Abstract
During development, neural progenitors undergo temporal patterning as they age to sequentially generate differently fated progeny. Temporal patterning of neural progenitors is relatively well-studied in Drosophila. Temporal cascades of transcription factors or opposing temporal gradients of RNA-binding proteins are expressed in neural progenitors as they age to control the fates of the progeny. The temporal progression is mostly driven by intrinsic mechanisms including cross-regulations between temporal genes, but environmental cues also play important roles in certain transitions. Vertebrate neural progenitors demonstrate greater plasticity in response to extrinsic cues. Recent studies suggest that vertebrate neural progenitors are also temporally patterned by a combination of transcriptional and post-transcriptional mechanisms in response to extracellular signaling to regulate neural fate specification. In this review, we summarize recent advances in the study of temporal patterning of neural progenitors in Drosophila and vertebrates. We also discuss the involvement of epigenetic mechanisms, specifically the Polycomb group complexes and ATP-dependent chromatin remodeling complexes, in the temporal patterning of neural progenitors.
Keywords: Temporal patterning, Neural progenitors, Temporal Transcription Factor cascade, Epigenetic regulation
Graphical Abstract:
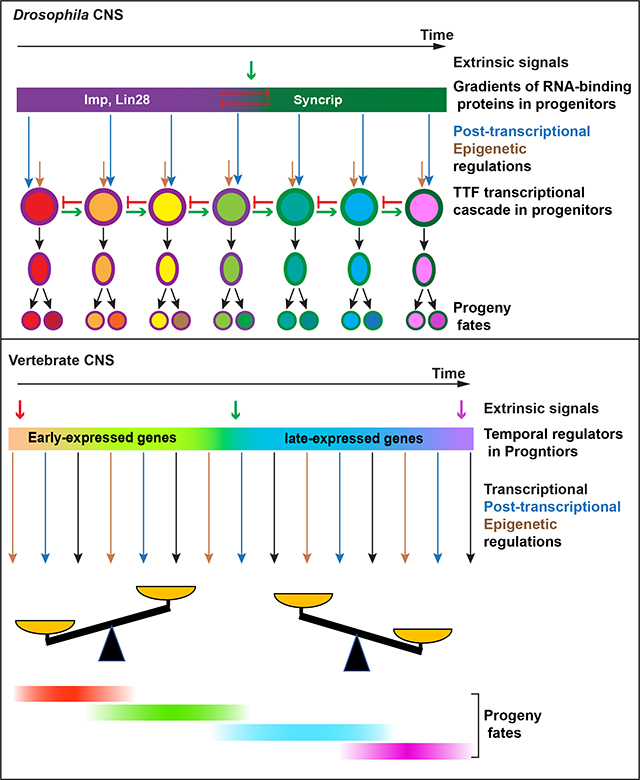
1. Introduction
A key question in developmental biology is how a small pool of progenitor cells generate the great diversity of cell types that comprise a multicellular organism. This question has inspired research over many decades and has led to the description of developmental patterning programs. In the developing nervous system, integration of spatial and temporal patterning of neural progenitors greatly diversifies neural progeny fates [reviewed in (Guillemot, 2007; Lin and Lee, 2012; Allan and Thor, 2015; Azzarelli, Hardwick and Philpott, 2015; Doe, 2017; Holguera and Desplan, 2018; Sagner and Briscoe, 2019)]. This review will focus on recent developments in the study of temporal patterning mechanisms of neural progenitors in Drosophila and vertebrate model systems. Temporal patterning refers to the developmental process in which neural progenitors change over time and sequentially generate differentiated progeny with distinct identities that correlate with the birth order (Pearson and Doe, 2004). Temporal patterning can be driven by both progenitor cell intrinsic mechanisms and environmental signals that impact progenitor gene expression.
Although the molecular mechanisms controlling temporal patterning are incompletely understood, there have been considerable progresses in this field. Studies have suggested that regulation of temporal patterning may involve transcriptional and epigenetic mechanisms as well as post-transcriptional mechanisms. Historically transcriptional events underlying temporal patterning have been most widely demonstrated: different subsets of transcription factors are expressed in neural progenitors as they age and the transcriptional changes over time direct their post-mitotic progeny to differentiate into distinct neuron subtypes. As with changes in patterns of transcription factor expression, the complement of post-transcriptional regulators including microRNAs, RNA-binding proteins and translational repressors in neural progenitors also evolves with developmental time, providing an additional layer of regulation. Finally changes in epigenetic landscapes of neural progenitors over their development have been shown to have important consequences for regulating temporal patterning in both Drosophila and in vertebrates.
In this review we summarize our existing knowledge of temporal patterning mechanisms. While we primarily focus on transcriptional and epigenetic mechanisms underlying temporal patterning due to the availability of a vast body of work in this area, we also briefly discuss the current understanding of the roles played by post-transcriptional regulatory mechanisms. Taken together we hope this will provide a unifying picture of the broad range of processes at work for specifying cell identities by temporal patterning in developing nervous systems.
2. Temporal patterning of Drosophila neuroblasts
2.1. Introduction of Drosophila neuroblasts
Drosophila neural progenitors called neuroblasts (NBs) exhibit three main modes of asymmetric divisions classified as type 0, type I and type II, based on the number of post-mitotic progeny (neurons or glia) generated at each division [reviewed in (Walsh and Doe, 2017)]. A type 0 division generates a self-renewed neuroblast and one post-mitotic progeny. A type I division generates a self-renewed neuroblast and a daughter Ganglion Mother Cell (GMC). The GMC in turn divides once to produce two postmitotic daughters. A type II neuroblast divides asymmetrically multiple times to self-renew and to generate a series of intermediate progenitor cells (INPs). Each INP then undergoes between 4 to 6 asymmetric divisions forming one GMC and a self-renewed INP at each division until it exits from the cell cycle (Walsh and Doe, 2017).
Temporal patterning of neuroblasts is studied in different parts of the Drosophila nervous system. Here we will focus on the embryonic and larval ventral nerve cord (VNC), larval central brain (including type I NBs in the mushroom body and antennal lobes, and type II NBs), and the larval optic-lobe medulla [reviewed in (Homem and Knoblich, 2012; Li, Chen and Desplan, 2013; Doe, 2017; Lee, 2017; Walsh and Doe, 2017; Maurange, 2020)]. The Drosophila VNC is analogous to the vertebrate spinal cord. In the developing VNC there are 30 neuroblasts per hemi-segment that obtain their specific lineage identity based on spatial patterning. At the embryo to larva transition, some embryonic NBs commit to apoptosis or exit the cell cycle, while others enter into quiescence to be reactivated later in the larval stages and generate ~90% of the neurons that constitute the adult CNS (Doe, 2017). Most NBs in the VNC are type I, although some switch to type 0 at the end of their lineage (Baumgardt et al., 2014). In the larval central brain, among the approximately100 type I NBs per lobe, the antennal lobe and the mushroom body neuroblasts are well characterized and they have long lineages (Lee, 2017). Three antennal lobe NBs sequentially give rise to neurons that comprise the antennal lobe, while the four mushroom body NBs generate mushroom body neurons in a defined order (Lee, 2017). There are also eight type II NBs per lobe in the larval central brain, including six dorsal medial (DM1-6) and two dorsal lateral (DL1-2) neuroblasts. Type II neuroblasts have expanded lineages and are capable of additional diversity beyond what is achievable with Type I division alone (Walsh and Doe, 2017). In the larval optic lobe, medulla NBs are sequentially converted from neuroepithelia cells (NEs) as a neurogenesis wave spreads through the NE. As a result NBs of different ages are aligned on a spatial axis with the younger neuroblasts positioned laterally and the older neuroblasts medially along the medulla, making it easier to identify age-related temporal changes (Yasugi et al., 2008; Li et al., 2013; Suzuki et al., 2013). Most medulla NBs are type I NBs except the first division of medulla tip NBs is type 0 (Bertet et al., 2014).
2.2. Temporal patterning of Drosophila neuroblasts
Studies of temporal patterning in Drosophila have demonstrated two prominent mechanisms. The first is the transcription factor cascade mechanism where a series of Temporal Transcription Factors (TTFs) are expressed sequentially in neuroblasts and specify distinct neuron subtypes over the duration of each transcription factor expression. The second is a post-transcriptional mechanism where two mutually opposing gradients of RNA-binding proteins are expressed in neuroblasts as they age. The two mechanisms can act together or separately to control the sequential generation of different neural fates [reviewed in (Homem and Knoblich, 2012; Li, Chen and Desplan, 2013; Doe, 2017; Lee, 2017; Walsh and Doe, 2017; Miyares and Lee, 2019; Maurange, 2020)]. In this section we discuss specific examples of temporal patterning in Drosophila nervous system.
The first TTF cascade in embryonic VNC NBs.
Pioneering studies in the embryonic VNC NBs identified the first TTF cascade in Drosophila, where Hunchback (Hb), Krüppel (Kr), Nubbin/Pdm2 (Pdm), Castor (Cas) and Grainy head (Grh) are sequentially expressed in NBs, and are required for the sequential specification of different neural fates (Kambadur et al., 1998; Brody and Odenwald, 2000; Isshiki et al., 2001; Grosskortenhaus et al., 2005; Grosskortenhaus, Robinson and Doe, 2006; Baumgardt et al., 2009) (Figure 1A). How this TTF cascade controls progeny fates was characterized in detail for several NB lineages, and small variations of temporal patterning are present in different lineages [Reviewed in (Doe, 2017)]. Subsequently, temporal regulators that are different from the embryonic VNC TTFs were identified in other parts of the fly nervous system.
Figure 1. Temporal patterning of Drosophila neuroblasts.
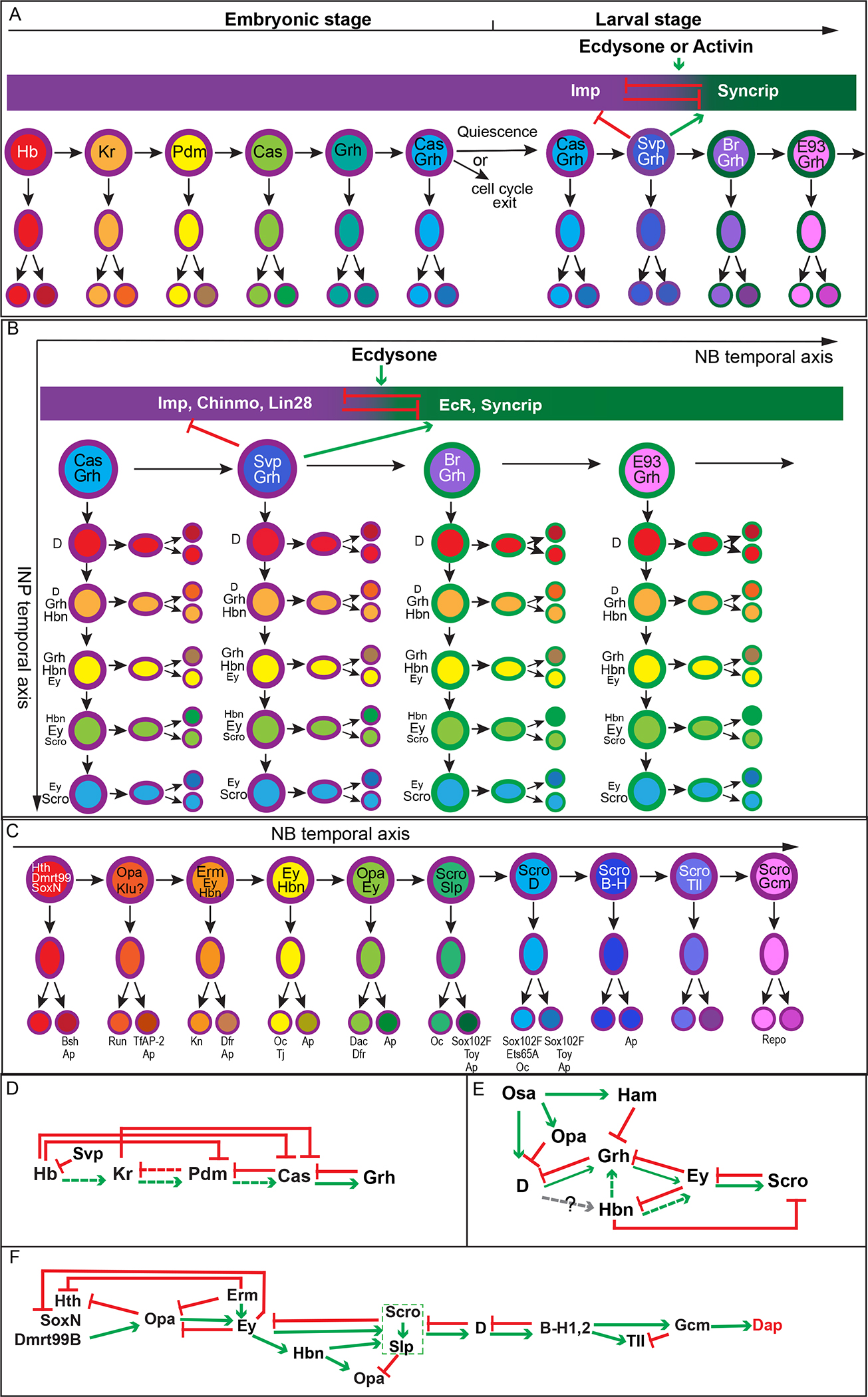
(A) Temporal patterning in embryonic and larval stage VNC and central brain type I neuroblasts. The embryonic VNC NBs sequentially express Hb, Kr, Pdm, Cas and Grh, which control the birth-order dependent progeny fate specification. Usually the two daughters of a GMC adopt different fates due to Notch dependent binary fate choices. This is a simplified and generalized model, and there are slight variations of temporal patterning in different NB linages (Doe, 2017). In certain NB lineages, there is another stage in which Cas and Grh are co-expressed. At the embryo to larva transition, some NBs exit the cell cycle, while others enter quiescence and become re-activated at the larval stage. Early larval TTFs Cas and Svp are required for the transition between RNA-binding proteins Imp and Syncrip that form opposing gradients. In at least some NBs, the transition is regulated by Ecdysone or Activin signaling. Late larval TTFs in the Syncrip window include Br and E93 is some NB lineages. (B) Temporal patterning in type II neuroblast lineages. In addition to the NB temporal axis, there is a second INP temporal axis. INPs sequentially express D, Grh, Hbn, Ey and Scro as they undergo self-renewing asymmetric divisions to generate GMCs which divide to produce neurons. Combinatorial temporal patterning greatly expands neural diversity. (C) Temporal patterning in medulla neuroblasts. Medulla neuroblasts sequentially express TTFs that control the sequential generation of different cell fates through regulating the expression of neuronal transcription factors. Hth, SoxN and Dmrt99B start their expression in the neuroepithelium, and each of them is required for the expression of Bsh in neurons. Opa is expressed in two waves in NBs and possibly serve as TTF for two different temporal stages. Opa is required for the generation of neurons expressing Run, TfAP-2, or Dac + Dfr (also known as Vvl). Erm and Ey are required for the generation of neurons expressing Kn, or Ap+Dfr. Hbn is required for the generation of neurons expressing Oc and Tj. Scro and Slp are required for the expression of Sox102F; D is required for the expression of Ets65A; and finally Gcm is required for the switch to gliogenesis and cell cycle exit. (D)(E)(F) Cross-regulatory interactions in Drosophila TTF cascades. Green pointed arrows indicate activation, and red flat-headed arrows indicate repression. Arrows with dashed lines indicate that a certain cross-regulation is sufficient but not required for the transition. (D) Cross-regulatory interactions in the embryonic VNC TTF cascade. (E) Regulation of temporal progression in the INP TTF cascade of type II NB lineages. Epigenetic regulators Osa, Ham and a transcription factor Opa are required to regulate the temporal progression in addition to cross-regulations between TTFs. (F) Cross-regulatory interactions in the medulla TTF cascade. Dashed rectangles around Scro and Slp indicates that Ey is required for the activation of both Slp and Scro; while both Slp and Scro are required to repress Ey.
