Abstract
The Bone Morphogenetic Protein (BMP) signaling pathway has established roles in early embryonic morphogenesis, particularly in the epiblast. More recently, however, it has also been implicated in development of extraembryonic lineages, including trophectoderm (TE), in both mouse and human. In this review, we will provide an overview of this signaling pathway, with a focus on BMP4, and its role in emergence and development of TE in both early mouse and human embryogenesis. Subsequently, we will build on these in vivo data and discuss the utility of BMP4-based protocols for in vitro conversion of primed vs. naïve pluripotent stem cells (PSC) into trophoblast, and specifically into trophoblast stem cells (TSC). PSC-derived TSC could provide an abundant, reproducible, and ethically acceptable source of cells for modeling placental development.
Keywords: Bone Morphogenetic Protein, Trophectoderm, Inner cell mass, Epiblast, Cytotrophoblast, Trophoblast stem cells, Primed pluripotency, Naïve pluripotency
Introduction
The Bone Morphogenetic Protein 4 (BMP4) is a secreted growth factor of the Transforming Growth Factor beta (TGFB also known as TGFβ) super-family. While initially described for its role in bone formation, BMP signaling has been implicated in the development, maintenance, and regeneration of numerous tissues and organs. It plays a role in various developmental processes, such as cell growth, apoptosis, and differentiation. In this review, we will discuss the role of BMP4 in the emergence of the trophoblast lineage from human pluripotent stem cells (PSC), in the context of the current knowledge of its role in early embryo development. While the focus of this controversy has surrounded human PSC, we will also discuss literature from other species, particularly mouse, where evidence supports a similar role for this ligand in expansion of pluripotency. Although the focus of this review is on signaling pathways mediated through BMP4, as the growth factor most commonly used in these experiments, we will briefly discuss related ligands in this family, but refer the reader to the following reviews for a broader overview of BMP signaling [1–4].
A brief overview of signaling pathways directed by BMP4
BMP4 is a growth factor activating a specific portion of the TGFB signaling pathway (Fig. 1), which is comprised of the BMP and TGFB/INHBA (also known as Activin) branches. There are 20 bone fide BMP ligands, some of which are referred to as Growth and Differentiation Factors (GDFs), classified in sub-groups based on sequence similarity and affinities for specific receptors [3, 4]. BMP/GDF receptors are transmembrane serine/threonine kinases, separated into five type I (ALK1/ACVRL1, ALK2/ACVR1, ALK3/BMPR1A, ALK4/ACVR1B, and ALK6/BMPR1B), and three type II (BMPR2, ACVR2A, and ACVR2B) receptors, with some type II receptors in common with the TGFB/Activin branch (Fig. 1) [2]. BMP2 and BMP4 show higher affinity for the type I receptors, ALK3/BMPR1A and ALK6/BMPR1B, and type II receptor, BMPR2, although binding to ACVR2A and ACVR2B has also been reported [2, 3]. BMP4 binds type I receptor homodimers that subsequently recruit type II homodimers forming a tetramer complex. From this initial interaction, two types of signaling pathways can be activated, referred to as “canonical”, or SMAD-dependent, and “non-canonical”, or SMAD-independent. In the “canonical” pathway, recruitment of type II receptors, with their constitutive kinase activity, allows the phosphorylation of Type I receptors at serine/threonine residues, which in turn use their kinase catalytic activities to phosphorylate receptor-regulated SMAD (R-SMAD) 1/5/8 proteins. Activated SMAD-1/5/8 bind to SMAD4, which is common to both branches of the signaling pathway, and translocate into the nucleus where they mediate the transcription of BMP-specific target genes, either directly, by binding to SMAD-binding elements (SBE), or indirectly, through interactions with other DNA-binding transcription factors and histone-modifying factors [5].
Fig. 1.
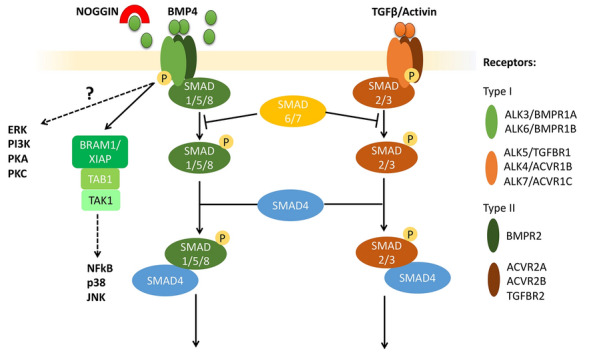
BMP and TGFB/INHBA (Activin) branches of the TGFB signaling pathway. Ligands of the TGFB super-family activate Type I and Type II receptors, transmembrane serine/threonine kinases, classified into five main type I (ALK3/BMPR1A, ALK4/ACVR1B, ALK5/TGFBR1, ALK6/BMPR1B, and ALK7/ACVR1C), and three type II (BMPR2, ACVR2A, and ACVR2B) receptors. BMP4 specifically binds homodimers of ALK3/BMPR1A or ALK6/BMPR1B, which subsequently recruit type II homodimers forming a tetramer complex. This interaction leads to the activation of both the “canonical”, or SMAD-dependent, and the “non-canonical”, or SMAD-independent, signaling pathways. In the canonical pathway, recruitment of type II receptors leads to the phosphorylation of Type I receptors, which then phosphorylate receptor-regulated SMAD (R-SMAD) 1/5/8 proteins. Activated SMAD-1/5/8 binds to SMAD4 and translocates into the nucleus, where it mediates the transcription of BMP-specific target genes. The “non-canonical” pathway activates various signaling cascades in a SMAD-independent manner, including those involving ERK, TAK1-p38, PI3K/AKT, and PKC. The SMAD-independent activation of TAK1, upstream of p38, JNK, and NFkB, is known to be mediated by TAB1 (TAK-binding protein 1) via the complex with BRAM1 (BMP Receptor Associated Molecule 1) or XIAP (X-linked inhibitor-of-apoptosis protein). NOGGIN is a well-known inhibitor of the BMP signaling pathway, acting by sequestering ligands and preventing their interaction with receptors. TGFB and Activin bind to specific Type I and II receptors, which phosphorylate R-SMAD 2/3. Inhibitory SMAD 6/7 interacts with both branches by preventing phosphorylation of R-SMADs
BMP signaling can also trigger additional signaling pathways. This includes pathways involving β-catenin (CTNNB1)-dependent WNT and NOTCH [6, 7]. BMP and TGFB-activated SMADs also regulate the biosynthesis and processing of a number of microRNAs [8, 9]. In addition, the “non-canonical” pathway activates various signaling cascades in a SMAD-independent manner, including those involving ERK, TAK1-p38, PI3K/AKT, and PKC [10, 11] (Fig. 1). In turn, these signaling pathways have downstream effects, with only a portion operating through transcriptional changes. The SMAD-independent activation of TAK1, upstream of p38, JNK, and NFkB, is known to be mediated by TAB1 (TAK-binding protein 1) via the complex with BRAM1 (BMP Receptor Associated Molecule 1) or XIAP (X-linked inhibitor-of-apoptosis protein) [10]. However, overall, the exact mechanisms activating other SMAD-independent pathways are poorly understood, though they mostly follow activation of type I receptors and are likely cell-context dependent [11].
BMP4 signaling is finely tuned by different proteins both in the intracellular and extracellular space [12, 13]. In the cytoplasm, inhibitory SMAD (I-SMADs) proteins 6 and 7 prevent activation of SMAD1/5/8 while promoting their interaction with SMAD ubiquitination regulatory factor (SMURF) proteins, thus mediating SMAD degradation and inhibiting SMAD-dependent pathways [1]. In the extracellular compartment, co-receptors, such as Betaglycan (TGFBR3, a cell surface proteoglycan), promote ligand/receptor interaction, thus enhancing BMP signaling, while BAMBI, a cell-membrane pseudo-receptor, sequesters ligands from the extracellular environment [1, 2]. Moreover, there are 15 known secreted natural antagonists, classified in 3 sub-groups based on the size of the cystine knot motif [2]. One of the most studied is NOGGIN, whose crystal structure with BMP7 has revealed a butterfly configuration that blocks the ligand epitopes responsible for binding both type I and type II receptors, thus working as a ligand trap [14]. NOGGIN, as well as other antagonists, are often induced downstream of BMP signaling as a form of negative feedback to mitigate excessive BMP activation. However, these inhibitors show ligand-specific affinities as, for example, BMP4 appears to be significantly more susceptible to inhibition by NOGGIN than BMP10 [15]. Finally, various synthetic small molecule inhibitors have been designed to block specific portions of the TGFB signaling cascade. These include dorsomorphin, LDN-193189, and DMH1, which inhibit the kinase activity of BMPR1A/ALK3 and BMPR1B/ALK6 on the BMP branch [16–18], A83-01 targeting ALK5 on the TGFβ/Activin arm [19], and 5Z-7-oxozeaenol and TAKinhib blocking the TAK1-mediated non-canonical pathway [20–22]. Many of the naturally occurring and synthetic agonists and inhibitors mentioned above have been used in context of both embryonic and trophoblast stem cells, as well as embryos, to probe the role of BMP signaling in trophoblast lineage specification and differentiation; some of these data are described in sections below.
BMP signaling in early embryonic development
BMP signaling has long been known to play a key role in early embryonic morphogenesis. In particular, BMP2 and BMP4 have been identified as the two key morphogens in the early mouse embryo (Fig. 2). In addition to cardiac development, BMP2 is required for closure of the proamniotic canal and proper patterning of amnion and chorion, and, interestingly, despite a 92% homology, BMP4 cannot replace BMP2 in this setting [23]. BMP4, on the other hand, has distinct roles in early mouse embryogenesis, with null embryos lacking primordial germ cells (PGC) and allantois [24], but also showing defects in gastrulation and mesoderm formation [25]. Initially, it is the BMP4 produced by extraembryonic ectoderm (ExEc, or trophectoderm-derived cells of the embryo) that is needed for establishment of both PGC and allantois; however, PGC localization/survival, allantois differentiation, and post-implantation embryonic development all require BMP4 that is derived from extraembryonic mesoderm (ExM) [26, 27]. ExEc-derived BMP4 also contributes to extraembryonic endoderm (ExEn, or yolk sac) patterning, contributing specifically to formation of anterior visceral endoderm [28–30].
Fig. 2.
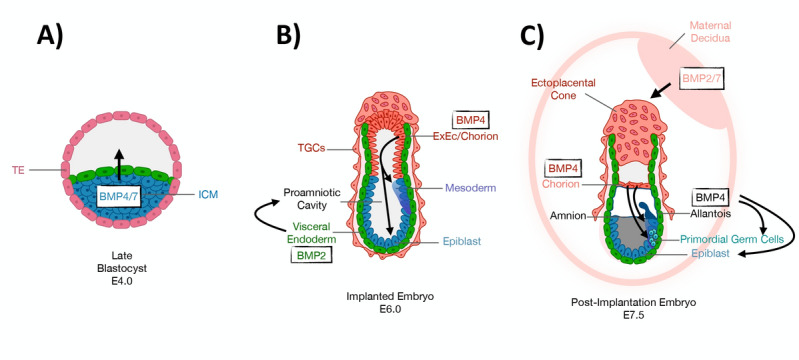
BMP signaling in early mouse embryonic morphogenesis. A Late blastocyst stage (E4.0), with BMP4/7 ligands arising from the inner cell mass (ICM, blue) and signaling toward the Trophectoderm (TE, pink), so-called “inside-out” (ICM-to-TE) BMP signaling prior to implantation. B Egg cylinder stage (E6.0) mouse embryo, showing the switch to “outside-in” BMP signaling in the peri-implantation period. Mesoderm induction is triggered by BMP4 signaling from the chorion/extraembryonic ectoderm (ExEc, dark orange) to the epiblast (blue). BMP2 originating from the visceral endoderm (green) drives the formation of the proamniotic cavity. C Post-implantation stage (E7.5) embryo with continued outside-in BMP signaling. BMP4, derived from the chorion (light red) continues to signal to the epiblast (blue), driving emergency of primordial germ cells (PGC, light green) and extraembryonic mesoderm/allantois (black) formation. BMP2 drives proper patterning of amnion (black) and chorion (pink). BMP2/7 from the surrounding decidua (light pink) signal to the ectoplacental cone (pink), regulating implantation
The above clearly suggest a role for BMP signaling in early development and patterning, particularly of the epiblast; however, there is also significant data supporting a role for this signaling pathway in patterning of the extraembryonic compartments, including trophectoderm (TE) (Fig. 2). As mentioned above, BMP2-null embryos fail to close the proamniotic canal, leading to abnormal patterning of both the amnion and chorion [23] and absence of chorioallantoic fusion [31]. Both BMP2 and -4 signal through BMPR1A, whose loss results in peri-implantation lethality [32]. While this phenotype has been mostly attributed to absence of mesoderm, a closer evaluation, particularly of the expression of EOMES, which in the mouse is a marker of both mesoderm and ExEc, reveals loss of both these compartments in the null embryo [32]. This latter paper also clearly shows active BMP signaling (in the form of phosphorylated SMAD1/5/8) within the ExEc compartment of the egg cylinder-stage mouse embryo [32]. Loss of SMAD1, a BMP signaling effector, also affects the patterning of ExEc, leading to paradoxical expansion of this compartment as shown by EOMES and BMP4 expression, suggesting a role for BMP signaling in initiation of differentiation [33].
There is also a significant body of evidence pointing to a possible role for BMP4 signaling in the pre-implantation embryo (Fig. 2). Specifically, Graham et al. [34] have shown that in the first wave of mouse embryonic asymmetric cell divisions (the 16-cell stage), the outer cells, which are progenitors of TE, are enriched for BMPR2 and show phosphorylation of SMAD1, while the inner cell mass (ICM) cells express BMP4 and -7. Inhibition of BMP signaling, using a dominant-negative form of either SMAD4 or BMPR2, or through small molecular inhibitors (including dorsomorphin and oxozeanol), led to the disruption of both TE and the primitive endoderm (PrE) [34]. Conversely, Home et al. [35] have shown that in vitro treatment of mouse embryos with BMP4 causes the embryos to arrest at the morula stage. BMP4 appeared to induce nuclear localization of the transcription factor TEAD4, leading to a TE-specific transcriptional program, including induction of CDX2 expression in the ICM [35]. Finally, Rivron et al. [36] used “Blastoids,” formed from combination of mESC and mTSC, to identify a role for mESC-derived BMP4 in TE morphogenesis, mediated through the BMP-inducible gene KLF6 within mTSC. These results suggest a role for inside-out (ICM-to-TE) BMP signaling in the correct development of TE, prior to implantation, with a subsequent switch to outside-in (from TE-derived ExEc to ICM-derived epiblast) BMP signaling in the post-implantation period, leading to PGC and mesoderm induction. While the latter processes have been significantly better studied, the former deserves more attention, in the context of both mouse and human embryonic development.
