Abstract
In addition to maintaining the GenBank® nucleic acid sequence database, the National Center for Biotechnology Information (NCBI) provides data analysis and retrieval and resources that operate on the data in GenBank and a variety of other biological data made available through NCBI’s Web site. NCBI data retrieval resources include Entrez, PubMed, LocusLink and the Taxonomy Browser. Data analysis resources include BLAST, Electronic PCR, OrfFinder, RefSeq, UniGene, Database of Single Nucleotide Polymorphisms (dbSNP), Human Genome Sequencing pages, GeneMap’99, Davis Human–Mouse Homology Map, Cancer Chromosome Aberration Project (CCAP) pages, Entrez Genomes, Clusters of Orthologous Groups (COGs) database, Retroviral Genotyping Tools, Cancer Genome Anatomy Project (CGAP) pages, SAGEmap, Online Mendelian Inheritance in Man (OMIM) and the Molecular Modeling Database (MMDB). Augmenting many of the Web applications are custom implementations of the BLAST program optimized to search specialized data sets. All of the resources can be accessed through the NCBI home page at: http://www.ncbi.nlm.nih.gov
INTRODUCTION
The National Center for Biotechnology Information (NCBI) at the National Institutes of Health was created in 1988 to develop information systems in the field of molecular biology. In addition to maintaining the GenBank® (1) nucleic acid sequence database, for which data is submitted directly by the scientific community, NCBI provides data analysis and retrieval and resources that operate on GenBank data and a variety of other biological data made available through NCBI.
The data accessible from NCBI’s home page (http://www.ncbi.nlm.nih.gov ) runs the gamut from short sequences representative of parts of genes, such as expressed sequence tags (ESTs), through complete genomic sequences, such as the 21 complete microbial genomes in GenBank today, to phenotypic descriptions such as those found in Online Mendelian Inheritance in Man (OMIM). Protein structures are linked to sequence data through the Molecular Modeling Database (MMDB). To access this data, NCBI offers powerful retrieval and search tools, such as Entrez and BLAST. NCBI also offers an array of computational resources to aid in the analysis of each type of data. For this overview, the NCBI suite of database resources is grouped into seven categories: database retrieval systems, sequence similarity search programs, resources for analysis of gene-level sequences, resources for chromosomal sequences, resources for genome-scale analysis, resources for the analysis of gene expression and phenotypes, and resources for protein structure and modeling. Table 1 provides an at-a-glance summary of these resources.
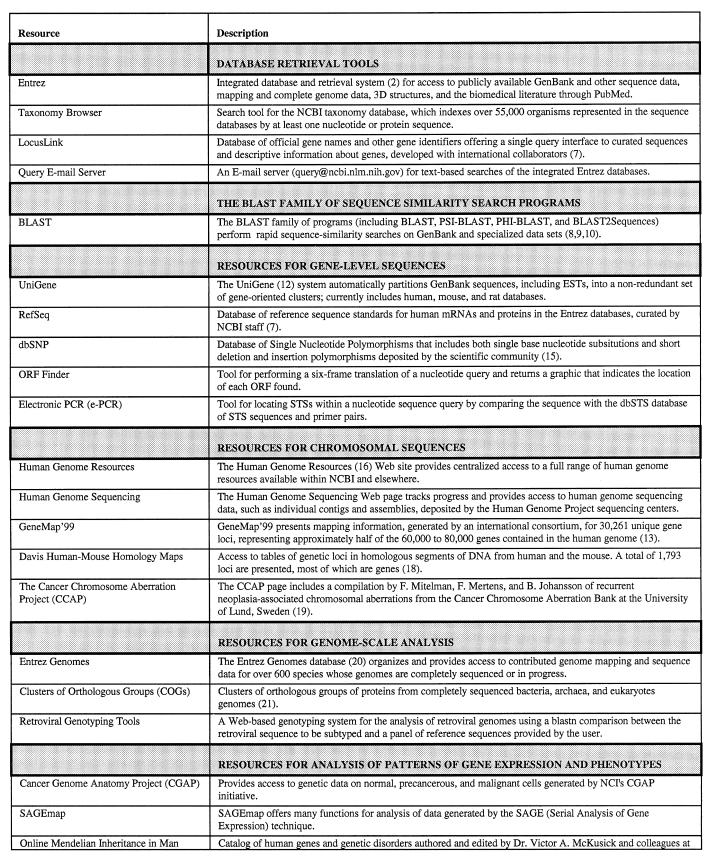
Table 1. A summary of selected Web-based data resources, in addition to GenBank, provided by the NCBI.
