Abstract
The construction of three-dimensional multi-modal tissue maps provides an opportunity to spur interdisciplinary innovations across temporal and spatial scales through information integration. While the preponderance of effort is allocated to the cellular level and explore the changes in cell interactions and organizations, contextualizing findings within organs and systems is essential to visualize and interpret higher resolution linkage across scales. There is a substantial normal variation of kidney morphometry and appearance across body size, sex, and imaging protocols in abdominal computed tomography (CT). A volumetric atlas framework is needed to integrate and visualize the variability across scales. However, there is no abdominal and retroperitoneal organs atlas framework for multi-contrast CT. Hence, we proposed a high-resolution CT retroperitoneal atlas specifically optimized for the kidney organ across non-contrast CT and early arterial, late arterial, venous and delayed contrast-enhanced CT. We introduce a deep learning-based volume interest extraction method by localizing the 2D slices with a representative score and crop within the range of the abdominal interest. An automated two-stage hierarchal registration pipeline is then performed to register abdominal volumes to a high-resolution CT atlas template with DEEDS affine and non-rigid registration. To generate and evaluate the atlas framework, multi-contrast modality CT scans of 500 subjects (without reported history of renal disease, age: 15–50 years, 250 males & 250 females) were processed. PDD-Net with affine registration achieved the best overall mean DICE for portal venous phase multi-organs label transfer with the registration pipeline (0.540 ± 0.275, p < 0.0001 Wilcoxon signed-rank test) comparing to the other registration tools. It also demonstrated the best performance with the median DICE over 0.8 in transferring the kidney information to the atlas space. DEEDS perform constantly with stable transferring performance in all phases average mapping including significant clear boundary of kidneys with contrastive characteristics, while PDD-Net only demonstrates a stable kidney registration in the average mapping of early and late arterial, and portal venous phase. The variance mappings demonstrate the low intensity variance in the kidney regions with DEEDS across all contrast phases and with PDD-Net across late arterial and portal venous phase. We demonstrate a stable generalizability of the atlas template for integrating the normal kidney variation from small to large, across contrast modalities and populations with great variability of demographics. The linkage of atlas and demographics provided a better understanding of the variation of kidney anatomy across populations.
Keywords: Retroperitoneal atlas, Deep learning, Multi-contrast computed tomography, Medical image registration, Kidney atlas
1. Introduction
Physiological and metabolic processes are performed in parallel in the human body. Complicated relationships between cells are required for proper organ function but are challenging to analyze. Extensive studies mapping the organization and molecular profiles of cells within tissues or organs are needed across the human body. While the majority of efforts are distributed to the cellular and molecular perspectives [1], generalizing information from cell to organ level is essential to provide a better understanding of the functionality and linkage across scales [2]. The use of computed tomography provides an opportunity to contextualize the anatomical characteristics of organs and systems in the human body. In addition, by creating a generalizable framework with integration of micro-scale information and system-scale information, this will provide clinicians and researchers the ability to visualize the complex organization of tissues from a cellular to tissue level.
Abdominal CT provides information about abdominal organs at a system scale. Contrast enhancement demonstrates additional anatomical and structural details of organs and neighboring vessels by injecting a contrast agent before imaging procedures. Five different contrast phases are typically generated corresponding to the timing of the contrast agent in the imaging cycle: 1) non-contrast, 2) early arterial, 3) late arterial, 4) portal venous and 5) delayed. The intensity range of organs fluctuates across the contrast enhanced imaging cycle and the variation of intensity helps to capture and specify contextual features of each specific organ. The kidneys, which are located retroperitoneally, also have challenges in imaging. From the anatomical information provided from the contrast enhanced CT of large clinical cohorts, healthy kidney morphometry and appearance may vary. An atlas reference framework is needed to generalize the anatomical and contextual features across the variations in sex, body size, and imaging protocols. However, due to the large variability in anatomy and morphology of various abdominal and retroperitoneal organs, generating a standard reference template for each of these organs is still challenging and no atlas framework for abdominal or retroperitoneal organs is currently publicly available.
Creating an atlas for particular anatomical regions has widely been used with magnetic resonance imaging (MRI). Extensive efforts are allocated in multiple perspectives of brain atlas with brain MRI. Kuklisova-Murgasova et al. generated multiple atlases for early developing babies with age ranging from 29 to 44 weeks using affine registration [3], while Shi et al. proposed an infant brain atlas using unbiased group-wise registration with three varying scanning time points of brain MRI from 56 males and 39 females normal infants [4]. Unbiased spatiotemporal 4-dimensional MRI atlas and time-variable longitudinal MRI atlas for infant brain were also proposed by Ali et al. [5]. with diffeomorphic deformable registration with kernel regression in age, and by Yuyao et al. using patch-based registration in spatial-temporal wavelet domain respectively [6]. Apart from generating atlas framework for normal brain, Rajashekar et al. generated two high-resolution normative disease-related brain atlases in FLAIR MRI and non-contrast CT modalities, to investigate lesion-related diseases such as stroke and multiple sclerosis in elderly population [7]. Meanwhile, limited studies have proposed creating a standard reference framework for abdominal and retroperitoneal organs. Development of abdominal and retroperitoneal organ atlases is challenging across multi-modality images (CT and MRI) due to the limited robustness of the registration methods and significant morphology variations in multiple organs associated with patients’ demographics [8].
To generate an atlas framework with stable context transfer, previous works have sought to improve the registration performance and the conventional frameworks perform spatial alignments between one image to another following with a global affine transformation and deformable transformation [9–11]. Significant effort has been invested in brain imaging by optimizing a regularized deformation field to match the moving image to a single fixed target with classical approach such as b-spline based deformation [12], discrete optimization [13,14], Demons [15], and Symmetric normalization [10]. Apart from the traditional approaches, learning-based methods provide an opportunity to reduce the time of inference and extract meaningful feature representation for deformation prediction. Voxelmorph provides an opportunity to learn a generalizable function to compute deformation field with unsupervised setting and localize the deformation with few millimeters [16,17]. For abdominal imaging, significant variation in body size and shape of organs are demonstrated with the several centimeters and large deformation for organs interest is needed to transfer the anatomical context to match the morphology of the fixed image. Zhao et al. proposed a recursive a cascaded network that use the organ labels to crop the region of interest (ROI) of specific organs and progressively refine the intermediate registered image to the fixed image space with multiple cascaded models [18]. Additionally, Yan et al. proposed a deep learning framework to compute bounding boxes that localize different organ of interest and use the organ-corresponding volumetric patches for non-rigid registration [19]. To further extend the abdominal registration with complete abdominal volumetric scans, Heinrich et al. proposed a probabilistic dense displacement network to adapt the large anatomical difference with organ label supervision [20]. This network aims to improve the robustness of the registration performance in the abdominal regions with limited label guidance and capture the deformation with lightweight feature representation.
In this work, we present a contrast-preserving CT retroperitoneal atlas framework, optimized for healthy kidney organs with contrast-characterized variability and the generalizable features across a large population of clinical cohorts as shown in Fig. 1. Specifically, as to reduce the failure rate of transferring the morphological and contrastive characteristics of kidney organs, we initially extracted the abdominal-to-retroperitoneal volume of interest with a similar field of view to the atlas target image, using a deep neural network called body part regression (BPR) [21]. 2D slices of the CT volume assessed with BPR model and generate a value ranging from −12 to +12, corresponding to the upper lung region and the pelvis region respectively in the body. By limiting the range of values for both abdominal and retroperitoneal regions, each CT volume is cropped and excludes other regions apart from the abdomen and retroperitoneum, such as the lung and pelvis. A two-stage hierarchical registration pipeline is then performed, registering the extracted volume interest to the high-resolution atlas target with traditional metric-based [22,23] and deep learning based [20,24] method respectively across all contrast phases. To ensure the stability and the variation localized in the atlas template, average and variance mappings across the multi-contrast registered output is computed to demonstrate a better understanding of anatomical details of kidney organs across different contrasts. Overall, our main contributions are summarized as:
We constructed the first multi-contrast CT healthy kidney atlas framework for the public usage domain.
We proposed metric-based and deep learning-based framework optimizing for kidney organs with corresponding contrast phase, and generalized the anatomical context of kidneys with significant variation of morphological and contrastive characteristics across demographics and imaging protocols.
We evaluate the generalizability of the atlas template by transferring the atlas target label to the 13 organs well-annotated CT space with inverse transformation. An unlabeled multi-contrast phase CT cohort is used to compute average and variance mapping to demonstrate the effectiveness and stability of the proposed atlas framework. Our proposed atlas framework demonstrates a stable transfer ability in both left and right kidneys with median Dice above 0.8.
The average template generated, and the associated 13 organs labels will be public for usage through HuBMAP.
Fig. 1.
