Abstract
Endometriosis is a common inflammatory disorder in women of reproductive age due to an abnormal endometrial immune environment and is associated with infertility. This study aimed to systematically understand the endometrial leukocyte types, inflammatory environment, and impaired receptivity at single-cell resolution. We profiled single-cell RNA transcriptomes of 138 057 endometrial cells from endometriosis patients (n = 6) and control (n = 7), respectively, using 10x Genomics platform. We found that one cluster of epithelial cells that expressed PAEP and CXCL14 was mostly from the control during the window of implantation (WOI). This epithelial cell type is absent in the eutopic endometrium during the secretory phase. The proportion of endometrial immune cells decreased in the secretory phase in the control group, whereas the cycle variation of total immune cells, NK cells, and T cells was absent in endometriosis. Endometrial immune cells secreted more IL-10 in the secretory phase than in the proliferative phase in the control group; the opposite trend was observed in endometriosis. Proinflammatory cytokines levels in the endometrial immune cells were higher in endometriosis than in the control group. Trajectory analysis revealed that the secretory phase epithelial cells decreased in endometriosis. Ligand–receptor analysis revealed that 11 ligand–receptor pairs were upregulated between endometrial immune and epithelial cells during WOI. These results provide new insights into the endometrial immune microenvironment and impaired endometrial receptivity in infertile women with minimal/mild endometriosis.
Keywords: single-cell RNA sequence, endometriosis-associated infertility, endometrial immune cells, endometrial receptivity
We systematically revealed the eutopic endometrial single-cell transcriptome immune landscape in minimal/mild endometriosis and compared it with that of disease-free controls. Our study provides new insights into the eutopic endometrial inflammatory microenvironment and defective endometrial receptivity in patients with endometriosis-associated infertility. Our results give clues into immunopathogenesis of endometriosis endometriosis-associated infertility.
Graphical Abstract
Graphical Abstract.
Introduction
Endometriosis is characterized by endometrial-like tissue outside the normal uterine environment and affects approximately 10% of reproductive-aged women and 30–50% of infertile women [1]. The pathogenesis of endometriosis-related infertility involves distorted pelvic anatomy, decreased oocyte quality, and an inhospitable endometrial environment for embryo nidation. In addition to altered steroid hormone signaling, the eutopic endometrial immune status in endometriosis, involving lymphocyte activation, antigen presentation, cytokine induction, and inflammation, contributes to adverse reproductive outcomes [2, 3].
Most endometrial immune cells are tissue-resident, and the population changes throughout the menstrual cycle. Endometrial immune cells, expressing the leukocyte common antigen surface marker (CD45), account for 10–20% of all endometrial cells and are increased in the secretory phase. T cells represent the majority of the endometrial immune cells in the proliferative phase; however, approximately 70–80% of total endometrial leukocytes are uterine natural killer cells (uNK) in the mid–late secretory phase [4]. T cells (CD3+), uNK cells (CD56+), and macrophages (CD68+) are abundant endometrial leukocytes. In addition, dendritic cells (ITGAX+), B cells (CD20+), mast cells (MS4A2), and neutrophils are endometrial immune cells. Dynamic changes in the endometrial immune cells maintain the homeostasis; aberrant population and function of the endometrial immune cells lead to an inhospitable environment for embryo implantation [5]. Previous studies have found an altered proinflammatory state and absent cycle variation of endometrial immune cells in endometriosis than in those without. Macrophages polarize from M2 (anti-inflammatory phenotype) to M1 (proinflammatory phenotype) in the eutopic endometrium of patients with endometriosis than in the control group [6–9]. Hey-Cunningham et al. found that the proportion of regulated T cells (Tregs) do not change between the proliferative and secretory phases, whereas in women without endometriosis, Tregs significantly decreased in the secretory phase [10].
Although a number of studies have supported an altered inflammatory immune environment in endometriosis, most have used flow cytometry and immunohistochemistry to define endometrial immune cells. These two methods can only focus on limited immune cell types in the eutopic endometrium, rather than on the complete endometrial immune landscape. Cellular components in the endometrial immune niche and inflammatory profiles have not yet been systematically determined. Recently, two studies used microarray and RNA sequencing databases in public repositories to analyze the endometrial immune environment in endometriosis [6, 11]. However, the resolution of microarray or standard RNA sequencing cannot compete and is not comparable to single-cell RNA sequencing (scRNA-seq), particularly for immune cell phenotyping [12]. To our knowledge, there is only one single-cell transcriptome analysis of eutopic endometrial immune cells in endometriosis. This study identified immune cell heterogeneity among endometriosis lesions, eutopic endometria, and normal endometria; the number and state of T cells, NK cells, and macrophages were different between the ectopic lesions and eutopic endometria [13]. However, this study only focused on the proliferative phase of the eutopic endometrium and stages III–IV of endometriosis (ovarian endometriosis). Transcriptome meta-analysis revealed that a pro-inflammatory profile is predominant in minimal/mild (stages I–II) endometriosis compared to moderate/severe (stages III–IV) endometriosis [6]. Moreover, this study emphasized the fibroblastic trajectory analysis and ligand-receptor analysis between fibroblasts and immune cells. Epithelial cells play a central role in endometrial receptivity during the window of implantation (WOI), and several markers, including PAEP and CXCL14, are expressed during WOI [14]. Therefore, we chose infertile women with minimal/mild endometriosis and investigated the eutopic immune environment, epithelial cell trajectory variation, and communication with immune cells throughout the menstrual cycle.
We aimed to comprehensively classify the transcriptional profiles of immune cell types in cyclic eutopic endometria in patients with minimal/mild endometriosis using scRNA-seq. Furthermore, we compared the trajectory and cell–cell communication analyses between patients with endometriosis and disease-free controls, in the WOI and other phases of the menstrual cycle. Our results revealed a detailed molecular and cellular immune map of eutopic endometrium in patients with minimal/mild endometriosis across the menstrual cycle. Additionally, it provided a better understanding of the endometrial immune environment and decreased endometrial receptivity in minimal/mild endometriosis.
Materials and methods
Study samples
This study was approved by the Ethics Committee of West China Second University Hospital of Sichuan University. A written informed consent was obtained from all participants. Endometrial samples were collected using Pipelle sampling from infertile women with minimal/mild endometriosis (ASRM classification) during hysteroscopy-laparoscopy [15]. The endometriosis-free controls were multipara women without an infertility history, who underwent laparoscopic surgery for benign ovarian cysts; their endometria were sampled using the Pipelle technique. Participants over the age of 35 years, or those with irregular menstruation (cycle <21 or >35 days), current or previous intrauterine disease (uterine adhesion, endometrial polyps, endometrial tuberculosis, and chronic endometritis), pelvic inflammatory disease, endocrine diseases (polycystic ovary syndrome, hyperthyroidism, hypothyroidism, Cushing’s syndrome, etc.), adenomyosis, malignancy, or exposure to steroid hormones within 3 months were excluded from the study. The infertile women in the endometriosis group whose partners presented with abnormal semen, or who had blocked fallopian tubes and other infertility-inducing diseases were excluded from the study. The samples obtained were immediately transferred to the laboratory for analysis.