DATABASE RETRIEVAL TOOLS
Entrez
Entrez (2) is an integrated database retrieval system that accesses DNA and protein sequence data, genome mapping, population sets, protein structures from MMDB (3) and the biomedical literature via PubMed, with embedded links to the NCBI taxonomy. The sequences in Entrez, especially protein sequences, are obtained from a variety of database sources [including GenBank protein translations, Protein Identification Resource (4), SWISS-PROT (5), Protein Research Foundation, Protein Data Bank (6) and RefSeq (7)], and therefore include more sequence data than GenBank alone. PubMed includes primarily the 10 million references and abstracts in MEDLINE®, with links to the full-text of nearly 500 journals available on the Web.
Entrez provides text searching of sequence or bibliographic records by simple Boolean queries, plus extensive links to related information. Some links are simple cross-references, for example, from a sequence to the abstract of the paper in which it was reported, from a protein sequence and its corresponding DNA sequence, or to alignments with other sequences. Other links are based on computed similarities among the sequences or MEDLINE abstracts. The resulting pre-computed document ‘neighbors’ allow rapid access for browsing groups of related records. A project called LinkOut was recently implemented to expand the range of external links from individual database records to related outside services, including organism-specific genome databases.
The Taxonomy Browser
The NCBI taxonomy database indexes over 55 000 organisms that are represented in the sequence databases with at least one nucleotide or protein sequence. The Taxonomy Browser can be used to view the taxonomic position or retrieve sequence and structural data for a particular organism or group of organisms. Searches of the NCBI taxonomy may be made on the basis of whole or partial organism names, and direct links to organisms commonly used in biological research are also provided. The Taxonomy Browser can also be used to display the number of nucleic acid sequences, protein sequences, and protein structures available for organisms included in the branch. From the data display for a particular organism, one can retrieve and download the sequence data for that organism, or protein 3D structure data if available.
LocusLink
The LocusLink database of official gene names and other gene identifiers, described elsewhere in this issue (7), was developed at NCBI in conjunction with several international collaborators, offers a single query interface to curated sequences and descriptive information about genes.
The QUERY Email server
The QUERY Email server (query@ncbi.nlm.nih.gov ) performs text-based searches of the integrated Entrez databases. In addition to the records that match the search query, the pre-computed document neighbors for retrieved records are also accessible. Various output formats, such as FASTA for sequence data, are available.
THE BLAST FAMILY OF SEQUENCE-SIMILARITY SEARCH PROGRAMS
The most frequent type of analysis performed on GenBank data is the search for sequences similar to a query sequence. NCBI offers the BLAST family of search programs (8,9) for this purpose. NCBI’s Web interface to the standard BLAST 2.0 program accepts a sequence or accession number as the input query. The search for similarity, performed using an identity matrix for blastn (nucleotide) searches and a PAM or BLOSUM scoring matrix for protein searches, results in a set of gapped alignments, with links to the full document records. Each BLAST alignment is accompanied by an alignment score and a measure of statistical significance, called the Expectation Value, for judging the quality of the alignment. Web BLAST also provides a graphical overview of the alignments, which are color-coded by alignment score and clearly show the extent and quality of the sequence similarities detected by BLAST, as well as the disposition of gaps in the alignments.
The default databases searched by BLAST are the non-redundant (nr) nucleotide and protein databases constructed from the Entrez databases. Several pre-defined specialized databases or subsets may also be searched, and searches may be restricted to sequences from a particular organism. Customized BLAST pages allow a nucleotide query against any combination of 21 complete and 40 incomplete microbial genomes, or against the genomes of malaria-associated pathogens.
Specialized versions of BLAST are also offered to facilitate other approaches to protein similarity searching. Position Specific Iterated BLAST (PSI-BLAST) (9) initially performs a conventional BLAST search to produce alignments from which it constructs a position specific profile. Subsequent BLAST iterations use this profile matrix in place of the initial query and scoring matrix to find similarities in a database. Pattern Hit Initiated BLAST (PHI-BLAST) (10) takes as input both a peptide query sequence and a peptide pattern, or motif, found within the peptide query sequence. The motif specifies an obligatory match between query and database sequences, about which optimal local alignments are constructed. Another variant, ‘BLAST2Sequences’ (11), can display the similarity between two DNA or peptide sequences by producing a dot-plot representation of the alignments it reports.
Basic BLAST 2.0 searches can also be performed by Email through the address: blast@ncbi.nlm.nih.gov . Documentation can be obtained by sending the word ‘help’ to the server address.
