Abstract
Late-onset Alzheimer’s disease is a prevalent age-related polygenic disease that accounts for 50–70% of dementia cases. Currently, only a fraction of the genetic variants underlying Alzheimer’s disease have been identified. Here we show that increased sample sizes allowed for identification of seven previously unidentified genetic loci contributing to Alzheimer’s disease. This study highlights microglia, immune cells, and protein catabolism as relevant to late-onset Alzheimer’s disease, while identifying and prioritizing previously unidentified genes of potential interest. We anticipate that these results can be included in larger meta-analyses of Alzheimer’s disease to identify further genetic variants which contribute to Alzheimer’s pathology.
Introduction
Dementia has an age- and sex- standardized prevalence of ~7.1% in Europeans1, with Alzheimer’s disease (AD) being the most common form of dementia (50–70% of cases)2. AD is pathologically characterized by the presence of amyloid-beta plaques and tau neurofibrillary tangles in the brain3. Most patients are diagnosed with AD after the age of 65, termed late onset AD (LOAD), while only 1% of the AD cases have an early onset (before the age of 65)3. Based on twin studies, the heritability of LOAD is estimated to be ~60–80%4,5, suggesting that a large proportion of individual differences in LOAD risk is driven by genetics. The heritability of LOAD is spread across many genetic variants; however, Zhang et al. (2020)6 suggested that LOAD is more of an oligogenic than polygenic disorder due to the large effects of APOE variants. Zhang et al. (2020) and Holland et al. (2021)7 predicted there to be ~100–10,000 causal variants contributing to LOAD; however, only a fraction have been identified. Increasing the sample size of GWAS studies will improve the statistical power to identify the missing causal variants and may highlight additional disease mechanisms. In combination with increasing samples, it is beneficial to use different approaches to identify rare and private variation to help identify additional causal variants and increase understanding of disease mechanisms; however, we deem this to be out of the scope of the current analysis.
The largest previous GWAS of LOAD, identified 29 risk loci from 71,880 (46,613 proxy) cases and 383,378 (318,246 proxy) controls8. Our current study expands this to include 90,338 (46,613 proxy) cases and 1,036,225 (318,246 proxy) controls. The recruitment of LOAD cases can be difficult due to the late age of onset, so proxy cases can allow for the inclusion of younger individuals by estimating their risk of LOAD using parental status. Proxy cases and controls were defined based on known parental LOAD status weighted by parental age (Supplementary Note). In the current study, we identified 38 loci, including seven loci that have not been reported previously. Functional follow-up analyses implicated tissues, cell types, and genes of interest through tissue and cell type enrichment, colocalization, and statistical fine-mapping. This study highlights microglia, immune cells, and protein catabolism as relevant to LOAD while identifying previously unidentified genes of potential interest.
Results
Genome-wide inferences
We meta-analyzed data from 13 cohorts, totaling 1,126,563 individuals (Supplementary Table 1). The inflation factors and linkage disequilibrium score (LDSC) regression9 intercepts of each dataset are reported in Supplementary Table 2. The liability-scale SNP heritability was estimated by LDSC regression9 to be 0.031 (SE=0.0062) given a population prevalence of 0.05 (UK Biobank (UKB) data excluded). This estimate is low but similar to the estimates obtained in a previous GWAS meta-analyses (Jansen8: h2l=0.055,SE=0.0099; Lambert10: h2=0.069, SE=0.013). The LDSC intercept was 1.022 (SE=0.013), the inflation factor (l) for the meta-analysis was 1.11, and the sample size adjusted inflation factor (l1000)11 was 1.007. The genetic correlation12 between proxy LOAD and case-control LOAD was 0.83 (SE=0.21, P=6.61×10−5). Separate Manhattan plots for the LOAD proxy data and the case-control LOAD data are available in Supplementary Figures 1, 2. Across 855 external phenotypes in LDhub13, two significant genetic correlations with the meta-analysis results were observed, both of which were identified in previous studies of LOAD (Supplementary Note, Supplementary Table 3).
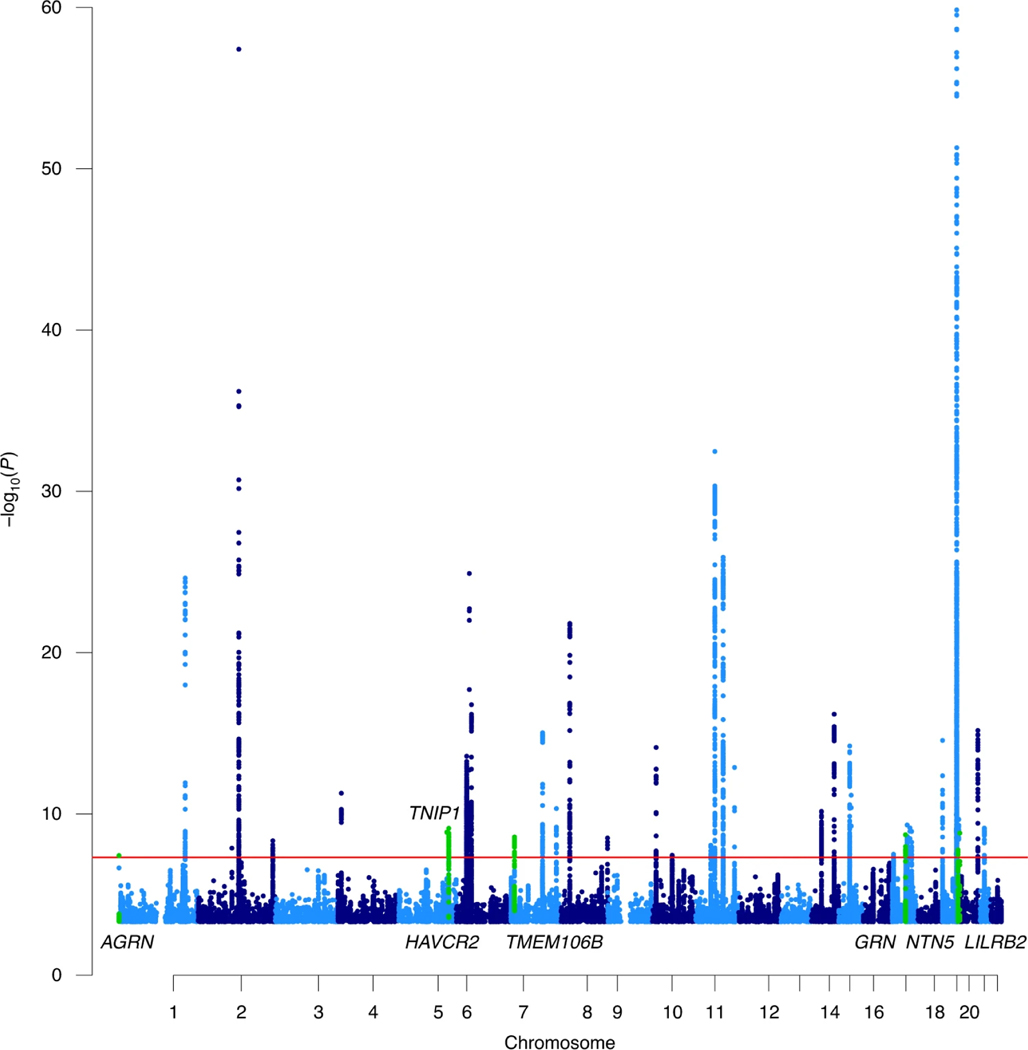
The meta-analysis identified 3,915 significant (P< 5×10−8) variants across 38 independent loci (Table 1, Figure 1). Of those 38 loci, seven have not shown associations with LOAD in previous GWAS, and five of those loci have not been associated with any form of dementia (AGRN, TNIP1, HAVCR2, NTN5, LILRB2). The lead variant effect estimates and significance values per dataset for each locus are reported in Supplementary Table 4. We largely replicated the loci identified in Jansen et al. (2019)8, however 7 loci were not found to be genome-wide significant in this study, five of those were just below significance and two were driven by rare variants (largely) not included in this study (Supplementary Note, Supplementary Table 5). However, we successfully replicated all the significant loci in Kunkle et al. (2019)14 (Supplementary Table 6).