Cell isolation
Endometrial tissues were washed in phosphate-buffered saline three times and then sheared into tiny pieces. Tissue pieces were digested with 2 mg/ml collagenase IV (Sigma) in Dulbecco’s modified Eagle’s medium (DMEM)/F12 1:1 (Gibco) at 37°C for 40 min to generate a single-cell suspension. The suspension was passed through 40-μm cell sieves, and the cell concentration was adjusted to 106/ml through dilution or centrifugation. The cell viability was quantified using the trypan blue exclusion method. When the suspension’s cell viability exceeded 85%, the sample was loaded onto a Chromium Single Cell Controller (10x Genomics).
scRNA-seq
The 10x-Genomics Single Cell 3 kit v2 was used to capture 6000–10 000 cells per sample according to the manufacturer’s instructions (https://www.10xgenomics.com). Single-cell and gel beads with 16 bp barcodes and 10 bp unique molecular identifiers (UMIs) were generated to gel bead-in-emulsions (GEMs) on a Chromium Controller, which were broken down after reverse transcription; the resultant cDNA with barcodes were amplified by PCR for library construction. Libraries were sequenced on an Illumina HiSeq PE 150 platform with a minimum coverage of 50 000–100 000 raw reads per cell.
scRNA-seq data analysis
Raw reads were demultiplexed to FASTQ files using “mkfastq application” from Cell Ranger (v3.0.0; 10x Genomics). STAR [16] was used to align FASTQ files with the human genome reference. The gene-barcode matrix, which contained valid cell barcodes and transcript UMI counts, was generated using Cell Ranger Count. The gene-barcode matrices of each sample were concatenated to one matrix and normalized to the same sequencing depth using the Cell Ranger “aggr” pipeline. We applied the Seurat R package [17] to process the gene barcode matrix for quality control. The analysis was performed according to the package’s user guidelines. Briefly, we filtered genes that were expressed in less than 3 cells and retained cells that had at least 200 expressed genes. In addition, cells with more than 10% of mitochondrial genes were removed to remove broken cells.
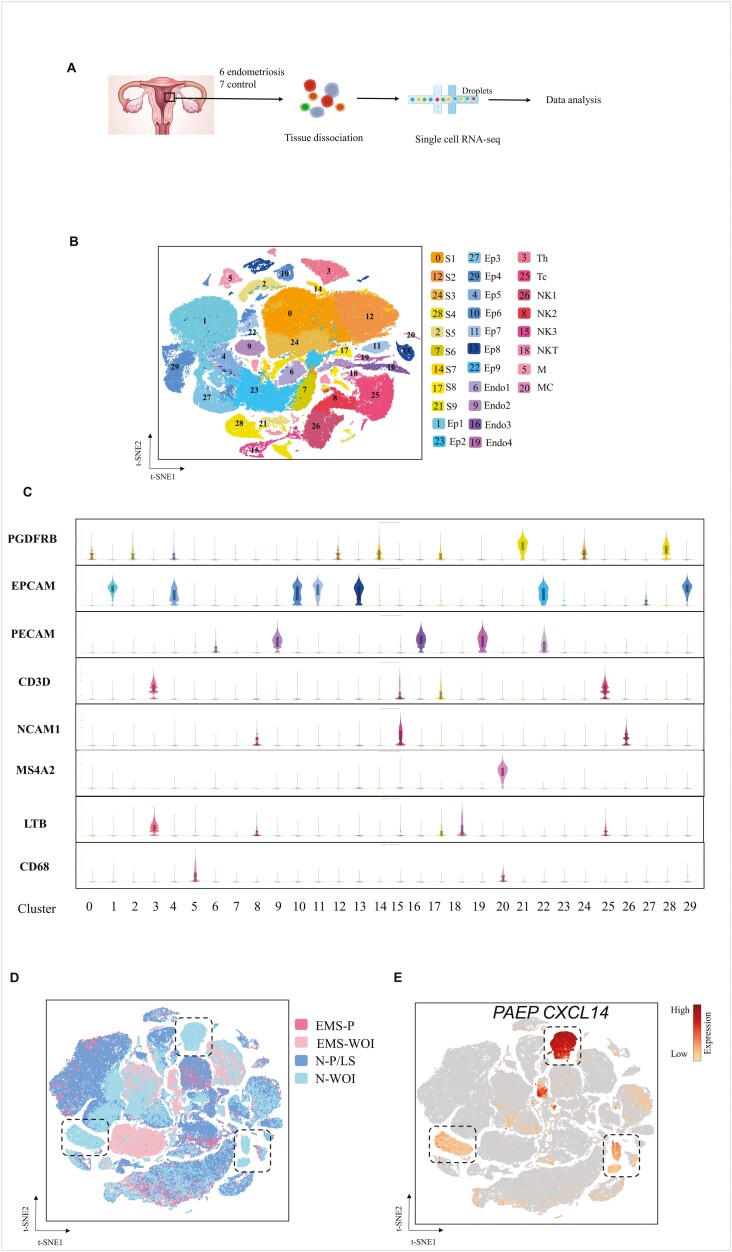
The filtered gene barcode matrix was input into the Seurat pipeline. First, the data matrices from different samples were normalized. To avoid bath differences, we used the canonical correlation analysis (CCA) method to integrate the database. Next, principal component analysis (PCA) was performed on the variable genes, and statistically significant principal components were used as inputs for the graph-based approach with the Louvain algorithm for unsupervised cluster cells. Dimensionality reduction and visualization were performed using the t-distributed stochastic neighbor embedding (t-SNE) algorithm, and cell clusters were visualized in two dimensions. Genes enriched in each cluster, were used to identify cluster markers. Cells expressing PTPRC/CD45 were selected for re-clustering through Seurat, using an approach similar to that described above, to define immune cell subtypes (Fig. 1A).
Figure 1:
Single-cell RNA-sequence of endometrial cells from the endometriosis and control groups. (A) Workflow schematic. (B) t-SNE projection of 138 057 endometrial cells from six patients with endometriosis and seven controls, identifying 30 clusters. (C) Violin plots of the representative marker genes expression levels in 30 clusters. The y-axis shows the log-scale normalised read count. (D) t-SNE projection of endometrial cells colored based on their sample origin. (E) Expression patterns of PAEP and CXCL14 projected on the t-SNE plot.The expression level refers to the color stripe on the right. S: stromal cell; Ep: epithelial cell; Endo: endothelial cell; Th: T-helper cell; Tc: T-cytotoxic cell; NK: natural killer cell; M: macrophage; MC: mast cells
Developmental trajectory analysis
The R package Monocle 3 [18] was used to order cells in pseudotime based on changes in cell gene marker expression. Significantly variable genes were used to construct the pseudotime trajectory, and the UMI count was used as a covariate in the tree construction. The trajectory was visualized in reduced dimensional space through the t-SNE.