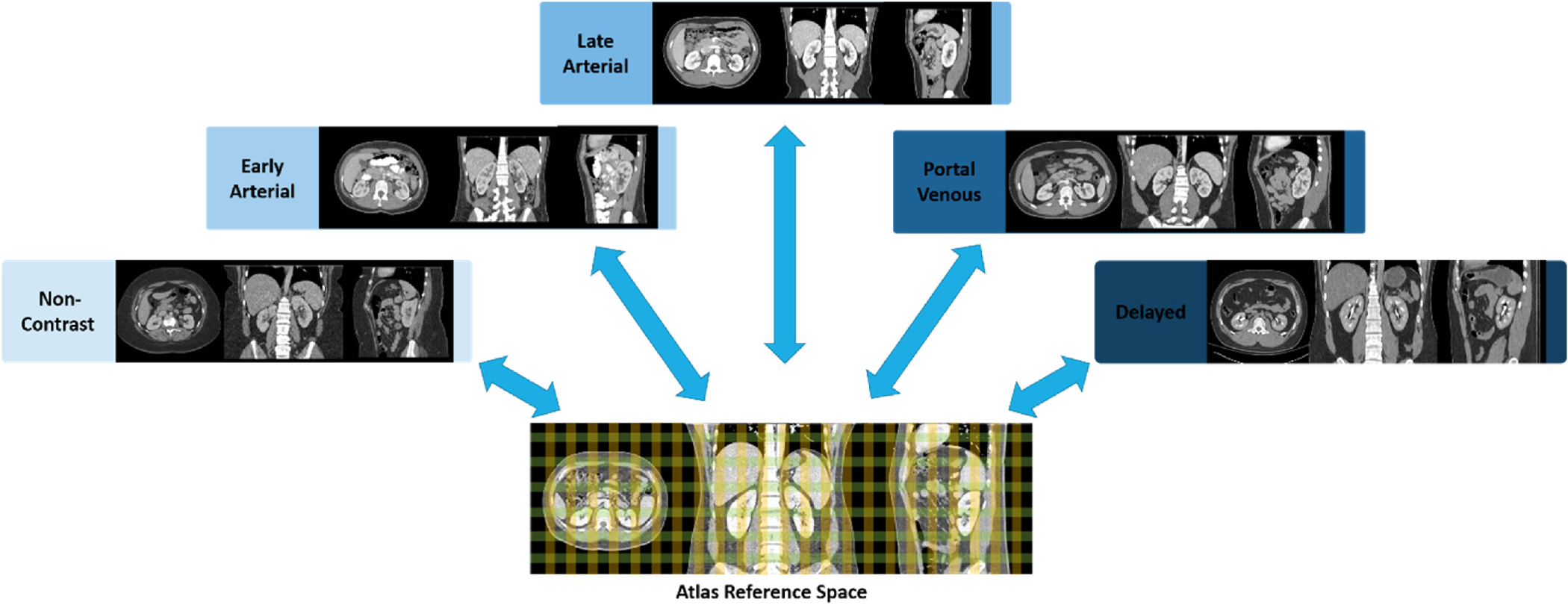
Illustration of multi-contrast phase CT atlas. The color grid in the three-dimensional atlas space represents the defined spatial reference for the abdominal-to-retroperitoneal volume of interest and localizes abdominal and retroperitoneal organs with each contrast phase characteristics. Blue arrows represent the bi-directional transformation across the atlas target defined spatial reference and the original source image space.
2. Materials and methods
Fig. 2 presents an overview of the complete pipeline for generating the kidney atlas framework. The volume of interest is first extracted with a deep learning-based BPR algorithm to obtain a similar field of view with the atlas target image, increasing the stability of registration in the abdominal body and kidneys. Here, we define the stability as the atlas not changing with randomized subjects and measure the stability with the mean or variance mapping of the atlas template. The integration of deep learning-based volume of interest extraction and classical registration provided an opportunity to reduce the subjectivity of choosing the field of view between source and target image, and increase the robustness of the image registration across the clinical cohorts.
Fig. 2.
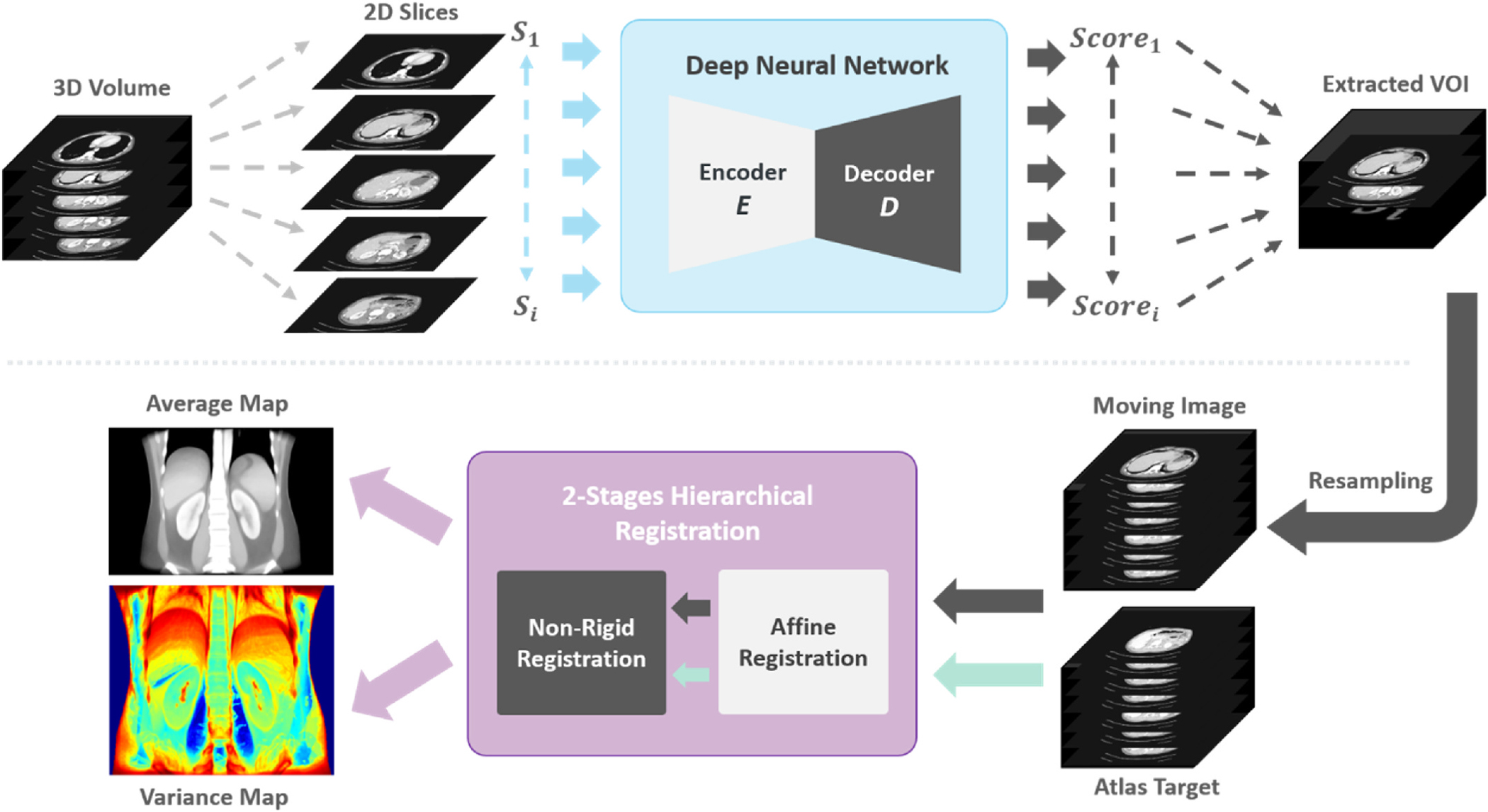
The overview of the complete pipeline to create kidney atlas template is illustrated. The input volume is initially cropped to a similar field of view with the atlas target. The extracted volumes of interest are resampled to the same resolution and dimension of the atlas template and performed 2-stages hierarchical registration. The successfully registered scans are finally used to compute the average template and the variance maps.
2.1. Dataset of studies
We evaluated the stability of our kidney atlas with a large cohort of multi-contrast unlabeled CT and a public portal venous contrast phase multi-organ labeled dataset. All CT data are unpaired and collected from different cohorts. With the use of both labeled and unlabeled datasets, we conducted comprehensive qualitative and quantitative evaluations for generalizing cross-modality information of kidney organs in CT.
