Abstract
We recently reported that the ER stress kinase PERK regulates ER-mitochondria appositions and ER– plasma membrane (ER-PM) contact sites, independent of its canonical role in the unfolded protein response. PERK regulation of ER-PM contacts was revealed by a proximity biotinylation (BioID) approach and involved a dynamic PERK-Filamin A interaction supporting the formation of ER-PM contacts by actin-cytoskeleton remodeling in response to depletion of ER-Ca2+ stores. In this report, we further interrogated the PERK BioID interactome by validating through co-IP experiments the interaction between PERK and two proteins involved in Ca2+ handling and ER-mitochondria contact sites. These included the vesicle associated membrane (VAMP)-associated proteins (VAPA/B) and the main ER Ca2+ pump sarcoplasmic/endoplasmic reticulum Ca ATPase 2 (SERCA2). These data identify new putative PERK interacting proteins with a crucial role in membrane contact sites and Ca2+ signaling further supporting the uncanonical role of PERK in Ca2+ signaling through membrane contact sites (MCSs).
Keywords: cell biology, endoplasmic reticulum, ER stress, sarco/endoplasmic reticulum Ca2+-ATPase (SERCA), mitochondrial associated membranes (MAM)
Introduction
The endoplasmic reticulum (ER) is a major site of protein folding in the cell, handling roughly one-third of all cellular proteins and all proteins destined for transport towards the plasma membrane (PM) or extracellular matrix (Brodsky & Wojcikiewicz, 2009). To cope with this demand, the ER has evolved to possess an intricate folding machinery and an ideal protein folding environment. However, when the ER can no longer match cellular protein folding demand, the resulting accumulation of unfolded proteins causes ER stress (Ron & Walter, 2007). The unfolded protein response (UPR) consists of the activation of a conserved signal transduction pathway, ultimately eliciting a transcriptional program that operates as a principal safeguard against the loss of ER homeostasis caused by ER stress. The UPR is launched by the activation of three ER membrane proteins. The ER stress kinase PKR-like endoplasmic reticulum kinase (PERK) is one of these three mediators and is activated upon ER stress (Ron & Walter, 2007). In homeostatic conditions, PERK is kept inactive through the binding of the ER chaperone BiP to its luminal domain, and is activated upon its release, resulting in the oligomerization of PERK, followed by its autophosphorylation. Activated PERK is then able to phosphorylate eukaryotic initiation factor 2 alpha (eIF2α), leading to a protein translation pause and giving the ER folding machinery time to deal with its protein burden (Ron & Walter, 2007).
In our previous studies (van Vliet et al., 2017; Verfaillie et al., 2012), we uncovered that independent of its UPR function, PERK moonlights at the ER-mitochondria contacts and aids apoptotic cell death by the transfer of ROS signals from the ER to the mitochondria. More recently, we showed that the activation of PERK can occur independently of its luminal domain and canonical ER stress, instead of being activated by a rise in cytosolic Ca2+.
To uncover the roles of PERK we carried out an unbiased proximity biotinylation (BioID) screen, using the promiscuous biotinylation enzyme BirA* tagged to PERK on its cytosolic side (Roux et al., 2012; van Vliet et al., 2017). Using this technique, we discovered the cytoskeletal protein Filamin A (FLNA) as a novel PERK interactor. The Ca2+ -mediated PERK-FLNA axis was found to be required to support the formation of ER-PM contacts and store-operated Ca2+ entry (SOCE) (van Vliet et al., 2017). These findings together hint at a broader cross-talk between ER stress proteins and the regulation of membrane contact sites (MCSs). However, many hits uncovered through our BioID experiment warrant further investigation in order to get a better picture of the potential additional roles of PERK. In this report, we reveal the full dataset of PERK interacting proteins identified through BioID analysis. We further confirmed by IP/Co-IP analysis, the physical interaction between PERK and sarcoplasmic/endoplasmic reticulum Ca ATPase 2 (SERCA2) and VAMP-associated protein A / B (VAPA/B) and show that for these interactions, PERK kinase activity is dispensable.
Results and Discussion
The PERK Proximity Interactome
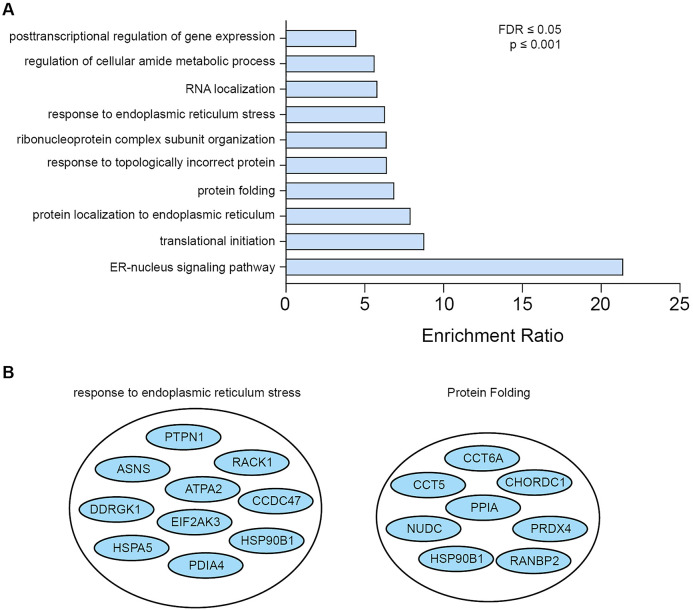
We generated a C-terminally tagged PERK-BirA* construct that was well expressed in HEK293-T cells and showed the expected ER localization (van Vliet et al., 2017). Our BioID approach was designed and performed to closely match the original study reporting BioID (Roux et al., 2012). Similar to that study, our approach relied on the expression of PERK-BirA* in HEK293-T cells, using mock-transfected parental HEK293-T cells treated in the same way (50 µM biotin for 24 h), as a control. Only biotinylated protein hits identified by LC-MS/MS in the streptavidin pulldown from PERK-BirA* transfected cells and not from mock-transfected cells were taken into consideration as putative interaction partners. Table 1 shows a list of PERK proximity interactors showing no spectral counts in the pulldown from mock-transfected cells. Interestingly, while our previous research focused on one of the most prominent hits, Filamin A, which we further validated functionally, the BioID dataset showed several other potential interacting proteins (Table 1), ranging from more proteins involved in actin cytoskeleton maintenance (cofilin, profilin,...), proteins involved in ER trafficking (syntaxin 5, coatomer subunits), proteins involved in membrane contact sites (VAPA/B, E-Syt1, junctophilin) to proteins linked with metabolism (ATP-citrate synthase, acyl-protein thioesterase 2). Interestingly, PERK has been linked recently with metabolic regulatory networks (Balsa et al., 2019; Moncan et al., 2021; Sorge et al., 2020), although whether these potential interactors uncovered by our BioID are relevant remains to be tested. To gain more insight into the various groups of proteins in the dataset, we performed a gene ontology analysis using WEB-based GEne SeT AnaLysis Toolkit (http://www.webgestalt.org/, Figure 1). This analysis yielded a set of enriched biological processes in our dataset. As a validation of our approach, two of these groups are linked to PERK's main role in protein folding and ER stress response (Figure 1A and B).
Table 1.
List of identified proteins (Scaffold, FDR < 1%) resulting from the BioID interactome screen using PERK-BirA as bait. Protein hits were only detected using PERK-BirA as bait and not in control. Parental cells are shaded in yellow. Relative quantification of proteins is based on spectral counts (‘Total spectra’). Only proteins with atleast 2 exclusive unique peptides per protein are listed.
| Molecular weight | Quantitative value (total spectra) | Exclusive unique peptide count | Quantitative value (total spectra) | Exclusive unique peptide count | Protein identification probability | Protein identification probability | |||
|---|---|---|---|---|---|---|---|---|---|
| Identified proteins (276) | Accession number | Alternate ID | PERK-BirA | PERK-BirA | Control | Control | PERK-BirA | Control | |
| Eukaryotic translation initiation factor 2-alpha kinase 3 OS = Homo sapiens OX = 9606 GN = EIF2AK3 PE = 1 SV = 3 | Q9NZJ5 | EIF2AK3 | 125 kDa | 415 | 70 | 0 | 0 | 100% | 0 |
| Filamin-A OS = Homo sapiens OX = 9606 GN = FLNA PE = 1 SV = 4 | P21333 | FLNA | 281 kDa | 94 | 66 | 0 | 0 | 100% | 0 |
| Coatomer subunit gamma-2 OS = Homo sapiens OX = 9606 GN = COPG2 PE = 1 SV = 1 | Q9UBF2 | COPG2 | 98 kDa | 40 | 26 | 0 | 0 | 100% | 0 |
| Lamina-associated polypeptide 2, isoforms beta/gamma OS = Homo sapiens OX = 9606 GN = TMPO PE = 1 SV = 2 | P42167 | TMPO | 51 kDa | 28 | 16 | 0 | 0 | 100% | 0 |
