ABSTRACT
Breast Cancer Gene 1 (BRCA1) is a tumour suppressor protein that modulates multiple biological processes including genomic stability and DNA damage repair. Although the main BRCA1 protein is well characterized, further proteomics studies have already identified additional BRCA1 isoforms with lower molecular weights. However, the accurate nucleotide sequence determination of their corresponding mRNAs is still a barrier, mainly due to the increased mRNA length of BRCA1 (~5.5 kb) and the limitations of the already implemented sequencing approaches. In the present study, we designed and employed a multiplexed hybrid sequencing approach (Hybrid-seq), based on nanopore and semi-conductor sequencing, aiming to detect BRCA1 alternative transcripts in a panel of human cancer and non-cancerous cell lines. The implementation of the described Hybrid-seq approach led to the generation of highly accurate long sequencing reads that enabled the identification of a wide spectrum of BRCA1 splice variants (BRCA1 sv.7 – sv.52), thus deciphering the transcriptional landscape of the human BRCA1 gene. In addition, demultiplexing of the sequencing data unveiled the expression profile and abundance of the described BRCA1 mRNAs in breast, ovarian, prostate, colorectal, lung and brain cancer as well as in non-cancerous human cell lines. Finally, in silico analysis supports that multiple detected mRNAs harbour open reading frames, being highly expected to encode putative protein isoforms with conserved domains, thus providing new insights into the complex roles of BRCA1 in genomic stability and DNA damage repair.
KEYWORDS: DNA repair, alternative splicing, nanopore sequencing, BRCA1, third-generation sequencing, mRNA transcripts
Introduction
Precursor mRNA (pre-mRNA) maturation is of utmost importance to the eukaryotic realm since nascent RNA must undergo extensive co- and/or post-transcriptional processing that involves 5′-end capping, splicing, and 3′-end polyadenylation, before being exported to the cytosol and translated into proteins [1,2]. In particular, splicing, the process of generating mature mRNAs by the removal of the intronic sequences and the subsequent joining of exons, has gained increasing attention especially due to the mechanism of alternative splicing (AS), in which different combinations of exonic sequences produce a variability of mRNA transcripts [3]. Thus, a single gene may give rise to multiple mRNAs with different functions, contributing to high protein diversity [4,5]. AS has a major impact on the features of the final conformation of the produced protein, as it affects functional domains by adding or deleting them, altering its stability, controls its localization and modifies its interactions with other proteins [6]. According to gene mapping analysis, almost 94% of the human genes undergo AS, therefore establishing the high phenotypic complexity in mammals [7,8]. Although AS is associated with cell differentiation, tissue and organ development as well as lineage determination [7], several studies underline its involution in specific tumour phenotypes [9–11]. AS mediates the differential expression of oncogenic and tumour suppressor genes, therefore the identification of alternative cancer-related mRNAs is a milestone in cancer research [12].
Datasets from comprehensive transcriptome analyses support that a plethora of tumour-associated genes lead to the generation of alternative transcripts that are implicated not only in carcinogenesis, but also in cancer progression and metastasis [13]. For instance, in multiple types of tumours, alternative transcripts of MET proto-oncogene (MET) contribute to the activation of MET and provide sensitivity to MET inhibitors [14]. As for Breast Cancer Gene 1 (BRCA1), it is considered to give rise to multiple alternatively spliced mRNAs [15]. In brief, BRCA1 was first cloned in 1994 [16], and after investigating adult and foetal tissues, it was found that alternative BRCA1 transcripts were present in both tissues [17]. Further studies have contributed to the detection of more than 100 BRCA1 alternative splicing events [18–20], however, sequencing of the full-length BRCA1 mRNAs has proven to be challenging [21].
BRCA1 is a 207 kDa tumour suppressor protein (1863 aa) that plays a significant role in genomic stability, DNA damage repair, cell cycle control and further critical physiological processes [22]. To maintain genomic integrity, via homologous recombination (HR) and non-homologous end-joining DNA repair (NHEJ), BRCA1 functions in the nucleus as part of vast multiprotein complexes [23]. Specifically, the protein contains a N-terminal RING domain, encoded by exons 2–5, which interacts with the BRCA1-Associated RING Domain protein 1 (BARD1), thus acquiring an E3 ligase activity [24]. It should be mentioned that both mutations in the RING E3 ligase activity and alterations in the interaction of BRCA1/BARD1 or BRCA1/UbcH5 (E2 ubiquitin-conjugating enzyme) can affect the tumour suppressor function of BRCA1 [25]. In addition, N-terminus harbours two Nuclear Export Signal (NES) regions, NES1 and NES2, respectively. NES1 includes the LECPICLEL segment (residues 22–30), whereas NES2 at positions 81–99 (QLVEELLKIICAFQLDTGL motif) promotes the BRCA1 exportation from the nucleus in a CRM1-dependent manner [26,27]. Furthermore, two Nuclear Localization Signal (NLS) sequences, encoded by exon 10, are located downstream of the NES regions and facilitate the transportation of BRCA1 from the cytosol to the nucleus when targeted by importin-a [28]. NLS1 is comprised of 7 residues (501-KLKRKRR-507) while NLS2 has 8 conserved aa (607-KKNRLRRK-614). Mutations in the NLS regions hinder interactions with importin-a hence confining BRCA1 in the cytosol and decreasing its tumour suppressor activity [29]. Furthermore, exons 10–15 are responsible for the translation of the serine cluster domain (SCD), located at positions 1280–1524, which contains an abundance of potential phosphorylation sites. In response to DNA damage, these sites get phosphorylated by ATM/ATR kinases, resulting in the recruitment of BRCA1 to double-strand break sites [28]. Moreover, SCD harbours an additional motif known as the coiled-coil domain (residues 1364–1437), that facilitates interactions with PALB2 [28]. The formed complex is charged of carrying out the DNA repair at break sites [30]. Of note, two tandem BRCA1 C-terminal (BRCT) repeats connected by a 23aa linker, constitute the BRCT domains, namely BRCT1 (residues 1646–1736) and BRCT2 (residues 1760–1859), which are crucial for its tumour suppressor, DNA repair and transcriptional regulation functions [31]. More precisely, phosphoprotein interactions between BRCA1 and proteins phosphorylated by ATM/ATR kinases in response to DNA damage are regulated by the BRCT domains [28,32].
Although functional studies at a protein level are still a barrier, isoforms with a lower molecular weight than the main BRCA1, have already been described [33]. According to previous studies, 3 additional BRCA1 isoforms have been identified and characterized as BRCA1a, BRCA1b and BRCA1-IRIS [15]. In detail, BRCA1a is encoded by a transcript variant that includes a truncated version of exon 10 (tr10) and has a molecular weight of approximately 110 kDa. BRCA1a lacks both the binding domains for specific proteins, such as Rb, p53, Rad50, c-Myc, γ-tubulin and angiopoietin-1 and the NLS motifs leading to cytoplasmic localization of the protein [34,35]. In a similar manner, BRCA1b (110 kDa) lacks the same binding domains and motifs as BRCA1a and is encoded by an alternative BRCA1 mRNA that further lacks exons 8 and 9 [34–36]. As for BRCA1-IRIS, its corresponding mRNA includes exons 1–10 and produces an isoform of 1,399 aa with abolished BRCT domains [15,37]. It is worth mentioning that although BRCA1-IRIS retains the RING finger at the N-terminus, the isoform does not interact with BARD1 and is exclusively chromatin-associated.
Even though several protein isoforms have already been characterized, the nucleotide sequences of their corresponding mRNAs still remain unclear, due to existing limitations in sequencing of full-length mRNA targets that exceed 1Kb. In the GenBank’s GRCh38 Annotation Release 110, a total of 6 curated mRNAs have been characterized, which consist of the main BRCA1 transcript (BRCA1 v.1, accession number: NM_007294.4) and 5 alternative splice variants (BRCA1 v.2 – v.6, accession numbers: NM_007300.4, NM_007297.4, NM_007298.3, NM_007299.4 and NR_027676.2, accordingly). Based on the current update in the database, the mRNA structure of these 6 annotated mRNAs has been subjected to minor modifications (Supplementary Figure S1). Briefly, BRCA1 v.2, v.3 and v.6 have a similar mRNA structure with the main transcript, bearing subtle splicing differentiations. On the other hand, BRCA1 v.4 and v.5 contain a significantly truncated exon 10 (tr10) and therefore they both encode protein isoforms with considerably decreased molecular weight. It should be mentioned that despite BRCA1 v.3 and v.6 are shown to utilize alternative ATG sites, no experimental validation of their functionality has yet been confirmed.
