Sulfur homeostasis is of vital importance for plant health and human nutrition. Sulfur nutrition strongly impacts plant resilience and nutritional quality traits in agricultural crop production. Recent discoveries provide dynamic perspectives on biological processes and mechanistic models delineating biochemical diversity and genetic regulation of plant sulfur metabolism. Invited reviews and articles in this Special Issue highlight current knowledge and ongoing research in sulfur biology.
In the sulfur cycle in nature, plants are producers of the essential amino acids cysteine (Cys) and methionine (Met), both of which are incorporated into storage proteins serving as a dietary source consumed by animals (Takahashi et al., 2011). Sulfur-containing metabolites and cofactors play pivotal roles in both abiotic and biotic stress mitigation processes, manifesting the importance of sulfur nutrition in plant resilience: for example, they contribute to redox signaling, heavy metal detoxification, and chemical defense against pathogen attack and herbivory in the environment. These topics were part of discussions at the 12th International Plant Sulfur Workshop held in London, Ontario, Canada in 2022, and are the basis for this Special Issue.
Plants monitor the environment and translate key information on external stimuli to intrinsic signals to alter gene expression, protein function, and protein stability, and take correct actions against the environmental stressors. We find intricate but tight connections between sulfur metabolism and cell signaling. Sulfide (S2–) and O-acetylserine (OAS), the substrates for Cys biosynthesis, modulate formation of the Cys synthase complex and sulfate assimilation (Hell and Wirtz, 2011; Sun et al., 2021; Apodiakou and Hoefgen, 2023; Wirtz et al., 2023) (Box 1). S2– and OAS can also modulate the activity of the target of rapamycin (TOR) complex which optimizes plant growth (Dong et al., 2017). Glutathione (γ-glutamylcysteinylglycine; GSH), the tripeptide synthesized from Cys, and Fe–S clusters impact the redox status (Caubrière et al., 2023; Ito and Ohkama-Ohtsu, 2023; Iven et al., 2023; Pedroletti et al., 2023). Sulfation of metabolites and peptides generates 3’-phosphoadenosine-5’-phosphate (PAP), a retrograde signaling molecule communicating messages relevant to a wide spectrum of abiotic stress factors (e.g. excess light, drought, and oxidative stress) (Chan et al., 2019). Biosynthesis and subcellular transport dynamics of these metabolite signals continue to represent intriguing questions about cell signaling over time and space.
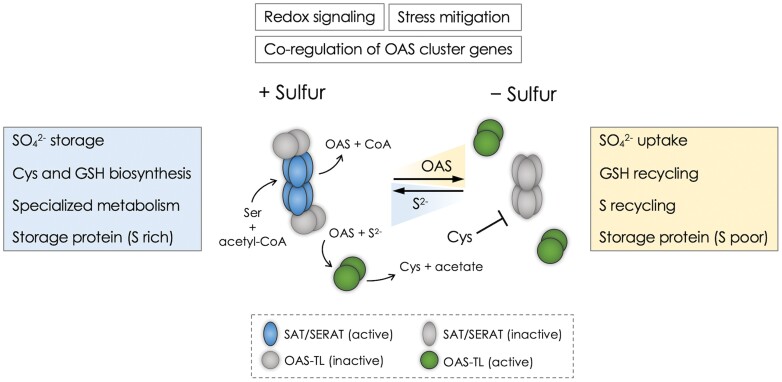
Box 1. Simplified model illustrating dynamic assembly of Cys synthase complex and regulation of sulfur metabolism in plants.
Under sulfur-replete conditions (+Sulfur), S2– promotes the assembly of the Cys synthase complex. Ser acetyltransferase (SAT/SERAT) hexamers and O-acetylserine(thiol)lyase (OAS-TL) dimers form this multienzyme complex. SAT/SERAT hexamers within the Cys synthase complex (blue) are free from feedback inhibition by Cys and remain active to synthesize OAS from Ser. OAS-TL dimers within the Cys synthase complex are catalytically inactive (gray). Free OAS-TL dimers (green) catalyze synthesis of Cys from OAS and S2–. Under sulfur-deficient conditions (–Sulfur), S2– gradually becomes unavailable and OAS accumulates in the cell. OAS promotes dissociation of the Cys synthase complex. SAT/SERAT hexamers dissociated from the complex are inactive (gray) because they are sensitive to feedback inhibition by Cys. Illustration modified based on work by Hell and Wirtz (2011).
Boxes highlight responses to changes in sulfur conditions: under sulfur-replete conditions (left), plants accumulate SO42– and synthesize Cys, GSH, and sulfur-containing specialized metabolites. These metabolites contribute to maintaining the redox status and stress mitigation. The seed storage protein composition will become sulfur rich. Under sulfur-deficient conditions (right), plants activate SO42– uptake and promote sulfur recycling. Sulfur-rich seed storage proteins will become less abundant and replaced by sulfur-poor storage proteins. The OAS cluster genes are co-regulated and play important roles in these processes.