With respect to the human embryo, significantly less has been done compared to the mouse, for multiple reasons, until recently. The advent of single cell RNA sequencing (scRNAseq) platforms, as well as the development of optimized conditions for extended culture of the human embryo [37, 38], have led to an explosion of research in this area. Data from these studies have also pointed to an enrichment of BMP signaling components in the TE of the human embryo, including expression of BMPR1/2 and SMAD1/5 [39]. However, to-date, no direct evaluation of BMP signaling on intact human embryos has been done.
BMP signaling in post-implantation placenta and at the maternal–fetal interface
BMP ligands and receptors have also been studied in the embryo-endometrium interface. BMP2 is required for decidualization of the endometrial stroma in mice [40], while BMP7 ablation causes defects in endometrial receptivity and implantation [41]. Uterine-specific knockout (achieved through use of Pgr-Cre mice) of both BMPR1A and BMPR2 result in female infertility, the former due to defects in endometrial receptivity and embryo attachment, and the latter due to abnormalities in decidualization and placentation [42, 43]. Similarly, in human, BMP2 protein has been detected in decidualized human endometrial stromal cells, with exogenous administration significantly enhancing decidualization in vitro [44]. This process is mediated through BMPR1A and SMAD1/5, and inhibited by dorsomorphin treatment [45]. BMP7 has also been implicated in implantation, with mutations in the gene identified in patients experiencing recurrent pregnancy loss [46].
BMP ligands and receptors have not been studied in detail in the mouse or human placenta. However, using a mouse STEMbryo model, where mouse embryonic and trophoblast stem cells have been combined to generate an embryo-like structure, Zernicka-Goetz’ group has confirmed SMAD1 signaling within the TE component of the STEMbryo [47] and more recently shown that BMP signaling is required to maintain this compartment [48]. In the human placenta, BMP2 has been identified in both decidua and trophoblast of the first trimester placenta and implicated in trophoblast function [49].
BMP signaling in pluripotent and trophoblast stem cells
BMP signaling components are expressed in both mouse and human pluripotent stem cells (PSC) [50, 51], emphasizing the potential for these cells to respond to BMPs either in an autocrine or paracrine manner. The effect of BMP signaling in these cells, the core of this review, is discussed further below.
Significantly less is known about BMP signaling in trophoblast stem cells (TSC). Au et al. [52] recently studied this in both mouse and human TSC, finding that both express various BMP ligands, including BMP4, BMP7, and low levels of BMP2, the receptors BMPR1A and BMPR2, and the BMP effector proteins SMAD1 and SMAD5 [52]. Interestingly, the transcription factors GATA2 and -3, which are implicated in TSC self-renewal, have been shown to directly regulate BMP4 expression in mouse TSC [53]. BMP signaling is highest in the undifferentiated state, decreasing with differentiation [52]. Mouse and human TSC show variable responses to BMP inhibition in the undifferentiated state: the non-canonical inhibitor 5Z-7-oxozeaenol has the strongest effect, downregulating TSC stemness markers and upregulating differentiation markers in the mouse, with significantly more modest effects in human TSC [52]. Interestingly, however, continuous treatment of both mouse and human TSC with BMP4 in the presence of differentiation-inducing media blunted the terminal differentiation process, affecting both labyrinthine and junctional zone markers equally in mouse TSC, but with a stronger moderating effect on EVT (rather than STB) differentiation in human TSC [52]. These data are consistent with a role for autocrine BMP signaling within the trophoblast compartment of the post-implantation embryo, as also recapitulated by the mouse STEMbryo model [48].
Established functions of BMP signaling in pluripotent stem cells
In this section, we will describe the more established roles of BMP signaling in pluripotent stem cells (PSC), including both embryonic (ESC) and induced pluripotent stem cells (iPSC). For the most part, we will discuss findings from both mouse and human PSC, though at times, PSC from other species will also be discussed.
The role of BMP4 in maintaining pluripotency: derivation of primed vs. naïve PSC
The original culture of mouse ESCs employed irradiated mouse embryonic fibroblasts (MEF) to provide a feeder layer and a medium supplemented with fetal bovine serum (FBS) as a source of other necessary factors for maintaining the pluripotent state [54]. Over time, this formula was simplified, following the discovery that the main active component controlling self-renewal in this complex medium was leukemia inhibitory factor (LIF), a cytokine supplied by MEFs [55]. Later, it was discovered that FBS could be replaced by adding either BMP4, BMP2, or GDF6 [56], although the former became the reagent of choice, presumably because of its availability rather than rational choice based on known physiology. Without LIF present, however, BMP4 can neither promote self-renewal nor prevent spontaneous differentiation of mESC. The role of BMP4 in enhancing stemness has been attributed both to its ability to upregulate Inhibitor of differentiation (ID) proteins, which are a family of negative transcriptional regulators, and to inhibit mitogen activated protein kinase (MAPK) pathways [57]. In particular, BMP4 appears to help preserve the potential of the ESC for multilineage differentiation, possibly through regulating their epigenetic state [58]. In other words, BMP/SMAD/ID and LIF/STAT3 signaling in these so-called naïve state mESC may have more to do with protecting pluripotency than maintaining self-renewal [59]. Indeed, it was found that mESC could be kept as self-renewing entities, without the external presence of either LIF, BMP4 or any other extrinsic factor, as long as their propensity for differentiation was minimized by inclusion of inhibitors of MAPK and glycogen synthase kinase 3 (so-called “2i medium”) [59]. These cells exhibit phenotypic uniformity in culture and have a higher pluripotent potential than the ESC grown on more complex media. They appear to exist in a self-sufficient ground state, now most commonly referred to as naïve pluripotency [60].
When ESC were first generated from monkey [61] and human [62] blastocysts by Thomson and his colleagues in the late 1990s, it was evident that they differed markedly from mESC in a number of respects. Most obviously, their colonies were more flattened, and the cells could not be readily propagated using trypsin dispersal, needing instead to be passaged in small clumps. They had no requirement for LIF but exhibited self-renewal with minimal differentiation when cultured on a complex medium supplemented with serum and a feeder layer of irradiated MEFs. Over time, defined media were formulated that supplied basic FGF (FGF2), and Activin A (a homodimer of INHBA), which could be substituted by the related cytokines NODAL and TGFB1 [63–66]. These human ESC (hESC) expressed some of the same core transcription factors, including SOX2, POU5F1, and NANOG, as the mouse counterparts [67], but there were also major differences in their transcriptome profiles. The hESC were clearly pluripotent, based on their ability to form well-differentiated teratomas in immunodeficient mice and to differentiate along the three major germ-line lineages both in standard 2D-cultures [68] and when aggregated into embryoid bodies [69]. One curious feature, however, was that under certain differentiation protocols, namely exposure to BMP4 and certain of its relatives, they released human chorionic gonadotrophin (hCG) into the culture medium and displayed a set of marker genes characteristic of placental trophoblast [70]. Human ESC also appeared to spontaneously differentiate into hCG-secreting cells, both in 2D culture [71] and within 3D embryoid bodies [72, 73]. We will address the role of BMP4 in trophoblast emergence from pluripotent stem cells in a subsequent section below, as it has been a subject of considerable controversy.
The conundrum raised by the apparent existence of two kinds of pluripotent stem cells, one in mouse and the other in primates, was at least partially solved when mouse cells showing many of the phenotypic features of hESC were isolated from post-implantation epiblast of mouse embryos [74, 75]. These cells have been named primed-type mESC or mEpiSC on the basis of their origin from epiblast, rather than inner cell mass (ICM). Like hESC, mEpiSCs showed no requirement for LIF but had a similar dual dependency on Activin A-related growth factors and FGF2, and possessed similar transcriptomes. Unlike naïve-type mESC, when injected into blastocyst-stage embryos, mEpiSC were incapable of colonizing the ICM and contributing to chimeric offspring; nevertheless, it was subsequently shown that mEpiSC could, in fact, contribute to chimeric offspring when injected into post-implantation stage embryos, from which they originate [76]. mEpiSC also exhibit differences in epigenetic characteristics, including an overall higher degree of DNA methylation, as well as X-chromosome inactivation, compared to naïve state cells [77]. It was soon demonstrated that the ground/naïve state and primed/epiblast states of mESC were in fact interconvertible, if appropriate selective culture conditions were employed [78]. More recently, homologous human naïve/ground state ESCs have been created [79, 80]; however, in both species, a phased progression between the two pluripotency states has been demonstrated, indicating that neither is a fixed configuration [81].
The role of BMP4 in early differentiation of embryonic lineages
The more established roles for BMP signaling in differentiation of pluripotent stem cells have been extrapolated from studies in the mouse embryo. As described above, this signaling pathway has been most well-studied in context of induction of mesendoderm [82], particularly the hematopoietic lineage [83], and specification of primordial germ cells [84]. Its role has also been well-described in derivation of epidermal lineage cells (keratinocytes) from pluripotent stem cells [85]. The role of BMP4 in induction of these lineages and associated protocols, starting with both mouse and human pluripotent stem cells, have been reviewed elsewhere [86, 87] and thus will not be discussed here in detail. However, several points will be mentioned here, due to the direct relevance to the discussion of trophoblast induction below. First, as correctly pointed out by Bernardo et al. [88] CDX2 is expressed in both early trophoblast and mesendoderm precursor cells, and as such, should not be used on its own to indicate trophoblast specification. In fact, there is not a single gene, but rather, a constellation of genes and functions that should be used to confirm trophoblast identity. However, the selection of such “trophoblast-associated” genes/gene networks should be based on the species being probed: as an example, Bernardo et al., wrongly claimed that the absence of EOMES in BMP4-treated hPSC is consistent with their non-trophoblast identity. While EOMES is required for establishment of both early trophoblast and mesoderm in the mouse [89], it is completely absent from early human trophoblast, both pre- and post-implantation [39, 90]. Third, if there is one lesson to be drawn from these numerous studies on the various functions of BMP signaling, it is that the effect of this growth factor on pluripotent stem cells is highly context-specific. Some of these cell- and environment-specific contexts have been described, both in vivo and in vitro; the most relevant of these to trophoblast induction is the simultaneous presence of other growth factors, particularly FGF and WNT signaling, which will be discussed further below.
In addition to the aforementioned lineages, the role of BMP signaling in amnion formation and patterning deserves mention here, as multiple groups have claimed this to be the identity of BMP4-treated primed hPSC (see below). As discussed above, BMP2 is required for proper patterning of amnion and chorion [23]. However, in vitro, hPSCs arranged in three-dimensional structures in Geltrex under pluripotency conditions upregulate expression of BMP2, -4, and -7, and self-organize into a squamous cyst-like structure identified as amnion [91]. This process is interrupted in the presence of SMAD-dependent BMP inhibitors, suggesting induction of endogenous BMP signaling as the primary mechanism of amniogenesis [91]. The same group has developed a more elegant, microfluidic-based device, for modeling hPSC-derived amniogenesis, in which the hPSC-derived cyst, exposed to BMP4 only on one side, forms an asymmetrically patterned disc, with a single layer of flattened amnion-like cells on the BMP4-exposed side and a stratified epithelium resembling epiblast on the non-BMP4-exposed side [92]. Interestingly, many of the markers used to confirm amnion identity in this setting are also markers of human villous cytotrophoblast (vCTB), including TP63, GATA2 and -3, and KRT18 [91–93]; however, at least in the context of the microfluidic device, in addition to the flattened morphology of the cells, induction of PGC-like cells at the junction of the amnion-like and epiblast-like regions of the cyst, and subsequent induction of mesoderm within the epiblast-like region of the cyst, confirm the ability to recapitulate peri-gastrulation events, including amniogenesis, by asymmetric application of BMP4 to hPSC [92]. Nevertheless, this still leaves open the problem of distinguishing trophoblast and amnion identity in the more commonly used two-dimensional context of BMP4-treated hPSC. In such analyses, the distinct developmental origins of the amnion should be kept in mind: specifically, that amnion formation occurs within the peri-gastrulation mouse embryo, but in the pre-gastrulation primate embryo [94]. While there is some single cell RNAseq data for amnion from cynomolgus monkey embryos [95], the datasets most commonly used for distinguishing these lineages in the human have been those of Xiang et al. [96], from extended culture human embryos, and Roost et al. [97], from first and second trimester human amnion. However, both datasets require further validation, particularly as Xiang et al., identified very few such cells (2 cells at day 12 and 11 cells at day 14 of cultured human embryos) and Roost et al.’s study lacked morphologic confirmation of their collected tissues.
Role of BMP4 in induction of trophoblast from pluripotency
There is a large amount of literature in this area, and therefore, we will first discuss data from mouse, and subsequently, human PSC. In each section, we will discuss potential mechanisms through which BMP4 may be mediating trophoblast induction, limitations of the experimental design, and future studies needed to address these limitations and resolve discrepancies between different studies.
BMP4-mediated trophoblast differentiation of mouse PSC
TE specification in the mouse occurs exclusively in the pre-implantation stage, when a combination of signals establishing cell polarity drives the asymmetric division of 8-cell-stage blastomeres [98], leading to specification of inside vs. outside cells. The outside cells turn off Hippo signaling, leading to induction of Cdx2 and TE specification [99]. It is generally accepted that TE specification in the mouse is completed by the blastocyst stage, as very few mESC contribute to TE-derived portions of the placenta when injected into blastocysts [100, 101]. One elegant study has used single cell RNAseq to better define lineage segregation in the mouse embryo, concluding that some ICM cells in the early blastocyst retain the ability to generate TE, and that cellular plasticity in mouse blastomeres is, in fact, linked to the window of sensitivity to Hippo signaling [102].
