Abstract
Black Veterans have higher a incidence of localized and metastatic prostate cancer compared to White Veterans yet are underrepresented in reports of frequencies of somatic and germline alterations. This retrospective analysis of somatic and putative germline alterations was conducted in a large cohort of Veterans with prostate cancer (N = 835 Black, 1613 White) who underwent next generation sequencing through the VA Precision Oncology Program, which facilitates molecular testing for Veterans with metastatic cancer. No differences were observed in gene alterations for FDA approved targetable therapies (13.5% in Black Veterans vs. 15.5% in White Veterans, P = .21), nor in any potentially actionable alterations (25.5% vs. 28.7%, P =.1). Black Veterans had higher rates of BRAF (5.5% vs. 2.6%, P < .001) alterations, White Veterans TMPRSS2 fusions (27.2% vs. 11.7%, P < .0001). Putative germline alteration rates were higher in White Veterans (12.0% vs. 6.1%, P < .0001). Racial disparities in outcome are unlikely attributable to acquired somatic alterations in actionable pathways.
Keywords: metastatic prostate cancer, actionable alterations, next-generation sequencing, disparities, race
Black veterans have higher incidence of prostate cancer than White veterans but are underrepresented in reports of frequencies of somatic and germline alterations. This retrospective analysis of somatic and putative germline alterations was conducted in a large cohort of veterans with prostate cancer who underwent next-generation sequencing through the VA Precision Oncology Program. No differences were observed in actionable alterations, suggesting disparities in prostate cancer outcomes are not attributable to acquired somatic alterations in actionable pathways.
Veterans self-identifying as Black/African American have a higher incidence of both localized (M0) and de novo metastatic (M1) PCa compared to White Veterans.1 Next Generation Sequencing (NGS) is recommended for all Veterans with metastatic PCa to identify actionable alterations targetable with approved therapies. Prior studies comparing racial differences in the frequency of somatic alterations in patients with metastatic PCa are mixed, though the representation of Black patients has been low. We evaluate differences in actionable genomic profiles between Black and White Veterans with metastatic PCa in a cohort with the largest total and proportional representation of Black patients in the published literature.
We retrospectively analyzed Veterans who underwent clinical tumor genomic sequencing for a submitted diagnosis of PCa as part of the National Precision Oncology Program (NPOP) sequencing initiative2 between January 2015 and February 2022. Race and ethnicity were self-reported by Veterans. Race categories in this analysis were Black/African American and White. For Black Veterans, Hispanic and non-Hispanic ethnicities were included. For White Veterans, only non-Hispanic ethnicities were included. FFPE tissue submitted for sequencing included prostate biopsies, prostatectomy specimens, and prostate cancer metastases. Foundation Medicine, Personalis, and Personal Genome Diagnostics (PGDx) platforms were deployed for sequencing, but only commonly reported genes were considered in this analysis. When multiple specimens from the same patient were sequenced, only the most recently sequenced specimen was analyzed. Alterations were defined as “likely oncogenic/oncogenic” according to described criteria (Supplementary Methods, Supplementary Tables S1, S2).
Likely oncogenic/oncogenic alterations were categorized as “FDA Approved,” (alterations in genes targetable by drugs with FDA-approved indications in metastatic PCa. Supplementary Table S3) or “Any Actionable,” (alterations in genes targetable by any drug), including targeted agents with FDA approval for another indication. In addition, putative germline alterations were defined as oncogenic/likely oncogenic variants if variant allele frequency exceeded 30% (VAF >30%) in genes identified by the ESMO Precision Medicine Working Group.3 Fisher’s exact test and Chi squared test compared differences among White versus Black Veterans.
NGS results from 2448 Veterans were available for analysis, 1613 (66%) White and 835 (34%) Black (Table 1, Supplementary Table S4). The cohort was balanced for vital status, but Black Veterans were younger, more likely to be unmarried, Medicaid eligible, and reside in an urban setting, and less likely to report Agent Orange exposure.