| Keratin, type I cytoskeletal 16 OS = Homo sapiens OX = 9606 GN = KRT16 PE = 1 SV = 4 | P08779 | KRT16 | 51 kDa | 26 | 4 | 0 | 0 | 100% | 0 |
| Kinectin OS = Homo sapiens OX = 9606 GN = KTN1 PE = 1 SV = 1 | Q86UP2 | KTN1 | 156 kDa | 24 | 23 | 0 | 0 | 100% | 0 |
| RuvB-like 1 OS = Homo sapiens OX = 9606 GN = RUVBL1 PE = 1 SV = 1 | Q9Y265 | RUVBL1 | 50 kDa | 20 | 13 | 0 | 0 | 100% | 0 |
| E3 SUMO-protein ligase RanBP2 OS = Homo sapiens OX = 9606 GN = RANBP2 PE = 1 SV = 2 | P49792 | RANBP2 | 358 kDa | 20 | 19 | 0 | 0 | 100% | 0 |
| 78 kDa glucose-regulated protein OS = Homo sapiens GN = HSPA5 PE = 1 SV = 2 | P11021| GRP78_HUMAN | HSPA5 | 72 kDa | 16 | 13 | 0 | 0 | 100% | 0 |
| Double-strand break repair protein MRE11 OS = Homo sapiens OX = 9606 GN = MRE11 PE = 1 SV = 3 | P49959 | MRE11 | 81 kDa | 15 | 14 | 0 | 0 | 100% | 0 |
| Zinc finger CCCH-type antiviral protein 1 OS = Homo sapiens OX = 9606 GN = ZC3HAV1 PE = 1 SV = 3 | Q7Z2W4 | ZC3HAV1 | 101 kDa | 14 | 10 | 0 | 0 | 100% | 0 |
| Src substrate cortactin OS = Homo sapiens OX = 9606 GN = CTTN PE = 1 SV = 2 | Q14247 | CTTN | 62 kDa | 14 | 12 | 0 | 0 | 100% | 0 |
| Bifunctional glutamate/proline--tRNA ligase OS = Homo sapiens OX = 9606 GN = EPRS PE = 1 SV = 5 | P07814 | EPRS | 171 kDa | 12 | 11 | 0 | 0 | 100% | 0 |
| Lamin-B receptor OS = Homo sapiens OX = 9606 GN = LBR PE = 1 SV = 2 | Q14739 | LBR | 71 kDa | 11 | 7 | 0 | 0 | 100% | 0 |
| Transgelin-2 OS = Homo sapiens OX = 9606 GN = TAGLN2 PE = 1 SV = 3 | P37802 | TAGLN2 | 22 kDa | 10 | 7 | 0 | 0 | 100% | 0 |
| eIF-2-alpha kinase activator GCN1 OS = Homo sapiens OX = 9606 GN = GCN1 PE = 1 SV = 6 | Q92616 | GCN1 | 293 kDa | 10 | 10 | 0 | 0 | 100% | 0 |
| UBX domain-containing protein 4 OS = Homo sapiens OX = 9606 GN = UBXN4 PE = 1 SV = 2 | Q92575 | UBXN4 | 57 kDa | 10 | 6 | 0 | 0 | 100% | 0 |
| Staphylococcal nuclease domain-containing protein 1 OS = Homo sapiens OX = 9606 GN = SND1 PE = 1 SV = 1 | Q7KZF4 | SND1 | 102 kDa | 9 | 9 | 0 | 0 | 100% | 0 |
| Vesicle-associated membrane protein-associated protein A OS = Homo sapiens OX = 9606 GN = VAPA PE = 1 SV = 3 | Q9P0L0 | VAPA | 28 kDa | 8 | 4 | 0 | 0 | 100% | 0 |
| Histone H1.5 OS = Homo sapiens OX = 9606 GN = HIST1H1B PE = 1 SV = 3 | P16401 | HIST1H1B | 23 kDa | 7 | 3 | 0 | 0 | 100% | 0 |
| Sodium/potassium-transporting ATPase subunit alpha-1 OS = Homo sapiens OX = 9606 GN = ATP1A1 PE = 1 SV = 1 | P05023 | ATP1A1 | 113 kDa | 7 | 7 | 0 | 0 | 100% | 0 |
| Eukaryotic translation initiation factor 5 OS = Homo sapiens OX = 9606 GN = EIF5 PE = 1 SV = 2 | P55010 | EIF5 | 49 kDa | 7 | 6 | 0 | 0 | 100% | 0 |
| Cytoskeleton-associated protein 4 OS = Homo sapiens OX = 9606 GN = CKAP4 PE = 1 SV = 2 | Q07065 | CKAP4 | 66 kDa | 7 | 7 | 0 | 0 | 100% | 0 |
| Protein ELYS OS = Homo sapiens OX = 9606 GN = AHCTF1 PE = 1 SV = 3 | Q8WYP5 | AHCTF1 | 253 kDa | 7 | 7 | 0 | 0 | 100% | 0 |
| MKL/myocardin-like protein 2 OS = Homo sapiens OX = 9606 GN = MKL2 PE = 1 SV = 3 | Q9ULH7 | MKL2 | 118 kDa | 7 | 7 | 0 | 0 | 100% | 0 |
| Neuroblast differentiation-associated protein AHNAK OS = Homo sapiens OX = 9606 GN = AHNAK PE = 1 SV = 2 | Q09666 | AHNAK | 629 kDa | 7 | 7 | 0 | 0 | 93% | 0 |
| Protein disulfide-isomerase A4 OS = Homo sapiens OX = 9606 GN = PDIA4 PE = 1 SV = 2 | P13667 | PDIA4 | 73 kDa | 6 | 5 | 0 | 0 | 100% | 0 |
| Coronin-1B OS = Homo sapiens OX = 9606 GN = CORO1B PE = 1 SV = 1 | Q9BR76 | CORO1B | 54 kDa | 6 | 6 | 0 | 0 | 100% | 0 |
| Splicing factor, proline- and glutamine-rich OS = Homo sapiens OX = 9606 GN = SFPQ PE = 1 SV = 2 | P23246 | SFPQ | 76 kDa | 6 | 4 | 0 | 0 | 100% | 0 |
| Profilin-1 OS = Homo sapiens OX = 9606 GN = PFN1 PE = 1 SV = 2 | P07737 | PFN1 | 15 kDa | 6 | 6 | 0 | 0 | 100% | 0 |
| Eukaryotic translation initiation factor 4 gamma 2 OS = Homo sapiens OX = 9606 GN = EIF4G2 PE = 1 SV = 1 | P78344 | EIF4G2 | 102 kDa | 6 | 6 | 0 | 0 | 100% | 0 |
| Coiled-coil domain-containing protein 47 OS = Homo sapiens OX = 9606 GN = CCDC47 PE = 1 SV = 1 | Q96A33 | CCDC47 | 56 kDa | 6 | 6 | 0 | 0 | 100% | 0 |
| Endoplasmin OS = Homo sapiens OX = 9606 GN = HSP90B1 PE = 1 SV = 1 | P14625 | HSP90B1 | 92 kDa | 6 | 6 | 0 | 0 | 100% | 0 |
| Eukaryotic translation initiation factor 4B OS = Homo sapiens OX = 9606 GN = EIF4B PE = 1 SV = 2 | P23588 | EIF4B | 69 kDa | 6 | 6 | 0 | 0 | 100% | 0 |
| Multifunctional protein ADE2 OS = Homo sapiens OX = 9606 GN = PAICS PE = 1 SV = 3 | P22234 | PAICS | 47 kDa | 6 | 5 | 0 | 0 | 100% | 0 |
| Torsin-1A-interacting protein 1 OS = Homo sapiens OX = 9606 GN = TOR1AIP1 PE = 1 SV = 2 | Q5JTV8 | TOR1AIP1 | 66 kDa | 6 | 6 | 0 | 0 | 100% | 0 |
| Threonylcarbamoyladenosine tRNA methylthiotransferase OS = Homo sapiens OX = 9606 GN = CDKAL1 PE = 1 SV = 1 | Q5VV42 | CDKAL1 | 65 kDa | 6 | 5 | 0 | 0 | 100% | 0 |
| Receptor of activated protein C kinase 1 OS = Homo sapiens OX = 9606 GN = RACK1 PE = 1 SV = 3 | P63244 | RACK1 | 35 kDa | 6 | 5 | 0 | 0 | 100% | 0 |
| Membrane-associated progesterone receptor component 2 OS = Homo sapiens OX = 9606 GN = PGRMC2 PE = 1 SV = 1 | O15173 | PGRMC2 | 24 kDa | 5 | 3 | 0 | 0 | 100% | 0 |
| Chromodomain-helicase-DNA-binding protein 4 OS = Homo sapiens OX = 9606 GN = CHD4 PE = 1 SV = 2 | Q14839 | CHD4 | 218 kDa | 5 | 5 | 0 | 0 | 100% | 0 |
| Afadin OS = Homo sapiens OX = 9606 GN = AFDN PE = 1 SV = 3 | P55196 | AFDN | 207 kDa | 5 | 5 | 0 | 0 | 100% | 0 |
| Adapter molecule crk OS = Homo sapiens OX = 9606 GN = CRK PE = 1 SV = 2 | P46108 | CRK | 34 kDa | 5 | 5 | 0 | 0 | 100% | 0 |
| Nucleophosmin OS = Homo sapiens OX = 9606 GN = NPM1 PE = 1 SV = 2 | P06748 | NPM1 | 33 kDa | 5 | 5 | 0 | 0 | 100% | 0 |
| Sarcoplasmic/endoplasmic reticulum calcium ATPase 2 OS = Homo sapiens OX = 9606 GN = ATP2A2 PE = 1 SV = 1 | P16615 | ATP2A2 | 115 kDa | 5 | 5 | 0 | 0 | 100% | 0 |
| Peroxiredoxin-4 OS = Homo sapiens OX = 9606 GN = PRDX4 PE = 1 SV = 1 | Q13162 | PRDX4 | 31 kDa | 5 | 2 | 0 | 0 | 100% | 0 |
| Eukaryotic translation initiation factor 4 gamma 1 OS = Homo sapiens OX = 9606 GN = EIF4G1 PE = 1 SV = 4 | Q04637 | EIF4G1 | 175 kDa | 5 | 4 | 0 | 0 | 100% | 0 |
| Cytoskeleton-associated protein 5 OS = Homo sapiens OX = 9606 GN = CKAP5 PE = 1 SV = 3 | Q14008 | CKAP5 | 226 kDa | 4 | 4 | 0 | 0 | 100% | 0 |
| PEST proteolytic signal-containing nuclear protein OS = Homo sapiens OX = 9606 GN = PCNP PE = 1 SV = 2 | Q8WW12 | PCNP | 19 kDa | 4 | 2 | 0 | 0 | 98% | 0 |
| Desmoplakin OS = Homo sapiens OX = 9606 GN = DSP PE = 1 SV = 3 | P15924 | DSP | 332 kDa | 4 | 4 | 0 | 0 | 100% | 0 |
| Ubiquitin-like modifier-activating enzyme 1 OS = Homo sapiens OX = 9606 GN = UBA1 PE = 1 SV = 3 | P22314 | UBA1 | 118 kDa | 4 | 4 | 0 | 0 | 100% | 0 |
| Fatty aldehyde dehydrogenase OS = Homo sapiens OX = 9606 GN = ALDH3A2 PE = 1 SV = 1 | P51648 | ALDH3A2 | 55 kDa | 4 | 4 | 0 | 0 | 100% | 0 |
| Creatine kinase B-type OS = Homo sapiens OX = 9606 GN = CKB PE = 1 SV = 1 | P12277 | CKB | 43 kDa | 4 | 4 | 0 | 0 | 100% | 0 |
| Syntaxin-5 OS = Homo sapiens OX = 9606 GN = STX5 PE = 1 SV = 2 | Q13190 | STX5 | 40 kDa | 4 | 4 | 0 | 0 | 100% | 0 |
| Rab-like protein 3 OS = Homo sapiens OX = 9606 GN = RABL3 PE = 1 SV = 1 | Q5HYI8 | RABL3 | 26 kDa | 4 | 3 | 0 | 0 | 100% | 0 |
| Regulator of chromosome condensation OS = Homo sapiens OX = 9606 GN = RCC1 PE = 1 SV = 1 | P18754 | RCC1 | 45 kDa | 4 | 4 | 0 | 0 | 100% | 0 |
| Ran GTPase-activating protein 1 OS = Homo sapiens OX = 9606 GN = RANGAP1 PE = 1 SV = 1 | P46060 | RANGAP1 | 64 kDa | 4 | 4 | 0 | 0 | 100% | 0 |
| Synaptobrevin homolog YKT6 OS = Homo sapiens OX = 9606 GN = YKT6 PE = 1 SV = 1 | O15498 | YKT6 | 22 kDa | 4 | 3 | 0 | 0 | 100% | 0 |
| Cold shock domain-containing protein E1 OS = Homo sapiens OX = 9606 GN = CSDE1 PE = 1 SV = 2 | O75534 | CSDE1 | 89 kDa | 4 | 4 | 0 | 0 | 100% | 0 |