Once segregated, the mouse epiblast signals to the TE through FGF4, leading to expansion of polar TE, the TE cells closest to the epiblast [103]. In fact, mouse trophoblast stem cells (mTSC) were first derived, from either mouse pre-implantation blastocysts or early post-implantation trophoblast, by using a combination of FGF4, heparin (which stabilizes FGF4), and a feeder layer (or feeder-conditioned media), later shown to be providing activin/TGFB [104, 105]. Unlike mouse ESC, which are mostly excluded from trophoblast compartment of the mouse placenta [100, 101], mTSCs contribute solely to this compartment when injected into blastocyst-stage embryos [104]. Since then, more defined conditions have been established for mTSC derivation and self-renewal, although differentiation of these cells still requires a serum-containing medium [106]. Both the standard and defined (TX) media have been used in maintaining primary TSC and TSC reprogrammed from mouse fibroblasts, using a combination of trophoblast-associated transcription factors Eomes, Tfap2c, and Gata3, and either Ets2 [107] or Myc [108].
mESC conversion to mTSC was first described through knockdown of Pou5f1 (a.k.a.Oct4) [109], and later, through forced overexpression of Cdx2, confirmed by the ability of these cells to contribute to the trophoblast portion of the placenta in vivo [110]. Subsequently, trophoblast-like cells were shown to arise from mESC by culture on various extracellular matrices, including gelatin [111] and collagen IV [112], as well as by BMP4 treatment of mESC cultured in defined media on laminin [113]. Interestingly, culture of mESC on laminin was shown to first induce an epiblast-like state, based on both morphology and upregulation of Fgf5, with upregulation of trophoblast-associated gene expression (including Gata3, Dlx3, and Mash2) following BMP4 treatment [113]. As mentioned above, when mouse epiblast stem cells (mEpiSC) were first derived from post-implantation embryos, they were described as being more akin to human ESC, based on their ability to make trophoblast-like cells in response to BMP4 [74, 75]. However, the trophoblast identity of these cells, whether derived from mESC (cultured in serum + LIF media) or mEpiSC, remained questionable, as they were only ever characterized based on in vitro assessment, including morphology and gene expression, or at best, by hemorrhagic tumor formation assay in immunocompromised mice; chimeric contribution to the trophoblast compartment in vivo was never published.
Since the identification of naïve (mESC cultured in 2i + LIF) vs. primed (mESC cultured in serum + LIF, or mEpiSC cultured in FGF2/activin) pluripotency, multiple groups have reported some level of heterogeneity in these different culture conditions, including different proportions of cells with some level of “totipotency.” The latter has been variously defined, as cells which transiently de-repress murine endogenous retroviral (MERVL) elements, thus resembling cells in the two-cell (2C) stage of the mouse embryo [114], and/or cells which induce the endoderm marker Hex [115]. In these studies, such cells showed an ability to give rise to TSC in vitro, but also to contribute to trophectoderm in vivo [114, 115].
Recently, several groups have identified a role for BMP4 in primed-to-naïve conversion of mESC, proposing several mechanisms, including enhancing responsiveness of mEpiSC to LIF [116], increasing the activity of autotaxin, an extracellular LPA-producing enzyme [117], and chromatin remodeling [118]. At least one group has expanded this work, showing that in 3D cultures BMP4 is even able to induce mEpiSC to convert to a 2C-like state, as defined by activation of the MERVL reporter, and to give rise to implantation-competent blastocyst-like structures that contain trophoblast-like cells [119, 120]. Nevertheless, these cysts fail to give rise to bona fide mTSC, and do not progress much beyond an egg cylinder-like stage in vivo [119]. Interesting, a recent small molecule screen of media additives for generation of synthetic embryos (STEMbryos), made from a combination of mouse iPSC and TSC, identified BMP4 as a factor that best promoted formation of morphologically high-grade structures [121].
Still, to-date, both the ability of mESC to be fully reprogrammed into mTSC without genetic manipulation and the potential role of BMP4 in this process, as well as in early embryogenesis, remain to be fully elucidated. Most recently, even the studies showing expanded potential/totipotency of mESC under specific conditions [122, 123], have been challenged by Posfai et al. [124], who have suggested more stringent criteria for defining totipotency. Specifically, they suggest including a morula aggregation assay with all such cells, followed by assessment of the developing embryo at blastocyst, egg cylinder, and post-chorioallantoic fusion-stage placenta, for contribution to TE-derived structures [124]. However, given the required technical expertise and low efficiency of these assays, even with wild-type mTSC [104, 125], identification of a set of totipotency-associated genes and/or development of ancillary assays, such as stem cell aggregation into “STEMbryos” [47], may help standardize such assessment, not just in mouse, but also in the human embryo.
BMP4-mediated trophoblast differentiation of human PSC
Unlike the mouse, TE specification in human is delayed, as shown by the ability of outer cells in pre-implantation embryos to contribute to the ICM when repositioned to the center of the embryo [126]. At least partly due to these, and other, differences between early development of mouse and human embryos [127], derivation of bona fide human TSC (hTSC) did not quickly follow mouse TSC derivation. These differences converged on the required culture conditions, with the major discrepancy being in FGF signaling within the early embryo [128, 129]. hTSC can be isolated from both TE at the blastocyst stage and first trimester vCTB, under conditions that promote EGF and WNT and inhibit TGFB signaling, without any FGF signaling [129]. This publication has been a huge step forward for studies on human placental development, although such studies still have serious limitations due to restricted availability of embryos and early gestation placental tissues, as well as associated ethical objections and technical constraints. In addition, since the protocol is mainly applied to tissues where pregnancy has been interrupted early in gestation, it cannot be used to generate TSC linked to particular placental disease states, such as preeclampsia, which cannot be readily diagnosed until relatively late in gestation. To avoid the limitations of accessibility and to obtain TSC lines from more diverse genetic sources, the use of hPSC, including both embryonic and induced pluripotent stem cells, is a must. In fact, it has recently become possible to derive TSC from hPSC, including from both primed and naïve-type hPSC (discussed below, see Table 1). Additionally, most recently, two groups have reported reprogramming somatic cells directly to TSC [130, 131], a topic that is briefly addressed below, but further detailed in Theunissen and David’s review in this issue. In this section, we will provide a historic review of hPSC conversion to trophoblast through use of different BMP4-based protocols, focusing mostly on primed hPSC as starting material, while also addressing the controversy surrounding the role of BMP signaling in trophoblast emergence.
Table 1.
TSC derivation studies by the PSC types
| PSC type | Approaches with BMP4 addition | No addition of BMP4 |
|---|---|---|
| Naïve PSC conversion to TSC | (Io, Kabata et al. 2021) | (Guo, Stirparo et al. 2021) |
| (Dong, Beltcheva et al. 2020) | ||
| (Cinkornpumin, Kwon et al. 2020) | ||
| EPSC conversion to TSC | – | (Gao, Nowak-Imialek et al. 2019) |
| Primed PSC conversion to TSC | (Wei, Wang et al. 2021) | (Li, Kurosawa et al. 2019) |
| (Mischler, Karakis et al. 2021) | ||
| (Soncin et al. 2022) | ||
| (Jang et al. 2022) |
Trophoblast differentiation of primed hPSC: before there were naïve hPSCs…
The first publication showing BMP4-induced differentiation of hPSC came from the Thomson lab, in which this growth factor was applied to cultures in the presence of feeder-conditioned media. After several days, they noted emergence of multinucleated cells producing the pregnancy hormone, hCG, resembling syncytiotrophoblast (STB) [70]. Soon, a flurry of publications from multiple independent groups followed [132, 133], including one showing differentiation into, not just STB-, but also extravillous trophoblast (EVT)-like cells, mononuclear cells with HLA-G expression [134]. Subsequent studies documented formation of a cytotrophoblast (CTB) precursor-like cell, co-expressing CDX2 and TP63, prior to terminal differentiation into STB- and EVT-like cells [135]. TP63, a transcription factor, which, at least within the human placenta, is specific to CTB [136], was also shown to be required for BMP4-induced emergence of subsequent STB and EVT [135]. Further validation of this model emerged from the ability to recapitulate hypoxia-inducible factor-dependent differentiation of human EVT and the STB differentiation defect of Trisomy 21-associated CTB, using this BMP4-based system [137].
As time progressed, BMP4-based protocols for trophoblast induction of hPSC evolved. In the initial study by Xu et al. [70], it was noted that FGF2 appeared to counteract trophoblast formation, an observation confirmed in numerous later studies (reviewed in [132, 138]), including in Vallier et al. [139] and Yu et al. [140], who used a chemically defined medium to avoid the complicating effects of feeder cells and serum on differentiation. While the combination of FGF2 and BMP4 resulted in prolonged NANOG expression and up-regulation of primitive streak gene markers, especially T (BRACHYURY), as well as mesoderm and endoderm marker genes, BMP4 alone, even at concentrations as low as 10 ng/ml, still favored expression of trophoblast lineage markers [139, 141]. Accordingly, most researchers seeking to follow trophoblast emergence no longer include FGF2 in the medium once BMP4 is added, although it should be emphasized that this precaution does not exclude a contribution from endogenously produced FGF2, which can be counteracted by providing FGF signaling inhibitors, such as PD173074 (Fig. 3) [140, 142]. Likewise, any autocrine/paracrine effects of Activin A signaling can be minimized pharmacologically, for example, with SB431542 [143, 144] or A83-01, thus potentiating the effects of BMP4 [141, 145, 146] (Fig. 3). It has also become clear that when BMP4 and FGF2 are provided together, the extent of emergence of trophoblast and the contribution to mesoderm depends on the relative concentrations of the two growth factors. By contrast, a defined medium supplemented with BMP4, FGF2 and high concentrations of Activin A mainly generates endoderm [88], while the same medium formulated for minimal Activin A signaling gives rise predominantly to mesoderm [88]. Additionally, Kurek et al. [147] found that BMP4 induction of mesoderm, but not trophoblast, requires WNT signaling, and, as such, WNT signaling inhibitors, such as IWP2 can be used to improve trophoblast emergence from primed hESC. Horii et al. [148] subsequently developed a two-step hPSC trophoblast differentiation protocol, in which the first step (CTB induction) occurs in basal medium containing only BMP4 and IWP2 (Fig. 3). Importantly, as discussed above, early cell fate decisions in primed hPSC appear to be controlled by the same signaling pathways as in mEpiSC [139], suggesting that cells of both species may be able to generate trophoblast in response to BMP4.
Fig. 3.
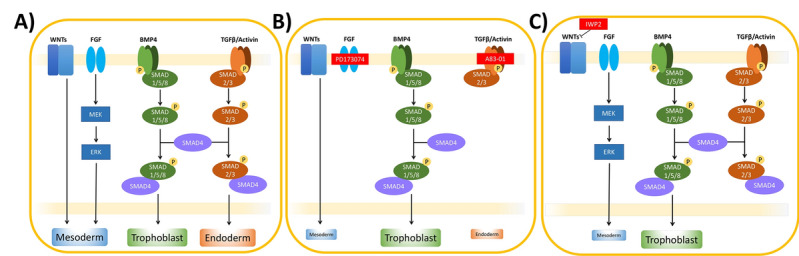
BMP4-based protocols for trophoblast induction of human pluripotent stem cells (hPSC). A In hPSC, activation of WNT, FGF, BMP, and TGFB pathways is known to drive differentiation into various lineages, including mesoderm, endoderm, and trophoblast. B In the so-called “BAP” protocol [141], exclusive differentiation into the trophoblast lineage is obtained by BMP4 treatment, concurrent with the inhibitors PD173074 and A83-01, targeting the FGF and TFGB pathways, respectively. C In the first step of the two-step protocol developed by Horii et al. [148], exclusive trophoblast differentiation is obtained by combination of BMP4 and IWP2, which interferes with WNT secretion
This context-dependent effect of BMP4 has also been reflected in multiple separate studies by Warmflash, Brivanlou, and colleagues, who first showed that circular micropatterns of hPSC exposed to BMP4 produced an outer ring of TE, with inner circles of mesendoderm and ectoderm (reviewed in [149]). This differentiation pattern is dependent on BMP4-induced generation of waves of WNT and NODAL signaling, which move toward the colony center, with duration of these signals controlling mesoderm induction, and duration of BMP signaling controlling TE induction [150]. Precise titration of hPSC down to a few cells has shown that, when only 1–8 cells are present within a colony, BMP4 treatment exclusively produces trophoblast-like cells, as defined by CDX2 and GATA3 expression, while this treatment in standard culture hPSC leads to a mixture of trophoblast-like and mesodermal cells [151]. Interestingly, differentiation into mesoderm could be modulated based on cell density, and completely prevented using inhibitors of activin/nodal signaling or high concentrations of BMP4, indicating a “community effect” controlling stem cell fate [151], highly analogous to cell fate decisions within the embryo.
Yang et al. [145] and Amita et al. [141] have shown that, in order for primed hPSC to differentiate fully to trophoblast, the presence of BMP4 is only necessary for the first 24 h; thereafter, differentiation toward trophoblast is guided by eliminating FGF2 and Activin A signaling. Between 24 and 36 h, the treated cells also transiently expressed CDX2 [145], a gene associated with trophoblast stem cells [104] and trophectoderm emergence in the mouse embryo [152], albeit with a less clear role in human TE [149]. These observations suggest that the BMP4-exposed cells may briefly pass through a stage in which the colonies are enriched in TSC-like cells. Li et al. [135] also identified TP63/CDX2 double-positive cells as early as 3 days after starting BMP4 treatment of hPSC, and were able to develop a two-step protocol whereby the vCTB-like cells obtained at the end of this first “step” could be replated and differentiated into STB- and EVT-like cells in the second “step” [137]. In an attempt to isolate such a stem cell population, Yang et al. [145] dissociated the BMP4-exposed colonies with trypsin and plated them out on a gelatin substratum, conditions that should not have supported primed hPSC growth. However, despite clear phenotypic differences between them and the initiating parental ESC, these “BMP-primed” cells still retained all the classical features of pluripotency rather than corresponding to trophoblast. In particular, they formed well-differentiated teratomas that included a trophoblast component when transplanted into immunocompromised mice [145]. The low FGF2 requirement of these cells, their high plating efficiency after trypsin dispersion, and their uniform NANOG expression, also clearly distinguished them from classical primed ESC. Although not named as such, these cells likely correspond to what has since become known as expanded potential stem cells or EPSC [122, 123, 153, 154], cells which are considered to be balanced in a metastable state somewhat intermediate between primed and naïve type ESC and poised to differentiate efficiently on multiple lineages given the right prompts. Their existence suggests that short-term BMP4 priming and inhibition of FGF/Activin A signaling are sufficient to launch primed ESC along the path for trophoblast emergence. This has also been confirmed by a study in which short exposures (no more than 24 h) of primed-type ESC to BMP4 triggered upregulation of a trophoblastic gene network circuit involving GATA2/GATA3/TFAP2A/TFAP2C in primed hESC [155], prior to the cells progressing to a differentiated trophoblast state. These so-called “trophectoderm four” (TEtra) genes are induced even before upregulation of CDX2 and TP63, and appear to be involved in both suppression of pluripotency and induction of a trophoblast-specific program [155].