Table 1.
Characteristics of patients with prostate cancer who underwent tumor sequencing.
| Characteristics | White (n = 1613) | Black (n = 835) | P-value | ||
|---|---|---|---|---|---|
| Number | % | Number | % | ||
| Vital status as of 3/15/2022 | |||||
| Deceased | 661 | 41.0 | 310 | 37.1 | .0673 |
| Lab | |||||
| Foundation medicine | 1349 | 83.6 | 692 | 82.9 | .0024 |
| Personalis | 89 | 5.5 | 73 | 8.7 | |
| PGDX | 175 | 10.8 | 70 | 8.4 | |
| Age | |||||
| Mean (SD) | 73.2 (7.6) | 69.6 (8.1) | |||
| Category (age group at index) | |||||
| 18-49 | * | * | * | * | <.0001 |
| 50-54 | 16 | 1.0 | 19 | 2.3 | |
| 55-59 | 47 | 2.9 | 45 | 5.4 | |
| 60-64 | 101 | 6.3 | 140 | 16.8 | |
| 65-69 | 216 | 13.4 | 187 | 22.4 | |
| 70-74 | 534 | 33.1 | 218 | 26.1 | |
| 75-79 | 403 | 25.0 | 126 | 15.1 | |
| 80 to older | 292 | 18.1 | 97 | 11.6 | |
| Marital status | |||||
| Other | 699 | 43.3 | 505 | 60.5 | <.0001 |
| Married | 914 | 56.7 | 330 | 39.5 | |
| Residential urban/rural | |||||
| Urban | 1025 | 63.5 | 716 | 85.7 | <.0001 |
| Rural | 588 | 36.5 | 119 | 14.3 | |
| Medicaid eligible | |||||
| No | 1598 | 99.1 | 813 | 97.4 | .0024 |
| Yes | 15 | 0.9 | 22 | 2.6 | |
| Type of VHA benefits | |||||
| Non-service-connected veteran | 36 | 2.2 | 21 | 2.5 | .0016 |
| 10%-39% | 191 | 11.8 | 100 | 12.0 | |
| 40%-59% | 59 | 3.7 | 25 | 3.0 | |
| 60%-89% | 98 | 6.1 | 89 | 10.7 | |
| 90+ | 1229 | 76.2 | 600 | 71.9 | |
| Potential exposure to agent orange | |||||
| No | 1210 | 75.0 | 743 | 89.0 | <.0001 |
| Yes | 403 | 25.0 | 92 | 11.0 | |
| Branch of service | |||||
| Other or missing | 292 | 18.1 | 105 | 12.6 | <.0001 |
| Air force | 191 | 11.8 | 105 | 12.6 | |
| Army | 688 | 42.7 | 431 | 51.6 | |
| Coast guard | 12 | 0.7 | * | * | |
| Marine corps | 154 | 9.5 | 97 | 11.6 | |
| Navy | 276 | 17.1 | 96 | 11.5 | |
| Service period | |||||
| Other or unknown | * | * | * | * | <.0001 |
| Pre-Korean (WWI, WWII, and Pre-Korean) | 12 | 0.7 | * | * | |
| Korean | 96 | 6.0 | 37 | 4.4 | |
| Post-Korean | 75 | 4.6 | 14 | 1.7 | |
| Vietnam | 1191 | 73.8 | 499 | 59.8 | |
| Post-Vietnam | 148 | 9.2 | 176 | 21.1 | |
| Persian Gulf War | 89 | 5.5 | 104 | 12.5 | |
*Less than 11 patients were observed. Statistical comparisons by Chi squared test.
