Abstract
It is now clear that the homeobox motif is well conserved across metazoan phyla. It has been established experimentally that a subset of genes containing this motif plays key roles in the orchestration of gene expression during development. Auto- and cross-regulatory functional interactions join homeobox genes into genetic networks. We have developed a specialized database HOX-Pro in order to arrange all available data on structure, function, phylogeny and evolution of Hox genes, Hox clusters and Hox networks. Its primary location is http://www.iephb.nw.ru/hoxpro . The database is also mirrored at http://www.mssm.edu/molbio/hoxpro . The HOX-Pro database is aimed at: (i) analysis and classification of regulatory and coding regions in diverse homeobox and related genes; (ii) comparative analysis of organization of ‘Hox-based’ genetic networks in the sea urchin Strongylocentrotus purpuratus, the fruit fly Drosophila melanogaster and the mouse Mus musculus; and (iii) analysis of phylogeny and evolution of homeobox genes and clusters.
INTRODUCTION
The investigation of metazoan genes involved in transcriptional control requires the creation of specialized databases. These must contain exhaustive information about the structure–function relationships of regulatory regions and the interactions of the protein products involved in transcriptional regulation (1,2). As a further complication, for developmentally important genes, function must be represented at multiple scales ranging from the molecular to the organismal.
In recent years it has been demonstrated that the protein products of a relatively small group of genes is of particular importance in controlling the expression of a much wider set of target genes, and through them the overall course of development and morphogenesis. An important subset of these genes contains a 180 bp structural motif known as the ‘homeobox’, and among these an important subset occurs in conserved clusters (‘Hox complexes’) on the chromosome. Members of Hox complexes are of particular importance in specifying the overall animal body plan, and have been the objects of intensive study. For these reasons, the homeobox containing genes are a natural choice for the subject matter of a database concerned with gene function in development at multiple levels. A further advantage of using the homeobox as a criterion for inclusion in the database is that its presence is an unambiguous sequence-based criterion that indicates a developmentally relevant gene, although not all such genes contain a homeobox domain.
The project which we describe here is called the ‘Homeobox Gene Promoter Regions DataBase’ (HOX Pro DB). Its aim is to integrate the molecular aspects of modern developmental biology by utilizing the information pathways that run from sequence data to developing organs and tissues. The long-term goal of HOX Pro is the reconstruction and prediction of functional genetic regulatory pathways from all relevant biological assays. These include not only sequence data but also information about protein binding, expression patterns, and so on.
GENERAL DESCRIPTION OF HOX Pro
A database of transcriptional regulatory networks
The HOX Pro database has been developed in order to describe the ensembles of homeobox containing genes which control embryogenesis (3). It contains a broad spectrum of information including images, diagrams and animations. Currently this amounts to ~600 html pages together with 300 images which contain information on 200 genes and 90 promoters, in turn linked to maps of 13 HOX clusters and nine genetic networks. Graphical representation of Hox clusters and Hox-based networks is accomplished by means of flow diagrams, JavaScript animations and Java applets. This permits the clear representation of gene interactions in the Hox gene ensembles and facilitates navigation in the database (4).
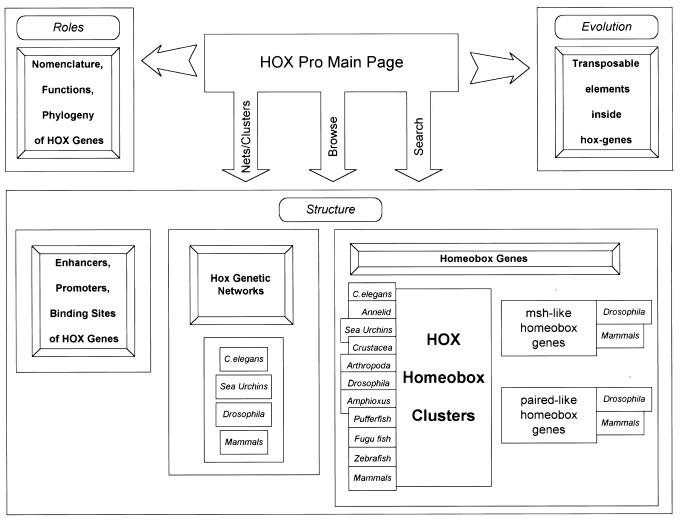
The HOX Pro database contains data on the structural and functional organization of the transcriptional regulatory machinery of homeobox and functionally related genes, as shown in Figure 1. The hierarchical organization of transcription regulation of metazoan genes is incorporated into the database schema. HOX Pro includes a hypertext description of the mechanisms of homeobox gene activation as well as the functional characteristics of proteins encoded by homeobox-containing and functionally related genes. HOX Pro also contains links to other databases such as TRANSFAC, COMPEL, EPD, EMBL, GeNet, FlyBase and The Interactive Fly.
Figure 1.
Block diagram of HOX Pro database organization.