Challenges to the trophoblast identity of BMP4-treated primed hPSC: before and after derivation of naïve hPSCs
The identity of these BMP4-treated primed hPSC was challenged, even before naïve hPSC were derived, with many investigators dismissing the possibility that primed cells have the potential to convert to trophoblast, largely on the grounds that these cells represent a post-implantation epiblast stage, one beyond that of TE specification in the embryo [88, 156]. Instead, there have been different hypotheses regarding the identity of these cells, with the first one being mesoderm, particularly based on the induction of BRACHYURY/T, an early mesoderm marker, shortly after initiation of BMP4 treatment [88]. However, the conditions used by this group were optimized for mouse EpiSC, not hPSC, thus resulting in formation of a BRACHYURY/CDX2 double-positive intermediate, which is not seen in other BMP-treated primed hPSC [150]. In addition, as mentioned above, induction of mesoderm in this setting can be prevented by stringently omitting FGF2[141, 145], reducing cell density, using inhibitors of activin/nodal signaling or high concentrations of BMP4 [151], or combining BMP4 treatment with a WNT inhibitor, such as IWP2[147, 148].
With advent of conditions for derivation of naïve hPSC, two independent groups soon converted these cells to TSC using media developed by Okae et al. [129] (Table 1). The first used naïve hPSC adapted to the 5i media (comprising the five inhibitors, PD0325901, IM-12, WH-4-023, SB590885, Y27632, along with LIF and Activin A) and subsequently passaged in the Okae TSC media multiple times [157], while the second used naïve hPSC cultured in PXGL (PD0325901, XAV939, Gö6983 and LIF) media with subsequent culture in TSC medium followed by sorting for ITGA2+ cells [158]. Both groups compared their “transdifferentiation” protocol between naïve and primed hPSC and showed that the former were significantly more efficient in derivation of such TSC [157, 158]; this is perhaps not surprising as conversion to the naïve state results in significant epigenetic remodeling, including significant loss of DNA methylation, in the form of loss of imprinting and X chromosome reactivation [159], and changes in the chromatin landscape [80]. More recently, two other groups provided a stepwise protocol for conversion of naïve hPSC, first to TE, and subsequently, to TSC. The first group, Guo et al. [160], also started with cells in PXGL media, subsequently removing all but PD0325901 and adding the ALK5 inhibitor A83-01 to induce the TE lineage over 2–3 days; they also showed that neither the addition of BMP2 nor the canonical BMP inhibitor, LDN-193189, affected TE induction in this setting [160]. Conversely, the second group, Io et al., who started with cells in yet another naïve-type media (t2iLGö, composed of PD0325901, CHIR99021, LIF and Gö6983), used a TE induction media containing BMP4, A83-01, and PD0325901 [161], a formulation not unlike that used in the BAP conversion of primed type hPSC to trophoblast [141]. BMP4 was only needed for 24 h, but, notably, removal of any of these three components (including BMP4) from the media significantly reduced the efficiency of TE induction (from ~ 50% down to 2–4%) [161]. Taken together, the role of BMP signaling in TE induction from naïve hPSC requires further study.
With the exception of Dong et al., all the above studies compared the transcriptomes of their naïve hPSC-derived TE to that of BMP4-treated primed hPSC, and concluded that, despite upregulation of many of the same genes and mostly because of co-expression of some “amnion-specific” genes, the latter have a differentiation trajectory ultimately leading towards amnion rather than trophoblast [158, 160, 161]. The identification of such “amnion-specific” genes comes from transcriptome data from either cynomolgus monkey amnion [95], or human data, the two main sources being the highly limited number of amniotic cells from extended culture human embryos [96], or the highly variable first and second trimester human amnion cells [97], all of which lack validation. A recent re-analysis of Xiang et al.’s extended culture human embryo single cell transcriptome data [96] found misclassification of several lineages, including amnion [162]. To discover a gene set that distinguishes amnion and TE, evaluation of more extended culture human embryos (containing a larger number of amniotic epithelial cells), as well as purified amnion epithelial cells (separated from underlying amniotic mesenchyme and chorionic trophoblast) and vCTB from early first trimester placentas, is needed to rigorously compare the transcriptomes of these cell types, with subsequent validation of potential lineage-specific genes. Even in the absence of such data, however, such co-expression of trophoblast- and amnion-associated genes in primed hPSC treated with a short course of BMP4, does not exclude trophoblast as the ultimate identity of the differentiated cells. A recent comparison of these short-term BMP4-treated hESC to the transcriptome of the cynomolgus monkey embryo found these cells to more closely resemble monkey amnion than TE, but maintained the possibility that the cells pass through an amnion intermediate prior to developing into mature STB and EVT [162]. Another group has recently come to the same conclusion, identifying a transient population of “nascent amnion” that precedes formation of STB in the context of endogenous activation of BMP signaling in primed hPSC [163]. Whether there is in fact a common precursor to amnion and TE within the human embryo, or whether at some point in gestation these two cell types can be interconverted, remains to be seen.
Conversion of primed hPSC to TSC
Nonetheless, there are now several published reports that bona fide TSC, with the expected transcriptome and protein identifiers and ability to differentiate to more advanced lineages, including STB and EVT, can arise from primed-type hPSC [164–167], including a manuscript by our group [168] (Table 1). With one exception, a study reporting derivation of TSC from iPSC-derived cysts using a micro-mesh technique [166], BMP4 was used to initiate conversions from prime-type hPSC to TSC, albeit with different culture medium formulations. One study used BMP4 combined with TGFB inhibitor SB43152 and S1PR3 agonist CYM-5541 in E7 medium for 3 days followed by switching to either Okae’s TSC medium, or to a medium (termed TM4) which contained the S1PR3 agonist. While both media formulations led to formation of TSC, TM4 led specifically to formation of a CDX2/TP63 double-positive TSC population, which the authors propose to be akin to an early TE progenitor [164]. Another study used the Okae TSC medium applied directly to hPSC, but learned that supplementation of this media with BMP4 for 5 days significantly improved efficiency of TSC derivation [165]. Our own study [168] applied the Okae TSC medium to cells treated with BMP4/IWP2 for 4 days, corresponding to the end of the first step of our previously described “two-step” trophoblast differentiation protocol [148].
While primed hPSC-TSC closely resemble primary hTSC, at the transcriptome level as well as functionally based on their ability to differentiate into STB and EVT in vitro [164, 165], the extent to which they have undergone epigenetic reprogramming remains to be fully addressed. Both Wei et al. [165] and [168] have demonstrated hypomethylation of the ELF5 promoter, and resulting expression of this gene, in primed hPSC-derived TSC, as occurs in primary hTSC [129] and early gestation human placenta [169]. However, the expression of another set of DNA-methylation-sensitive genes, the C19 miRNA cluster, located in a maternally imprinted region of chromosome 19 and known to be highly expressed in human trophoblast [170], remains low. While some members of the C19 miRNA cluster are upregulated during the induction from primed hPSC to TSC, it is unclear whether their expression reaches the same levels as they do in primary hTSC [165, 168]. Nevertheless, to-date, no specific function for these genes has been described in hTSC, and thus the significance of their lower than expected expression in primed hPSC-TSC remains questionable. It should also be noted that naïve hPSC-derived TSC also showed some aberrations in their methylome, specifically at some imprinted loci, with resulting alterations in gene expression [158]. Given that culture in naïve conditions is known to result in widespread loss of imprinting [171, 172], and the importance of many imprinted genes in human placental function (reviewed in [173]), future studies using iPSC to model placental disease should carefully choose the method of TSC conversion and evaluate the DNA methylome in the process.
Early insights into possible mechanisms of primed hPSC conversion to TSC
While much remains to be explored in context of naïve and primed pluripotency and TSC derivation from the respective pluripotent state, it should be noted that hPSC lines and cultures are highly heterogeneous [174]. With the advent of single cell RNAseq, this heterogeneity has been probed even within single cultures and single lines, resulting in the discovery of a population that is “intermediate” between naïve and primed state and has a unique transcriptome [175]. Interestingly, upregulated genes within this intermediate state include many trophoblast-associated genes, including GATA2, GATA3, and VGLL1 [175], the latter being a transcriptional co-factor uniquely expressed in vCTB of early gestation human placenta [90].
Recently, a similar “intermediate” has been identified during somatic cell reprogramming. Specifically, two groups have generated human induced trophoblast stem cells (iTSC) by reprogramming fibroblasts with the standard transcription factors used for iPSC generation, namely POU5F1, KLF4, SOX2 and MYC, also known as OSKM [130, 131]. Following introduction of these factors, which is most often done using non-integrating, non-replicating vectors, such as Sendai virus, into the somatic cell, the cells are often cultured in hPSC media until pluripotent colonies emerge, usually after about 4 weeks from vector introduction [176, 177]. The above two independent studies reported the isolation of iTSC by providing a medium that selects for TSC at about day 21, i.e., a week or so earlier than PSC emergence [130, 131]. Liu et al. also revealed molecular trajectories for cells undergoing various routes of reprogramming using a combination of single cell and single nucleus RNAseq performed on different days, and showed that pathways to the primed and naïve state diverged late in the second week of reprogramming [131]. Importantly, cells expressing the transcription factors GATA2, GATA3, as well as TFAP2C (broken circle in Fig. 4A) and with other features of TE formed a secondary tract shortly after the branch point to primed pluripotency had emerged. These genes were only transiently upregulated in the path to the primed state and subsequently become silenced (Fig. 4B). By contrast, their expression was partially retained in naïve-type cells [131]. The switch to TSC-supportive media was able to “catch” this transitory intermediate state, stabilizing it as TSC, rather than returning it to the naïve state [131]. Upregulation of the TE-associated genes appeared to be an inherent and essential part of naïve state reprogramming, because knockdown of either GATA2 or TFAP2C in the initiating fibroblasts impaired iPSC generation [131]. Interestingly, endogenous BMP4 expression provided a similar pattern to that of the TE signature genes during reprogramming (Fig. 4B), raising the intriguing idea that BMP signaling may be required for such reprogramming, if not into the pluripotent state then at least for the TSC state. This could also explain a role for exogenous BMP4 in trophoblast emergence from primed pluripotency. Specifically, BMP4 may not necessarily be promoting primed-to-naïve conversion as discussed above for mEpiSC, but instead promoting conversion to this “intermediate” state between primed and naïve pluripotency, allowing for easier capture of the TSC state.
Fig. 4.
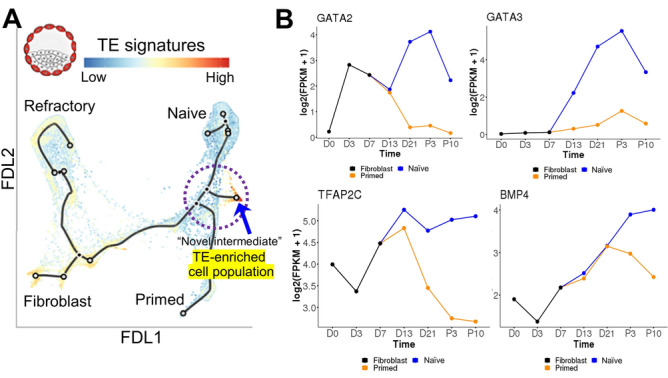
Roadmap for reprogramming human cells. A Either primed or naïve reprogramming trajectories, starting from fibroblast transduced with POU5F1, SOX2, KLF4, and MYC (OSKM). The single cell RNA sequencing (scRNAseq) data were visualized by force-directed layout (FDL), overlaid with in vivo trophectoderm (TE) signatures in color. Red shows high TE signature enrichment as exemplified with an inserted blastocyst drawing. Broken circle represents the intermediate stage of reprogramming.
Modified from Fig. 3A in [131]. B GATA2, GATA3, TFAP2C, and BMP4 gene expressions during reprogramming to naïve and primed human pluripotency prepared from interactive online tool (http://hrpi.ddnetbio.com/) by [131]. Gene expressions are shown in log-transformed log2 (FPKM + 1) and days (D) or passage numbers (P) from the OSKM transduction. Black, blue, and orange lines represent gene expression in fibroblast, naïve (t2iLGoY), and primed medium, respectively
Perhaps a similar process is at play during induction of “iBlastoids,” blastocyst-like structures containing epiblast, primitive endoderm (hypoblast), and an outer layer of TE. To date, these structures have been derived from extended potential stem cells (EPSC), cells in process of being reprogrammed (~ day 21 post-transduction, or “day-21 iPSC”), or naïve hPSC, but not primed hPSC. While naïve PSC appear able to generate blastoids without BMP4 supplementation [154, 178, 179], blastoid formation from EPSC [180, 181] and from day-21 iPSC [182] require BMP4 supplementation. Both of these EPSC-blastoid studies used slightly different protocols to offset an otherwise dearth of TE cells in the assembled blastoid. While Sozen et al. [180] combined BMP4 and ALK inhibitor A83-01 to promote TE differentiation, Fan et al. [181] mixed two different populations of EPSC, the first of which had been pretreated for 3 days with BMP4 to ensure TE fate transition, with the second untreated cell population providing the precursors of epiblast and hypoblast. Liu et al. [182] also created blastoids by placing ~ 100 cells of day 21-iPSCs in an AggreWell in the presence of BMP4-supplemented iBlastoid medium, where after aggregate formation, they could be cultured for a further 6 days to yield blastocyst-like structures. Both the Sozen et al., and Liu et al., protocols reported higher efficiency rates (7.2% and 5.8–18%, respectively) than Fan et al. (1.9%), likely due to addition of the ALK inhibitor A83-01, which has been shown to be necessary, both for derivation of TE [160, 161], and for establishment of hTSC [129]. While methods for generating blastoids without BMP4 have reported efficiency rates above 70% [178, 179], these studies report efficiencies solely based on formation of a cavitated structure without verifying proper development of the three lineages using immune-localization, such as was done by Sozen et al. [180] (7.2%). It should be noted that a recent pre-print manuscript by Zhao et al. [183] has challenged the conclusions of Liu et al. [182], indicating that the TE component of the iBlastoids is most likely amnion and not trophoblast, based on expression of so-called amnion-associated genes, GABRP and ISL1. However, as discussed above, a consensus list of amnion marker transcripts or proteins has yet to be attained; additionally, GABRP is readily detected in STB of first trimester placental tissue as demonstrated by immunohistochemistry [184]. Thus, as discussed above, such conclusions must await validation of a true set of amnion (vs. trophoblast)-specific markers.
In summary, there is a significant body of evidence, both for the ability of primed hPSC to give rise to trophoblast and for the role of BMP4 signaling in this process. The exact mechanisms, however, including whether this conversion occurs through an intermediate with similarities to both amnion and TE, remain to be elucidated.