Clinical Multi-Contrast Abdominal CT Cohort:
A large clinical cohort of multi-contrast CT was employed for abdominal and retroperitoneal organs registration. In total, 2000 patients’ de-identified CT data were initially retrieved in de-identified form from ImageVU with the approval of Institutional Review Board (IRB). In these 2000 patients, since some had renal disease, criteria in ICD-9 codes and age range from 18 to 50 years old were set and applied to extract scans with healthy kidneys from all subjects. 720 subjects out of 2000 were identified after quality assessment and extract the corresponding contrast phase abdominal CT scans, which included 290 unlabeled CT volumes in total with: 1) non-contrast: 50 vol, 2) early arterial: 30, 3) late arterial: 80 vol, 4) portal venous: 100 vol, 5) delayed: 30 volumes. All CT volumes are initially reoriented to standard orientation before further processing [25]. BPR was performed to each modality volumes and obtain the similar field of view with the atlas target. They were then resampled to the same resolution and dimensions with the atlas target for performing registration pipeline. We aim to adapt a generalized atlas framework for localizing the anatomical and contextual characteristics of kidney organs across multi-contrast.
Multi-Organ Labeled Portal Venous Abdominal CT Cohort:
We used a separate healthy clinical cohort with 100 portal venous contrast phase abdominal CT volumes and 20 of the volumetric scans are the testing scans in the 2015 MICCAI Multi-Atlas Abdomen Labeling challenge. The ground truth labels of 13 multiple organs are provided including: 1) spleen, 2) right kidney, 3) left kidney, 4) gall bladder, 5) esophagus, 6) liver, 7) stomach, 8) aorta, 9) inferior vena cava (IVC), 10) portal splenic vein (PSV), 11) pancreas, 12) right adrenal gland (RAD), 13) left adrenal gland (LAD), with which we conduct label transfer on this dataset to evaluate the generalizability and stability of the atlas template. In order to reduce the number of failed registrations to the atlas target, BPR is performed on abdominal and retroperitoneal regions only with soft-tissue window. To evaluate the atlas framework, the inverse transformation was applied to the multi-organ atlas label, and labels were transferred back to the spatial space of each portal venous phase CT.
High Resolution Single Subject Atlas Template:
We choose the single subject atlas template with several conditions: 1) high-resolution characteristics, 2) significant appearance in kidney morphology and 3) clear kidney boundary with contrast. The atlas template is provided by Human Biomolecular Atlas Program (HuBMAP) with high resolution setting of 0.8 × 0.8 × 0.8 and healthy condition. The dimension of the atlas subject is 512 × 512 × 434 with 13 Organs annotated by joint label fusion from the registered subjects. The volumetric measure of both left and right kidneys are ~256 cc.
2.2. Deep body part regression network
The use of deep learning in the medical imaging domain contributed to a great increase in automatic models for classification and segmentation. Due to the shift of various domains and the variation of imaging protocols, medical images usually present with different visual appearances and fields of view. The goal of the body part regression network (BPR) is to narrow the difference of field of view between the source images and the atlas target image for reducing the failure rate of registration. Formally, given an unlabeled dataset from the moving image domain, and a labeled dataset from the atlas target domain, we aim to crop the original volume of interest to an approximate field of view with the atlas target . The obtained volume of interest only consists of abdomen regions and is resampled to the same voxel resolution as the atlas target.
Tang et al. proposed a self-supervised method to predict a continuous score for each axial slice of CT volumes as the normalized body coordinate value without any labels [21,26]. The self-supervised model predicts scores in the range of −12 to +12 and each body part region corresponds well to an approximate score (e.g., −12: upper chest, −5: diaphragm/upper liver, 4: lower retroperitoneum, 6: pelvis). Linear regression is performed to correct the discontinuity of the predicted score, and we use the regressed output as the self-supervised label to train a new refined model. Both the atlas target image and the unlabeled dataset are input into the well-trained model and compute scores for each slice of the volume. To extract the abdominal-to-retroperitoneal regions only for each dataset, we limit the slices with a range of scores from −5 to 5 and crop the slices that are out of this range. All unlabeled datasets are then enforced to have a closer field of view to the atlas target image .
2.3. Two-stage hierarchal metric-based registration pipeline
The metric-based registration pipeline is composed of 2 hierarchal stages: 1) affine registration and 2) non-rigid registration. Dense displacement sampling (DEEDS) is a 3D medical registration tool with a discretized sampling space that has been shown to yield a great performance in abdominal and retroperitoneal organs registration, is used for both affine non-rigid registration in this pipeline [8,22,23]. The DEEDS affine registration is first performed to initially align both moving images and the atlas target to preserve 12° of freedom of transformation and provide a prior definition of the spatial context and each affine component. An affine transformation matrix is generated as the output and become the second stage non-rigid registrations’ input. The DEEDS non-rigid registration is refined with the spatial context as the local voxel-wise correspondence with its specific similarity metric, which will be illustrated below. Five different scale levels are used with grid spacing ranging from 8 to 4 voxels to extract patches and displacement search radii from six to two steps between 5 and 1 voxels [8,22,23,27]. Deformed scans with the displacement data from selecting control points is generated and transfer the source image space voxel information to the atlas target space after deformation. To ensure the stability of the atlas generated, all successfully deformed scans are averaged and variance mapping is used to visualize the intensity fluctuation and variation around the abdominal body and kidney organs.
The similarity metric defined in DEEDS registration tool is self-similarity context with the patches extracted from moving images. Such a similarity metric aims to find a similar context around neighboring voxels in patches [22]. The self-similarity metric S is optimizing a distance function between the image patches extracted from the moving image . A function is computed to estimate both the noise in local and global perspectives. Meanwhile, a certain number of neighborhood relations is also defined as to determine the kinds of self-similarities in the neighborhood. As extracting an image patch with a center at , the measurement calculation of the self-similarity can be demonstrated as follows:
| (1) |
where is defined as the center of another patch from one of the neighborhood . This similarity metric helps to avoid the negative influence of image artifacts or random noise from the central patch extracted and prevent a direct adverse effect in calculation. Twelve distances between pairwise patches are calculated within six neighborhoods and concentrate in extracting the contextual neighboring information, instead of the direct shape representation.
2.4. Experimental settings
In the preprocessing stage, the BPR model with U-Net like architecture is pretrained with a total of 230,625 2D slices from a large-scale cohorts of 1030 whole body CT scans from the public domain [26]. The portal venous multi-organ labeled cohorts are used only for external validation. The pretrained model is trained from scratch and optimized with Adam using learning rate of 0.0001. The batch size is 4 for end-to-end training.
For the atlas generation process, we investigate multiple registration methods including traditional registration tools and deep learning registration methods to compare the robustness of registration across all contrast phases scans in qualitative and quantitative perspective. We performed comprehensive analysis which included ANTS [8,10], NIFTYREG (NIFTYR) [8,28] and DEEDS [8,22,23,27] as the traditional medical image registration tools, while VoxelMorph [16] and PDD-Net [20] are used as deep learning-based registration framework on our portal venous dataset. For metric-based registration, the preprocessed moving scans are upsampled to the same resolution with the preprocessed atlas target and directly performed hierarchical affine and deformable registration in high resolution setting. For deep learning-based method, we downsampled the moving scans and the atlas target to a resolution of 1.5 × 1.5 × 1.5 with dimension 192 × 160 × 256. Affine registration is performed with NiftyReg to coarse align the moving scans and the atlas fixed target before non-linear mapping prediction with deep network pipelines. VoxelMorph is trained with the multi-organ labeled portal venous scans as the moving scans and the atlas target as the fixed scans in unsupervised setting with 100 epochs and learning rate of 0.0001. We use the pretrained model of PDD-Net from [20] and directly predict a 4D displacement field for non-linear transformation. The pretrained model of PDD-Net is trained with 3-fold cross validations on 10 contrast-enhanced CT scans of the VISCERAL3 training data with learning rate of 0.01 for 1500 iterations [29]. The training process of both VoxelMorph and PDD-Net are optimized end-to-end with Adam and batch size of 1. The predicted deformation fields from deep learning based framework are finally upsampled to the atlas template resolution and compute deformation warp with non-linear transformation in high resolution setting.
For ablation studies, we further investigate the effectiveness on the BPR preprocessing and located the affect of the significant difference of field of view for registration with variance mapping. As there is significant intensity variation across each organ interests in each phase, we investigate the effect of domain shift for the robustness of the registration pipeline and the label transfer of non-contrast phase dataset is further evaluated to demonstrate the registration stability with significant domain shift across metric-based to deep learning-based framework.
2.5. Evaluation metrics
We employed three commonly used metrics to evaluate the similarity between the prediction label from automatic models and the original ground truth label: 1) Dice score, 2) mean surface distance (MSD), and 3) Hausdorff distance (HD).
The definition of Dice is to measure the overlapping of volume between the segmentation label prediction and the ground truth segmentation label:
Experimental results are measured by accuracy score.
| (3) |
where is the predicted label from models and is the original ground truth segmentation label, while || is the L1 norm operation.