Temporal regulators in larval VNC and central brain NBs.
In the larval stage, temporal patterning continues in the postembryonic neuroblasts of the VNC and central brain (including both type I and type II NBs). In addition to TTFs, two RNA-binding proteins, IGF-II mRNA-binding protein (Imp) and Syncrip (Syp), are expressed in opposing temporal gradients in both type I and type II NBs, with Imp expression being the highest in the youngest NBs and gradually diminishing as NBs age, and Syncrip being expressed in gradually increasing concentrations peaking in the oldest NBs (Liu et al., 2015; Ren et al., 2017; Syed, Mark and Doe, 2017; Yang et al., 2017) (Figure 1A,B). The two proteins post-transcriptionally regulate target genes including chronologically inappropriate morphogenesis (chinmo), encoding a BTB-zinc finger transcription factor, to determine neural fates (Zhu et al., 2006; Liu et al., 2015). TTFs and RNA binding proteins co-operate in the regulation of temporal patterning. In type II and some type I larval NBs, early stage NBs express RNA-binding proteins Imp and Lin-28, as well as early larval TTFs Cas, Seven-up (Svp) and Chinmo (Maurange, Cheng and Gould, 2008; Benito-Sipos et al., 2011; Bayraktar and Doe, 2013; Liu et al., 2015; Narbonne-Reveau et al., 2016; Syed, Mark and Doe, 2017). Svp triggers the switch from Imp to Syncrip by activating the expression of the Ecdysone receptor (EcR), enabling neuroblasts to respond to the hormone ecdysone (Ren et al., 2017; Syed, Mark and Doe, 2017). EcR is also required for the sequential expression of late TTFs including Broad (Br) and Eip93 in the Syncrip window (Syed, Mark and Doe, 2017). Although type I and type II larval NBs follow a largely similar temporal patterning scheme, there are variations in TTF composition in different lineages. In some type I NB lineages including those of the mushroom body and antennal lobe, Chinmo and Br proteins are not expressed in NBs (Zhu et al., 2006), and the Imp to Syncrip transition was found to be dependent on activin signaling with ligands coming from glia in mushroom body NBs (Marchetti and Tavosanis, 2019; Rossi and Desplan, 2020). In addition, one of the embryonic TTFs Kr was found to define one out of 40 temporal fates in the antennal lobe neuroblast lineage (Kao et al., 2012). In summary, in larval VNC and central brain neuroblasts, temporal patterning of neural fates is achieved by a combination of patterning transcription factors and gradients of RNA-binding proteins. This system also demonstrates how cell-intrinsic programs enable neuroblasts to respond to cell extrinsic signals like ecdysone which in turn control subsequent stages of temporal progression.
INP temporal cascade in type II NB lineages.
In addition to temporal patterning of type II NBs, the INPs undergo a second temporal patterning program. The INP temporal patterning axis acts in combination with the temporal patterning of type II NBs to further expand neural diversity (Bayraktar and Doe, 2013; Ren et al., 2017) (Figure 1B).Three transcription factors, Dichaete (D), Grh, and Eyeless (Ey), identified through antibody screening, were shown to form a temporal cascade in INPs and control their sequential generation of differently fated progeny (Bayraktar and Doe, 2013) (Figure 1B). Recently, using a combination of single-cell RNA-seq (scRNA-seq) and a new technique, NanoDam (a modified Dam-ID approach using tag-recognizing nanobodies fused to Dam to profile binding sites of endogenously tagged known TTFs in the genome), a study identified two novel TTFs, Homeobrain (Hbn) and Scarecrow (Scro), as part of the INP temporal cascade (Tang et al., 2021) (Figure 1B). Therefore, the current INP temporal cascade is D->Grh/Hbn->Ey->Scro (Figure 1B).
A TTF cascade in medulla neuroblast.
In the larval optic lobe medulla neuroblasts, six sequentially expressed TTFs were identified through antibody screening: Homothorax (Hth), Klumpfuss (Klu), Eyeless (Ey), Sloppy paired (Slp), Dichaete (D), and Tailless (Tll) (Li et al., 2013; Suzuki et al., 2013)(Figure 1C). They control the sequential generation of different medulla neurons through regulating the expression of neuronal transcription factors. Loss of Hth, Ey, Slp, or D caused loss of the corresponding neuronal transcription factors (Li et al., 2013; Suzuki et al., 2013; Naidu et al., 2020). Loss of Klu caused a general defect in neuroblast development, and precluded examination of neural fates (Suzuki et al., 2013). NBs at the posterior tips of the outer proliferation center (OPC) use a slightly different TTF cascade from the main medulla, which starts with Distal-less (Dll) instead of Hth, and stops at the D stage (Bertet et al., 2014). Recently, two scRNA-seq studies using all larval optic lobe cells or FACS-sorted medulla neuroblasts respectively, identified overlapping and complementary lists of novel TTFs in the Drosophila medulla temporal cascade: SoxNeuro (SoxN), Doublesex-Mab related 99B (Dmrt99B), Odd paired (Opa), Earmuff (Erm, dFezf2), Hbn, Scro, BarH1, BarH2 and Glial cells missing (Gcm) (Konstantinides et al., 2021; Zhu et al., 2021). The majority of these novel TTFs were shown to be each required for the expression of certain neuronal transcription factors or glia markers, thus together they control the sequential generation of different progeny fates (Konstantinides et al., 2021; Zhu et al., 2021) (Figure 1C).
2.3. Control of temporal transitions
The temporal progression of these TTF cascades were shown to be mostly driven by intrinsic mechanisms mainly the cross-regulation between temporal genes, but specific mechanisms may differ case by case. In the Hb->Kr->Pdm->Cas TTF cascade, misexpression experiments showed that mis-expression of one TTF is sufficient to activate the next TTF in the pathway and repress the “next plus one” TTF (Isshiki et al., 2001) (Figure 1D). Loss of Hb, Kr, or Pdm causes the corresponding fates to be skipped, but does not block temporal progression, with the exception that Cas is required to repress Pdm and activate Grh, while Grh is required to repress Cas (Isshiki et al., 2001; Grosskortenhaus, Robinson and Doe, 2006; Maurange, Cheng and Gould, 2008; Baumgardt et al., 2009) (Figure 1D). These cross-regulations suggest that temporal progression can be driven by either an activator-relay timer (in which the next TTF is switched on when the activator level is increased above a certain threshold ) or a repressor-decay timer (in which the next TTF is switched on once a repressor decays below a certain threshold) (Averbukh et al., 2018). Computational analysis suggests that the decay timer is more robust than the relay timer (Averbukh et al., 2018). Experimental studies showed that loss of Hb does not affect the expression of the next TTF Kr, but advances the induction timing of the next plus one TTF Pdm; similarly, kr mutant does not affect Pdm expression, but advanced Cas induction; also pdm mutant didn’t have much effect on Cas induction (Averbukh et al., 2018). Thus theoretical analysis in combination with evidence from experiments suggest that in this TTF cascade, the repressor-decay mechanism is dominant in driving the temporal transitions (Averbukh et al., 2018). In addition, the Hb to Kr transition requires a “switching factor”, Svp, which represses Hb expression (Kanai, Okabe and Hiromi, 2005) (Figure 1D). Switching factors regulate temporal transitions, but may not act directly to specify neural fates.
In the medulla TTF cascade, there are both similarities and differences in the cross-regulations as compared with the temporal patterning programs of the VNC. With the addition of several new TTFs in the cascade, extensive cross-regulations were identified among TTFs that generally follow the same rule: one TTF is required to activate the next, and repress the previous TTF (Li et al., 2013; Suzuki et al., 2013; Konstantinides et al., 2021; Zhu et al., 2021). For the most part, loss of one TTF leads to loss of the next TTF, and blocks temporal progression. Thus, in the most part of the medulla temporal cascade, the expression of one TTF is dependent on the expression of its activator (Li et al., 2013; Suzuki et al., 2013; Konstantinides et al., 2021; Zhu et al., 2021) (Figure 1F). Therefore, the activator relay mechanism is at least necessary to drive most of the medulla TTF cascade progression. However, the repressor-decay mechanism may also shape the kinetics of the progression, and it will be interesting to identify such possible regulatory interactions. Furthermore, within this comprehensive list of temporal genes, the cross-regulations are more complex than a simple linear cascade, and this complexity can increase the number of temporal windows marked by combinations of TTFs which can further diversify neural fates (Figure 1F). Since regulatory interactions between TTFs have mostly been inferred from analyses of TTF mutant phenotypes, it is important to note that we cannot conclude whether each interaction is direct transcriptional regulation (one TTF directly regulates the transcription of another TTF by binding to its enhancer) or indirect regulation involving intermediate factors. To examine whether they are direct transcriptional regulation, future studies will need to identify regulatory elements controlling expression of TTFs, profile the binding of TTFs genome wide and examine expressions of TTFs after mutagenizing binding sites of their regulators.
In the INP temporal cascade, the D->Grh/Hbn->Ey->Scro transitions also require cross-regulations including feedforward activation and feedback repression. Among them, Hbn is sufficient to activate Grh and Ey; Ey is required and sufficient to repress Hbn and Grh, and activate Scro; Scro is in turn required and sufficient to repress Ey; Finally Hbn is necessary and sufficient to repress Scro (the next plus one TTF) (Bayraktar and Doe, 2013; Tang et al., 2021) (Figure 1B,E). In addition, temporal transitions also require epigenetic regulators (see section 5) and a switching factor Odd paired (Opa), which represses D, and allow the transition to the Grh stage (Abdusselamoglu et al., 2019) (Figure 1E). Interestingly, Opa, D, Ey, Hbn, and Scro participate in temporal patterning of both INPs and medulla neuroblasts (Tang et al., 2021). The cross-regulations between Ey and Scro seems conserved in both cascades, but cross-regulations for other members appear different based on these studies (Konstantinides et al., 2021; Tang et al., 2021; Zhu et al., 2021) (Figure 1,E,F).
In summary, the temporal progression in Drosophila temporal gene cascades is mainly driven by intrinsic mechanisms such as cross-regulations and switching factors, but extrinsic signals also play important roles, as shown by the regulation of the Imp to Syncrip transition by ecdysone signaling (Ren et al., 2017; Syed, Mark and Doe, 2017) or activin signaling (Rossi and Desplan, 2020).
3. Temporal patterning of vertebrate neural progenitors
There is accumulating evidence that vertebrate neural progenitors also undergo the temporal patterning process. In this review, we will focus on recent studies in vertebrate retinal progenitor cells (RPCs), cortical progenitors also called apical progenitors (APs) or apical radial glia (aRGs), and neural progenitors in the spinal cord.