BMP4 signaling in early embryonic development and pluripotent stem cells from comparative animal models
The involvement of BMP4 and its relatives on the development of the extra-embryonic compartment in species other than human and mouse has been less-extensively explored. Studies in early development of rabbit embryos that employed in-situ hybridization showed spatio-temporal expression of BMP2 and BMP4 distinct from that observed in the mouse [185]. BMP4 was first observed in pre-gastrulation embryos (stage 2) across the embryonic disc and at the border between embryonic and extra-embryonic compartments, increasing in the gastrulating embryo but never detected in the extra-embryonic compartment [185]. Expression of BMP2 started earlier in a ring-like domain at the border between the embryonic disc and the extra-embryonic compartment, persisting in that region and in the nascent mesoderm at the onset of gastrulation [185]. BMP2 expression was detected in both epiblast and hypoblast cells of the embryonic compartment and in the yolk sac lining the trophoblast compartment [185]. However, without evaluation of BMP signaling activity and knowledge of the spatio-temporal localization of BMP receptors, it is difficult to implicate BMP signaling, including possible effects on TE [185]. BMP4 treatment of rabbit ESCs, which are of the primed type [186], caused differentiation into epithelial-like cells expressing trophoblast-associated markers, including CDX2, EOMES, HAND1, and GCM1 [187]. These cells were able to self-renew in vitro for over 60 passages in an FGF- and TGFB-dependent manner, could be differentiated into both relaxin (RLN)-secreting syncytiotrophoblast [188] and invasive trophoblast giant cells in vitro, and could contribute to placentas of chimeric embryos in vivo. Therefore, they were classified as bona fide rabbit trophoblast stem cells [187], suggesting a potential role for BMP4 signaling in trophoblast emergence in rabbit ESC.
Bovine embryos are a widely used model to study early embryonic development and have been used to examine BMP signaling in early development [189]. BMP receptors BMPR1A and BMPR2, ligands BMP2 and BMP7, and effector proteins SMAD1 and SMAD5, have all been detected by qRT-PCR during pre-implantation development, from the two-cell stage to early blastocyst [190, 191]. In addition, both BMP2 and BMP4, as well as the BMP response machinery, were expressed within the elongated peri-implantation embryos (day 17 post-insemination) [192], and BMP4 expression has also been detected in extra-embryonic tissues post-implantation [193]. Treatment of bovine early pre-implantation embryos with BMP2, BMP4, or BMP5 differentially affected embryo development: BMP2 treatment caused no changes in rates of cleavage and blastocyst formation but increased expression of both NANOG and CDX2 in the blastocysts [190]. BMP4 treatment decreased blastocyst formation and hatching rate of in vitro fertilized (IVF), but not parthenogenically activated (PA) embryos; but, surprisingly, NOGGIN, an antagonist of BMP signaling, had a similar effect, along with a lower rate of cleavage of both PA and IVF embryos. No change in the number of total cells was observed in both BMP4- and NOGGIN-treated blastocysts but a decrease in the number of POU5F1+ cells was observed with both treatments [194, 195]. BMP5 treatment increased the rate of blastocyst formation without affecting cleavage, and increased the expression of POU5F1 and NANOG, but not SOX2 [191]. While there may be potential ligand-specific effects, these various results are likely also due to differences in medium/culture composition, as well as developmental time and duration of ligand treatment. Finally, addition of exogenous BMP4 boosted trophoblast cell line derivation from bovine blastocysts [196]. Altogether, these data suggest a role for BMP4 signaling in the peri-implantation bovine embryo, possibly including trophoblast emergence.
Studies on porcine embryos showed expression of BMP4 initially in a ring-like pattern between the epiblast and the TE of the peri-elongation embryos and subsequently in the posterior epiblast at the site of mesoderm formation during gastrulation [197]. Similar to the rabbit, expression of BMP2 preceded that of BMP4, and was stronger in epiblast cells than in TE [197]. BMPR2 was detected in the TE of pre-elongation embryos, but not in the embryonic compartment. BMP activity, detected by phosphorylation of SMAD1/5/8, was seen concentrated in the TE and extra-embryonic mesoderm of the peri-elongation embryos, suggesting a paracrine signal of BMP2/4 from the embryonic compartment towards the TE [197]. With respect to pluripotency, it is notable that, to maintain porcine ESC in a naïve-like state, inhibitors of BMP and TGFB appear to be required, in addition to inhibitors of MAPK and GSKβ signaling, suggesting that BMP4 has a negative influence on maintaining naïve pluripotency, likely promoting differentiation [198]. These data strongly support a role for BMP signaling in TE emergence in the porcine embryo.
In the developing equine placenta, the chorionic girdle, a ring of trophoblast that circles the spherical preimplantation conceptus, beginning at about day 30, contains terminally differentiated, binucleated trophoblast cells, which produce equine chorionic gonadotropin and other hormones, while the chorion harbors trophoblast progenitor. One study has shown that the chorionic girdle expresses higher levels of the BMP receptors, BMPR1A and BMPR2, compared to the chorion, which, in turn, expresses BMP4. This might suggest that a paracrine BMP signaling, from the chorion, promotes differentiation/maintenance of the terminally differentiated cells in the chorionic girdle [199]. Indeed, phospho-SMAD 1/5/8 was detected between day 27 and 34 of embryonic development, with a peak at day 31, the time of initiation of binucleated cell differentiation. Moreover, treatment with exogenous BMP4 in vitro increased the number of binucleated cells in the chorionic girdle suggesting a role in promoting the terminal differentiation of trophoblast cells in equids [199].
Finally, data on the role of BMP signaling in non-human primate embryo development are scarce. Nakamura and colleagues [200] performed single-cell analysis of both pre- and post-implantation rhesus monkey embryos and identified BMP4 expression in the gene cluster of gastrulating cells. Following extended culture conditions for human embryos, Ma and colleagues [95] performed single-cell analysis on cynomologus monkey blastocysts, cultured up to 20 (days post-fertilization). They observed three clusters of POU2F1+/SOX2+/BMP4+ cells, which they identified as potential monkey amnion cells. However, both studies focused on the epiblast, and not TE-derived, compartments. Finally, treatment of cynomolgus monkey ESCs with BMP4 did not up-regulate trophoblast markers, instead initiating primitive endoderm differentiation [201]. Therefore, further studies will be required to investigate the role of BMP signaling in TE/trophoblast specification in primates.
Summary and future studies
While much has been done to probe the role of BMP signaling in the peri-implantation period, significant questions remain, particularly with respect to the role of this pathway, both in establishment and development of the trophoblast compartment, as well as in reprogramming toward the trophoblastic fate. While the mouse provides a highly manipulatable system, particularly allowing assessment of contribution to chimeras which can develop in vivo, species-specific differences must be kept in mind, and hence, the need for further development of stem cell-based blastocyst-like structures, including iBlastoids and STEMbryos, which can be applied to human pluripotent stem cells.
Acknowledgements
Not applicable.
Author contributions
All authors contributed to writing the first drafts of different sections of this review article, as well as to generation of Figures and Tables. RMR and MMP revised and edited the manuscript. All authors subsequently read and approved the final manuscript.
Funding
This work was supported by the National Institutes of Health: R01HD089534 (M.M.P.), R01HD094937 and R21AI145071 (R.M.R.), and R01HD096260 (F.S.). J.R.O. was supported through a grant from the American Society for Reproductive Medicine.
Availability of data and material
Not applicable.
Declarations
Conflict of interests
The authors have no relevant financial or non-financial interests to disclose.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Sieber C, Kopf J, Hiepen C, Knaus P. Recent advances in BMP receptor signaling. Cytokine Growth Factor Rev. 2009;20:343–355. doi: 10.1016/J.CYTOGFR.2009.10.007. [DOI] [PubMed] [Google Scholar]
- 2.Bragdon B, Moseychuk O, Saldanha S, et al. Bone Morphogenetic Proteins: a critical review. Cell Signal. 2011;23:609–620. doi: 10.1016/J.CELLSIG.2010.10.003. [DOI] [PubMed] [Google Scholar]
- 3.Yadin D, Knaus P, Mueller TD. Structural insights into BMP receptors: specificity, activation and inhibition. Cytokine Growth Factor Rev. 2016;27:13–34. doi: 10.1016/j.cytogfr.2015.11.005. [DOI] [PubMed] [Google Scholar]
- 4.García de Vinuesa A, Abdelilah-Seyfried S, Knaus P, et al. BMP signaling in vascular biology and dysfunction. Cytokine Growth Factor Rev. 2016;27:65–79. doi: 10.1016/J.CYTOGFR.2015.12.005. [DOI] [PubMed] [Google Scholar]
- 5.Ross S, Hill CS. How the Smads regulate transcription. Int J Biochem Cell Biol. 2008;40:383–408. doi: 10.1016/J.BIOCEL.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 6.Hussein SM, Duff EK, Sirard C. Smad4 and β-catenin Co-activators functionally interact with lymphoid-enhancing factor to regulate graded expression of Msx2 *. J Biol Chem. 2003;278:48805–48814. doi: 10.1074/JBC.M305472200. [DOI] [PubMed] [Google Scholar]
- 7.Itoh F, Itoh S, Goumans MJ, et al. Synergy and antagonism between Notch and BMP receptor signaling pathways in endothelial cells. EMBO J. 2004;23:541–551. doi: 10.1038/SJ.EMBOJ.7600065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blahna MT, Hata A. Smad-mediatedregulation of microRNA biosynthesis. FEBS Lett. 2012;586:1906. doi: 10.1016/J.FEBSLET.2012.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davis BN, Hilyard AC, Lagna G, Hata A. SMAD proteins control DROSHA-mediated microRNA maturation. Nature. 2008;454:56. doi: 10.1038/NATURE07086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nohe A, Keating E, Knaus P, Petersen NO. Signal transduction of bone morphogenetic protein receptors. Cell Signal. 2004;16:291–299. doi: 10.1016/J.CELLSIG.2003.08.011. [DOI] [PubMed] [Google Scholar]
- 11.Zhang YE. Non-Smad pathways 128 Non-Smad pathways in TGF-β signaling. Cell Res. 2009;19:128–139. doi: 10.1038/cr.2008.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brazil DP, Church RH, Surae S, et al. BMP signalling: agony and antagony in the family. Trends Cell Biol. 2015;25:249–264. doi: 10.1016/j.tcb.2014.12.004. [DOI] [PubMed] [Google Scholar]
- 13.Walsh DW, Godson C, Brazil DP, Martin F. Extracellular BMP-antagonist regulation in development and disease: tied up in knots. Trends Cell Biol. 2010;20:244–256. doi: 10.1016/j.tcb.2010.01.008. [DOI] [PubMed] [Google Scholar]
- 14.Groppe J, Greenwald J, Wiater E, et al. Structural basis of BMP signalling inhibition by the cystine knot protein Noggin. Nature. 2002;420:636–642. doi: 10.1038/NATURE01245. [DOI] [PubMed] [Google Scholar]
- 15.Lichtner B, Knaus P, Lehrach H, Adjaye J. BMP10 as a potent inducer of trophoblast differentiation in human embryonic and induced pluripotent stem cells. Biomaterials. 2013;34:9789–9802. doi: 10.1016/J.BIOMATERIALS.2013.08.084. [DOI] [PubMed] [Google Scholar]
- 16.Ali JL, Lagasse BJ, Minuk AJ, et al. Differential cellular responses induced by dorsomorphin and LDN-193189 in chemotherapy-sensitive and chemotherapy-resistant human epithelial ovarian cancer cells. Int J Cancer. 2015;136:E455–E469. doi: 10.1002/IJC.29220. [DOI] [PubMed] [Google Scholar]
- 17.Hao J, Ho JN, Lewis JA, et al. In vivo structure—activity relationship study of dorsomorphin analogues identifies selective VEGF and BMP inhibitors. ACS Chem Biol. 2010;5:245–253. doi: 10.1021/cb9002865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hao J, Lee R, Chang A, et al. DMH1, a small molecule inhibitor of BMP type I receptors, suppresses growth and invasion of lung cancer. PLoS ONE. 2014;9:e90748. doi: 10.1371/JOURNAL.PONE.0090748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tojo M, Hamashima Y, Hanyu A, et al. The ALK-5 inhibitor A-83-01 inhibits Smad signaling and epithelial-to-mesenchymal transition by transforming growth factor-beta. Cancer Sci. 2005;96:791–800. doi: 10.1111/J.1349-7006.2005.00103.X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ninomiya-Tsuji J, Kajino T, Ono K, et al. A resorcylic acid lactone, 5Z–7-oxozeaenol, prevents inflammation by inhibiting the catalytic activity of TAK1 MAPK kinase kinase. J Biol Chem. 2003;278:18485–18490. doi: 10.1074/jbc.M207453200. [DOI] [PubMed] [Google Scholar]
- 21.Wu J, Powell F, Larsen NA, et al. Mechanism and in vitro pharmacology of TAK1 inhibition by (5Z)-7-oxozeaenol. ACS Chem Biol. 2013;8:643–650. doi: 10.1021/CB3005897. [DOI] [PubMed] [Google Scholar]
- 22.Totzke J, Gurbani D, Raphemot R, et al. Takinib, a selective TAK1 inhibitor, broadens the therapeutic efficacy of TNF-α inhibition for cancer and autoimmune disease. Cell Chem Biol. 2017;24:1029–1039.e7. doi: 10.1016/J.CHEMBIOL.2017.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang H, Bradley A. Mice deficient for BMP2 are nonviable and have defects in amnion/chorion and cardiac development. Development. 1996;122:2977–2986. doi: 10.1242/dev.122.10.2977. [DOI] [PubMed] [Google Scholar]
