Highlights
-
•
To deal with missing modalities for brain tumor segmentation, we propose a novel framework TISS-Net that obtains the synthesized target modality and segmentation of brain tumors end-to-end.
-
•
We propose a novel dual-task-regularized generator that simultaneously obtains a synthesized target modality and a coarse segmentation, which is further regularized by a tumor-aware synthesis loss with perceptibility regularization.
-
•
A dual-task segmentor to predict a refined segmentation and error in the coarse segmentation simultaneously, where a consistency between these two predictions is introduced for regularization.
-
•
Experiments on two public brain tumor datasets showed that our method outperformed state-of-the-art image synthesis-based segmentation methods.
Keyword: Brain tumor, Image synthesis, Segmentation, Deep learning, Regularization
Abstract
Accurate segmentation of brain tumors from medical images is important for diagnosis and treatment planning, and it often requires multi-modal or contrast-enhanced images. However, in practice some modalities of a patient may be absent. Synthesizing the missing modality has a potential for filling this gap and achieving high segmentation performance. Existing methods often treat the synthesis and segmentation tasks separately or consider them jointly but without effective regularization of the complex joint model, leading to limited performance. We propose a novel brain Tumor Image Synthesis and Segmentation network (TISS-Net) that obtains the synthesized target modality and segmentation of brain tumors end-to-end with high performance. First, we propose a dual-task-regularized generator that simultaneously obtains a synthesized target modality and a coarse segmentation, which leverages a tumor-aware synthesis loss with perceptibility regularization to minimize the high-level semantic domain gap between synthesized and real target modalities. Based on the synthesized image and the coarse segmentation, we further propose a dual-task segmentor that predicts a refined segmentation and error in the coarse segmentation simultaneously, where a consistency between these two predictions is introduced for regularization. Our TISS-Net was validated with two applications: synthesizing FLAIR images for whole glioma segmentation, and synthesizing contrast-enhanced T1 images for Vestibular Schwannoma segmentation. Experimental results showed that our TISS-Net largely improved the segmentation accuracy compared with direct segmentation from the available modalities, and it outperformed state-of-the-art image synthesis-based segmentation methods.
1. Introduction
Brain and other Central Nervous System (CNS) tumors are one of the most common types of cancers, with an estimated incidence of 29.9 per million per year among adults, and approximately one-third of them are malignant [24]. As an example, gliomas that originate in glial cells constitute 80% of malignant primary brain tumors. High-Grade Gliomas (HGG) have a median survival rate of two years or less, while Low-Grade Gliomas (LGG) are less aggressive with a relatively promising prognosis [23]. In contrast, Vestibular Schwannoma (VS) is a benign tumor caused by the abnormal proliferation of schwann cells on the outside of the vestibulocochlear nerve that connects the brain to the ear. The incidence of VS is increasing in recent years and has been estimated to be 14 to 20 cases per million per year [27].
Currently, Magnetic Resonance Imaging (MRI) is an important tool for diagnosis and treatment management of brain tumors due to its good contrast for soft tissues. Especially, segmentation of the tumor structure from MRI plays a critical role in accurate volumetric measurement and 3D modeling of the tumors that is required by tumor growth detection and surgical planning. As manual segmentation is time-consuming, labor-intensive and subject to inter-observer and intra-observer variations, automatic segmentation is highly desirable in clinical practice. Usually, accurate automatic segmentation requires multi-modal scanning or contrast-enhanced imaging to visualize the entire tumor or tumor subregions. For example, state-of-the-art glioma segmentation methods typically require four modalities [22], [6], including T1-weighted, contrast enhanced T1-weighted (ceT1), T2-weighted and Fluid Attenuation Inversion Recovery (FLAIR) imaging. T1 and ceT1 mostly highlight the tumor core region (without peri-tumoral edema), and T2 and FLAIR provide a better contrast for the whole tumor region (with peri-tumoral edema). Specifically, FLAIR images show hyperintensity signal abnormality in peri-tumoral edema surrounding the main mass lesion that generally represents infiltrative edema.
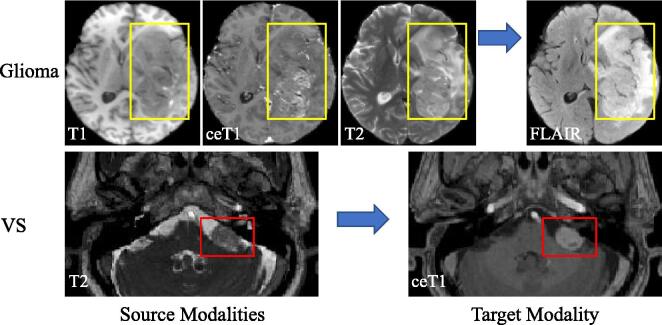
In clinical practice, since obtaining multiple sequences is time-consuming and expensive, some modalities may be missing [18], [20], which leads to challenges for the segmentation task. Fig. 1 shows two examples of such cases. In the first example of glioma, the segmentation task often involves T1, T2, ceT1 and FLAIR images, and the segmentation accuracy of the whole tumor would be largely reduced when FLAIR is not available. In the second example of VS, high-resolution T2-weighted MRI is commonly used for imaging, but it suffers from a low contrast between the tumor and the background. To improve the visibility of the tumor for accurate assessment, Radiologists may use gadolinium contrast agents for ceT1 MRI scanning, which makes the tumor boundary easier to recognize. Despite the fact that the performance of automatic VS segmentation from ceT1 can be comparable to that of manual segmentation [27], ceT1 scanning requires the use of gadolinium contrast agents that raise concerns on potentially harmful cumulative side-effect, leading to a demand on segmentation of VS with only T2 images being available.
Fig. 1.
Examples of source and target modalities for glioma and Vestibular Schwannoma (VS) segmentation. The bounding boxes highlight the segmentation targets in the images. Note that the tumor is less visible in the source modalities.
To tackle these problems, synthesizing a missing target modality from one or more available modalities for the downstream segmentation has attracted increasing attentions recently [8], [36]. Traditionally, researchers have used dictionary learning [13] and random forest [17] methods for this purpose. But they usually focus on a low-level pixel-wise optimization for synthesis and can hardly obtain realistic images at a high level. Recently, Convolutional Neural Networks (CNNs) and Generative Adversarial Networks (GANs) have been proposed for more realistic synthesis, such as generating high-dose Positron Emission Tomography (PET) images conditioned on low-dose PET images [33] and generating FLAIR images from T1 images for brain tumor segmentation [39], [20]. Such methods typically use a generator to obtain the synthesized image, a discriminator to encourage realistic synthesis, and a segmentation CNN taking the synthesized image as input to obtain the segmentation result. The synthesis in these works was not well optimized for segmentation due to that the image generation model and segmentation model were trained independently, which may limit the final segmentation performance.
To overcome this issue, end-to-end medical image synthesis and segmentation methods have been proposed recently [36]. However, it remains challenging to achieve accurate segmentation results from the synthesized images due to the following reasons: First, there is a domain gap between the synthesised and real target modalities, leading the segmentation based on synthesised images to be inferior to segmentation with real target modalities [32]. Second, an end-to-end synthesis and segmentation model becomes more complex and much deeper than independent models and it has a higher risk of over-fitting, which requires more effective regularization methods to keep the performance during testing. However, regularization of the end-to-end model has rarely been explored in-depth in existing works.
The contribution of this work is threefold. First, to deal with missing modality for brain tumor segmentation, we propose a novel brain Tumor Image Synthesis and Segmentation Network (TISS-Net) based on a cascaded dual-task architecture for end-to-end training and inference, where the synthesis and segmentation models are learned synergistically with several novel high-level regularization strategies. Second, we introduce segmentation-aware target-modality image synthesis, where a coarse segmentation is used as an auxiliary task to regularize the synthesis task, and a tumor-aware synthesis loss with perceptibility regularization is introduced to generate segmentation-friendly images in the missing modality. Thirdly, we propose a novel error-prediction consistency loss for improving the segmentation performance, where the dual-task segmentor uses two branches to predict a fine segmentation and errors in the coarse segmentation simultaneously, and a consistency between these two predictions is introduced as a regularization for better segmentation performance. The dual-task generator and dual-task segmentor are trained end-to-end so that they are adaptive to each other for high segmentation performance. We extensively evaluated our method on FLAIR image synthesis for glioma whole tumor segmentation and ceT1 image synthesis for Vestibular Schwannoma segmentation. Experimental results show that our method outperformed several state-of-the-art deep learning-based image synthesis and segmentation methods.
2. Related Works
2.1. Brain Tumor Segmentation
Brain tumor segmentation from multi-modal images has made great advances based on the development of CNNs [25], [14]. They have achieved better performance than traditional methods using hand-crafted features [22]. Some techniques such as attention mechanism have proven effective for improving performance for glioma segmentation [44], [21] and Vestibular Schwannoma (VS) segmentation [27]. To deal with brain tumors in multiple scales, Zhou et al. [46] used atrous convolution feature pyramid to keep high spatial resolution, and Ye et al. [38] introduced a dense neural network with parallel pathways at different scales. Sun et al. [29] used a multi-pathway architecture to effectively extract features from multi-modal MRI images. Hu et al. [11] proposed mutual ensemble learning to enable knowledge exchange between networks and let them teach each other for better performance. Coarse to fine architectures have also shown their potential in glioma segmentation [42], [4]. They often perform well when the images have a good contrast or multi-sequence images are used [31], and the segmentation accuracy is limited when the image has a low contrast or some of the multiple modalities are missing [18].