Ligand–receptor interaction analysis
The CellPhone DB v.2.0 [19] python package was used to perform ligand–receptor interaction analysis. Single-cell transcriptomic data of endometrial epithelial cells (PECAM+) during WOI (PAEP+, CXCL14+) or other phases (PAEP−, CXCL14−), T-cells (Th1 (CD4+ and IFNG+), Th2 (CD4+ and GATA3+), Th17 (CD4+ and RORC+), Treg (CD4+ and FOXP3+), cytotoxic T-cells (Tc, CD8+), NK-cells (NCAM+), macrophages (M1 [CD68+ and CD86+] and M2 [CD68+ and MRC1+]), dendritic cells (ITGAX+), mast cells (MS4A2), and B-cells (CD20+) were entered into the CellPhone DB. The expression of receptors in one cell type and the corresponding ligands expressed in another cell type were identified. The P-value for each receptor–ligand pair among cluster–cluster interactions was computed using a null distribution. We selected significantly different expressed interactions (P < 0.01) during the WOI and other phases in endometriosis and control group, and visualized them in dot plots.
Results
Single-cell transcriptomic profiles of endometrial cells
We acquired single-cell transcriptome profiles from 13 endometrial samples (six patients with endometriosis [EMS] and four disease-free controls [N]) and further divided them into proliferative and secretory phases based on their menstrual cycle and endometrial pathology (Supplementary Table S1). Using a 10x Genomics Chromium system, 138 057 endometrial cells were captured. All data matched a median depth of 29 760–87 537 reads/cell and 1519–3470 genes/cell (Supplementary Table S2).
Cell partitioning via t-SNE analysis using Seurat identified 30 cell clusters (Fig. 1B). Based on known cell type-specific markers, we annotated nine clusters of stromal cells (S, PDGFRB+), nine of epithelial cells (Ep, EPCAM+), four of endothelial cells (Endo, PECAM+), and eight of immune cells (CD3D for T-cells, NCAM1 for NK-cells, CD68 for macrophages, LTB for Th cells, and MS4A2 for mast cells) (Fig. 1C).
Epithelial cells accounted for 27.60% and 33.2% of cells in the control and endometriosis groups, respectively. There were 49.36% and 34.1% stromal cells in the control and endometriosis groups, respectively. The proportion of endothelial and immune cells was 8.43% and 15.43% in the control group, respectively, and 6.4% and 26.3% in the endometriosis group, respectively. The most obvious difference was observed in the stromal and immune cells between the two groups. Cells from 13 endometria were distributed evenly in all clusters, while there was some variation in epithelial cells between the endometriosis and control groups in the WOI phase (Fig. 1D). PAEP and CXCL14 were specific markers for epithelial cells that were selected in dotted boxes, and these epithelial cells were almost entirely from control WOI phase (Fig. 1D and 1F). CXCL14 and PAPE are luminal epithelial markers for entry into the WOI phase, and the CXCL14 expression notably declines in the late secretory phase [14]. We speculated that epithelial cells from endometriosis in the mid-secretory phase might exhibit defective marker expression, affecting endometrial receptivity.
Cycle variation and cytokine expression in eutopic endometrial immune cells in endometriosis
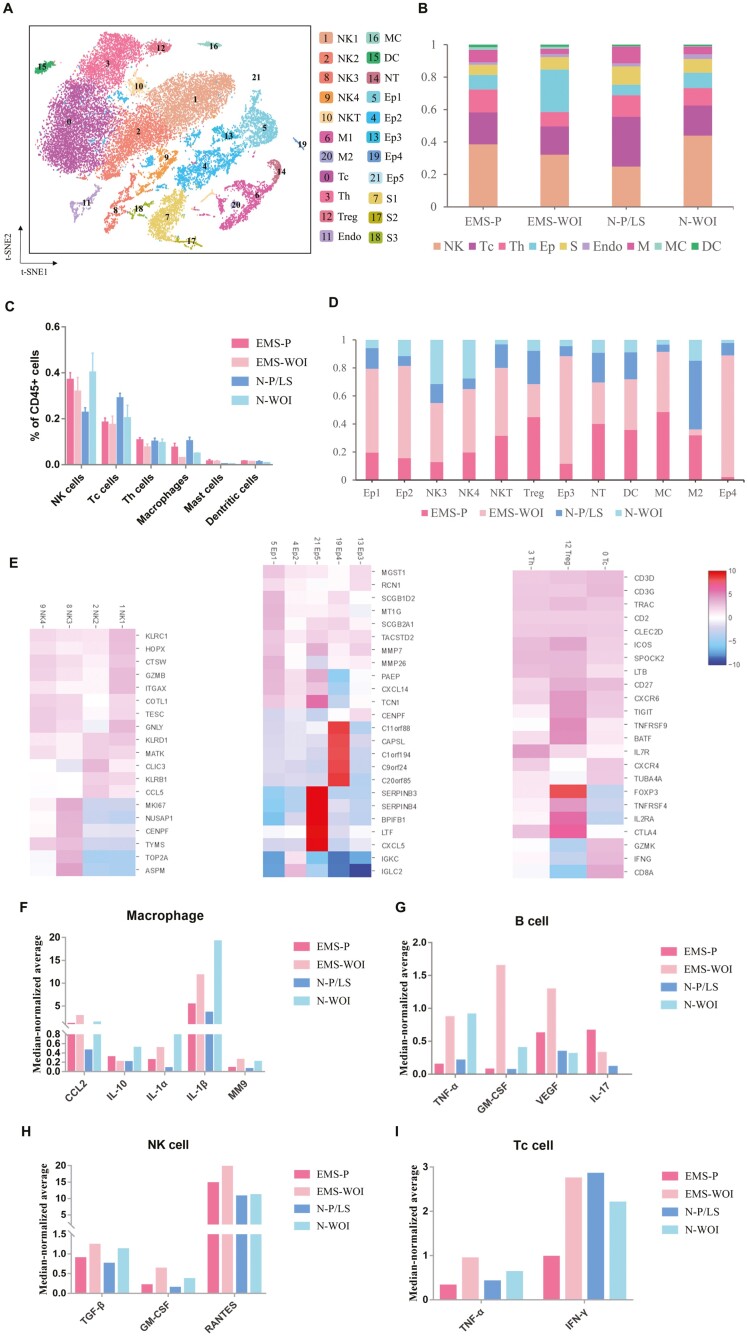
To further characterize the immune environment of endometria, we selected CD45/PTPRC+ endometrial cells and applied Seruat to normalize and re-cluster the gene expression. We identified 22 clusters of immune cells including NK-cells, Tc-cells, Th cells, dendritic cells, macrophages, mast cells, and neutrophils. Unexpectedly, there were nine clusters of CD45+ epithelial, stromal, and endothelial cells (Fig. 2A). The average percentage of immune cells (PTPRC/CD45+) in the endometriosis eutopic endometrium was 26.78% and 24.33% in the proliferative and mid-secretory phases, respectively; higher than that in the endometriosis-free controls (17.11% and 12.43%, respectively). The increased CD45+ leukocyte cells in the secretory phase endometriosis eutopic endometrium indicated an inflammatory environment during the WOI.