RESOURCES FOR GENE-LEVEL SEQUENCES
UniGene
To manage the redundancy of the EST datasets and facilitate the extraction of information, NCBI has developed UniGene (12), a system for automatically partitioning GenBank sequences, including ESTs, into a non-redundant set of gene-oriented clusters. There are currently three UniGene databases, for human, mouse and rat. UniGene starts with entries in the Primate (or Rodent) division of GenBank, combines these with ESTs of the specific organism, and creates clusters of sequences that share virtually identical 3′ untranslated regions (3′ UTRs). Each UniGene cluster contains sequences that represent a unique gene, and is linked to related information, such as the tissue types in which the gene is expressed and the map location of the gene. For the human UniGene database, over 1.5 million human ESTs in GenBank have been reduced 18-fold in number to approximately 83 000 sequence clusters. In a similar fashion, the mouse and rat ESTs have been organized as 25 000 and 27 000 clusters, respectively. The human UniGene collection has been effectively used as a source of mapping candidates for the construction of a human gene map (13). In this case, the 3′ UTRs of genes and ESTs are converted to sequence-tagged sites (STSs) that are then placed on physical maps and integrated with pre-existing genetic maps of the genome. The UniGene collection has also been used as a source of unique sequences for the fabrication of ‘chips’ for the large-scale study of gene expression (14). UniGene databases are updated weekly with new EST sequences, and bimonthly with newly characterized sequences. UniGene clusters may be searched in several ways, including gene name, chromosome, cDNA library, accession number and ordinary text words. They may also be downloaded by FTP.
RefSeq database of reference sequence standards
The References Sequence (RefSeq) database, described elsewhere in this issue (7), provides reference sequences for human mRNAs and proteins.
Database of Single Nucleotide Polymorphisms (dbSNP)
The database of Single Nucleotide Polymorphisms (dbSNP), described elsewhere in this issue (15), serves as a repository for both single base nucleotide substitutions and short deletion and insertion polymorphisms that are deposited by the research community.
ORF Finder
An early step in the analysis of a nucleic acid sequence suspected of containing a gene is to perform a search for Open Reading Frames (ORFs). ORF Finder performs a six-frame translation of a nucleotide query and returns a graphic that indicates the location of each ORF found. Restrictions on the size of the ORFs returned may be set by the user. The sequences of predicted protein products can be submitted directly for BLAST similarity searching.
Electronic PCR
PCR-based assays for STSs can be used for gene identification and mapping. Electronic PCR (e-PCR) is a tool for locating STSs within a nucleotide sequence by comparing the query against the dbSTS database of STS sequences and primer pairs. The e-PCR application accepts either an accession number or sequence as input, and returns a table of links to matching dbSTS records as well as the primer pairs used to amplify each STS identified.
RESOURCES FOR CHROMOSOMAL SEQUENCES
Collection of Human Genome Resources
The Human Genome Resources (16) page provides centralized access to a full range of human genome resources available from NCBI and elsewhere, including core research resources for human genes, sequences, maps, genetic variations and gene expression described below. It also includes the Genes and Disease site, which has general synopses of over 60 diseases of genetic origin.
Human Genome Sequencing
The Human Genome Sequencing site shows chromosome-specific progress of the human sequencing project, provides access to individual contigs and assemblies, and offers chromosome-specific BLAST searches. Links to contributing genome sequencing centers are also provided. Sequence data may be downloaded by contig or chromosome.
GeneMap’99
An international consortium was formed in 1994 to construct a human gene map by determining the locations of ESTs relative to a framework of well-characterized genetic markers (17). The current version of this map is the radiation hybrid map, GeneMap’99 (13). GeneMap’99 features 30 261 unique gene loci representing approximately half of the 60 000–80 000 genes thought to be contained in the human genome. Mapped genes and STSs can be searched by marker name, gene symbol, accession number or UniGene identifier.
The Davis Human–Mouse Homology Maps
NCBI provides the Web access to the Davis Human–Mouse Homology Maps (18), which are tables of genetic loci in homologous segments of DNA from human and the mouse. A total of 1793 loci are presented, most of which are genes. The homology maps are linked to GeneMap’99, OMIM, and the Mouse Genome Database at The Jackson Laboratory.
The Cancer Chromosome Aberration Project (CCAP)
The CCAP service is an initiative of the National Cancer Institute (NCI) and NCBI. The data includes a compilation by F. Mitelman, F. Mertens and B. Johansson of recurrent neoplasia-associated chromosomal aberrations from the Cancer Chromosome Aberration Bank at the University of Lund, Sweden (19). Also provided are bacterial artificial chromosome (BAC) human chromosome mapping data provided through CCAP’s fluorescent in situ hybridization (FISH) effort.