| Vesicle-associated membrane protein-associated protein B/C OS = Homo sapiens OX = 9606 GN = VAPB PE = 1 SV = 3 | O95292 | VAPB | 27 kDa | 4 | 2 | 0 | 0 | 99% | 0 |
| Vigilin OS = Homo sapiens OX = 9606 GN = HDLBP PE = 1 SV = 2 | Q00341 | HDLBP | 141 kDa | 4 | 4 | 0 | 0 | 99% | 0 |
| Microtubule-associated protein 4 OS = Homo sapiens OX = 9606 GN = MAP4 PE = 1 SV = 3 | P27816 | MAP4 | 121 kDa | 4 | 3 | 0 | 0 | 100% | 0 |
| TATA-binding protein-associated factor 2N OS = Homo sapiens OX = 9606 GN = TAF15 PE = 1 SV = 1 | Q92804 | TAF15 | 62 kDa | 3 | 2 | 0 | 0 | 84% | 0 |
| Peptidyl-prolyl cis-trans isomerase A OS = Homo sapiens OX = 9606 GN = PPIA PE = 1 SV = 2 | P62937 | PPIA | 18 kDa | 3 | 3 | 0 | 0 | 100% | 0 |
| CAD protein OS = Homo sapiens OX = 9606 GN = CAD PE = 1 SV = 3 | P27708 | CAD | 243 kDa | 3 | 3 | 0 | 0 | 100% | 0 |
| Synapse-associated protein 1 OS = Homo sapiens OX = 9606 GN = SYAP1 PE = 1 SV = 1 | Q96A49 | SYAP1 | 40 kDa | 3 | 3 | 0 | 0 | 100% | 0 |
| Leucine-rich repeat-containing protein 59 OS = Homo sapiens OX = 9606 GN = LRRC59 PE = 1 SV = 1 | Q96AG4 | LRRC59 | 35 kDa | 3 | 3 | 0 | 0 | 100% | 0 |
| WW domain-containing oxidoreductase OS = Homo sapiens OX = 9606 GN = WWOX PE = 1 SV = 1 | Q9NZC7 | WWOX | 47 kDa | 3 | 3 | 0 | 0 | 99% | 0 |
| D-3-phosphoglycerate dehydrogenase OS = Homo sapiens OX = 9606 GN = PHGDH PE = 1 SV = 4 | O43175 | PHGDH | 57 kDa | 3 | 3 | 0 | 0 | 100% | 0 |
| 40S ribosomal protein S3 OS = Homo sapiens OX = 9606 GN = RPS3 PE = 1 SV = 2 | P23396 | RPS3 | 27 kDa | 3 | 3 | 0 | 0 | 100% | 0 |
| Voltage-dependent anion-selective channel protein 2 OS = Homo sapiens OX = 9606 GN = VDAC2 PE = 1 SV = 2 | P45880 | VDAC2 | 32 kDa | 3 | 3 | 0 | 0 | 100% | 0 |
| Band 4.1-like protein 3 OS = Homo sapiens OX = 9606 GN = EPB41L3 PE = 1 SV = 2 | Q9Y2J2 | EPB41L3 | 121 kDa | 3 | 3 | 0 | 0 | 100% | 0 |
| Junctophilin-1 OS = Homo sapiens OX = 9606 GN = JPH1 PE = 1 SV = 2 | Q9HDC5 | JPH1 | 72 kDa | 3 | 2 | 0 | 0 | 100% | 0 |
| Emerin OS = Homo sapiens OX = 9606 GN = EMD PE = 1 SV = 1 | P50402 | EMD | 29 kDa | 3 | 2 | 0 | 0 | 100% | 0 |
| Thioredoxin-dependent peroxide reductase, mitochondrial OS = Homo sapiens OX = 9606 GN = PRDX3 PE = 1 SV = 3 | P30048 | PRDX3 | 28 kDa | 3 | 3 | 0 | 0 | 100% | 0 |
| Eukaryotic translation initiation factor 5B OS = Homo sapiens OX = 9606 GN = EIF5B PE = 1 SV = 4 | O60841 | EIF5B | 139 kDa | 3 | 3 | 0 | 0 | 99% | 0 |
| Clathrin heavy chain 1 OS = Homo sapiens OX = 9606 GN = CLTC PE = 1 SV = 5 | Q00610 | CLTC | 192 kDa | 3 | 3 | 0 | 0 | 99% | 0 |
| Stromal interaction molecule 1 OS = Homo sapiens OX = 9606 GN = STIM1 PE = 1 SV = 3 | Q13586 | STIM1 | 77 kDa | 3 | 2 | 0 | 0 | 98% | 0 |
| Splicing factor 3B subunit 3 OS = Homo sapiens OX = 9606 GN = SF3B3 PE = 1 SV = 4 | Q15393 | SF3B3 | 136 kDa | 3 | 3 | 0 | 0 | 96% | 0 |
| Nuclear migration protein nudC OS = Homo sapiens OX = 9606 GN = NUDC PE = 1 SV = 1 | Q9Y266 | NUDC | 38 kDa | 3 | 3 | 0 | 0 | 93% | 0 |
| Proliferation marker protein Ki-67 OS = Homo sapiens OX = 9606 GN = MKI67 PE = 1 SV = 2 | P46013 | MKI67 | 359 kDa | 3 | 2 | 0 | 0 | 84% | 0 |
| Chloride channel CLIC-like protein 1 OS = Homo sapiens OX = 9606 GN = CLCC1 PE = 1 SV = 1 | Q96S66 | CLCC1 | 62 kDa | 3 | 3 | 0 | 0 | 99% | 0 |
| Far upstream element-binding protein 2 OS = Homo sapiens OX = 9606 GN = KHSRP PE = 1 SV = 4 | Q92945 | KHSRP | 73 kDa | 3 | 3 | 0 | 0 | 96% | 0 |
| Clathrin interactor 1 OS = Homo sapiens OX = 9606 GN = CLINT1 PE = 1 SV = 1 | Q14677 | CLINT1 | 68 kDa | 3 | 3 | 0 | 0 | 91% | 0 |
| Eukaryotic translation initiation factor 2A OS = Homo sapiens OX = 9606 GN = EIF2A PE = 1 SV = 3 | Q9BY44 | EIF2A | 65 kDa | 3 | 3 | 0 | 0 | 61% | 0 |
| Junction plakoglobin OS = Homo sapiens OX = 9606 GN = JUP PE = 1 SV = 3 | P14923 | JUP | 82 kDa | 3 | 3 | 0 | 0 | 40% | 0 |
| Pre-mRNA-splicing factor ATP-dependent RNA helicase DHX15 OS = Homo sapiens OX = 9606 GN = DHX15 PE = 1 SV = 2 | O43143 | DHX15 | 91 kDa | 2 | 2 | 0 | 0 | 100% | 0 |
| T-complex protein 1 subunit zeta OS = Homo sapiens OX = 9606 GN = CCT6A PE = 1 SV = 3 | P40227 | CCT6A | 58 kDa | 2 | 2 | 0 | 0 | 100% | 0 |
| Cytoplasmic dynein 1 heavy chain 1 OS = Homo sapiens OX = 9606 GN = DYNC1H1 PE = 1 SV = 5 | Q14204 | DYNC1H1 | 532 kDa | 2 | 2 | 0 | 0 | 100% | 0 |
| 60S ribosomal protein L15 OS = Homo sapiens OX = 9606 GN = RPL15 PE = 1 SV = 2 | P61313 | RPL15 | 24 kDa | 2 | 2 | 0 | 0 | 100% | 0 |
| Dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit STT3B OS = Homo sapiens OX = 9606 GN = STT3B PE = 1 SV = 1 | Q8TCJ2 | STT3B | 94 kDa | 2 | 2 | 0 | 0 | 100% | 0 |
| Protein SGT1 homolog OS = Homo sapiens OX = 9606 GN = SUGT1 PE = 1 SV = 3 | Q9Y2Z0 | SUGT1 | 41 kDa | 2 | 2 | 0 | 0 | 100% | 0 |
| Protein 4.1 OS = Homo sapiens OX = 9606 GN = EPB41 PE = 1 SV = 4 | P11171 | EPB41 | 97 kDa | 2 | 2 | 0 | 0 | 100% | 0 |
| Heterogeneous nuclear ribonucleoprotein D0 OS = Homo sapiens OX = 9606 GN = HNRNPD PE = 1 SV = 1 | Q14103 | HNRNPD | 38 kDa | 2 | 2 | 0 | 0 | 100% | 0 |
| Chloride intracellular channel protein 1 OS = Homo sapiens OX = 9606 GN = CLIC1 PE = 1 SV = 4 | O00299 | CLIC1 | 27 kDa | 2 | 2 | 0 | 0 | 100% | 0 |
| Signal recognition particle subunit SRP68 OS = Homo sapiens OX = 9606 GN = SRP68 PE =1 SV= 2 | Q9UHB9 | SRP68 | 71 kDa | 2 | 2 | 0 | 0 | 100% | 0 |
| Signal recognition particle 54 kDa protein OS = Homo sapiens OX= 9606 GN= SRP54 PE = 1 SV = 1 | P61011 | SRP54 | 56 kDa | 2 | 2 | 0 | 0 | 100% | 0 |
| Polyadenylate-binding protein 1 OS = Homo sapiens OX = 9606 GN = PABPC1 PE = 1 SV = 2 | P11940 | PABPC1 | 71 kDa | 2 | 2 | 0 | 0 | 100% | 0 |
| T-complex protein 1 subunit epsilon OS = Homo sapiens OX = 9606 GN = CCT5 PE = 1 SV = 1 | P48643 | CCT5 | 60 kDa | 2 | 2 | 0 | 0 | 100% | 0 |
| C-1-tetrahydrofolate synthase, cytoplasmic OS = Homo sapiens OX = 9606 GN = MTHFD1 PE = 1 SV = 3 | P11586 | MTHFD1 | 102 kDa | 2 | 2 | 0 | 0 | 100% | 0 |
| Voltage-dependent anion-selective channel protein 3 OS = Homo sapiens OX = 9606 GN = VDAC3 PE = 1 SV = 1 | Q9Y277 | VDAC3 | 31 kDa | 2 | 2 | 0 | 0 | 100% | 0 |
| RNA-binding protein 26 OS = Homo sapiens OX = 9606 GN = RBM26 PE = 1 SV = 3 | Q5T8P6 | RBM26 | 114 kDa | 2 | 2 | 0 | 0 | 99% | 0 |
| Ran-binding protein 3 OS = Homo sapiens OX = 9606 GN = RANBP3 PE = 1 SV = 1 | Q9H6Z4 | RANBP3 | 60 kDa | 2 | 2 | 0 | 0 | 99% | 0 |
| 60S ribosomal protein L10 OS = Homo sapiens OX = 9606 GN = RPL10 PE = 1 SV = 4 | P27635 | RPL10 | 25 kDa | 2 | 2 | 0 | 0 | 99% | 0 |
| Protein RCC2 OS = Homo sapiens OX = 9606 GN = RCC2 PE = 1 SV = 2 | Q9P258 | RCC2 | 56 kDa | 2 | 2 | 0 | 0 | 98% | 0 |
| Extended synaptotagmin-1 OS = Homo sapiens OX = 9606 GN = ESYT1 PE = 1 SV = 1 | Q9BSJ8 | ESYT1 | 123 kDa | 2 | 2 | 0 | 0 | 97% | 0 |
| PC4 and SFRS1-interacting protein OS = Homo sapiens OX = 9606 GN = PSIP1 PE = 1 SV = 1 | O75475 | PSIP1 | 60 kDa | 2 | 2 | 0 | 0 | 94% | 0 |
| SAFB-like transcription modulator OS = Homo sapiens OX = 9606 GN = SLTM PE = 1 SV = 2 | Q9NWH9 | SLTM | 117 kDa | 2 | 2 | 0 | 0 | 92% | 0 |
| Probable rRNA-processing protein EBP2 OS = Homo sapiens OX = 9606 GN = EBNA1BP2 PE = 1 SV = 2 | Q99848 | EBNA1BP2 | 35 kDa | 2 | 2 | 0 | 0 | 80% | 0 |
| Cysteine and histidine-rich domain-containing protein 1 OS = Homo sapiens OX = 9606 GN = CHORDC1 PE = 1 SV = 2 | Q9UHD1 | CHORDC1 | 37 kDa | 2 | 2 | 0 | 0 | 79% | 0 |
| ATP-dependent RNA helicase DDX39A OS = Homo sapiens OX = 9606 GN = DDX39A PE = 1 SV = 2 | O00148 (+1) | DDX39A | 49 kDa | 2 | 2 | 0 | 0 | 100% | 0 |
| Heterogeneous nuclear ribonucleoprotein H3 OS = Homo sapiens OX = 9606 GN = HNRNPH3 PE = 1 SV = 2 | P31942 | HNRNPH3 | 37 kDa | 2 | 2 | 0 | 0 | 100% | 0 |
| Peroxisomal multifunctional enzyme type 2 OS = Homo sapiens OX = 9606 GN = HSD17B4 PE = 1 SV = 3 | P51659 | HSD17B4 | 80 kDa | 2 | 2 | 0 | 0 | 98% | 0 |
| Eukaryotic translation initiation factor 3 subunit D OS = Homo sapiens OX = 9606 GN = EIF3D PE = 1 SV = 1 | O15371 | EIF3D | 64 kDa | 2 | 2 | 0 | 0 | 96% | 0 |