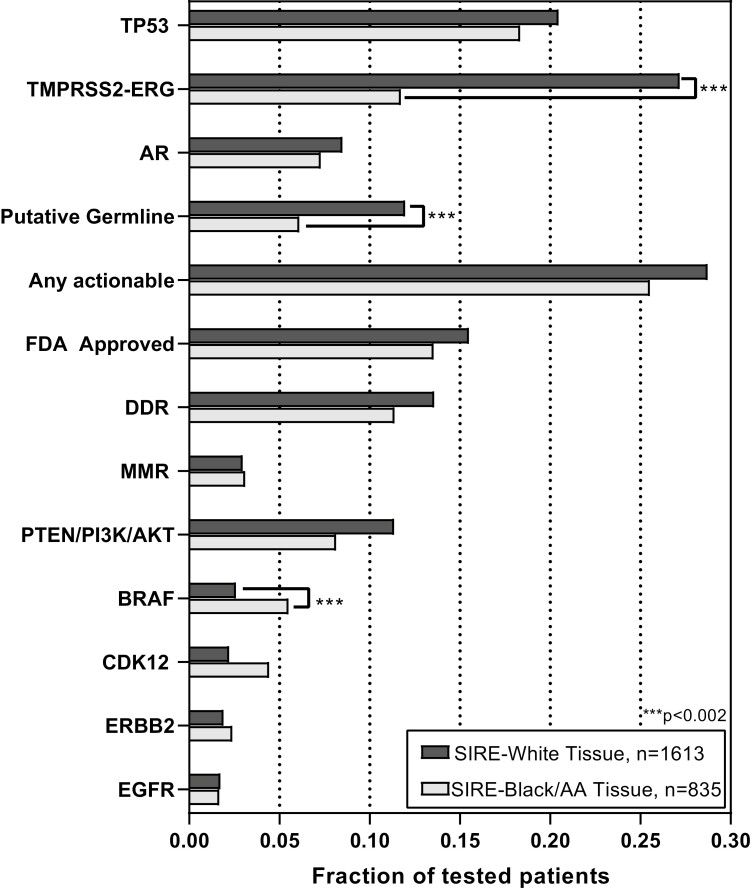
White Veterans had higher rates of TMPRSS2-ERG fusions (27.2% vs. 11.7% of tested patients, P < .001). No differences were observed in TP53 or AR alteration rates (Fig. 1; Table 2). Oncogenic alteration rates were not different in the “FDA Approved” gene category for White vs. Black Veterans (15.5% vs. 13.7, P = 0.2), respectively, owing to similar rates of alterations in DDR genes, MMR genes, TMB status, and MSI status. Similarly, there was no difference in the rates of “Any Actionable” alterations (28.7% vs. 25.5%, P = 0.1). Black Veterans displayed higher rates of BRAF (5.5% vs. 2.6%, P < .001) oncogenic alterations. White Veterans had higher rates of putative germline alterations (12.0% vs. 6.1%, P < .001) compared to Black Veterans.
Figure 1.
Alteration profiles in Black vs. White Veterans with metastatic prostate cancer. Alteration rates for the listed genes or gene groupings according to the fraction of total patients tested for that particular gene. “MMR” deficiency was defined as oncogenic alterations (short variants or copy number loss) in MLH1, MSH2, MSH6 or PMS2; MSI-High status on report; or TMB-High status on report. “DDR” deficiency was defined as oncogenic alterations (short variants or copy number loss) in ATM, BARD1, BRCA1, BRCA2, BRIP1, CHEK1, CHEK2, FANCA, FANCL, NBN, PALB2, RAD51, RAD51B, RAD51C, RAD51D, or RAD54L. “Any Actionable” further included oncogenic alterations in BRAF, PTEN, PI3KCA, AKT, CDK12, ERBB2, EGFT, IDH1, CHKN2A, NTRK1/2/3, ALK, FGFR1/2/3, KIT, PDGFRA, RET, and ROS1. Short variants in TP53, AR, and TMPRSS2 fusions were not considered actionable but reported. Tabulation of this data is available in Supplementary Table S5. ***Refers to the Bonferroni corrected significance threshold P < .002.
Table 2.