Table 1:
The 38 genomic risk loci identified from 90,338 (46,613 proxy) cases and 1,036,225 (318,246 proxy) controls. The P-values were identified through a meta-analysis (two-sided test) of summary statistics generated by linear/logistic regressions (two-sided test) and were not adjusted for multiple testing. The previously unidentified loci are highlighted in bold. The genes were assigned based on colocalization results, fine-mapping results, and previous literature.
| Genomic Locus | Gene | Position (GRCh37) | Lead variant | A1 | A1 frequency | P | N |
|---|---|---|---|---|---|---|---|
| 1 | AGRN | 1:985377 | rs113020870 | T | 0.0041 | 3.83×10 −8 | 776379 |
| 2 | CR1 | 1:207750568 | rs679515 | C | 0.82 | 2.42×10−25 | 762176 |
| 3 | NCK2 | 2:106235428 | rs115186657 | C | 0.0035 | 1.33×10−8 | 727537 |
| 4 | BIN1 | 2:127891427 | rs4663105 | C | 0.41 | 3.92×10−58 | 1078540 |
| 5 | INPPD5 | 2:234082577 | rs7597763 | C | 0.45 | 4.65×10−9 | 819541 |
| 6 | CLNK | 4:11014822 | rs4504245 | G | 0.79 | 5.23×10−12 | 1080458 |
| 7 | TNIP1 | 5:150432388 | rs871269 | T | 0.32 | 1.37×10 −9 | 1089904 |
| 8 | HAVCR2 | 5:156526331 | rs6891966 | G | 0.77 | 7.91×10 −10 | 1089230 |
| 9 | HLA-DRB1 | 6:32583813 | rs1846190 | A | 0.30 | 2.66×10−14 | 754040 |
| 10 | TREM2 | 6:40942196 | rs187370608 | G | 0.997 | 1.26×10−25 | 791668 |
| 11 | CD2AP | 6:47552180 | rs9369716 | T | 0.27 | 1.70×10−17 | 1052285 |
| 12 | TMEM106B | 7:12268758 | rs5011436 | C | 0.41 | 2.70×10−9 | 1123678 |
| 13 | ZCWPW1/NYAP1 | 7:99932049 | rs7384878 | T | 0.69 | 9.41×10−16 | 1084138 |
| 14 | EPHA1-AS1 | 7:143104331 | rs3935067 | G | 0.62 | 4.69×10−11 | 1117025 |
| 15 | CLU | 8:27466315 | rs1532278 | T | 0.39 | 1.57×10−22 | 1126563 |
| 16 | SHARPIN | 8:145108151 | rs61732533 | G | 0.95 | 3.14×10−9 | 1122653 |
| 17 | USP6NL/ECHDC3 | 10:11718713 | rs7912495 | G | 0.46 | 7.68×10−15 | 1120367 |
| 18 | CCDC6 | 10:61738152 | rs7902657 | T | 0.54 | 3.68×10−8 | 1126388 |
| 19 | MADD/SPI1 | 11:47380340 | rs3740688 | T | 0.54 | 8.78×10−9 | 1123185 |
| 20 | MS4A4A | 11:60021948 | rs1582763 | G | 0.62 | 3.40×10−33 | 1125804 |
| 21 | PICALM | 11:85800279 | rs561655 | G | 0.35 | 1.24×10−26 | 1126563 |
| 22 | SORL1 | 11:121435587 | rs11218343 | T | 0.96 | 1.33×10−13 | 1125100 |
| 23 | FERMT2 | 14:53298853 | rs7146179 | G | 0.89 | 6.99×10−11 | 1089904 |
| 24 | RIN3 | 14:92938855 | rs12590654 | G | 0.67 | 6.63×10−17 | 1116967 |
| 25 | ADAM10 | 15:59057023 | rs602602 | T | 0.70 | 6.22×10−15 | 1124268 |
| 26 | APH1B | 15:63569902 | rs117618017 | T | 0.13 | 7.00×10−12 | 889854 |
| 27 | SCIMP/RABEP1 | 17:4969940 | rs7209200 | T | 0.33 | 3.18×10−8 | 1125637 |
| 28 | GRN | 17:42442344 | rs708382 | T | 0.61 | 1.98×10 −9 | 1125622 |
| 29 | ABI3 | 17:47450775 | rs28394864 | G | 0.54 | 4.90×10−10 | 1084218 |
| 30 | TSPOAP1-AS1 | 17:56409089 | rs2632516 | G | 0.54 | 7.46×10−10 | 1082451 |
| 31 | ACE | 17:61545779 | rs6504163 | T | 0.61 | 1.23×10−9 | 1083145 |
| 32 | ABCA7 | 19:1050874 | rs12151021 | G | 0.68 | 2.81×10−15 | 1082434 |
| 33 | APOE | 19:45411941 | rs429358 | T | 0.84 | <1.0×10−300 | 1126190 |
| 34 | NTN5 | 19:49213504 | rs2452170 | G | 0.47 | 1.72×10 −8 | 1088626 |
| 35 | CD33 | 19:51737991 | rs1354106 | G | 0.37 | 2.21×10−10 | 716038 |
| 36 | LILRB2 | 19:54825174 | rs1761461 | C | 0.49 | 1.56×10 −9 | 1116336 |
| 37 | CASS4 | 20:54995699 | rs6069737 | T | 0.083 | 6.73×10−16 | 1087703 |
| 38 | APP | 21:27520931 | rs2154482 | T | 0.44 | 7.66×10−10 | 1124606 |
Bold rows indicate previously unidentified loci
Figure 1:
A Manhattan plot of the meta-analysis results highlighting 38 loci, including 7 previously unidentified regions. Only variants with a P< 0.0005 are displayed. The APOE region cannot be fully observed because the y-axis is limited to the top variant in the second most significant locus, -log10(1×10−60), in order to display the less significant variants. The red line represents genome wide significance (5×10−8). The P-values were identified through a meta-analysis (two-sided test) of summary statistics generated by linear/logistic regressions (two-sided test) and were not adjusted for multiple testing. The previously unidentified loci are highlighted in green and indicated by the assigned gene name. The TNIP1/HAVCR2 regions and the NTN5/LILRB2 regions are close enough together that they cannot be visually distinguished at this scale but are different genomic risk loci.