The rendered surface is another perspective which used to evaluate result. The 3-dimensional coordinates of vertices were initially extracted from both the prediction label and the ground truth label. The average distance and the Hausdorff distance between the sets of vertices coordinate were calculated as follows:
| (4) |
| (5) |
where and represents the vertices coordinates of prediction label and ground truth label respectively, while avg refers to average, and sup and inf refers to the greatest lower bound and least upper bound of the distance function measure Dist.
3. Results
Registrations were performed to the atlas target image with the preprocessed cropped volume of interest. We performed five registration methods with portal venous cohorts to ensure the highest accuracy for the localization of both left and right kidneys. The quantitative representation of each organ in the registered output was then demonstrated in terms of Dice score, MSD and HD, and illustrated the distribution of the performance across the multi-organ labeled portal venous cohort with Fig. 3 and Table 1. As shown in Table 1, the deep registration pipeline PDD-Net with affine registration from NiftyReg achieved the best overall mean Dice across all 13 organs. From the demonstration of Fig. 3, NiftyReg demonstrated a better performance in registering liver organ comparing to DEEDS, while ANTs performed registration with inferiority across all organs comparing to the other two methods. Apart from comparing with metric-based methods, we also performed deep learning registration baseline with VoxelMorph and characterize the efficiency of the registration with deep networks. The performance of PDD-Net outperforms VoxelMorph with 24.2% Dice across 13 organs by training from scratch with both with or without initial affine registration scenario. In terms of optimizing for kidney organs with the atlas template, the Dice score of both left and right kidneys are separately computed to obtain the ability of kidneys localization with the atlas template. Both PDD-Net and DEEDS have a comparable performance in the kidney regions and outperforms the registration performance of kidney with NiftyReg and ANTs. PDD-Net demonstrate significant improvement in right kidneys registrations, while DEEDS perform better with a small extent in transferring the left kidney regions. NiftyReg and ANTs illustrated the lack of generalizability of transferring kidney organs and computed significant variance of Dice across all registrations. The reduction of variance and population of outliers are shown from boxplots with PDD-Net, leading to a significant improvement of Dice score, MSD, and HD for both left and right kidneys comparing to the other two methods. The Wilcoxon signed-rank test showed that PDD-Net was significantly better (p < 0.0001) than all other methods in Dice [30]. The median Dice of both transferred left and right kidney using DEEDS and PDD-Net are above 0.8, while it is a significant boost comparing with the other registration pipelines.
Fig. 3.
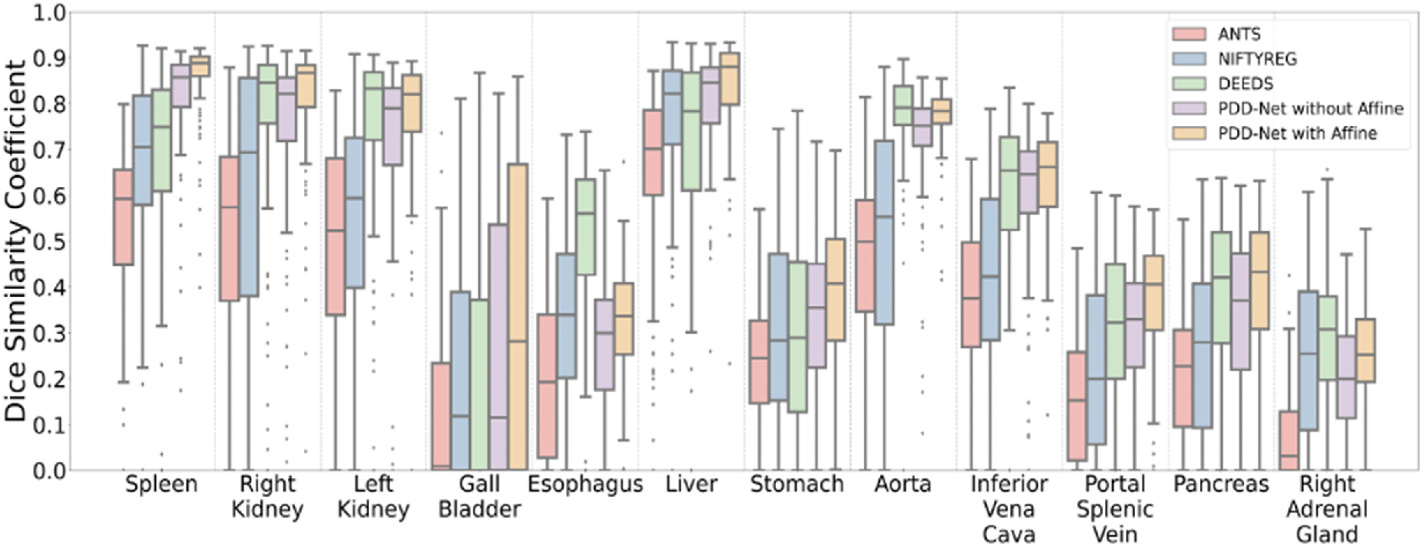
The quantitative representation of label transfer with multi-organ portal venous dataset are demonstrated that PDD-Net with affine registration outperforms the other four traditional methods in an organ-wise manner. Significant increase of DICE is also demonstrated with median Dice score over 0.8 in the transferring result of both left and right kidneys using PDD-Net with improving outliers.
Table 1.
Evaluation metric on 100 Portal Venous registration for three registration methods on 13 Organs (Mean ± STD).
| Methods | Dice Score | MSD (mm) | HD (mm) |
|---|---|---|---|
|
| |||
| ANTS (A) | 0.246 ± 0.224 | 12.8 ± 10.2 | 60.2 ± 47.5 |
| NIFTYR (A) | 0.270 ± 0.222 | 13.1 ± 11.1 | 62.1 ± 50.1 |
| DEEDS (A) | 0.200 ± 0.199 | 19.6 ± 18.2 | 80.9 ± 59.3 |
| ANTS | 0.319 ± 0.252 | 10.1 ± 9.00 | 51.6 ± 45.3 |
| NIFTYR | 0.406 ± 0.279 | 10.9 ± 13.8 | 55.0 ± 51.8 |
| DEEDS | 0.496 ± 0.284 | 8.52 ± 17.1 | 41.6 ± 51.2 |
| VoxelMorph without A | 0.335 ± 0.275 | 12.5 ± 10.9 | 34.9 ± 21.7 |
| VoxelMorph with A | 0.435 ± 0.283 | 9.27 ± 9.47 | 30.8 ± 23.5 |
| PDD-Net without A | 0.486 ± 0.286 | 8.47 ± 9.87 | 31.0 ± 25.0 |
| PDD-Net with A * | 0.540 ± 0.275 | 6.92 ± 8.73 | 28.5 ± 25.4 |
Note that p < 0.0001 with Wilcoxon signed-rank test
A: affine registration only.
Apart from the quantitative result, we compare the qualitative representations of DEEDS and PDD-Net registrations across multiple contrast phases and shown in terms of average template and variance mapping in Fig. 4 and Fig. 5. The average mapping of each contrast phase was then computed with all registered contrast-corresponding volumes. With DEEDS, the contrast and anatomical context of kidney regions in each phase are stably transferred to the atlas space, while other organs’ regions such as liver and spleen, are demonstrated with blurry appearance. With PDD-Net, the contextual representations of the kidney regions are demonstrated with sharper appearance comparing with the DEEDS average template in early arterial, late arterial and portal venous phase. The anatomy of kidney sub-structure and renal-related vessels can appear with sharpness in the average template of early arterial and late arterial phase. However, the non-contrast and delayed phase template demonstrate an unclear structure of the kidney anatomy with PDD-Net. To further ensure the stability of transferring the kidneys’ anatomical information, variance maps of each contrast phase template with corresponding registration methods are also computed to demonstrate the voxel variability of each organ across the clinical cohort. With DEEDS registration, the small variation in the kidney is illustrated with a color range from yellow to green, while significant variation in voxels is shown near the diaphragm region and the color range from orange to red indicated the highly deformed variability across the registered outputs. For PDD-Net, the boundary around the kidney regions demonstrated significant variation across each contrast phase, while the intensity variance is small in the renal cortex regions in late arterial and portal venous phase. The variance map of non-contrast and delayed phase demonstrate the significant variability across the contrast intensity and the kidney anatomy, which correlated the instability of the registration of both phase subject scans. Overall, contrast phase context can be stability preserved with DEEDS in the kidney regions and PDD-Net demonstrated a higher robustness of transferring the kidney findings with sharp appearance in late arterial and portal venous phase.
Fig. 4.