3.1. Temporal patterning of vertebrate retinal progenitor cells
In the vertebrate retina, major cell types are born in a stereotypical but overlapping order, with retinal ganglion cells first and Müller glia last [reviewed in (Cepko, 2014)] (Figure 2A). In vivo lineage tracing studies indicate that RPCs are multipotent and give rise to distinct cell types, but the clone size and composition are highly variable (Cepko, 2014). Current view is that the temporal progression of RPCs is largely driven by intrinsic mechanisms, but extrinsic cues including feedback signals from progeny or stochastic effects may also regulate RPC division patterns and progeny fates [reviewed in (Cepko, 2014; Javed and Cayouette, 2017)]. How are the different cell types generated by temporal patterning of RPCs? Several studies examined whether orthologs of Drosophila VNC TTFs have roles in the specification of different cell types. Ikzf1 (Ikaros), the ortholog of Drosophila Hb, is expressed in all early RPCs but not in late RPCs and is sufficient to confer competence to generate early-born neurons including RGCs, horizontal cells, and amacrine cells when mis-expressed in late RPCs. Loss of Ikzf1 causes a reduction of early-born cell types (Elliott et al., 2008) (Figure 2A). Subsequently, Casz1, the ortholog of Drosophila Castor, was found to be expressed in mid-/late-stage RPCs, and bias RPC output by promoting mid-/late-born cell fates such as rods and bipolar cells at the expense of early-born cell fates (Mattar et al., 2015) (Figure 2A). Recently, Pou2f1/Pou2f2, the homologs of Drosophila TTF Pdm, were found to be expressed in early RPCs. Misexpression of Pou2f1 or Pou2f2 in late RPCs promotes ectopic cone cell generation, and loss of Pou2f2 causes a reduction in cone and horizontal cells (Javed et al., 2020) (Figure 2A). Some cross-regulations were identified among these factors: mis-expression of Ikzf1 in E14 retina increases the expression level of Pou2f1/2, while mis-expression of Pou2f1 in P0 retina decreases the level of CasZ1 expression (Javed et al., 2020). In addition to these three homologs of Drosophila TTFs, a Fox domain transcription factor, Foxn4, was identified as another temporal identity factor, that biases PRCs to generate mid-stage cell fates (horizontal, amacrine, cone, and rod cells) (Liu et al., 2020) (Figure 2A).
Figure 2. Temporal patterning of vertebrate neural progenitors.
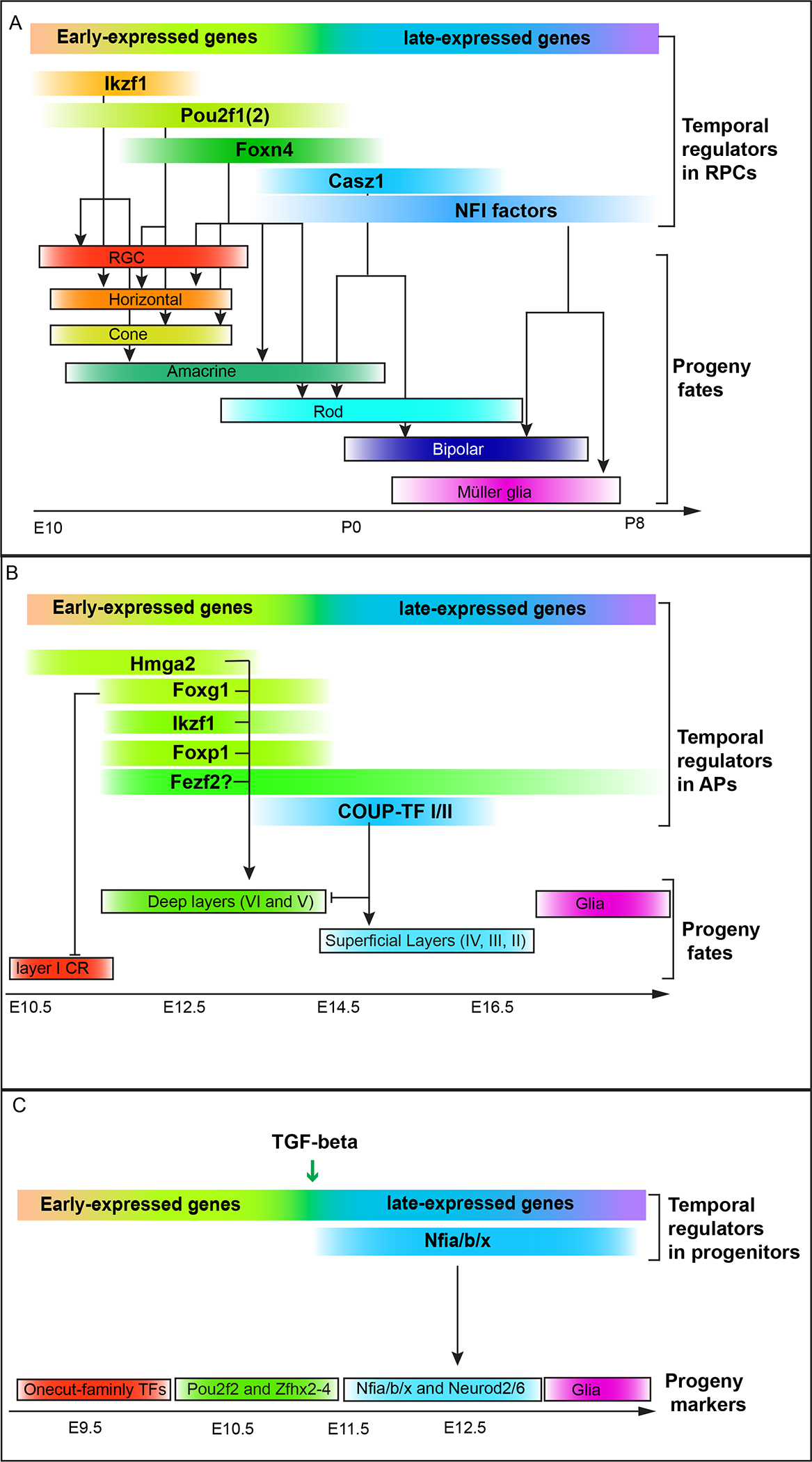
(A) Temporal patterning in mouse retinal progenitors (RPCs). Five transcription factors temporally expressed in mouse RPCs have been shown to promote the production of different retinal cell types. In addition, scRNA-sequencing identified sets of genes differentially expressed in early vs. late RPCs that can be candidate temporal regulators. (B) Temporal patterning in mouse cortical progenitors (APs). Several transcription factors show temporal expression patterns in APs and regulate temporal specification of progeny fates. Foxg1 is required to repress layer I neuron generation, and promote deep layer neuron fates. In addition, Hmga2, Ikzf1, Foxp1 and Fezf2 were all shown to be required for deep layer neuron fates. COUP-TF I/II are required for the switch from generating deep layer neurons to generating superficial layer neurons. Furthermore, scRNA-sequencing identified groups of genes differentially expressed in early vs. late APs that can be candidate temporal regulators. (C) Temporal patterning in vertebrate spinal cord. Neurons born at different times express different transcription factors. A list of temporal genes were shown to be differentially expressed in early vs. late progenitors. Among them, Nfi factors were shown to be required for the generation of Neurod2 expressing neurons. The temporal transition in neural progenitors require TGF-β signaling.
Finally, single-cell RNA-seq studies of mouse and human retina across different developmental stages revealed the whole developmental trajectories of RPCs and differentiated cell types (Clark et al., 2019; Lu et al., 2020). According to transcriptional profiles, RPCs are clearly segregated into early primary RPCs and late primary PRCs, as well as the corresponding early neurogenic and late neurogenic progenitors that express neurogenic bHLH genes and connect primary RPCs to differentiated cell types (Clark et al., 2019). Differentially expressed genes along the RPC pseudotime including genes in different signaling pathways (FGF, Wnt, Notch and TGF-β pathways) as well as transcription factors (Foxp1, Six6, Otx1, Zic1, Six3, Hopx, Sox8, Sox9, Sox11 and Nfi factors among others) are promising candidates to regulate RPC competence and cell fate specification (Clark et al., 2019). Among them, the NFI transcription factors (Nfia/b/x) were confirmed to be expressed in late RPCs. In triple mutants of Nfia/b/x, the latest-born cell fates (bipolar interneuron and Müller glia) are almost completely lost, and RPC failed to stop proliferation, showing that these factors control the latest-born cell fate and the cell-cycle exit (Clark et al., 2019) (Figure 2A).
In addition to transcriptional regulation, post-transcriptional mechanisms involving microRNAs also play important roles in temporal transitions, but different sets of microRNAs were shown to be involved in Xenopus and mouse retinogenesis [reviewed in (Cepko, 2014)].
Although homologs of three Drosophila TTFs were found to regulate temporal patterning in vertebrate RPCs, there appear to be significant differences in their roles between Drosophila neuroblasts and vertebrate RPCs. Drosophila neuroblasts have discrete temporal windows defined by the differential expression of TTFs, and a TTF is usually required and sufficient for the fate specification of the neural types born in this TTF stage: loss of a given TTF often leads to a nearly complete loss of the cell fates it controls. In contrast, vertebrate retinal cell types are generated in overlapping intervals, and are broadly classified into early-born cell types and late-born cell types. Loss of a temporal regulator in vertebrate RPCs often causes modest changes in early-born or late-born cell types. It is possible that multiple mechanisms or factors act partially redundantly or in combinations to control temporal patterning. Functional characterization of candidate genes and pathways identified in the scRNA-seq studies will help to elucidate the temporal patterning networks. It is also possible that within each major cell type, different subtypes are generated at different time points, and identification of subtype markers will greatly facilitate the study of temporal patterning if this is the case.
3.2. Temporal patterning of mammalian cortical progenitors
Sequential generation of different neuron fates in the mammalian neocortex
The ordered manner of neuron generation in the mammalian CNS is well studied in the mouse neocortex, which is organized into six layers composed of different neural subtypes [reviewed in (Jacob, Maurange and Gould, 2008; Okano and Temple, 2009; Greig et al., 2013; Oberst, Agirman and Jabaudon, 2019)]. The apical progenitors (APs) located in the ventricular zone (VZ) produce these neural subtypes in a defined order directly or through intermediate progenitors (IPs). At E10.5, the Cajal-Retzius cells are generated first and make up layer I. From E11.5 to E16.5, layer II-VI neurons are generated in an inside-out pattern, with early-born neurons occupying deep layers (layers VI and V), and late-born neurons occupying superficial layers (layers IV and II/III) (McConnell, 1995). Clonal analysis demonstrated that the majority of APs are multipotent and generate both deep layer and superficial layer neurons sequentially, and at around E17.5, a defined fraction of APs transition to gliogenesis while others exit the cell cycle (Gao et al., 2014). A recent study showed that individual lineages are extremely heterogeneous, and suggested probabilistic decisions based on the combination of internal competence and extrinsic cues (Llorca et al., 2019; Llorca and Marín, 2021).
Intrinsic and extrinsic control of temporal patterning
Early heterochronic transplantation experiments in ferrets suggest that the sequential neural fate specification is controlled by a combination of intrinsic competence and extrinsic cues, and the competence to generate early-born neurons is gradually lost (McConnell, 1988; McConnell and Kaznowski, 1991; Desai and McConnell, 2000). Recently, it was demonstrated in mice that late APs retain the plasticity to revert back to previous temporal states in response to environmental cues, but the IPs are committed progenitors (Oberst et al., 2019). Multiple extrinsic cues have been shown to influence the cell divisions and temporal progression of APs, which have been reviewed in detail (Reillo et al., 2017; Kawaguchi, 2019; Llorca and Marín, 2021). Most notably are the feedback signals from the progeny within the lineage. Cortical progenitors cultured in vitro recapitulate the sequential generation from deep layer neurons to superficial layer neurons, suggesting that temporal transitions depend on cell-intrinsic and environmental factors generated within the clonal lineage (Shen et al., 2006; Gaspard et al., 2008). In these culture conditions, the progeny are able to contact the progenitor. When cultured APs are maintained at one-cell state, they have more limited changes in temporal gene expression compared to those cultured in neurospheres (Okamoto et al., 2016). These results together suggest that feedback signals generated from the progeny are important for temporal progression, consistent with previous reports that ablating newly-born deep layer neurons until E14.5 delays the onset of upper layer neuron generation (Toma et al., 2014). In addition to feedback signaling, extrinsic cues such as Wnt signaling (Oberst et al., 2019) and Sonic hedgehog (SHH) signaling (Zhang et al., 2020) have been shown to influence the temporal progression of APs. The extensive regulation of temporal patterning by extrinsic cues does not preclude intrinsic temporal patterning mechanisms. The intrinsic states can modulate the response to extrinsic cues, and extrinsic cues can also regulate the expression of temporal patterning factors. For example, it has been shown that APs become increasingly hyperpolarized as they age, and the hyperpolarization of APs promotes temporal progression to later fates through the inhibition of extrinsic Wnt signaling (Vitali et al., 2018).