2.2. Segmentation with Missing Modality
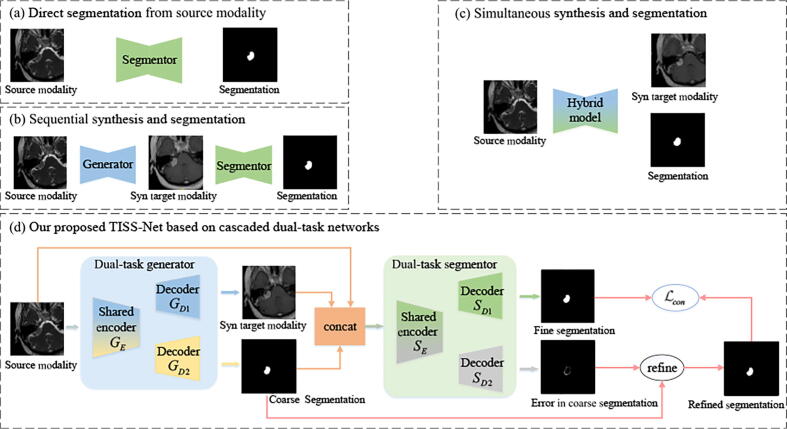
Segmentation with missing modality is a challenging problem in medical image analysis. It is often challenging to achieve satisfactory results when performing segmentation directly on the remaining available (source) modalities [32], [40], as depicted in Fig. 2(a). There have been several approaches to handle the problem of missing modalities. One popular approach is Domain Adaptation (DA), which transfers models trained in a source domain (i.e., one modality) to a target domain (i.e., another modality)[9]. The DA methods usually train a model with annotated source-domain images and unannotated target-domain images, and then use target-domain images for inference. For example, Dou et al. [7] proposed a GAN-based method to align the features between source and target domains for adaptation. Zhu et al. [47] proposed a boundary-weighted domain adaptive neural network to improve the performance of prostate MR image segmentation by considering the boundary information between the source and target domains. Zhong et al. [43] proposed joint image and feature adaptive attention-aware networks to alleviate the domain shift for cross-modality semantic segmentation. Liu et al. [19] also used collaborative adaptations from both image and feature perspectives in a supervised learning framework. Other techniques such as pseudo label-based methods [34] and disentanglement [37] have also been proposed for cross-modality domain adaptation. In HeMIS [10], a common feature space was learned to represent different modalities, and it was used to perform down-stream segmentation or classification tasks.
Fig. 2.
Illustration of existing pipelines (a-c) and our proposed framework (d) for segmentation of brain tumors with missing modalities. This figure takes the VS case as an example. Our proposed TISS-Net based on cascaded dual-task networks deals with the synthesis and segmentation tasks end-to-end. A dual-task generator obtains a synthetic target-modality image and a coarse segmentation mask simultaneously. It is followed by a dual-task segmentor obtaining a fine segmentation and errors in the coarse segmentation simultaneously, and a consistency loss between them (i.e, ) is proposed for regularization. Note that the generator is further regularized by a tumor-aware synthesis loss with perceptibility regularization detailed in Section 3.2.
Knowledge Distillation (KD) has also been proposed to deal with missing modalities. Hu et al. [12] proposed to use generalized knowledge distillation to transfer knowledge from a teacher network trained with multi-modal images that are registered [28] to a mono-modal student. A similar framework was proposed by Chen et al. [2], where both pixel-level and image-level distillation are leveraged for better knowledge transfer. In addition, synthesizing the missing modality based on available modalities for segmentation is appealing [45], as the synthesized image can provide additional important features to improve the performance and the result is more explainable [36]. The synthesis-based methods are detailed in the following.
2.3. Medical Image Synthesis for Improved Segmentation
Synthesis-based segmentation methods can be briefly summarized as two categories: 1) sequential synthesis and segmentation where the two models are trained independently or end-to-end; 2) simultaneous synthesis and segmentation where a hybrid model is used to obtain the synthesized target modality and segmentation jointly. Fig. 2(b) and (c) illustrate the workflow of these two categories, respectively.
Most existing works follow the sequential image synthesis and segmentation workflow. For example, Luo et al. [20] first generated the missing modality based on an edge-preserving generator, and then segmented the target with the synthesized modality, where the image generation model and segmentation model were independently optimized during training and cascaded during testing. However, dealing with the synthesis and segmentation independently may restrict the segmentation performance. To overcome this problem, end-to-end synthesis and segmentation methods have been increasingly employed recently. For example, Xu et al. [36] proposed progressive sequential casual GANs (PSCGAN) to simultaneously synthesize a contrast-enhanced image and segment tissues related to diagnosis of ischemic heart disease. However, as the synthesis and segmentation models are cascaded, the whole pipeline has a risk of over-fitting and it’s performance is limited if without effective regularization [36].
Compared with sequential synthesis and segmentation, simultaneous synthesize and segmentation takes a better advantage of the inter-dependency between these two tasks. In such methods, a model takes the available modalities as input, and gives target modality and segmentation result simultaneously, where the implicit constraints between synthesis and segmentation is used as a regularization. For example, Bahrami et al. [1] jointly learned two parallel CNNs for 7T MR image reconstruction and brain tissue segmentation from 3T MR images. Sun et al. [30] proposed a unified network for simultaneous compressed sensing MRI reconstruction and brain tissue segmentation, where a high-quality MRI synthesis network and a segmentation model share the encoder and use independent decodes to get the outputs. However, in such methods, the synthesized image was not further used to guide the segmentation process, which may limit the segmentation accuracy.
3. Methods
Fig. 2(d) is an overview of our TISS-Net for end-to-end target modality synthesis and brain tumor segmentation, where only partial modalities are given and the real target modality is not available at test time. Both the target-modality generator and brain tumor segmentor have two branches with a shared encoder for different tasks that can regularize each other. The dual-task generator obtains a synthesized target-modality image and a coarse segmentation simultaneously, and it is further regularized by a tumor-aware perceptibility loss. The segmentor takes the coarse segmentation, the input modalities and the synthesized modality as input, and uses one branch to directly predict a fine segmentation, and another branch to predict errors in the coarse segmentation for refinement. As the two branches are designed to obtain the final segmentation of the same target using different mechanisms, we impose a consistency between these two branches as an additional regularization to achieve better performance.
3.1. Dual-Task Generator for Image Synthesis and Coarse Segmentation
Let denote the input image with the available source modalities, and denote the corresponding target modality of the same subject. We use y to denote the segmentation ground truth. Our dual-task generator G takes as input, and simultaneously obtains a synthesized target-modality image and a coarse segmentation . G is composed of a shared encoder and two decoders: to obtain and to obtain , respectively. Compared with using two different networks to obtain and sequentially or independently, our dual-task generator with a shared encoder can save network parameters and the synthesis and coarse segmentation branches are regularized by each other.
Theoretically, G can be implemented by any image-in and image-out CNNs. In this work, we use a 2.5D U-Net [27] as the backbone for the glioma and VS segmentation from 3D volumes due to the following reasons. First, VS images have high inter-plane resolution and low through-plane resolution, and a 2.5D network combining 2D and 3D convolutions has been shown more effective than standard 3D networks [27]. Second, for 3D volumes with isotropic resolutions, 2.5D networks can achieve a good trade-off between model complexity, receptive field and GPU memory with competitive performance [31].
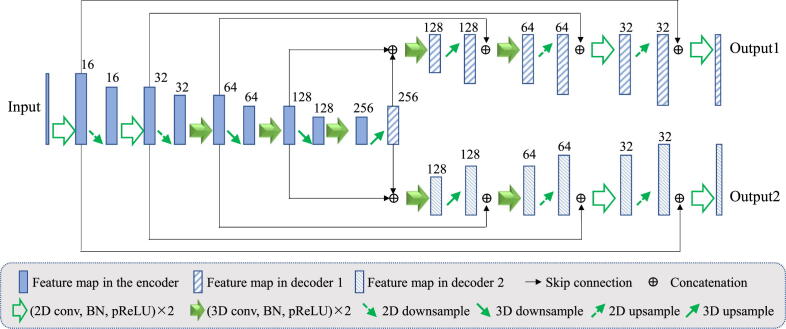
The original 2.5D U-Net [27] has an encoder-decoder structure with five resolution levels, where the two highest resolution levels use 2D convolutions and the other three resolution levels use 3D convolutions. We add another decoder with the same structure as the existing decoder with skip connections to obtain the dual-branch network, which is denoted as 2.5D DB-Net and illustrated in Fig. 3. The two branches are trained to obtain and respectively. The loss function for coarse segmentation branch is defined as a standard Dice loss , and the loss for the synthesis branch is detailed in the following.
Fig. 3.
Details of the 2.5D dual-branch network structure used in this work. It includes one encoder and two decoders, where the two highest resolution levels use 2D convolutions and the other three resolution levels use 3D convolutions. Channel numbers are shown on the top of feature maps.