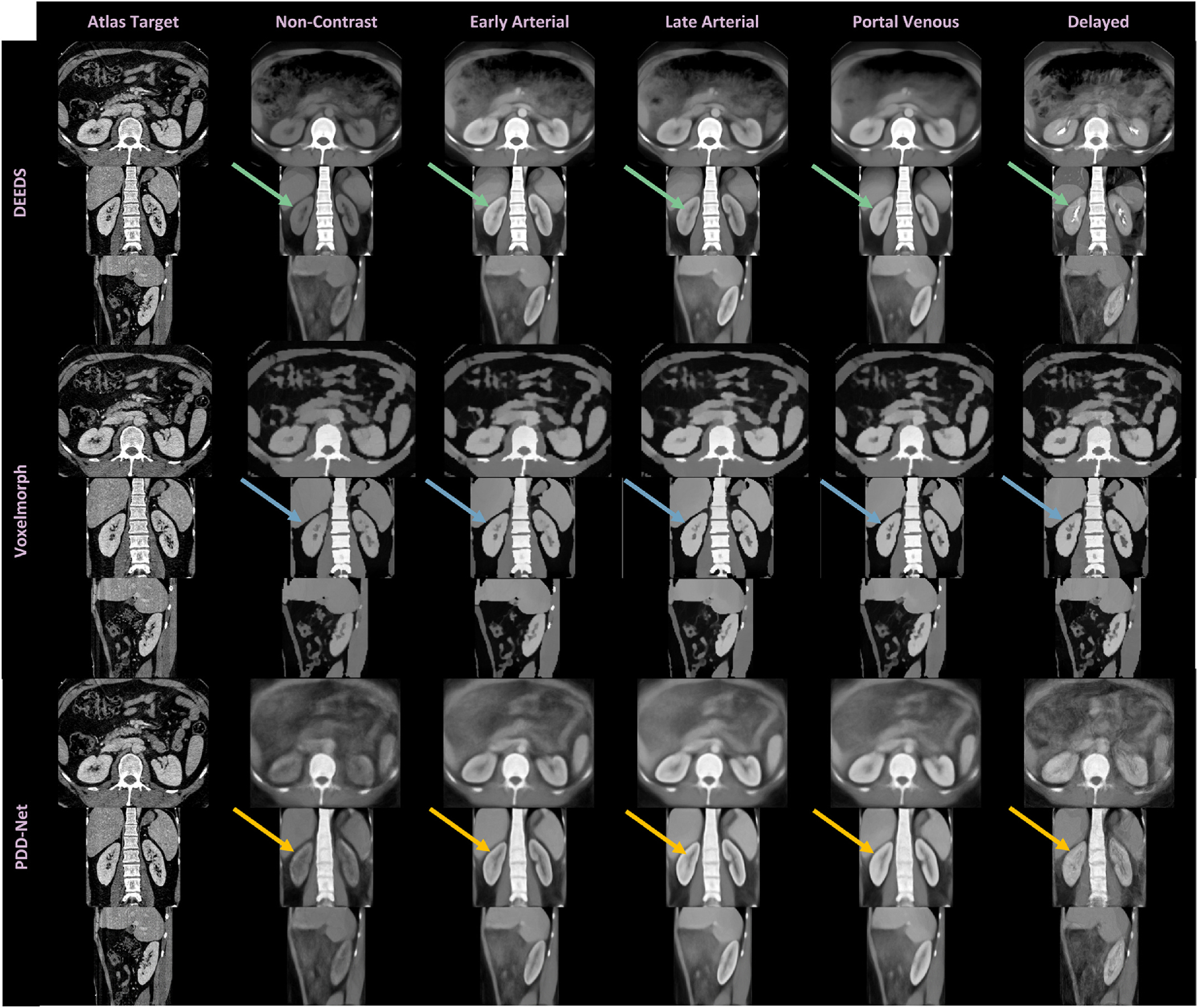
We investigate the registration stability across all contrast phase with average mapping. The metric-based registration representative DEEDS demonstrates a stable transfer of the kidney contextual findings across all phases, while the deep learning representation PDD-Net illustrates sharp appearance in the kidney sub-structural context in contrast-enhanced phase such as late arterial and portal venous, but with unstable registration appearance in non-contrast and delayed phase. We additionally compare the average mapping from the VoxelMorph. It is limited to transfer the sub-structural contrast characteristics and preserve the boundary of kidney organs well compared with DEEDS and PDD-Net. (See arrows).
Fig. 5.
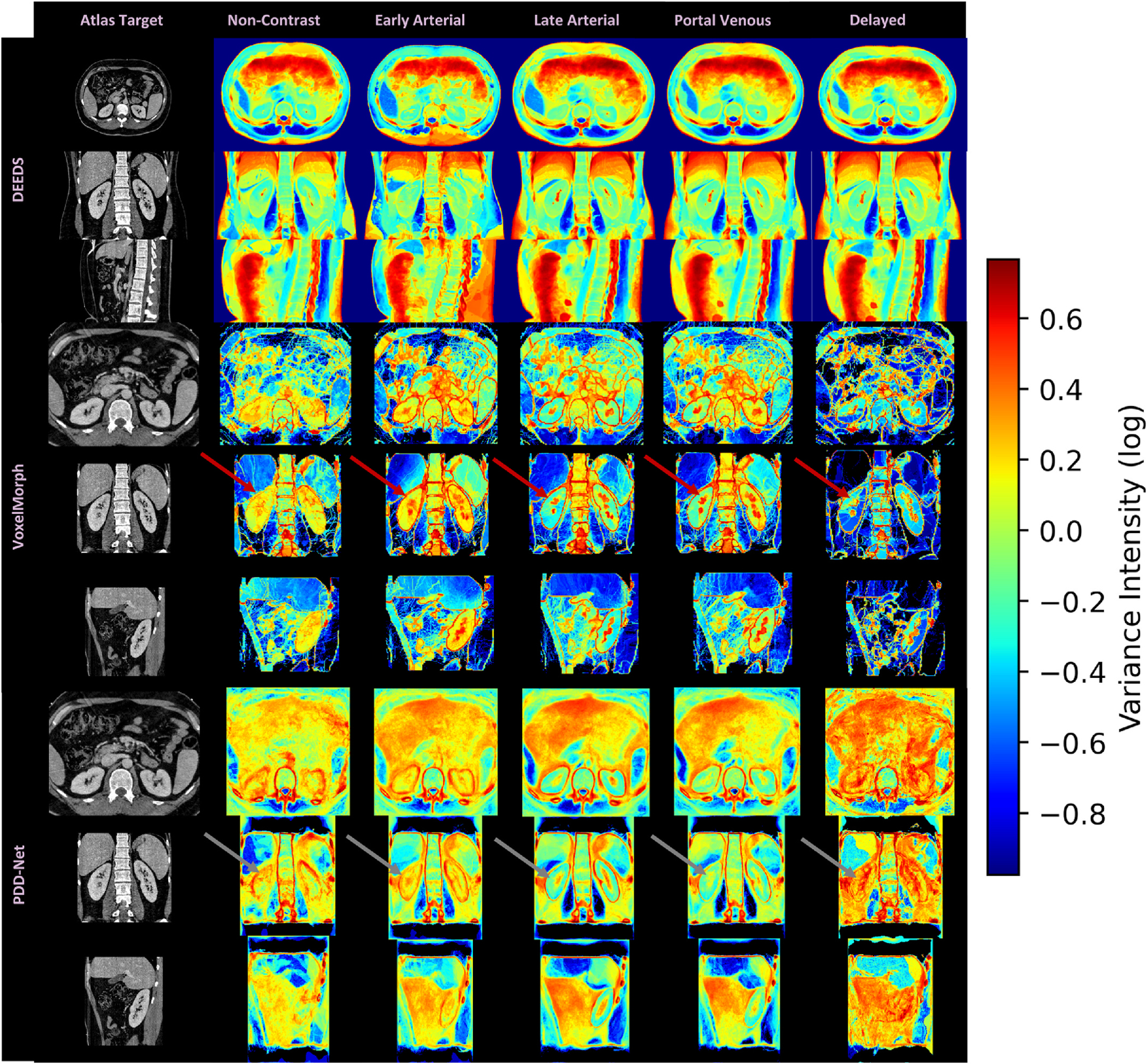
We further evaluate the intensity variance across the registration outputs with the average template in each contrast phase. The variance mapping of DEEDS demonstrates the kidney context transferal with stability and the variance value near the kidney region is ~0–0.15, while significant variance are localized in the boundary of the kidney region with the variance mapping of PDD-Net and VoxelMorph. For late arterial and portal venous phase, PDD-Net well preserved the core context of the renal cortex region. However, unstable registrations are demonstrated with the high variance value in kidney with non-contrast and delayed phase mapping (see arrows), which match the blurry appearance of kidney regions in the average mapping.