- 24.Lawson KA, Dunn NR, Roelen BA, et al. Bmp4 is required for the generation of primordial germ cells in the mouse embryo. Genes Dev. 1999;13:424–436. doi: 10.1101/gad.13.4.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Winnier G, Blessing M, Labosky PA, Hogan BL. Bone morphogenetic protein-4 is required for mesoderm formation and patterning in the mouse. Genes Dev. 1995;9:2105–2116. doi: 10.1101/gad.9.17.2105. [DOI] [PubMed] [Google Scholar]
- 26.Fujiwara T, Dunn NR, Hogan BL. Bone morphogenetic protein 4 in the extraembryonic mesoderm is required for allantois development and the localization and survival of primordial germ cells in the mouse. Proc Natl Acad Sci USA. 2001;98:13739–13744. doi: 10.1073/pnas.241508898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fujiwara T, Dehart DB, Sulik KK, Hogan BLM. Distinct requirements for extra-embryonic and embryonic bone morphogenetic protein 4 in the formation of the node and primitive streak and coordination of left-right asymmetry in the mouse. Development. 2002;129:4685–4696. doi: 10.1242/dev.129.20.4685. [DOI] [PubMed] [Google Scholar]
- 28.Soares ML, Haraguchi S, Torres-Padilla M-E, et al. Functional studies of signaling pathways in peri-implantation development of the mouse embryo by RNAi. BMC Dev Biol. 2005;5:28. doi: 10.1186/1471-213X-5-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Soares ML, Torres-Padilla M-E, Zernicka-Goetz M. Bone morphogenetic protein 4 signaling regulates development of the anterior visceral endoderm in the mouse embryo. Dev Growth Differ. 2008;50:615–621. doi: 10.1111/j.1440-169X.2008.01059.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cho LTY, Wamaitha SE, Tsai IJ, et al. Conversion from mouse embryonic to extra-embryonic endoderm stem cells reveals distinct differentiation capacities of pluripotent stem cell states. Development. 2012;139:2866–2877. doi: 10.1242/dev.078519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ying Y, Zhao GQ. Cooperation of endoderm-derived BMP2 and extraembryonic ectoderm-derived BMP4 in primordial germ cell generation in the mouse. Dev Biol. 2001;232:484–492. doi: 10.1006/dbio.2001.0173. [DOI] [PubMed] [Google Scholar]
- 32.Di-Gregorio A, Sancho M, Stuckey DW, et al. BMP signalling inhibits premature neural differentiation in the mouse embryo. Development. 2007;134:3359–3369. doi: 10.1242/dev.005967. [DOI] [PubMed] [Google Scholar]
- 33.Tremblay KD, Dunn NR, Robertson EJ. Mouse embryos lacking Smad1 signals display defects in extra-embryonic tissues and germ cell formation. Development. 2001;128:3609–3621. doi: 10.1242/dev.128.18.3609. [DOI] [PubMed] [Google Scholar]
- 34.Graham SJL, Wicher KB, Jedrusik A, et al. BMP signalling regulates the pre-implantation development of extra-embryonic cell lineages in the mouse embryo. Nat Commun. 2014;5:5667. doi: 10.1038/ncomms6667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Home P, Saha B, Ray S, et al. Altered subcellular localization of transcription factor TEAD4 regulates first mammalian cell lineage commitment. Proc Natl Acad Sci USA. 2012;109:7362–7367. doi: 10.1073/pnas.1201595109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rivron NC, Frias-Aldeguer J, Vrij EJ, et al. Blastocyst-like structures generated solely from stem cells. Nature. 2018;557:106–111. doi: 10.1038/s41586-018-0051-0. [DOI] [PubMed] [Google Scholar]
- 37.Shahbazi MN, Jedrusik A, Vuoristo S, et al. Self-organization of the human embryo in the absence of maternal tissues. Nat Cell Biol. 2016;18:700–708. doi: 10.1038/ncb3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Deglincerti A, Croft GF, Pietila LN, et al. Self-organization of the in vitro attached human embryo. Nature. 2016;533:251–254. doi: 10.1038/nature17948. [DOI] [PubMed] [Google Scholar]
- 39.Blakeley P, Fogarty NME, Del Valle I, et al. Defining the three cell lineages of the human blastocyst by single-cell RNA-seq. Development. 2015;142:3613. doi: 10.1242/dev.131235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee KY, Jeong J-W, Wang J, et al. Bmp2 is critical for the murine uterine decidual response. Mol Cell Biol. 2007;27:5468–5478. doi: 10.1128/MCB.00342-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Monsivais D, Clementi C, Peng J, et al. BMP7 induces uterine receptivity and blastocyst attachment. Endocrinology. 2017;158:979–992. doi: 10.1210/en.2016-1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Monsivais D, Clementi C, Peng J, et al. Uterine ALK3 is essential during the window of implantation. Proc Natl Acad Sci USA. 2016;113:E387–395. doi: 10.1073/pnas.1523758113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nagashima T, Li Q, Clementi C, et al. BMPR2 is required for postimplantation uterine function and pregnancy maintenance. J Clin Invest. 2013;123:2539–2550. doi: 10.1172/JCI65710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stoikos CJ, Harrison CA, Salamonsen LA, Dimitriadis E. A distinct cohort of the TGFbeta superfamily members expressed in human endometrium regulate decidualization. Hum Reprod. 2008;23:1447–1456. doi: 10.1093/humrep/den110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang Y, Zhu H, Chang H-M, Leung PCK. ALK3-SMAD1/5 signaling mediates the BMP2-induced decrease in PGE2 production in human endometrial stromal cells and decidual stromal cells. Front Cell Dev Biol. 2020;8:573028. doi: 10.3389/fcell.2020.573028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pape ME, Kim KH. Effect of tumor necrosis factor on acetyl-coenzyme A carboxylase gene expression and preadipocyte differentiation. Mol Endocrinol. 1988;2:395–403. doi: 10.1210/mend-2-5-395. [DOI] [PubMed] [Google Scholar]
- 47.Harrison SE, Sozen B, Christodoulou N, et al. Assembly of embryonic and extraembryonic stem cells to mimic embryogenesis in vitro. Science. 2017;356:eaal1810. doi: 10.1126/science.aal1810. [DOI] [PubMed] [Google Scholar]
- 48.Sozen B, Demir N, Zernicka-Goetz M. BMP signalling is required for extra-embryonic ectoderm development during pre-to-post-implantation transition of the mouse embryo. Dev Biol. 2021;470:84–94. doi: 10.1016/j.ydbio.2020.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yi Y, Zhu H, Klausen C, et al. Dysregulated BMP2 in the placenta may contribute to early-onset preeclampsia by regulating human trophoblast expression of extracellular matrix and adhesion molecules. Front Cell Dev Biol. 2021;9:768669. doi: 10.3389/fcell.2021.768669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sato N, Sanjuan IM, Heke M, et al. Molecular signature of human embryonic stem cells and its comparison with the mouse. Dev Biol. 2003;260:404–413. doi: 10.1016/s0012-1606(03)00256-2. [DOI] [PubMed] [Google Scholar]
- 51.Yabe S, Alexenko AP, Amita M, et al. Comparison of syncytiotrophoblast generated from human embryonic stem cells and from term placentas. Proc Natl Acad Sci USA. 2016;113:E2598–2607. doi: 10.1073/pnas.1601630113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Au J, Requena DF, Rishik H, et al. Role of autocrine bone morphogenetic protein signaling in trophoblast stem cells. Biol Reprod. 2021 doi: 10.1093/biolre/ioab213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Home P, Kumar RP, Ganguly A, et al. Genetic redundancy of GATA factors in the extraembryonic trophoblast lineage ensures the progression of preimplantation and postimplantation mammalian development. Development. 2017;144:876–888. doi: 10.1242/dev.145318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chambers I, Smith A. Self-renewal of teratocarcinoma and embryonic stem cells. Oncogene. 2004;23:7150–7160. doi: 10.1038/sj.onc.1207930. [DOI] [PubMed] [Google Scholar]
- 55.Smith AG, Heath JK, Donaldson DD, et al. Inhibition of pluripotential embryonic stem cell differentiation by purified polypeptides. Nature. 1988;336:688–690. doi: 10.1038/336688a0. [DOI] [PubMed] [Google Scholar]
- 56.Ying QL, Nichols J, Chambers I, Smith A. BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell. 2003;115:281–292. doi: 10.1016/s0092-8674(03)00847-x. [DOI] [PubMed] [Google Scholar]
- 57.Qi X, Li T-G, Hao J, et al. BMP4 supports self-renewal of embryonic stem cells by inhibiting mitogen-activated protein kinase pathways. Proc Natl Acad Sci USA. 2004;101:6027–6032. doi: 10.1073/pnas.0401367101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gomes Fernandes M, Dries R, Roost MS, et al. BMP-SMAD signaling regulates lineage priming, but is dispensable for self-renewal in mouse embryonic stem cells. Stem Cell Reports. 2016;6:85–94. doi: 10.1016/j.stemcr.2015.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ying Q-L, Wray J, Nichols J, et al. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nichols J, Smith A. Naive and primed pluripotent states. Cell Stem Cell. 2009;4:487–492. doi: 10.1016/j.stem.2009.05.015. [DOI] [PubMed] [Google Scholar]
- 61.Thomson JA, Kalishman J, Golos TG, et al. Pluripotent cell lines derived from common marmoset (Callithrix jacchus) blastocysts. Biol Reprod. 1996;55:254–259. doi: 10.1095/biolreprod55.2.254. [DOI] [PubMed] [Google Scholar]
- 62.Thomson JA, Itskovitz-Eldor J, Shapiro SS, et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 63.Amit M, Itskovitz-Eldor J. Derivation and maintenance of human embryonic stem cells. Methods Mol Biol. 2006;331:43–53. doi: 10.1385/1-59745-046-4:43. [DOI] [PubMed] [Google Scholar]
- 64.Amit M, Itskovitz-Eldor J. Sources, derivation, and culture of human embryonic stem cells. Semin Reprod Med. 2006;24:298–303. doi: 10.1055/s-2006-954939. [DOI] [PubMed] [Google Scholar]
- 65.James D, Levine AJ, Besser D, Hemmati-Brivanlou A. TGFbeta/activin/nodal signaling is necessary for the maintenance of pluripotency in human embryonic stem cells. Development. 2005;132:1273–1282. doi: 10.1242/dev.01706. [DOI] [PubMed] [Google Scholar]
- 66.Vallier L, Alexander M, Pedersen RA. Activin/Nodal and FGF pathways cooperate to maintain pluripotency of human embryonic stem cells. J Cell Sci. 2005;118:4495–4509. doi: 10.1242/jcs.02553. [DOI] [PubMed] [Google Scholar]
- 67.Wei CL, Miura T, Robson P, et al. Transcriptome profiling of human and murine ESCs identifies divergent paths required to maintain the stem cell state. Stem Cells. 2005;23:166–185. doi: 10.1634/stemcells.2004-0162. [DOI] [PubMed] [Google Scholar]
- 68.Odorico JS, Kaufman DS, Thomson JA. Multilineage differentiation from human embryonic stem cell lines. Stem Cells. 2001;19:193–204. doi: 10.1634/stemcells.19-3-193. [DOI] [PubMed] [Google Scholar]
- 69.Itskovitz-Eldor J, Schuldiner M, Karsenti D, et al. Differentiation of human embryonic stem cells into embryoid bodies compromising the three embryonic germ layers. Mol Med. 2000;6:88–95. doi: 10.1007/BF03401776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xu R-H, Chen X, Li DS, et al. BMP4 initiates human embryonic stem cell differentiation to trophoblast. Nat Biotechnol. 2002;20:1261–1264. doi: 10.1038/nbt761. [DOI] [PubMed] [Google Scholar]
- 71.Ezashi T, Das P, Roberts RM. Low O2 tensions and the prevention of differentiation of hES cells. Proc Natl Acad Sci USA. 2005;102:4783–4788. doi: 10.1073/pnas.0501283102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Golos TG, Pollastrini LM, Gerami-Naini B. Human embryonic stem cells as a model for trophoblast differentiation. Semin Reprod Med. 2006;24:314–321. doi: 10.1055/s-2006-952154. [DOI] [PubMed] [Google Scholar]
- 73.Harun R, Ruban L, Matin M, et al. Cytotrophoblast stem cell lines derived from human embryonic stem cells and their capacity to mimic invasive implantation events. Hum Reprod. 2006;21:1349–1358. doi: 10.1093/humrep/del017. [DOI] [PubMed] [Google Scholar]
- 74.Tesar PJ, Chenoweth JG, Brook FA, et al. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature. 2007;448:196–199. doi: 10.1038/nature05972. [DOI] [PubMed] [Google Scholar]
- 75.Brons IGM, Smithers LE, Trotter MWB, et al. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature. 2007;448:191–195. doi: 10.1038/nature05950. [DOI] [PubMed] [Google Scholar]
- 76.Huang Y, Osorno R, Tsakiridis A, Wilson V. In Vivo differentiation potential of epiblast stem cells revealed by chimeric embryo formation. Cell Rep. 2012;2:1571–1578. doi: 10.1016/j.celrep.2012.10.022. [DOI] [PubMed] [Google Scholar]
- 77.Takahashi S, Kobayashi S, Hiratani I. Epigenetic differences between naïve and primed pluripotent stem cells. Cell Mol Life Sci. 2018;75:1191–1203. doi: 10.1007/s00018-017-2703-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hanna JH, Saha K, Jaenisch R. Pluripotency and cellular reprogramming: facts, hypotheses, unresolved issues. Cell. 2010;143:508–525. doi: 10.1016/j.cell.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Guo G, von Meyenn F, Santos F, et al. Naive Pluripotent Stem Cells Derived Directly from Isolated Cells of the Human Inner Cell Mass. Stem Cell Reports. 2016;6:437–446. doi: 10.1016/j.stemcr.2016.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Theunissen TW, Powell BE, Wang H, et al. Systematic identification of culture conditions for induction and maintenance of naive human pluripotency. Cell Stem Cell. 2014;15:471–487. doi: 10.1016/j.stem.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Weinberger L, Ayyash M, Novershtern N, Hanna JH. Dynamic stem cell states: naive to primed pluripotency in rodents and humans. Nat Rev Mol Cell Biol. 2016;17:155–169. doi: 10.1038/nrm.2015.28. [DOI] [PubMed] [Google Scholar]
- 82.Czyz J, Wobus A. Embryonic stem cell differentiation: the role of extracellular factors. Differentiation. 2001;68:167–174. doi: 10.1046/j.1432-0436.2001.680404.x. [DOI] [PubMed] [Google Scholar]
- 83.Sadlon TJ, Lewis ID, D’Andrea RJ. BMP4: its role in development of the hematopoietic system and potential as a hematopoietic growth factor. Stem Cells. 2004;22:457–474. doi: 10.1634/stemcells.22-4-457. [DOI] [PubMed] [Google Scholar]
- 84.Sybirna A, Wong FCK, Surani MA. Genetic basis for primordial germ cells specification in mouse and human: conserved and divergent roles of PRDM and SOX transcription factors. Curr Top Dev Biol. 2019;135:35–89. doi: 10.1016/bs.ctdb.2019.04.004. [DOI] [PubMed] [Google Scholar]