RESOURCES FOR GENOME-SCALE ANALYSIS
Entrez Genomes
The Entrez Genomes database (20) provides access to genomic data contributed by the scientific community for over 600 species whose sequencing and mapping is complete or in progress, including 21 complete microbial genomes. It offers a variety of integrated views for whole genomes, complete chromosomes, contiged sequence maps. Each genome is represented as a clickable graphic for quick navigation to more detailed representations of smaller regions. The detailed views depict ORFs, each of which has a set of alignments to similar proteins, highlighted by taxonomic kingdom.
For each genome, a Coding Regions function displays the location of each coding region, length of the product, GenBank identification number for the protein sequence, and name of the protein product. An RNA Genes function lists the location and gene names for ribosomal and transfer RNA genes.
The TaxTables function, available for all complete microbial genomes in GenBank, compares the proteome of an organism to those for organisms from the three taxonomic kingdoms of life, Archaea, Eubacteria and Eukaryota. The taxonomic distribution of the homologs across the three kingdoms can be displayed at various levels of detail. For each translation product in the genome, the TaxTable lists the most similar protein from each of the three kingdoms.
Chromosome mapping data for human and other eukaryotes includes genetic, cytogenetic, physical and sequence maps that have been integrated and graphically represented to show common markers. Each chromosome can be viewed in its entirety or explored in progressively greater detail.
Clusters of Orthologous Groups (COGs)
The COGs Web page, described elsewhere in this issue (21), presents a compilation of orthologous groups of proteins from completely sequenced organisms representing phylogenetically distant clades.
Retroviral genotyping tools
Genotyping retrovirus sequences has a number of implications for attempts at characterization of viral genetic diversity, tracking of the epidemics, and in the case of HIV-1, with an impact on the vaccine development. NCBI has developed a Web-based genotyping tool for the analysis of retroviral genomes. The genotyping method employs a blastn comparison between the retroviral sequence to be subtyped and a panel of reference sequences provided by the user. An HIV-1-specific subtyping tool uses a set of reference sequences taken from the principle HIV-1 variants. The subtyping panel includes complete genomic references for the nine clades (A–J) of group M, as well as for the groups O and N. Because retroviruses are highly recombinogenic, a substantial number of HIV-1 genomes are mosaics of subtypes. The tool helps detect HIV-1 recombinant genomes by performing local blastn comparisons over a sliding window of size and step values set by the user.
RESOURCES FOR ANALYSIS OF PATTERNS OF GENE EXPRESSION AND PHENOTYPES
The Cancer Genome Anatomy Project (CGAP)
The CGAP service provides access to genetic data on normal, precancerous and malignant cells generated by the NCI’s CGAP initiative. CGAP cDNA library information may be retrieved by text words using an interactive Library Browser, by selecting from a summary table that organizes the libraries by gene, clone, tissue type, method of sample preparation and stage of tumor development, or by UniGene Cluster ID (via the GeneExpress interface). Expression profiles of cDNA libraries may be compared using either the Digital Differential Display (DDD) tool or the xProfiler. CGAP also includes a directory of tumor suppressor genes and oncogenes.
SAGEmap
Serial Analysis of Gene Expression (SAGE) refers to a technique for taking a snapshot of the messenger RNA population of a cell to obtain a quantitative measure of gene expression. NCBI’s SAGEmap service implements many functions useful in the analysis of SAGE data. A tag-to-gene function maps a SAGE tag to one or more UniGene clusters. The reciprocal gene-to-tag function maps a UniGene cluster ID to the SAGE tags found within the cluster. SAGEmap can also construct a user-configurable table of data comparing one group of SAGE libraries with another. Groups may be chosen for inclusion in the table on the basis of several expression criteria specified by the user. SAGEMap is updated weekly, immediately following the update of UniGene.
Online Mendelian Inheritance in Man (OMIM)
NCBI provides Web access to the OMIM database, a catalog of human genes and genetic disorders authored and edited by Dr Victor A. McKusick at The Johns Hopkins University (22). The database contains information on disease phenotypes and genes, including extensive descriptions, gene names, inheritance patterns, map locations and gene polymorphisms. OMIM currently contains 10 820 entries, including data on 7597 established gene loci and 659 phenotypic descriptions, and is heavily linked to associated records in Entrez.