| Tyrosine-protein phosphatase non-receptor type 1 OS = Homo sapiens OX = 9606 GN = PTPN1 PE = 1 SV = 1 | P18031 | PTPN1 | 50 kDa | 2 | 2 | 0 | 0 | 92% | 0 |
| Jupiter microtubule associated homolog 2 OS = Homo sapiens OX = 9606 GN = JPT2 PE = 1 SV = 1 | Q9H910 | JPT2 | 20 kDa | 2 | 2 | 0 | 0 | 90% | 0 |
| U5 small nuclear ribonucleoprotein 200 kDa helicase OS = Homo sapiens OX = 9606 GN = SNRNP200 PE = 1 SV = 2 | O75643 | SNRNP200 | 245 kDa | 2 | 2 | 0 | 0 | 89% | 0 |
| PDZ and LIM domain protein 5 OS = Homo sapiens OX = 9606 GN = PDLIM5 PE = 1 SV = 5 | Q96HC4 | PDLIM5 | 64 kDa | 2 | 2 | 0 | 0 | 86% | 0 |
| Coatomer subunit alpha OS = Homo sapiens OX = 9606 GN = COPA PE = 1 SV = 2 | P53621 | COPA | 138 kDa | 2 | 2 | 0 | 0 | 86% | 0 |
| Activator of 90 kDa heat shock protein ATPase homolog 1 OS = Homo sapiens OX = 9606 GN = AHSA1 PE = 1 SV = 1 | O95433 | AHSA1 | 38 kDa | 2 | 2 | 0 | 0 | 76% | 0 |
| Asparagine synthetase [glutamine-hydrolyzing] OS = Homo sapiens OX = 9606 GN = ASNS PE = 1 SV = 4 | P08243 | ASNS | 64 kDa | 2 | 2 | 0 | 0 | 69% | 0 |
| Transitional endoplasmic reticulum ATPase OS = Homo sapiens OX = 9606 GN = VCP PE = 1 SV = 4 | P55072 | VCP | 89 kDa | 2 | 2 | 0 | 0 | 62% | 0 |
| RNA cytidine acetyltransferase OS = Homo sapiens OX = 9606 GN = NAT10 PE = 1 SV = 2 | Q9H0A0 | NAT10 | 116 kDa | 2 | 2 | 0 | 0 | 54% | 0 |
| Protein arginine N-methyltransferase 5 OS = Homo sapiens OX = 9606 GN = PRMT5 PE = 1 SV = 4 | O14744 | PRMT5 | 73 kDa | 2 | 2 | 0 | 0 | 38% | 0 |
| 60S ribosomal protein L38 OS = Homo sapiens OX = 9606 GN = RPL38 PE = 1 SV = 2 | P63173 | RPL38 | 8 kDa | 2 | 2 | 0 | 0 | 37% | 0 |
| Angiomotin OS = Homo sapiens OX = 9606 GN = AMOT PE = 1 SV = 1 | Q4VCS5 | AMOT | 118 kDa | 2 | 2 | 0 | 0 | 29% | 0 |
| Segment polarity protein dishevelled homolog DVL-2 OS = Homo sapiens OX = 9606 GN = DVL2 PE = 1 SV = 1 | O14641 | DVL2 | 79 kDa | 2 | 2 | 0 | 0 | 21% | 0 |
| DDRGK domain-containing protein 1 OS = Homo sapiens OX = 9606 GN = DDRGK1 PE = 1 SV = 2 | Q96HY6 | DDRGK1 | 36 kDa | 2 | 2 | 0 | 0 | 41% | 0 |
| Calponin-3 OS = Homo sapiens OX = 9606 GN = CNN3 PE = 1 SV = 1 | Q15417 | CNN3 | 36 kDa | 2 | 2 | 0 | 0 | 19% | 0 |
| Synaptosomal-associated protein 29 OS = Homo sapiens OX = 9606 GN = SNAP29 PE = 1 SV = 1 | O95721 | SNAP29 | 29 kDa | 2 | 2 | 0 | 0 | 16% | 0 |
| T-complex protein 1 subunit theta OS = Homo sapiens OX = 9606 GN = CCT8 PE = 1 SV = 4 | P50990 | CCT8 | 60 kDa | 58 | 32 | 1 | 1 | 100% | 55% |
| Fatty acid synthase OS = Homo sapiens OX = 9606 GN = FASN PE = 1 SV = 3 | P49327 | FASN | 273 kDa | 32 | 27 | 1 | 1 | 100% | 82% |
| Protein LYRIC OS = Homo sapiens OX = 9606 GN = MTDH PE = 1 SV = 2 | Q86UE4 | MTDH | 64 kDa | 24 | 15 | 1 | 1 | 100% | 11% |
| Plasminogen activator inhibitor 1 RNA-binding protein OS = Homo sapiens OX = 9606 GN = SERBP1 PE = 1 SV = 2 | Q8NC51 | SERBP1 | 45 kDa | 12 | 8 | 1 | 1 | 100% | 77% |
| Ubiquitin-40S ribosomal protein S27a OS = Homo sapiens OX = 9606 GN = RPS27A PE = 1 SV = 2 | P62979 | RPS27A | 18 kDa | 16 | 7 | 1 | 1 | 100% | 15% |
| Heterogeneous nuclear ribonucleoprotein L OS = Homo sapiens OX = 9606 GN = HNRNPL PE = 1 SV = 2 | P14866 | HNRNPL | 64 kDa | 9 | 9 | 1 | 1 | 100% | 94% |
| Poly(rC)-binding protein 1 OS = Homo sapiens OX = 9606 GN = PCBP1 PE = 1 SV = 2 | Q15365 | PCBP1 | 37 kDa | 7 | 5 | 1 | 1 | 100% | 98% |
| Eukaryotic initiation factor 4A-I OS = Homo sapiens OX = 9606 GN = EIF4A1 PE = 1 SV = 1 | P60842 | EIF4A1 | 46 kDa | 6 | 6 | 1 | 1 | 100% | 81% |
| Heterogeneous nuclear ribonucleoprotein A3 OS = Homo sapiens OX = 9606 GN = HNRNPA3 PE = 1 SV = 2 | P51991 | HNRNPA3 | 40 kDa | 5 | 5 | 1 | 1 | 100% | 98% |
| GTP-binding nuclear protein Ran OS = Homo sapiens OX = 9606 GN = RAN PE = 1 SV = 3 | P62826 | RAN | 24 kDa | 5 | 5 | 1 | 1 | 100% | 93% |
| ADP/ATP translocase 2 OS = Homo sapiens OX = 9606 GN = SLC25A5 PE = 1 SV = 7 | P05141 | SLC25A5 | 33 kDa | 4 | 4 | 1 | 1 | 100% | 92% |
| Splicing regulatory glutamine/lysine-rich protein 1 OS = Homo sapiens OX = 9606 GN = SREK1 PE = 1 SV = 1 | Q8WXA9 | SREK1 | 59 kDa | 4 | 4 | 1 | 1 | 100% | 47% |
| 60S ribosomal protein L12 OS = Homo sapiens OX = 9606 GN = RPL12 PE = 1 SV = 1 | P30050 | RPL12 | 18 kDa | 3 | 2 | 1 | 1 | 100% | 76% |
| Fructose-bisphosphate aldolase A OS = Homo sapiens OX = 9606 GN = ALDOA PE = 1 SV = 2 | P04075 | ALDOA | 39 kDa | 4 | 4 | 1 | 1 | 100% | 20% |
| 60S ribosomal protein L3 OS = Homo sapiens OX = 9606 GN = RPL3 PE = 1 SV = 2 | P39023 | RPL3 | 46 kDa | 5 | 4 | 1 | 1 | 100% | 8% |
| Heterogeneous nuclear ribonucleoprotein H OS = Homo sapiens OX = 9606 GN = HNRNPH1 PE = 1 SV = 4 | P31943 | HNRNPH1 | 49 kDa | 3 | 3 | 1 | 1 | 100% | 98% |
| Desmoglein-1 OS = Homo sapiens OX = 9606 GN = DSG1 PE = 1 SV = 2 | Q02413 | DSG1 | 114 kDa | 2 | 2 | 1 | 1 | 100% | 97% |
| 40S ribosomal protein S2 OS = Homo sapiens OX = 9606 GN = RPS2 PE = 1 SV = 2 | P15880 | RPS2 | 31 kDa | 2 | 2 | 1 | 1 | 100% | 96% |
| Dermcidin OS = Homo sapiens OX = 9606 GN = DCD PE = 1 SV = 2 | P81605 | DCD | 11 kDa | 2 | 2 | 1 | 1 | 100% | 94% |
| 40S ribosomal protein S16 OS = Homo sapiens OX = 9606 GN = RPS16 PE = 1 SV = 2 | P62249 | RPS16 | 16 kDa | 2 | 2 | 1 | 1 | 100% | 92% |
| Heterogeneous nuclear ribonucleoprotein M OS = Homo sapiens OX = 9606 GN = HNRNPM PE = 1 SV = 3 | P52272 | HNRNPM | 78 kDa | 2 | 2 | 1 | 1 | 100% | 92% |
| Serine/arginine-rich splicing factor 7 OS = Homo sapiens OX = 9606 GN = SRSF7 PE = 1 SV = 1 | Q16629 | SRSF7 | 27 kDa | 3 | 3 | 1 | 1 | 99% | 77% |
| 40S ribosomal protein S15a OS = Homo sapiens OX = 9606 GN = RPS15A PE = 1 SV = 2 | P62244 | RPS15A | 15 kDa | 3 | 3 | 1 | 1 | 100% | 74% |
| T-complex protein 1 subunit alpha OS = Homo sapiens OX = 9606 GN = TCP1 PE = 1 SV = 1 | P17987 | TCP1 | 60 kDa | 3 | 3 | 1 | 1 | 100% | 56% |
| Pre-mRNA-processing factor 40 homolog A OS = Homo sapiens OX = 9606 GN = PRPF40A PE = 1 SV = 2 | O75400 | PRPF40A | 109 kDa | 3 | 3 | 1 | 1 | 99% | 50% |
| Glyceraldehyde-3-phosphate dehydrogenase OS = Homo sapiens OX = 9606 GN = GAPDH PE = 1 SV = 3 | P04406 | GAPDH | 36 kDa | 2 | 2 | 1 | 1 | 100% | 31% |
| RNA-binding protein 39 OS = Homo sapiens OX = 9606 GN = RBM39 PE = 1 SV = 2 | Q14498 | RBM39 | 59 kDa | 3 | 3 | 1 | 1 | 99% | 28% |
| Probable ATP-dependent RNA helicase DDX46 OS = Homo sapiens OX = 9606 GN = DDX46 PE = 1 SV = 2 | Q7L014 | DDX46 | 117 kDa | 3 | 3 | 1 | 1 | 100% | 8% |
| Filaggrin-2 OS = Homo sapiens OX = 9606 GN = FLG2 PE = 1 SV = 1 | Q5D862 | FLG2 | 248 kDa | 3 | 3 | 1 | 1 | 85% | 23% |
| Cofilin-1 OS = Homo sapiens OX = 9606 GN = CFL1 PE = 1 SV = 3 | P23528 | CFL1 | 19 kDa | 3 | 3 | 1 | 1 | 100% | 7% |
| Heterogeneous nuclear ribonucleoprotein A/B OS = Homo sapiens OX = 9606 GN = HNRNPAB PE = 1 SV = 2 | Q99729 | HNRNPAB | 36 kDa | 2 | 2 | 1 | 1 | 98% | 98% |
| Pre-mRNA-processing-splicing factor 8 OS = Homo sapiens OX = 9606 GN = PRPF8 PE = 1 SV = 2 | Q6P2Q9 | PRPF8 | 274 kDa | 4 | 4 | 1 | 1 | 85% | 31% |
| Elongation factor Tu, mitochondrial OS = Homo sapiens OX = 9606 GN = TUFM PE = 1 SV = 2 | P49411 | TUFM | 50 kDa | 9 | 8 | 2 | 2 | 100% | 95% |
| RNA-binding protein FUS OS = Homo sapiens OX = 9606 GN = FUS PE = 1 SV = 1 | P35637 | FUS | 53 kDa | 11 | 6 | 2 | 2 | 100% | 30% |
| Peroxiredoxin-2 OS = Homo sapiens OX = 9606 GN = PRDX2 PE = 1 SV = 5 | P32119 | PRDX2 | 22 kDa | 6 | 3 | 2 | 1 | 100% | 73% |
| ADP-ribosylation factor-like protein 6-interacting protein 4 OS = Homo sapiens OX = 9606 GN = ARL6IP4 PE = 1 SV= 2 | Q66PJ3 | ARL6IP4 | 45 kDa | 6 | 4 | 2 | 1 | 100% | 98% |
| Nucleolar RNA helicase 2 OS = Homo sapiens OX = 9606 GN = DDX21 PE= 1 SV = 5 | Q9NR30 | DDX21 | 87 kDa | 7 | 7 | 2 | 2 | 100% | 96% |
| Fibronectin OS = Homo sapiens OX = 9606 GN = FN1 PE = 1 SV = 4 | P02751 | FN1 | 263 kDa | 4 | 3 | 2 | 2 | 100% | 100% |