Comparison of rates of genomic alterations between White and Black Veterans.
| SIRE-White | SIRE-Black/AA | P-value (8) | % change (W-B) | |||
|---|---|---|---|---|---|---|
| n | % of 1613 | n | % of 835 | |||
| TP53 | 331 | 20.52 | 154 | 18.44 | .2181 | 2.1 |
| TMPRSS2-ERG | 438 | 27.15 | 98 | 11.74 | <.0001* | 15.4 |
| DDR (1) | 219 | 13.58 | 95 | 11.38 | .1264 | 2.2 |
| PTEN/PI3K/AKT | 183 | 11.35 | 68 | 8.14 | .0138 | 3.2 |
| AR | 137 | 8.49 | 61 | 7.31 | .3481 | 1.2 |
| BRAF | 42 | 2.60 | 46 | 5.51 | .0005* | −2.9 |
| PTEN | 97 | 6.01 | 39 | 4.6 | .1925 | 1.3 |
| CDK12 | 36 | 2.23 | 37 | 4.43 | .0036 | −2.2 |
| MMRd (2) | 48 | 2.98 | 26 | 3.11 | .901 | −0.1 |
| ERBB2 | 31 | 1.92 | 20 | 2.40 | .457 | −0.5 |
| EGFR | 28 | 1.74 | 14 | 1.68 | 1 | 0.1 |
| CDKN2A | 14 | 0.87 | <11 | <1.3 | N/A | 0.0 |
| FGFR (3) | <11 | <0.7 | <11 | <1.3 | N/A | −0.5 |
| IDH1 | <12 | <0.7 | <11 | <1.3 | N/A | −0.2 |
| PDGFRA | <13 | <0.7 | <11 | <1.3 | N/A | 0.0 |
| RET | 12 | 0.74 | <11 | <1.3 | N/A | 0.5 |
| ROS1 | <11 | <0.7 | <11 | <1.3 | N/A | −0.2 |
| NTRK (4) | <11 | <0.7 | <11 | <1.3 | N/A | 0.1 |
| ALK | <11 | <0.7 | <11 | <1.3 | N/A | 0.1 |
| KIT | 13 | 0.81 | <11 | <1.3 | N/A | 0.7 |
| ABL1 | <11 | <0.7 | <11 | <1.3 | N/A | 0.3 |
| Any actionable (5) | 463 | 28.70 | 213 | 25.51 | .0953 | 3.2 |
| FDA approved (6) | 250 | 15.50 | 113 | 13.53 | .2079 | 2.0 |
| Putative Germline (7) | 193 | 11.97 | 51 | 6.11 | <.0001* | 5.9 |
(1) DDR: LP/P alteration in ATM, BARD1, BRCA1, BRCA2, BRIP1, CHEK1, CHEK2, FANCA, FANCL, NBN, PALB2, RAD51, RAD51B RAD51C, RAD51D, RAD54L.
(2) MMR deficiency = LP/P alteration in MLH1, MSH2, MSH6, PMS2 and/or MSI-H and/or TMB-H.
(3) FGFR: LP/P alteration in FGFR2, FGFR3.
(4) NTRK: LP/P alteration in NTRK1, NTRK2, NTRK3.
(5) Any actionable: LP/P alteration in DDR, MMRd, ABL1, AKT, BRAF, CDK12, ERBB2, EGFR, IDH1, CDKN2A, NTRK, ALK, FGFR, KIT, PDGFRA, PI3KCA, PTEN, RET, ROS1.
(6) FDA approved: LP/P alteration in MMR deficiency + DDR.
(7) Putative germline = LP/P alteration with VAF>30% in ESMO guideline gene.
(8) *Refers to the Bonferroni corrected value significance level P < .002.
To our knowledge, this cohort has the largest total and proportional representation of Black men with PCa who underwent NGS to date. Defined narrowly or broadly, actionable alterations do not differ between Black and White Veterans, suggesting current therapeutic strategies based on actionable alterations are likely equally beneficial for Black and White patients. Additionally, differences in somatic alterations are likely insufficient to explain the race-based disparities in the incidence and outcomes of metastatic PCa.