3.2. Tumor-Aware Synthesis Loss with Perceptibility Regularization
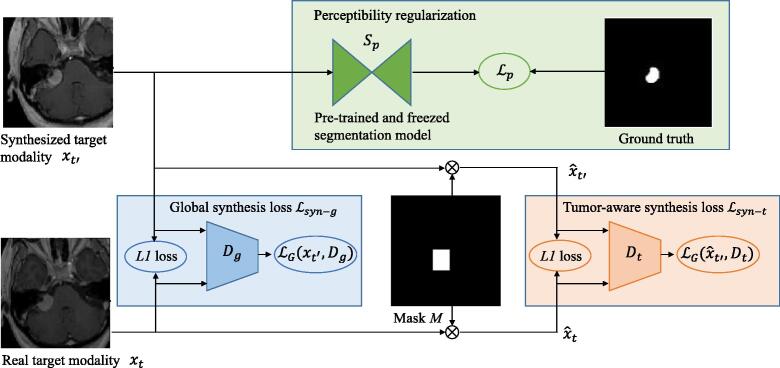
Most existing image synthesis methods define a global synthesis loss to supervise quality of the overall image [15], [20], which may not ensure a high synthesis quality around the tumor region and lead to low performance in the down-stream tumor segmentation task. To address this problem, in addition to the widely used global synthesis loss, we introduce a tumor-aware synthesis loss that highlights the synthesis quality around the tumor and a perceptibility regularization to reduce high-level domain gap between the synthesized and real target-modality images.
Global Synthesis Loss: The global synthesis loss in typical image synthesis methods [15] is formulated as a combination of an term and an adversarial term:
| (1) |
where is weight for the term. is a global discriminator implemented by PatchGANs [15] to recognize and as real or fake, respectively, and means the concatenation operation. The generator G is trained to fool the discriminator to obtain realistic outputs, and the corresponding loss is:
| (2) |
And the adversarial loss function for discriminator is:
| (3) |
Tumor-Aware Synthesis Loss: To improve the synthesis quality around the tumor region, we introduce a tumor-focused discriminator for training. Let M denote a binary mask around the tumor according to the bounding box of y, we multiply and by M respectively, and the corresponding masked results are denoted as and respectively. is defined as:
| (4) |
where is a weight for the term, and is a tumor-focused local discriminator to recognize the masked images and as real or fake, respectively. Similarly to Eq. (2) and Eq. (3), we replace and by and respectively to define the generator’s local adversarial loss and the local discriminator’s loss , respectively.
Perceptibility Regularization: As good low-level synthesis quality measurement such as SSIM and PSNR may not necessarily lead to a high segmentation performance due to the high-level semantic gap between and [32], we introduce a perceptibility loss to encourage a segmentation model trained with real target-modality images to keep high performance on the synthesized images with parameters freezed, which makes synthesized and real target-modality images have similar semantic features. Let denote a segmentation model pre-trained with real target-modality images and freezed during training of G, we aim to generate so that performs well on . In this paper, is implemented by the 2.5D U-Net [27], and the perceptibility regularization is:
| (5) |
Note that the gradient of is back-propagated to the generator G, rather than that is freezed.
Overall Synthesis Loss: As shown in Fig. 4, our proposed synthesis loss is a combination of the global synthesis loss, the tumor-aware synthesis loss and the perceptibility regularization:
| (6) |
where is weight for the perceptibility regularization.
Fig. 4.
Illustration of our proposed synthesis loss. Global synthesis loss encourages good quality in the entire image as a whole, and tumor-aware synthesis loss highlights the synthesis quality around the tumor region. The perceptibility loss encourages segmentation-friendly synthesis so that (a segmentation model trained with real target modality images and then freezed) performs well on the synthesized target modality image, leading to minimized semantic gap between synthesized and real target modality images.
3.3. Multi-Task Segmentor with Error-Prediction Consistency
With the synthesized target modality image and the coarse segmentation , we concatenate them with the original input image and denote the concatenation result as . Then is sent to the following segmentation network that can leverage the information from the synthesized missing modality and coarse segmentation to obtain better segmentation results than just using the available modalities for segmentation.
To obtain a fine segmentation considering that a coarse segmentation has been incorporated into , there are two basic approaches: one is to predict the fine segmentation directly [16], and the other is to first predict the error information in [35] and then combine the error information with to obtain a refined segmentation. Differently from existing works using only one of these two predictions, we take advantages of both of them, and add a consistency between these two predictions as a regularization to improve the robustness. Therefore, we use a dual-task structure again to implement the fine segmentation network.
Similarly to the dual-branch generator G, our dual-task fine segmentor S has a shared encoder and two decoders, where the first decoder directly obtains a fine segmentation and the second decoder predicts the probability of errors (denoted as ) in and then assembles and to obtain a refined segmentation (foreground probability map) :
| (7) |
where when a pixel in is 0 (1), a high corresponding value in leads to a high (low) value, indicating that this pixel should have a high probability of being the foreground (background) after refinement.
As both and can represent the new segmentation refined from , there should be a consistency between them. Therefore we define a consistency loss as:
| (8) |
The entire loss function for S is defined as:
| (9) |
where measures the difference between the fine prediction and the segmentation ground truth y, and measures the difference between and mis-segmentations in . Both and are implemented by Dice loss. Note that in the binary segmentation task of this paper, instead of predicting under-segmentation and over-segmentation respectively [35] that introduces extra difficulty due to extremely severe class-imbalance, our error prediction indicating whether a pixel value in equals to that in y is simpler to train.
3.4. Overall Loss Function
The overall loss function for training our dual-task generator G and dual-task fine segmentor S is summarized as:
| (10) |
where is the synthesis loss, is the coarse segmentation loss and is loss for the fine segmentor S. are weighting coefficients for and , respectively. The overall loss function for training the discriminators and is summarized as:
| (11) |
With Eq. (10) and Eq. (11), the generator G and segmentor S are trained end-to-end (i.e., the gradient of segmentation loss flows back to the synthesis network), so that they are adaptive to each other for the synthesis and segmentation tasks.
4. Experiments and Results
We validated our proposed TISS-Net for target-modality synthesis and brain tumor segmentation in two applications: 1) synthesizing FLAIR images using T1, T2 and ceT1 images for whole glioma segmentation, and 2) VS tumor segmentation based on synthesizing ceT1 images from T2 MR images.
4.1. Implementation Details
All the experiments were implemented with PyTorch, using an Ubuntu 20.04 Desktop with an Intel i9-10940X CPU and an NVIDIA GeForce RTX 2080Ti GPU. Both G and S used the dual-branch network structure illustrated in Fig. 3 based on the backbone of a 2.5D U-Net [27]. and were based on 167070 PatchGANs [15], as they have been demonstrated with higher performance than only letting the discriminator output a scalar to judge the entire synthetic image as a whole. For both glioma and VS segmentation tasks, we used a batch size of 2, and the patch size was 16128128. Adam optimizer was used for training, and the learning rate for was initialized to in the first 100 epochs and linearly decayed to 0 in the following 150 epochs. was also implemented by the 2.5D U-Net and pre-trained with the target modality. The learning rate for was initialized to that was halved when no performance improvement was observed on the validation set for 30 consecutive epochs. Note that the parameters of was freezed when training TISS-Net. The hyper-parameter setting was and according to the optimal performance on the validation set. For the dual-branch segmentor S, we used prediction in the first branch () as the segmentation result during inference.
To evaluate low-level synthesis quality, we used Peak Signal to Noise Ratio (PSNR) and Structural Similarity Index (SSIM). These two metrics were calculated both globally (i.e., in the entire image region) and locally (i.e., around the ground truth tumor). In addition, we evaluated high-level synthesis quality based on perceptability, which was measured by the performance of on synthesized images. A high perceptability indicates a high semantic similarity between synthesized and real target-modality images. For quantitative evaluation of segmentation performance, we reported Dice, Average Symmetric Surface Distance (ASSD) and 95 Hausdorff distance (HD95) between segmentation results and the ground truth tumor masks.
4.2. Glioma Segmentation from Multimodal MR Images with Absence of FLAIR
4.2.1. Data
We used the Multimodal Brain Tumor Segmentation Challenge (BraTS) 2020 training set for experiments [22]. In this dataset, spatially aligned four 3D MRI modalities (T1, ceT1, T2 and FLAIR) of 369 patients with resolution 1.0 mm 1.0 mm 1.0 mm and in-plane size 240240 were annotated into 3 heterogeneous histological sub-regions by expert raters: peri-tumoral edema, necrotic core and non-enhancing tumor core and enhancing tumor core. As FLAIR images provide a high contrast for the edema region and thus important for the whole tumor segmentation, we investigate synthesizing FLAIR images from T1, T2 and ceT1 for whole tumor segmentation. We randomly selected images from 258, 37 and 74 patients for training, validation and testing, respectively. For preprocessing, each image was manually cropped along the z-axis centered on the tumor. The intensity values were normalized to the range of [-1, 1] for each modality, respectively.