| Serine/arginine-rich splicing factor 1 OS = Homo sapiens OX = 9606 GN = SRSF1 PE = 1 SV = 2 | Q07955 | SRSF1 | 28 kDa | 5 | 4 | 2 | 2 | 100% | 66% |
| Serine/threonine-protein kinase PRP4 homolog OS = Homo sapiens OX = 9606 GN = PRPF4B PE = 1 SV = 3 | Q13523 | PRPF4B | 117 kDa | 4 | 4 | 2 | 2 | 100% | 100% |
| Nucleolar protein 58 OS = Homo sapiens OX = 9606 GN = NOP58 PE = 1 SV = 1 | Q9Y2X3 | NOP58 | 60 kDa | 4 | 4 | 2 | 2 | 100% | 100% |
| Alpha-enolase OS = Homo sapiens OX = 9606 GN = ENO1 PE = 1 SV = 2 | P06733 | ENO1 | 47 kDa | 4 | 3 | 2 | 2 | 100% | 98% |
| Histone H2A type 1 OS = Homo sapiens OX = 9606 GN = HIST1H2AG PE = 1 SV = 2 | P0C0S8 (+6) | HIST1H2AG | 14 kDa | 2 | 2 | 2 | 2 | 100% | 100% |
| Serine/arginine-rich splicing factor 2 OS = Homo sapiens OX = 9606 GN = SRSF2 PE = 1 SV = 4 | Q01130 | SRSF2 | 25 kDa | 2 | 2 | 2 | 2 | 100% | 100% |
| 60S ribosomal protein L36a OS = Homo sapiens OX = 9606 GN = RPL36A PE = 1 SV = 2 | P83881 (+1) | RPL36A | 12 kDa | 2 | 2 | 2 | 2 | 99% | 93% |
| Poly(rC)-binding protein 2 OS = Homo sapiens OX = 9606 GN = PCBP2 PE = 1 SV = 1 | Q15366 | PCBP2 | 39 kDa | 4 | 1 | 2 | 2 | 40% | 41% |
| Proliferation-associated protein 2G4 OS = Homo sapiens OX = 9606 GN = PA2G4 PE = 1 SV = 3 | Q9UQ80 | PA2G4 | 44 kDa | 4 | 4 | 2 | 2 | 100% | 71% |
| 40S ribosomal protein S26 OS = Homo sapiens OX = 9606 GN = RPS26 PE = 1 SV = 3 | P62854 | RPS26 | 13 kDa | 2 | 2 | 2 | 1 | 100% | 67% |
| Keratinocyte proline-rich protein OS = Homo sapiens OX = 9606 GN = KPRP PE = 1 SV = 1 | Q5T749 | KPRP | 64 kDa | 2 | 2 | 2 | 2 | 73% | 85% |
| ATP-dependent RNA helicase DDX3X OS = Homo sapiens OX = 9606 GN = DDX3X PE = 1 SV = 3 | O00571 | DDX3X | 73 kDa | 2 | 2 | 2 | 2 | 100% | 27% |
| ATP-citrate synthase OS = Homo sapiens OX = 9606 GN = ACLY PE = 1 SV = 3 | P53396 | ACLY | 121 kDa | 5 | 5 | 2 | 2 | 100% | 16% |
| 60S ribosomal protein L4 OS = Homo sapiens OX = 9606 GN = RPL4 PE = 1 SV = 5 | P36578 | RPL4 | 48 kDa | 3 | 3 | 2 | 2 | 100% | 15% |
| 60S ribosomal protein L7a OS = Homo sapiens OX = 9606 GN = RPL7A PE = 1 SV = 2 | P62424 | RPL7A | 30 kDa | 2 | 2 | 2 | 1 | 92% | 10% |
| 60S ribosomal protein L8 OS = Homo sapiens OX = 9606 GN = RPL8 PE = 1 SV = 2 | P62917 | RPL8 | 28 kDa | 1 | 1 | 2 | 2 | 83% | 98% |
| Uncharacterized protein NKAPD1 OS = Homo sapiens OX = 9606 GN = NKAPD1 PE = 1 SV = 2 | Q6ZUT1 | NKAPD1 | 34 kDa | 2 | 2 | 2 | 2 | 60% | 66% |
| Glutathione S-transferase P OS = Homo sapiens OX = 9606 GN = GSTP1 PE = 1 SV = 2 | P09211 | GSTP1 | 23 kDa | 1 | 1 | 2 | 2 | 71% | 46% |
| Interleukin enhancer-binding factor 3 OS = Homo sapiens OX = 9606 GN = ILF3 PE = 1 SV = 3 | Q12906 | ILF3 | 95 kDa | 0 | 0 | 2 | 2 | 0 | 100% |
| Non-histone chromosomal protein HMG-14 OS = Homo sapiens OX = 9606 GN = HMGN1 PE = 1 SV = 3 | P05114 | HMGN1 | 11 kDa | 1 | 1 | 2 | 2 | 9% | 31% |
| ATP-dependent RNA helicase A OS = Homo sapiens OX = 9606 GN = DHX9 PE = 1 SV = 4 | Q08211 | DHX9 | 141 kDa | 20 | 15 | 3 | 2 | 100% | 98% |
| Peroxiredoxin-1 OS = Homo sapiens OX = 9606 GN = PRDX1 PE = 1 SV = 1 | Q06830 | PRDX1 | 22 kDa | 13 | 11 | 3 | 3 | 100% | 100% |
| Retinitis pigmentosa 9 protein OS = Homo sapiens OX = 9606 GN = RP9 PE = 1 SV = 2 | Q8TA86 | RP9 | 26 kDa | 11 | 8 | 3 | 3 | 100% | 100% |
| Non-POU domain-containing octamer-binding protein OS = Homo sapiens OX = 9606 GN = NONO PE = 1 SV = 4 | Q15233 | NONO | 54 kDa | 12 | 9 | 3 | 2 | 100% | 100% |
| L-lactate dehydrogenase B chain OS = Homo sapiens OX = 9606 GN = LDHB PE = 1 SV = 2 | P07195 | LDHB | 37 kDa | 7 | 5 | 3 | 2 | 100% | 100% |
| Vimentin OS = Homo sapiens OX = 9606 GN = VIM PE = 1 SV = 4 | P08670 | VIM | 54 kDa | 7 | 6 | 3 | 2 | 100% | 100% |
| 40S ribosomal protein S3a OS = Homo sapiens OX = 9606 GN = RPS3A PE = 1 SV = 2 | P61247 | RPS3A | 30 kDa | 3 | 3 | 3 | 3 | 100% | 100% |
| THO complex subunit 4 OS = Homo sapiens OX = 9606 GN = ALYREF PE = 1 SV = 3 | Q86V81 | ALYREF | 27 kDa | 2 | 2 | 3 | 2 | 100% | 100% |
| Histone H1x OS = Homo sapiens OX = 9606 GN = H1FX PE = 1 SV = 1 | Q92522 | H1FX | 22 kDa | 3 | 2 | 3 | 2 | 100% | 100% |
| 40S ribosomal protein S14 OS = Homo sapiens OX = 9606 GN = RPS14 PE = 1 SV = 3 | P62263 | RPS14 | 16 kDa | 2 | 2 | 3 | 2 | 100% | 97% |
| 60S ribosomal protein L13 OS = Homo sapiens OX = 9606 GN = RPL13 PE = 1 SV = 4 | P26373 | RPL13 | 24 kDa | 2 | 2 | 3 | 2 | 100% | 100% |
| Serum albumin OS = Homo sapiens OX = 9606 GN = ALB PE = 1 SV = 2 | P02768 | ALB | 69 kDa | 2 | 1 | 3 | 3 | 96% | 100% |
| Elongation factor 1-gamma OS = Homo sapiens OX = 9606 GN = EEF1G PE = 1 SV = 3 | P26641 | EEF1G | 50 kDa | 1 | 1 | 3 | 3 | 97% | 100% |
| 60S ribosomal protein L10a OS = Homo sapiens OX = 9606 GN = RPL10A PE = 1 SV = 2 | P62906 | RPL10A | 25 kDa | 1 | 1 | 3 | 2 | 41% | 98% |
| ATP synthase subunit alpha, mitochondrial OS = Homo sapiens OX = 9606 GN = ATP5F1A PE = 1 SV = 1 | P25705 | ATP5F1A | 60 kDa | 1 | 1 | 3 | 3 | 8% | 100% |
| Heat shock protein HSP 90-alpha OS = Homo sapiens OX = 9606 GN = HSP90AA1 PE = 1 SV = 5 | P07900 | HSP90AA1 | 85 kDa | 12 | 9 | 4 | 3 | 100% | 100% |
| Pyruvate kinase PKM OS = Homo sapiens OX = 9606 GN = PKM PE = 1 SV = 4 | P14618 | PKM | 58 kDa | 15 | 15 | 4 | 4 | 100% | 100% |
| Heat shock protein HSP 90-beta OS = Homo sapiens OX = 9606 GN = HSP90AB1 PE = 1 SV = 4 | P08238 | HSP90AB1 | 83 kDa | 10 | 4 | 4 | 1 | 100% | 8% |
| Splicing factor, arginine/serine-rich 19 OS = Homo sapiens OX = 9606 GN = SCAF1 PE = 1 SV = 3 | Q9H7N4 | SCAF1 | 139 kDa | 8 | 7 | 4 | 3 | 100% | 100% |
| Heterogeneous nuclear ribonucleoprotein K OS = Homo sapiens OX = 9606 GN = HNRNPK PE = 1 SV = 1 | P61978 | HNRNPK | 51 kDa | 5 | 5 | 4 | 4 | 100% | 100% |
| Trypsin-1 OS = Homo sapiens OX = 9606 GN = PRSS1 PE = 1 SV = 1 | P07477 | PRSS1 | 27 kDa | 4 | 2 | 4 | 2 | 100% | 100% |
| 40S ribosomal protein S30 OS = Homo sapiens OX = 9606 GN = FAU PE = 1 SV = 1 | P62861 | FAU | 7 kDa | 3 | 1 | 4 | 2 | 97% | 100% |
| DNA topoisomerase 1 OS = Homo sapiens OX = 9606 GN = TOP1 PE = 1 SV = 2 | P11387 | TOP1 | 91 kDa | 6 | 5 | 4 | 4 | 100% | 97% |
| 60S ribosomal protein L7 OS = Homo sapiens OX = 9606 GN = RPL7 PE = 1 SV = 1 | P18124 | RPL7 | 29 kDa | 4 | 3 | 4 | 3 | 100% | 100% |
| 40S ribosomal protein S8 OS = Homo sapiens OX = 9606 GN = RPS8 PE = 1 SV = 2 | P62241 | RPS8 | 24 kDa | 3 | 3 | 4 | 2 | 100% | 100% |
| 60S ribosomal protein L5 OS = Homo sapiens OX = 9606 GN = RPL5 PE = 1 SV = 3 | P46777 | RPL5 | 34 kDa | 1 | 1 | 4 | 2 | 95% | 80% |
| Ribosomal RNA processing protein 1 homolog B OS = Homo sapiens OX = 9606 GN = RRP1B PE = 1 SV = 3 | Q14684 | RRP1B | 84 kDa | 0 | 0 | 4 | 4 | 0 | 100% |
| Elongation factor 2 OS = Homo sapiens OX = 9606 GN = EEF2 PE = 1 SV = 4 | P13639 | EEF2 | 95 kDa | 31 | 19 | 5 | 5 | 100% | 100% |
| Tubulin beta chain OS = Homo sapiens OX = 9606 GN = TUBB PE = 1 SV = 2 | P07437 | TUBB | 50 kDa | 16 | 11 | 5 | 4 | 100% | 100% |
| Tubulin alpha-1B chain OS = Homo sapiens OX = 9606 GN = TUBA1B PE = 1 SV = 1 | P68363 (+2) | TUBA1B | 50 kDa | 13 | 8 | 5 | 4 | 100% | 100% |
| L-lactate dehydrogenase A chain OS = Homo sapiens OX = 9606 GN = LDHA PE = 1 SV = 2 | P00338 | LDHA | 37 kDa | 7 | 6 | 5 | 5 | 100% | 100% |
| E3 ubiquitin-protein ligase RBBP6 OS = Homo sapiens OX = 9606 GN = RBBP6 PE = 1 SV = 1 | Q7Z6E9 | RBBP6 | 202 kDa | 11 | 10 | 5 | 5 | 100% | 100% |
| Protein LLP homolog OS = Homo sapiens OX = 9606 GN = LLPH PE = 1 SV = 1 | Q9BRT6 | LLPH | 15 kDa | 7 | 4 | 5 | 3 | 100% | 100% |
| Probable ATP-dependent RNA helicase DDX17 OS = Homo sapiens OX = 9606 GN = DDX17 PE = 1 SV = 2 | Q92841 | DDX17 | 80 kDa | 4 | 3 | 5 | 4 | 100% | 100% |
| Protein SREK1IP1 OS = Homo sapiens OX = 9606 GN = SREK1IP1 PE = 1 SV = 1 | Q8N9Q2 | SREK1IP1 | 18 kDa | 4 | 3 | 5 | 3 | 100% | 100% |
| RNA-binding motif protein, X chromosome OS = Homo sapiens OX = 9606 GN = RBMX PE = 1 SV = 3 | P38159 | RBMX | 42 kDa | 8 | 6 | 5 | 4 | 100% | 97% |