Transcriptional priming and post-transcriptional regulation
Recent studies have revealed the importance of transcriptional priming and post-transcriptional regulation in cortical neuron fate specification [reviewed in (Albert and Huttner, 2018; Hoye and Silver, 2021)]. APs express mRNAs of genes that are only translated and functioning in IPs or neurons, including general markers for IPs and neurons, and transcriptional specifiers of both deep and superficial layer neurons, a phenomenon termed transcriptional priming (Florio et al., 2015; Telley et al., 2016; Nowakowski et al., 2017; Yoon et al., 2017; Zahr et al., 2018). Several post-transcriptional mechanisms were shown to prevent the protein expression of these genes in APs, including m6A modification of mRNA molecules (Yoon et al., 2017), microRNAs [reviewed in (Rajman and Schratt, 2017; Albert and Huttner, 2018)] and translational repression complexes (Zahr et al., 2018). Of particular interest to temporal patterning, some mRNAs, including the transcriptional regulators that specify superficial versus deep layer fates, were shown to be co-expressed in APs, and associate with a Pum2/4E-T translational repression complex (Zahr et al., 2018). Depletion of this complex caused abnormal co-expression of deep layer neuron specification proteins in newborn superficial layer neurons (Zahr et al., 2018). A model was proposed that APs are transcriptionally primed and competent to make different cortical neuron subtypes; and that extracellular signals then regulate mRNA interactions with translational repression complexes to specify neural fates (Zahr et al., 2018). It will be interesting to further examine how transcriptional specifiers of deep vs. superficial layer neurons are selectively and differentially regulated by translational repression complexes at different stages. Since microRNAs binding to mRNA 3’UTRs can recruit protein complexes including 4E-T and 4EHP to suppress cap-dependent mRNA translation (Jafarnejad et al., 2018), and Pumilio can also work together with miRNAs (Kedde et al., 2010), it is possible that miRNAs may provide some specificity.
Recently it was found that three miRNAs, miR-128/miR-9 and let-7, form opposing temporal gradients in APs (decreasing for miR-128/miR-9, and increasing for let-7), and they are required for the sequential generation of deep and superficial layer neurons (Shu et al., 2019). One of let-7’s targets, transcription factor Hmga2, is highly expressed in young APs, and gradually lost in late-stage APs (Nishino et al., 2008; Shu et al., 2019; Telley et al., 2019). Overexpression of Hmga2 in late-stage APs shifts the distribution of their progeny from superficial to deep-layers, while knocking it down in early stage APs causes a weak shift of progeny fate to superficial layers (Shu et al., 2019). In addition to Hmga2, two RNA-binding proteins, Imp-1 (another target of let-7) and Lin-28a (an activator of Imp-1) are also highly expressed in early-stage neural progenitors, and decline in late stages (Nishino et al., 2008, 2013; Sagner et al., 2020; Konstantinides et al., 2021). They were previously shown to be required for the expansion of fetal neural stem cells (Nishino et al., 2008, 2013). Thus it appears that the temporal gradients of RNA-binding proteins Lin-28 and Imp observed in Drosophila neuroblasts is conserved in vertebrate neural progenitors. It will be interesting to test whether they function in the deep layer neural fate specification.
Transcriptional regulation in APs
The extensive post-transcriptional regulation involved in cortical neuron specification does not preclude transcriptional regulation in APs. Studies have revealed that several transcription factors temporally expressed in APs function in the specification of neurons in different layers. Forkhead Box G1 (Foxg1), the mammalian ortholog of Slp, is expressed in APs as they are transiting from generating layer I Cajal-Retzius cells to making deep layer neurons (Hanashima et al., 2004). Loss of Foxg1 caused prolonged generation of Cajal-Retzius cells; and delayed activation of Foxg1 is sufficient to promote the generation of deep layer neurons, suggesting that it is required and sufficient for this temporal transition (Hanashima et al., 2004; Kumamoto et al., 2013) (Figure 2B). Foxg1 acts on a transcriptional network, in which cross-repression between layer-enriched TFs including Fezf2, Ctip2, Satb2, and Tbr1, allows specification of deep versus superficial layer subtypes (Fame, MacDonald and Macklis, 2011; Srinivasan et al., 2012; Hanashima and Toma, 2015). Foxg1 was shown to repress Tbr1, which then allows de-repression of Fezf2 (Toma et al., 2014). Most of these TFs are only detected in postmitotic neurons at the protein level, but Fezf2 is expressed in both APs and deep layer neurons. Studies showed that Fezf2 is expressed in early APs and is required for the specification of deep layer neurons, and that over-expression of Fezf2 in late APs is sufficient to produce supernumerary deep layer neurons (Chen, Schaevitz and McConnell, 2005; Chen et al., 2008), making it a good candidate as a TTF. However, a recent study showed that Fezf2 is still expressed in late APs when they are generating upper-layer neurons and glia, and suggested that the expression level of Fezf2 matters (Guo et al., 2013) (Figure 2B). Whether Fezf2 is a TTF depends on whether it is functioning in APs for temporal specification, or only functioning in post-mitotic neurons. Another transcription factor LHX2 that is expressed in all APs, initially functions as a cortical selector gene, and then has a second role in repressing the expression of Fezf2 (Muralidharan et al., 2017). Loss of LHX2 causes upregulation of Fezf2 expression in APs and an increase in layer 5 neurons specified by Fezf2, while overexpression of LHX2 causes the opposite phenotype (Muralidharan et al., 2017). LHX2 appears to function in APs, because loss of LHX2 in post-mitotic neurons only did not have the same phenotype (Zembrzycki et al., 2015). This suggests that the Fezf2 level may indeed matters in neural progenitors.
In addition, the ortholog of Drosophila Hb, Ikzf1(Ikaros), was found to function in mammalian early APs to promote deep layer neuron fates (Alsiö et al., 2013), similar to its role in early RPCs. Ikzf1(Ikaros) is expressed in early stage APs, and sustained Ikaros expression results in prolonged generation of early-born deep-layer neurons and delayed production of late-born upper-layer neurons (Alsiö et al., 2013) (Figure 2B). COUP-TFI and II, the orthologs of Drosophila switching factor Svp, are co-expressed in the ventricular zone starting from E12.5 and become diminished at E16.5. Knocking-down both of them caused sustained neurogenesis and prolonged generation of early-born deep layer neurons, suggesting that COUP-TFI and II are involved in the transition from generating deep layer neurons to superficial layer neurons (Naka et al., 2008) (Figure 2B). It will be interesting to examine whether the role of COUP-TFI and II in this switch is through down-regulation of Ikzf1(Ikaros), which would be analogous to the regulatory relationships in Drosophila (Figure 2B). A recent study showed that Foxg1 directly represses the transcription of COUP-TFI by binding to its enhancer. As Foxg1 expression is down-regulated, COUP-TFI expression is de-repressed and promotes the transition to the layer IV neuron fate (Hou, Miyoshi and Hanashima, 2019) (Figure 2B). In a sense this might be analogous to the feedforward repression that one TTF represses the next plus one TTF in the Drosophila embryonic TTF cascade.
Recently, another Forkhead box TF, Foxp1, was shown to be expressed at high levels in early APs and promote deep-layer neuron production (Pearson et al., 2020). Foxp1 expression then decreases during the transition to the superficial-layer neurogenesis, and sustained Foxp1 expression extends the deep layer neuron production period into postnatal life (Pearson et al., 2020) (Figure 2B).
Finally, the presence of temporal regulation of transcription in APs was clearly demonstrated by single-cell transcriptomics studies (Okamoto et al., 2016; Yuzwa et al., 2017; Telley et al., 2019). A core set of temporally patterned genes are sequentially expressed in APs of different ages, with genes involved in cell cycle regulation and nucleus/chromatin–related processes prominent in the early stage, and genes involved in susceptibility to environmental signals increasing later-on (Telley et al., 2019). These age-dependent temporally patterned genes are transmitted to the progeny of APs as “seeds” of initial neural identity, and on the basis of which a largely conserved neural differentiation program and environmental cues drive the neurons to their final identities (Telley et al., 2019). One of the candidate transcription factors highly expressed in early stage APs is Hmga2, which we have discussed in the previous sub-section.
In summary, mammalian cortex APs are temporally patterned by a combination of transcriptional and post-transcriptional mechanisms as well as environmental cues. Although a number of transcriptional regulators were identified, it is still not clear whether they form a temporal cascade in neural progenitors. ScRNA-seq studies of APs from different developmental stages revealed dynamic temporal changes in transcriptome, and suggested groups of genes rather than single genes function in temporal patterning. Further investigation of differentially expressed genes will help to reveal the temporal patterning mechanisms.
3.3. Temporal patterning of neural progenitors in the vertebrate spinal cord
In the vertebrate spinal cord, studies have demonstrated generation of different neural subtypes at different times from the same neural progenitors (Sockanathan and Jessell, 1998; Müller et al., 2002; Tripodi, Stepien and Arber, 2011; Benito-Gonzalez and Alvarez, 2012; Stam et al., 2012; Luxenhofer et al., 2014; Hayashi et al., 2018; Deska-Gauthier et al., 2020). Recently, scRNA-sequencing studies of developing mammalian spinal cord suggested the existence of a global temporal patterning scheme in neural progenitors (Delile et al., 2019; Sagner et al., 2020). Across all spatial domains of the spinal cord, neurons born at the same embryonic stages express the same sets of transcription factors: Onecut family TFs for earliest born neurons, Pou2f2 and Zfhx2-4 for neurons born in intermediate stages, and Nfia/b/x and Neurod2/6 for late-born neurons (Delile et al., 2019; Sagner et al., 2020). Furthermore, this temporal code for neurons is likely to be conserved in other regions of the nervous system, including the retina and different brain regions (Sapkota et al., 2014; Clark et al., 2019; Javed et al., 2020; Sagner et al., 2020). On the progenitor level, a group of transcription factors or RNA binding proteins (including Hmga2, Nr6a1, Sox9, Npas3, Zbtb20, Nfi factors, Hopx and Lin28a/b among others) show largely consistent temporal expression patterns in all spatial domains of the neural tube in aging neural progenitors (Sagner et al., 2020). A number of them, including Sox9, Nfi factors and Hopx, are also among the top differentially expressed genes in RPCs (Clark et al., 2019). Among these temporally-expressed candidate genes, Nfi factors were shown to be required for the generation of late-born neural types (expressing Neurod2) and the transition to gliogenesis in the spinal cord, consistent with the results in the retina (Deneen et al., 2006; Kang et al., 2012; Matuzelski et al., 2017; Clark et al., 2019). The transition from early to late temporal program was shown to be in part regulated by TGFβ signaling: blocking or ectopic activation of TGFβ signaling pathway affects the speed of the temporal changes, and this was proposed to be a possible mechanism of feedback regulation from newborn neurons (Sagner et al., 2020).
These studies suggested the possibility of a global temporal patterning scheme operating in all parts of the vertebrate nervous system, and some common candidate genes were shown to have conserved temporal expression patterns in neural progenitors. Among them, Nfi factors have been demonstrated to play conserved roles in the specification of late-born fates. However, for other candidate temporal genes, further studies are required to examine which temporal factors in neural progenitors control the expression of neuronal transcription factors that constitute the temporal code for neurons. Some of the previously characterized temporal regulators in the retina or cortex were not included in the list of candidate genes for the spinal cord. It is possible that although a common temporal patterning program including a number of genes may function globally, different parts of the nervous system may also deploy specific temporal patterning programs.
4. Connections between cell cycle progression to temporal patterning in flies and vertebrates
During neurogenesis, neuroepithelial cells initially undergo symmetrical proliferative divisions to increase the stem cell pool, and then they make the transition to become asymmetrically dividing neural progenitors. Division modes of neural progenitors are also temporally regulated, and the involvement of cell cycle length, signaling pathways especially the dynamic Notch signaling, have been reviewed elsewhere (Egger, Gold and Brand, 2011; Kawaguchi, 2019). In this section, we will focus on recent studies exploring any possible links between cell cycle progression and the expression of temporal regulators controlling progeny fate specification.
Since neural progenitors need to generate a defined number of progeny through cell divisions at the same time as they express each of the temporal patterning transcription factors, investigators have long questioned whether counting the cell cycles serves as an intrinsic timer determining how long a TTF is expressed and when its expression is to be terminated (Grosskortenhaus et al., 2005). Early studies in Drosophila embryonic VNC neuroblasts showed that the first temporal transition ( Hb to Kr ) requires cytokinesis, but all later temporal transitions progress normally in G2-arrested neuroblasts (Grosskortenhaus et al., 2005). The switching factor Svp is necessary for the Hb to Kr transition, and the requirement of cytokinesis for this transition is because the nuclear export of svp mRNA (and hence its efficient translation) is dependent on mitosis (Mettler, Vogler and Urban, 2006). In neuroblasts of Drosophila larval VNC, the temporal transition from Imp /Castor/Chinmo to Syncrip/Broad has also been linked to cell cycle progression. Delaying the G1 to S transition caused the majority of neuroblasts to remain at the Chinmo stage. In contrast, delaying the G2 to M transition did not affect the temporal progression (Van Den Ameele and Brand, 2019). Since this temporal transition also requires the switching factor Svp, it will be interesting to examine whether the same mechanism as in the embryonic neuroblasts is involved.