4.2.2. Ablation Study of the Synthesis Method
To evaluate the performance of our segmentation-aware target-modality image synthesis, we first ignore the segmentor in TISS-Net (i.e., equals to setting to 0), and conducted an ablation study to investigate effectiveness of each of our proposed losses for the synthesis: tumor-aware synthesis loss , perceptibility regularization and using the coarse segmentation branch (i.e., ) as regularization for the synthesis task. The baseline method was using global synthesis loss only. The quantitative evaluation results of different synthesis loss configurations are shown in Table 1. The segmentation performance (perceptibility) of with freezed parameters when applied to the synthesized images was used to measure the domain similarity between synthesized and real FLAIR images, where a higher perceptibility indicates closer high-level semantic appearance.
Table 1.
Quantitative comparison between different input and loss functions for FLAIR image synthesis for whole glioma segmentation. Perceptibility means the performance of applying (i.e., a segmentation model pre-trained with real FLAIR images and then freezed) to the synthesized images for segmentation.
| Input | Loss functions |
Synthesis quality |
Perceptibility |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Global SSIM | Local SSIM | Global PSNR | Local PSNR | Dice (%) | ASSD (mm) | HD95 (mm) | |||||
| T1, ceT1, T2 | ✓ | 0.730.05 | 0.500.09 | 22.722.07 | 19.851.84 | 83.2010.00 | 2.412.23 | 10.0812.01 | |||
| T1, ceT1, T2 | ✓ | ✓ | 0.750.09 | 0.520.12 | 22.992.39 | 20.012.14 | 84.668.09 | 2.061.73 | 8.0311.06 | ||
| T1, ceT1, T2 | ✓ | ✓ | 0.710.08 | 0.490.08 | 22.522.36 | 19.402.09 | 84.248.43 | 2.252.12 | 9.8214.10 | ||
| T1, ceT1, T2 | ✓ | ✓ | ✓ | 0.730.05 | 0.500.11 | 22.632.47 | 19.532.14 | 85.747.99 | 1.841.29 | 6.726.17 | |
| T1, ceT1, T2 | ✓ | ✓ | ✓ | ✓ | 0.740.09 | 0.500.12 | 22.762.53 | 19.752.05 | 86.097.58 | 1.761.00 | 6.705.92 |
| T2 | ✓ | ✓ | ✓ | ✓ | 0.650.06 | 0.370.09 | 21.142.12 | 18.002.13 | 83.719.84 | 2.281.93 | 8.9410.28 |
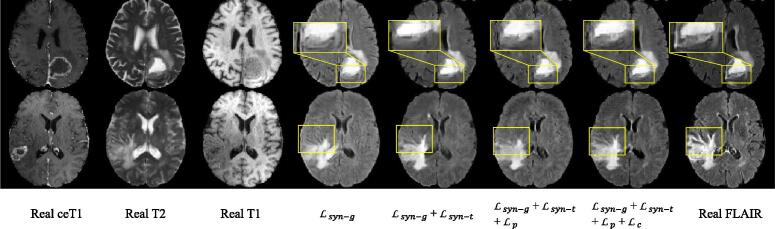
In Table 1, it can be observed that our tumor-aware synthesis loss helps to improve the image quality in terms of SSIM and PSNR, as well as the perceptibility. improves the perceptibility of the whole tumor, due to that makes the image synthesis aware of the segmentation, which alleviates the high-level semantic domain shift between real and synthesized FLAIR images. We found that and did not improve the SSIM and PSNR scores. This is mainly due to that these metrics only measure the low-level image quality and may not be directly related to the semantic segmentation task. In contrast, and are designed to enhance high-level semantic information related to segmentation in the image, and they are optimized for better segmentation accuracy, rather than low-level pixel intensity similarity between synthesized and real images. However, for perceptibility measurement, the baseline method obtained an average Dice of 83.20%, and introducing and improved it to 84.66% and 85.74%, respectively. Additionally using to regularize the synthesis task with the coarse segmentation branch further improved the average Dice to 86.09%. The results show that our proposed synthesis method that combines and outperformed the other variants in synthesizing segmentation-friendly FLAIR images of glioma. In addition, a visual comparison between synthesized FLAIR images obtained by different loss functions are shown in Fig. 5. It can be observed that the proposed method obtained better local contrast and structure details around the tumor region than the other variants.
Fig. 5.
Visual comparison of different loss functions for synthesizing FLAIR images of glioma.
In addition, to demonstrate the effectiveness of combining all the three available modalities as input, we conducted an experiment with only using T2 images as input. The results in the last row of Table 1 shows that removing T1 and ceT1 from the network input led the average Dice value to drop from 86.09% to 83.71%, which shows the importance of leveraging all the available modalities for synthesizing the missing modality for segmentation, as also demonstrated in previous works [18].
4.2.3. Effectiveness of Fine Segmentation using Error-Prediction Consistency
To further investigate the effectiveness of the proposed dual-task fine segmentor based on error-prediction consistency, we compared it with three variants: 1) only using the fine segmentation decoder (), without the error prediction branch; 2) error prediction branch only () without fine segmentation and thus without consistency loss; 3) predicting fine segmentation and error in the coarse segmentation simultaneously ( and ) but without consistency regularization. Quantitative results are shown in Table 2. It can observed that when taking a concatenation of source-modality image , synthesized target-modality image and coarse segmentation as input, using one of and leads to an average Dice of 86.95 and 86.81 respectively. Combining and together improved the score to 87.17, and introducing the consistency loss further improved the score to 87.55, which outperformed the other variants. We also compared these variants when using a concatenation of and as input of the segmentor. The results in Table 2 show that our proposed error-prediction consistency strategy still performed better than the other three variants.
Table 2.
Quantitative evaluation results of different inputs and loss functions for segmentor S in whole glioma segmentation. denotes our synthesized FLAIR image, is the input multi-modal image with absence of FLAIR, and is the coarse segmentation. The last two sections are based on a concatenation of these images as input. denotes significant improvement from as input based on a paired t-test (p-value 0.05).
| Input | Training loss |
Segmentation Performance |
||||
|---|---|---|---|---|---|---|
| Dice (%) | ASSD (mm) | HD95 (mm) | ||||
| ✓ | 84.549.62 | 2.322.12 | 10.7515.86 | |||
| ✓ | 86.336.85 | 1.700.91 | 6.405.13 | |||
| ✓ | 86.768.11 | 1.721.15 | 6.686.69 | |||
| ✓ | 86.628.39 | 1.701.11 | 6.576.93 | |||
| ✓ | ✓ | 86.817.86 | 1.701.12 | 6.817.86 | ||
| ✓ | ✓ | ✓ | 87.157.41 | 1.600.84 | 5.834.75 | |
| ✓ | 86.958.02 | 1.671.07 | 6.366.07 | |||
| ✓ | 86.818.75 | 1.691.27 | 6.556.88 | |||
| ✓ | ✓ | 87.178.56 | 1.631.16 | 6.346.78 | ||
| ✓ | ✓ | ✓ | 87.557.62 | 1.530.85 | 5.674.92 | |
We also investigated only using as the input for the fine segmentor (i.e., is not applicable), and it can be observed that this method obtained better results than direct segmentation from , but its performance is much lower than that of our method, as shown in Table 2. In addition, for our error-prediction consistency segmentor, we compared and their average in Table 3. It shows that the three results are very close to each other, and the average Dice difference between and was only 0.28 (p-value 0.05).
Table 3.
Quantitative comparison between and their average obtained by our error-prediction consistency segmentor for whole glioma segmentation.
| Results Used | Dice (%) | ASSD (mm) | HD95 (mm) |
|---|---|---|---|
| 87.557.62 | 1.530.85 | 5.674.92 | |
| 87.277.91 | 1.601.08 | 6.336.28 | |
| average | 87.477.56 | 1.591.04 | 6.318.20 |
4.2.4. Hyper-parameter Analysis
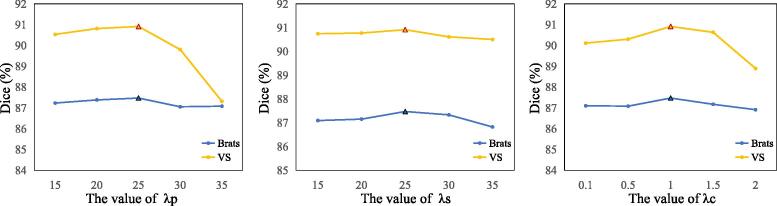
Our method has three main hyper-parameters related to the proposed loss function: for the perceptibility regularization loss (), for the coarse segmentation loss (), and for the fine segmentor loss (). We conducted ablation experiments on the validation set to investigate the sensitivity of these hyper-parameters. The results are presented in Fig. 6, which shows that our method performs the best when , and . It can be found that the performance our method is relatively not sensitive to and when they are in the range of [15], [25].
Fig. 6.
Performance of our method with different hyper-parameter values on the validation set of Brats dataset and VS dataset.
4.2.5. Comparison with State-of-the-art Methods
We further compared our framework with different types of existing methods for the synthesis and segmentation task: 1) separated synthesis and segmentation. We respectively used Pix2pix [15] and PGAN [3] for synthesizing FLAIR based on T1, ceT1 and T2 images, and then trained a 2.5D U-Net [27] to segment whole glioma from the concatenation of available and synthesized modalities. These two steps were trained separately. 2) End-to-end synthesis and segmentation. We used the methods of Wang et al. [32], PSCGAN [36] and UAGAN [41] for this purpose, respectively. As these methods were originally proposed for 2D images, we replaced their 2D CNN-based backbones with the 2.5D U-Net [27] respectively. All these methods were otherwise trained in the same way as the original papers. As UAGAN [41] had reported their results on BraTS dataset, we also list their reported results, which is denoted as UAGAN [41]. We found that our re-implementation of UAGAN had a better performance than the original paper, mainly due to the different data split and preprocessing methods.