Figure 2:
Endometrial immune cell re-clustering in different menstrual phases and cytokine expression. (A) t-SNE projection of endometrial PTPRC+ cells from six patients with endometriosis and seven controls, identifying 22 clusters. (B) Bar plot representation of the relative frequency of different immune cell subclusters across the menstrual cycle. (C) Bar plot representation of the percentage of different immune cell subtypes. Data represent the mean ± SEM. (D) Bar plot representation of the relative ratio of the four groups (EMS-P, EMS-WOI, N-P/LS, and N-WOI) in 12 immune cell clusters. (E) Heatmap of gene expression in NK-cells, epithelial cells, T cells. (G–I) Cytokines expressed in endometrial immune cells. The y-axis shows the single-cell median-normalized average expression. S: stromal cell; Ep: epithelial cell; endo: endothelial cell; Th: T helper cell; Tc, Treg, regulatory T cell; T cytotoxic cell; NK: natural killer cell; M: macrophage; MC: mast cells; DC: dendritic cells; EMS-P: endometriosis in the proliferative phase; EMS-WOI: endometriosis in the window of implantation phase; N-P: control in the proliferative phase; N-P/LS: control in the proliferative phase and late secretory phase; N-S: control in the secretory phase; N-WOI: control in the window of implantation phase
In the control group, T-cells represented the majority of the immune cells in the proliferative phase and decreased in the secretory phase. NK-cells comprised the majority of leukocytes in the secretory phase. The T- and NK-cell distributions across the menstrual cycle are consistent with those reported in previous studies [20–22]. In eutopic endometria of endometriosis, NK-cells were abundant in the proliferative phase and decreased in the secretory phase. NK cells are necessary for implantation, and the decreased NK-cells in the secretory phase of endometriosis probably contribute to defective endometrial receptivity [23]. One group of epithelial cells expressed CD45 and accounted for approximately 30% of the secretory phase of endometriosis. Tc-cells sharply changed from the proliferative to the secretory phase in the normal endometrium but varied slightly throughout the menstrual cycle in endometriosis. Tc-cells exhibit antitumor and antimicrobial effects via the release of cytotoxins. The proportion of Tc cells is reportedly higher in the eutopic endometria of women with endometriosis than in endometriosis-free controls; the elevated number of CD8+ T cells is related to endometriosis-associated infertility [11, 24]. The proportion of mast cells was higher in endometriosis than in control. Mast cells reportedly play a role in endometriosis pathology [25], and the abnormal number of mast cells in our results may provide evidence for this viewpoint. In summary, the cycle variation and inflammatory state of immune cells were disordered in endometriosis, which demonstrated its chronic inflammatory niche [26] (Fig. 2B and C). Endometrial immune cells foster immunological changes to establish tolerance during embryo implantation and early pregnancy, and abnormal immune cell regulation across the menstrual cycle in endometriosis could probably influence the establishment of immune tolerance [5, 27].
Four clusters of NK-cells shared the expression of granzymes B and pore-forming protein (GNLY). NK3 specifically expressed TOP2A and MKI67, which are associated with apoptosis and the cell cycle, and has recently been described in chronic neutropenia [28]. These NK-cells tended to be immature. NK3 was primarily detected in the secretory phase of endometriosis (42.00%) (Fig. 2D). In five clusters of CD45+ epithelial cells, Ep4 highly expressed C20orf85, C11orf88, and C1orf194, which are markers of ciliated epithelial cells in the endometrium [14]. SERPINB3 and SERPINB4 are maker genes for Ep5, these two genes have been reported to contribute to inflammatory disease and cancer [29]. And CD45+ epithelial cells mostly originate from the WOI phase of endometriosis (Fig. 2D and E). IL-2RA and CTLA4 were markers of endometrial Tregs, and Tc was marked by IFNG and GZMK (Fig. 2E). Twelve clusters of CD45+ cells showed an uneven distribution between endometriosis and the control; eleven clusters mostly came from endometriosis, including CD45+ epithelial cells, NK3, NK4, NKT, Tregs, mast cells, dendritic cells, and neutrophils, whereas most M2 cells were from the control group (Fig. 2D).
CD45+ cells were divided into four groups: endometriosis in the proliferative phase, endometriosis in the WOI phase, control in the proliferative/late secretory phase, and control in the WOI phase. We then selected common cytokines expressed by endometrial leukocytes and counted their median-normalized average expression in each cluster at the transcription level. We found that CCL2, IL-10, IL-1α, IL-1β, and MMP9 were expressed at the highest levels in macrophages; TNF-α, VEGF, and IL-17 were mostly secreted in B-cells; TGF-β and RANTES were expressed in NK cells; IFN-γ was expressed in Tc cells (Supplementary Fig. S1). IL-10 alone had an anti-inflammatory effect, and it was upregulated in the control secretory phase but reduced in the endometriosis secretory phase (Fig. 2F). Other proinflammatory cytokines mostly had the same cycle variation in both the endometriosis and control groups, with relatively higher or similar expression levels in the endometriosis group (Fig. 2F–I). These results also suggest a disordered inflammatory environment in the eutopic endometria of endometriosis.
Trajectory analysis of endometrial immune and epithelial cells in the endometriosis and control groups across the menstrual cycle.
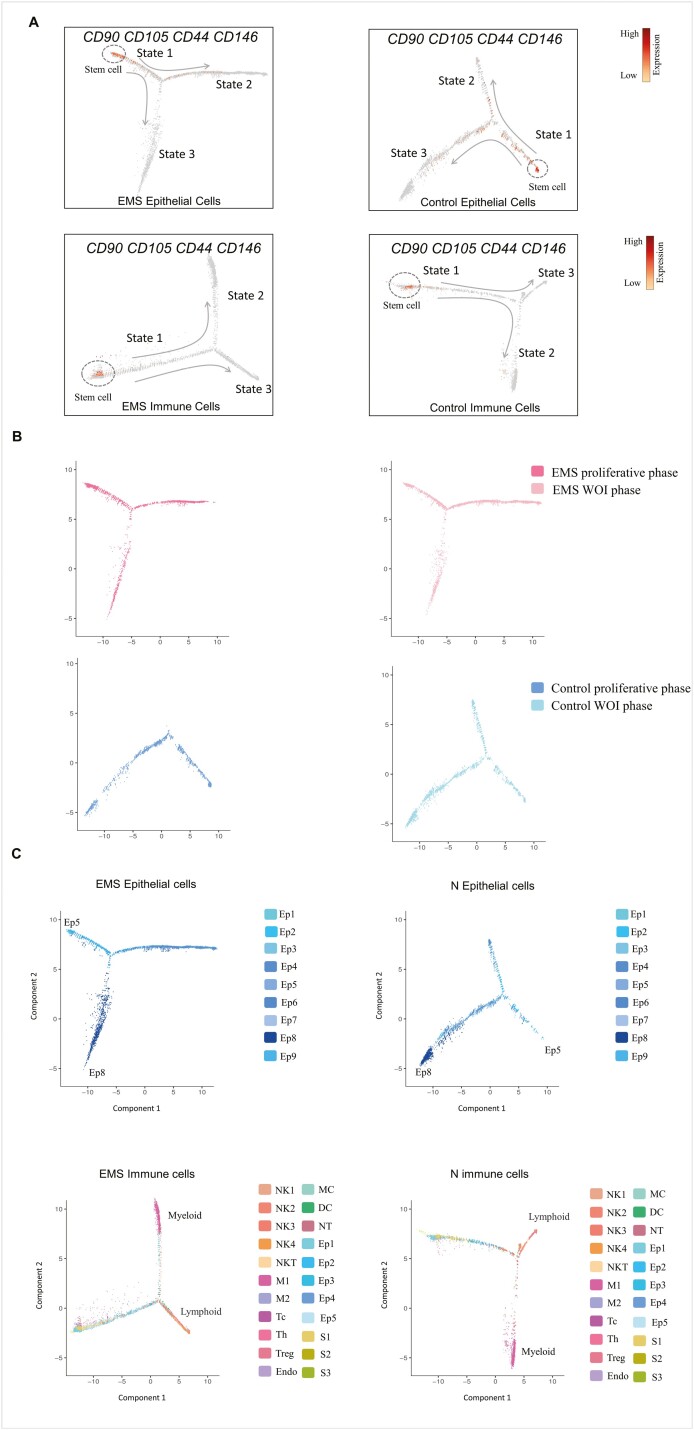
We used Monocle to order endometrial immune and epithelial cells in pseudotime to investigate the regulation of endometrial cells [18]. CD90/THY1 (a mesenchymal marker), CD105/ENG (endoglin), CD44 (endometrial stromal marker), and CD146/MCAM (hematopoietic marker) were used as endometrial stem cells to ensure the beginning of the trajectory [30]. The trajectories of epithelial cells and immune cells from the endometriosis group were different from that of the control. The trajectory was divided into three states: cells before the branching point were in the initial state (state 1), intermediate state cells were in state 2, and cells in state 3 were relatively mature at pseudotime (Fig. 3A).