4. Discussion
In this study, we constructed a healthy kidney atlas for five different contrast phases CT and generalized the kidney anatomical context across population demographics and the variation of contrast characteristics. High variance score is located near the diaphragm regions and the boundary of the abdominal body (see Fig. 5). Such variability is contributed to the large deformation of the lower and upper boundary of the volume interest and transfer specific organs’ contextual information to other organs’ anatomical locations. The low variance score in both the left and right kidney region indicated the stable registration of structural information, as the kidney organs are localized in the middle region of the volume interest with contrast. The surface rendering of the kidney across the morphological sizes visualizes the generalizability of the atlas template across shape variability (see Fig. 5). We adapt a 2D color-space checkerboard to visualize the deformation on the surface. The color of each box in the checkerboard pattern changes both horizontally and vertically. The color-boxes in atlas space are equivalent to that in the inverse original space. The smoothnesss of the registration is qualitatively defined by the movement of the checkerboard pattern. If there is significant movement of the colored pattern, there is significant deformation on that particular regions and the smoothness of deformation field is low. From Fig. 7, stable deformation is demonstrated across small to large kidneys.
Fig. 7.
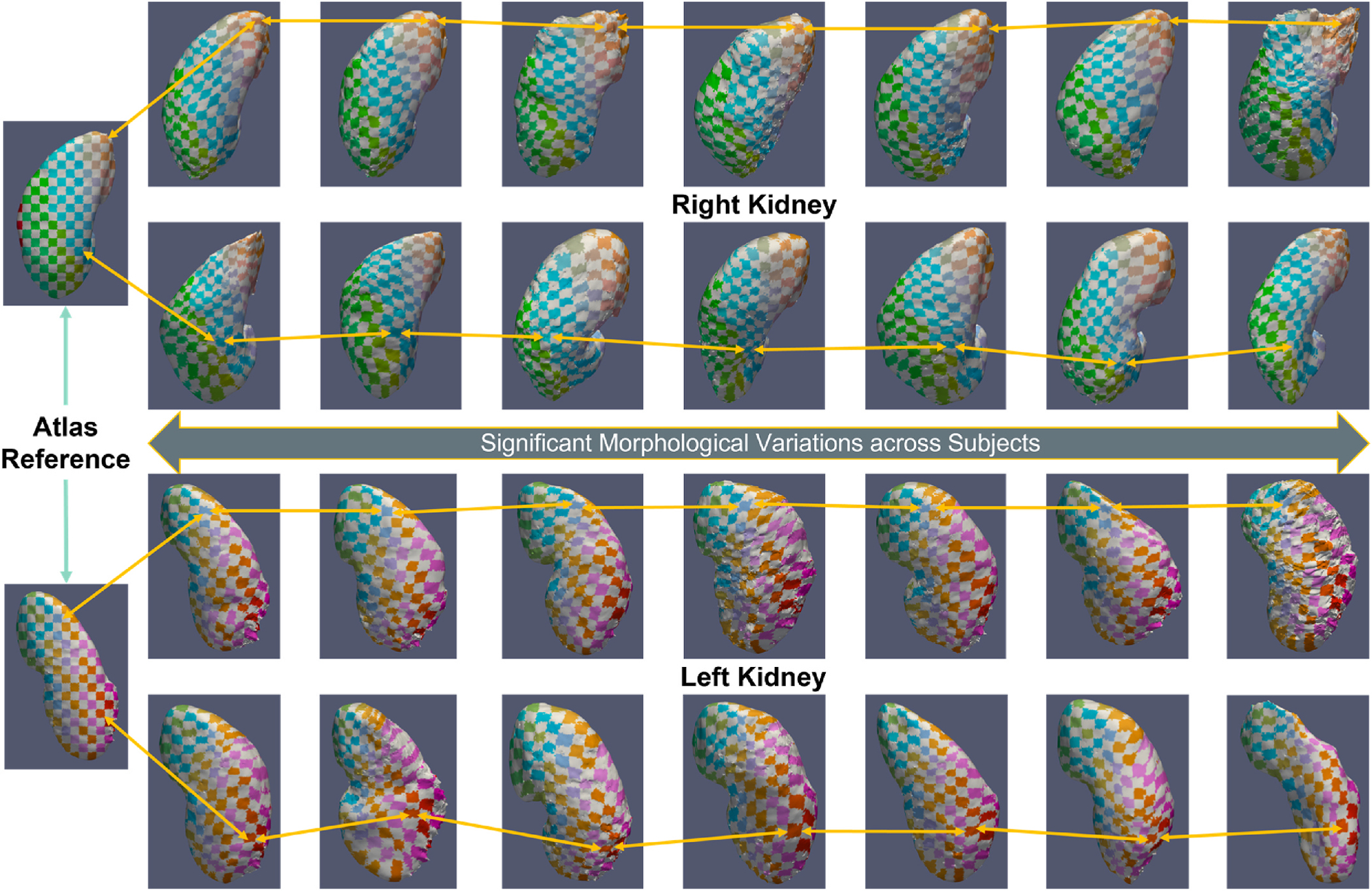
The surface rendering of the registered kidney with significant morphological variation are also illustrated. The 2D checkerboard pattern with arrows demonstrates the correspondence of the deformation from atlas space to the moving image space. Astable deformation across the change in volumetric morphology of kidney (100 cc–308 cc) are demonstrated with the deformed checkerboard.
The high-resolution characteristics of the atlas template preserve highly detailed voxel-wise information across all organs. DEEDS provided an overall performance with mean Dice of 0.50 [8]. However, the DEEDS performance cannot provide accurate measures towards the organ of interests due to high sensitivity of field of view, leading to significant deformation. With the use of PDD-Net [20], it yields the best performance with mean Dice of 0.54 across all organs.The sharpness of the organ interests’ structure such as liver and spleen become more appealing and the substructure context of kidneys are also demonstrated with stability across the contrast-enhanced phase such as late arterial and portal venous. Furthermore, the extraction time of each computing registered output is signficantly slower than metric-based method (DEEDs), as demonstrated in Table 5. Learning-based approaches demonstrate the significant advantage of efficiency in image registration. In addition to the ablation study, we evaluate the effectiveness of the BPR preprocessing for abdominal registration with DEEDS. As abdominal registration is sensitive to the similarity of the field of view between the fixed scans and the moving scans, the BPR algorithm provides an opportunity to extract approximate ranges within the abdominal region and allows a certain extent of deformation, while limited field of view is generated by cropping the ROI of the organs with the segmentation masks and may lead to over-deformation. We aim to increase the successful rate of registration with the BPR algorithm with a proportional increase of label transfer performance. Significant improvement is demonstrated with the use of BPR in Tables 2 and 3 for both volumetric segmentation using 3D U-Net as network architecture [31] and image registration respectively. The cropped abdominal interest reduces the over-deformation of organs towards the lung or pelvis region. The variance in registration performance is also reduced with the decrease of outliers in Fig. 6 and demonstrate the effect reduction by the significant field of view variability. Furthermore, Table 4 demonstrates the efficiency of using BPR and each scan only needs ~30 secs to compute the anatomical score and use it as guidance to crop the region of abdominal interest. Among the average template of each contrast phase in Fig. 4, the portal venous phase average template provides a sharper and smoother abdominal body comparing to that of the other four phases. However, the portal venous phase average template is not suitable to be an unbiased template for registration. The small tissue voxels are difficult to represent and cannot provide sufficient accurate information for registration. Large population of clinical cohorts may need to be used to obtain a higher confidence level of voxel representation that is as sharp as the atlas target image (see Figs. 4 and 5).
Table 5.
Time consumption of metric-based & deep learning-based registration methods.
| Methods | Extraction Time per Scan (Sec) |
|---|---|
|
| |
| DEEDS (A) | 170.105 |
| DEEDS (D) | 591.349 |
| PDD-Net | 7.43 |
A: affine registration, D: deformable registration.
Table 2.
Segmentation Performance Evaluation with/out Body Part Regression with 13 Organs (Mean Dice ± STD).
| Organs | Without BPR | With Ke et al. | With BPR |
|---|---|---|---|
|
| |||
| Spleen | 0.928 ± 0.0213 | 0.946 ± 0.0224 | 0.956 ± 0.0201 |
| Right Kidney | 0.899 ± 0.0254 | 0.904 ± 0.0263 | 0.932 ± 0.0217 |
| Left Kidney | 0.889 ± 0.0201 | 0.904 ± 0.0321 | 0.924 ± 0.0212 |
| Gall Bladder | 0.539 ± 0.201 | 0.540 ± 0.211 | 0.559 ± 0.189 |
| Esophagus | 0.748 ± 0.0547 | 0.758 ± 0.0753 | 0.770 ± 0.602 |
| Liver | 0.946 ± 0.0194 | 0.954 ± 0.0215 | 0.960 ± 0.0146 |
| Stomach | 0.830 ± 0.0421 | 0.840 ± 0.0601 | 0.855 ± 0.0375 |
| Aorta | 0.906 ± 0.0345 | 0.912 ± 0.0392 | 0.920 ± 0.0233 |
| Inferior Vena Cava | 0.807 ± 0.0397 | 0.815 ± 0.0462 | 0.830 ± 0.0219 |
| Portal Splenic Vein | 0.622 ± 0.135 | 0.638 ± 0.143 | 0.716 ± 0.0947 |
| Pancreas | 0.677 ± 0.103 | 0.717 ± 0.109 | 0.742 ± 0.0901 |
| Right Adrenal Glands | 0.632 ± 0.113 | 0.640 ± 0.131 | 0.652 ± 0.0823 |
| Average Dice | 0.785 ± 0.0675 | 0.797 ± 0.0764 | 0.818 ± 0.0564* |
Note that p < 0.0001 with Wilcoxon signed-rank test
between Ke et al. and BPR.