Thiols in action
Soil contaminants, such as arsenic (As), cadmium (Cd), lead (Pb), and mercury (Hg), are significant threats to global food security, deteriorating plant performance and human health. Plants exposed to these toxic metals and metalloids detoxify them through conjugation, sequestration, and avoidance mechanisms (Clemens and Ma, 2016). In this Special Issue, Sun et al. (2023) introduce As and Cd detoxification mechanisms, addressing what specific conjugants and transporters are involved and how they are mechanistically related to sulfur metabolism. Their recent work on the rice arsenite-tolerant mutant astol1 and the causal dominant mutation which eliminates the catalytic activity of chloroplast-localizing O-acetylserine(thiol)lyase (OAS-TL) highlights the importance of the proposed sulfur-sensing function of the Cys synthase complex in As tolerance (Sun et al., 2021). Their findings suggest the assembly of the Cys synthase complex—stabilized and activated to function as Ser acetyltransferase (SAT or SERAT) in the mutant—promotes sulfate uptake, reductive sulfur assimilation, and subsequent Cys and GSH biosynthesis, thus contributing to As detoxification (Sun et al., 2021).
These toxic metals and metalloids are detrimental to plant health and thus may impact general cellular and organellar responses. Iven et al. (2023) extend discussion on how GSH and PC pools alter in the event of plant exposure to Cd and influence the ethylene and reactive oxygen species (ROS)-mediated signaling networks. Ramification of stresses imposed by Cd appears to involve ethylene and ROS signaling communicating with mitochondrial retrograde signaling, unfolded protein response, and autophagy. The authors further introduce potential actions of S2– in plant Cd responses. As the product of sulfate assimilation and Cys catabolism, the potential signaling function of this highly reactive small molecule draws increasing attention. The prevalence of its biological impact may well be explained by translational and post-translational regulations associated with protein persulfidation (Aroca et al., 2017). S2– impacts the activity states and assembly/disassembly of the Cys synthase complex (Hell and Wirtz, 2011), altering the GSH pool size and signaling networks governing cellular responses to the redox status (Box 1).
Ito and Ohkama-Ohtsu (2023) highlight GSH metabolism and recycling pathways influencing the GSH pool size. GSH is synthesized from Cys in the chloroplast and partially in the cytosol by two-step reactions catalyzed by γ-glutamylcysteine synthetase and GSH synthetase. GSH forms conjugates with xenobiotics and endogenous bioactive metabolites including phytoallexins. γ-Glutamylpeptidases subsequently process these conjugates in the cytosol and vacuoles. Aside from these canonical aspects, the authors’ recent findings point to the concept of sulfur economy: γ-glutamylpeptidase and γ-glutamyl cyclotransferase are differentially expressed to enable GSH recycling, and GSH serves for conjugating glucosinolate breakdown products when there is a demand for sulfur recycling under sulfur deficiency (Sugiyama et al., 2021; Ito et al., 2022).
Cys biosynthesis and metallocofactors
Cys biosynthesis is essential to protein synthesis in three subcellular compartments, the cytosol, chloroplasts, and mitochondria (Hell and Wirtz, 2011). SAT and OAS-TL exist in all three subcellular compartments. The rate of Cys biosynthesis depends on provision of two substrates—S2– from the sulfate assimilation pathway in the chloroplast and OAS which derives from the amino acid Ser. The Cys synthase complex, the multienzyme complex of SAT and OAS-TL, functions not only as the catalyst for OAS biosynthesis but also for sulfur sensing. The suggested action mechanism for sulfur sensing involves assembly of the enzyme complex promoted by S2– and its dissociation triggered by OAS (Hell and Wirtz, 2011) (Box 1).
Wirtz et al. (2023) present new data on this topic. By testing the impact of overexpression of enzymatically active and inactive forms of SAT in tobacco plants, they provide evidence that SAT overexpression enhances sulfate assimilation and Cys biosynthesis regardless of the SAT enzyme activity—the phenotype probably associated with stable Cys synthase complex formation encouraged in both the cytosol and chloroplasts. Interestingly, only the SAT overexpression in chloroplasts (with both the active and inactive forms of SAT) appears causative of chloroplast dysfunction and consequential downfall of the photosynthetic capacity. The impact of Cys overproduction on photosynthesis appears compartment specific and may require interacting partners specific to the Cys synthase complex in chloroplasts (Müller et al., 2017).
Sulfur-containing cofactors play pivotal roles in metabolism (Caubrière et al., 2023; Pedroletti et al., 2023). Fe–S clusters are undoubtedly the most prominent and indispensable units, as they are essential for the photosynthetic and respiratory electron transfer chains (Balk and Schaedler, 2014). They are also essential for key enzyme functions in nitrogen and sulfur assimilation pathways, and synthesis of other important cofactors, such as lipoic acid, biotin, and thiamine. The molybdenum cofactor (Moco) is another essential metallocofactor required for nitrogen assimilation as well as ureide biosynthesis, and it is also involved in sulfite oxidation and abscisic acid (ABA) biosynthesis. Thus, the Fe–S cluster and Moco biogenesis is of crucial importance to the photosynthesis and basic metabolic function.