Studies in mammalian cortical progenitors have also explored the possible connection between cell cycle and laminar fate specification for similar reasons (Okamoto et al., 2016). For this purpose, early APs were arrested in the cell cycle for two days by co-electroporation of a Cdk inhibitor and the intracellular domain of Notch1 (NICD) to maintain the undifferentiated state, and then allowed to re-enter the cell cycle by excision of the Cdk inhibitor and NICD transgenes (Okamoto et al., 2016). Such APs re-entering the cell cycle generated upper layer neurons, suggesting that the temporal transition to generate late-born neurons is independent of cell cycle when Notch signaling is provided (Okamoto et al., 2016). These APs can still receive extrinsic cues provided by normal cycling progenitors and their progeny. After isolated APs co-expressing the Cdk inhibitor and NICD were cultured in vitro as single-cell clones, they were allowed to re-enter the cell cycle. Staining of the progeny showed that the transitions of laminar fate potential occur at low frequency and incompletely suggesting the transition is controlled by both cell-autonomous and non-cell-autonomous mechanisms (Okamoto et al., 2016).
In summary, although specific temporal transitions may depend on the cell cycle/cytokinesis, counting the number of cell cycles is unlikely to be a universal timer controlling all temporal transitions.
An alternative model postulates that the length of the cell cycle especially the G1 phase in cycling progenitors may drive neural differentiation or regulate cell fate (Calegari and Huttner, 2003; Decembrini et al., 2009; Hardwick et al., 2015; Kawaguchi, 2019). In Xenopus retina, it was shown that the expression of a set of microRNAs is related to the cell cycle speed, and is upregulated in faster-cycling early RPCs, and downregulated in slower-cycling late RPCs, and that these miRNAs inhibit the later-born neural fates by repressing their targets (Decembrini et al., 2009). According to this model, critical temporal fate determinants may accumulate or degrade at only certain phases of the cell cycle, hence the length of these phases will determine whether the concentration of these determinants will be above or below a certain threshold (Calegari and Huttner, 2003; Decembrini et al., 2009). In the mammalian cortex, neurogenic APs also increase their cell cycle length as they age, specifically a ~50% increase in the cell cycle length between E12 and E15 (Telley et al., 2019). However, whether the cell cycle length has a role in the temporal specification of cortical neural fates remains to be investigated.
The exact coordination between cell cycle progression/progeny generation and temporal patterning might also be explained by co-regulation of temporal patterning factors and cell cycle genes by certain mechanisms, including post-transcriptional m6A modification of their mRNAs (Yoon et al., 2017), and transcriptional mechanisms that remain to be identified. On the other hand, regulation of cell cycle genes by temporal patterning factors may also coordinate the neuroblast proliferation and temporal progression. In the Drosophila VNC, a set of early temporal factors and a set of late temporal factors have been shown to regulate key cell-cycle genes differently, and thus control the neuroblast proliferation, transitions in the neuroblast division modes and the final cell-cycle exit (Bahrampour et al., 2017).
5. Epigenetic mechanisms regulating temporal patterning.
Epigenetic mechanisms including DNA methylation, histone and chromatin modifications, chromatin remodeling, and 3D genome architecture are required for the regulation, maintenance and inheritance of transcriptional patterns, and have been shown to play important roles in the regulation of neural development [reviewed in (Yao et al., 2016; Sokpor et al., 2017, 2018; Albert and Huttner, 2018; Yoon et al., 2018; Seritrakul and Gross, 2019)]. In this review, we will focus on recent findings on the roles of Polycomb group (PcG) complexes and ATP-dependent chromatin remodeling complexes in the regulation of temporal patterning of neural progenitors in both Drosophila and vertebrates.
5.1. Involvement of PcG complexes in temporal patterning.
The Polycomb group proteins (PcGs) are organized into two complexes: Polycomb repressive complex 1 (PRC1) and Polycomb repressive complex 2 (PRC2). PRC2 and PRC1 catalyze trimethylation of histone H3 at lysine 27 (H3K27me3), and mono-ubiquitination of histone 2A at lysine 119 (H2AK119ub1), respectively, and mainly function in transcriptional repression [reviewed in (Aranda, Mas and Di Croce, 2015)]. The underlying molecular mechanism is under intensive investigation, and may include histone-modification dependent direct blocking of transcription, and histone modification independent induction of chromatin compaction [reviewed in (Aranda, Mas and Di Croce, 2015; Geng and Gao, 2020)]. PcGs may regulate many targets in neuronal development, and thus loss of PcG function often display composite or different phenotypes depending on the experimental conditions.
During cortical lineage progression, there are extensive H3K27me3 changes at the promoters of transcription factors involved in neural fate specification in neural progenitors at major developmental transitions, suggesting that PcGs have important roles in the regulation of gene expression during neocortical development (Albert et al., 2017). PcG complex proteins have been shown to be required for the timely termination of temporal factor expression. For example, Ring1B, an essential component of PRC1, was shown to be required for the timely down-regulation of Fezf2 expression in APs of the mouse neocortex (Morimoto-Suzki et al., 2014). H3K27me3 modification and Ring1B binding increase at the promoter of Fezf2 as its expression decreases in APs. Deletion of Ring1B in APs at the time when layer V to II neurons are born, caused prolonged expression of Fezf2 and increased generation of deep-layer neurons, while deletion of Ring1B in postmitotic neurons did not have the same phenotype (Morimoto-Suzki et al., 2014). These data also suggest that Fezf2 level matters in APs, and support that Fezf2 is a TTF functioning in neural progenitors. Another example is at the neurogenic to gliogenic transition, PRCs are required to repress the expression of Neurogenin (Ngn) 1 and Ngn2, which suppress gliogenesis (Hirabayashi et al., 2009). Deletion of PRC1 or PRC2 components in late stage APs resulted in a prolonged neurogenic phase and delayed gliogenesis (Hirabayashi et al., 2009). PcG proteins could also be involved in preventing precocious expression of temporal factors before their scheduled expression period. Deletion of Ezh2, the histone methyltransferase of PRC2, or Eed, a regulatory subunit of PRC2, before the onset of neurogenesis, caused precocious neurogenesis and accelerated temporal progression, resulting in a shortened neurogenic period and greatly reduced the thickness of neocortex (Pereira et al., 2010; Telley et al., 2019).
PRCs also regulate temporal patterning in vertebrate retinal progenitors. In Xenopus retina, PRCs were shown to be crucial for the initiation of neural differentiation, and loss of PRC2 function caused loss of most retinal neural types and a precocious transition to gliogenesis (Aldiri et al., 2013). In mouse retina, PRCs are required for the TTF Casz1 to promote the rod photoreceptor production and to prevent precocious transition to gliogenesis (Mattar et al., 2020). Loss of Casz1 or PRC components caused reduction of rod photoreceptor production and precocious gliogenesis. Further, loss of PRC components reversed the suppression of gliogenesis caused by mis-expression of Casz1(Mattar et al., 2020). These data showed that PRCs are also required downstream of a TTF to control temporal specification.
In Drosophila, there is also accumulating evidence that PRCs regulate temporal patterning. In the VNC, PRCs were shown to be necessary and sufficient to restrict motor neuron specific competence windows in several neuroblast lineages that transition from producing motor neurons to interneurons (Touma, Weckerle and Cleary, 2012). Loss of PRC function extended the competence window of neuroblasts to respond to the TTF Kr to generate motor neurons, while PRC gain of function caused premature loss of competence to produce motor neurons (Touma, Weckerle and Cleary, 2012). In the Drosophila brain, PRCs were shown to associate with genes encoding TTFs of the medulla temporal cascade using Targeted DamID (TaDa) (Marshall and Brand, 2017). This study classified PcG-associated chromatin into two groups: PcG mixed and PcG repressive domains, with PcG mixed domains also associated with RNA Pol II and Brahma (Brm, a chromatin remodeling protein). Since the experiment was done on a mixed population of neuroblasts of all ages, genes within the PcG mixed chromatin state are likely to be in the repressed state (associated with PRC) in some neuroblasts, but in the active state (associated with RNA RNA Pol II and Brm) in other neuroblasts (Marshall and Brand, 2017). Specifically, genes encoding medulla TTFs Ey, Slp1, Hth and Tll were found to be within the PcG mixed domains, suggesting that PcG complexes might also be involved in the regulation of TTF expression in the Drosophila medulla (Marshall and Brand, 2017).
In summary, it is clear that PRCs play important roles in the temporal patterning of neural progenitors in both flies and vertebrates, although the specific phenotypes of PRC component mutants may vary because PRCs can have different targets at different stages. How PRCs target specific genes at different temporal stages is still under investigation, but may involve interactions with specific transcription factors, non-coding RNAs and other chromatin factors (Albert and Huttner, 2018).
5.2. Involvement of ATP-dependent chromatin remodeling complexes in temporal patterning of neural progenitors
There are four major families of chromatin remodeling complexes: SWI/SNF family, ISWI family, NuRD/Mi-2/CHD family, and INO80 family, that function extensively in nervous system development [reviewed in (Sokpor et al., 2018)]. We will be focusing on recent evidence that the SWI/SNF family and NuRD/Mi-2/CHD family chromatin remodeling complexes are involved in the temporal patterning of neural progenitors.
SWI/SNF chromatin remodeling complexes.
In mammals, there are two SWI/SNF chromatin remodeling complexes called BAF and PBAF (BRG-/BRM-Associated Factors and Polybromo-Associated BAF respectively) [reviewed in (Ho and Crabtree, 2010; Hodges, Kirkland and Crabtree, 2016)]. The BAF complex contains one of two ATPase subunits Brahma or BRG1 (Brahma-related Gene 1) and signature subunit ARID1A/B, while PBAF contains the BRG1 ATPase, ARID2 and PBRM1. The BAF and PBAF complexes share some core components called BAFs (Ho and Crabtree, 2010; Tang, Nogales and Ciferri, 2010; Hodges, Kirkland and Crabtree, 2016). In Drosophila the two complexes are called BAP and PBAP (Brahma Associated Protein and Polybromo-associated BAP respectively) complexes, and they share the same ATPase subunit Brahma, as well as other core components. The signature subunit of BAP complex is Osa, the ortholog of ARID1A/B, while PBAP contains Polybromo and BAP170 but lacks Osa (Mohrmann et al., 2004). The SWI/SNF family of chromatin remodeling complexes function by destabilizing histone-DNA interactions using the energy from ATP hydrolysis, leading to nucleosome rearrangement and increased accessibility for transcription factor binding to activate transcription (Becker and Workman, 2013; Kingston and Tamkun, 2014; Hota and Bruneau, 2016).
In vertebrates, genes encoding subunits of the complexes had undergone expansions to form gene families, and tissue or cell-type specific BAF complexes have been reported [reviewed in (Ho and Crabtree, 2010; Sokpor et al., 2018)]. For example, BAF complex in neural progenitors (npBAF) contains BAF45a/d and BAF53a that are required for proliferation, while BAF in postmitotic neurons (nBAF) contain the alternative BAF45b/c and BAF53b subunits (Lessard et al., 2007; Ho and Crabtree, 2010). Different BAF subunit composition may also regulate temporal patterning in neural progenitors. In cortical progenitors at the VZ, BAF170 has a temporal expression pattern: high during early neurogenesis, lost during late neurogenesis, and reappearing at the beginning of gliogenesis (Tuoc et al., 2013). During early neurogenesis, BAF170 competes with BAF155 in npBAF, and represses Pax6 target genes that regulate the generation of IPs and late APs that produce upper-layer neurons. Conditional deletion of BAF170 promotes indirect neurogenesis through IPs and generation of significantly more upper layer neurons, while mis-expression of BAF-170 has the opposite phenotype (Tuoc et al., 2013). In another study, Brg1 expression was shown to be upregulated in cortical progenitors after E13, where it is required for the maintenance of neural progenitors and for the transition from neurogenesis to gliogenesis (Matsumoto et al., 2006).
The BAP complex in Drosophila has been shown to initiate temporal patterning by activating TTF expression in neural progenitors. In the Drosophila type II NB lineages, the signature subunit of the BAP complex, Osa (ortholog of ARID1), is required to initiate the temporal patterning of INPs and prevent their dedifferentiation (Eroglu et al., 2014; Abdusselamoglu et al., 2019). Osa directly binds near the transcription start site of temporal genes, and is required for the expression of both TTF D and its repressor Opa, but activation of Opa has a slower kinetics. After Opa reaches a high level, it represses D, and allows the expression of the next TTF, Grh (Eroglu et al., 2014; Abdusselamoglu et al., 2019) (Figure 1E). Osa also directly activates the expression of Hamlet (Ham), which belongs to a histone methyltransferase family homologous to vertebrate Prdm3/Evi1 and Prdm16. Ham is then required to repress Grh and limit neuroblast proliferation (Eroglu et al., 2014). Thus the BAP complex has essential roles in every step of the INP temporal cascade.
In summary, there is accumulating evidence that the BAF and BAP complexes regulate temporal patterning of neural progenitors in vertebrates and Drosophila, respectively. In contrast to Drosophila, vertebrate BAF complexes can have different compositions in different tissue/cell types, providing a certain degree of specificity. In both Drosophila and vertebrates, different pioneering factors or initiating factors may interact dynamically with chromatin remodeling complexes to provide specificity to the regulation (Swinstead et al., 2016).
NuRD/Mi-2/CHD family chromatin remodeling complexes.