Tissue type, cell type, and gene set enrichment
MAGMA tissue specificity analysis15 identified spleen (PBonferroni=0.034) as the only Genotype-Tissue Expression (GTEx) tissue where expression of the MAGMA genes was significantly associated (Supplementary Figure 3, Supplementary Table 7). However, this tissue was slightly above the significance threshold (PBonferroni= 0.054) when the larger APOE region (GRCh37: 19:40000000–50000000) was excluded (Supplementary Table 7). Spleen was also significant in the previous MAGMA tissue specificity analysis performed in Jansen et al. (2019)8 and is a known contributor to immune function. To investigate enrichment at the cell type level, FUMA cell type analysis16 was performed with a collection of cell types in mouse brain, human brain, and human blood tissue. Six single-cell (scRNA-seq) datasets were significantly associated, after multiple testing correction, with the expression of LOAD-associated genes (Supplementary Figure 4, Supplementary Table 8). Microglia was the only significant cell type in all six independent scRNA-seq datasets. We confirm previously observed enrichment for non-human microglial cells8, and report additional similar enrichments in human microglia. Four of these enrichments remained significant after exclusion of the larger APOE region suggesting that genomic regions outside of these two play a substantial role in the microglia finding. A combination of the cell type and tissue specificity results identifies microglia and immune tissues as potential experimental models for identifying the contribution of LOAD-associated genes towards LOAD pathogenesis.
MAGMA gene set analysis15 identified 25 Gene Ontology biological processes (Supplementary Table 9) that were significantly enriched, after multiple testing correction, for LOAD-associated variants. Subsequent conditional gene set analyses confirmed independent association of four out of these 25 gene-sets, reflecting the role of LOAD-associated genes in amyloid and tau plaque formation, protein catabolism of plaques, immune cell recruitment, and glial cells (Supplementary Table 9). The exclusion of the larger APOE region resulted in the loss of 5 significant gene-sets related to amyloid beta clearance, phospholipid efflux, cholesterol transport, protein lipid interactions, and tau binding, and the gain of 2 significant gene-sets related to tau degradation and astrocyte activation (Supplementary Table 9). Conditional gene-set analysis, with the larger APOE region excluded, identified 4 independent gene-sets related to astrocyte activation, immune cell recruitment, amyloid catabolism, and neurofibrillary tangles. The gene-set related to glial cells was still significant after removal of the APOE region, but was not identified as an independent gene-set, which suggests that this association can be explained by the APOE region in addition to another significant independent gene-set. Largely, the themes highlighted in the gene-set analysis are robust to the exclusion of the APOE region. Our gene-set analysis identified the same themes as Jansen et al. (2019)8 and further identified significant gene-sets involved in immune cell recruitment and neuronal cell types.
Gene prioritization
As expected, the genomic risk loci identified in this study were enriched for active chromatin and variant annotations relating to gene function (Supplementary Note). We performed functional follow-up (colocalization and fine-mapping) to further dissect the genomic risk loci to identify potential disease drivers. Functional mapping of variants to genes based on position and expression quantitative trait loci (eQTL) information from brain and immune tissues/cells identified 989 genes which mapped to one of the 38 genomic risk loci (Supplementary Table 10). These mapped genes were annotated with the drugs which target them based on information from DrugBank17.
Due to linkage disequilibrium (LD) and the inability to distinguish true causal variants from variants in LD, many of the mapped genes may be functionally irrelevant to LOAD. In order to highlight potentially relevant genes, eQTL data from immune tissues, brain, and microglia were colocalized with the genomic risk loci using Coloc18. We used the 19 successful colocalizations (Supplementary Table 11) for nine genes (TNIP1, MADD, APH1B, GRN, AC004687.2, ACE, NTN5, CD33, and CASS4) to prioritize genes in those loci. Statistical fine-mapping with susieR was additionally performed to narrow down the associated region (Supplementary Table 12). The statistical fine-mapping required an external reference panel, which limits the interpretation of the findings, so only high confidence variants (posterior inclusion probability (PIP) in a credible set >0.95) will be considered in gene prioritization. Gene prioritization of the previously unidentified loci and a description of colocalization and fine-mapping evidence for previously identified loci is available in the Supplementary Note. Some of the most interesting findings for the previously unidentified loci are highlighted below.
The lead variant of locus 7 (rs871269; P=1.37×10−9; minor allele frequency (MAF) =0.34) is located in an intron of TNIP1 (Supplementary Figure 5) and maps to GPX3, TNIP1, and SLC36A1 based on eQTLs within blood tissue. The lead variant is supported by a few variants with suggestive signal (rs34294852; P=1.05×10−6) but none of these variants are in LD (R2>0.1) in the 1000 Genomes (1KG) European (EUR) population. However, these variants are in moderate/low LD with the lead variant (R2=0.2–0.6) in the 1KG East Asian (EAS) and American populations. This suggests that the 1KG EUR reference panel does not accurately represent the LD structure of our data at this locus. The fine-mapping results from susieR identified the lead variant as the only variant with high posterior probability of inclusion (PIP>0.99). However, the association signal in this locus colocalized with a nearby suggestive variant (rs34294852; R2=0.29 in 1KG EAS), this variant is an eQTL for TNIP1 in blood tissue (TwinsUK). Support from previous literature is sparse; however, TNIP1 has the most support of the three genes. TNIP1 contributes to hyperinflammation and has been previously identified in an autoimmune GWAS19. TNIP1 was included in a transcription module regulated by Bcl3 in mouse microglia20 where this module was implicated in prolonged exposure to inflammation and aging of microglia. The gene encoding Bcl3 (BCL3) was found to be significantly associated with cerebrospinal fluid amyloid-beta1–42 peptide after conditioning for APOE21 and was observed as upregulated in the postmortem brain of LOAD patients22. Further investigation into this locus in non-European populations may yield more support for the lead variant and improve the fine-mapping analysis.
The lead variant of locus 8 (rs6891966; P=7.91×10−10) is located in an intron of HAVCR2 (Supplementary Figure 6). HAVCR1 and TIMD4 also map to this region based on brain eQTLs (PsychENCODE). HAVCR2 was significantly differentially expressed in bulk brain tissue of LOAD patients compared to controls23. HAVCR2 is preferentially expressed in aged microglia24, was included as one of the top 100 enriched transcripts in brain and microglia, and was included in a cluster of transcripts which are involved in sensing endogenous ligands and microbes25. The protein encoded by HAVCR2 (Havcr2) has been suggested to bind to phosphatidylserine on cell surfaces to mediate apoptosis26 and to interact with amyloid precursor protein27. TIMD4 is another gene in this region which encodes a protein (TIM-4) with a similar function to Havcr2; it binds to phosphatidylserine on cell surfaces to mediate apoptosis and microglia without TIM-4 receptors have reduced apoptotic clearance28. Follow-up experimental work would be useful to determine the role that these genes play within LOAD.
Locus 12 and locus 28 have been previously associated with dementia29 but not within a previous LOAD GWAS. The lead variant in locus 12 (rs5011436; P=2.7×10−9) is an intron variant in TMEM106B (Supplementary Figure 7). A nearby exonic variant (rs3173615; R2=0.976 in 1KG EUR; P=6.61×10−9) with a CADD score of 21.2 has been discussed as the association signal driving variant in frontotemporal dementia (FTD) by causing decreased transmembrane protein 106B (the protein encoded by TMEM106B) abundance through increased protein degradation30. TMEM106B was also found to be significantly differentially expressed in bulk brain tissue of LOAD patients compared to controls23. The lead variant in locus 28 (rs708382; P=1.98×10−9) is an upstream variant of FAM171A2 (Supplementary Figure 8). Interestingly, the protein (integrin alpha-IIb) encoded by a nearby gene (ITGA2B) is a target for Abciximab, an antibody which inhibits platelet aggregation and is used to estimate concentrations of coated-platelets31. In patients with mild cognitive impairments, elevated coated-platelet levels are linked to increased risk of LOAD progression. However, the association signal in this locus colocalized with an eQTL for GRN in brain tissue (ROSMAP and BrainSeq) with the lead variant identified as the colocalized variant. GRN is also a known FTD gene32 and has the most evidence for being the causal gene in the region. The association signals in locus 12 and locus 28 do not appear to be primarily driven by the UKB data (Supplementary Note) which suggests that the associations of the known FTD genes are not driven by the proxy phenotype. These results suggest that TMEM106B and GRN are not solely contributing to FTD, but also to LOAD, implying that their biological implications might be related to protein clearance mechanisms rather than the involvement in specific disease-related protein aggregates.