Representation of networks. The main principles of data presentation in HOX Pro are as follows. The genetic cluster and genetic network maps form the basis for information structuring in HOX Pro. The genetic network diagrams are represented in a form of a directed graph, in which each gene is represented as a node. Each node is represented by a symbol denoting its broad structural and functional class (homeobox containing, controller or target, etc.).
Representation of genes. Each gene entry contains information on the gene’s function, key features of its encoded protein, expression pattern, regulatory interactions (upstream and downstream genes) as well as links to other databases. The regulatory element entry in HOX Pro contains data on the organism source, bibliographical data, regulatory element sequence and coordinates of sites for transcription factor binding, as well as key words and definitions. Gene interaction entries hold information on the mechanism of gene interaction with experimental lines of evidence supporting the named mechanism.
Taxonomic coverage. HOX Pro holds information on Hox clusters and networks in different organisms: the sea urchin Strongylocentrotus purpuratus, the fruit fly Drosophila melanogaster, and several vertebrates including teleost fishes, chicken, mouse and human. It contains up to six types of data: genetic cluster maps, network maps, gene entries, gene sequence entries, regulatory region entries and bibliography. In addition, the database contains comparative schemes for HOX clusters in a larger set of organisms including Caenorbabditis elegans, Drosophila, Amphioxus, pufferfish, zebrafish and certain annelids, sea urchins, crustaceans, arthropods and mammals.
The trimodal HOX Pro user interface. Three entrance html pages are provided allowing the user to browse the database, to search the database, and to work with genetic cluster or network maps. While browsing HOX Pro the user moves sequentially from the page containing the list of database sections to genetic clusters and genetic network maps. Each gene within a map is linked to a gene entry. This holds hypertext links to data on gene sequence, regulatory regions, gene interactions and a bibliography. By selecting a gene name in the map the user gets detailed information about the gene and mechanisms of its regulation.
Another entrance page into HOX Pro enables the user to work with gene clusters or network maps. Interactive cluster diagrams present a physical map of a Hox cluster. Genetic network maps are depicted as directed graphs. These enable the user to find out which genes regulate a particular gene as well as which genes are its regulatory targets. Each gene in the cluster or network diagram is hyperlinked to its gene entry so that the user can retrieve all the information about a gene of interest by following the hypertext links in the database.
The HOX Pro graphical user interface, written in Java, allows exploration and visualization of the HOX Pro database through the Internet. It includes tools for automated generation of gene network diagrams, visualization filters, as well as tools for data navigation. These include interactive images within the diagram, online help, interactive cross references within this database, and references to other databases (4).
To make HOX Pro as easy to use as possible, we use JavaScript to orient the user by means of dialogue boxes. These windows open automatically when a user connects with a particular html page and contain, depending on the page, either a brief database structure map or a map of a Hox/Hom cluster or network. Such help pages assist users who are novices to the database structure. In addition, all 200 gene title pages include a thumbnail image of the network and/or cluster which contains the given gene. The thumbnail is linked to the interactive diagram of the network or cluster.
Data model for description of gene regulatory regions
The formulation of an appropriate data model is a central problem in database design. This is particularly acute when constructing a database concerned with transcriptional regulation. One recurrent characteristic of gene regulatory regions is their modular structure (5) and recognizable levels of hierarchy (1). The lowest level in the regulatory hierarchy corresponds to a binding site for a particular ligand, which binds that ligand with a certain affinity.
The HOX Pro database contains descriptions of more than 200 binding sites on the promoters of invertebrate and vertebrate homeobox containing genes. Currently the HOX Pro database contains entries characterizing 190 cis-regulatory elements of Drosophilidae homeotic genetic networks and 30 cis-regulatory elements of vertebrate hox genetic networks.
The second level of the hierarchy consists of the composite response elements. These contain two to three closely situated binding sites for different transcription factors. Composite response elements act as entire units due to specific protein–protein interactions between the respective transcription factors (6). As a rule a composite element is formed from adjacent or partially overlapping sites.
Promoters and enhancers lie at the next level of the transcriptional regulatory hierarchy, above composite elements. (Although ‘promoter’ sometimes refers to the entire regulatory region of a gene, in much of the literature and in Hox DB it refers to the regulatory part of the gene that is responsible for the basal level of transcription.) Promoters consist of several composite elements and/or individual binding sites for transcription factors. They include the region of transcription start and the adjacent upstream region of several tens of nucleotides. Cis-regulatory elements of a promoter provide the formation of the minimal transcription initiating complex and subsequent formation of a complete transcription complex (7). The HOX Pro DB contains sequences and description of promoter regions from 12 Drosophilidae HOM-C genes, 63 vertebrate Hox-cluster genes, 12 eve-like and seven msh-like vertebrate genes.
Enhancers also consist of composite elements and/or single binding sites. They modulate the level of transcription depending on the type of tissue, developmental stage, stage of the cell cycle, induction by hormones or other molecular signals. An enhancer can act over many kilobases 3′ or 5′ from the transcription start site, possibly from within an intron, and its activity does not depend on its orientation.