Figure 3:
Trajectory analysis of epithelial and immune cells in endometria from the endometriosis and control groups. (A) The single-cell trajectory was predicted by Monocle 3 and visualized by t-SNE. Three states were identified in the endometriosis and control groups based on cell distribution. Using CD90, CD105, CD44, and CD146 as markers of endometrial stem cells, arrows represent the differentiation pathway. (B) The endometrial epithelial cell trajectories in the endometriosis and control groups from the proliferative to the WOI phase. (C) The subcluster locations of endometrial epithelial and immune cells in the endometriosis and control trajectories.
In this study, we focused on the distribution of epithelial and immune cells during the menstrual cycle. We found that epithelial cells in the proliferative phase were mostly in states 1 and 3, while epithelial cells in the secretory phase were distributed in all three states. Epithelial cells in state 2 were of a specific cell type in the secretory phase, and there were fewer state 2 cells in the endometriosis group than in the control group (Fig. 3B). The distribution of immune cells in the proliferative and secretory phases was similar between the two groups (Supplementary Fig. S2).
Epithelial cells of Ep5 expressed LRRC75A, TFPI2, and N-cadherin on the initiation of the epithelial cell trajectory, which were relatively progenitor epithelial cell clusters. The end of the state 3 branch was Ep8; they were ciliated epithelial cells that expressed C20orf85 and RSPH1 [14] (Figs. 1B and 3C). The immune cells that were initiated were CD45+ epithelial cells, stromal cells, and endothelial cells. Myeloid immune cells (macrophages, mast cells, neutrophils) were primarily in state 2 of the immune cell trajectories, while lymphoid immune cells (NK cells, T cells) were distributed in state 3 (Figs. 2A and 3C).
Ligand–receptor analysis of endometrial epithelial cells and immune cells
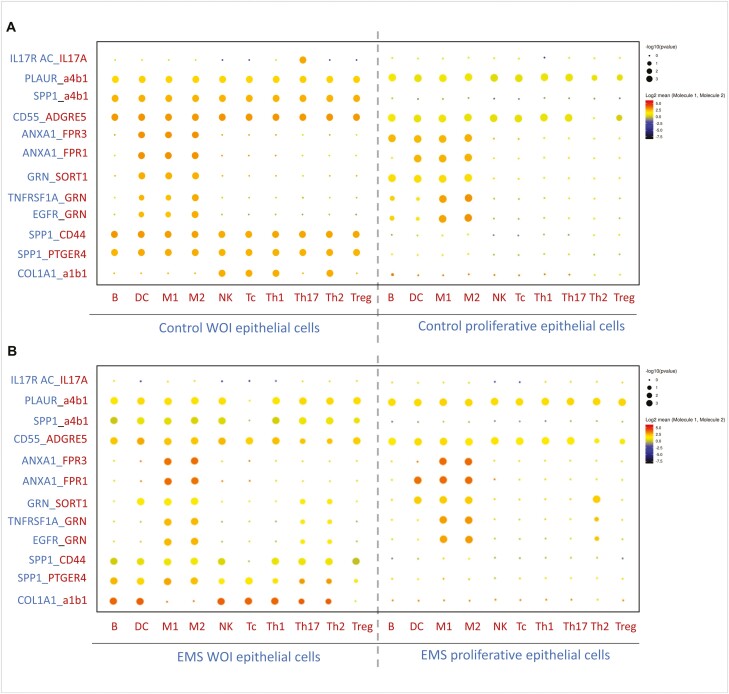
We used Cellphone DB [19] to identify ligand–receptor pairs among epithelial cells and subtypes of immune cells (B cells, DC, M1, M2, NK cells, Tc, Th1, Th2, Th17, and Treg). We compared epithelial cells expressing PAEP and CXCL14 in the WOI with those in the proliferative phase of the control, proliferative phase of endometriosis, and WOI phase of endometriosis. We found 11 pairs that were highly expressed in epithelial cells and immune cells in the WOI group and were expressed relatively poorly in the other groups. COL1A1_a1b1 was the only pair specifically expressed in the WOI phase of endometriosis between epithelial and immune cells (Fig. 4A and B).
Figure 4:
Ligand–receptor analysis of endometrial cells in the endometriosis and control groups. (A) Dot plot of the predicted interactions of endometrial epithelial cells with immune cell subtypes during the window of implantation and in the proliferative phase of controls. (B) Dot plot of the predicted interactions of endometrial epithelial cells with immune cell subtypes in the secretory phase and proliferative phase in endometriosis. P values are indicated by circle size. The expression levels of all the interacting genes are indicated by color stripe on the right
IL17 is a proinflammatory cytokine that is mostly secreted by Th17 cells; it has been reported to participate in defective endometrial receptivity [31]. Our data showed for the first time that the IL17_IL-17 receptor was expressed significantly in the epithelial and Th17 cells during WOI, indicating that an appropriate inflammatory response is beneficial for embryo implantation. Secreted phosphoprotein 1 (SPP1), also known as osteopontin (OPN), is a marker of endometrial receptivity secreted by epithelial cells [32]. Our results showed that SPP1 and its receptor pairs (SPP1_a4b1, SPP1_CD44, and SPP1_PTGER4) were highly expressed in epithelial cells and immune cells during the WOI, relatively less expressed in the endometriosis WOI phase, and almost not expressed in the two proliferative phases.
The annexin A1, (ANXA1)-formyl peptide receptor (FPR) system, is a potent mediator of the inflammatory response [33]. Our ligand–receptor analysis revealed its upregulated expression among epithelial cells, B cells, dendritic cells, and macrophages during WOI, and it may participate in the immune microenvironment balance of the endometrium. Defective expression of ANXA1_FPRs in the WOI phase of endometriosis could be the cause of immune disorders in the endometrial environment. Three pairs of ligands–receptors associated with (GRN) were highly expressed during WOI (GRN_SORT1, TNFSF1A_GRN, and EGFR_GRN). GRN is a growth factor with anti-inflammatory properties, and one study found decreased protein levels in eutopic endometria in patients with endometriosis compared with controls [34]. GRN has been reported to play a role in implantation, and we found defective GRN expression between epithelial and immune cells in the WOI phase of endometriosis. Collagen, an extracellular matrix (ECM), and integrins are involved in the interaction between embryos and the endometrium, which governs implantation success. One study reported significantly increased expression of integrin α1β1 during a spontaneous abortion and increased adhesion to collagen I [35]. Our study found that COL1A1_integrin α1β1 was upregulated in endometriosis during the WOI phase, and abnormalities in ECM and integrin may play a role in decreasing endometrial receptivity in endometriosis.