Recently, more than 300 records that describe alternative transcripts of BRCA1 have been curated by NCBI. However, they are derived from in silico analysis and obsolete datasets that were submitted in 2002 (accession number: AC135721), whereas there is still no experimental verification of their existence and their expression levels by newer scientific reports. Moreover, according to the Ensembl database, 4 additional cryptic exons are distributed between the annotated exons. Briefly, cryptic exon 1 (C1) and cryptic exon 2 (C2) are located between the second and third exon, whereas cryptic exon 3 (C3) and cryptic exon 4 (C4) are located between the third and fourth exon.
Our research aims to investigate the transcriptional landscape of BRCA1 gene through nanopore sequencing, a state-of-the-art technology that enables the detection of full-length transcripts. For this purpose, an in-house developed multiplexed hybrid sequencing approach (Hybrid-seq), based on nanopore and semi-conductor sequencing methodologies, was designed and employed for the detection and expression profiling of BRCA1 splice variants in major human malignancies as well as in human non-cancerous cell lines. Through the implementation of the Hybrid-seq approach that led to the generation of highly accurate sequencing reads, we highlight the existence of a wide spectrum of BRCA1 mRNA splice variants (BRCA1 sv.7 – sv.52) and assess their expression levels among the investigated human tissues. Finally, in silico analysis of the identified BRCA1 mRNA transcripts supports the existence of several protein-coding mRNAs harbouring open reading frames (ORFs), suggesting the translation of putative protein isoforms.
Materials and methods
Human cell line culture
The present work was accomplished using a total of 25 human cell lines (23 cancerous and 2 non-cancerous), which were the following: MCF-7, SK-BR-3, BT-20, MDA-MB-231, MDA-MB-468, BT-474 (Breast/ductal adenocarcinoma), OVCAR-3, SK-OV-3, ES-2, MDAH-2774 (Ovarian cancer), PC-3, DU 145, LNCaP (Prostate cancer), Caco-2, DLD-1, HT-29, HCT 116, SW 620 (Colorectal cancer), A549 (Lung adenocarcinoma), U-87 MG, U-251 MG, H4, SH-SY5Y (Brain cancer), HEK-293 (Human embryonic kidney), HaCaT (Human keratinocytes). All cell lines were propagated based on the American Type Culture Collection (ATCC) guidelines.
Total RNA isolation, mRNA enrichment and first-strand cDNA synthesis
The isolation of total RNA from each human cell line was employed using the TRIzol Reagent (Ambion™, Thermo Fisher Scientific Inc., Waltham, MA, USA). The purity and the concentration of each RNA sample was assessed spectrophotometrically at 260 and 280 nm, using BioSpec-nano Micro-volume UV-Vis Spectrophotometer (Shimadju, Kyoto, Japan). Finally, the downstream mRNA enrichment from 5 μg of each total RNA sample was carried out with NEBNext® Poly(A) mRNA Magnetic Isolation Module (New England Biolabs, Inc).
To generate first-strand cDNAs, reverse transcription (RT) was implemented in reaction volumes of 20 μl using 100 ng of mRNA from each cell line as template, an oligo-dT20 as primer and Maxima H Minus Reverse Transcriptase (Invitrogen™, Thermo Fisher Scientific Inc.). Briefly, the initial cDNA synthesis mixtures included 12.5 μl poly(A)+ RNA and 1 μl oligo-dT20 (10 μM) and was incubated at 65°C for 5 min in a hot-lid Veriti 96-Well Fast Thermal Cycler (Applied Biosystems™). In the next step, the cDNA synthesis mixtures were completed by adding 4 μl 5× RT Buffer, 1 μl dNTP mix (10 mM each), 0.5 μl (20 U) RNaseOUT inhibitor (Invitrogen™, Thermo Fisher Scientific Inc.) and 1 μl (200 U) Maxima H Minus Reverse Transcriptase (Invitrogen™, Thermo Fisher Scientific Inc.). The first-strand cDNAs were synthesized by incubating the RT reaction mixtures at 50°C for 30 min, whereas a heat inactivation step was followed at 85°C for 5 min. The quality of the derived cDNA samples was evaluated by the amplification of the human housekeeping gene GAPDH (Glyceraldehyde 3-phosphate dehydrogenase). Finally, the cDNA samples were mixed equimolarly based on the tissue of origin/type of malignancy (breast, ovarian, prostate, colorectal, lung and brain cancer as well as non-cancerous cell lines) to create 7 distinct cDNA pools that were used as templates for the downstream PCR-based assays.
Specific amplification of BRCA1 mRNA transcripts
A touchdown PCR-based assay, using two gene-specific primers (GSPs) was optimized and employed for the specific amplification of BRCA1 mRNA transcripts. A forward GSP (F: 5΄ – GGATTTATCTGCTCTTCGCGTT − 3΄) was designed to anneal near the ATG site and was used along with a reverse GSP (R: 5΄ – GCTACACTGTCCAACACCCA − 3΄) designed to target the last annotated exon of the main BRCA1 v.1.
The amplification of BRCA1 transcript variants was implemented in reaction volumes that included 5 μl of 10× LA PCR Buffer ll (Mg2+ plus), 8 μl of dNTP mix (2.5 mM each), 1 μl of each GSP (10 μM), 0.5 μl (2.5 U) of TaKaRa LA Taq polymerase and sterile distilled water to a final volume of 50 μl. The applied thermal protocol was performed in a Veriti 96-Well Fast Thermal Cycler (Applied Biosystems™) and included an initial denaturation step at 94°C for 3 min, 35 cycles of 94°C for 30 sec, 65°C (auto-ΔTa: −0.3°C/cycle) for 30 sec, 72°C for 6 min, and a final extension step at 72°C for 10 min. The specificity and yield of the PCR assays were evaluated with electrophoresis in agarose gels. The produced amplicons were purified using the NucleoSpin® Gel and PCR Clean-up kit (Macherey-Nagel GmbH & Co. KG, Duren, Germany) and then were quantified with the Qubit® DNA HS Assay Kit (Invitrogen™, Thermo Fisher Scientific Inc.).
Targeted Hybrid-seq approach for the generation of highly accurate long reads
An initial amount of 1 μg from each purified amplicon was used for the preparation of 7 DNA-seq barcoded libraries for nanopore sequencing, which corresponded to amplified BRCA1 mRNAs from breast cancer, ovarian cancer, prostate cancer, colorectal cancer, lung cancer, brain cancer and non-cancerous cell lines (HEK-293 and HaCaT). The Ligation Sequencing Kit (SQK-LSK109, ONT) and the Native Barcoding Expansion Kit 1–12 (EXP-NBD104, ONT) were used for the construction of the barcoded libraries, following the guidelines of the manufacturer. During the library preparation, the end repair step was completed with NEBNext® Ultra™ II End Repair/dA-Tailing Module (New England Biolabs, Inc), the adapter ligation was conducted with the use of Quick T4 Ligase (New England Biolabs, Inc) and the clean-up steps were performed with the Agencourt AMPure XP beads (Beckman Coulter, Brea, CA, USA). Each barcoded library was mixed equimolarly to create a final DNA-seq library that was loaded on a FLO-MIN106D flow cell with R9.4.1 chemistry and a nanopore sequencing run was performed on a MinION Mk1C sequencer (Oxford Nanopore Technologies Ltd, ONT, Oxford, UK).
To implement the described Hybrid-seq approach, the same starting material was exploited as template for the creation of DNA-seq libraries for next-generation sequencing. The library preparation workflow was performed with the Ion Xpress™ Plus Fragment Library Kit (Ion Torrent™, Thermo Fisher Scientific Inc.) and included an initial enzymatic fragmentation step, the adapter ligation and nick-repair, as well as the purification of the ligated DNA. A bead-based size selection (target fragment ~ 400bp) was followed to enrich the library for the desired fragments, using the KAPA Pure Beads (Kapa Biosystems Inc.) in the recommended dsDNA: beads ratio. In the next step, the quantification of the NGS libraries was performed on an ABI 7500 Fast Real-Time PCR System (Applied Biosystems™) with the Ion Library TaqMan™ Quantitation Kit (Ion Torrent™). Finally, the Ion PGM™ Hi-Q™ View OT2 kit (Ion Torrent™) was used for the template preparation and enrichment, whereas semi-conductor sequencing was employed on the Ion Personal Genome Machine™ (PGM™) platform with the Ion PGM™ Hi-Q™ View Sequencing kit.