The regulatory regions of Hox clusters and functionally related genes consist of several elements of the previously mentioned levels (separate cis-elements, composite elements, enhancers and promoters) and occupy relatively large regions of DNA. Certain Hox clusters are known to act as a large, high order transcriptional regulation unit. To systematically analyze this we have started to accumulate DNA sequences for the whole clusters. Currently the HOX Pro database includes 14 complete intergenic sequences for vertebrate Hox clusters. Also, there are two completely sequenced clusters: D.Melanogaster BX-C (>330 kb) and human HOX A (~228 kb). The regulatory regions of some genes are located at extremely long distances from the transcription start site. For example, there are at least five control elements of the D.melanogaster Ubx gene scattered at 100 kb upstream and downstream the transcription start (8,9).
In conclusion, we are able to represent the highest level of the regulatory hierarchy, namely the modular cis-regulatory organization of developmentally expressed genes. These genes are modular in organization: specific, separable fragments of the cis-regulatory DNA each containing multiple transcription factor target sites that execute particular regulatory subfunctions. Cis-regulatory modules are thought to be the units of developmental transcription control, and also of evolution, in the assembly of transcription control systems (5).
DISCUSSION
The genomic sequencing projects for model organisms have led to a rapid growth of biological information (10–12), much of it accessible in databases. These databases have been designed specifically for the storage, processing and retrieval of molecular biology data. At present neither analysis of results, nor planning of experiments are possible without utilizing these databases.
Now, at the beginning of the ‘post-genomic era’, when bio-medical research shifts from identifying genes to characterization of their function, the design of databases containing functional information has become crucial. HOX Pro contains functional information on the mechanisms of gene action in embryogenesis, in a manner similar to other functional information databases. However the distinctive feature of HOX Pro is its particular model for information presentation, which is based on the concept of genetic ensembles (clusters and networks), the hierarchical nature of transcriptional regulation and comparative evolutionary approaches. Such a database structure enables end users to retrieve information on the functional organization and evolutionary conservation of a whole ensemble of interacting genes. Another distinctive feature of the HOX Pro is its user-friendly decentralized architecture, so that from any particular html page the user can see the main content of the database.
One of the difficulties in the design and use of a database of this type is that for some genes there are detailed data at the molecular level on the structure of their regulatory regions, including binding sites for transcription factors, while for other genes this data is not available. Beyond the question of the scale of the data (molecular, cellular, organismal, etc.), there is the issue of how to treat inferential or derived data. Such data might include the analysis of mutations in gene regulatory regions, functional relationships with other genes, and so on. Often such data is dependent of the interpretation of particular experiments that may be reinterpreted in light of new information.
A flexible data representation scheme in HOX Pro allows these difficulties to be circumvented. In accordance with the available experimental data, the structure of the transcriptional regulatory regions of some genes is given in a highly structured format, while regulation of the expression of others is described in a freeform manner with hypertext, figures and tables.
Acknowledgments
ACKNOWLEDGEMENTS
A.S. was supported by RFBR 98-04-49422, INTAS 97-30950, and by CRDF GAP project RB0-685 from NIH RO1 RR07801-07. J.R. and T.B. were supported by NIH RO1 RR07801-07 and ONR N0014-97-1-0422.
REFERENCES
- 1.Kel A.E., Ponomarenko,M.P., Likhachev,E., Orlov,Yu.L., Ischenko,I.V., Milanesi,L. and Kolchanov,N.A. (1993) Comput. Appl. Biosci., 9, 617–627. [DOI] [PubMed] [Google Scholar]
- 2.Kanehisa M. (1996) Sci. Technol. Jpn, 59, 34–38. [Google Scholar]
- 3.Spirov A.V. (1996) J. Evol. Biochem. Physiol. [St.-Petersburg], 32, 556–568. [Google Scholar]
- 4.Serov V.N., Spirov,A.V. and Samsonova,M.G. (1998) Bioinformatics, 14, 546–547. [DOI] [PubMed] [Google Scholar]
- 5.Kirchhamer C.V., Yuh,C.H. and Davidson,E.H. (1996) Proc. Natl Acad. Sci. USA, 93, 9322–9328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kel O.V., Romaschenko,A.G., Kel,A.E., Wingender,E. and Kolchanov,N.A. (1995) Nucleic Acids Res., 23, 4097–4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buratowski S. (1994) Cell, 77, 1–3. [DOI] [PubMed] [Google Scholar]
- 8.Bienz M. and Tremml,G. (1988) Nature, 333, 576–578. [DOI] [PubMed] [Google Scholar]
- 9.Muller J. and Bienz,M. (1992) EMBO J., 11, 3653–3661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schuler G.D., Boguski,M.S., Stewart,E.A., Stein,L.D., Gyapay,G., Rice,K., White,R.E., Rodriguez-Tome,P., Aggarwal,A., Bajorek,E., Bentolila,S. et al. (1996) Science, 274, 540–546. [PubMed] [Google Scholar]
- 11.Lander E.S. (1996) Science, 274, 536–539. [DOI] [PubMed] [Google Scholar]
- 12.Nowak R. (1995) Science, 270, 368–369. [DOI] [PubMed] [Google Scholar]