Table 3.
Registration Performance Evaluation with/out Body Part Regression with 13 Organs (Mean Dice ± STD).
| Organs | Without BPR | With BPR |
|---|---|---|
|
| ||
| Spleen | 0.642 ± 0.200 | 0.701 ± 0.180 |
| Right Kidney | 0.696 ± 0.235 | 0.766 ± 0.203 |
| Left Kidney | 0.696 ± 0.246 | 0.746 ± 0.215 |
| Gall Bladder | 0.191 ± 0.280 | 0.196 ± 0.279 |
| Esophagus | 0.464 ± 0.178 | 0.524 ± 0.155 |
| Liver | 0.650 ± 0.222 | 0.714 ± 0.194 |
| Stomach | 0.277 ± 0.211 | 0.303 ± 0.213 |
| Aorta | 0.729 ± 0.144 | 0.771 ± 0.113 |
| Inferior Vena Cava | 0.552 ± 0.181 | 0.621 ± 0.141 |
| Portal Splenic Vein | 0.306 ± 0.165 | 0.310 ± 0.173 |
| Pancreas | 0.387 ± 0.172 | 0.385 ± 0.174 |
| Right Adrenal Gland | 0.280 ± 0.157 | 0.289 ± 0.153 |
| Left Adrenal Gland | 0.260 ± 0.156 | 0.272 ± 0.157 |
| Average Dice | 0.472 ± 0.196 | 0.523 ± 0.181* |
Note that p < 0.0001 with Wilcoxon signed-rank test
between registration without BPR and that with BPR.
Fig. 6.
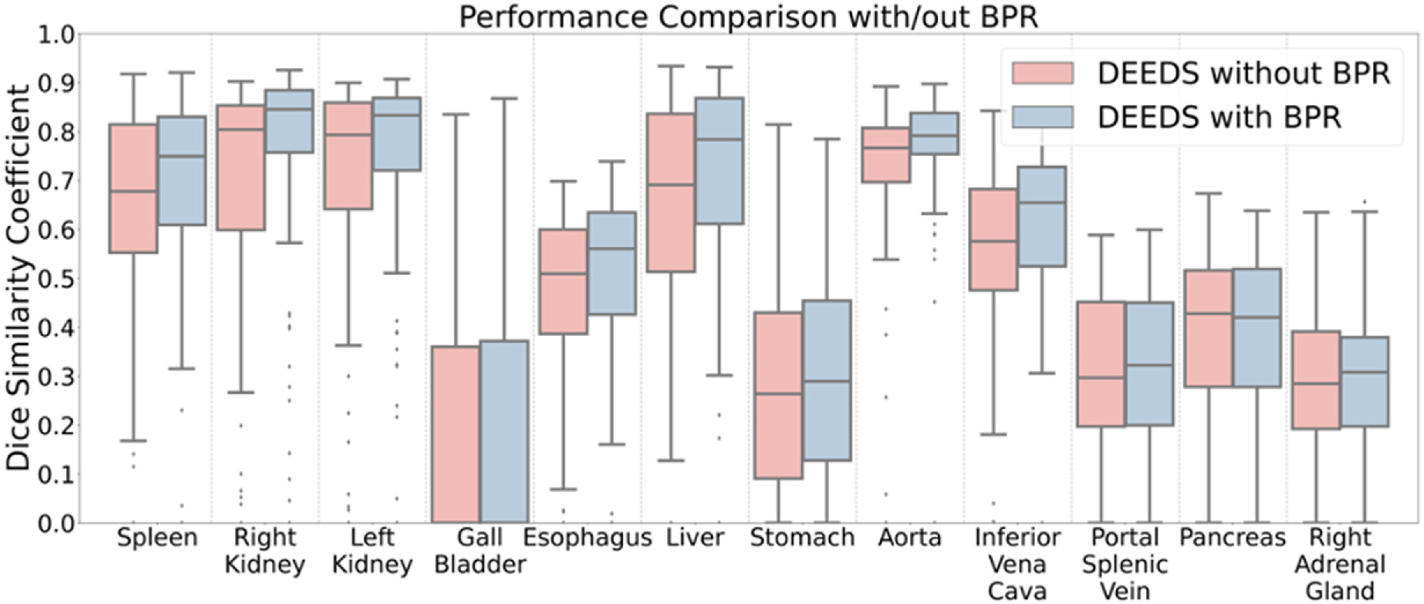
We evaluate the failure rate of the registration with/out using BPR. The use of BPR reduce the number of outliers in some of organs, especially for right and left kidneys.
Table 4.
Time consumption of preprocessing and sampling data samples.
| Methods | Extraction Time per Scan (Sec) |
|---|---|
|
| |
| Apply BPR | 30.237 |
| Downsampling for Deep Learning Registration | 13.111 |
| Upsampling to Atlas Resolution | 124.071 |
Initially, one of the bottlenecks of providing a stable anatomical information transfer for organs is the registration method (see Table 1). The registration performance of kidneys may be affected by the secondary targets such as liver and spleen. Further optimization of the registration pipeline is needed for reducing the possibility of significant deformation. Instead of relying on similarity metric (mutual information, cross correlation, Hamming distances of the self-similarity context) as the loss function [15,32], a learning-based method may be another promising direction to learn a registration function and predict the registration field for the moving images [20,33]. However, most of the proposed learning-based pipeline is focused on the brain [16,34]. PDD-Net provide an opportunity to reduce the gap for robust abdominal region registration with deep neural networks to adapt large deformation field. However, the significant improvement of the registration is limited with contrast-enhanced dataset such as late arterial phase and portal venous phase scans as shown in Fig. 4. We further perform the evaluation in registration performance with non-contrast dataset as Table 6 and a significant decrease of robustness in registration is illustrated with PDD-Net. As the pretrained model of PDD-Net is trained with limited portal venous dataset, the significant of domain shift contribute to the adverse performance of the registration. Multi-Modality registration with deep learning approaches can be the next step to contribute a robust generation of atlas with abdominal organs.
Table 6.
Evaluation metric on 50 Non-Contrast registration for three registration methods on 13 Organs (Mean ± STD).
| Methods | Dice Score | MSD (mm) | HD (mm) |
|---|---|---|---|
|
| |||
| DEEDS * | 0.485 ± 0.275 | 9.45 ± 18.2 | 43.4 ± 48.5 |
| PDD-Net with A | 0.278 ± 0.223 | 12.4 ± 15.6 | 59.4 ± 45.1 |
Note that p < 0.0001 with Wilcoxon signed-rank test
A: affine registration.
The kidney atlas template also provides contributions in the segmentation of other abdominal and retroperitoneal organs. The high-quality atlas multi-organ label can be transferred with the inverse transformation and use to provide accurate measures for abdominal and retroperitoneal organs. Also, high quality labels can help perform training strategies to innovate automatic learning-based model. Huo et al. proposed a whole brain segmentation using spatially localized network tiles with the atlas-transferred label [35]. Dong et al. proposed left ventricle segmentation network, which integrate the ventricle atlas at echocardiogram into learning framework and provides consistency constraints with atlas label to perform accurate segmentations [36]. Bai et al. presented a population study of relating the phenome-wide association to the function of cardiac and aortic structures using machine-learning-based segmentation pipeline [37]. Clinical validation and phenotypic analysis can be performed and reveal biomarkers of specific organs in certain conditions such as disease pathogenesis, with high-quality segmentation labels. Further exploration can be investigated in the abdominal and retroperitoneal domain with the use of the high-quality atlas label.
5. Conclusion
This manuscript presents a healthy kidney atlas to generalize the contrastive and morphological characteristics across patients with significant variability in demographics and imaging protocols. Specifically, the healthy kidney atlas provides a stable reference standard for both left and right kidney organs in 3-dimensional space to transfer kidney information using an adapted registration pipeline. Significant variance on the field of view and the organ shape can be focused as the optimization parameters to reduce the possibility of failure registration. Potential future exploration with the use of the atlas template can be further investigated in both engineering and clinical perspectives, to provide better understandings and measures towards the kidneys.