Post-processing and bioinformatics analysis
The basecalling, demultiplexing, adapter trimming and QC analysis of the raw nanopore sequencing datasets were completed with Guppy [38]. After the analysis, the nanopore sequencing reads were separated into two folders, the ‘pass’ and the ‘fail’, based on their quality scores. Only the reads existing in the ‘pass’ folder were used for the investigation of the BRCA1 transcriptional profile and the identification of potential novel mRNA transcripts. In the next step, a long-read polishing step was implemented with hybrid error correction algorithms that utilized both the raw nanopore sequencing data and the short-read NGS datasets to produce highly accurate long reads, which were used for the downstream bioinformatics analysis for BRCA1. The polished long-reads from each barcoded nanopore sequencing library were aligned to the human reference genome (GRCh38) with Minimap2 [39], whereas Integrative Genomics Viewer (IGV) enabled the visualization of the successfully mapped reads [40].
Expression analysis of the detected BRCA1 transcripts
Demultiplexing of the polished nanopore sequencing reads enabled not only the identification of the existing BRCA1 transcript variants in each barcoded library, but also the evaluation of their expression levels. For the assessment of each BRCA1 transcript’s abundance in each library and the estimation of the read per million (RPM) values, only the sequencing reads that covered the entire coding sequence (from the translation initiation site until the last exon) were taken into consideration. Furthermore, reads derived from random fragments during the library preparation workflow were excluded from the analysis since they fail to represent a unique variant. The identification of the existing BRCA1 splice variants in each dataset was carried out with our in-house developed algorithm ‘ASDT’ [41], whereas specialized algorithms for long-read alternative isoform quantification were employed for the abundancy estimation of each mRNA splice variant in each barcoded library [42,43].
ORF evaluation of the identified BRCA1 mRNA transcripts
Following the identification of BRCA1 transcriptional profile, an in silico ORF query step was carried out for each identified mRNA transcript. Since all BRCA1 splice variants described in the present study share the exon 2 that harbours the annotated initiation codon, we used the ExPASy’s translate tool [44] to investigate the potential ORFs that are derived from this annotated initiation codon. The ORF for each mRNA was further analysed using the mammalian nonsense-mediated mRNA decay (NMD) rule and was tested for the existence of any conserved BRCA1 domains. According to this rule, transcript variants harbouring ORFs in which a termination codon resides more than 50–55 nt upstream of the last exon junction are most likely subjected to NMD [45]. Finally, the Gene Ontology (GO) terms with significant prediction scores for each putative BRCA1 isoform were obtained by DeepGOWeb [46].
Results
Investigation of BRCA1 splicing through a targeted Hybrid-seq approach
Briefly, a touchdown PCR-based assay using two GSPs was developed and optimized for the specific amplification of BRCA1 splice variants in cDNA pools from six human malignancies (breast, ovarian, prostate, colorectal, lung and brain cancer) and non-cancerous human cell lines. The derived PCR amplicons were used as input for the construction of long-read-barcoded libraries that were sequenced on a MinION Mk1C sequencer. To obliterate the high-error rates of nanopore sequencing, NGS libraries from the same PCR amplicons were created and the short-read data was used for error correction of the acquired nanopore sequencing reads with read polishing algorithms. The combination of long- and short-read sequencing approaches (Hybrid-seq) led to the generation of polished long reads, which were aligned to GRCh38 for the study of BRCA1 transcriptional profile. Analysis of the aligned datasets was carried out with bioinformatics tools specialized for splicing detection and transcript quantification. Ultimately, the applied in silico pipeline that included ORF query algorithms and GO term prediction tools indicated the existence of mRNAs that are highly expected to encode putative BRCA1 isoforms, which merit further investigation.
Long-read sequencing unveils the transcriptional landscape of BRCA1
The acquired sequencing datasets included long reads covering the entire length of the produced amplicons from the annotated ‘ATG site’ till the last exon of the gene. Bioinformatics analysis using the in-house developed ‘ASDT’ algorithm and visualization of the successfully aligned sequencing reads clearly confirmed a wide variety of previously described as well as novel splicing events and validated the existence of all 4 cryptic exons that appear in the Ensembl database in several alternative splice variants (Supplementary Figures S2 & S3).
Overall, in the present study we identified a total of 46 BRCA1 mRNAs (BRCA1 sv.7 – sv.52) (Supplementary Table S1). The detected transcripts BRCA1 sv.7 – sv.52 were deposited in the GenBank® database, under the accession numbers OP156938 – OP156983, respectively. In brief, BRCA1 sv.7 – sv.37 are generated from a variety of splicing patterns and cassette exon combinations, whereas 15 transcripts (BRCA1 sv.38 – v.52) include the aforementioned cryptic exons (Figure 1 and Supplementary Figure S4). Additionally, the major annotated BRCA1 mRNA transcripts (BRCA1 v.1 – v.6) were detected in all investigated human malignancies as well as in non-cancerous human cells.
Figure 1.
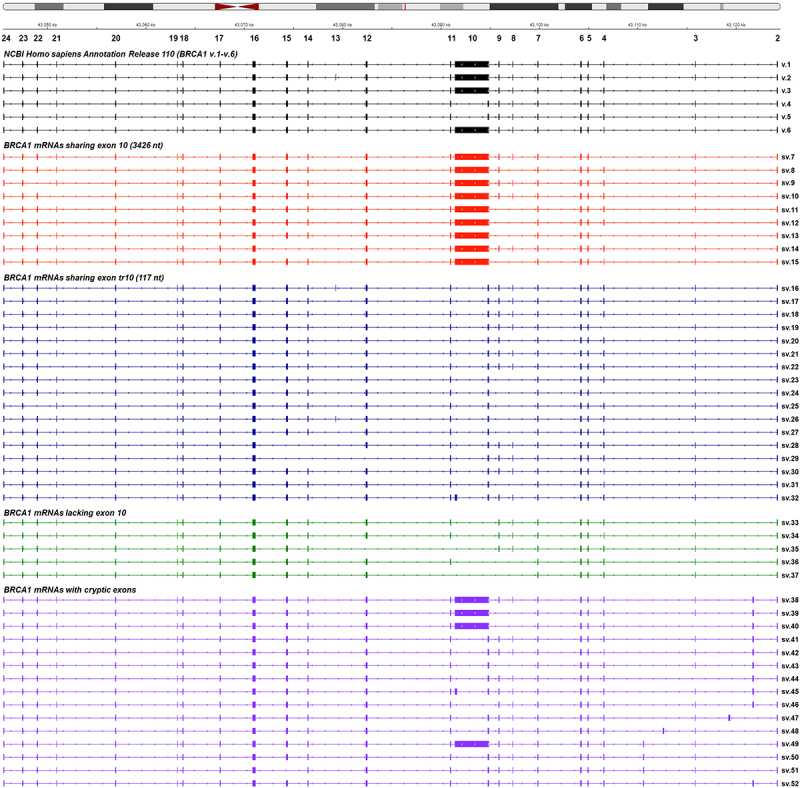
Visualization of the detected BRCA1 splice variants aligned to the reference genome GRCh38 with IGV. The annotated BRCA1 mRNAs existing in the GenBank’s Annotation Release 110 are shown in black colour. For visual purposes, each group of BRCA1 mRNAs is demonstrated with a different colour.
Interestingly, additional new splicing events corresponding to novel transcripts were detected only in the barcoded library of the brain tumour cell lines and therefore could be characterized as ‘brain-specific’ mRNAs (Supplementary Figure S5). However, in contrast to the rest findings of the present study, these transcripts fail to be detected in other tumour cell lines or even the human non-cancerous cell lines that were investigated. Taking into consideration their absence from non-cancerous cells, it is possible that these findings are rather a collection of splicing errors or even cell line-specific genomic variants that can make adverse effects on the alignment of sequencing reads. In our effort to preclude the latter, additional sequencing experiments were carried out for each brain cancer cell line separately (U-87 MG, U-251 MG, H4, SH-SY5Y). The derived results showed that five mRNAs were present in more than one brain cell line (BRCA1 sv.54, sv.56, sv.58, sv.59 and sv.60), thus do not represent cell line-specific genomic variants (Supplementary data.pdf). Still however, the ambiguity on whether these mRNAs are functional and possess regulatory roles is rather increased.