- 85.Aberdam D. Derivation of keratinocyte progenitor cells and skin formation from embryonic stem cells. Int J Dev Biol. 2004;48:203–206. doi: 10.1387/ijdb.15272386. [DOI] [PubMed] [Google Scholar]
- 86.Li Z, Chen Y-G. Functions of BMP signaling in embryonic stem cell fate determination. Exp Cell Res. 2013;319:113–119. doi: 10.1016/j.yexcr.2012.09.016. [DOI] [PubMed] [Google Scholar]
- 87.Sui L, Bouwens L, Mfopou JK. Signaling pathways during maintenance and definitive endoderm differentiation of embryonic stem cells. Int J Dev Biol. 2013;57:1–12. doi: 10.1387/ijdb.120115ls. [DOI] [PubMed] [Google Scholar]
- 88.Bernardo AS, Faial T, Gardner L, et al. BRACHYURY and CDX2 mediate BMP-induced differentiation of human and mouse pluripotent stem cells into embryonic and extraembryonic lineages. Cell Stem Cell. 2011;9:144–155. doi: 10.1016/j.stem.2011.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Russ AP, Wattler S, Colledge WH, et al. Eomesodermin is required for mouse trophoblast development and mesoderm formation. Nature. 2000;404:95–99. doi: 10.1038/35003601. [DOI] [PubMed] [Google Scholar]
- 90.Soncin F, Khater M, To C, et al. Comparative analysis of mouse and human placentae across gestation reveals species-specific regulators of placental development. Development. 2018 doi: 10.1242/dev.156273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Shao Y, Taniguchi K, Gurdziel K, et al. Self-organized amniogenesis by human pluripotent stem cells in a biomimetic implantation-like niche. Nature Mater. 2017;16:419–425. doi: 10.1038/nmat4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zheng Y, Xue X, Shao Y, et al. Controlled modelling of human epiblast and amnion development using stem cells. Nature. 2019;573:421–425. doi: 10.1038/s41586-019-1535-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Shao Y, Taniguchi K, Townshend RF, et al. A pluripotent stem cell-based model for post-implantation human amniotic sac development. Nat Commun. 2017;8:208. doi: 10.1038/s41467-017-00236-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Dobreva MP, Pereira PNG, Deprest J, Zwijsen A. On the origin of amniotic stem cells: of mice and men. Int J Dev Biol. 2010;54:761–777. doi: 10.1387/ijdb.092935md. [DOI] [PubMed] [Google Scholar]
- 95.Ma H, Zhai J, Wan H, et al. In vitro culture of cynomolgus monkey embryos beyond early gastrulation. Science. 2019 doi: 10.1126/science.aax7890. [DOI] [PubMed] [Google Scholar]
- 96.Xiang L, Yin Y, Zheng Y, et al. A developmental landscape of 3D-cultured human pre-gastrulation embryos. Nature. 2020;577:537–542. doi: 10.1038/s41586-019-1875-y. [DOI] [PubMed] [Google Scholar]
- 97.Roost MS, van Iperen L, Ariyurek Y, et al. KeyGenes, a tool to probe tissue differentiation using a human fetal transcriptional atlas. Stem Cell Rep. 2015;4:1112–1124. doi: 10.1016/j.stemcr.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Plusa B, Frankenberg S, Chalmers A, et al. Downregulation of Par3 and aPKC function directs cells towards the ICM in the preimplantation mouse embryo. J Cell Sci. 2005;118:505–515. doi: 10.1242/jcs.01666. [DOI] [PubMed] [Google Scholar]
- 99.Nishioka N, Inoue K, Adachi K, et al. The Hippo signaling pathway components Lats and Yap pattern Tead4 activity to distinguish mouse trophectoderm from inner cell mass. Dev Cell. 2009;16:398–410. doi: 10.1016/j.devcel.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 100.Beddington RS, Robertson EJ. An assessment of the developmental potential of embryonic stem cells in the midgestation mouse embryo. Development. 1989;105:733–737. doi: 10.1242/dev.105.4.733. [DOI] [PubMed] [Google Scholar]
- 101.Nagy A, Gócza E, Diaz EM, et al. Embryonic stem cells alone are able to support fetal development in the mouse. Development. 1990;110:815–821. doi: 10.1242/dev.110.3.815. [DOI] [PubMed] [Google Scholar]
- 102.Posfai E, Petropoulos S, de Barros FRO, et al. Position- and Hippo signaling-dependent plasticity during lineage segregation in the early mouse embryo. Elife. 2017;6:e22906. doi: 10.7554/eLife.22906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gasperowicz M, Natale DRC. Establishing three blastocyst lineages—then what? Biol Reprod. 2011;84:621–630. doi: 10.1095/biolreprod.110.085209. [DOI] [PubMed] [Google Scholar]
- 104.Tanaka S, Kunath T, Hadjantonakis AK, et al. Promotion of trophoblast stem cell proliferation by FGF4. Science. 1998;282:2072–2075. doi: 10.1126/science.282.5396.2072. [DOI] [PubMed] [Google Scholar]
- 105.Erlebacher A, Price KA, Glimcher LH. Maintenance of mouse trophoblast stem cell proliferation by TGF-beta/activin. Dev Biol. 2004;275:158–169. doi: 10.1016/j.ydbio.2004.07.032. [DOI] [PubMed] [Google Scholar]
- 106.Kubaczka C, Senner C, Araúzo-Bravo MJ, et al. Derivation and maintenance of murine trophoblast stem cells under defined conditions. Stem Cell Rep. 2014;2:232–242. doi: 10.1016/j.stemcr.2013.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kubaczka C, Senner CE, Cierlitza M, et al. Direct induction of trophoblast stem cells from murine fibroblasts. Cell Stem Cell. 2015;17:557–568. doi: 10.1016/j.stem.2015.08.005. [DOI] [PubMed] [Google Scholar]
- 108.Benchetrit H, Herman S, van Wietmarschen N, et al. Extensive nuclear reprogramming underlies lineage conversion into functional trophoblast stem-like cells. Cell Stem Cell. 2015;17:543–556. doi: 10.1016/j.stem.2015.08.006. [DOI] [PubMed] [Google Scholar]
- 109.Niwa H, Miyazaki J, Smith AG. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet. 2000;24:372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- 110.Niwa H, Toyooka Y, Shimosato D, et al. Interaction between Oct3/4 and Cdx2 determines trophectoderm differentiation. Cell. 2005;123:917–929. doi: 10.1016/j.cell.2005.08.040. [DOI] [PubMed] [Google Scholar]
- 111.Peng S, Hua J, Cao X, Wang H. Gelatin induces trophectoderm differentiation of mouse embryonic stem cells. Cell Biol Int. 2011;35:587–591. doi: 10.1042/CBI20100452. [DOI] [PubMed] [Google Scholar]
- 112.Schenke-Layland K, Angelis E, Rhodes KE, et al. Collagen IV induces trophoectoderm differentiation of mouse embryonic stem cells. Stem Cells. 2007;25:1529–1538. doi: 10.1634/stemcells.2006-0729. [DOI] [PubMed] [Google Scholar]
- 113.Hayashi Y, Furue MK, Tanaka S, et al. BMP4 induction of trophoblast from mouse embryonic stem cells in defined culture conditions on laminin. In Vitro Cell Dev Biol-Animal. 2010;46:416–430. doi: 10.1007/s11626-009-9266-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Macfarlan TS, Gifford WD, Driscoll S, et al. Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature. 2012;487:57–63. doi: 10.1038/nature11244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Morgani SM, Canham MA, Nichols J, et al. Totipotent embryonic stem cells arise in ground-state culture conditions. Cell Rep. 2013;3:1945–1957. doi: 10.1016/j.celrep.2013.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Onishi K, Tonge PD, Nagy A, Zandstra PW. Local BMP-SMAD1 signaling increases LIF receptor-dependent STAT3 responsiveness and primed-to-naive mouse pluripotent stem cell conversion frequency. Stem Cell Rep. 2014;3:156–168. doi: 10.1016/j.stemcr.2014.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kime C, Sakaki-Yumoto M, Goodrich L, et al. Autotaxin-mediated lipid signaling intersects with LIF and BMP signaling to promote the naive pluripotency transcription factor program. Proc Natl Acad Sci USA. 2016;113:12478–12483. doi: 10.1073/pnas.1608564113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Yu S, Zhou C, Cao S, et al. BMP4 resets mouse epiblast stem cells to naive pluripotency through ZBTB7A/B-mediated chromatin remodelling. Nat Cell Biol. 2020;22:651–662. doi: 10.1038/s41556-020-0516-x. [DOI] [PubMed] [Google Scholar]
- 119.Kime C, Kiyonari H, Ohtsuka S, et al. Induced 2C expression and implantation-competent blastocyst-like cysts from primed pluripotent stem cells. Stem Cell Reports. 2019;13:485–498. doi: 10.1016/j.stemcr.2019.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Tomoda K, Hu H, Sahara Y, et al. Reprogramming epiblast stem cells into pre-implantation blastocyst cell-like cells. Stem Cell Rep. 2021;16:1197–1209. doi: 10.1016/j.stemcr.2021.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Guo J, Wang P, Sozen B, et al. Machine learning-assisted high-content analysis of pluripotent stem cell-derived embryos in vitro. Stem Cell Reports. 2021;16:1331–1346. doi: 10.1016/j.stemcr.2021.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Yang J, Ryan DJ, Wang W, et al. Establishment of mouse expanded potential stem cells. Nature. 2017;550:393–397. doi: 10.1038/nature24052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Yang Y, Liu B, Xu J, et al. Derivation of pluripotent stem cells with in vivo embryonic and extraembryonic potency. Cell. 2017;169:243–257.e25. doi: 10.1016/j.cell.2017.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Posfai E, Schell JP, Janiszewski A, et al. Defining totipotency using criteria of increasing stringency. Cell Biol. 2020;33:991. doi: 10.1038/s41556-020-00609-2. [DOI] [PubMed] [Google Scholar]
- 125.Takahashi Y, Dominici M, Swift J, et al. Trophoblast stem cells rescue placental defect in SOCS3-deficient mice. J Biol Chem. 2006;281:11444–11445. doi: 10.1074/jbc.C600015200. [DOI] [PubMed] [Google Scholar]
- 126.De Paepe C, Cauffman G, Verloes A, et al. Human trophectoderm cells are not yet committed. Hum Reprod. 2013;28:740–749. doi: 10.1093/humrep/des432. [DOI] [PubMed] [Google Scholar]
- 127.Gerri C, Menchero S, Mahadevaiah SK, et al. Human embryogenesis: a comparative perspective. Annu Rev Cell Dev Biol. 2020;36:411–440. doi: 10.1146/annurev-cellbio-022020-024900. [DOI] [PubMed] [Google Scholar]
- 128.Kunath T, Yamanaka Y, Detmar J, et al. Developmental differences in the expression of FGF receptors between human and mouse embryos. Placenta. 2014;35:1079–1088. doi: 10.1016/j.placenta.2014.09.008. [DOI] [PubMed] [Google Scholar]
- 129.Okae H, Toh H, Sato T, et al. Derivation of human trophoblast stem cells. Cell Stem Cell. 2018;22:50–63.e6. doi: 10.1016/j.stem.2017.11.004. [DOI] [PubMed] [Google Scholar]
- 130.Castel G, Meistermann D, Bretin B, et al. Induction of human trophoblast stem cells from somatic cells and pluripotent stem cells. Cell Rep. 2020;33:108419. doi: 10.1016/j.celrep.2020.108419. [DOI] [PubMed] [Google Scholar]
- 131.Liu X, Ouyang JF, Rossello FJ, et al. Reprogramming roadmap reveals route to human induced trophoblast stem cells. Nature. 2020;586:101–107. doi: 10.1038/s41586-020-2734-6. [DOI] [PubMed] [Google Scholar]
- 132.Roberts RM, Ezashi T, Sheridan MA, Yang Y. Specification of trophoblast from embryonic stem cells exposed to BMP4. Biol Reprod. 2018;99:212–224. doi: 10.1093/biolre/ioy070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Horii M, Touma O, Bui T, Parast MM. Modeling human trophoblast, the placental epithelium at the maternal fetal interface. Reproduction. 2020;160:R1–R11. doi: 10.1530/REP-19-0428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Das P, Ezashi T, Schulz LC, et al. Effects of fgf2 and oxygen in the bmp4-driven differentiation of trophoblast from human embryonic stem cells. Stem Cell Res. 2007;1:61–74. doi: 10.1016/j.scr.2007.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Li Y, Moretto-Zita M, Soncin F, et al. BMP4-directed trophoblast differentiation of human embryonic stem cells is mediated through a ΔNp63+ cytotrophoblast stem cell state. Development. 2013;140:3965–3976. doi: 10.1242/dev.092155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Lee Y, Kim K-R, McKeon F, et al. A unifying concept of trophoblastic differentiation and malignancy defined by biomarker expression. Hum Pathol. 2007;38:1003–1013. doi: 10.1016/j.humpath.2006.12.012. [DOI] [PubMed] [Google Scholar]
- 137.Horii M, Li Y, Wakeland AK, et al. Human pluripotent stem cells as a model of trophoblast differentiation in both normal development and disease. Proc Natl Acad Sci USA. 2016;113:E3882–3891. doi: 10.1073/pnas.1604747113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Ezashi T, Telugu BPVL, Roberts RM. Model systems for studying trophoblast differentiation from human pluripotent stem cells. Cell Tissue Res. 2012;349:809–824. doi: 10.1007/s00441-012-1371-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Vallier L, Touboul T, Chng Z, et al. Early cell fate decisions of human embryonic stem cells and mouse epiblast stem cells are controlled by the same signalling pathways. PLoS ONE. 2009;4:e6082. doi: 10.1371/journal.pone.0006082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Yu P, Pan G, Yu J, Thomson JA. FGF2 sustains NANOG and switches the outcome of BMP4-induced human embryonic stem cell differentiation. Cell Stem Cell. 2011;8:326–334. doi: 10.1016/j.stem.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Amita M, Adachi K, Alexenko AP, et al. Complete and unidirectional conversion of human embryonic stem cells to trophoblast by BMP4. Proc Natl Acad Sci USA. 2013;110:E1212–1221. doi: 10.1073/pnas.1303094110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Sudheer S, Bhushan R, Fauler B, et al. FGF inhibition directs BMP4-mediated differentiation of human embryonic stem cells to syncytiotrophoblast. Stem Cells Dev. 2012;21:2987–3000. doi: 10.1089/scd.2012.0099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Wu Z, Zhang W, Chen G, et al. Combinatorial signals of activin/nodal and bone morphogenic protein regulate the early lineage segregation of human embryonic stem cells. J Biol Chem. 2008;283:24991–25002. doi: 10.1074/jbc.M803893200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Erb TM, Schneider C, Mucko SE, et al. Paracrine and epigenetic control of trophectoderm differentiation from human embryonic stem cells: the role of bone morphogenic protein 4 and histone deacetylases. Stem Cells Dev. 2011;20:1601–1614. doi: 10.1089/scd.2010.0281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Yang Y, Adachi K, Sheridan MA, et al. Heightened potency of human pluripotent stem cell lines created by transient BMP4 exposure. Proc Natl Acad Sci USA. 2015;112:E2337–2346. doi: 10.1073/pnas.1504778112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Koel M, Võsa U, Krjutškov K, et al. Optimizing bone morphogenic protein 4-mediated human embryonic stem cell differentiation into trophoblast-like cells using fibroblast growth factor 2 and transforming growth factor-β/activin/nodal signalling inhibition. Reprod Biomed Online. 2017;35:253–263. doi: 10.1016/j.rbmo.2017.06.003. [DOI] [PubMed] [Google Scholar]