Our findings are consistent with a prior study,4 which evaluated a mixed population of 861 metastatic and non-metastatic men with PCa. Though only 250 Black men were included in their cohort, they found a similar proportion of actionable alterations between White and Black patients. A separate comparative analysis5 evaluated sequencing data from 165 Black men and similarly uncovered no differences in rates of DDR alterations. Also consistent with our findings, TMPRSS2 rearrangements were less common in patients of African ancestry compared to those of European ancestry, a finding that has been recapitulated across multiple studies.5-7 In contrast, another study reported greater rates of actionable alterations in the metastatic tumors of 71 Black men compared to those of 801 White men.8 It is possible that the low representation of Black men in that analysis contributed to an over-emphasis of the rates of potentially actionable alterations.
The almost two-fold difference in putative germline variants by race is striking. Annotation databases lack representation of men of African ancestry making it difficult to properly annotate and identify germline variants in this population. However, given that most putative germline variants were truncating variants, differential inclusion in annotation databases is unlikely explain this difference entirely.
Limitations include the lack of annotation to distinguish if sequenced tissue was from metastases versus primary tissue, the potential phenotypic heterogeneity of this metastatic cohort, and our inability to correlate corresponding clinicopathologic data such as tumor burden and castrate sensitivity with genomic findings. Importantly, our primary analysis of actionable mutation rates should not be affected by different proportions of primary versus metastatic tissue because DDR alterations and MSI are truncal alterations.9-11 While sequencing was accomplished using multiple platforms, our analysis was restricted to commonly reported genes across all platforms to maintain a robust analysis. We feel that the benefits of reporting genomic sequencing data with a high total and proportional representation of Black patients outweigh these limitations, given the critical need to understand race-based drivers of PCa disparities.
Our work adds the largest NGS analysis of Black patients with PCa and suggests that clinically actionable alteration rates do not vary with race. Thus, racial disparities in outcome are unlikely attributable to acquired somatic alterations in currently known actionable pathways, though we cannot exclude the possibility that unknown actionable pathways play a role. We echo others12 in underscoring the importance of diverse and equitable representation in studies seeking to characterize cancer genomic profiles. This is especially true in the context of a relatively lower mutation rate of PCa, where conclusions from comparative studies with low Black representation may provide an incomplete picture of the true tumor mutational landscape in this population. Ultimately, we support genomic sequencing efforts for Veterans with advanced PCa in service of improving patient selection for targeted therapies and simultaneously encourage complementary strategies that address modifiable cancer risk factors, healthcare access, social determinants of health, and other interactions with genomics to address disparities in cancer outcomes.
Supplementary Material
Contributor Information
Luca F Valle, Veterans Affairs Greater Los Angeles Healthcare System, Los Angeles, CA, USA; Department of Radiation Oncology, David Geffen School of Medicine at the University of California Los Angeles, Los Angeles, CA, USA.
Nicholas G Nickols, Veterans Affairs Greater Los Angeles Healthcare System, Los Angeles, CA, USA; Department of Radiation Oncology, David Geffen School of Medicine at the University of California Los Angeles, Los Angeles, CA, USA; UCLA Jonsson Comprehensive Cancer Center, Los Angeles, CA, USA; Department of Urology, David Geffen School of Medicine at the University of California, Los Angeles, CA, USA.
Ryan Hausler, Department of Veterans Affairs Informatics and Computing Infrastructure, Department of Veterans Affairs Salt Lake City Health Care System, Salt Lake City, UT, USA; Division of Hematology/Oncology, Department of Medicine, Perelman School of Medicine at the University of Pennsylvania, Philadelphia, PA, USA.
Patrick R Alba, Department of Veterans Affairs Informatics and Computing Infrastructure, Department of Veterans Affairs Salt Lake City Health Care System, Salt Lake City, UT, USA; Division of Epidemiology, Department of Internal Medicine, University of Utah School of Medicine, Salt Lake City, UT, USA.