| Putative RNA-binding protein Luc7-like 2 OS = Homo sapiens OX = 9606 GN = LUC7L2 PE = 1 SV = 2 | Q9Y383 | LUC7L2 | 47 kDa | 2 | 2 | 5 | 4 | 99% | 100% |
| 60S ribosomal protein L37 OS = Homo sapiens OX = 9606 GN = RPL37 PE = 1 SV = 2 | P61927 | RPL37 | 11 kDa | 0 | 0 | 5 | 2 | 0 | 100% |
| Heat shock 70 kDa protein 1A OS = Homo sapiens OX = 9606 GN = HSPA1A PE = 1 SV = 1 | P0DMV8 (+1) | HSPA1A | 70 kDa | 20 | 16 | 6 | 6 | 100% | 100% |
| Heat shock cognate 71 kDa protein OS = Homo sapiens OX = 9606 GN = HSPA8 PE = 1 SV = 1 | P11142 | HSPA8 | 71 kDa | 18 | 8 | 6 | 4 | 100% | 100% |
| Heterogeneous nuclear ribonucleoproteins A2/B1 OS = Homo sapiens OX = 9606 GN = HNRNPA2B1 PE = 1 SV = 2 | P22626 | HNRNPA2B1 | 37 kDa | 16 | 11 | 6 | 4 | 100% | 100% |
| Histone H4 OS = Homo sapiens OX = 9606 GN = HIST1H4A PE = 1 SV = 2 | P62805 | HIST1H4A | 11 kDa | 9 | 7 | 6 | 5 | 100% | 100% |
| AP-3 complex subunit delta-1 OS = Homo sapiens OX = 9606 GN = AP3D1 PE = 1 SV = 1 | O14617 | AP3D1 | 130 kDa | 9 | 7 | 6 | 4 | 100% | 100% |
| Nucleolin OS = Homo sapiens OX = 9606 GN = NCL PE = 1 SV = 3 | P19338 | NCL | 77 kDa | 8 | 8 | 6 | 6 | 100% | 100% |
| A-kinase anchor protein 17A OS = Homo sapiens OX = 9606 GN = AKAP17A PE = 1 SV = 2 | Q02040 | AKAP17A | 81 kDa | 11 | 8 | 6 | 6 | 100% | 100% |
| 60S ribosomal protein L6 OS = Homo sapiens OX = 9606 GN = RPL6 PE = 1 SV = 3 | Q02878 | RPL6 | 33 kDa | 2 | 2 | 6 | 4 | 95% | 100% |
| Heterogeneous nuclear ribonucleoprotein A1 OS = Homo sapiens OX = 9606 GN = HNRNPA1 PE = 1 SV = 5 | P09651 | HNRNPA1 | 39 kDa | 16 | 11 | 7 | 4 | 100% | 100% |
| La-related protein 7 OS = Homo sapiens OX = 9606 GN = LARP7 PE = 1 SV = 1 | Q4G0J3 | LARP7 | 67 kDa | 6 | 6 | 7 | 6 | 100% | 100% |
| G patch domain-containing protein 4 OS = Homo sapiens OX = 9606 GN = GPATCH4 PE = 1 SV = 2 | Q5T3I0 | GPATCH4 | 50 kDa | 5 | 5 | 7 | 6 | 100% | 100% |
| NF-kappa-B-activating protein OS = Homo sapiens OX = 9606 GN = NKAP PE = 1 SV = 1 | Q8N5F7 | NKAP | 47 kDa | 8 | 6 | 8 | 5 | 100% | 100% |
| 60S ribosomal protein L23a OS = Homo sapiens OX = 9606 GN = RPL23A PE = 1 SV = 1 | P62750 | RPL23A | 18 kDa | 8 | 6 | 8 | 4 | 100% | 100% |
| Histone H2B type 1-K OS = Homo sapiens OX = 9606 GN = HIST1H2BK PE = 1 SV = 3 | O60814 (+8) | HIST1H2BK | 14 kDa | 7 | 5 | 8 | 4 | 100% | 100% |
| Histone H3.1 OS = Homo sapiens OX = 9606 GN = HIST1H3A PE = 1 SV = 2 | P68431 (+3) | HIST1H3A | 15 kDa | 6 | 4 | 8 | 4 | 100% | 100% |
| Transcription initiation factor TFIID subunit 3 OS = Homo sapiens OX = 9606 GN = TAF3 PE = 1 SV = 1 | Q5VWG9 | TAF3 | 104 kDa | 6 | 6 | 8 | 7 | 100% | 100% |
| Keratin, type I cytoskeletal 17 OS = Homo sapiens OX = 9606 GN = KRT17 PE = 1 SV = 2 | Q04695 | KRT17 | 48 kDa | 12 | 3 | 9 | 2 | 100% | 76% |
| Heterogeneous nuclear ribonucleoproteins C1/C2 OS = Homo sapiens OX = 9606 GN = HNRNPC PE = 1 SV = 4 | P07910 | HNRNPC | 34 kDa | 9 | 6 | 9 | 4 | 100% | 100% |
| Hornerin OS = Homo sapiens OX = 9606 GN = HRNR PE = 1 SV = 2 | Q86YZ3 | HRNR | 282 kDa | 12 | 9 | 10 | 7 | 100% | 100% |
| Heterogeneous nuclear ribonucleoprotein U OS = Homo sapiens OX = 9606 GN = HNRNPU PE = 1 SV = 6 | Q00839 | HNRNPU | 91 kDa | 17 | 12 | 11 | 6 | 100% | 100% |
| Serine/arginine-rich splicing factor 11 OS = Homo sapiens OX = 9606 GN = SRSF11 PE = 1 SV = 1 | Q05519 | SRSF11 | 54 kDa | 11 | 8 | 11 | 6 | 100% | 100% |
| Serine/arginine repetitive matrix protein 2 OS = Homo sapiens OX = 9606 GN = SRRM2 PE = 1 SV = 2 | Q9UQ35 | SRRM2 | 300 kDa | 15 | 7 | 11 | 6 | 100% | 100% |
| Poly [ADP-ribose] polymerase 1 OS = Homo sapiens OX = 9606 GN = PARP1 PE = 1 SV = 4 | P09874 | PARP1 | 113 kDa | 29 | 22 | 12 | 12 | 100% | 100% |
| Elongation factor 1-alpha 1 OS = Homo sapiens OX = 9606 GN = EEF1A1 PE = 1 SV = 1 | P68104 (+1) | EEF1A1 | 50 kDa | 21 | 14 | 13 | 9 | 100% | 100% |
| Nucleolar protein 56 OS = Homo sapiens OX = 9606 GN = NOP56 PE = 1 SV = 4 | O00567 | NOP56 | 66 kDa | 16 | 11 | 13 | 10 | 100% | 100% |
| U2 snRNP-associated SURP motif-containing protein OS = Homo sapiens OX = 9606 GN = U2SURP PE = 1 SV = 2 | O15042 | U2SURP | 118 kDa | 25 | 19 | 14 | 10 | 100% | 100% |
| Guanine nucleotide-binding protein-like 3 OS = Homo sapiens OX = 9606 GN = GNL3 PE = 1 SV = 2 | Q9BVP2 | GNL3 | 62 kDa | 9 | 7 | 16 | 9 | 100% | 100% |
| Actin, cytoplasmic 1 OS = Homo sapiens OX = 9606 GN = ACTB PE = 1 SV = 1 | P60709 (+1) | ACTB | 42 kDa | 22 | 12 | 17 | 9 | 100% | 100% |
| Multiple myeloma tumor-associated protein 2 OS = Homo sapiens OX = 9606 GN = MMTAG2 PE = 1 SV = 1 | Q9BU76 | MMTAG2 | 29 kDa | 21 | 13 | 17 | 8 | 100% | 100% |
| Keratin, type II cytoskeletal 5 OS = Homo sapiens OX = 9606 GN = KRT5 PE = 1 SV = 3 | P13647 | KRT5 | 62 kDa | 27 | 15 | 20 | 11 | 100% | 100% |
| Histone H1.0 OS = Homo sapiens OX = 9606 GN = H1F0 PE = 1 SV = 3 | P07305 | H1F0 | 21 kDa | 16 | 9 | 20 | 9 | 100% | 100% |
| Nucleolar protein of 40 kDa OS = Homo sapiens OX = 9606 GN = ZCCHC17 PE = 1 SV = 1 | Q9NP64 | ZCCHC17 | 28 kDa | 19 | 9 | 20 | 10 | 100% | 100% |
| 60S ribosomal protein L29 OS = Homo sapiens OX = 9606 GN = RPL29 PE = 1 SV = 2 | P47914 | RPL29 | 18 kDa | 10 | 2 | 21 | 5 | 100% | 100% |
| Keratin, type I cytoskeletal 14 OS = Homo sapiens OX = 9606 GN = KRT14 PE = 1 SV = 4 | P02533 | KRT14 | 52 kDa | 32 | 16 | 22 | 7 | 100% | 100% |
| Protein SON OS = Homo sapiens OX = 9606 GN = SON PE = 1 SV = 4 | P18583 | SON | 264 kDa | 15 | 15 | 23 | 17 | 100% | 100% |
| Keratin, type II cytoskeletal 6A OS = Homo sapiens OX = 9606 GN = KRT6A PE = 1 SV = 3 | P02538 | KRT6A | 60 kDa | 30 | 8 | 25 | 7 | 100% | 100% |
| Transcription termination factor 1 OS = Homo sapiens OX = 9606 GN = TTF1 PE = 1 SV = 3 | Q15361 | TTF1 | 103 kDa | 31 | 23 | 25 | 17 | 100% | 100% |
| Treacle protein OS = Homo sapiens OX = 9606 GN = TCOF1 PE = 1 SV = 3 | Q13428 | TCOF1 | 152 kDa | 26 | 17 | 27 | 17 | 100% | 100% |
| Methylcrotonoyl-CoA carboxylase subunit alpha, mitochondrial OS = Homo sapiens OX = 9606 GN = MCCC1 PE = 1 SV = 3 | Q96RQ3 | MCCC1 | 80 kDa | 50 | 25 | 40 | 19 | 100% | 100% |
| Lysine-rich nucleolar protein 1 OS = Homo sapiens OX = 9606 GN = KNOP1 PE = 1 SV = 1 | Q1ED39 | KNOP1 | 52 kDa | 61 | 33 | 42 | 23 | 100% | 100% |
| Arginine and glutamate-rich protein 1 OS = Homo sapiens OX = 9606 GN = ARGLU1 PE = 1 SV = 1 | Q9NWB6 | ARGLU1 | 33 kDa | 29 | 17 | 46 | 20 | 100% | 100% |
| Cell growth-regulating nucleolar protein OS = Homo sapiens OX = 9606 GN = LYAR PE = 1 SV = 2 | Q9NX58 | LYAR | 44 kDa | 50 | 24 | 50 | 23 | 100% | 100% |
| Keratin, type I cytoskeletal 9 OS = Homo sapiens OX = 9606 GN = KRT9 PE = 1 SV = 3 | P35527 | KRT9 | 62 kDa | 98 | 34 | 54 | 19 | 100% | 100% |
| Keratin, type I cytoskeletal 10 OS = Homo sapiens OX = 9606 GN = KRT10 PE = 1 SV = 6 | P13645 | KRT10 | 59 kDa | 60 | 27 | 54 | 24 | 100% | 100% |
| Keratin, type II cytoskeletal 2 epidermal OS = Homo sapiens OX = 9606 GN = KRT2 PE = 1 SV = 2 | P35908 | KRT2 | 65 kDa | 64 | 29 | 60 | 24 | 100% | 100% |
| Histone H1.2 OS = Homo sapiens OX = 9606 GN = HIST1H1C PE = 1 SV = 2 | P16403 | HIST1H1C | 21 kDa | 72 | 2 | 69 | 2 | 100% | 100% |
| Acetyl-CoA carboxylase 1 OS = Homo sapiens OX = 9606 GN = ACACA PE = 1 SV = 2 | Q13085 | ACACA | 266 kDa | 41 | 33 | 69 | 45 | 100% | 100% |
| Histone H1.4 OS = Homo sapiens OX = 9606 GN = HIST1H1E PE = 1 SV = 2 | P10412 | HIST1H1E | 22 kDa | 74 | 13 | 75 | 18 | 100% | 100% |
| Keratin, type II cytoskeletal 1 OS = Homo sapiens OX = 9606 GN = KRT1 PE = 1 SV = 6 | P04264 | KRT1 | 66 kDa | 135 | 45 | 94 | 31 | 100% | 100% |
| Propionyl-CoA carboxylase alpha chain, mitochondrial OS = Homo sapiens OX = 9606 GN = PCCA PE = 1 SV = 4 | P05165 | PCCA | 80 kDa | 117 | 49 | 114 | 45 | 100% | 100% |
| Pyruvate carboxylase, mitochondrial OS = Homo sapiens OX = 9606 GN = PC PE = 1 SV = 2 | P11498 | PC | 130 kDa | 122 | 54 | 121 | 49 | 100% | 100% |
Figure 1.