Nucleosome Remodeling Deacetylase (NuRD) complexes have CHD3 (CHD: chromodomain-helicase-DNA binding) or CHD4 as core catalytic ATPase components, which directly bind to the histone deacetylases HDAC1 and HDAC2, and a number of DNA-binding proteins [reviewed in (Sokpor et al., 2018)]. NuRD complexes promote transcriptional repression, and the molecular mechanism involves eviction of transcriptional activators and RNAPolII due to CHD4-dependent chromatin remodeling, and subsequent maintenance of silencing which requires both nucleosome remodeling and HDAC activity (Liang et al., 2017). The subunit composition of this complex is also highly variable in different tissue /cell types (Sokpor et al., 2018). NuRD was found to regulate temporal patterning in vertebrate retinal and cortical progenitors. In mouse retina RPCs, TTF Casz1 physically interacts with NuRD complex, which subsequently recruits PRCs, and both complexes are required for Casz1 to promote rod photoreceptor fate and suppress gliogenesis (Mattar et al., 2021). Here the NuRD complex is acting downstream of a TTF to regulate progeny fate. The NuRD complex may also regulate the expression of temporal patterning factors. In mouse cortical progenitors, NuRD complex proteins physically interact with Lhx2, and bind to the transcription start site or the distal enhancer of Fezf2 gene, and active chromatin marks in these regions are increased with loss of Lhx2 (Muralidharan et al., 2017). These data suggest that Lhx2 recruits the NuRD complex to remove active chromatin marks in the Fezf2 gene to repress its expression. Since PRCs are also involved in the repression of Fezf2 (Morimoto-Suzki et al., 2014), PRCs and NuRD might also act together in this case. In another study, deletion of MBD3, a structural component of the NuRD complex, affects the division modes of Pax6+ APs resulting in a reduction of basal IPs and neurons (Knock et al., 2015). Loss of MBD3 also compromised proper differentiation of upper-layer neurons, causing them to co-express deep-layer and upper-layer neuronal markers. Analysis of global gene expression patterns in neural progenitors suggests that temporal transitions in gene expression do not occur normally with loss of MBD3 (Knock et al., 2015). Thus, studies in mammalian retinal and cortical progenitors suggest that the NuRD complex have important roles in temporal patterning. In Drosophila, it was reported that the NuRD complex has a role in preventing de-differentiation of progeny by decommissioning stem-cell enhancers (Zacharioudaki, Falo Sanjuan and Bray, 2019), but whether the complex plays a role in temporal patterning has not yet been examined to our knowledge.
6. Conclusions
The last few years have seen great advances in the study of temporal patterning of neural progenitors in model organisms ranging from Drosophila to vertebrates. This has been aided to a great extent by advancements in single cell multi-omics technologies especially single-cell RNA sequencing, which has brought to light transcriptional heterogeneities between neural progenitors of different ages that when combined with environmental signals can nudge their progeny along different developmental trajectories. In the Drosophila neuroblasts, sequential expression of TTFs regulated mainly by cross-regulations and modulated by extrinsic signals is a central mechanism by which temporal patterning of neural fates is achieved. Recent discoveries of opposing temporal gradients of RNA-binding proteins in Drosophila neuroblasts that regulate the expressions of patterning transcription factors appears to be one other critical mechanism for achieving temporal patterning. Thus, both transcriptional and post-transcriptional mechanisms may act in concert to differentiate the largely invariant neuron lineages in Drosophila.
In contrast to the deterministic roles played by TTFs in flies, mechanisms of temporal patterning in vertebrates are less clearly defined. As in flies, vertebrate neurons are generated in a stereotypical order. However, neurogenesis in vertebrates demonstrates far greater plasticity in response to environmental signals than in flies. Although a number of temporal-expressed transcription factors have been shown to function in neural progenitors to regulate neuron fate specification, loss or gain of function of individual candidate regulators often results in relatively modest phenotypes. ScRNA-sequencing studies on vertebrate retinal, cortical and spinal cord neural progenitors revealed changes in the expression of cohorts of genes in early vs. late neural progenitors, suggesting that groups of genes rather than single genes function in temporal patterning in vertebrates (Clark et al., 2019; Telley et al., 2019; Sagner et al., 2020). Further characterization of these temporal genes will help to elucidate the temporal patterning network in greater details. Additionally, transcriptional priming in combination with post-transcriptional mechanisms also play important roles in temporal fate specification. Epigenetic changes facilitated by chromatin modifiers and remodelers contribute to the dynamic transcriptome of neural progenitors in both Drosophila and vertebrates by either regulating temporal factor expression or acting downstream of temporal factors. In the future, new techniques including advanced single-cell multi omics methods such as single cell RNA-seq and ATAC-seq (Lopes, Magrinelli and Telley, 2020) as well as high resolution imaging technologies for visualizing higher order genome organization in single cells (Lakadamyali and Cosma, 2020) will continue to facilitate our research on the temporal patterning of neural progenitors and neuron fate specification across different animal phyla.
Highlights.
TTF cascades and gradients of RNA-binding proteins regulate temporal patterning in Drosophila neuroblasts
Vertebrate neural progenitors also undergo temporal patterning
Single-cell RNA-sequencing is a powerful tool to study temporal patterning
Chromatin modifiers and remodelers regulate temporal patterning of neural progenitors
Acknowledgements
The authors are supported by the National Institutes of Health (Grant 1 R01 EY026965-01A1).
Footnotes
The authors declare no competing or financial interests.
References:
- Abdusselamoglu MD et al. (2019) ‘The transcription factor odd-paired regulates temporal identity in transit- amplifying neural progenitors via an incoherent feed-forward loop’, eLife. doi: 10.7554/eLife.46566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert M et al. (2017) ‘Epigenome profiling and editing of neocortical progenitor cells during development’, The EMBO Journal. doi: 10.15252/embj.201796764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert M and Huttner WB (2018) ‘Epigenetic and transcriptional pre-patterning-An emerging theme in cortical neurogenesis’, Frontiers in Neuroscience. doi: 10.3389/fnins.2018.00359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldiri I et al. (2013) ‘Polycomb repressive complex PRC2 regulates Xenopus retina development downstream of Wnt/β-catenin signaling’, Development (Cambridge). doi: 10.1242/dev.088096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allan DW and Thor S (2015) ‘Transcriptional selectors, masters, and combinatorial codes: Regulatory principles of neural subtype specification’, Wiley Interdisciplinary Reviews: Developmental Biology. doi: 10.1002/wdev.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsiö JM et al. (2013) ‘Ikaros promotes early-born neuronal fates in the cerebral cortex’, Proceedings of the National Academy of Sciences of the United States of America. doi: 10.1073/pnas.1215707110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Den Ameele J and Brand AH (2019) ‘Neural stem cell temporal patterning and brain tumour growth rely on oxidative phosphorylation’, eLife. doi: 10.7554/eLife.47887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aranda S, Mas G and Di Croce L (2015) ‘Regulation of gene transcription by Polycomb proteins’, Science Advances. doi: 10.1126/sciadv.1500737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Averbukh I et al. (2018) ‘A repressor-decay timer for robust temporal patterning in embryonic drosophila neuroblast lineages’, eLife. doi: 10.7554/eLife.38631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzarelli R, Hardwick LJA and Philpott A (2015) ‘Emergence of neuronal diversity from patterning of telencephalic progenitors’, Wiley Interdisciplinary Reviews: Developmental Biology. doi: 10.1002/wdev.174. [DOI] [PubMed] [Google Scholar]
- Bahrampour S et al. (2017) ‘Neural Lineage Progression Controlled by a Temporal Proliferation Program’, Developmental Cell. doi: 10.1016/j.devcel.2017.10.004. [DOI] [PubMed] [Google Scholar]
- Baumgardt M et al. (2009) ‘Neuronal Subtype Specification within a Lineage by Opposing Temporal Feed-Forward Loops’, Cell. doi: 10.1016/j.cell.2009.10.032. [DOI] [PubMed] [Google Scholar]
- Baumgardt M et al. (2014) ‘Global Programmed Switch in Neural Daughter Cell Proliferation Mode Triggered by a Temporal Gene Cascade’, Developmental Cell. doi: 10.1016/j.devcel.2014.06.021. [DOI] [PubMed] [Google Scholar]
- Bayraktar OA and Doe CQ (2013) ‘Combinatorial temporal patterning in progenitors expands neural diversity’, Nature. doi: 10.1038/nature12266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker PB and Workman JL (2013) ‘Nucleosome remodeling and epigenetics’, Cold Spring Harbor Perspectives in Biology. Cold Spring Harb Perspect Biol, 5(9). doi: 10.1101/cshperspect.a017905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benito-Gonzalez A and Alvarez FJ (2012) ‘Renshaw cells and ia inhibitory interneurons are generated at different times from p1 progenitors and differentiate shortly after exiting the cell cycle’, Journal of Neuroscience. J Neurosci, 32(4), pp. 1156–1170. doi: 10.1523/JNEUROSCI.3630-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benito-Sipos J et al. (2011) ‘Seven up acts as a temporal factor during two different stages of neuroblast 5–6 development’, Development. doi: 10.1242/dev.070946. [DOI] [PubMed] [Google Scholar]
- Bertet C et al. (2014) ‘Temporal patterning of neuroblasts controls notch-mediated cell survival through regulation of hid or reaper’, Cell. doi: 10.1016/j.cell.2014.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brody T and Odenwald WF (2000) ‘Programmed transformations in neuroblast gene expression during Drosophila CNS lineage development’, Developmental Biology. doi: 10.1006/dbio.2000.9829. [DOI] [PubMed] [Google Scholar]
- Calegari F and Huttner WB (2003) ‘An inhibition of cyclin-dependent kinases that lengthens, but does not arrest, neuroepithelial cell cycle induces premature neurogenesis’, Journal of Cell Science. doi: 10.1242/jcs.00825. [DOI] [PubMed] [Google Scholar]
- Cepko C (2014) ‘Intrinsically different retinal progenitor cells produce specific types of progeny’, Nature Reviews Neuroscience. doi: 10.1038/nrn3767. [DOI] [PubMed] [Google Scholar]
- Chen B et al. (2008) ‘The Fezf2-Ctip2 genetic pathway regulates the fate choice of subcortical projection neurons in the developing cerebral cortex’, Proceedings of the National Academy of Sciences of the United States of America. doi: 10.1073/pnas.0804918105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Schaevitz LR and McConnell SK (2005) ‘Fezl regulates the differentiation and axon targeting of layer 5 subcortical projection neurons in cerebral cortex’, Proceedings of the National Academy of Sciences of the United States of America. doi: 10.1073/pnas.0508732102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark BS et al. (2019) ‘Single-Cell RNA-Seq Analysis of Retinal Development Identifies NFI Factors as Regulating Mitotic Exit and Late-Born Cell Specification’, Neuron. doi: 10.1016/j.neuron.2019.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decembrini S et al. (2009) ‘MicroRNAs couple cell fate and developmental timing in retina’, Proceedings of the National Academy of Sciences of the United States of America. doi: 10.1073/pnas.0909167106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delile J et al. (2019) ‘Single cell transcriptomics reveals spatial and temporal dynamics of gene expression in the developing mouse spinal cord’, Development (Cambridge). Company of Biologists Ltd, 146(12). doi: 10.1242/dev.173807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deneen B et al. (2006) ‘The Transcription Factor NFIA Controls the Onset of Gliogenesis in the Developing Spinal Cord’, Neuron. Neuron, 52(6), pp. 953–968. doi: 10.1016/j.neuron.2006.11.019. [DOI] [PubMed] [Google Scholar]
- Desai AR and McConnell SK (2000) ‘Progressive restriction in fate potential by neural progenitors during cerebral cortical development’, Development. [DOI] [PubMed] [Google Scholar]
- Deska-Gauthier D et al. (2020) ‘The temporal neurogenesis patterning of spinal p3–V3 interneurons into divergent subpopulation assemblies’, Journal of Neuroscience. Society for Neuroscience, 40(7), pp. 1440–1452. doi: 10.1523/JNEUROSCI.1518-19.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doe CQ (2017) ‘ Temporal Patterning in the Drosophila CNS ‘, Annual Review of Cell and Developmental Biology. doi: 10.1146/annurev-cellbio-111315-125210. [DOI] [PubMed] [Google Scholar]
- Egger B, Gold KS and Brand AH (2011) ‘Regulating the balance between symmetric and asymmetric stem cell division in the developing brain’, Fly. Taylor and Francis Inc., 5(3), pp. 237–241. doi: 10.4161/fly.5.3.15640. [DOI] [PubMed] [Google Scholar]