Structural analysis unravels a comprehensive catalogue of BRCA1 transcripts
Based on the acquired sequencing data, BRCA1 sv.7 – sv.37 are derived from various cassette exon(s) events between the annotated exons. These transcripts can be categorized into three different groups in terms of their exon/intron boundaries; splice variants sharing the ‘full-length’ exon 10 (3426 nt) that is present in the main mRNA (BRCA1 sv.7 – sv.15), splice variants that include the exon tr10 (BRCA1 sv.16 – sv.32) and splice variants that lack the entire sequence of exon 10, namely BRCA1 sv.33 – sv.37 (Figure 1).
Most mRNAs of the first group that contain the ‘full-length’ exon 10 harbour ORFs and therefore are predicted to encode BRCA1 isoforms. In specific, BRCA1 sv.7, sv.9, sv.11, sv.13 and sv.14 share the same initiation codon with the main BRCA1 v.1 and hence they can be characterized as the most promising protein-coding mRNAs (Figure 2). On the contrary, ORF query analysis showed that BRCA1 sv.8, sv.10, sv.12 and sv.15 have premature termination codons (PTCs) and are candidates for the nonsense-mediated mRNA decay (NMD) pathway.
Figure 2.

BRCA1 splice variants bearing the ‘full-length’ exon 10 (BRCA1 sv.7 – sv.15). Exons are shown as boxes and introns as lines. The numbers inside the boxes and above the lines represent the length of each exon and intron, accordingly. Black boxes denote the coding region of the main BRCA1 v.1. Dark blue boxes are used to indicate the coding region of the splice variants that utilize the annotated initiation site of exon 2, while light blue is used to demonstrate non-coding RNAs with PTCs and untranslated regions.
The 17 transcripts of the second group (BRCA1 sv.16 – sv.32) have two common features; the truncated exon 10 (tr10) and a variety of splicing events between annotated exons (Figure 3). Due to the existence of tr10, these splice variants are more similar to the annotated BRCA1 v.4 and v.5, and as a consequence, the primary aa sequence of their putative protein isoforms resemble to isoforms encoded by BRCA1 v.4 and v.5 (Supplementary Figure S1). For instance, the subtle difference between BRCA1 v.4 and sv.16 is the existence of the additional exon 13 at the nucleotide sequence of BRCA1 sv.16, whereas BRCA1 v.4 and sv.17 only differ in a nucleotide triplet at exon 14. Besides BRCA1 sv.18, sv.20 – sv.22, sv.27 and sv.29 – sv.31 that contain PTCs and therefore are most likely non-coding RNAs, all the remaining transcripts of this group harbour ORFs and are predicted to encode truncated isoforms due to the existence of exon skipping events in their respective mRNA sequences (Figure 3).
Figure 3.

BRCA1 splice variants sharing the truncated version of exon 10 (BRCA1 sv.16 – sv.32). Exons are shown as boxes and introns as lines. The numbers inside the boxes and above the lines represent the length of each exon and intron, accordingly. Black boxes denote the coding region of the annotated BRCA1 v.4. Dark blue boxes are used to indicate the coding region of the splice variants that utilize the annotated initiation site of exon 2, while light blue is used to demonstrate non-coding RNAs with PTCs and untranslated regions.
The next group consists of 5 alternative transcripts that completely lack exon 10 (BRCA1 sv.33 – sv.37). The absence of exon 10 involves the splicing of either exon 7 or 9 with exons downstream of 10, thus leading to the generation of significantly shorter mRNA splice variants as compared to the main BRCA1 v.1 (Figure 4). In silico analysis revealed that BRCA1 sv.33, sv.35 and sv.36 are expected to be protein-coding, sharing the annotated start and stop codon with BRCA1 v.1, whereas BRCA1 sv.34 and sv.37 represent non-coding transcripts.
Figure 4.
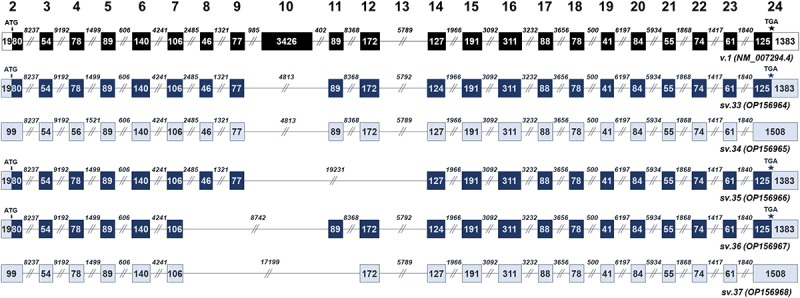
BRCA1 splice variants lacking the entire sequence of exon 10 (BRCA1 sv.33 – sv.37). Exons are shown as boxes and introns as lines. The numbers inside the boxes and above the lines represent the length of each exon and intron, accordingly. Black boxes denote the coding region of the main BRCA1 v.1. Dark blue boxes are used to indicate the coding region of the splice variants that utilize the annotated initiation site of exon 2, while light blue is used to demonstrate non-coding RNAs with PTCs and untranslated regions.
Interestingly, bioinformatics analysis also revealed the expression of 15 additional BRCA1 mRNAs that include cryptic exons (Supplementary Figure S3). More precisely, a total of 9 transcripts (BRCA1 sv.38 – sv.46) share the cryptic exon C1, which is located between the annotated exons 2 and 3 (Figure 5). The cryptic exon C2, which resides between exons 2 and 3, was documented only in BRCA1 sv.47, while C3 was uniquely detected in BRCA1 sv.48. Additionally, our results confirm that exon C4, which is also located between exons 3 and 4, is present in 3 BRCA1 mRNAs (BRCA1 sv.49 – sv.51). Finally, in the mRNA sequence of BRCA1 sv.52, both C1 and C4 are detected. The existence of the described cryptic exons leads to the formation of PTCs, suggesting that this group of mRNAs represent non-coding RNAs (Figure 5).
Figure 5.
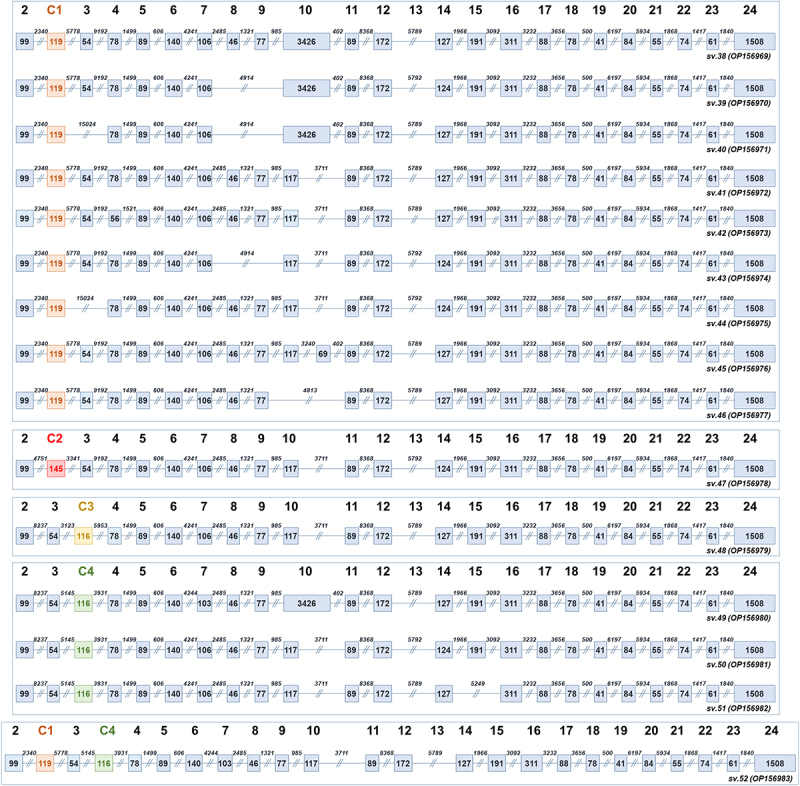
Structural demonstration of the BRCA1 transcripts that contain the cryptic exons (C1-C4) described in the present study. Exons are shown as boxes and introns as lines. The numbers inside the boxes and above the lines represent the length of each exon and intron, accordingly. Light blue is used to demonstrate non-coding splice variants with PTCs. For visual purposes, the boxes that correspond to the cryptic exons are shown in different colour.