Discussion
Inflammatory, chemical, immunologic, epigenetic, and genetic changes have been discovered in eutopic endometria of patients with endometriosis compared to healthy women [36]. However, current studies usually focus on specific markers or pathways, one type of endometrial cell culture in vitro, or one immune cell subtype assessed using immunochemistry or flow cytometry. The overall landscape of the endometrial immune environment in endometriosis remains poorly understood. Endometrial immune cells include complex subtypes and dynamic changes across the menstrual cycle. The endometrial immune cells in endometriosis, and the cytokines secreted by them, contribute to the hallmarks of the disease pathophysiology [37]. We generated a single-cell transcriptome analysis of endometrial cells to better understand the mechanism of pathogenesis of this disease and to decipher the endometrial receptivity and immune environment of the eutopic endometrium in endometriosis.
We found that one cluster of epithelial cells that expressed endometrial receptivity markers (PAPE and CXCL14) came from the control group in the mid-secretory phase. These results were consistent with previous quantitative immunohistochemical and scRNA-seq studies [14, 38]. Furthermore, our data identified for the first time that this type of epithelial cell was absent in eutopic endometria of patients with endometriosis in the secretory phase. We reclustered CD45+ endometrial cells, divided these cells into proliferative and secretory phases, and compared the control with endometriosis. We first revealed the disorder of endometrial leukocyte fluctuation throughout the menstrual cycle using single-cell resolution and whole-tissue specimens, which were more accurate and closer to physiological status [4]. Interestingly, we found that some epithelial, stromal, and endothelial cells expressed CD45. Our trajectory analysis revealed that these cells initiated an immune cell trajectory. This indicates that these cells are probably relatively immature progenitor cells in the endometrium. Our database reflected the cytokine secretion of endometrial immune cells and revealed the inflammatory status of eutopic endometrium in endometriosis, which results in defective endometrial receptivity [26].
Trajectory analysis of the epithelial cells revealed that one state of cells specifically existed in the secretory phase of the endometrium. The number of cells in this state was significantly lower in the endometriosis group than that in the control group. This suggests that abnormal differentiation of epithelial cells in the secretory phase may contribute to defective endometrial receptivity in endometriosis. Another branch of epithelial cells ends with ciliated epithelial cells. Previous studies have reported two main lineages of endometrial epithelial cells, secretory and ciliated, across the menstrual cycle [39]. Our trajectory analysis using the census algorithm supports this conclusion. Except for one group of CD45+ epithelial cells, stromal cells, and endothelial cells mentioned above, immune cell trajectory analysis showed that mast cells, neutrophils, and macrophage were distributed in different branches from NK cells and T cells. These results indicate that endometrial immune cells differentiated lineage including myeloid and lymphoid-like peripheral blood.
Crosstalk between endometrial epithelial and immune cells influences endometrial physiology and homeostasis and is also involved in the development of endometriosis [40]. We identified extensive ligand–receptor pairs between endometrial immune and epithelial cells. We found that intercellular interactions during WOI were more active than those during the proliferative phase in the control and endometriosis groups during all menstrual cycles. The crosstalk between epithelial cells and trophoblasts influences embryo implantation [41]. Our ligand–receptor analysis demonstrated that epithelial and immune cell crosstalk during WOI is crucial for endometrial receptivity. These bioinformatics data provide clues for further analysis of the defective endometrial receptivity of endometriosis.
Our study had some limitations, such as the small sample size and lack of validation experiments. Further studies are needed to demonstrate the conclusions of our bioinformatic analysis, like in situ validation of receptor-ligand pairs analysis. Therefore, more evidence is needed to clearly state the pathogenesis of endometriosis-associated infertility.
In this study, we systematically revealed the eutopic endometrial single-cell transcriptome immune landscape in endometriosis and compared it with that of disease-free controls. Our trajectory and receptor–ligand analysis will be useful for deciphering endometrial cell differentiation rules and endometrial intercellular crosstalk between epithelial cells and the immune microenvironment. This database will provide an essential resource for understanding the eutopic endometrial inflammatory environment and defective endometrial receptivity in patients with endometriosis-associated infertility.
Supplementary Material
Acknowledgements
We thank for data analysis suggestions by You Duan, Key Laboratory of Birth Defects and Related Diseases of Women and Children of Ministry of Education.
Abbreviations
- ANXA1
annexin A1
- CCA
canonical correlation analysis
- CCL
chemokine(C-C motif) ligand
- DC
dendritic cells
- DMEM
Dulbecco’s modified Eagle’s medium
- ECM
extracellular matrix
- FPR
formyl peptide receptor
- GEMs
gel bead-in-emulsions
- IFN
interferon
- IL
interleukin
- MC
mast cells
- OPN
osteopontin
- PCA
principal component analysis
- SPP1
Secreted phosphoprotein 1
- Tc
cytotoxic T cells
- Th
T helper cells
- TNF
tumor necrosis factor
- Treg
regulatory T cells
- t-SNE
t-distributed stochastic neighbor embedding
- UMIs
unique molecular identifiers
- uNK
Uterine natural killer cell
- VEGF
vascular endothelial growth factor
- WOI
window of implantation
Contributor Information
Xin Huang, Division of Reproductive Medicine, West China Second University Hospital of Sichuan University, Chengdu, Sichuan, China; Key Laboratory of Birth Defects and Related Diseases of Women and Children of Ministry of Education, Chengdu, Sichuan, China; NHC Key Laboratory of Chronobiology (Sichuan University), Chengdu, Sichuan, China.
Lukanxuan Wu, Division of Reproductive Medicine, West China Second University Hospital of Sichuan University, Chengdu, Sichuan, China; Key Laboratory of Birth Defects and Related Diseases of Women and Children of Ministry of Education, Chengdu, Sichuan, China; NHC Key Laboratory of Chronobiology (Sichuan University), Chengdu, Sichuan, China.
Tianjiao Pei, Division of Reproductive Medicine, West China Second University Hospital of Sichuan University, Chengdu, Sichuan, China; Key Laboratory of Birth Defects and Related Diseases of Women and Children of Ministry of Education, Chengdu, Sichuan, China; NHC Key Laboratory of Chronobiology (Sichuan University), Chengdu, Sichuan, China.
Dong Liu, Division of Reproductive Medicine, West China Second University Hospital of Sichuan University, Chengdu, Sichuan, China; Key Laboratory of Birth Defects and Related Diseases of Women and Children of Ministry of Education, Chengdu, Sichuan, China.
Chang Liu, Division of Reproductive Medicine, West China Second University Hospital of Sichuan University, Chengdu, Sichuan, China; Key Laboratory of Birth Defects and Related Diseases of Women and Children of Ministry of Education, Chengdu, Sichuan, China.