(A) Gene ontology analysis of the protein hits discovered using PERK-BirA* obtained using WEB-based GEne SeT AnaLysis toolkit (http://www.webgestalt.org/). The minimum number of IDs in each category was required to be 5, the maximum was 2000, the false-discovery rate (FDR) correction used was Benjamini and Hochberg (BH). The categories were first ranked based on FDR and after that the topmost significant categories were selected. 10 categories of biological processes were identified, many related to the function of PERK. The enrichment ratio indicates the ratio between the fraction of proteins belonging to each biological process in our dataset over the fraction of these proteins expected If our dataset was completely random. (B) Two specific categories are highlighted, response to endoplasmic reticulum stress, and protein folding, indicating an enrichment in the dataset of proteins and processes linked to PERK's role in the unfolded protein response and ER stress.
Considering our previous and ongoing studies, we decided to focus on validating the interactions of PERK with proteins with a known tethering role and/or with a relevant function in membrane contact sites and ER Ca2+ homeostasis. Two hits that were interesting in this regard were SERCA1/2 and VAPA/B, found at positions 47 and 62, respectively, on our list (Table 1).
PERK Interaction With VAPB
The isoforms VAPA and B are members of a small VAP protein family and have broadly similar structures and functions (Murphy & Levine, 2016). They are tail-anchored ER membrane proteins that are central to the formation of MCSs. VAPs act as tethers for a growing group of proteins, including protein tyrosine phosphatase interacting protein 51 (PTPIP51), StAR Related Lipid Transfer Domain Containing 3 (STARD3), oxysterol-binding protein (OSBP), Nir2, and Sorting nexin 2 (SNX2), among others (Alpy et al., 2013; Amarilio et al., 2005; De Vos et al., 2012; Dong et al., 2016; Wyles et al., 2002). VAPB is an important mediator of both ER-mitochondria contact sites and ER-plasma membrane contact sites, where it can interact with various proteins responsible for tethering and lipid trafficking at these contact sites, including PTPIP51 on mitochondria and Nir2 at the ER-PM contacts. Given the role of PERK in modulating both ER-mitochondria and indirectly ER-PM contacts, we then explored the possibility that VAPA/B is a bona fide interacting partner of PERK.
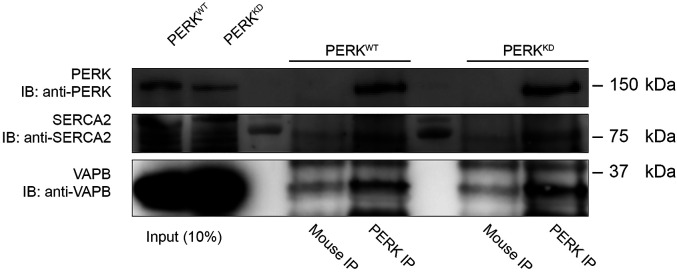
In our dataset, we picked up unique peptides for both VAPA and VAPB, indicating that both proteins might interact with PERK. Because VAPB has traditionally been the more studied of the two at MCS, and because both proteins are highly similar, for simplicity, here we focused on VAPB. We tested the possible interaction between VAPB and PERK by co-immunoprecipitation (co-IP) experiments and successfully confirmed that immunoprecipitation of wild type PERK-myc (PERKWT) expressed in HEK293-T cells, pulled down VAPB (Figures 2 and S2). In previous studies, we showed that the function of PERK required at the ER-mitochondria and ER-PM contacts was independent of its kinase function (van Vliet et al., 2017; Verfaillie et al., 2012). Here, to investigate whether PERK's kinase activity was dispensable for PERK-VAPB interaction, we performed the IP in cells expressing either a PERKWT or a kinase dead mutant of PERK (PERKKD). Interestingly, the expression of the PERKKD mutant yielded no difference in VAPB binding (Figures 2 and S2). Together these observations suggest that PERK interacts with VAPB constitutively, and in the absence of a signal, evoking its UPR activation. It is possible that VAPB could recruit PERK to specific MCS upon certain stresses, or vice versa; further research would be needed to validate this possibility.
Figure 2.
Identification of VAPB and SERCA2 as putative PERK interactors. HEK293-T cells were transfected with myc-tagged expression vectors for PERK WT (PERKWT) and PERK Kinase dead (PERKKD). After 48 h of transfection, PERKWT and PERKKD were immunoprecipitated (PERK IP) and interactors were detected using antibodies against VAPB and SERCA2 by immunoblot. Input was 10% of the total protein amount used for the IP (50 µg of protein was loaded as an input versus 500 µg of total protein used for the IP). Unspecific anti-mouse antibody was used as a negative control (Mouse IP). Data show are representative of N = 3 (VAPB), N = 2 (SERCA2) biologically independent experiments.
Proteins that interact with VAPs usually do so through a specific domain, termed a FFAT motif (two phenylalanines (FF) in an acidic tract) that binds the major sperm protein domain (MSP) on VAP (Murphy & Levine, 2016). However, using a previously published algorithm (Murphy & Levine, 2016) we did not find any robust FFAT motif in the PERK primary sequence. This suggests that the PERK-VAPB interaction may involve either another FFAT containing protein or may be mediated through another binding in cis, as they are both ER membrane proteins. This would leave the MSP of VAPB free to bind FFAT motifs in other proteins. A recent publication has explored the human “VAPome” in a systematic way using BioID and found that PERK was a hit for at least VAPA (Cabukusta et al., 2020). This same study also reports the putative interaction between PERK and MOSDP1 and 3. These proteins are VAP-like but bind to a slight variation of the FFAT motif, the FFNT motif (two phenylalanines (FF) in a neutral tract). Their analysis indicates that PERK contains an FFNT motif at location 283 of the protein, which is located in the lumen of the ER. This study, along with ours, does give support for a functional role between PERK and VAPA/B and other MCS proteins.
PERK Interaction With SERCA2
SERCA1/2 are ATPases located in the ER membrane that pump Ca2+ ions from the cytosol into the ER lumen, counteracting the Ca2+ leak from various sources (translocon, inositol 1,4,5-trisphosphate receptor) and maintaining a high level of Ca2+ in the ER lumen with a steady-state Ca2+ concentration of approximately 1 mM (de la Fuente et al., 2013).
In contrast to SERCA2, no unique peptides for SERCA1 could be identified in the BioID data set, so the presence of the latter isoform in the PERK interactome could not be proven. Neither SERCA1 nor SERCA2 have been detected at the ER-mitochondria contact sites, but a truncated version of SERCA1, S1 T (truncated after amino acid 395) has been shown to play a role at ER-mitochondria contact sites (Chami et al., 2008). As with VAPB, we confirmed the interaction of SERCA2 with PERK through co-IP, which was again detected independently of its kinase activity (Figures 2 and S2). Our previous results indicated that PERK is strongly activated by increases in cytosolic Ca2+ levels, and we speculated that this is linked to the depletion of ER luminal Ca2+ leading to ER stress, as shown also in a recent study (Preissler et al., 2020). Furthermore, we reported that PERK is involved in SOCE, by regulating the ER-PM contacts through its binding to FLNA (van Vliet et al., 2017). SOCE is induced by ER Ca2+ depletion, which leads to oligomerization of STIM1 and its translocation to the PM where it interacts with the PM localized ORAI1 protein. The interaction of STIM1 with ORAI1 then allows Ca2+ to enter the cytosol from the extracellular medium through the opening of the ORAI1 channel (Zhang et al., 2005). PERK's interaction with SERCA2 is an intriguing link since, although ORAI1 allows Ca2+ to enter the cytosol, the refilling of ER-Ca2+ store requires the activity of the Ca2+ pump SERCA. A previous study has indicated a close relationship between STIM1/ORAI1 driven SOCE and SERCA (Courjaret & Machaca, 2014). We can then speculate that PERK may be located close to the site of STIM1/ORAI1 contact, playing a part in the mechanism that ensures Ca2+ entering the cytosol is rapidly internalized into the ER lumen through SERCA2. Further study is needed to investigate a possible role of PERK in regulating SERCA2 activity, and therefore, influencing this mechanism.
Conclusions and Limitations of the Study
In this report, we show a BioID dataset obtained by tagging the promiscuous biotin ligase BirA* to PERK, in order to map its close interactors. We highlight and validate two of these, VAPB and SERCA2, which lead us to further speculate about the role of PERK in ER MCS formation and Ca2+ signaling.
Our study was performed when BioID was just emerging as a tool, and at the time of our planning, only the original study had been published (Roux et al., 2012). We, therefore, closely modeled our experimental setup on this study. It is important to note when interpreting this dataset that later studies using BioID (and derivatives of BioID like TurboID, where the BirA* ligase has been mutated to biotinylate at a much higher rate), have implemented more stringent controls to detect false positives (inherent with using BioID as an approach to screen protein-protein interactions). For example, the initial study reporting BioID used only parental cells (mock-transfected) as a control, while some recent studies used BirA* (or TuboID) not tagged to their protein of interest to control for non-specific biotinylation events (Szczesniak et al., 2021; Vermehren-Schmaedick et al., 2021; Zhang et al., 2019). Our reasoning for using only parental cells as control was that by using free BirA* as a control, there was a risk that this enzyme might biotinylate bona fide hits randomly, leading to false negatives. Since the initial study reporting BioID, the original authors have published numerous extensive updates on how to set up a BioID study, incorporating more appropriate or bespoke controls and comparing the different new techniques (May et al., 2020; May & Roux, 2019; Roux et al., 2018; Sears et al., 2019). In summary, we have identified 129 potential interacting partners of PERK, of which we have tested and confirmed two, SERCA2 and VAPB. We hope that this dataset can yield new insights concerning PERK and cellular signaling.
Materials and Methods
Cell Lines and Transfection:
HEK293-T cells have been maintained in Dulbecco's modified Eagle's medium containing 4.5 g/l glucose and 0.11 g/l sodium pyruvate and supplemented with 2 mM glutamine, 100 units/ml penicillin, 100 μg/ml streptomycin, and 10% fetal bovine serum (FBS) (all added, AA medium). Cells were transiently transfected with different PERK constructs encoding PERKK618A (Addgene plasmid 21815) or PERKFL (Addgene plasmid 21814), both myc tagged, using Trans-IT X2 transfection reagent (Mirus Bio LLC, Science Dr.Madison, WI USA).
BioID, Biotinylation Assay, and Mass Spectrometry
BioID was performed as described previously (Roux et al., 2012), with minor modifications. Overexpression of PERK-BioID was achieved by transiently transfecting Hek293-T cells using X-tremegene 9 (Roche, Germany). A 6 μl X-tremegene9 was mixed with a 2 μg PERK-BioID plasmid and dispersed on cultured cells. A 30 h post-transfection medium containing 50 μM biotin was added to the cells for 24 h. After incubation with biotin, cells were lysed in 1 mL lysis buffer (50 mM Tris, pH 7.4, 500 mM NaCl, 0.4% SDS, 5 mM EDTA, 1 mM DTT, and 1x complete protease inhibitor [Roche]) and sonicated. Triton X-100 was added to a 2% final concentration. After further sonication, an equal volume of 4 °C 50 mM Tris (pH 7.4) was added before additional sonication (subsequent steps at 4 °C) and centrifugation at 16,000 g. Supernatants were incubated with 600 μl Dynabeads (50% slurry) (MyOne Streptavidin C1; Invitrogen) overnight. Beads were collected and washed twice for 8 min at 25 °C (all subsequent steps at 25 °C) in 1 mL wash buffer 1 (2% SDS in dH2O). This was repeated once with wash buffer 2 (0.1% deoxycholate, 1% Triton X-100, 500 mM NaCl, 1 mM EDTA, and 50 mM HEPES, pH 7.5), once with wash buffer 3 (250 mM LiCl, 0.5% NP-40, 0.5% deoxycholate, 1 mM EDTA, and 10 mM Tris, pH 8.1), and twice with wash buffer 4 (50 mM Tris, pH 7.4, and 50 mM NaCl). For western blot analysis, bound proteins were removed from the magnetic beads with 50 μl of Laemmli SDS-sample buffer saturated with biotin at 98 °C. For mass spectrometry analysis, beads were washed repeatedly in MQ water containing 50 mM ammonium bicarbonate (AmBic) before being incubated for 30 min at 37 °C in 50 mM AmBic containing 5 mM DTT. Beads were further washed in 50 mM AmBic and incubated with 25 mM iodoacetamide in the dark for 30 min at 37 °C. After reduction/alkylation, beads were washed in 50 mM AmBic and incubated with 1:20 (w/w) modified trypsin (Pierce) ON at 37 °C in 50 mM AmBic containing 5% acetonitrile. After removal of beads by magnetic separation, formic acid was added to the peptide solution (to 2%) before desalting by C18 Micro Spin Columns (Harvard Apparatus). The resulting peptide mixture was analyzed by nano LC-MS on a hybrid quadrupole-orbitrap mass spectrometer (Q Exactive, Thermo Fisher Scientific). Peptides were identified by MASCOT 2.2 (Matrix Science) using the SwissProt database (taxonomy Homo sapiens, 20231 entries) adopting the following MASCOT search parameters: trypsin/P, two missed cleavages allowed, variable modification oxidation (M), fixed modification carbamidomethylation (C). The mascot was searched with a fragment ion mass tolerance of 0,02 Da and a parent ion mass tolerance of 10 ppm.