- Elliott J et al. (2008) ‘Ikaros Confers Early Temporal Competence to Mouse Retinal Progenitor Cells’, Neuron. doi: 10.1016/j.neuron.2008.08.008. [DOI] [PubMed] [Google Scholar]
- Eroglu E et al. (2014) ‘SWI/SNF complex prevents lineage reversion and induces temporal patterning in neural stem cells’, Cell. doi: 10.1016/j.cell.2014.01.053. [DOI] [PubMed] [Google Scholar]
- Fame RM, MacDonald JL and Macklis JD (2011) ‘Development, specification, and diversity of callosal projection neurons’, Trends in Neurosciences. doi: 10.1016/j.tins.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florio M et al. (2015) ‘Human-specific gene ARHGAP11B promotes basal progenitor amplification and neocortex expansion’, Science. doi: 10.1126/science.aaa1975. [DOI] [PubMed] [Google Scholar]
- Gao P et al. (2014) ‘Deterministic progenitor behavior and unitary production of neurons in the neocortex’, Cell. doi: 10.1016/j.cell.2014.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspard N et al. (2008) ‘An intrinsic mechanism of corticogenesis from embryonic stem cells’, Nature. doi: 10.1038/nature07287. [DOI] [PubMed] [Google Scholar]
- Geng Z and Gao Z (2020) ‘Mammalian prc1 complexes: Compositional complexity and diverse molecular mechanisms’, International Journal of Molecular Sciences. MDPI AG, pp. 1–18. doi: 10.3390/ijms21228594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greig LC et al. (2013) ‘Molecular logic of neocortical projection neuron specification, development and diversity’, Nature Reviews Neuroscience. doi: 10.1038/nrn3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosskortenhaus R et al. (2005) ‘Regulation of temporal identity transitions in drosophila neuroblasts’, Developmental Cell. doi: 10.1016/j.devcel.2004.11.019. [DOI] [PubMed] [Google Scholar]
- Grosskortenhaus R, Robinson KJ and Doe CQ (2006) ‘Pdm and Castor specify late-born motor neuron identity in the NB7–1 lineage’, Genes and Development. doi: 10.1101/gad.1445306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillemot F (2007) ‘Spatial and temporal specification of neural fates by transcription factor codes’, Development. doi: 10.1242/dev.006379. [DOI] [PubMed] [Google Scholar]
- Guo C et al. (2013) ‘Fezf2 expression identifies a multipotent progenitor for neocortical projection neurons, astrocytes, and oligodendrocytes’, Neuron. doi: 10.1016/j.neuron.2013.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanashima C et al. (2004) ‘Foxg1 Suppresses Early Cortical Cell Fate’, Science. doi: 10.1126/science.1090674. [DOI] [PubMed] [Google Scholar]
- Hanashima C and Toma K (2015) ‘Switching modes in corticogenesis: Mechanisms of neuronal subtype transitions and integration in the cerebral cortex’, Frontiers in Neuroscience. doi: 10.3389/fnins.2015.00274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardwick LJA et al. (2015) ‘Cell cycle regulation of proliferation versus differentiation in the central nervous system’, Cell and Tissue Research. doi: 10.1007/s00441-014-1895-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi M et al. (2018) ‘Graded Arrays of Spinal and Supraspinal V2a Interneuron Subtypes Underlie Forelimb and Hindlimb Motor Control’, Neuron. Cell Press, 97(4), pp. 869–884.e5. doi: 10.1016/j.neuron.2018.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirabayashi Y et al. (2009) ‘Polycomb Limits the Neurogenic Competence of Neural Precursor Cells to Promote Astrogenic Fate Transition’, Neuron. doi: 10.1016/j.neuron.2009.08.021. [DOI] [PubMed] [Google Scholar]
- Ho L and Crabtree GR (2010) ‘Chromatin remodelling during development’, Nature. doi: 10.1038/nature08911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges C, Kirkland JG and Crabtree GR (2016) ‘The many roles of BAF (mSWI/SNF) and PBAF complexes in cancer’, Cold Spring Harbor Perspectives in Medicine. doi: 10.1101/cshperspect.a026930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holguera I and Desplan C (2018) ‘Neuronal specification in space and time’, Science. doi: 10.1126/science.aas9435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homem CCF and Knoblich JA (2012) ‘Drosophila neuroblasts: A model for stem cell biology’, Development (Cambridge). doi: 10.1242/dev.080515. [DOI] [PubMed] [Google Scholar]
- Hota SK and Bruneau BG (2016) ‘ATP-dependent chromatin remodeling during mammalian development’, Development (Cambridge). doi: 10.1242/dev.128892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou PS, Miyoshi G and Hanashima C (2019) ‘Sensory cortex wiring requires preselection of short- and long-range projection neurons through an Egr-Foxg1-COUP-TFI network’, Nature Communications. doi: 10.1038/s41467-019-11043-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoye ML and Silver DL (2021) ‘Decoding mixed messages in the developing cortex: translational regulation of neural progenitor fate’, Current Opinion in Neurobiology. doi: 10.1016/j.conb.2020.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isshiki T et al. (2001) ‘Drosophila neuroblasts sequentially express transcription factors which specify the temporal identity of their neuronal progeny’, Cell. doi: 10.1016/S0092-8674(01)00465-2. [DOI] [PubMed] [Google Scholar]
- Jacob J, Maurange C and Gould AP (2008) ‘Temporal control of neuronal diversity: Common regulatory principles in insects and vertebrates?’, Development. doi: 10.1242/dev.016931. [DOI] [PubMed] [Google Scholar]
- Jafarnejad SM et al. (2018) ‘Translational control of ERK signaling through miRNA/4EHP-directed silencing’, eLife. doi: 10.7554/eLife.35034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javed A et al. (2020) ‘Pou2f1 and Pou2f2 cooperate to control the timing of cone photoreceptor production in the developing mouse retina’, Development (Cambridge). Company of Biologists Ltd, 147(18). doi: 10.1242/dev.188730. [DOI] [PubMed] [Google Scholar]
- Javed A and Cayouette M (2017) ‘Temporal progression of retinal progenitor cell identity: Implications in cell replacement therapies’, Frontiers in Neural Circuits. doi: 10.3389/fncir.2017.00105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kambadur R et al. (1998) ‘Regulation of POU genes by castor and hunchback establishes layered compartments in the Drosophila CNS’, Genes and Development. doi: 10.1101/gad.12.2.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang P et al. (2012) ‘Sox9 and NFIA Coordinate a Transcriptional Regulatory Cascade during the Initiation of Gliogenesis’, Neuron. Neuron, 74(1), pp. 79–94. doi: 10.1016/j.neuron.2012.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao CF et al. (2012) ‘Hierarchical Deployment of Factors Regulating Temporal Fate in a Diverse Neuronal Lineage of the Drosophila Central Brain’, Neuron. doi: 10.1016/j.neuron.2011.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi A (2019) ‘Temporal patterning of neocortical progenitor cells: How do they know the right time?’, Neuroscience Research. doi: 10.1016/j.neures.2018.09.004. [DOI] [PubMed] [Google Scholar]
- Kedde M et al. (2010) ‘A Pumilio-induced RNA structure switch in p27–3′2 UTR controls miR-221 and miR-222 accessibility’, Nature Cell Biology. doi: 10.1038/ncb2105. [DOI] [PubMed] [Google Scholar]
- Kingston RE and Tamkun JW (2014) ‘Transcriptional regulation by trithorax-group proteins’, Cold Spring Harbor Perspectives in Biology. doi: 10.1101/cshperspect.a019349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knock E et al. (2015) ‘The methyl binding domain 3/nucleosome remodelling and deacetylase complex regulates neural cell fate determination and terminal differentiation in the cerebral cortex’, Neural Development. doi: 10.1186/s13064-015-0040-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konstantinides N et al. (2021) ‘A comprehensive series of temporal transcription factors in the fly visual system’, bioRxiv. Cold Spring Harbor Laboratory, p. 2021.06.13.448242. doi: 10.1101/2021.06.13.448242. [DOI] [Google Scholar]
- Kumamoto T et al. (2013) ‘Foxg1 coordinates the switch from nonradially to radially migrating glutamatergic subtypes in the neocortex through spatiotemporal repression’, Cell Reports. doi: 10.1016/j.celrep.2013.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakadamyali M and Cosma MP (2020) ‘Visualizing the genome in high resolution challenges our textbook understanding’, Nature Methods. doi: 10.1038/s41592-020-0758-3. [DOI] [PubMed] [Google Scholar]
- Lee T (2017) ‘Wiring the Drosophila Brain with Individually Tailored Neural Lineages’, Current Biology. doi: 10.1016/j.cub.2016.12.026. [DOI] [PubMed] [Google Scholar]
- Lessard J et al. (2007) ‘An Essential Switch in Subunit Composition of a Chromatin Remodeling Complex during Neural Development’, Neuron. doi: 10.1016/j.neuron.2007.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X et al. (2013) ‘Temporal patterning of Drosophila medulla neuroblasts controls neural fates’, Nature. Nature Publishing Group, 498(7455), pp. 456–462. doi: 10.1038/nature12319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Chen Z and Desplan C (2013) ‘Temporal patterning of neural progenitors in drosophila’, in Current Topics in Developmental Biology. doi: 10.1016/B978-0-12-396968-2.00003-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Z et al. (2017) ‘A high-resolution map of transcriptional repression’, eLife. eLife Sciences Publications Ltd, 6. doi: 10.7554/eLife.22767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S and Lee T (2012) ‘Generating neuronal diversity in the Drosophila central nervous system’, Developmental Dynamics. doi: 10.1002/dvdy.22739. [DOI] [PubMed] [Google Scholar]
- Liu S et al. (2020) ‘Foxn4 is a temporal identity factor conferring mid/late-early retinal competence and involved in retinal synaptogenesis’, Proceedings of the National Academy of Sciences of the United States of America. doi: 10.1073/pnas.1918628117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z et al. (2015) ‘Opposing intrinsic temporal gradients guide neural stem cell production of varied neuronal fates’, Science. doi: 10.1126/science.aad1886. [DOI] [PubMed] [Google Scholar]
- Llorca A et al. (2019) ‘A stochastic framework of neurogenesis underlies the assembly of neocortical cytoarchitecture’, eLife. doi: 10.7554/eLife.51381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorca A and Marín O (2021) ‘Orchestrated freedom: new insights into cortical neurogenesis’, Current Opinion in Neurobiology. doi: 10.1016/j.conb.2020.09.004. [DOI] [PubMed] [Google Scholar]
- Lopes A, Magrinelli E and Telley L (2020) ‘Emerging roles of single-cell multi-omics in studying developmental temporal patterning’, International Journal of Molecular Sciences. doi: 10.3390/ijms21207491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y et al. (2020) ‘Single-Cell Analysis of Human Retina Identifies Evolutionarily Conserved and Species-Specific Mechanisms Controlling Development’, Developmental Cell. Cell Press, 53(4), pp. 473–491.e9. doi: 10.1016/j.devcel.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luxenhofer G et al. (2014) ‘MicroRNA-9 promotes the switch from early-born to late-born motor neuron populations by regulating Onecut transcription factor expression’, Developmental Biology. Academic Press Inc., 386(2), pp. 358–370. doi: 10.1016/j.ydbio.2013.12.023. [DOI] [PubMed] [Google Scholar]
- Marchetti G and Tavosanis G (2019) ‘Modulators of hormonal response regulate temporal fate specification in the Drosophila brain’, PLoS Genetics. doi: 10.1371/journal.pgen.1008491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall OJ and Brand AH (2017) ‘Chromatin state changes during neural development revealed by in vivo cell-type specific profiling’, Nature Communications. doi: 10.1038/s41467-017-02385-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto S et al. (2006) ‘Brg1 is required for murine neural stem cell maintenance and gliogenesis’, Developmental Biology. doi: 10.1016/j.ydbio.2005.10.044. [DOI] [PubMed] [Google Scholar]
- Mattar P et al. (2015) ‘A conserved regulatory logic controls temporal identity in mouse neural progenitors’, Neuron. doi: 10.1016/j.neuron.2014.12.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattar P et al. (2020) ‘A Casz1 - NuRD complex regulates temporal identity transitions in neural progenitors’, bioRxiv. doi: 10.1101/2020.02.11.944470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattar P et al. (2021) ‘A Casz1–NuRD complex regulates temporal identity transitions in neural progenitors’, Scientific Reports. Nature Research, 11(1). doi: 10.1038/s41598-021-83395-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matuzelski E et al. (2017) ‘Transcriptional regulation of Nfix by NFIB drives astrocytic maturation within the developing spinal cord’, Developmental Biology. Elsevier Inc., 432(2), pp. 286–297. doi: 10.1016/j.ydbio.2017.10.019. [DOI] [PubMed] [Google Scholar]
- Maurange C (2020) ‘Temporal patterning in neural progenitors: From Drosophila development to childhood cancers’, DMM Disease Models and Mechanisms. doi: 10.1242/dmm.044883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurange C, Cheng L and Gould AP (2008) ‘Temporal Transcription Factors and Their Targets Schedule the End of Neural Proliferation in Drosophila’, Cell. doi: 10.1016/j.cell.2008.03.034. [DOI] [PubMed] [Google Scholar]
- McConnell SK (1988) ‘Fates of visual cortical neurons in the ferret after isochronic and heterochronic transplantation’, Journal of Neuroscience. doi: 10.1523/jneurosci.08-03-00945.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell SK (1995) ‘Constructing the cerebral cortex: Neurogenesis and fate determination’, Neuron. doi: 10.1016/0896-6273(95)90168-X. [DOI] [PubMed] [Google Scholar]