Table 4 shows a quantitative comparison between these methods. For the existing separated synthesis and segmentation methods, PGAN [3] outperformed Pix2Pix [15], and their average Dice values were 85.79% and 84.67%, respectively. Among the existing end-to-end synthesis and segmentation methods, PSCGAN [36] outperformed the others, with an average Dice of 86.45%. Our end-to-end cascaded dual-task framework obtained an average Dice of 87.55%, which outperformed the existing methods.
Table 4.
Quantitative comparison of different synthesis-based and synthesis-free methods for whole glioma segmentation. and denote separated and end-to-end image synthesis and segmentation respectively. denotes synthesis-free methods for the segmentation task. Bold font highlights the best values obtained by synthesis-based methods. Results with no significant difference from the upper bound are denoted by *, according to a paired t-test (p-value 0.05).
| Methods | Dice (%) | ASSD (mm) | HD95 (mm) |
|---|---|---|---|
| Pix2Pix [15] | 84.677.93 | 2.262.09 | 9.12 11.97 |
| PGAN [3] | 85.797.73 | 1.901.29 | 7.217.53 |
| PSCGAN [36] | 86.458.04 | 1.991.59 | 8.629.62 |
| Wang et al. [32] | 83.778.48 | 2.411.70 | 9.9711.26 |
| UAGAN[41] | 81.552.96 | 2.530.29 | not reported |
| UAGAN [41] | 86.278.10 | 1.961.37 | 8.048.47 |
| w/o FLAIR | 84.549.62 | 2.322.12 | 10.7515.86 |
| Real FLAIR | 87.498.51 | 1.571.71 | 5.694.62 |
| Ours | 87.557.62 | 1.530.85 | 5.674.92 |
| Upper bound | 88.6510.01 | 1.501.89 | 5.544.69 |
We also trained a segmentation model based on 2.5D U-Net only using the available source-modality images, which is denoted as “w/o FLAIR”. The same network structure trained and tested with real FLAIR images only is denoted as “Real FLAIR”, as shown in Table 4. We can observe that our framework outperformed these two methods. It should be noted that compared with “w/o FLAIR” that directly uses source-modality images for training and testing, our method significantly improved the average Dice from 84.54 to 87.55. For comparison, we also segmented the whole tumor from a complete set of the four modalities, and the average Dice was 88.65, which serves as a upper bound for our synthesis-based segmentation. There is no significant difference between our result and the upper bound (p-value = 0.17 0.05 for Dice, p-value = 0.77 0.05 for ASSD and p-value = 0.83 0.05 for HD95).
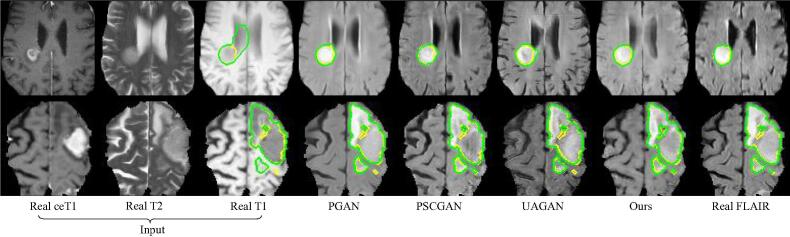
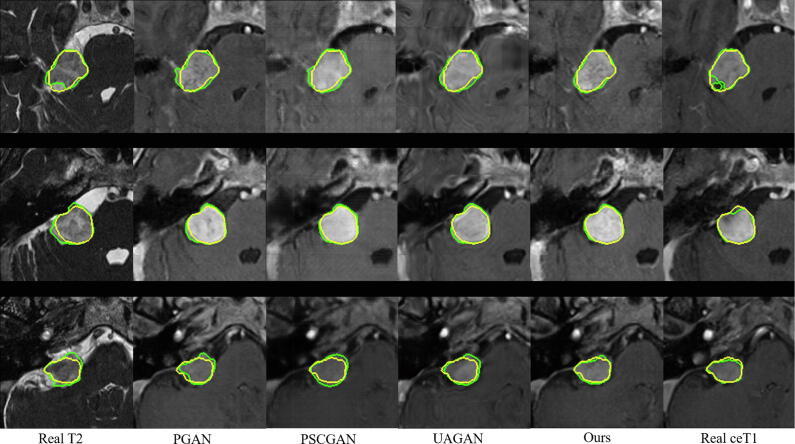
Fig. 7 shows a visual comparison between our method and the top three existing methods according to Table 4, i.e., PGAN [3], PSCGAN [36] and UAGAN [41]. It can be observed that the images synthesized by PGAN [3] are fuzzier than those of the other methods. The results of PSCGAN [36] have a different structure compared with the ground truth in some local regions, and UAGAN [41] introduced some artifacts. In contrast, our method leads to a better image quality, and its segmentation accuracy is also higher than that of the compared methods.
Fig. 7.
Visual comparison of different methods for glioma FLAIR image synthesis and segmentation. Yellow and green curves show the ground truth and segmentation results, respectively. Direct segmentation from the input source-modality images (T1, T2 and ceT1) is shown on the T1 image. Columns 4–7 show the segmentation results on the synthesized FLAIR images.
4.3. Segmentation of Vestibular Schwannoma
4.3.1. Data
We further used a public VS dataset for experiments [26]. In this dataset, spatially aligned T2 and ceT1 MR images of 242 patients with VS were acquired with in-plane resolution around 0.4 mm0.4 mm, in-plane size 512512 and slice thickness 1.5 mm. Manually segmented results by an experienced neurosurgeon and physicist were used as the ground truth by consensus [26]. We randomly selected images from 169, 24 and 49 patients for training, validation and testing, respectively. For preprocessing, each 3D volume was cropped by a cubic box centered on the tumor with 256, 128 and 40 pixels along the width, height and depth dimensions, respectively. The intensity values for each modality were normalized to the range of [-1, 1]. Here we treat T2 as the available source modality and ceT1 as the target modality to synthesize.
4.3.2. Ablation Study of the Synthesis Method
To demonstrate the effectiveness of our segmentation-aware ceT1 image synthesis, in this experiment we ignore segmentor (i.e., setting to 0), and compared different combinations of and , where is the baseline of using the global synthesis loss only. Note that was pre-trained on the ceT1 images and then freezed before training TISS-Net. To evaluate the domain similarity between synthesized and real ceT1 images, we measured the segmentation performance (perceptibility) of when applied to the synthesized ceT1 images, where a higher perceptibility indicates that they have closer high-level semantic appearance.
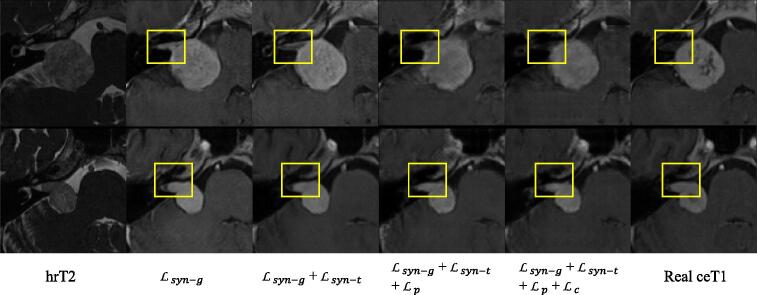
The quantitative evaluation results are shown in Table 5. It can be observed that our tumor-aware synthesis loss improved the image quality in terms of SSIM and PSNR, as well as the perceptibility (from 83.26% to 86.03% in terms of average Dice). Despite that did not improve the SSIM and PSNR scores that measure low-level image quality, it improved the perceptibility, showing its effectiveness in reducing the high-level semantic domain shift between real and synthesized ceT1 images. Our method of combining and achieved higher perceptibility (87.03% in average Dice) than the other variants, showing its effectivness in synthesizing segmentation-friendly ceT1 images of VS. Fig. 8 presents a visual comparison between synthesized ceT1 images of VS obtained by different loss functions. It can be observed that leads to an improved image quality in the tumor region, and when combining and , the image contrast and local structure in the synthesised ceT1 image is closer to those in the real ceT1 image than results of the other variants.
Table 5.
Quantitative evaluation results of different loss functions for ceT1 image synthesis and their effect on VS segmentation. Perceptibility means the performance of applying (i.e., a segmentation model pre-trained with real ceT1 images and freezed) to the synthesized images for segmentation.
| Loss functions |
Synthesis quality |
Perceptibility |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Global SSIM | Local SSIM | Global PSNR | Local PSNR | Dice (%) | ASSD (mm) | HD95(mm) | ||||
| ✓ | 0.600.04 | 0.680.09 | 23.061.33 | 22.711.92 | 83.2612.85 | 0.890.75 | 3.323.91 | |||
| ✓ | ✓ | 0.650.04 | 0.720.10 | 23.701.64 | 23.421.80 | 86.037.50 | 0.870.69 | 2.221.45 | ||
| ✓ | ✓ | 0.600.04 | 0.670.11 | 23.061.38 | 23.342.02 | 84.669.78 | 0.720.50 | 2.512.80 | ||
| ✓ | ✓ | ✓ | 0.600.04 | 0.710.09 | 23.371.41 | 22.832.20 | 86.577.50 | 0.550.20 | 1.701.07 | |
| ✓ | ✓ | ✓ | ✓ | 0.620.04 | 0.710.10 | 23.061.59 | 23.332.02 | 87.037.50 | 0.550.22 | 1.651.32 |
Fig. 8.