Scaffold 4 (Proteome Software Inc.) was used to validate MS/MS-based peptide and protein identifications. Peptide and protein identifications were accepted to achieve an FDR less than 10%. Standard protein grouping was adopted. The presence of at least 2 exclusive unique peptides per protein was required.
Immunoprecipitation
After 48 h of transfection with selected plasmids cells, they were collected through scraping and lysed in lysis buffer (1% CHAPS, 100 mM KCl, 150 mM NaCl, 1x protease inhibitor (Pierce Protease Inhibitor Tablets, Thermo Fisher Scientific Inc.)) for 30 min at 4 °C. Cells were centrifuged at 13.000 g for 15 min to remove debris and unbroken cells. From the supernatant, 500 μg of proteins were combined with primary antibodies overnight (ON) at 4 °C, against PERK and using a non-specific Mouse IgG as a control. Protein-antibody complexes were captured by the addition of Protein AG magnetic beads (Pierce) for 1.5 h at room temperature (RT). Protein AG magnetic beads with captured protein-antibody complexes were washed three times with lysis buffer. Proteins were eluted with sample buffer (62.5 μM Tris-HCl, 10% glycerol, 2% SDS, 1x protease inhibitor, 1x phosphatase inhibitor (Pierce phosphatase Inhibitor Tablets, Thermo Fisher Scientific Inc.) in MQ water) and loaded on a gel for western blot analysis.
Western Blotting
Samples were separated by SDS-PAGE on the Criterion system (Bio-Rad Laboratories, Hercules, CA, USA) on a 4%–12% Bis-TRIS gel and electrophoretically transferred to Protran 2 μm-pored nitrocellulose paper (PerkinElmer, Wellesley, MA, USA). The blots were blocked for 1 h at RT in TBS-T buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 0.1% Tween-20) containing 5% nonfat dry milk and then incubated with selected antibody solutions. Samples were processed and enhanced chemiluminescence using Pierce ECL Western Blotting Substrate was used for western blot detection and membranes were scanned using the Bio-Rad Chemidoc Imager (Bio-Rad Laboratories N.V.3, Winninglaan, Temse, Belgium).
Antibodies
Antibodies used were mouse monoclonal anti-c-Myc (Sigma, Cat# M4439), Control normal mouse IgG (sc-2025), anti-ATP2A2/SERCA2 (Cell signaling 388S), anti-VAPB (Invitrogen, PA5-53023), and an HRP-based detection using as a secondary antibody, the Veriblot antibody (#ab131366, Abcam).
Footnotes
Author Contributions: Experiment design, analysis, and manuscript writing: MLS, PA, ARVV. Performing and analyzing western blots: MLS. Mass spectrometry analysis: RD, EW. Project and funding: PA.
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This study was supported by the EOS consortium (grant number 30837538, FAF-F/2018/1252, G049817N, G070115N, G076617N).
ORCID iDs: Derua https://orcid.org/0000-0002-1784-0677
AR van Vliet https://orcid.org/0000-0003-4729-4379
References
- Alpy F., Rousseau A., Schwab Y., Legueux F., Stoll I., Wendling C., Spiegelhalter C., Kessler P., Mathelin C., Rio M. C., Levine T. P., Tomasetto C. (2013). STARD3 Or STARD3NL and VAP form a novel molecular tether between late endosomes and the ER. Journal of Cell Science, 126(Pt 23), 5500–5512. 10.1242/jcs.139295 [DOI] [PubMed] [Google Scholar]
- Amarilio R., Ramachandran S., Sabanay H., Lev S. (2005). Differential regulation of endoplasmic reticulum structure through VAP-Nir protein interaction. Journal of Biological Chemistry, 280(7), 5934–5944. 10.1074/jbc.M409566200 [DOI] [PubMed] [Google Scholar]
- Balsa E., Soustek M. S., Thomas A., Cogliati S., Garcia-Poyatos C., Martin-Garcia E., Jedrychowski M., Gygi S. P., Enriquez J. A., Puigserver P. (2019). ER and nutrient stress promote assembly of respiratory chain supercomplexes through the PERK-eIF2alpha axis. Molecular Cell, 74(5), 877–90e6. 10.1016/j.molcel.2019.03.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky J. L., Wojcikiewicz R. J. (2009). Substrate-specific mediators of ER associated degradation (ERAD). Current Opinion in Cell Biology, 21(4), 516–521. 10.1016/j.ceb.2009.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabukusta B., Berlin I., van Elsland D. M., Forkink I., Spits M., de Jong A. W. M., Akkermans Jjll, Wijdeven R. H. M., Janssen G. M. C, van Veelen P. A., Neefjes J. (2020). Human VAPome analysis reveals MOSPD1 and MOSPD3 as membrane contact site proteins interacting with FFAT-related FFNT motifs. Cell Reports, 33(10), 108475. 10.1016/j.celrep.2020.108475 [DOI] [PubMed] [Google Scholar]
- Chami M., Oules B., Szabadkai G., Tacine R., Rizzuto R., Paterlini-Brechot P. (2008). Role of SERCA1 truncated isoform in the proapoptotic calcium transfer from ER to mitochondria during ER stress. Molecular Cell, 32(5), 641–651. 10.1016/j.molcel.2008.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courjaret R., Machaca K. (2014). Mid-range Ca2+ signalling mediated by functional coupling between store-operated Ca2+ entry and IP3-dependent Ca2+ release. Nature Communications, 5, 3916. 10.1038/ncomms4916 [DOI] [PubMed] [Google Scholar]
- de la Fuente S., Fonteriz R. I., Montero M., Alvarez J. (2013). Ca2+ homeostasis in the endoplasmic reticulum measured with a new low-Ca2+-affinity targeted aequorin. Cell Calcium, 54(1), 37–45. 10.1016/j.ceca.2013.04.001 [DOI] [PubMed] [Google Scholar]
- De Vos K. J., Morotz G. M., Stoica R., Tudor E. L., Lau K. F., Ackerley S., Warley A., Shaw C. E., Miller C. C. (2012). VAPB Interacts with the mitochondrial protein PTPIP51 to regulate calcium homeostasis. Human Molecular Genetics, 21(6), 1299–1311. 10.1093/hmg/ddr559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong R., Saheki Y., Swarup S., Lucast L., Harper J. W., De Camilli P. (2016). Endosome-ER contacts control actin nucleation and retromer function through VAP-dependent regulation of PI4P. Cell, 166(2), 408–423. 10.1016/j.cell.2016.06.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- May D. G., Roux K. J. (2019). BioID: A method to generate a history of protein associations. Methods in Molecular Biology, 2008, 83–95. 10.1007/978-1-4939-9537-0_7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- May D. G., Scott K. L., Campos A. R., Roux K. J. (2020). Comparative application of BioID and TurboID for protein-proximity biotinylation. Cells, 9(5). 10.3390/cells9051070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moncan M., Mnich K., Blomme A., Almanza A., Samali A., Gorman A. M. (2021). Regulation of lipid metabolism by the unfolded protein response. Journal of Cellular and Molecular Medicine, 25(3), 1359–1370. 10.1111/jcmm.16255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy S. E., Levine T. P. (2016). VAP, a Versatile access point for the endoplasmic Reticulum: Review and analysis of FFAT-like motifs in the VAPome. Biochimica et Biophysica Acta, 1861(8 Pt B), 952–961. 10.1016/j.bbalip.2016.02.009 [DOI] [PubMed] [Google Scholar]
- Preissler S., Rato C., Yan Y., Perera L. A., Czako A., Ron D. (2020). Calcium depletion challenges endoplasmic reticulum proteostasis by destabilising BiP-substrate complexes. Elife, 9. 10.7554/eLife.62601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron D., Walter P. (2007). Signal integration in the endoplasmic reticulum unfolded protein response. Nature Reviews Molecular Cell Biology, 8(7), 519–529. 10.1038/nrm2199 [DOI] [PubMed] [Google Scholar]
- Roux K. J., Kim D. I., Burke B., May D. G. (2018). BioID: A screen for protein-protein interactions. Current Protocols in Protein Science, 91, 19231–192315. 10.1002/cpps.51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux K. J., Kim D. I., Raida M., Burke B. (2012). A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. Journal of Cell Biology, 196(6), 801–810. 10.1083/jcb.201112098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sears R. M., May D. G., Roux K. J. (2019). BioID as a tool for protein-proximity labeling in living cells. Methods in Molecular Biology, 2012, 299–313. 10.1007/978-1-4939-9546-2_15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorge S., Theelke J., Yildirim K., Hertenstein H., McMullen E., Muller S., Altburger C., Schirmeier S., Lohmann I. (2020). ATF4-induced Warburg metabolism drives over-proliferation in drosophila. Cell Reports, 31(7), 107659. 10.1016/j.celrep.2020.107659 [DOI] [PubMed] [Google Scholar]
- Szczesniak L. M., Bonzerato C. G., Wojcikiewicz R. J. H. (2021). Identification of the Bok interactome using proximity labeling. Frontiers in Cell and Developmental Biology, 9, 689951. 10.3389/fcell.2021.689951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Vliet A. R., Giordano F., Gerlo S., Segura I., Van Eygen S., Molenberghs G., Rocha S., Houcine A., Derua R., Verfaillie T., Vangindertael J., De Keersmaecker H., Waelkens E., Tavernier J., Hofkens J., Annaert W., Carmeliet P., Samali A., Mizuno H., Agostinis P. (2017). The ER stress sensor PERK coordinates ER-plasma membrane contact site formation through interaction with filamin-A and F-actin remodeling. Molecular Cell, 65(5), 885–99e6. 10.1016/j.molcel.2017.01.020 [DOI] [PubMed] [Google Scholar]
- Verfaillie T., Rubio N., Garg A. D., Bultynck G., Rizzuto R., Decuypere J. P., Piette J., Linehan C., Gupta S., Samali A., Agostinis P. (2012). PERK Is required at the ER-mitochondrial contact sites to convey apoptosis after ROS-based ER stress. Cell Death & Differentiation, 19(11), 1880–1891. 10.1038/cdd.2012.74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermehren-Schmaedick A., Huang J. Y., Levinson M., Pomaville M. B., Reed S., Bellus G. A., Gilbert F., Keren B., Heron D., Haye D., Janello C., Makowski C., Danhauser K., Fedorov L. M., Haack T. B., Wright K. M., Cohen M. S. (2021). Characterization of PARP6 function in knockout mice and patients with developmental delay. Cells, 10(6). 10.3390/cells10061289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyles J. P., McMaster C. R., Ridgway N. D. (2002). Vesicle-associated membrane protein-associated protein-A (VAP-A) interacts with the oxysterol-binding protein to modify export from the endoplasmic reticulum. Journal of Biological Chemistry, 277(33), 29908–29918. 10.1074/jbc.M201191200 [DOI] [PubMed] [Google Scholar]
- Zhang S. L., Yu Y., Roos J., Kozak J. A., Deerinck T. J., Ellisman M. H., Stauderman K. A., Cahalan M. D. (2005). STIM1 Is a Ca2+ sensor that activates CRAC channels and migrates from the Ca2+ store to the plasma membrane. Nature, 437(7060), 902–905. 10.1038/nature04147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Song G., Lal N. K., Nagalakshmi U., Li Y., Zheng W., Huang P. J., Branon T. C., Ting A. Y., Walley J. W., Dinesh-Kumar S. P. (2019). TurboID-based proximity labeling reveals that UBR7 is a regulator of N NLR immune receptor-mediated immunity. Nature Communications, 10(1), 3252. 10.1038/s41467-019-11202-z [DOI] [PMC free article] [PubMed] [Google Scholar]