The lead variant of locus 36 (rs1761461, P= 1.56×10−9) is an intergenic variant upstream of LILRA5 (Supplementary Figure 9). The lead variant is an eQTL for LILRA5, LILRP2, LILRB1, LILRA4 in GTEx whole blood. These genes encode a family of transmembrane glycoproteins which mediate immune activation33. LILRB5, LILRA5, and LILRB2 were significantly differentially expressed in bulk brain tissue of LOAD patients compared to controls23. Interestingly, LILRB2 is a nearby gene in the same family and encodes a protein (leukocyte immunoglobulin like receptor B2) known to inhibit axonal regeneration and to contribute to LOAD through amyloid binding33. The role of LILRB2 in LOAD has been investigated in mouse models and results suggest that drug targeting this gene could be a beneficial treatment approach34. While prioritizing this region to a single gene is difficult, the LILR family appears to be the most likely candidate for explaining the association signal.
Discussion
We performed a large GWAS for LOAD, including 1,126,563 individuals, and identified 38 LOAD-associated loci, including seven previously unidentified loci. The data included both clinical cases and proxy cases, defined based on parental LOAD status, a strategy that was validated previously by us8 and others35. Through gene set analysis, tissue and single cell specificity analysis, colocalization, and fine-mapping, this study highlighted additional biological routes that connect genetic variants to LOAD pathology. These functional analyses all implicated immune cells and microglia as cells of interest which provided genetic support to the current understanding of LOAD pathology36. The seven previously unidentified loci were functionally annotated and fine-mapped to help narrow down candidate causal genes. Two of the previously unidentified loci have been previously associated with frontotemporal dementia (FTD)29. This signal is not driven by the non-medically verified LOAD cases in the UKB proxy LOAD data (Supplementary Note), which suggests that this region is pleiotropic for FTD or contains separate causal variants within the same LD blocks.
A recent study7 produced a power curve for LOAD using a model which accounts for large and small effect variants. This model was based on summary statistics from a previous GWAS of LOAD10. A sample size of 2.2 million is predicted to identify 80% of genetic variance on chromosome 19 and a sample size of 7.8 million is predicted to identify 80% genetic variance outside of chromosome 19. The effective sample size35 of our meta-analysis was ~169,608, so based on previous power estimates our study was powered to explain ~6% of genetic variance outside of chromosome 19 and 58.9% of genetic variance on chromosome 19 (Supplementary Figure 10). We demonstrated that an increased sample size in a GWAS meta-analysis approach allowed for identification of previously unidentified loci; however, Holland et al. (2021)7 also predicted there to be approximately 300 large effect causal variants contributing to LOAD. These large effect variants (and small effect rare variants) are unlikely to be identified through traditional GWAS approaches focusing on common variants. Larger sample size GWAS approaches should be complemented with rare variant, copy number variant (CNV), and private variant discovery in order to identify the remaining causal variants.
Future work focusing on fine-mapping, generating larger QTL databases in more specific cells types, and incorporating other ancestries will improve the interpretability of associated loci. Our colocalization analysis identified a candidate causal gene in 9 of the 38 loci and we expect that larger and more specific QTL datasets will improve the number of successful colocalization. Yao et al. (2020)37 highlighted a need for higher sample size eQTL discovery and suggested that genes with smaller effect eQTLs are more likely to be causal for common traits. The identification of human microglia, but not bulk brain tissue, as a cell/tissue type of interest in this study supported a finding in a recent single-cell epigenomic study38, which showed that investigating individual cell types will be more fruitful than bulk brain tissue for understanding the route from variant to LOAD pathology.
One important goal for LOAD GWAS is the identification of medically actionable information that can help in diagnosis or treatment in all populations. This study was limited in the ability to identify causal genes and in the applicability to non-European populations. Further study in non-European populations will improve the equity of genetic information and also help with fine-mapping of associated regions. Larger sample sizes of GWAS, epigenomic studies, and eQTL studies in all populations will improve identification and explanation of additional LOAD loci while increasing the applicability of these findings to a larger group of individuals. This could be accomplished by a push for facilitating data-sharing and global collaboration within the field of Alzheimer’s disease genetics. The current work provided genetic support for the role of immune cells and microglia in LOAD, identified previously unidentified LOAD-associated regions, prioritized causal genes of interest, and highlighted the importance of collaboration to discern the biological process that mediate LOAD pathology.
Methods
Dataset Processing
Quality Control and Meta-analysis
The data from the participants in this study were obtained from freely available summary statistics and from genotype level data. Additional cohorts were obtained since our previous analysis8 (as well as an increased deCODE sample); these cohorts contain 12,968 additional cases and 488,616 additional controls. An overview of the cohorts is available in Supplementary Table 1. Informed consent was obtained from all participants and we complied with all relevant ethical regulations. Full description of each dataset, the quality control (QC) procedures, and the analysis protocol are available in the Supplementary Note. In short, each dataset underwent initial QC, imputation, logistic/linear regression with at least sex and principal components as covariates, and post-regression QC of the summary statistics using EasyQC39. If necessary, the data were converted to build GRCh37 before QC using the UCSC LiftOver tool40. During post-regression QC, each dataset was matched to the Haplotype Reference Consortium (HRC) or 1KG reference panel and variants with absolute allele frequency differences > 0.2 compared to the reference panel were removed. Variants with an imputation quality score < 0.8, minor allele count (MAC) < 6, N < 30, or absolute beta or SE > 10 were removed. Low minor allele frequency (MAF) variants were removed; low MAF41 was defined as . All datasets were meta-analyzed using mv-GWAMA (https://github.com/Kyoko-wtnb/mvGWAMA), a sample size weighted method previously developed in Jansen et al. (2019)8. The option to account for overlapping individuals was not utilized because no datasets were expected to contain overlapping samples and the estimates of overlapping samples (genetic covariance intercepts) were unreliable due to low heritability of the datasets. The effective sample size of the full meta-analysis for power estimates was calculated by assuming the individuals in the UKB proxy data with phenotype values <1 are controls and >=1 are cases.
Genomic risk loci definition
We used FUMA v1.3.6a42 (http://fuma.ctglab.nl) to annotate and functionally map variants included in the meta-analysis. Genomic risk loci were defined around significant variants (<5×10−8); the genomic risk loci included all variants correlated (R2>0.6) with the most significant variant. The correlation estimates were defined using 1KG European reference information43. The 1KG European reference panel was chosen over the UKB44 10K reference panel because the meta-analysis included individuals from a range of European ancestries and this diversity would be better reflected in the 1KG European sample than the primarily British UKB sample. Genomic risk loci within 250 Kb of each other are incorporated into the same locus. Previously unidentified genomic risk loci are loci which do not overlap with variants identified as significant in previous studies of LOAD8,10,45–50. Regional plots were generated using LocusZoom51 and 1KG reference information.