Visual comparison between synthesized ceT1 images of VS obtained by different loss functions.
4.3.3. Effectiveness of Fine Segmentation using Error-Prediction Consistency
We further investigated the effectiveness of our dual-task fine segmentor based on error-prediction consistency. We compared it with three variants: 1) only using the fine segmentation decoder (), without the error prediction branch; 2) error prediction branch only () without fine segmentation and thus without consistency loss; 3) predicting fine segmentation and error in the coarse segmentation simultaneously ( and ) but without consistency regularization.
Quantitative comparison between these methods for VS segmentation are shown in Table 6. It can be observed that when taking a concatenation of the source-modality image , the synthesized ceT1 image and coarse segmentation as input, using one of and leads to an average Dice of 88.39 and 88.25 respectively. Combining and together improved the average Dice to 88.87, and introducing the consistency loss further improved the average Dice to 89.46, which outperformed the other variants. We also compared these variants when using a concatenation of and as input of the segmentor. The results in the second section of Table 6 show that our proposed error-prediction consistency strategy still performed better than the other three variants.
Table 6.
Quantitative evaluation results of different inputs and loss functions for segmentor S in VS segmentation. denotes our synthesized ceT1 image, is the input T2 image, and is the coarse segmentation. The last two sections are based on a concatenation of these images as input. denotes significant improvement from as input based on a paired t-test (p-value 0.05).
| Input | Training loss |
Segmentation Performance |
||||
|---|---|---|---|---|---|---|
| Dice (%) | ASSD (mm) | HD95 (mm) | ||||
| ✓ | 86.0014.79 | 0.710.53 | 1.961.42 | |||
| ✓ | 87.206.22 | 0.510.20 | 1.540.72 | |||
| ✓ | 88.316.44 | 0.450.11 | 1.430.55 | |||
| ✓ | 88.364.67 | 0.470.13 | 1.400.49 | |||
| ✓ | ✓ | 89.037.12 | 0.460.15 | 1.400.56 | ||
| ✓ | ✓ | ✓ | 89.335.89 | 0.440.16 | 1.350.69 | |
| ✓ | 88.397.08 | 0.450.15 | 1.350.61 | |||
| ✓ | 88.256.29 | 0.470.17 | 1.410.84 | |||
| ✓ | ✓ | 88.874.51 | 0.430.15 | 1.320.59 | ||
| ✓ | ✓ | ✓ | 89.465.49 | 0.420.15 | 1.310.59 | |
We also investigated only using the synthesized ceT1 image as the input for the fine segmentor (where is not applicable), and found that it obtained better results than direct segmentation from T2 images. Its average Dice was 87.20%, compared with 89.33% obtained by using a concatenation of and and 89.46% obtained by using a concatenation of and .
4.3.4. Comparison with State-of-the-art Methods
Additionally, we compared our framework with different types of existing methods for the synthesis and segmentation task: 1) Separated synthesis and segmentation. We respectively used Pix2pix [15] and PGAN [3] for the synthesis step, and trained a 2.5D U-Net [27] to segment VS from a concatenation of T2 and synthesized ceT1 images. These two steps were trained separately. 2) End-to-end synthesis and segmentation. We used the methods of Wang et al. [32], PSCGAN [36] and UAGAN [41] for this purpose, respectively. As these methods were originally proposed for 2D images, we replaced their 2D CNN-based backbones with the 2.5D U-Net [27] respectively. All these methods were otherwise trained in the same way as the original papers.
Table 7 shows a quantitative comparison between these methods. Our method achieved an average Dice of 89.46%, compared with 83.75% of Pix2Pix [15], 82.90% by PGAN [3], 85.89% by Wang et al. [32], 84.65% by PSCGAN [36] and 87.30% by UAGAN [41], respectively. The results demonstrate that our cascaded dual-task framework outperformed the existing methods.
Table 7.
Quantitative comparison of different synthesis-based and synthesis-free methods for VS segmentation. and denote separated and end-to-end methods for synthesis and segmentation, respectively. denotes synthesis-free methods for the segmentation task. Bold font highlights the best values obtained by the synthesis-based methods. denotes significant improvement from “T2 only” based on a paired t-test (p-value 0.05).
| Methods | Dice (%) | ASSD (mm) | HD95 (mm) |
|---|---|---|---|
| Pix2Pix [15] | 83.7515.00 | 0.780.43 | 2.602.37 |
| PGAN [3] | 82.9012.31 | 1.191.09 | 2.995.52 |
| [32] | 85.896.50 | 0.530.14 | 1.540.59 |
| PSCGAN [36] | 84.6510.07 | 0.650.39 | 1.790.87 |
| UAGAN [41] | 87.308.65 | 0.500.43 | 1.650.62 |
| T2 only | 86.0014.79 | 0.710.53 | 1.961.42 |
| Real ceT1 | 92.803.83 | 0.310.13 | 1.090.24 |
| Ours | 89.465.49 | 0.420.15 | 1.310.59 |
We also trained a segmentation model based on 2.5D U-Net using the T2 images and real ceT1 images, respectively for comparison. It can be observed that our framework improved the average Dice from 86.00% to 89.46% compared with simply segmenting from T2 MRI and the improvement was significant based on a paired t-test (p-value0.05). Using real ceT1 images for training and testing achieved an average Dice of 92.80. Visual comparison in Fig. 9 also shows the better performance of our framework than the other synthesis-based methods and direct segmentation from the T2 images.
Fig. 9.
Visual comparison of different methods for VS image synthesis and segmentation. Yellow and green curves show the ground truth and segmentation results, respectively. Columns 2–5 show the segmentation results on the synthesized ceT1 images.
5. Discussion
Accurate segmentation of brain tumors relies on multi-modal images or high-contrast images, but the access to some modalities may be limited as it is expansive, time-consuming or faced with safety concerns with the use of contrast agents, which has been a crucial obstacle for developing deep learning methods for accurate segmentation of brain tumors. To alleviate these problems, we propose a new method for missing modality synthesis for better segmentation. Our proposed TISS-Net is a unification of simultaneous synthesis and segmentation through dual-task networks, coarse-to-fine segmentation and error-prediction consistency. Compared with typical synthesis followed by segmentation methods [39], [5], our framework is trained end-to-end, so that the synthesis and segmentation are adaptive to each other and it could obtain segmentation-friendly synthesis results. Differently from existing end-to-end methods for image synthesis [31], [36], we propose a cascaded dual-task architecture, and introduce several regularization strategies to improve the performance, i.e., simultaneous synthesis and coarse segmentation, perceptibility regularization and error-prediction consistency.
The general effectiveness of our method has been demonstrated on two different brain tumor segmentation tasks. For the whole glioma segmentation, our synthesis-based method achieved a performance that is comparable to segmentation from multi-modal images with FLAIR. However, we found that synthesizing ceT1 images of VS from a single modality of T2 is more challenging as shown in Table 7, and similar phenomenon had also been reported by previous works [18]. The main reason is that the input single-modality T2 image has a low contrast and contains limited information of the contrast agent. Introducing shape and contrast prior information could be a potential solution to further narrow the gap, which will be investigated in the future. Due to the memory limitation, we used 2.5D networks considering anisotropic resolution and to achieve a trade-off among patch size, 3D feature learning and GPU memory consumption. However, our method can also be extended with 3D networks.
This work also has some limitations. First, the cascaded networks with dual decoders increase the model complexity, and it has more parameters than methods using single-decoder networks or single-stage methods. Compared with Pix2Pix [15] and PGAN [3], our method increases the model size from 81.06 M to 126.96 M due to the auxiliary decoders under the the same backbone. On the VS dataset, the training time per epoch is 134 s, compared with 126 s of PGAN. However, at the testing stage, as only the first branch is used in the segmentor, our method has a similar inference time compared with existing methods, i.e., 0.09 s/case for PGAN and 0.11 s/case for TISS-Net, respectively. Second, in this work, we have investigated binary segmentation of brain tumors based on synthesis of a missing modality, and its effectiveness on multi-class segmentation tasks remains to be verified. In addition, this work only considered the synthesis of a single missing modality, and in some cases, multiple modalities might be missing. It is of interest to extend our method to deal with multiple missing modalities in the future.