Tori Anglin-Foote, Department of Veterans Affairs Informatics and Computing Infrastructure, Department of Veterans Affairs Salt Lake City Health Care System, Salt Lake City, UT, USA.
Cristina Perez, Department of Veterans Affairs Informatics and Computing Infrastructure, Department of Veterans Affairs Salt Lake City Health Care System, Salt Lake City, UT, USA.
Kosj Yamoah, Department of Radiation Oncology, H. Lee Moffitt Cancer Center & Research Institute, Tampa, FL, USA; James A. Haley Veterans’ Hospital, Tampa, FL, USA.
Brent S Rose, Department of Radiation Oncology, University of California, San Diego, CA, USA; Veterans Affairs San Diego Healthcare System, San Diego, CA.
Michael J Kelley, Duke University Medical Center, Durham, NC, USA; Department of Veteran Affairs Medical Center, Durham, NC, USA.
Scott L DuVall, Department of Veterans Affairs Informatics and Computing Infrastructure, Department of Veterans Affairs Salt Lake City Health Care System, Salt Lake City, UT, USA; Division of Epidemiology, Department of Internal Medicine, University of Utah School of Medicine, Salt Lake City, UT, USA.
Isla P Garraway, Veterans Affairs Greater Los Angeles Healthcare System, Los Angeles, CA, USA; UCLA Jonsson Comprehensive Cancer Center, Los Angeles, CA, USA; Department of Urology, David Geffen School of Medicine at the University of California, Los Angeles, CA, USA.
Kara N Maxwell, Division of Hematology/Oncology, Department of Medicine, Perelman School of Medicine at the University of Pennsylvania, Philadelphia, PA, USA; Corporal Michael J. Crescenz Veterans Affairs Medical Center, Philadelphia, PA, USA; Abramson Cancer Center, Perelman School of Medicine at the University of Pennsylvania, Philadelphia, PA, USA.
Julie A Lynch, Department of Veterans Affairs Informatics and Computing Infrastructure, Department of Veterans Affairs Salt Lake City Health Care System, Salt Lake City, UT, USA; Division of Epidemiology, Department of Internal Medicine, University of Utah School of Medicine, Salt Lake City, UT, USA; Department of Nursing and Health Sciences, University of Massachusetts, Boston, MA, USA.
Acknowledgments
Research support for this study was generously provided by the Department of Veterans Affairs (VA) Informatics and Computing Infrastructure (VINCI) to Put VA Data to Work for Veterans (VA ORD 22-D4V, VA HSR RES 13-457, PRA, CP, SLD, JAL), The National Institutes of Health (K08CA215312, KNM; 5P50CA092131, R01 PAR-20-077, IPG); US Department of Defense (W81XWH211075, IPG; W81XWH-19-1-0435, KY); Prostate Cancer Foundation (PCF17CHAL04, IPG; 20YOUNG02, KNM; 18VALO10, KY, NGN), Burroughs Wellcome Foundation (1017184, KNM); Basser Center for BRCA at the University of Pennsylvania (KNM); Jean Perkins Foundation (IPG), STOP Cancer Foundation (IPG), and VA ORD CSR&D (NGN). LV was also a Recipient of the Robert A. Winn Diversity in Clinical Trials Career Development Award, funded by Bristol Myers Squibb Foundation.
Conflict of Interest
Nicholas G. Nickols reported research support from Lantheus, Janssen, and Bayer, and consulting for PrimeFour outside the scope of the present work. Michael J. Kelley reported research support from Novartis, AstraZeneca, Bristol-Myers Squibb, Regeneron, and Genentech. Scott L. DuVall and Julie A Lynch reported research support from Alnylam Pharmaceuticals, Inc, Astellas Pharma, Inc, AstraZeneca Pharmaceuticals LP, Biodesix, Boehringer Ingelheim International GmbH, Celgene Corporation, Eli Lilly and Company, Genentech Inc., Gilead Sciences Inc., GlaxoSmithKline PLC, Innocrin Pharmaceuticals Inc., IQVIA Inc., Janssen Pharmaceuticals, Inc., Kantar Health, MDxHealth, Merck & Co., Inc., Myriad Genetic Laboratories, Inc., Novartis International AG, and Parexel International Corporation through the University of Utah or Western Institute for Veteran Research outside the submitted work. The other authors indicated no financial relationships.