Bin Luo, Division of Reproductive Medicine, West China Second University Hospital of Sichuan University, Chengdu, Sichuan, China; Key Laboratory of Birth Defects and Related Diseases of Women and Children of Ministry of Education, Chengdu, Sichuan, China.
Li Xiao, Division of Reproductive Medicine, West China Second University Hospital of Sichuan University, Chengdu, Sichuan, China; Key Laboratory of Birth Defects and Related Diseases of Women and Children of Ministry of Education, Chengdu, Sichuan, China.
Yujing Li, Division of Reproductive Medicine, West China Second University Hospital of Sichuan University, Chengdu, Sichuan, China; Key Laboratory of Birth Defects and Related Diseases of Women and Children of Ministry of Education, Chengdu, Sichuan, China; NHC Key Laboratory of Chronobiology (Sichuan University), Chengdu, Sichuan, China.
Ruiying Wang, Division of Reproductive Medicine, West China Second University Hospital of Sichuan University, Chengdu, Sichuan, China; Key Laboratory of Birth Defects and Related Diseases of Women and Children of Ministry of Education, Chengdu, Sichuan, China; NHC Key Laboratory of Chronobiology (Sichuan University), Chengdu, Sichuan, China.
Yunwei Ouyang, Division of Reproductive Medicine, West China Second University Hospital of Sichuan University, Chengdu, Sichuan, China; Key Laboratory of Birth Defects and Related Diseases of Women and Children of Ministry of Education, Chengdu, Sichuan, China.
Huili Zhu, Division of Reproductive Medicine, West China Second University Hospital of Sichuan University, Chengdu, Sichuan, China; Key Laboratory of Birth Defects and Related Diseases of Women and Children of Ministry of Education, Chengdu, Sichuan, China.
Wei Huang, Division of Reproductive Medicine, West China Second University Hospital of Sichuan University, Chengdu, Sichuan, China; Key Laboratory of Birth Defects and Related Diseases of Women and Children of Ministry of Education, Chengdu, Sichuan, China; NHC Key Laboratory of Chronobiology (Sichuan University), Chengdu, Sichuan, China.
Ethics Approval
The study was approved by the Ethics Committee of West China second University Hospital of Sichuan University (Grant number:2021028).
Patient Consent
Written informed consents were obtained from all participants.
Conflict of Interests
The authors have no conflicts of interest to declare in relation to this work.
Funding
This study was funded by the National Natural Science Foundation of China (Grant number: 82071625), National Key R&D Program of China (Grant number: 2017YCF1001200), Fundamental Research Funds for the Central Universities (Grant number: SCU2019C4198), and International Science and Technology Cooperation Project of Chengdu (Grant number: 2017-GH02-000060-HZ).
Data Availability
The datasets generated and analyzed during the current study are available in the NCBI’s Gene Expression Omnibus (accession code GSE214411).
Author Contributions
X.H. implemented research and analyzed data, drafted the manuscript; L.W. designed the research and revised the manuscript; T.P., D.L., C.L. collected samples and analyzed data; B.L., Y.L., R.W. collected and prepared samples; L.X., H.Z., Y.O. conceived and supervised the research; W.H. designed the research, revised the manuscript and approved the final vision.
References
- 1. Zondervan KT, Becker CM, Missmer SE. Endometriosis.. New Engl J Med 2020, 382, 1244–56. [DOI] [PubMed] [Google Scholar]
- 2. Khine YM, Taniguchi F, Harada T.. Clinical management of endometriosis-associated infertility. Reprod Med Biol 2016, 15, 217–25. doi: 10.1007/s12522-016-0237-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lee JY, Lee M, Lee SK.. Role of endometrial immune cells in implantation. Clin Exp Reprod Med 2011, 38, 119–25. doi: 10.5653/cerm.2011.38.3.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Vallve-Juanico J, Houshdaran S, Giudice LC.. The endometrial immune environment of women with endometriosis. Hum Reprod Update 2019, 25, 565–92. doi: 10.1093/humupd/dmz018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kitazawa J, Kimura F, Nakamura A, Morimune A, Takahashi A, Takashima A, et al. Endometrial immunity for embryo implantation and pregnancy establishment. Tohoku J Exp Med 2020, 250, 49–60. doi: 10.1620/tjem.250.49. [DOI] [PubMed] [Google Scholar]
- 6. Poli-Neto OB, Meola J, Rosa-e-Silva JC, Tiezzi D.. Transcriptome meta-analysis reveals differences of immune profile between eutopic endometrium from stage I-II and III-IV endometriosis independently of hormonal milieu. Sci Rep 2020, 10, 313. doi: 10.1038/s41598-019-57207-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Li M-Z, Wu Y-H, Ali M, Wu X-Q, Nie M-F.. Endometrial stromal cells treated by tumor necrosis factor-alpha stimulate macrophages polarized toward M2 via interleukin-6 and monocyte chemoattractant protein-1. J Obstet Gynaecol Res 2020, 46, 293–301. doi: 10.1111/jog.14135. [DOI] [PubMed] [Google Scholar]
- 8. Takebayashi A, Kimura F, Kishi Y, Ishida M, Takahashi A, Yamanaka A, et al. Subpopulations of macrophages within eutopic endometrium of endometriosis patients. Am J Reprod Immunol 2015, 73, 221–31. doi: 10.1111/aji.12331. [DOI] [PubMed] [Google Scholar]
- 9. Vallve-Juanico J, Santamaria X, Vo KC, Houshdaran S, Giudice LC.. Macrophages display proinflammatory phenotypes in the eutopic endometrium of women with endometriosis with relevance to an infectious etiology of the disease. Fertil Steril 2019, 112, 1118–28. doi: 10.1016/j.fertnstert.2019.08.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hey-Cunningham AJ, Riaz A, Fromm PD, Kupresanin F, Markham R, McGuire HM.. Circulating and endometrial regulatory t-cell and related populations in endometriosis and infertility: endometriosis is associated with blunting of endometrial cyclical effects and reduced proportions in moderate-severe disease. Reprod Sci 2021, 29, 229–42. doi: 10.1007/s43032-021-00658-4. [DOI] [PubMed] [Google Scholar]
- 11. Wu X-G, Chen J-J, Zhou H-L, Wu Y, Lin F, Shi J, et al. Identification and validation of the signatures of infiltrating immune cells in the eutopic endometrium endometria of women with endometriosis. Front Immunol 2021, 12, 671201. doi: 10.3389/fimmu.2021.671201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ranzoni AM, Strzelecka PM, Cvejic A.. Application of single-cell RNA sequencing methodologies in understanding haematopoiesis and immunology. Essays Biochem 2019, 63, 217–25. doi: 10.1042/EBC20180072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ma J, Zhang L, Zhan H, Mo Y, Ren Z, Shao A, et al. Single-cell transcriptomic analysis of endometriosis provides insights into fibroblast fates and immune cell heterogeneity. Cell Biosci 2021, 11, 125. doi: 10.1186/s13578-021-00637-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wang W, Vilella F, Alama P, Moreno I, Mignardi M, Isakova A, et al. Single-cell transcriptomic atlas of the human endometrium during the menstrual cycle. Nat Med 2020, 26, 1644–53. doi: 10.1038/s41591-020-1040-z. [DOI] [PubMed] [Google Scholar]