Heritability and genetic correlation
Linkage disequilibrium score (LDSC) regression9 (https://github.com/bulik/ldsc) was used to estimate the liability scale heritability of the non-proxy LOAD meta-analysis (UKB data excluded). The non-proxy LOAD meta-analysis (43,725 cases and 717,979 controls) was performed in the same way as the full meta-analysis described above. The UKB data (N=364,859) was excluded because LDSC liability scale heritability estimates are sensitive to sample prevalence and the UKB data was generated with a continuous phenotype and therefore a sample prevalence could not be perfectly estimated if the UKB data was included. Heritability estimates were converted to a liability scale using the LOAD population prevalence of 0.05 and a sample prevalence of 0.0574041885. LDSC12 was also used to determine the genetic correlation between a meta-analysis of the non-proxy LOAD datasets and the UKB proxy LOAD dataset. Pre-calculated LD scores for LDSC were derived from the 1KG European reference population (https://data.broadinstitute.org/alkesgroup/LDSCORE/eur_w_ld_chr.tar.bz2). Heritability and genetic correlation estimates were calculated using HapMap3 variants only. Further genetic correlations were determined using the full meta-analysis and LDhub13 (http://ldsc.broadinstitute.org/), where all 855 traits were tested using the HapMap3 variants (http://ldsc.broadinstitute.org/static/media/w_hm3.noMHC.snplist.zip). The heritability estimate of Lambert et al. (2013)10 summary statistics was obtained from LDhub.
Gene-based and gene-set analyses
Genome-wide gene association analysis was performed using MAGMA v1.0815 (http://ctg.cncr.nl/software/magma). All variants in the GWAS outside of the MHC region (GRCh37: 6:28,477,797–33,448,354) that positionally map within one of the 19,019 protein coding genes were included to estimate the significance value of that gene. Genes were considered significant if the P-value was <0.05 after Bonferroni correction for 19,019 genes. All MAGMA analyses utilized 1KG43 LD information. MAGMA gene-set analysis was performed where variants map to 15,496 gene-sets from the MSigDB v7.0 database52. Gene-sets were considered significant if the P-value was <0.05 after Bonferroni correction for the number of tested gene-sets. Forward selection of significantly associated gene-sets was performed using MAGMA v1.08 conditional analysis53. Initially the most significant gene-set was selected as a covariate and the remaining gene-sets were analyzed. The most significant gene-set from this conditional analysis was added as a covariate in addition to the previous gene-set and a new analysis was run. This process was repeated until no gene-set met the significance threshold (PBonferroni<0.05). MAGMA tissue specificity analysis was performed in FUMA using 30 general tissue type gene expression profiles (from GTEx v8). Tissues were considered significant if the P-value was < 0.05 after Bonferroni correction for 30 tissues.
FUMA cell type specificity analysis16 utilises the MAGMA gene association results to identify cell types enriched in expression of trait associated genes. We focused on brain and immune related cell types with the inclusion of pancreas as a control, therefore selecting the following scRNA-seq datasets: Allen_Human_LGN_level154, Allen_Human_LGN_level254, Allen_Human_MTG_level154, Allen_Human_MTG_level254, DroNc_Human_Hippocampus55, DroNc_Mouse_Hippocampus55, GSE104276_Human_Prefrontal_cortex_all_ages56, GSE67835_Human_Cortex57, GSE81547_Human_Pancreas58, Linnarsson_GSE101601_Human_Temporal_cortex59, MouseCellAtlas_all60, PBMC_10x_68k61, and PsychENCODE_Adult62. Within-dataset corrected results were reported to indicate which single cells are most likely to be disease relevant. The gene-based and gene-set analyses were also performed without the larger APOE region (19:40000000–50000000).
Gene mapping
The individual genomic risk loci were mapped to genes using FUMA v1.3.6a42 using positional mapping and eQTL mapping. For positional mapping, all variants within 10Kb of a gene in the genomic risk locus were assigned to that gene. For eQTL mapping, variants were mapped to genes based on significant eQTL interactions in a collection of immune and brain tissues. Brain tissue eQTLs were used due to importance of brain tissue in LOAD pathology and immune tissue/cell eQTLs were used for gene mapping because MAGMA tissue specificity analysis highlighted immune tissues as tissues of interest. The brain and immune tissues eQTLs used for mapping were: Alasoo naive macrophage63, BLUEPRINT monocyte64, BLUEPRINT neutrophil64, BLUEPRINT T-cell64, BrainSeq Brain65, CEDAR B-cell66, CEDAR monocyte, CEDAR neutrophil66, CEDAR T-cell66, Fairfax B-cell67, Fairfax naive monocyte68, GENCORD T-cell69, Kasela CD4 T-cell70, Kasela CD8 T-cell70, Lepik Blood71, Naranbhai neutrophil72, Nedelec macrophage73, Quach monocyte74, Schwartzentruber sensory neuron75, TwinsUK blood76, PsychENCODE brain62, eQTLGen blood cis and trans77, BloodeQTL blood78, BIOS Blood79, xQTLServer blood80, CommonMind Consortium brain81, BRAINEAC brain82, GTEX v8 lymphocytes, brain, spleen, and whole blood. The genes which mapped to previously unidentified loci were searched in a database (https://diegomscoelho.github.io/AD-IsoformSwitch/index.html)23 to identify if they were differential expressed in bulk brain tissue of LOAD patients compared to controls.
Colocalization
All variants within 1.5 Mb of the lead variant of each genomic risk loci were used in the colocalization analysis. The GWAS data and eQTL data were trimmed so that all variants overlap. Colocalization was performed per gene using coloc.abf from the Coloc R package18. Default priors were used for prior probability of association with the GWAS data and eQTL data. The prior probability of colocalization was set as 1×10−6 as recommended83. Nominal P, sample size, and minor allele frequency from the GWAS data and eQTL data were used in all the colocalization analyses. Colocalizations with a posterior probability > 0.8 were considered successful colocalizations. eQTL data from all tissues except microglia were obtained from the eQTL catalogue84. The microglia data were obtained from Young et al. (2019)85.
Fine-mapping
Fine-mapping was performed with susieR v0.9.186 on all variants within 1.5 Mb of the lead variant of each genomic risk loci. The APOE and HLA-DRB1 (MHC) regions were excluded from fine-mapping due to the complicated LD structure. The sample size of the fine-mapping reference panel should be proportional to the sample size of the data being fine-mapped. A good-sized reference panel is 10% to 20% the sample size of the data87. UKB data were used as a reference panel for the fine-mapping because it had the largest sample size of the available reference panels and was the only available European reference panel to fulfill the criteria for a good-sized reference panel. The reference panel was ~10% the size of the GWAS data. An LD matrix was generated using 100,000 individuals in R v3.4.388. The 100,000 individuals were chosen for each locus as the top 100,000 people with the most genotyped variants in the locus in order to maintain the highest number of variants in the fine-mapping. Only the top 100,000 were chosen for computational feasibility and in order to maintain as many variants as possible while having a large reference panel. The meta-analysis data was trimmed to match the variants included in the LD reference. The maximum number of causal variants in the region was set to 10. The susieR credible sets are reported in Supplementary Table 12. The allele frequency in the UKB data and meta-analysis data of all the variants in the fine-mapping analyses were compared to identify outliers. No variants included in the confidence set or credible set had an allele frequency difference > 0.2.