THE MOLECULAR MODELING DATABASE
FOR FURTHER INFORMATION
Most of the resources described here include documentation, other explanatory material, and references to collaborators and data sources on the respective Web sites. Several tutorials are also offered under the Education link from NCBI’s home page. A Site Map provides a comprehensive table of NCBI resources, and the What’s New feature announces new and enhanced resources. Additional tools to guide users to NCBI’s growing array of services are also being developed. A user support staff is available to answer questions at info@ncbi.nlm. nih.gov
REFERENCES
- 1.Benson D.A., Karsch-Mizrachi,I., Lipman,D.J., Ostell,J., Rapp,B.A. and Wheeler,D.L. (2000) Nucleic Acids Res., 28, 15–18 (this issue). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schuler G.D., Epstein,J.A., Ohkawa,H. and Kans,J.A. (1996) Methods Enzymol., 266, 141–162. [DOI] [PubMed] [Google Scholar]
- 3.Wang Y., Addess,K.J., Geer,L., Madej,T., Marchler-Bauer,A., Zimmerman,D. and Bryant,S.H. (2000) Nucleic Acids Res., 28, 243–245 (this issue). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barker W.C., Garavelli,J.S., McGarvey,P.B., Marzec,C.R., Orcutt,B.C., Srinivarsarao,G.Y., Yeh,L.S., Ledley,R.S., Mewes,H.W., Pfeiffer,F. et al. (1999) Nucleic Acids Res., 27, 39–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bairoch A. and Apweiler,R. (1999) Nucleic Acids Res., 27, 44–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berman H.M., Westbrook,J., Feng,Z., Gilliland,G., Bhat,T.N., Weissig,H., Shindyalov,I.N. and Bourne,P.E. (2000) Nucleic Acids Res., 28, 235–242 (this issue). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maglott D., Katz,K., Sicotte,H. and Pruitt,K. (2000) Nucleic Acids Res., 28, 126–128 (this issue). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Altschul S.E., Gish,W., Miller,W., Myers,E.W. and Lipman,D.J. (1990) J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 9.Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Miller,W. and Lipman,D.J. (1997) Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Z., Schaffer,A.A., Miller,W., Madden,T.L., Lipman,D.J., Koonin,E.V. and Altschul,S.F. (1998) Nucleic Acids Res., 26, 3986–3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tatusova T.A. and Madden,T.L. (1999) FEMS Microbiol. Lett., 174, 247–250. [DOI] [PubMed] [Google Scholar]
- 12.Schuler G.D. (1997) J. Mol. Med., 75, 694–698. [DOI] [PubMed] [Google Scholar]
- 13.Deloukas P., Schuler,G.D., Gyapay,G., Beasley,E.M., Soderlund,C., Rodriguez-Tome,P., Hui,L., Matise,T.C., McKusick,K.B., Beckmann,S. et al. (1998) Science, 282, 744–746. [DOI] [PubMed] [Google Scholar]
- 14.Ermolaeva O., Rastogi,M., Pruitt,K.D., Schuler,G.D., Bittner,M.L., Chen,Y., Simon,R., Meltzer,P., Trent,J.M. and Boguski,M.S. (1998) Nature Genet., 20, 19–23. [DOI] [PubMed] [Google Scholar]
- 15.Smigielski E.M., Sirotkin,K., Ward,M. and Sherry,S.T. (2000) Nucleic Acids Res., 28, 352–355 (this issue). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jang W., Chen,H.C., Sicotte,H. and Schuler,G.D. (1999) Trends Genet., 15, 284–286. [DOI] [PubMed] [Google Scholar]
- 17.Schuler G.D., Boguski,M.S., Stewart,E.A., Stein,L.D., Gyapay,G., Rice,K., White,R.E., Rodriguez-Tome,P., Aggarwal,A., Bajorek,E. et al. (1996) Science, 274, 540–546. [PubMed] [Google Scholar]
- 18.DeBry R.W. and Seldin,M.F. (1996) Genomics, 33, 337–351. [DOI] [PubMed] [Google Scholar]
- 19.Mitelman F., Mertens,F. and Johansson,B. (1997) Nature Genet., 15, 417–474. [DOI] [PubMed] [Google Scholar]
- 20.Tatusova T., Karsch-Mizrachi,I. and Ostell,J. (1999) Bioinformatics, 15, 536–543. [DOI] [PubMed] [Google Scholar]
- 21.Tatusov R.L., Galperin,M.Y., Natale,D.A. and Koonin,E.V. (2000) Nucleic Acids Res., 28, 33–36 (this issue). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McKusick V.A. (1998) Mendelian Inheritance in Man. Catalogs of Human Genes and Genetic Disorders, 12th edition. Johns Hopkins University Press, Baltimore.