Nanopore sequencing captures the expression levels of BRCA1 mRNAs
Besides the detection of BRCA1 mRNA transcripts, demultiplexing of the acquired Hybrid-seq datasets provided new insights into the expression levels of each transcript among the investigated barcoded libraries (Supplementary Tables S2 & S3). Most of the described BRCA1 mRNAs were identified in all the barcoded libraries and hence they demonstrate broad expression patterns. Regarding the annotated transcripts, BRCA1 v.1 and v.4 exhibited similar expression levels in all barcoded libraries (Supplementary Figure S6). As for the rest annotated BRCA1 mRNAs, not only they were detected in significantly lower read counts as compared to v.1 and v.4, but also demonstrated diverse expression levels among the investigated barcoded libraries, which may be an indication of their diverging roles in each cell type.
Among the detected BRCA1 mRNAs, 8 transcripts (BRCA1 sv.11, sv.13, sv.18, sv.19, sv.23, sv.30, sv.38 and sv.41) emerged as the most abundant in terms of RPM values, and hence could be characterized as major BRCA1 splice variants (Figure 6). Interestingly, BRCA1 sv.23 exhibits a wide expression profile and significantly higher expression levels in brain cancer cells (Supplementary Figure S6). In addition, BRCA1 sv.14 was detected only in lung cancer cells indicating a rather tissue-specific role. Moreover, analysis revealed that the abundance of most BRCA1 transcripts displays a wide range of differentiation among the panel of human cell lines that was investigated. For instance, BRCA1 sv.16 is represented by higher read counts in ovarian, prostate, colorectal and lung cancer, compared to breast and brain cancer. Similarly, the expression levels of BRCA1 sv.33 are notably lower in the malignant tumours of prostate and lung (Supplementary Figure S6). In addition, sequencing data revealed that the abundancy of 8 BRCA1 transcripts (BRCA1 sv.9, sv.20, s.26, sv.40, sv.42, sv.44, sv.47 and sv.51) is significantly decreased as compared to BRCA1 v.1 and v.4, as they are hardly detectable among the same libraries Supplementary Figure S6).
Figure 6.
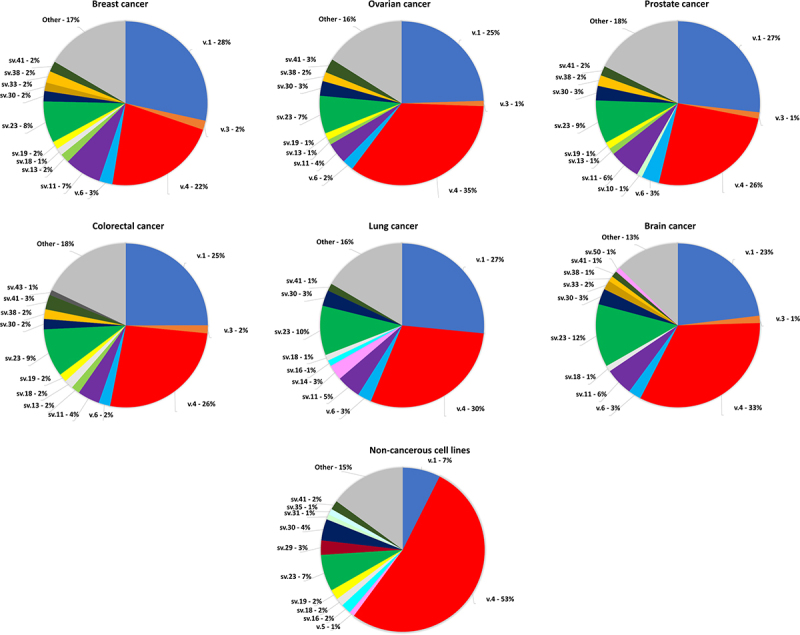
Pie charts demonstrating the % abundance of the major BRCA1 transcripts in the investigated nanopore sequencing datasets. Only transcripts with >1% abundancy are shown, whereas splice variants with < 1% are denoted as other and their aggregated abundancy is shown.
Despite the increased number of BRCA1 mRNA transcripts that were detected in the investigated malignancies, most of them are generated by exon skipping events. Apart from the alternative 5’ alternative splice site of exon 10 that creates mRNAs bearing exon tr10 (117 nt), the overwhelming mechanism that generates this wide spectrum of transcripts is cassette exon(s), whereas it should be mentioned that no intron retentions, alternative 3’ alternative splice sites or mutually exclusive exons were detected.
In silico characterization of the putative BRCA1 isoforms
The derived BRCA1 isoforms can be categorized in two distinct groups regarding their similarity to the main BRCA1 protein or their homology to the isoform encoded by BRCA1 v.4. The putative protein isoforms of the mRNAs of the first group that include the ‘full-length’ exon 10 (3426 nt) maintain both the vital domains (RING, SCD and BRCT) and the protein’s exportation and localization signals (Figure 7A). Of note, the putative isoforms encoded by BRCA1 sv.9 and sv.14 are predicted to encompass the RING domain that enables interactions with BARD1, but BRCA1 sv.9 has a truncated BRCT2 domain whereas BRCA1 sv.14 possesses an altered SCD region. As far as BRCA1 sv.11 is concerned, its alleged produced protein conserves the entire aa sequences that constitute the aforementioned domains. The rest coding mRNAs of this group possess a canonical C-terminal that comprises the entire sequence of both SCD and BRCT domains. However, skipping events in their mRNA sequences can lead to alterations in the N-terminal, hence a truncated RING region (Figure 7B).
Figure 7.
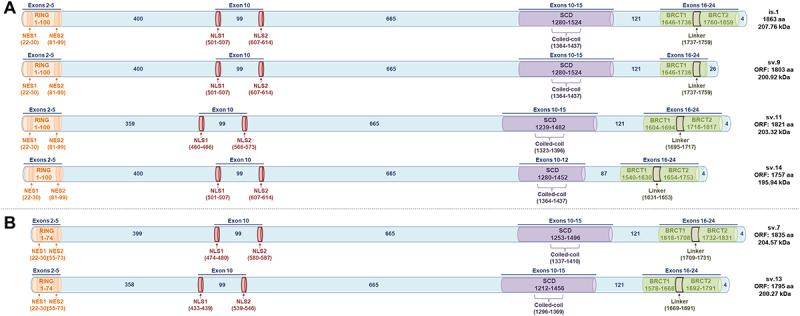
Putative BRCA1 isoforms bearing NLS signals, encoded by the described BRCA1 splice variants of the present study. The conserved motifs and signals are exhibited in coloured barrels (RING, NLS, SCD and BRCT domains). (A) Putative isoforms encompassing the full RING domain. (B) Putative isoforms with truncated RING domains.
As for the group of transcripts that include the truncated exon tr10, their ORFs are similar to BRCA1 v.4. Consequently, despite featuring the conserved domains, they lack the NLS motifs (Figure 8). In the same manner, coding BRCA1 transcripts that completely lack exon 10 fail to encode the NLS motif even though they are expected to encode the rest crucial regions. Most of the alleged BRCA1 isoforms of this group maintain the entire RING region, the truncated SCD domain of 158 aa that is also present at the protein encoded by BRCA1 v.4 as well as the BRCT domains, but in some cases such as BRCA1 sv.25 and sv.28, the C-terminal is modified. On the contrary, an additional group of BRCA1 mRNAs that feature multiple cassette exon(s) events seems to encode proteins with critical alterations in their functional domains (Figure 8).
Figure 8.