- 147.Kurek D, Neagu A, Tastemel M, et al. Endogenous WNT signals mediate BMP-induced and spontaneous differentiation of epiblast stem cells and human embryonic stem cells. Stem Cell Rep. 2015;4:114–128. doi: 10.1016/j.stemcr.2014.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Horii M, Bui T, Touma O, et al. An improved two-step protocol for trophoblast differentiation of human pluripotent stem cells. Curr Protoc Stem Cell Biol. 2019 doi: 10.1002/cpsc.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Deglincerti A, Etoc F, Ozair MZ, Brivanlou AH. Self-organization of spatial patterning in human embryonic stem cells. Curr Top Dev Biol. 2016;116:99–113. doi: 10.1016/bs.ctdb.2015.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Chhabra S, Liu L, Goh R, et al. Dissecting the dynamics of signaling events in the BMP, WNT, and NODAL cascade during self-organized fate patterning in human gastruloids. PLoS Biol. 2019;17:e3000498. doi: 10.1371/journal.pbio.3000498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Nemashkalo A, Ruzo A, Heemskerk I, Warmflash A. Morphogen and community effects determine cell fates in response to BMP4 signaling in human embryonic stem cells. Development. 2017;144:3042–3053. doi: 10.1242/dev.153239. [DOI] [PubMed] [Google Scholar]
- 152.Strumpf D, Mao C-A, Yamanaka Y, et al. Cdx2 is required for correct cell fate specification and differentiation of trophectoderm in the mouse blastocyst. Development. 2005;132:2093–2102. doi: 10.1242/dev.01801. [DOI] [PubMed] [Google Scholar]
- 153.Gao X, Nowak-Imialek M, Chen X, et al. Establishment of porcine and human expanded potential stem cells. Nat Cell Biol. 2019;21:687–699. doi: 10.1038/s41556-019-0333-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Yu L, Wei Y, Sun H-X, et al. Derivation of intermediate pluripotent stem cells amenable to primordial germ cell specification. Cell Stem Cell. 2021;28:550–567.e12. doi: 10.1016/j.stem.2020.11.003. [DOI] [PubMed] [Google Scholar]
- 155.Krendl C, Shaposhnikov D, Rishko V, et al. GATA2/3-TFAP2A/C transcription factor network couples human pluripotent stem cell differentiation to trophectoderm with repression of pluripotency. Proc Natl Acad Sci USA. 2017;114:E9579–E9588. doi: 10.1073/pnas.1708341114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Lee CQE, Gardner L, Turco M, et al. What is trophoblast? A combination of criteria define human first-trimester trophoblast. Stem Cell Reports. 2016;6:257–272. doi: 10.1016/j.stemcr.2016.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Dong C, Beltcheva M, Gontarz P, et al. Derivation of trophoblast stem cells from naïve human pluripotent stem cells. Elife. 2020;9:e52504. doi: 10.7554/eLife.52504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Cinkornpumin JK, Kwon SY, Guo Y, et al. Naive human embryonic stem cells can give rise to cells with a trophoblast-like transcriptome and methylome. Stem Cell Rep. 2020;15:198–213. doi: 10.1016/j.stemcr.2020.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Guo G, von Meyenn F, Rostovskaya M, et al. Correction: epigenetic resetting of human pluripotency. Development. 2018;145:dev166397. doi: 10.1242/dev.166397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Guo G, Stirparo GG, Strawbridge SE, et al. Human naive epiblast cells possess unrestricted lineage potential. Cell Stem Cell. 2021;28:1040–1056.e6. doi: 10.1016/j.stem.2021.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Io S, Kabata M, Iemura Y, et al. Capturing human trophoblast development with naive pluripotent stem cells in vitro. Cell Stem Cell. 2021;28:1023–1039.e13. doi: 10.1016/j.stem.2021.03.013. [DOI] [PubMed] [Google Scholar]
- 162.Chhabra S, Warmflash A. BMP-treated human embryonic stem cells transcriptionally resemble amnion cells in the monkey embryo. Biol Open. 2021;10:bio058617. doi: 10.1242/bio.058617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Ohgushi M, Eiraku M. Cell-autonomous differentiation of human primed embryonic stem cells into trophoblastic syncytia through the nascent amnion-like cell state. Dev Biol. 2021;11:5877. [Google Scholar]
- 164.Mischler A, Karakis V, Mahinthakumar J, et al. Two distinct trophectoderm lineage stem cells from human pluripotent stem cells. J Biol Chem. 2021;296:100386. doi: 10.1016/j.jbc.2021.100386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Wei Y, Wang T, Ma L, et al. Efficient derivation of human trophoblast stem cells from primed pluripotent stem cells. Sci Adv. 2021;7:eabf4416. doi: 10.1126/sciadv.abf4416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Li Z, Kurosawa O, Iwata H. Establishment of human trophoblast stem cells from human induced pluripotent stem cell-derived cystic cells under micromesh culture. Stem Cell Res Ther. 2019;10:245. doi: 10.1186/s13287-019-1339-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Jang YJ, Kim M, Lee B-K, Kim J. Induction of human trophoblast stem-like cells from primed pluripotent stem cells. Proc Natl Acad Sci USA. 2022;119:e2115709119. doi: 10.1073/pnas.2115709119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Soncin F, Morey R, Bui T, Requena DF, Cheung VC, Kallol S, Kittle R, Jackson MG, Farah O, Dumdie J, Meads M, Pizzo D, Horii M, Fisch KM, Parast MM. Derivation of functional trophoblast stem cells from primed human pluripotent stem cells. Stem Cell Rep. 2022;17(1303):1317. doi: 10.1016/j.stemcr.2022.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Hemberger M, Udayashankar R, Tesar P, et al. ELF5-enforced transcriptional networks define an epigenetically regulated trophoblast stem cell compartment in the human placenta. Hum Mol Genet. 2010;19:2456–2467. doi: 10.1093/hmg/ddq128. [DOI] [PubMed] [Google Scholar]
- 170.Donker RB, Mouillet JF, Chu T, et al. The expression profile of C19MC microRNAs in primary human trophoblast cells and exosomes. Mol Hum Reprod. 2012;18:417–424. doi: 10.1093/molehr/gas013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Pastor WA, Chen D, Liu W, et al. Naive human pluripotent cells feature a methylation landscape devoid of blastocyst or germline memory. Cell Stem Cell. 2016;18:323–329. doi: 10.1016/j.stem.2016.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Theunissen TW, Friedli M, He Y, et al. Molecular criteria for defining the naive human pluripotent state. Cell Stem Cell. 2016;19:502–515. doi: 10.1016/j.stem.2016.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Nelissen ECM, van Montfoort APA, Dumoulin JCM, Evers JLH. Epigenetics and the placenta. Hum Reprod Update. 2011;17:397–417. doi: 10.1093/humupd/dmq052. [DOI] [PubMed] [Google Scholar]
- 174.Hayashi Y, Ohnuma K, Furue MK. Pluripotent stem cell heterogeneity. Adv Exp Med Biol. 2019;1123:71–94. doi: 10.1007/978-3-030-11096-3_6. [DOI] [PubMed] [Google Scholar]
- 175.Messmer T, von Meyenn F, Savino A, et al. Transcriptional heterogeneity in naive and primed human pluripotent stem cells at single-cell resolution. Cell Rep. 2019;26:815–824.e4. doi: 10.1016/j.celrep.2018.12.099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Polo JM, Anderssen E, Walsh RM, et al. A molecular roadmap of reprogramming somatic cells into iPS cells. Cell. 2012;151:1617–1632. doi: 10.1016/j.cell.2012.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Schwarz BA, Cetinbas M, Clement K, et al. Prospective isolation of poised iPSC intermediates reveals principles of cellular reprogramming. Cell Stem Cell. 2018;23:289–305.e5. doi: 10.1016/j.stem.2018.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Kagawa H, Javali A, Khoei HH, et al. Human blastoids model blastocyst development and implantation. Nature. 2022;601:600–605. doi: 10.1038/s41586-021-04267-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Yanagida A, Spindlow D, Nichols J, et al. Naive stem cell blastocyst model captures human embryo lineage segregation. Cell Stem Cell. 2021;28:1016–1022.e4. doi: 10.1016/j.stem.2021.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Sozen B, Jorgensen V, Weatherbee BAT, et al. Reconstructing aspects of human embryogenesis with pluripotent stem cells. Nat Commun. 2021;12:5550. doi: 10.1038/s41467-021-25853-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Fan Y, Min Z, Alsolami S, et al. Generation of human blastocyst-like structures from pluripotent stem cells. Cell Discov. 2021;7:81. doi: 10.1038/s41421-021-00316-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Liu X, Tan JP, Schröder J, et al. Modelling human blastocysts by reprogramming fibroblasts into iBlastoids. Nature. 2021;591:627–632. doi: 10.1038/s41586-021-03372-y. [DOI] [PubMed] [Google Scholar]
- 183.Zhao C, Reyes AP, Schell JP, et al. Reprogrammed blastoids contain amnion-like cells but not trophectoderm. Dev Biol. 2021;28:1016. [Google Scholar]
- 184.Karvas RM, McInturf S, Zhou J, et al. Use of a human embryonic stem cell model to discover GABRP, WFDC2, VTCN1 and ACTC1 as markers of early first trimester human trophoblast. Mol Hum Reprod. 2020;26:425–440. doi: 10.1093/molehr/gaaa029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Hopf C, Viebahn C, Püschel B. BMP signals and the transcriptional repressor BLIMP1 during germline segregation in the mammalian embryo. Dev Genes Evol. 2011 doi: 10.1007/s00427-011-0373-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Schmaltz-Panneau B, Jouneau L, Osteil P, et al. Contrasting transcriptome landscapes of rabbit pluripotent stem cells in vitro and in vivo. Anim Reprod Sci. 2014;149:67–79. doi: 10.1016/j.anireprosci.2014.05.014. [DOI] [PubMed] [Google Scholar]
- 187.Tan T, Tang X, Zhang J, et al. Generation of trophoblast stem cells from rabbit embryonic stem cells with BMP4. PLoS ONE. 2011;6:17124. doi: 10.1371/JOURNAL.PONE.0017124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Lee VH, Zhang SJ, Chang SMT, et al. In vitro transformation of rabbit cytotrophoblast cells into syncytiotrophoblast: stimulation of hormone secretion by progesterone and dibutyryl cyclic 3’,5’-adenosine monophosphate. Biol Reprod. 1995;52:868–877. doi: 10.1095/BIOLREPROD52.4.868. [DOI] [PubMed] [Google Scholar]
- 189.La Rosa I. Bone morphogenetic proteins in preimplantation embryos. Vitam Horm. 2015;99:223–248. doi: 10.1016/BS.VH.2015.04.001. [DOI] [PubMed] [Google Scholar]
- 190.Lee K-B, Folger JK, Rajput SK, Smith GW. Temporal regulation of mRNAs for select bone morphogenetic proteins (BMP), BMP receptors and their associated SMAD proteins during bovine early embryonic development: effects of exogenous BMP2 on embryo developmental progression. Reprod Biol Endocrinol. 2014;12:67. doi: 10.1186/1477-7827-12-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.García EV, Miceli DC, Rizo G, et al. Effect of early addition of bone morphogenetic protein 5 (BMP5) to embryo culture medium on in vitro development and expression of developmentally important genes in bovine preimplantation embryos. Theriogenology. 2015;84:589–599. doi: 10.1016/j.theriogenology.2015.04.018. [DOI] [PubMed] [Google Scholar]
- 192.Pennington KA, Ealy AD. The expression and potential function of bone morphogenetic proteins 2 and 4 in bovine trophectoderm. Reprod Biol Endocrinol. 2012 doi: 10.1186/1477-7827-10-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Degrelle SA, Lê Cao K-A, Heyman Y, et al. A small set of extra-embryonic genes defines a new landmark for bovine embryo staging. Reproduction. 2011;141:79–89. doi: 10.1530/REP-10-0174. [DOI] [PubMed] [Google Scholar]
- 194.La Rosa I. Bone morphogenetic proteins in preimplantation embryos. Vitamins and hormones. Cambridge: Academic Press; 2015. pp. 223–248. [DOI] [PubMed] [Google Scholar]
- 195.La Rosa I, Camargo LSA, Pereira MM, et al. Effects of bone morphogenic protein 4 (BMP4) and its inhibitor, Noggin, on in vitro maturation and culture of bovine preimplantation embryos. Reprod Biol Endocrinol. 2011;9:18. doi: 10.1186/1477-7827-9-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Suzuki Y, Koshi K, Imai K, et al. Bone morphogenetic protein 4 accelerates the establishment of bovine trophoblastic cell lines. Reproduction. 2011;142:733–743. doi: 10.1530/REP-11-0275. [DOI] [PubMed] [Google Scholar]
- 197.Valdez Magaña G, Rodríguez A, Zhang H, et al. Paracrine effects of embryo-derived FGF4 and BMP4 during pig trophoblast elongation. Dev Biol. 2014;387:15–27. doi: 10.1016/J.YDBIO.2014.01.008. [DOI] [PubMed] [Google Scholar]
- 198.Yuan Y, Park J, Tian Y, et al. A six-inhibitor culture medium for improving naïve-type pluripotency of porcine pluripotent stem cells. Cell Death Discov. 2019;5:104. doi: 10.1038/s41420-019-0184-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Cabrera-Sharp V, Read JE, Richardson S, et al. SMAD1/5 signaling in the early equine placenta regulates trophoblast differentiation and chorionic gonadotropin secretion. Endocrinology. 2014;155:3054–3064. doi: 10.1210/en.2013-2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Nakamura T, Okamoto I, Sasaki K, Yabuta Y, Iwatani C, Tsuchiya H, Seita Y, Nakamura S, Yamamoto T, Saitou M. A developmental coordinate of pluripotency among mice, monkeys and humans. Nature. 2016;537:57–62. doi: 10.1038/nature19096. [DOI] [PubMed] [Google Scholar]
- 201.Kobayashi M, Takada T, Takahashi K et al (2008) BMP4 induces primitive endoderm but not trophectoderm in monkey embryonic stem cells. Cloning Stem Cells 10:495–502. 10.1089/CLO.2008.0030. https://home.liebertpub.com/clo [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.