- McConnell SK and Kaznowski CE (1991) ‘Cell cycle dependence of laminar determination in developing neocortex’, Science. doi: 10.1126/science.1925583. [DOI] [PubMed] [Google Scholar]
- Mettler U, Vogler G and Urban J (2006) ‘Timing of identity: Spatiotemporal regulation of hunchback in neuroblast lineages of Drosophila by Seven-up and Prospero’, Development. doi: 10.1242/dev.02229. [DOI] [PubMed] [Google Scholar]
- Miyares RL and Lee T (2019) ‘Temporal control of Drosophila central nervous system development’, Current Opinion in Neurobiology. doi: 10.1016/j.conb.2018.10.016. [DOI] [PubMed] [Google Scholar]
- Mohrmann L et al. (2004) ‘Differential Targeting of Two Distinct SWI/SNF-Related Drosophila Chromatin-Remodeling Complexes’, Molecular and Cellular Biology. doi: 10.1128/mcb.24.8.3077-3088.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto-Suzki N et al. (2014) ‘The polycomb component Ring1B regulates the timed termination of subcerebral projection neuron production during mouse neocortical development’, Development (Cambridge). doi: 10.1242/dev.112276. [DOI] [PubMed] [Google Scholar]
- Müller T et al. (2002) ‘The homeodomain factor Lbx1 distinguishes two major programs of neuronal differentiation in the dorsal spinal cord’, Neuron. Elsevier, 34(4), pp. 551–562. doi: 10.1016/S0896-6273(02)00689-X. [DOI] [PubMed] [Google Scholar]
- Muralidharan B et al. (2017) ‘LHX2 interacts with the NuRD complex and regulates cortical neuron subtype determinants Fezf2 and Sox11’, Journal of Neuroscience. doi: 10.1523/JNEUROSCI.2836-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naidu VG et al. (2020) ‘Temporal progression of Drosophila medulla neuroblasts generates the transcription factor combination to control T1 neuron morphogenesis’, Developmental Biology. doi: 10.1016/j.ydbio.2020.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naka H et al. (2008) ‘Requirement for COUP-TFI and II in the temporal specification of neural stem cells in CNS development’, Nature Neuroscience. doi: 10.1038/nn.2168. [DOI] [PubMed] [Google Scholar]
- Narbonne-Reveau K et al. (2016) ‘Neural stem cell-encoded temporal patterning delineates an early window of malignant susceptibility in Drosophila’, eLife. doi: 10.7554/eLife.13463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino J et al. (2008) ‘Hmga2 Promotes Neural Stem Cell Self-Renewal in Young but Not Old Mice by Reducing p16Ink4a and p19Arf Expression’, Cell. Elsevier B.V., 135(2), pp. 227–239. doi: 10.1016/j.cell.2008.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino J et al. (2013) ‘A network of heterochronic genes including Imp1 regulates temporal changes in stem cell properties’, eLife. eLife Sciences Publications Ltd, 2013(2), p. 924. doi: 10.7554/eLife.00924.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowakowski TJ et al. (2017) ‘Spatiotemporal gene expression trajectories reveal developmental hierarchies of the human cortex’, Science. doi: 10.1126/science.aap8809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberst P et al. (2019) ‘Temporal plasticity of apical progenitors in the developing mouse neocortex’, Nature. doi: 10.1038/s41586-019-1515-6. [DOI] [PubMed] [Google Scholar]
- Oberst P, Agirman G and Jabaudon D (2019) ‘Principles of progenitor temporal patterning in the developing invertebrate and vertebrate nervous system’, Current Opinion in Neurobiology. doi: 10.1016/j.conb.2019.03.004. [DOI] [PubMed] [Google Scholar]
- Okamoto M et al. (2016) ‘Cell-cycle-independent transitions in temporal identity of mammalian neural progenitor cells’, Nature Communications. doi: 10.1038/ncomms11349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano H and Temple S (2009) ‘Cell types to order: temporal specification of CNS stem cells’, Current Opinion in Neurobiology. doi: 10.1016/j.conb.2009.04.003. [DOI] [PubMed] [Google Scholar]
- Pearson BJ and Doe CQ (2004) ‘Specification of temporal identity in the developing nervous system’, Annual Review of Cell and Developmental Biology. doi: 10.1146/annurev.cellbio.19.111301.115142. [DOI] [PubMed] [Google Scholar]
- Pearson CA et al. (2020) ‘Foxp1 Regulates Neural Stem Cell Self-Renewal and Bias Toward Deep Layer Cortical Fates’, Cell Reports. doi: 10.1016/j.celrep.2020.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira JD et al. (2010) ‘Ezh2, the histone methyltransferase of PRC2, regulates the balance between self-renewal and differentiation in the cerebral cortex’, Proceedings of the National Academy of Sciences of the United States of America. doi: 10.1073/pnas.1002530107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajman M and Schratt G (2017) ‘MicroRNAs in neural development: From master regulators to fine-tuners’, Development (Cambridge). doi: 10.1242/dev.144337. [DOI] [PubMed] [Google Scholar]
- Reillo I et al. (2017) ‘A complex code of extrinsic influences on cortical progenitor cells of higher mammals’, Cerebral Cortex. doi: 10.1093/cercor/bhx171. [DOI] [PubMed] [Google Scholar]
- Ren Q et al. (2017) ‘Stem Cell-Intrinsic, Seven-up-Triggered Temporal Factor Gradients Diversify Intermediate Neural Progenitors’, Current Biology. doi: 10.1016/j.cub.2017.03.047. [DOI] [PubMed] [Google Scholar]
- Rossi AM and Desplan C (2020) ‘Extrinsic activin signaling cooperates with an intrinsic temporal program to increase mushroom body neuronal diversity’, eLife. doi: 10.7554/eLife.58880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagner A et al. (2020) ‘Temporal patterning of the central nervous system by a shared transcription factor code’, bioRxiv. Cold Spring Harbor Laboratory, p. 2020.11.10.376491. doi: 10.1101/2020.11.10.376491. [DOI] [Google Scholar]
- Sagner A and Briscoe J (2019) ‘Establishing neuronal diversity in the spinal cord: A time and a place’, Development (Cambridge). doi: 10.1242/dev.182154. [DOI] [PubMed] [Google Scholar]
- Sapkota D et al. (2014) ‘Onecut1 and Onecut2 redundantly regulate early retinal cell fates during development’, Proceedings of the National Academy of Sciences of the United States of America. National Academy of Sciences, 111(39), pp. E4086–E4095. doi: 10.1073/pnas.1405354111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seritrakul P and Gross JM (2019) ‘Genetic and epigenetic control of retinal development in zebrafish’, Current Opinion in Neurobiology. doi: 10.1016/j.conb.2019.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Q et al. (2006) ‘The timing of cortical neurogenesis is encoded within lineages of individual progenitor cells’, Nature Neuroscience. doi: 10.1038/nn1694. [DOI] [PubMed] [Google Scholar]
- Shu P et al. (2019) ‘Opposing Gradients of MicroRNA Expression Temporally Pattern Layer Formation in the Developing Neocortex’, Developmental Cell. doi: 10.1016/j.devcel.2019.04.017. [DOI] [PubMed] [Google Scholar]
- Sockanathan S and Jessell TM (1998) ‘Motor neuron-derived retinoid signaling specifies the subtype identity of spinal motor neurons’, Cell. Elsevier B.V., 94(4), pp. 503–514. doi: 10.1016/S0092-8674(00)81591-3. [DOI] [PubMed] [Google Scholar]
- Sokpor G et al. (2017) ‘Chromatin remodeling BAF (SWI/SNF) complexes in neural development and disorders’, Frontiers in Molecular Neuroscience. doi: 10.3389/fnmol.2017.00243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokpor G et al. (2018) ‘ATP-dependent chromatin remodeling during cortical neurogenesis’, Frontiers in Neuroscience. doi: 10.3389/fnins.2018.00226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan K et al. (2012) ‘A network of genetic repression and derepression specifies projection fates in the developing neocortex’, Proceedings of the National Academy of Sciences of the United States of America. doi: 10.1073/pnas.1216793109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stam FJ et al. (2012) ‘Renshaw cell interneuron specialization is controlled by a temporally restricted transcription factor program’, Development. Development, 139(1), pp. 179–190. doi: 10.1242/dev.071134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T et al. (2013) ‘A temporal mechanism that produces neuronal diversity in the Drosophila visual center’, Developmental Biology. doi: 10.1016/j.ydbio.2013.05.002. [DOI] [PubMed] [Google Scholar]
- Swinstead EE et al. (2016) ‘Pioneer factors and ATP-dependent chromatin remodeling factors interact dynamically: A new perspective’, BioEssays. doi: 10.1002/bies.201600137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syed MH, Mark B and Doe CQ (2017) ‘Steroid hormone induction of temporal gene expression in drosophila brain neuroblasts generates neuronal and glial diversity’, eLife. doi: 10.7554/eLife.26287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang JLY et al. (2021) ‘NanoDam identifies novel temporal transcription factors conserved between the Drosophila central brain and visual system’, bioRxiv. Cold Spring Harbor Laboratory, p. 2021.06.07.447332. doi: 10.1101/2021.06.07.447332. [DOI] [Google Scholar]
- Tang L, Nogales E and Ciferri C (2010) ‘Structure and function of SWI/SNF chromatin remodeling complexes and mechanistic implications for transcription’, Progress in Biophysics and Molecular Biology. Prog Biophys Mol Biol, pp. 122–128. doi: 10.1016/j.pbiomolbio.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telley L et al. (2016) ‘Sequential transcriptional waves direct the differentiation of newborn neurons in the mouse neocortex’, Science. doi: 10.1126/science.aad8361. [DOI] [PubMed] [Google Scholar]
- Telley L et al. (2019) ‘Temporal patterning of apical progenitors and their daughter neurons in the developing neocortex’, Science. doi: 10.1126/science.aav2522. [DOI] [PubMed] [Google Scholar]
- Toma K et al. (2014) ‘The timing of upper-layer neurogenesis is conferred by sequential derepression and negative feedback from deep-layer neurons’, Journal of Neuroscience. doi: 10.1523/JNEUROSCI.2334-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touma JJ, Weckerle FF and Cleary MD (2012) ‘Drosophila Polycomb complexes restrict neuroblast competence to generate motoneurons’, Development. doi: 10.1242/dev.071589. [DOI] [PubMed] [Google Scholar]
- Tripodi M, Stepien AE and Arber S (2011) ‘Motor antagonism exposed by spatial segregation and timing of neurogenesis’, Nature. Nature, 479(7371), pp. 61–66. doi: 10.1038/nature10538. [DOI] [PubMed] [Google Scholar]
- Tuoc TC et al. (2013) ‘Chromatin Regulation by BAF170 Controls Cerebral Cortical Size and Thickness’, Developmental Cell. doi: 10.1016/j.devcel.2013.04.005. [DOI] [PubMed] [Google Scholar]
- Vitali I et al. (2018) ‘Progenitor Hyperpolarization Regulates the Sequential Generation of Neuronal Subtypes in the Developing Neocortex’, Cell. doi: 10.1016/j.cell.2018.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh KT and Doe CQ (2017) ‘Drosophila embryonic type II neuroblasts: Origin, temporal patterning, and contribution to the adult central complex’, Development (Cambridge). doi: 10.1242/dev.157826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CP et al. (2017) ‘Imp and Syp RNA-binding proteins govern decommissioning of Drosophila neural stem cells’, Development (Cambridge). doi: 10.1242/dev.149500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao B et al. (2016) ‘Epigenetic mechanisms in neurogenesis’, Nature Reviews Neuroscience. doi: 10.1038/nrn.2016.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasugi T et al. (2008) ‘Drosophila optic lobe neuroblasts triggered by a wave of proneural gene expression that is negatively regulated by JAK/STAT’, Development. doi: 10.1242/dev.019117. [DOI] [PubMed] [Google Scholar]
- Yoon KJ et al. (2017) ‘Temporal Control of Mammalian Cortical Neurogenesis by m6A Methylation’, Cell. doi: 10.1016/j.cell.2017.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon KJ et al. (2018) ‘Epigenetics and epitranscriptomics in temporal patterning of cortical neural progenitor competence’, Journal of Cell Biology. doi: 10.1083/jcb.201802117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuzwa SA et al. (2017) ‘Developmental Emergence of Adult Neural Stem Cells as Revealed by Single-Cell Transcriptional Profiling’, Cell Reports. doi: 10.1016/j.celrep.2017.12.017. [DOI] [PubMed] [Google Scholar]
- Zacharioudaki E, Falo Sanjuan J and Bray S (2019) ‘Mi-2/NuRD complex protects stem cell progeny from mitogenic Notch signaling’, eLife. doi: 10.7554/eLife.41637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahr SK et al. (2018) ‘A Translational Repression Complex in Developing Mammalian Neural Stem Cells that Regulates Neuronal Specification’, Neuron. doi: 10.1016/j.neuron.2017.12.045. [DOI] [PubMed] [Google Scholar]
- Zembrzycki A et al. (2015) ‘Postmitotic regulation of sensory area patterning in the mammalian neocortex by Lhx2’, Proceedings of the National Academy of Sciences of the United States of America. doi: 10.1073/pnas.1424440112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y et al. (2020) ‘Cortical Neural Stem Cell Lineage Progression Is Regulated by Extrinsic Signaling Molecule Sonic Hedgehog’, Cell Reports. doi: 10.1016/j.celrep.2020.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H et al. (2021) ‘A comprehensive temporal patterning gene network in Drosophila medulla neuroblasts revealed by single-cell RNA sequencing’, bioRxiv. Cold Spring Harbor Laboratory, p. 2021.06.12.448145. doi: 10.1101/2021.06.12.448145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S et al. (2006) ‘Gradients of the Drosophila Chinmo BTB-Zinc Finger Protein Govern Neuronal Temporal Identity’, Cell. doi: 10.1016/j.cell.2006.08.045. [DOI] [PubMed] [Google Scholar]