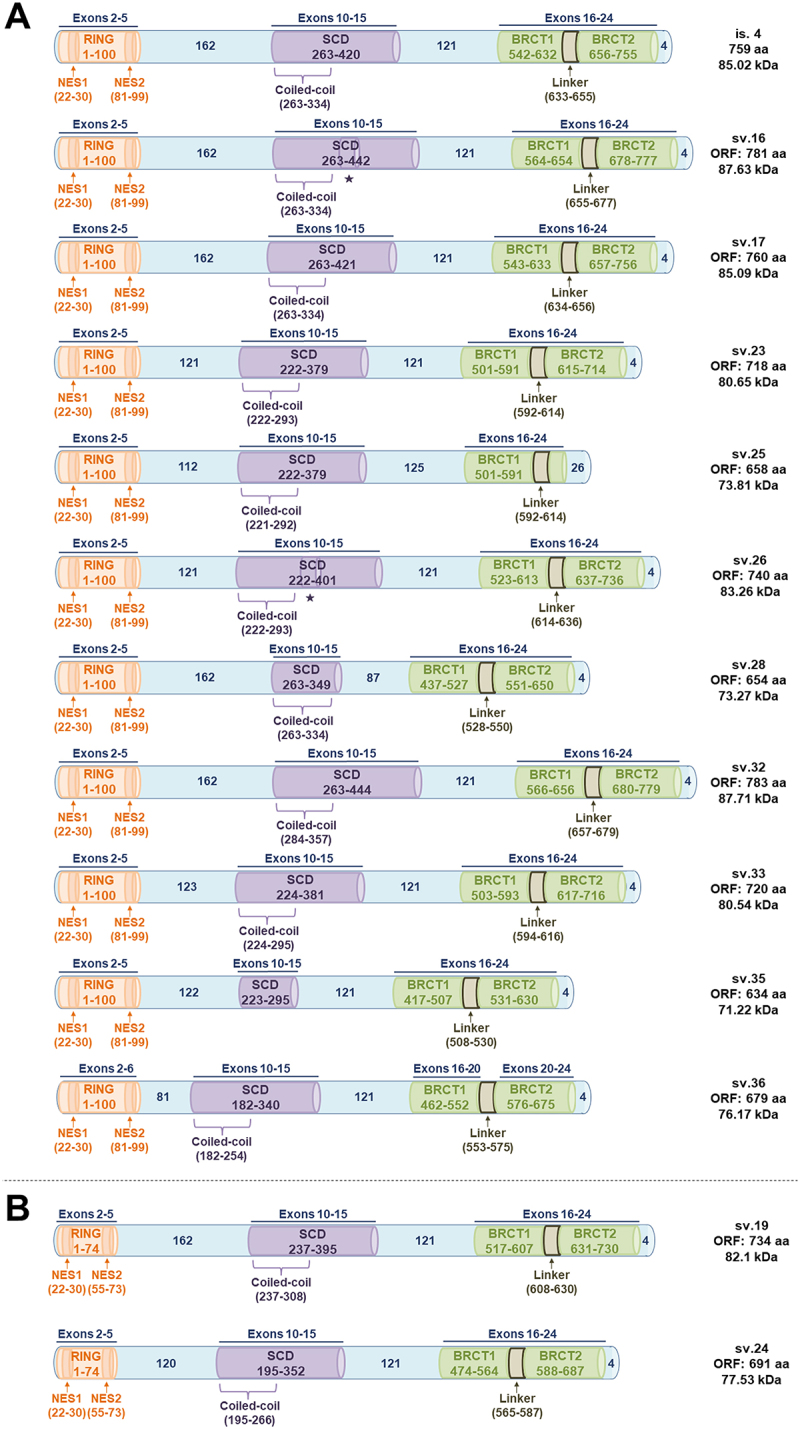
Putative BRCA1 isoforms lacking NLS signals, encoded by the described BRCA1 splice variants of the present study. The conserved motifs are exhibited in coloured barrels (RING, SCD and BRCT domains). Symbol * is shown below any protein region encoded by exon 13, which exists in the annotated BRCA1 v.2. (A) Putative isoforms encompassing the full RING domain. (B) Putative isoforms with truncated RING domains.
Discussion
It is well known that a great number of human genes exhibit differentiated mRNA expression profiles in malignant tissues, affecting therapy response and drug resistance depending on the cancer type [47,48]. Since long-read sequencing strategies showed up, our perception about transcriptomics has been radically changed [49]. Due to their innovative characteristics, long-read sequencing technologies have turned into the key method for the analysis of the transcriptome diversity, especially for genes with increased lengths, such as BRCA1. Although several studies have attempted to decipher the transcriptional landscape of BRCA1, its great complexity led to unsatisfactory results. In the present study, the implementation of our hybrid-seq approach and the generation of polished long reads highlighted a wide spectrum of expressed BRCA1 mRNA transcripts that comprise cassette exon events as well as cryptic exons and revealed their expression pattern in major human malignancies and non-cancerous cell lines. In detail, 31 BRCA1 mRNAs described in the present work (BRCA1 sv.7 – sv.37) originate from different combinations of the annotated exons, whereas 15 mRNAs (BRCA1 sv.38 – sv.52) contain the presented cryptic exons (C1 – C4).
A major point of the current study is that most of the described BRCA1 mRNAs were also detected in non-cancerous human cell lines. Over the last decade, many transcripts have been identified in cancer cells but since they are undetected in normal cell lines or tissues, they are called tumour-specific [50]. Under that prism, the existence of the presented BRCA1 mRNAs in non-cancerous cells firmly implies that they represent transcripts of the physiological alternative splicing mechanism and not splicing errors due to dysregulation of the splicing machinery. It is well known that transcribed RNAs are subjected to several regulatory mechanisms and modifications to end up being functional transcripts [51]. Nevertheless, even the recent advances in massive parallel sequencing, tumour transcriptome has not been fully decoded at transcript resolution. Recent scientific studies have indicated that from ~ 500,000 transcripts that were identified from RNA-seq data across more than 1000 cancer cell lines, almost half of them were unannotated [51]. Hence, the identification of BRCA1 mRNA splice variants that were detected only in brain cancer cell lines, being generated by a plethora of novel alternative donor and acceptor splice sites (Supplementary figure S5) is in accordance with the notion of these studies and further suggest that tumour transcriptome is far from being elucidated.
The potential protein-coding capacity of the identified BRCA1 mRNAs was investigated in silico based on whether they harbour ORFs that initiate from the common initiation codon that exists on the main mRNA BRCA1 v.1 (GenBank® accession number: NM_007294.4). Using the aforementioned strategy, 17 BRCA1 mRNAs were predicted as protein-coding, whereas the remaining represented prominent candidates for nonsense-mediated decay. However, several scientific reports support that eukaryotic ribosomes can recognize various translational initiation sites, thus translation of downstream ORFs is possible by either reinitiation or leaky scanning [52,53]. In fact, it has been shown that additional translated ORFs upstream and downstream of annotated ORFs are indeed functional, leading to the translation of protein isoforms from alternative start codons both in-frame and out-of-frame of annotated ORFs [54–59]. More importantly, RNAs previously considered non-coding, including long non-coding RNAs and circular RNAs, have been found to produce small functional proteins by previously ignored ORFs [60,61].
Our in silico ORF determination suggested that many of the described BRCA1 transcripts are rather coding mRNAs and share the crucial-conserved domains for protein’s interactions. Hence, we categorized the BRCA1 isoforms into two groups based on whether they are predicted to harbour the NLS signals. As for the first group that includes isoforms whose primary aa sequence is similar to the main protein encoded by BRCA1 v.1, Gene Ontology (GO) functional analysis’ results are in accordance with our in silico analysis. The putative isoforms encoded by BRCA1 sv.11 and sv.14 are highly probable to be co-localized with the main BRCA1 since they demonstrate similar GO prediction scores in terms of cellular compartmentalization (Figure 9). The rest members of this group are expected to possess significant truncations in their aa sequences that obviously affect their subcellular localization. In the same manner, the molecular functions of the putative proteins, apart from BRCA1 sv.9, sv.11 and sv.14 isoforms, may differ compared to BRCA1, since they lack aa residues that form the critical domains. Even though all of them are predicted to maintain protein’s DNA binding affinity, the lack of residues that constitute the RING domain leads to a truncated RING region that affects its transferase activity. The second group, that comprises isoforms similar to the protein encoded by BRCA1 v.4, is expected to exhibit a considerably differentiated cellular localization pattern. Briefly, the absence of the NLS signals affects their transportation into the nucleus and their involvement in formatting nuclear complexes. Moreover, the expected reduced molecular weight of the produced isoforms and hence, the possible alterations in the tertiary structure between these proteins and BRCA1, suggest that their transferase, hydrolase and peptidase activities are reduced (Figure 8).
Figure 9.
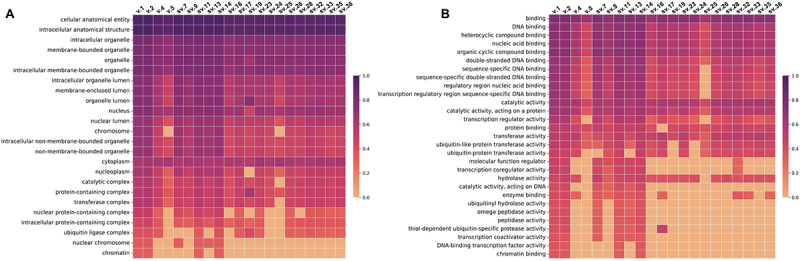
Heatmaps showing the DeepGOWeb scores for the main and putative new BRCA1 isoforms, which indicate how probable the predictions are regarding their (A) Cellular component and (B) Molecular function.