- 15. Schenken RS, Guzick DS.. Revised endometriosis classification: 1996. Fertil Steril 1997, 67, 815–6. doi: 10.1016/s0015-0282(97)81390-8. [DOI] [PubMed] [Google Scholar]
- 16. Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Butler A, Hoffman P, Smibert P, Papalexi E, Satija R.. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 2018, 36, 411–20. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Qiu X, Hill A, Packer J, Lin D, Ma Y-A, Trapnell C.. Single-cell mRNA quantification and differential analysis with Census. Nat Methods 2017, 14, 309–15. doi: 10.1038/nmeth.4150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Efremova M, Vento-Tormo M, Teichmann SA, Vento-Tormo RC.. Inferring cell-cell communication from combined expression of multi-subunit ligand-receptor complexes. Nat Protoc 2020, 15, 1484–506. [DOI] [PubMed] [Google Scholar]
- 20. Janosevic DR, Trandafilovic M, Krtinic D, Colovic H, Stevanovic JM, Dinic SP-T.. Endometrial immunocompetent cells in proliferative and secretory phase of normal menstrual cycle. Folia Morphol 2020, 79, 296–302. [DOI] [PubMed] [Google Scholar]
- 21. Mselle TF, Meadows SK, Eriksson M, Smith JM, Shen L, Wira CR, et al. Unique characteristics of NK cells throughout the human female reproductive tract. Clin Immunol 2007, 124, 69–76. doi: 10.1016/j.clim.2007.04.008. [DOI] [PubMed] [Google Scholar]
- 22. Yeaman GR, Guyre PM, Fanger MW, et al. Unique CD8(+) T cell-rich lymphoid aggregates in human uterine endometrium. J Leukocyte Biol 1997, 61, 427–35. [PubMed] [Google Scholar]
- 23. Huber WJ, III, Sauerbrun-Cutler M-T, Krueger PM, Sharma S.. Novel predictive and therapeutic options for better pregnancy outcome in frozen embryo transfer cycles. Am J Reprod Immunol 2021, 85, e13300. [DOI] [PubMed] [Google Scholar]
- 24. Poli-Neto OB, Carlos D, Favaretto Junior A, Rosa-e-Silva JC, Meola J, Tiezzi D.. Eutopic endometrium from women with endometriosis and chlamydial endometritis share immunological cell types and DNA repair imbalance: a transcriptome meta-analytical perspective. J Reprod Immunol 2021, 145, 103307. [DOI] [PubMed] [Google Scholar]
- 25. McCallion A, Nasirzadeh Y, Lingegowda H, Miller JE, Khalaj K, Ahn SH, et al. Estrogen mediates inflammatory role of mast cells in endometriosis pathophysiology. Front Immunol 2022, 13, 961599. doi: 10.3389/fimmu.2022.961599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lin Y-H, Chen Y-H, Chang H-Y, Au H-K, Tzeng C-R, Huang Y-H.. Chronic Niche inflammation in endometriosis-associated infertility: current understanding and future therapeutic strategies. Inter J Mol Sci. 2018, 19, 2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Murata H, Tanaka S, Okada H.. Immune tolerance of the human decidua. J Clin Med 2021, 10, 351. doi: 10.3390/jcm10020351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sohlberg E, Pfefferle A, Ask EH, et al. Perturbed NK-cell homeostasis associated with disease severity in chronic neutropenia. Blood 2022, 139, 704–16. [DOI] [PubMed] [Google Scholar]
- 29. Sun Y, Sheshadri N, Zong WX.. SERPINB3 and B4: From biochemistry to biology. Semin Cell Dev Biol 2017, 62, 170–7. doi: 10.1016/j.semcdb.2016.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tavakol S, Azedi F, Hoveizi E, Ai J, Joghataei MT.. Human endometrial stem cell isolation from endometrium and menstrual blood. Bio-Protocol 2018, 8, e2693. doi: 10.21769/BioProtoc.2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang W, Zhang H, Chen Z, et al. Endometrial TGF-beta, IL-10, IL-17 and autophagy are dysregulated in women with recurrent implantation failure with chronic endometritis. Reprod Biol Endocrinol 2019, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Frank JW, Seo H, Burghardt RC, Bayless KJ, Johnson GA.. ITGAV (alpha v integrins) bind SPP1 (osteopontin) to support trophoblast cell adhesion. Reproduction 2017, 153, 695–706. doi: 10.1530/REP-17-0043. [DOI] [PubMed] [Google Scholar]
- 33. Marmorato MP, Gimenes AD, Costa Andrade FE, Oliani SM, Gil CD.. Involvement of the annexin A1-Fpr anti-inflammatory system in the ocular allergy. Eur J Pharmacol 2019, 842, 298–305. [DOI] [PubMed] [Google Scholar]
- 34. Holzer I, Weber AM, Marshall A, Freis A, Jauckus J, Strowitzki T, et al. GRN, NOTCH3, FN1, and PINK1 expression in eutopic endometrium—potential biomarkers in the detection of endometriosis—a pilot study. J Assist Reprod Gen 2020, 7, 2723–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sohn JO, Park HJ, Kim SH, Kim MJ, Song HJ, Yun JI, et al. Integrins expressed on the surface of human endometrial stromal cells derived from a female patient experiencing spontaneous abortion. Hum Cell 2020, 33, 29–36. doi: 10.1007/s13577-019-00278-w. [DOI] [PubMed] [Google Scholar]
- 36. Lagana AS, Garzon S, Goette M, et al. The pathogenesis of endometriosis: molecular and cell biology insights. Inter J Mol Sci 2019, 20, 5615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Symons LK, Miller JE, Kay VR, Marks RM, Liblik K, Koti M, et al. The immunopathophysiology of endometriosis. Trends Mol Med 2018, 24, 748–62. doi: 10.1016/j.molmed.2018.07.004. [DOI] [PubMed] [Google Scholar]
- 38. Leach RE, Jessmon P, Coutifaris C, Kruger M, Myers ER, Ali-Fehmi R, et al.; Reproductive Medicine Network. High throughput, cell type-specific analysis of key proteins in human endometrial biopsies of women from fertile and infertile couples. Hum Reprod 2012, 27, 814–28. doi: 10.1093/humrep/der436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Garcia-Alonso L, Handfield L-F, Roberts K, Nikolakopoulou K, Fernando RC, Gardner L, et al. Mapping the temporal and spatial dynamics of the human endometrium in vivo and in vitro. Nat Genet 2021, 53, 1698–711. doi: 10.1038/s41588-021-00972-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Li T, Wang J, Guo X, Yu Q, Ding S, Xu X, et al. Possible involvement of crosstalk between endometrial cells and mast cells in the development of endometriosis via CCL8/CCR1. Biomed Pharmacother 2020, 129, 110476. doi: 10.1016/j.biopha.2020.110476. [DOI] [PubMed] [Google Scholar]
- 41. Karizbodagh MP, Rashidi B, Sahebkar A, Masoudifar A, Mirzaei H.. Implantation window and angiogenesis. J Cell Biochem 2017, 118, 4141–51. doi: 10.1002/jcb.26088. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and analyzed during the current study are available in the NCBI’s Gene Expression Omnibus (accession code GSE214411).