Functional enrichment of significantly associated regions
All enrichment analyses were performed using a Fisher’s exact test (fisher.test) implemented in R 4.0.188. The enrichment analyses compared all variants within the genomic risk loci (excluding the MHC region; GRCh37: 6:28,477,797–33,448,354) to all other variants present in the meta-analysis (excluding of the MHC region). Enrichment of active chromatin was performed using ROADMAP Core 15-state model annotation89 obtained from https://egg2.wustl.edu/roadmap/data/byFileType/chromhmmSegmentations/ChmmModels/coreMarks/jointModel/final/all.mnemonics.bedFiles.tgz . For each of the 127 cell types, all variants within the analysis were annotated with one of the 15 states using the R package Genomic Ranges90. All variants annotated with a state < 8 were defined as being within active chromatin. The enrichment of active chromatin within the specified region was performed for each of the cell types and the resulting P-values were corrected for 127 tests using Bonferroni correction. To perform enrichments of functional consequences, variants were annotated with ANNOVAR91 using ANNOVAR and FASTA sequences for all annotated transcripts in RefSeq Gene92. Enrichments were considered significant if the P-value was < 0.05 after Bonferroni correction for 11 functional consequences. The enrichment plots were generated using the R package ggplot293.
Statistics & Reproducibility
No statistical method was used to predetermine sample size, all available datasets were included in the meta-analysis. Exclusion of data was predetermined and based on quality control procedures outlined in the Supplementary Note. Phenotype values were assigned based on (parental) diagnoses so the experiments were not randomized. The investigators were not blinded to allocation during experiments and outcome assessment. Scientific findings were compared to findings from previous LOAD meta-analyses. Replication of previously identified loci is reported in the Main Text and Supplementary Note.
Data Availability Statement
Access to raw data can be requested via the Psychiatric Genomics Data Access portal https://www.med.unc.edu/pgc/shared-methods/open-source-philosophy/), UKBiobank (www.ukbiobank.ac.uk), or 23andMe. Restriction of raw data is to protect the privacy of participants. Summary statistics from IGAP (https://web.pasteur-lille.fr/en/recherche/u744/igap/igap_download.php) and Finngen (https://www.finngen.fi/en/access_results) can be obtained from their respective online portals. Summary statistics from the meta-analysis excluding 23andMe are available at https://ctg.cncr.nl/software/summary_statistics. Access to the full set including 23andMe results can be obtained after the approval from 23andMe is presented to the corresponding author. Approval can be obtained by completion of a Data Transfer Agreement. The Data Transfer Agreement exists to protect the privacy of 23andMe participants. Please visit https://research.23andme.com/dataset-access/ to initiate a request. Summary statistics of the primary microglia eQTLs are also available from EGA (Accession ID: EGAD00001005736). MSigDB gene-sets are available online (https://www.gsea-msigdb.org/gsea/msigdb/) and integrated in FUMA (https://fuma.ctglab.nl/).
Code Availability Statement
The code used to perform the analyses is available at https://github.com/dwightman/PGC-ALZ2. All software used in the analyses is freely available online.
Supplementary Material
Acknowledgements
Thank you to all the participants included in this study including the participants from Finngen, GR@CE, IGAP, UKB, DemGene, TwinGene, STSA, the Gothenburg H70 Birth Cohort Studies and Clinical AD from Sweden, ANMerge, BioVU, 23andMe, HUNT, and deCODE.
We thank the research participants from 23andMe who made this study possible. Members of the 23andMe Research Team are: Michelle Agee, Stella Aslibekyan, Elizabeth Babalola, Robert K. Bell, Jessica Bielenberg, Katarzyna Bryc, Emily Bullis, Briana Cameron, Daniella Coker, Gabriel Cuellar Partida, Devika Dhamija, Sayantan Das, Sarah L. Elson, Teresa Filshtein, Kipper Fletez-Brant, Pierre Fontanillas, Will Freyman, Pooja M. Gandhi, Barry Hicks, David A. Hinds, Karen E. Huber, Ethan M. Jewett, Yunxuan Jiang, Aaron Kleinman, Katelyn Kukar, Vanessa Lane, Keng-Han Lin, Maya Lowe, Marie K. Luff, Jennifer C. McCreight, Matthew H. McIntyre, Kimberly F. McManus, Steven J. Micheletti, Meghan E. Moreno, Joanna L. Mountain, Sahar V. Mozaffari, Priyanka Nandakumar, Elizabeth S. Noblin, Jared O’Connell, Aaron A. Petrakovitz, G. David Poznik, Morgan Schumacher, Anjali J. Shastri, Janie F. Shelton, Jingchunzi Shi, Suyash Shringarpure, Chao Tian, Vinh Tran, Joyce Y. Tung, Xin Wang, Wei Wang, Catherine H. Weldon, and Peter Wilton.
The authors would like to thank the participants of the Norwegian Dementia Genetics Network (DemGene). This work was supported by the Research Council of Norway (RCN; 248980, 248778, 223273), Norwegian Regional Health Authorities, Norwegian Health Association (22731), EU JPND: PMI-AD RCN 311993. National Institutes of Health, National Institute on Aging R01 AG08724, R01 AG17561, R01 AG028555, and R01 AG060470. DP was supported by the European Research Council advanced grant (grant no. ERC-2018AdG GWAS2FUNC 834057)
We thank the International Genomics of Alzheimer’s Project (IGAP) for providing summary results data for these analyses. The investigators within IGAP contributed to the design and implementation of IGAP and/or provided data but did not participate in analysis or writing of this report. IGAP was made possible by the generous participation of the control subjects, the patients, and their families. The i–Select chips was funded by the French National Foundation on Alzheimer’s disease and related disorders. EADI was supported by the LABEX (laboratory of excellence program investment for the future) DISTALZ grant, Inserm, Institut Pasteur de Lille, Université de Lille 2 and the Lille University Hospital. GERAD/PERADES was supported by the Medical Research Council (Grant n° 503480), Alzheimer’s Research UK (Grant n° 503176), the Wellcome Trust (Grant n° 082604/2/07/Z) and German Federal Ministry of Education and Research (BMBF): Competence Network Dementia (CND) grant n° 01GI0102, 01GI0711, 01GI0420. CHARGE was partly supported by the NIH/NIA grant R01 AG033193 and the NIA AG081220 and AGES contract N01–AG– 12100, the NHLBI grant R01 HL105756, the Icelandic Heart Association, and the Erasmus Medical Center and Erasmus University. ADGC was supported by the NIH/NIA grants: U01 AG032984, U24 AG021886, U01 AG016976, and the Alzheimer’s Association grant ADGC– 10–196728.
HZ has served at scientific advisory boards for Denali, Roche Diagnostics, Wave, Samumed, Siemens Healthineers, Pinteon Therapeutics and CogRx, has given lectures in symposia sponsored by Fujirebio, Alzecure and Biogen, and is a co-founder of Brain Biomarker Solutions in Gothenburg AB (BBS), which is a part of the GU Ventures Incubator Program (outside submitted work).
KB has served as a consultant, at advisory boards, or at data monitoring committees for Abcam, Axon, Biogen, JOMDD/Shimadzu. Julius Clinical, Lilly, MagQu, Novartis, Roche Diagnostics, and Siemens Healthineers, and is a co-founder of Brain Biomarker Solutions in Gothenburg AB (BBS), which is a part of the GU Ventures Incubator Program.
OAA is a consultant to HealthLytix, and received speaker’s honorarium from Lundbeck and Sunovion. All other authors declare no financial interests or potential conflicts of interest.
JBN is employed by Regeneron Pharmaceuticals, Inc.
TWM has received speaker’s honorarium from Roche.
Footnotes
Competing Interests Statement
All other authors declare no competing interests.