Acknowledgements
This research is supported by HuBMAP, NIH Common Fund and National Institute of Diabetes, Digestive and Kidney Diseases U54DK120058 (Spraggins), NSF CAREER 1452485, NIH 2R01EB006136, NIH 1R01EB017230 (Landman), and NIH R01NS09529. This study was in part using the resources of the Advanced Computing Center for Research and Education (ACCRE) at Vanderbilt University, Nashville, TN. The identified datasets used for the analysis described were obtained from the Research Derivative (RD), database of clinical and related data. ImageVU and RD are supported by the VICTR CTSA award (ULTR000445 from NCATS/NIH) and Vanderbilt University Medical Center institutional funding. ImageVU pilot work was also funded by PCORI (contract CDRN-1306–04869).
Footnotes
Declaration of competing interest
None declared.
References
- [1].Rozenblatt-Rosen O, Stubbington MJ, Regev A, Teichmann SA, The human cell atlas: from vision to reality, Nature News 550 (7677) (2017) 451. [DOI] [PubMed] [Google Scholar]
- [2].Consortium H, The human body at cellular resolution: the NIH Human Biomolecular Atlas Program, Nature 574 (7777) (2019) 187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Kuklisova-Murgasova M, et al. , A dynamic 4D probabilistic atlas of the developing brain, Neuroimage 54 (4) (2011) 2750–2763. [DOI] [PubMed] [Google Scholar]
- [4].Shi F, et al. , Infant brain atlases from neonates to 1-and 2-year-olds, PLoS One 6 (4) (2011) e18746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Gholipour A, et al. , A normative spatiotemporal MRI atlas of the fetal brain for automatic segmentation and analysis of early brain growth, Sci. Rep. 7 (1) (2017) 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Zhang Y, Shi F, Wu G, Wang L, Yap P-T, Shen D, Consistent spatial-temporal longitudinal atlas construction for developing infant brains, IEEE Trans. Med. Imag. 35 (12) (2016) 2568–2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Rajashekar D, et al. , High-resolution T2-FLAIR and non-contrast CT brain atlas of the elderly, Sci. Data 7 (1) (2020) 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Xu Z, et al. , Evaluation of six registration methods for the human abdomen on clinically acquired CT, IEEE (Inst. Electr. Electron. Eng.) Trans. Biomed. Eng. 63 (8) (2016) 1563–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Ashburner J, A fast diffeomorphic image registration algorithm, Neuroimage 38 (1) (2007) 95–113. [DOI] [PubMed] [Google Scholar]
- [10].Avants BB, Epstein CL, Grossman M, Gee JC, Symmetric diffeomorphic image registration with cross-correlation: evaluating automated labeling of elderly and neurodegenerative brain, Med. Image Anal. 12 (1) (2008) 26–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Balakrishnan G, Zhao A, Sabuncu MR, Guttag J, Dalca AV, An unsupervised learning model for deformable medical image registration, in: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 2018, pp. 9252–9260. [Google Scholar]
- [12].Rueckert D, Sonoda LI, Hayes C, Hill DL, Leach MO, Hawkes DJ, Nonrigid registration using free-form deformations: application to breast MR images, IEEE Trans. Med. Imag. 18 (8) (1999) 712–721. [DOI] [PubMed] [Google Scholar]
- [13].Dalca AV, Bobu A, Rost NS, Golland P, Patch-based discrete registration of clinical brain images, in: International Workshop on Patch-Based Techniques in Medical Imaging, Springer, 2016, pp. 60–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Glocker B, Komodakis N, Tziritas G, Navab N, Paragios N, Dense image registration through MRFs and efficient linear programming, Med. Image Anal. 12 (6) (2008) 731–741. [DOI] [PubMed] [Google Scholar]
- [15].Vercauteren T, Pennec X, Perchant A, Ayache N, Diffeomorphic demons: efficient non-parametric image registration, Neuroimage 45 (1) (2009) S61–S72. [DOI] [PubMed] [Google Scholar]
- [16].Balakrishnan G, Zhao A, Sabuncu MR, Guttag J, Dalca AV, Voxelmorph: a learning framework for deformable medical image registration, IEEE Trans. Med. Imag. 38 (8) (2019) 1788–1800. [DOI] [PubMed] [Google Scholar]
- [17].Dalca AV, Balakrishnan G, Guttag J, Sabuncu MR, Unsupervised learning of probabilistic diffeomorphic registration for images and surfaces, Med. Image Anal. 57 (2019) 226–236. [DOI] [PubMed] [Google Scholar]
- [18].Zhao S, Dong Y, Chang EI, Xu Y, Recursive cascaded networks for unsupervised medical image registration, in: Proceedings of the IEEE/CVF International Conference on Computer Vision, 2019, pp. 10600–10610. [Google Scholar]
- [19].Zhao Y.-q., Yang Z, Wang Y.-j., Zhang F, Yu L.-l., Wen X.-b., Target organ non-rigid registration on abdominal CT images via deep-learning based detection, Biomed. Signal Process Control 70 (2021) 102976. [Google Scholar]
- [20].Heinrich MP, Closing the gap between deep and conventional image registration using probabilistic dense displacement networks, in: International Conference on Medical Image Computing and Computer-Assisted Intervention, Springer, 2019, pp. 50–58. [Google Scholar]
- [21].Yan K, Lu L, Summers RM, Unsupervised body part regression via spatially self-ordering convolutional neural networks, in: 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018), IEEE, 2018, pp. 1022–1025. [Google Scholar]
- [22].Heinrich MP, Jenkinson M, Papież BW, Brady M, Schnabel JA, Towards realtime multimodal fusion for image-guided interventions using self-similarities, in: International Conference on Medical Image Computing and Computer-Assisted Intervention, Springer, 2013, pp. 187–194. [DOI] [PubMed] [Google Scholar]
- [23].Heinrich MP, Maier O, Handels H, Multi-modal Multi-Atlas Segmentation Using Discrete Optimisation and Self-Similarities, 1390, VISCERAL Challenge@ ISBI, 2015, p. 27. [Google Scholar]
- [24].Heinrich MP, Oktay O, Bouteldja N, OBELISK-Net: fewer layers to solve 3D multi-organ segmentation with sparse deformable convolutions, Med. Image Anal. 54 (2019) 1–9. [DOI] [PubMed] [Google Scholar]
- [25].Jenkinson M, Beckmann CF, Behrens TE, Woolrich MW, Smith SM, Fsl,” Neuroimage 62 (2) (2012) 782–790. [DOI] [PubMed] [Google Scholar]
- [26].Tang Y, et al. , Body Part Regression with self-supervision, IEEE Trans. Med. Imag. 40 (5) (2021) 1499–1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Heinrich MP, Jenkinson M, Brady M, Schnabel JA, MRF-based deformable registration and ventilation estimation of lung CT, IEEE Trans. Med. Imag. 32 (7) (2013) 1239–1248. [DOI] [PubMed] [Google Scholar]
- [28].Modat M, et al. , Fast free-form deformation using graphics processing units, Comput. Methods Progr. Biomed. 98 (3) (2010) 278–284. [DOI] [PubMed] [Google Scholar]
- [29].Jimenez-del-Toro O, et al. , Cloud-based evaluation of anatomical structure segmentation and landmark detection algorithms: VISCERAL anatomy benchmarks, IEEE Trans. Med. Imag. 35 (11) (2016) 2459–2475. [DOI] [PubMed] [Google Scholar]
- [30].Rosner B, Glynn RJ, Lee MLT, The Wilcoxon signed rank test for paired comparisons of clustered data, Biometrics 62 (1) (2006) 185–192. [DOI] [PubMed] [Google Scholar]
- [31].Çiçek Ö, Abdulkadir A, Lienkamp SS, Brox T, Ronneberger O, 3D U-Net: learning dense volumetric segmentation from sparse annotation, in: International Conference on Medical Image Computing and Computer-Assisted Intervention, Springer, 2016, pp. 424–432. [Google Scholar]
- [32].Urschler M, Werlberger M, Scheurer E, Bischof H, Robust optical flow based deformable registration of thoracic CT images, in: Medical Image Analysis for the Clinic: A Grand Challenge, 2010, pp. 195–204. [Google Scholar]
- [33].Mok TC, Chung AC, Large deformation diffeomorphic image registration with laplacian pyramid networks, in: International Conference on Medical Image Computing and Computer-Assisted Intervention, Springer, 2020, pp. 211–221. [Google Scholar]
- [34].de Vos BD, Berendsen FF, Viergever MA, Sokooti H, Staring M, Išgum I, A deep learning framework for unsupervised affine and deformable image registration, Med. Image Anal. 52 (2019) 128–143. [DOI] [PubMed] [Google Scholar]
- [35].Huo Y, et al. , 3D whole brain segmentation using spatially localized atlas network tiles, Neuroimage 194 (2019) 105–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Dong S, et al. , Deep atlas network for efficient 3d left ventricle segmentation on echocardiography, Med. Image Anal. 61 (2020) 101638. [DOI] [PubMed] [Google Scholar]
- [37].Bai W, et al. , A population-based phenome-wide association study of cardiac and aortic structure and function, Nat. Med. (2020) 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]