6. Conclusion
In conclusion, we propose a novel cascaded dual-task network TISS-Net to synthesize a missing modality for brain tumor segmentation given one or a set of available source modalities. To synthesize segmentation-friendly target-modality images, we employ a dual-branch network to predict the target modality and a coarse segmentation simultaneously, and propose a tumor-aware synthesis loss with perceptibility regularization that improves the image quality around the tumor region and reduces the high-level domain gap between synthesized and real target-modality images. For the final segmentation network, a consistency loss between fine segmentation and error prediction in the coarse segmentation is proposed for regularization. Experiments on glioma and VS images show that our TISS-Net outperformed state-of-the-art segmentation methods based on target-modality image synthesis, and it leads to significantly higher accuracy than segmentation from the original partial modalities. This work increases the accuracy of automated tumor assessment with a missing modality or without the need of gadolinium-based scanning that is associated with more time consumption or even potentially harmful side-effects of cumulative gadolinium contrast agent use. In the future, it is of interest to apply the proposed method for other types of target modalities and tissues, and investigate more efficient network structures for the synthesis and segmentation.
CRediT authorship contribution statement
Jianghao Wu: Methodology, Writing - original draft, Writing - review & editing. Dong Guo: Methodology. Lu Wang: Methodology. Shuojue Yang: Writing - original draft. Yuanjie Zheng: Conceptualization, Methodology. Jonathan Shapey: Supervision. Tom Vercauteren: Conceptualization, Supervision. Sotirios Bisdas: Supervision. Robert Bradford: Supervision. Shakeel Saeed: Conceptualization, Supervision. Neil Kitchen: Supervision, Conceptualization. Sebastien Ourselin: Conceptualization, Supervision. Shaoting Zhang: Methodology, Supervision. Guotai Wang: Supervision, Writing - review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
This work was supported by the National Natural Science Foundations of China [81771921, 61901084] funding, the Wellcome Trust [203145Z/16/Z; 203148/Z/16/Z; WT106882], and EPSRC [NS/A000050/1; NS/A000049/1] funding. TV is supported by a Medtronic/Royal Academy of Engineering Research Chair [RCSRF1819/7/34].
Biographies
Jianghao Wu is a graduate student at University of Electronic Science and Technology of China. His research interest is medical image analysis and computer vision.
Dong Guo is a graduate student at University of Electronic Science and Technology of China. His research interest is medical image synthesis and deep learning.
Lu Wang is a graduate student at University of Electronic Science and Technology of China. His research interest is medical image synthesis and deep learning.
Shuojue Yang is an undergraduate student at University of Electronic Science and Technology of China. His research interest is medical image analysis and computer vision
Yuanjie Zheng is a professor at Shandong Normal University. He received his PhD degree at Shanghai Jiao Tong University in 2006. His main research interests are artificial intelligence, computer vision and medical image analysis.
Jonathan Shapey is a clinical Senior Lecturer in Neurosurgery at King’s College London. Jonathan’s academic interest focuses on the application of medical technology and artificial intelligence to neurosurgery.
Tom Vercauteren is Professor of Interventional Image Computing at King’s College London since 2018 where he holds the Medtronic/Royal Academy of Engineering Research Chair in Machine Learning for Computer-assisted Neurosurgery. From 2014 to 2018, he was Associate Professor at UCL where he acted as Deputy Director for the Wellcome / EPSRC Centre for Interventional and Surgical Sciences (2017-18). He is a Columbia University and Ecole Polytechnique graduate and obtained his PhD from Inria in 2008.
Sotirios Bisdas is consultant neuroradiologist and MRI lead in the Department of Neuroradiology at the National Hospital for Neurology in London, senior lecturer in neuroradiology at the Institute of Neurology University College London, and professor of radiology at Eberhard Karls University in Tübingen, Germany. His expertise fields include advanced CT, intraoperative MRI, advanced and functional MRI and molecular MR-PET imaging in brain diseases.
Robert Bradford is Past Chair Brain/CNS tumour board North London Cancer Network and currently clinical director neurosciences at National Hospital for Neurology and Neurosurgery. His research interests are Clinical neuro-oncology, Stereotaxis and image-guided neurosurgery for brain tumours, and Management of acoustic neuromas.
Shakeel Saeed is a professor at University College Hospital. He is currently a leading surgeon and researcher in disorders of the ear, hearing, balance, facial nerve and skullbase. He was the President of the European Academy of Otology and Neurotology 2018-2021, and has Over 120 peer-reviewed publications.
Neil Kitchen is a consultant neurosurgeon at the National Hospital for Neurology and Neurosurgery (NHNN), Director of the Gamma Knife Unit and lead neurosurgeon for skull-base surgery. He studied medicine at St Bartholomew’s hospital and Cambridge University. Before moving to Cambridge he completed a BSc degree in the history of medicine at the Wellcome Institute, UCL.
Sebastien Ourselin is Head of the School of Biomedical Engineering & Imaging Sciences at King’s College London. His core skills are in medical image analysis, software engineering, and translational medicine. He is best known for his work on image registration and segmentation, its exploitation for robust image-based biomarkers in neurological conditions, as well as for his development of image-guided surgery systems.
Shaoting Zhang is a Professor at University of Electronic Science and Technology of China. He received PhD in Computer Science from Rutgers in 2011, M.S. from Shanghai Jiao Tong University in 2007, and B.E. from Zhejiang University in 2005. His research is on the interface of medical imaging informatics, computer vision and machine learning.
Guotai Wang is an Associate Professor at University of Electronic Science and Technology of China. He obtained his Bachelor and Master degree of Biomedical Engineering in Shanghai Jiao Tong University in 2011 and 2014 respectively. He then obtained his PhD degree of Medical and Biomedical Imaging in University College London in 2018. His research interests include medical image computing, computer vision and deep learning.
Communicated by Zidong Wang
Data availability
We used public datasets in this work
References
- 1.Bahrami K., Rekik I., Shi F., Shen D. International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer; 2017. Joint reconstruction and segmentation of 7T-like MR images from 3T MRI based on cascaded convolutional neural networks; pp. 764–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen C., Dou Q., Jin Y., Liu Q., Heng P.A. Learning with privileged multimodal knowledge for unimodal segmentation. IEEE Transactions on Medical Imaging. 2022;41:621–632. doi: 10.1109/TMI.2021.3119385. [DOI] [PubMed] [Google Scholar]
- 3.Dar S.U., Yurt M., Karacan L., Erdem A., Erdem E., Çukur T. Image synthesis in multi-contrast MRI with conditional generative adversarial networks. IEEE transactions on medical imaging. 2019;38:2375–2388. doi: 10.1109/TMI.2019.2901750. [DOI] [PubMed] [Google Scholar]
- 4.Ding Y., Zhang C., Cao M., Wang Y., Chen D., Zhang N., Qin Z. Tostagan: An end-to-end two-stage generative adversarial network for brain tumor segmentation. Neurocomputing. 2021;462:141–153. doi: 10.1016/j.neucom.2021.07.066. [DOI] [Google Scholar]
- 5.Dong X., Lei Y., Tian S., Wang T., Patel P., Curran W.J., Jani A.B., Liu T., Yang X. Synthetic MRI-aided multi-organ segmentation on male pelvic CT using cycle consistent deep attention network. Radiother. Oncol. 2019;141:192–199. doi: 10.1016/j.radonc.2019.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dorent R., Joutard S., Modat M., Ourselin S., Vercauteren T. International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer; 2019. Hetero-modal variational encoder-decoder for joint modality completion and segmentation; pp. 74–82. [Google Scholar]
- 7.Dou Q., Ouyang C., Chen C., Chen H., Heng P.A. Proceedings of International Joint Conference on Artificial Intelligence. 2018. Unsupervised cross-modality domain adaptation of convnets for biomedical image segmentations with adversarial loss; pp. 691–697. [Google Scholar]
- 8.Frangi A.F., Tsaftaris S.A., Prince J.L. Simulation and synthesis in medical imaging. IEEE transactions on medical imaging. 2018;37:673–679. doi: 10.1109/TMI.2018.2800298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guan H., Liu M. Domain adaptation for medical image analysis: A survey. IEEE Transactions on Biomedical Engineering. 2022;69:1173–1185. doi: 10.1109/TBME.2021.3117407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Havaei M., Guizard N., Chapados N., Bengio Y. MICCAI. Springer; 2016. Hemis: Hetero-modal image segmentation; pp. 469–477. [Google Scholar]