Another major issue that should be mentioned is the potential existence of mutations, especially insertions/deletions and nonsense mutations. Since BRCA1 represents a widely mutated tumour-suppressor gene, any coding mutations can potentially lead to cancer susceptibility. According to the Catalogue of Somatic Mutations in Cancer (COSMIC) database [62], there are several confirmed BRCA1 coding mutations that can affect variant abundance in cancers, most of which include nonsense, frameshift as well as missense mutations that alter key functional domains of the protein (Supplementary data). These mutations can trigger NMD, leading to reduced levels of functional BRCA1 protein and increased cancer risk. For instance, the BRCA1 185delAG mutation, which results in a frameshift and a PTC, has been shown to have reduced levels of mRNA and protein compared to the normal BRCA1 allele [63,64]. Similarly, BRCA1 5382insC mutation introduces a PTC and has also been associated with reduced mRNA and protein levels [65]. Variant calling in our nanopore sequencing datasets from each human malignancy confirmed the existence of several characterized BRCA1 coding mutations from COSMIC database (Supplementary data). Finally, any detected uncharacterized mutation in the coding region of BRCA1 demonstrated notably lower frequency and therefore is most-likely not significantly in terms of cancer susceptibility and increased cancer risk.
Besides the coding transcripts, in silico ORF query unveiled a group of long non-coding BRCA1 mRNAs that may represent tumour-related components in the DNA repair pathway. Multiple reports support that BRCA1 modulates cellular responses to damage through the activation of DNA repair associated complexes, while additional studies have highlighted several cancer-related lncRNAs as mediators in DNA damage [66–68]. For instance, the lncRNAs CUPID1 and CUPID2 regulate responses to DNA damage in breast cancer [69]. Taken together all these considerations, it can be assumed that the identified BRCA1 lncRNAs could be involved in the process of DNA repair although their specific role merits further investigation.
Overall, most of the identified transcripts of the current study may represent key regulatory factors in tumour growth, be involved in DNA damage response pathways and inhibit or promote BRCA1’s molecular functions. Especially, BRCA1 transcripts that are characterized by differentiated expression patterns among the examined human malignancies should be thoroughly investigated since they could lead to the detection of novel diagnostic and/or prognostic biomarkers or promising therapeutic targets.
Supplementary Material
Funding Statement
Part of this work was supported by Empirikeion Foundation, Athens, Greece.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The nucleotide sequences of the BRCA1 mRNA transcripts described in the present study (BRCA1 sv.7 – sv.52) have been deposited to the GenBank® database (https://www.ncbi.nlm.nih.gov/genbank/) and correspond to the accession numbers OP156938 – OP156983, accordingly. The datasets related to the present work are available in the Sequence Read Archive (SRA) repository, BioProject ID: PRJNA899869.
Author contributions
P.G.A. designed research, performed experiments, facilitated bioinformatics analysis, wrote the manuscript, prepared the original draft, reviewed the manuscript and coordinated the project; K.A. performed experiments, facilitated bioinformatics analysis, wrote the manuscript and prepared the original draft; M.A.B. facilitated bioinformatics analysis, wrote the manuscript and prepared the original draft; G.D. performed experiments, facilitated bioinformatics analysis and wrote the manuscript; G.N.D facilitated bioinformatics analysis; P.T. performed experiments and prepared the original draft; A.S. reviewed and edited the manuscript.
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/15476286.2023.2220210
References
- [1].Darnell JE Jr. Reflections on the history of pre-Mrna processing and highlights of current knowledge: a unified picture. RNA. 2013;19:443–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Nott A, Le Hir H, Moore MJ.. Splicing enhances translation in mammalian cells: an additional function of the exon junction complex. Genes Dev. 2004;18:210–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Bonnal SC, Lopez-Oreja I, Valcarcel J. Roles and mechanisms of alternative splicing in cancer - implications for care. Nat Rev Clin Oncol. 2020;17:457–474. [DOI] [PubMed] [Google Scholar]
- [4].Nilsen TW, Graveley BR. Expansion of the eukaryotic proteome by alternative splicing. Nature. 2010;463:457–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem. 2003;72:291–336. [DOI] [PubMed] [Google Scholar]
- [6].Lee SC, Abdel-Wahab O. Therapeutic targeting of splicing in cancer. Nat Med. 2016;22:976–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Wang ET, Sandberg R, Luo S, et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Johnson JM, Castle J, Garrett-Engele P, et al. Genome-wide survey of human alternative pre-Mrna splicing with exon junction microarrays. Science. 2003;302:2141–2144. [DOI] [PubMed] [Google Scholar]
- [9].Dvinge H, Kim E, Abdel-Wahab O, et al. RNA splicing factors as oncoproteins and tumour suppressors. Nat Rev Cancer. 2016;16:413–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Singh B, Eyras E. The role of alternative splicing in cancer. Transcription. 2017;8:91–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].da Silva MR, Moreira GA, da Silva RA G, et al. Splicing regulators and their roles in cancer biology and therapy. BioMed Res Int. 2015;2015:150514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Belluti S, Rigillo G, Imbriano C. Transcription factors in cancer: when alternative splicing determines opposite cell fates. Cells. 2020;9(3):760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Kahles A, Lehmann KV, Toussaint NC, et al. Comprehensive analysis of alternative splicing across tumors from 8,705 patients. Cancer Cell. 2018;34:211–24 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Frampton GM, Ali SM, Rosenzweig M, et al. Activation of MET via diverse exon 14 splicing alterations occurs in multiple tumor types and confers clinical sensitivity to MET inhibitors. Cancer Discov. 2015;5:850–859. [DOI] [PubMed] [Google Scholar]
- [15].Li D, Harlan-Williams LM, Kumaraswamy E, et al. BRCA1-No matter how you splice it. Cancer Res. 2019;79:2091–2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Miki Y, Swensen J, Shattuck-Eidens D, et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science. 1994;266:66–71. [DOI] [PubMed] [Google Scholar]
- [17].Orban TI, Olah E. Expression profiles of BRCA1 splice variants in asynchronous and in G1/S synchronized tumor cell lines. Biochem Biophys Res Commun. 2001;280:32–38. [DOI] [PubMed] [Google Scholar]
- [18].de Jong LC, Cree S, Lattimore V, et al. Nanopore sequencing of full-length BRCA1 mRNA transcripts reveals co-occurrence of known exon skipping events. Breast Cancer Res. 2017;19:127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Hojny J, Zemankova P, Lhota F, et al. Multiplex PCR and NGS-based identification of mRNA splicing variants: analysis of BRCA1 splicing pattern as a model. Gene. 2017;637:41–49. [DOI] [PubMed] [Google Scholar]
- [20].Davy G, Rousselin A, Goardon N, et al. Detecting splicing patterns in genes involved in hereditary breast and ovarian cancer. Eur J Hum Genet. 2017;25:1147–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Gabrieli T, Sharim H, Fridman D, et al. Selective nanopore sequencing of human BRCA1 by Cas9-assisted targeting of chromosome segments (CATCH). Nucleic Acids Res. 2018;46:e87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Fu X, Tan W, Song Q, et al. BRCA1 and breast cancer: molecular mechanisms and therapeutic strategies. Front Cell Dev Biol. 2022;10:813457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Bekker-Jensen S, Mailand N. Assembly and function of DNA double-strand break repair foci in mammalian cells. DNA Repair (Amst). 2010;9:1219–1228. [DOI] [PubMed] [Google Scholar]
- [24].Nelson AC, Holt JT. Impact of RING and BRCT domain mutations on BRCA1 protein stability, localization and recruitment to DNA damage. Radiat Res. 2010;174:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Greenberg RC. BRCA1, everything but the RING? Science. 2011;334:459–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Rodriguez JA, Henderson BR. Identification of a functional nuclear export sequence in BRCA1. J Biol Chem. 2000;275:38589–38596. [DOI] [PubMed] [Google Scholar]