References
- 1.Bacigalupo I.et al. A Systematic Review and Meta-Analysis on the Prevalence of Dementia in Europe: Estimates from the Highest-Quality Studies Adopting the DSM IV Diagnostic Criteria. J. Alzheimers. Dis 66, 1471–1481 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Winblad B.et al. Defeating Alzheimer’s disease and other dementias: a priority for European science and society. Lancet. Neurol 15, 455–532 (2016). [DOI] [PubMed] [Google Scholar]
- 3.DeTure MA & Dickson DW The neuropathological diagnosis of Alzheimer’s disease. Mol. Neurodegener 14, 32 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gatz M.et al. Heritability for Alzheimer’s disease: the study of dementia in Swedish twins. J. Gerontol. A. Biol. Sci. Med. Sci 52, M117–25 (1997). [DOI] [PubMed] [Google Scholar]
- 5.Gatz M.et al. Role of genes and environments for explaining Alzheimer disease. Arch. Gen. Psychiatry 63, 168–174 (2006). [DOI] [PubMed] [Google Scholar]
- 6.Zhang Q.et al. Risk prediction of late-onset Alzheimer’s disease implies an oligogenic architecture. Nat. Commun 11, 4799 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holland D.et al. The genetic architecture of human complex phenotypes is modulated by linkage disequilibrium and heterozygosity. Genetics 217, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jansen IE et al. Genome-wide meta-analysis identifies new loci and functional pathways influencing Alzheimer’s disease risk. Nat. Genet 51, 404–413 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bulik-Sullivan BK et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet 47, 291–295 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lambert J-C et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat. Genet 45, 1452–1458 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Bakker PIW et al. Practical aspects of imputation-driven meta-analysis of genome-wide association studies. Hum. Mol. Genet 17, R122–R128 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bulik-Sullivan B.et al. An atlas of genetic correlations across human diseases and traits. Nat. Genet 47, 1236–1241 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zheng J.et al. LD Hub: a centralized database and web interface to perform LD score regression that maximizes the potential of summary level GWAS data for SNP heritability and genetic correlation analysis. Bioinformatics 33, 272–279 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kunkle BW et al. Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Aβ, tau, immunity and lipid processing. Nat. Genet 51, 414–430 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Leeuw CA, Mooij JM, Heskes T.& Posthuma D.MAGMA: Generalized Gene-Set Analysis of GWAS Data. PLOS Comput. Biol 11, e1004219 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Watanabe K, Umićević Mirkov M, de Leeuw CA, van den Heuvel MP & Posthuma D.Genetic mapping of cell type specificity for complex traits. Nat. Commun 10, 3222 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wishart DS et al. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 46, D1074–D1082 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giambartolomei C.et al. Bayesian Test for Colocalisation between Pairs of Genetic Association Studies Using Summary Statistics. PLOS Genet. 10, e1004383 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shamilov R.& Aneskievich BJ TNIP1 in Autoimmune Diseases: Regulation of Toll-like Receptor Signaling. J. Immunol. Res 2018, 3491269 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cho CE et al. A modular analysis of microglia gene expression, insights into the aged phenotype. BMC Genomics 20, 164 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nho K.et al. Association analysis of rare variants near the APOE region with CSF and neuroimaging biomarkers of Alzheimer’s disease. BMC Med. Genomics 10, 29 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li X.et al. Systematic Analysis and Biomarker Study for Alzheimer’s Disease. Sci. Rep 8, 17394 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marques-Coelho D.et al. Differential transcript usage unravels gene expression alterations in Alzheimer’s disease human brains. npj Aging Mech. Dis 7, 2 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Olah M.et al. A transcriptomic atlas of aged human microglia. Nat. Commun 9, 539 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hickman SE et al. The microglial sensome revealed by direct RNA sequencing. Nat. Neurosci 16, 1896–1905 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nam KN et al. Effect of high fat diet on phenotype, brain transcriptome and lipidome in Alzheimer’s model mice. Sci. Rep 7, 4307 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oláh J.et al. Interactions of pathological hallmark proteins: tubulin polymerization promoting protein/p25, beta-amyloid, and alpha-synuclein. J. Biol. Chem 286, 34088– 34100 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mazaheri F.et al. Distinct roles for BAI1 and TIM-4 in the engulfment of dying neurons by microglia. Nat. Commun 5, 4046 (2014). [DOI] [PubMed] [Google Scholar]
- 29.Ciani M, Benussi L, Bonvicini C.& Ghidoni R.Genome Wide Association Study and Next Generation Sequencing: A Glimmer of Light Toward New Possible Horizons in Frontotemporal Dementia Research. Front. Neurosci 13, 506 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li Z.et al. The TMEM106B FTLD-protective variant, rs1990621, is also associated with increased neuronal proportion. Acta Neuropathol. 139, 45–61 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prodan CI et al. Coated-platelet levels and progression from mild cognitive impairment to Alzheimer disease. Neurology 76, 247–252 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Greaves CV & Rohrer JD An update on genetic frontotemporal dementia. J. Neurol 266, 2075–2086 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang J.et al. Leukocyte immunoglobulin-like receptors in human diseases: an overview of their distribution, function, and potential application for immunotherapies. J. Leukoc. Biol 102, 351–360 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cao Q.et al. Inhibiting amyloid-β cytotoxicity through its interaction with the cell surface receptor LilrB2 by structure-based design. Nat. Chem 10, 1213–1221 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu JZ, Erlich Y.& Pickrell JK Case-control association mapping by proxy using family history of disease. Nat. Genet 49, 325–331 (2017). [DOI] [PubMed] [Google Scholar]
- 36.Schwabe T, Srinivasan K.& Rhinn H.Shifting paradigms: The central role of microglia in Alzheimer’s disease. Neurobiol. Dis 143, 104962 (2020). [DOI] [PubMed] [Google Scholar]
- 37.Yao DW, O’Connor LJ, Price AL & Gusev A.Quantifying genetic effects on disease mediated by assayed gene expression levels. Nat. Genet 52, 626–633 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Corces MR et al. Single-cell epigenomic analyses implicate candidate causal variants at inherited risk loci for Alzheimer’s and Parkinson’s diseases. Nat. Genet 52, 1158–1168 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Winkler TW et al. Quality control and conduct of genome-wide association meta-analyses. Nat. Protoc 9, 1192–1212 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kuhn RM, Haussler D.& Kent WJ The UCSC genome browser and associated tools. Brief. Bioinform 14, 144–161 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ionita-Laza I, Lee S, Makarov V, Buxbaum JD & Lin X.Sequence Kernel Association Tests for the Combined Effect of Rare and Common Variants. Am. J. Hum. Genet 92, 841–853 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Watanabe K, Taskesen E, van Bochoven A.& Posthuma D.Functional mapping and annotation of genetic associations with FUMA. Nat. Commun 8, 1826 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Auton A.et al. A global reference for human genetic variation. Nature 526, 68–74 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sudlow C.et al. UK Biobank: An Open Access Resource for Identifying the Causes of a Wide Range of Complex Diseases of Middle and Old Age. PLOS Med. 12, e1001779 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu P-P, Xie Y, Meng X-Y & Kang J-S History and progress of hypotheses and clinical trials for Alzheimer’s disease. Signal Transduct. Target. Ther 4, 29 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marioni RE et al. GWAS on family history of Alzheimer’s disease. Transl. Psychiatry 8, 99 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Desikan RS et al. Genetic assessment of age-associated Alzheimer disease risk: Development and validation of a polygenic hazard score. PLOS Med. 14, e1002258 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jun G.et al. A novel Alzheimer disease locus located near the gene encoding tau protein. Mol. Psychiatry 21, 108–117 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.de Rojas I.et al. Common variants in Alzheimer's disease: Novel association of six genetic variants with AD and risk stratification by polygenic risk scores. medRxiv 19012021 (2020) doi: 10.1101/19012021. [DOI] [Google Scholar]