- 11.Hu J., Gu X., Gu X. Mutual ensemble learning for brain tumor segmentation. Neurocomputing. 2022;504:68–81. doi: 10.1016/j.neucom.2022.06.058. [DOI] [Google Scholar]
- 12.Hu M., Maillard M., Zhang Y., Ciceri T., La Barbera G., Bloch I., Gori P. MICCAI. Springer; 2020. Knowledge distillation from multi-modal to mono-modal segmentation networks; pp. 772–781. [Google Scholar]
- 13.Huang Y., Shao L., Frangi A.F. Cross-modality image synthesis via weakly coupled and geometry co-regularized joint dictionary learning. IEEE Transactions on Medical Imaging. 2017;37:815–827. doi: 10.1109/TMI.2017.2781192. [DOI] [PubMed] [Google Scholar]
- 14.Isensee F., Kickingereder P., Wick W., Bendszus M., Maier-Hein K.H. International MICCAI Brainlesion Workshop. Springer; 2018. No new-net; pp. 234–244. [Google Scholar]
- 15.Isola P., Zhu J.-Y., Zhou T., Efros A.A. Proceedings of the IEEE con- ference on Computer Vision and Pattern Recognition. 2017. Image-to-image translation with conditional adversarial networks; pp. 1125–1134. [Google Scholar]
- 16.Jiang Z., Ding C., Liu M., Tao D. International MICCAI Brainlesion Workshop. Springer; 2019. Two-stage cascaded U-Net: 1st place solution to BraTS challenge 2019 segmentation task; pp. 231–241. [Google Scholar]
- 17.Jog A., Carass A., Roy S., Pham D.L., Prince J.L. Random forest regression for magnetic resonance image synthesis. Medical image analysis. 2017;35:475–488. doi: 10.1016/j.media.2016.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee D., Moon W.-J., Ye J.C. Assessing the importance of magnetic resonance contrasts using collaborative generative adversarial networks. Nature Machine Intelligence. 2020;2:34–42. [Google Scholar]
- 19.Liu J., Xuan W., Gan Y., Zhan Y., Liu J., Du B. An end-to-end supervised domain adaptation framework for cross-domain change detection. Pattern Recognition. 2022;132 [Google Scholar]
- 20.Luo Y., Nie D., Zhan B., Li Z., Wu X., Zhou J., Wang Y., Shen D. Edge-preserving mri image synthesis via adversarial network with iterative multi-scale fusion. Neurocomputing. 2021;452:63–77. doi: 10.1016/j.neucom.2021.04.060. [DOI] [Google Scholar]
- 21.Mazumdar I., Mukherjee J. Fully automatic mri brain tumor segmentation using efficient spatial attention convolutional networks with composite loss. Neurocomputing. 2022;500:243–254. doi: 10.1016/j.neucom.2022.05.050. [DOI] [Google Scholar]
- 22.Menze B.H., Jakab A., Bauer S., Kalpathy-Cramer J., Farahani K., et al. The multimodal brain tumor image segmentation benchmark (BRATS) IEEE transactions on medical imaging. 2015;34:1993–2024. doi: 10.1109/TMI.2014.2377694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ohgaki H., Kleihues P. Population-based studies on incidence, survival rates, and genetic alterations in astrocytic and oligodendroglial gliomas. Journal of Neuropathology & Experimental Neurology. 2005;64:479–489. doi: 10.1093/jnen/64.6.479. [DOI] [PubMed] [Google Scholar]
- 24.Ostrom Q.T., Gittleman H., Truitt G., Boscia A., Kruchko C., Barnholtz-Sloan J.S. CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the united states in 2011–2015. Neuro-oncology. 2018;20 doi: 10.1093/neuonc/noy131. iv1–iv86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pereira S., Pinto A., Alves V., Silva C.A. Brain tumor segmentation using convolutional neural networks in MRI images. IEEE transactions on medical imaging. 2016;35:1240–1251. doi: 10.1109/TMI.2016.2538465. [DOI] [PubMed] [Google Scholar]
- 26.Shapey J., Kujawa A., Dorent R., Wang G., Dimitriadis A., Grishchuk D., Paddick I., Kitchen N., Bradford R., Saeed S.R., et al. Segmentation of vestibular schwannoma from MRI, an open annotated dataset and baseline algorithm. Scientific Data. 2021;8:1–6. doi: 10.1038/s41597-021-01064-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shapey J., Wang G., Dorent R., Dimitriadis A., Li W., Paddick I., Kitchen N., Bisdas S., Saeed S.R., Ourselin S., et al. An artificial intelligence framework for automatic segmentation and volumetry of vestibular schwannomas from contrast-enhanced T1-weighted and high-resolution T2-weighted MRI. Journal of Neurosurgery. 2019;134:171–179. doi: 10.3171/2019.9.JNS191949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Song X., Chao H., Xu X., Guo H., Xu S., Turkbey B., Wood B.J., Sanford T., Wang G., Yan P. Cross-modal attention for multi-modal image registration. Medical Image Analysis. 2022;82 doi: 10.1016/j.media.2022.102612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun J., Peng Y., Guo Y., Li D. Segmentation of the multimodal brain tumor image used the multi-pathway architecture method based on 3d fcn. Neurocomputing. 2021;423:34–45. doi: 10.1016/j.neucom.2020.10.031. [DOI] [Google Scholar]
- 30.Sun L., Fan Z., Ding X., Huang Y., Paisley J. International Conference on Information Processing in Medical Imaging. Springer; 2019. Joint CS-MRI reconstruction and segmentation with a unified deep network; pp. 492–504. [Google Scholar]
- 31.Wang G., Li W., Ourselin S., Vercauteren T. International MICCAI brainlesion workshop. Springer; 2017. Automatic brain tumor segmentation using cascaded anisotropic convolutional neural networks; pp. 178–190. [Google Scholar]
- 32.Wang G., Song T., Dong Q., Cui M., Huang N., Zhang S. Automatic ischemic stroke lesion segmentation from computed tomography perfusion images by image synthesis and attention-based deep neural networks. Medical Image Analysis. 2020;65 doi: 10.1016/j.media.2020.101787. [DOI] [PubMed] [Google Scholar]
- 33.Wang Y., Yu B., Wang L., Zu C., Lalush D.S., Lin W., Wu X., Zhou J., Shen D., Zhou L. 3D conditional generative adversarial networks for high-quality PET image estimation at low dose. NeuroImage. 2018;174:550–562. doi: 10.1016/j.neuroimage.2018.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu J., Gu R., Dong G., Wang G., Zhang S. 2022 IEEE 19th International Symposium on Biomedical Imaging (ISBI) IEEE; 2022. Fpl-uda: Filtered pseudo label-based unsupervised cross-modality adaptation for vestibular schwannoma segmentation; pp. 1–5. [Google Scholar]
- 35.Xie Y., Lu H., Zhang J., Shen C., Xia Y. International Conference on Medical Image Computing and Computer- Assisted Intervention. Springer; 2019. Deep segmentation-emendation model for gland instance segmentation; pp. 469–477. [Google Scholar]
- 36.Xu C., Xu L., Ohorodnyk P., Roth M., Chen B., Li S. Contrast agent-free synthesis and segmentation of ischemic heart disease images using progressive sequential causal gans. Medical Image Analysis. 2020;62 doi: 10.1016/j.media.2020.101668. [DOI] [PubMed] [Google Scholar]
- 37.Yang J., Dvornek N.C., Zhang F., Chapiro J., Lin M., Duncan J.S. International Conference on Medical Image Computing and Computer- Assisted Intervention. 2019. Unsupervised domain adaptation via disentangled representations: Application to cross-modality liver segmentation; pp. 255–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ye F., Zheng Y., Ye H., Han X., Li Y., Wang J., Pu J. Parallel pathway dense neural network with weighted fusion structure for brain tumor segmentation. Neurocomputing. 2021;425:1–11. doi: 10.1016/j.neucom.2020.11.005. [DOI] [Google Scholar]
- 39.Yu, B., Zhou, L., Wang, L., Fripp, J., & Bourgeat, P. (2018). cGAN based cross-modality MR image synthesis for brain tumor segmentation. In 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018) (pp. 626–630). IEEE.
- 40.Yu Q., Gao Y., Zheng Y., Zhu J., Dai Y., Shi Y. Crossover-net: Leveraging vertical-horizontal crossover relation for robust medical image segmentation. Pattern Recognition. 2021;113 [Google Scholar]
- 41.Yuan W., Wei J., Wang J., Ma Q., Tasdizen T. International Conference on Medical Image Computing and Computer- Assisted Intervention. Springer; 2019. Unified attentional generative adversarial network for brain tumor segmentation from multimodal unpaired images; pp. 229–237. [Google Scholar]
- 42.Zhang J., Zeng J., Qin P., Zhao L. Brain tumor segmentation of multi-modality mr images via triple intersecting u-nets. Neurocomputing. 2021;421:195–209. doi: 10.1016/j.neucom.2020.09.016. [DOI] [Google Scholar]
- 43.Zhong Q., Zeng F., Liao F., Liu J., Du B., Shang J.S. Joint image and feature adaptative attention-aware networks for cross-modality semantic segmentation. Neural Computing and Applications. 2023;35:3665–3676. [Google Scholar]
- 44.Zhou C., Ding C., Wang X., Lu Z., Tao D. One-pass multi-task networks with cross-task guided attention for brain tumor segmentation. IEEE Transactions on Image Processing. 2020;29:4516–4529. doi: 10.1109/TIP.2020.2973510. [DOI] [PubMed] [Google Scholar]
- 45.Zhou T., Canu S., Vera P., Ruan S. Feature-enhanced generation and multi-modality fusion based deep neural network for brain tumor segmentation with missing mr modalities. Neurocomputing. 2021;466:102–112. doi: 10.1016/j.neucom.2021.09.032. [DOI] [Google Scholar]
- 46.Zhou Z., He Z., Jia Y. Afpnet: A 3d fully convolutional neural network with atrous-convolution feature pyramid for brain tumor segmentation via mri images. Neurocomputing. 2020;402:235–244. doi: 10.1016/j.neucom.2020.03.097. [DOI] [Google Scholar]
- 47.Zhu Q., Du B., Yan P. Boundary-weighted domain adaptive neural network for prostate mr image segmentation. IEEE transactions on medical imaging. 2019;39:753–763. doi: 10.1109/TMI.2019.2935018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
We used public datasets in this work