- [27].Thompson ME, Robinson-Benion CL, Holt JT. An amino-terminal motif functions as a second nuclear export sequence in BRCA1. J Biol Chem. 2005;280:21854–21857. [DOI] [PubMed] [Google Scholar]
- [28].Clark SL, Rodriguez AM, Snyder RR, et al. Structure-function of the tumor suppressor BRCA1. Comput Struct Biotechnol J. 2012;1:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Chen CF, Li S, Chen Y, et al. The nuclear localization sequences of the BRCA1 protein interact with the importin-alpha subunit of the nuclear transport signal receptor. J Biol Chem. 1996;271:32863–32868. [DOI] [PubMed] [Google Scholar]
- [30].Sy SM, Huen MS, Chen J. PALB2 is an integral component of the BRCA complex required for homologous recombination repair. Proc Natl Acad Sci U S A. 2009;106:7155–7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Williams RS, Green R, Glover JN. Crystal structure of the BRCT repeat region from the breast cancer-associated protein BRCA1. Nat Struct Biol. 2001;8:838–842. [DOI] [PubMed] [Google Scholar]
- [32].Mohammad DH, Yaffe MB. 14-3-3 proteins, FHA domains and BRCT domains in the DNA damage response. DNA Repair (Amst). 2009;8:1009–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Orban TI, Olah E. Emerging roles of BRCA1 alternative splicing. Mol Pathol. 2003;56:191–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Yuli C, Shao N, Rao R, et al. Brca1a has antitumor activity in TN breast, ovarian and prostate cancers. Oncogene. 2007;26:6031–6037. [DOI] [PubMed] [Google Scholar]
- [35].Thakur S, Zhang HB, Peng Y, et al. Localization of BRCA1 and a splice variant identifies the nuclear localization signal. Mol Cell Biol. 1997;17:444–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Wilson CA, Payton MN, Elliott GS, et al. Differential subcellular localization, expression and biological toxicity of BRCA1 and the splice variant BRCA1-delta11b. Oncogene. 1997;14:1–16. [DOI] [PubMed] [Google Scholar]
- [37].ElShamy WM, Livingston DM. Identification of BRCA1-IRIS, a BRCA1 locus product. Nat Cell Biol. 2004;6:954–967. [DOI] [PubMed] [Google Scholar]
- [38].Wick RR, Judd LM, Holt KE. Performance of neural network basecalling tools for Oxford Nanopore sequencing. Genome Biol. 2019;20:129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Li H, Birol I. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 2018;34:3094–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14:178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Adamopoulos PG, Theodoropoulou MC, Scorilas A. Alternative splicing detection tool-a novel PERL algorithm for sensitive detection of splicing events, based on next-generation sequencing data analysis. Ann Transl Med. 2018;6:244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Holmqvist I, Backerholm A, Tian Y, et al. FLAME: long-read bioinformatics tool for comprehensive spliceome characterization. RNA. 2021;27:1127–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Tian L, Jabbari JS, Thijssen R, et al. Comprehensive characterization of single-cell full-length isoforms in human and mouse with long-read sequencing. Genome Biol. 2021;22:310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Artimo P, Jonnalagedda M, Arnold K, et al. ExPASy: sIB bioinformatics resource portal. Nucleic Acids Res. 2012;40:W597–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Nagy E, Maquat LE. A rule for termination-codon position within intron-containing genes: when nonsense affects RNA abundance. Trends Biochem Sci. 1998;23:198–199. [DOI] [PubMed] [Google Scholar]
- [46].Kulmanov M, Hoehndorf R. DeepGOPlus: improved protein function prediction from sequence. Bioinformatics. 2021;37:1187–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Wen J, Toomer KH, Chen Z, et al. Genome-wide analysis of alternative transcripts in human breast cancer. Breast Cancer Res Treat. 2015;151:295–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Bianchi JJ, Zhao X, Mays JC, et al. Not all cancers are created equal: tissue specificity in cancer genes and pathways. Curr Opin Cell Biol. 2020;63:135–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Amarasinghe SL, Su S, Dong X, et al. Opportunities and challenges in long-read sequencing data analysis. Genome Biol. 2020;21:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Guo W, Hu Z, Bao Y, et al. A LIN28B tumor-specific transcript in cancer. Cell Rep. 2018;22:2016–2025. [DOI] [PubMed] [Google Scholar]
- [51].Hu W, Wu Y, Shi Q, et al. Systematic characterization of cancer transcriptome at transcript resolution. Nat Commun. 2022;13:6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Kochetov AV, Ahmad S, Ivanisenko V, et al. uOrfs, reinitiation and alternative translation start sites in human mRnas. FEBS Lett. 2008;582:1293–1297. [DOI] [PubMed] [Google Scholar]
- [53].Orr MW, Mao Y, Storz G, et al. Alternative ORFs and small ORFs: shedding light on the dark proteome. Nucleic Acids Res. 2020;48:1029–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Meydan S, Marks J, Klepacki D, et al. Retapamulin-assisted ribosome profiling reveals the alternative bacterial proteome. Molecular Cell. 2019;74(3):481–493.e6. DOI: 10.1016/j.molcel.2019.02.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Hsu PY, Calviello L, Wu HL, et al. Super-resolution ribosome profiling reveals unannotated translation events in Arabidopsis. Proc Natl Acad Sci U S A. 2016;113:E7126–E35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Fritsch C, Herrmann A, Nothnagel M, et al. Genome-wide search for novel human uOrfs and N-terminal protein extensions using ribosomal footprinting. Genome Res. 2012;22:2208–2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Shell SS, Wang J, Lapierre P, et al. Leaderless transcripts and small proteins are common features of the mycobacterial translational landscape. PLoS Genet. 2015;11:e1005641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Brunet MA, Leblanc S, Roucou X. Reconsidering proteomic diversity with functional investigation of small ORFs and alternative ORFs. Exp Cell Res. 2020;393:112057. [DOI] [PubMed] [Google Scholar]
- [59].Wiesner T, Lee W, Obenauf AC, et al. Alternative transcription initiation leads to expression of a novel ALK isoform in cancer. Nature. 2015;526:453–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Ragan C, Goodall GJ, Shirokikh NE, et al. Insights into the biogenesis and potential functions of exonic circular RNA. Sci Rep. 2019;9:2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Furuno M, Kasukawa T, Saito R, et al. CDS annotation in full-length cDNA sequence. Genome Res. 2003;13:1478–1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Tate JG, Bamford S, Jubb HC, et al. COSMIC: the catalogue of somatic mutations in cancer. Nucleic Acids Res. 2019;47:D941–D7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Abeliovich D, Kaduri L, Lerer I, et al. The founder mutations 185delAG and 5382insC in BRCA1 and 6174delT in BRCA2 appear in 60% of ovarian cancer and 30% of early-onset breast cancer patients among Ashkenazi women. Am J Hum Genet. 1997;60:505–514. [PMC free article] [PubMed] [Google Scholar]
- [64].Wang F, Fang Q, Ge Z, et al. Common BRCA1 and BRCA2 mutations in breast cancer families: a meta-analysis from systematic review. Mol Biol Rep. 2012;39:2109–2118. [DOI] [PubMed] [Google Scholar]
- [65].Mullen P, Miller WR, Mackay J, et al. BRCA1 5382insC mutation in sporadic and familial breast and ovarian carcinoma in Scotland. Br J Cancer. 1997;75:1377–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Mylavarapu S, Das A, Roy M. Role of BRCA mutations in the modulation of response to platinum therapy. Front Oncol. 2018;8:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Jiang MC, Ni JJ, Cui WY, et al. Emerging roles of lncRNA in cancer and therapeutic opportunities. Am J Cancer Res. 2019;9:1354–1366. [PMC free article] [PubMed] [Google Scholar]
- [68].Zhou Y, Zhong Y, Wang Y, et al. Activation of p53 by MEG3 non-coding RNA. J Biol Chem. 2007;282:24731–24742. [DOI] [PubMed] [Google Scholar]
- [69].Betts JA, Moradi Marjaneh M, Al-Ejeh F, et al. Long noncoding RNAs CUPID1 and CUPID2 mediate breast cancer risk at 11q13 by modulating the response to DNA damage. Am J Hum Genet. 2017;101:255–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The nucleotide sequences of the BRCA1 mRNA transcripts described in the present study (BRCA1 sv.7 – sv.52) have been deposited to the GenBank® database (https://www.ncbi.nlm.nih.gov/genbank/) and correspond to the accession numbers OP156938 – OP156983, accordingly. The datasets related to the present work are available in the Sequence Read Archive (SRA) repository, BioProject ID: PRJNA899869.