Author Contributions
Conception/design: N.G.N., I.P.G., K.N.M., J.A.L. Provision of study material or patients: M.J.K., S.L.D., I.P.G., J.A.L. Collection and/or assembly of data: R.H., P.R.A., T.A.F., C.P., M.J.K., S.L.D., J.A.L. Data analysis and interpretation: L.F.V., N.G.N., R.H., P.R.A., T.A.F., C.P., K.Y., B.S.R., M.J.K., S.L.D., I.P.G., K.N.M., J.A.L. Manuscript writing: N.G.N., L.F.V., K.Y., B.S.R., M.J.K., I.P.G., K.N.M., J.A.L. Final approval of manuscript: All authors.
Data Availability
The data underlying this article cannot be shared publicly due to privacy.
References
- 1. Yamoah K, Lee KM, Awasthi S, et al. Racial and ethnic disparities in prostate cancer outcomes in the veterans affairs health care system. JAMA Netw Open. 2022;5:e2144027. 10.1001/jamanetworkopen.2021.44027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kelley MJ. VA National precision oncology program. Fed Pract Health Care Prof VA DoD PHS. 2020;37:S22-S27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mandelker D, Donoghue M, Talukdar S, et al. Germline-focused analysis of tumour-only sequencing: recommendations from the ESMO Precision Medicine Working Group. Ann Oncol Off J Eur Soc Med Oncol. 2019;30:1221-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Koga Y, Song H, Chalmers ZR, et al. Genomic profiling of prostate cancers from men with African and European ancestry. Clin Cancer Res Off J Am Assoc Cancer Res. 2020;26:4651-4660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Stopsack KH, Nandakumar S, Arora K, et al. Differences in prostate cancer genomes by self-reported race: contributions of genetic ancestry, modifiable cancer risk factors, and clinical factors. Clin Cancer Res Off J Am Assoc Cancer Res. 2022;28:318-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Schumacher FR, Basourakos SP, Lewicki PJ, et al. Race and genetic alterations in prostate cancer. JCO Precis Oncol. 2021;5:PO.21.00324. 10.1200/PO.21.00324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yuan J, Kensler KH, Hu Z, et al. Integrative comparison of the genomic and transcriptomic landscape between prostate cancer patients of predominantly African or European genetic ancestry. PLoS Genet. 2020;16:e1008641. 10.1371/journal.pgen.1008641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mahal BA, Alshalalfa M, Kensler KH, et al. Racial differences in genomic profiling of prostate cancer. N Engl J Med. 2020;383:1083-1085. 10.1056/NEJMc2000069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Schweizer MT, Yu EY.. “Matching” the “mismatch” repair-deficient prostate cancer with immunotherapy. Clin Cancer Res Off J Am Assoc Cancer Res. 2020;26:981-983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kolinsky MP, Niederhoffer KY, Kwan EM, et al. Considerations on the identification and management of metastatic prostate cancer patients with DNA repair gene alterations in the Canadian context. Can Urol Assoc J J Assoc Urol Can. 2022;16:132-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mateo J, Seed G, Bertan C, et al. Genomics of lethal prostate cancer at diagnosis and castration resistance. J Clin Invest. 2020;130:1743-1751. 10.1172/JCI132031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Arenas-Gallo C, Owiredu J, Weinstein I, et al. Race and prostate cancer: genomic landscape. Nat Rev Urol. 2022;19:547-561. 10.1038/s41585-022-00622-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article cannot be shared publicly due to privacy.