- 50.Schwartzentruber J.et al. Genome-wide meta-analysis, fine-mapping and integrative prioritization implicate new Alzheimer’s disease risk genes. Nat. Genet 53, 392–402 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pruim RJ et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics 26, 2336–2337 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liberzon A.et al. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 1, 417–425 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.de Leeuw CA, Stringer S, Dekkers IA, Heskes T.& Posthuma D.Conditional and interaction gene-set analysis reveals novel functional pathways for blood pressure. Nat. Commun 9, 3768 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hodge RD et al. Conserved cell types with divergent features in human versus mouse cortex. Nature 573, 61–68 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Habib N.et al. Massively parallel single-nucleus RNA-seq with DroNc-seq. Nat. Methods 14, 955–958 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhong S.et al. A single-cell RNA-seq survey of the developmental landscape of the human prefrontal cortex. Nature 555, 524–528 (2018). [DOI] [PubMed] [Google Scholar]
- 57.Darmanis S.et al. A survey of human brain transcriptome diversity at the single cell level. Proc. Natl. Acad. Sci. U. S. A 112, 7285–7290 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Enge M.et al. Single-Cell Analysis of Human Pancreas Reveals Transcriptional Signatures of Aging and Somatic Mutation Patterns. Cell 171, 321–330.e14 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hochgerner H.et al. STRT-seq-2i: dual-index 5’ single cell and nucleus RNA-seq on an addressable microwell array. Sci. Rep 7, 16327 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Han X.et al. Mapping the Mouse Cell Atlas by Microwell-Seq. Cell 172, 1091–1107.e17 (2018). [DOI] [PubMed] [Google Scholar]
- 61.Zheng GXY et al. Massively parallel digital transcriptional profiling of single cells. Nat. Commun 8, 14049 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang D.et al. Comprehensive functional genomic resource and integrative model for the human brain. Science 362, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Alasoo K.et al. Shared genetic effects on chromatin and gene expression indicate a role for enhancer priming in immune response. Nat. Genet 50, 424–431 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen L.et al. Genetic Drivers of Epigenetic and Transcriptional Variation in Human Immune Cells. Cell 167, 1398–1414.e24 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jaffe AE et al. Developmental and genetic regulation of the human cortex transcriptome illuminate schizophrenia pathogenesis. Nat. Neurosci 21, 1117–1125 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Momozawa Y.et al. IBD risk loci are enriched in multigenic regulatory modules encompassing putative causative genes. Nat. Commun 9, 2427 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fairfax BP et al. Genetics of gene expression in primary immune cells identifies cell type–specific master regulators and roles of HLA alleles. Nat. Genet 44, 502–510 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fairfax BP et al. Innate Immune Activity Conditions the Effect of Regulatory Variants upon Monocyte Gene Expression. Science (80-. ) 343, 1246949 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gutierrez-Arcelus M.et al. Passive and active DNA methylation and the interplay with genetic variation in gene regulation. Elife 2, e00523 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kasela S.et al. Pathogenic implications for autoimmune mechanisms derived by comparative eQTL analysis of CD4+ versus CD8+ T cells. PLOS Genet. 13, e1006643 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lepik K.et al. C-reactive protein upregulates the whole blood expression of CD59 - an integrative analysis. PLOS Comput. Biol 13, e1005766 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Naranbhai V.et al. Genomic modulators of gene expression in human neutrophils. Nat. Commun 6, 7545 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nédélec Y.et al. Genetic Ancestry and Natural Selection Drive Population Differences in Immune Responses to Pathogens. Cell 167, 657–669.e21 (2016). [DOI] [PubMed] [Google Scholar]
- 74.Quach H.et al. Genetic Adaptation and Neandertal Admixture Shaped the Immune System of Human Populations. Cell 167, 643–656.e17 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schwartzentruber J.et al. Molecular and functional variation in iPSC-derived sensory neurons. Nat. Genet 50, 54–61 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Buil A.et al. Gene-gene and gene-environment interactions detected by transcriptome sequence analysis in twins. Nat. Genet 47, 88–91 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Võsa U.et al. Unraveling the polygenic architecture of complex traits using blood eQTL metaanalysis. bioRxiv 447367 (2018) doi: 10.1101/447367. [DOI] [Google Scholar]
- 78.Westra H-J et al. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat. Genet 45, 1238–1243 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhernakova DV et al. Identification of context-dependent expression quantitative trait loci in whole blood. Nat. Genet 49, 139–145 (2017). [DOI] [PubMed] [Google Scholar]
- 80.Ng B.et al. An xQTL map integrates the genetic architecture of the human brain’s transcriptome and epigenome. Nat. Neurosci 20, 1418–1426 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fromer M.et al. Gene expression elucidates functional impact of polygenic risk for schizophrenia. Nat. Neurosci 19, 1442–1453 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ramasamy A.et al. Genetic variability in the regulation of gene expression in ten regions of the human brain. Nat. Neurosci 17, 1418–1428 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wallace C.Eliciting priors and relaxing the single causal variant assumption in colocalisation analyses. PLOS Genet. 16, e1008720 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kerimov N.et al. eQTL Catalogue: a compendium of uniformly processed human gene expression and splicing QTLs. bioRxiv 2020.01.29.924266 (2020) doi: 10.1101/2020.01.29.924266. [DOI] [Google Scholar]
- 85.Young AMH et al. A map of transcriptional heterogeneity and regulatory variation in human microglia. bioRxiv 2019.12.20.874099 (2019) doi: 10.1101/2019.12.20.874099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang G, Sarkar A, Carbonetto P.& Stephens M.A simple new approach to variable selection in regression, with application to genetic fine mapping. J. R. Stat. Soc. Ser. B (Statistical Methodol n/a, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Benner C.et al. Prospects of Fine-Mapping Trait-Associated Genomic Regions by Using Summary Statistics from Genome-wide Association Studies. Am. J. Hum. Genet 101, 539–551 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; (2017). [Google Scholar]
- 89.Kundaje A.et al. Integrative analysis of 111 reference human epigenomes. Nature 518, 317–330 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lawrence M.et al. Software for Computing and Annotating Genomic Ranges. PLOS Comput. Biol 9, e1003118 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wang K, Li M.& Hakonarson H.ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38, e164–e164 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.O’Leary NA et al. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 44, D733–D745 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wickham Hadley. ggplot2: Elegant Graphics for Data Analysis. (Springer-Verlag New York, 2016). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Access to raw data can be requested via the Psychiatric Genomics Data Access portal https://www.med.unc.edu/pgc/shared-methods/open-source-philosophy/), UKBiobank (www.ukbiobank.ac.uk), or 23andMe. Restriction of raw data is to protect the privacy of participants. Summary statistics from IGAP (https://web.pasteur-lille.fr/en/recherche/u744/igap/igap_download.php) and Finngen (https://www.finngen.fi/en/access_results) can be obtained from their respective online portals. Summary statistics from the meta-analysis excluding 23andMe are available at https://ctg.cncr.nl/software/summary_statistics. Access to the full set including 23andMe results can be obtained after the approval from 23andMe is presented to the corresponding author. Approval can be obtained by completion of a Data Transfer Agreement. The Data Transfer Agreement exists to protect the privacy of 23andMe participants. Please visit https://research.23andme.com/dataset-access/ to initiate a request. Summary statistics of the primary microglia eQTLs are also available from EGA (Accession ID: EGAD00001005736). MSigDB gene-sets are available online (https://www.gsea-msigdb.org/gsea/msigdb/) and integrated in FUMA (https://fuma.ctglab.nl/).