Cys desulfurase is the enzyme which is central to the extraction of sulfur from Cys, facilitating subsequent sulfur transfer or ‘sulfur trafficking’ in the Fe–S cluster and Moco biogenesis pathways. Caubrière et al. (2023) discuss functional diversity of Cys desulfurases in plants based on structural features and isoform-specific roles of this enzyme in three subcellular compartments, the cytosol, chloroplasts, and mitochondria. They further explore biological implication and possible evolutionary origins of Cys desulfurases coupling with Fe–S cluster assembly enzymes, sulfur trafficking enzymes, and thioredoxins involved in sulfide formation and redox regulations.
Fe–S cluster formation in mitochondria parallels its essentiality in chloroplasts. Pedroletti et al. (2023) provide detailed analyses of mitochondrial Fe–S cluster assembly mechanisms and machineries introducing sulfur to lipoic acid and biotin biosynthesis. They also mention possible impacts of loss of mitochondrial Fe–S clusters and target proteins and consequences of turnover of Fe–S cluster proteins. The article provides critical insights into the mitochondrial SAT function. Given the major contribution (~80%) of the mitochondrial SAT to the overall OAS production (Watanabe et al., 2008; Hell and Wirtz, 2011), scavenging of S2– released after disassembly of Fe–S clusters could rely on Cys biosynthesis locally in this organelle.
Impact of OAS and sulfur nutrition
OAS is arguably the compound gaining traction over the past few decades in this research area. While it serves as a substrate for Cys biosynthesis, it is also recognized categorically as a signaling molecule triggering dissociation of the Cys synthase complex (Box 1). OAS accumulates under sulfur deficiency because of the appearance of SAT and OASTL, the enzymes responsible for Cys biosynthesis, at the juncture of sulfur assimilation and Ser biosynthetic pathways. Along these lines, OAS has been studied as a diagnostic molecular marker for plant sulfur status and an effector modulating sulfur deficiency response.
Apodiakou and Hoefgen (2023) provide an overview of the ‘OAS cluster genes’ co-expressed or co-regulated by sulfur deficiency and OAS in Arabidopsis. The robustness of gene regulatory networks of OAS cluster genes shown in this work corroborates the earlier findings of co-regulation of sulfur-responsive genes by the SULFUR LIMITATION 1/ETHYLENE INSENSITIVE 3-LIKE 3 (SLIM1/EIL3) transcription factor (Maruyama-Nakashita et al., 2006; Aarabi et al., 2016). The additional evidence shown based on comparative transcriptome analyses and DNA affinity purification (DAP) sequencing for SLIM1/EIL3 further emphasizes the role of this master regulator in sulfur nutritional response. Proposed interactive contributions of other transcription factors for sulfur and OAS responses await further investigations (Rakpenthai et al., 2022).
Sulfur nutrition not only impacts Cys and Met biosynthesis, but also the ‘sulfur richness’ of seed storage proteins. Bonnot et al. (2023) discuss sulfur balancing mechanisms, the impact of abiotic stress factors on seed sulfur content and quality, and possible strategies for seed quality trait improvement. The authors address the importance of Met recycling through the S-methyl-Met cycle as a possible strategy to improve seed quality. They further mention the OAS cluster gene regulations in developing seeds of pea and wheat grains (Bonnot et al., 2020; Henriet et al., 2021), highlighting the impact of sulfur input and the role of the nuclear-localizing regulatory protein SULFUR DEFICIENCY INDUCED 1 (SDI1) in optimization of seed storage protein compositions, which extends the idea of sulfur homeostasis with this regulatory protein beyond its previously established function to inhibit the aliphatic glucosinolate biosynthesis in Arabidopsis (Aarabi et al., 2016, 2021).
Future directions
Sulfur metabolic enzymes are co-regulated to fulfill the demand for sulfur and stress mitigation. The concept of sulfur economy applies to balancing of primary and secondary metabolism, recycling of sulfur metabolites and cofactors, and alterations in seed storage protein compositions (Sugiyama et al., 2021; Bonnot et al., 2023; Ito and Ohkama-Ohtsu, 2023; Pedroletti et al., 2023). The assembly/disassembly dynamics of the Cys synthase complexes with the actions of S2– and OAS would impact sulfur metabolism and recycling pathways (Box 1). Beside the suggested direct impact of sulfur input and demands, movement of S2– and OAS across subcellular compartments may also be decisive factors for this sensing mechanism. Co-regulation of the OAS cluster genes further signifies the OAS actions in sulfur signaling. The potential modularity of SLIM1/EIL3 in transcriptional regulation reveals important questions about interactions between sulfur demand signaling and mechanisms that control basic cell functions.
Studying plant–microbe and plant–insect interactions may offer new research directions. Plants are capable of mounting resistance mechanisms against pathogens and herbivores. Sulfur-containing specialized metabolites may be part of these plant defense mechanisms, as they are pre-emptively synthesized, stored, and metabolized into active forms to fend off pathogens and herbivores (Jacoby et al., 2021). Pathogens, on the other hand, can deliver mechanisms to target sulfate transport and metabolism of host plant systems to compete for sulfur nutrients (Cernadas et al., 2014; Wang et al., 2022). Specialist insects co-evolved with host plants to evade plant defense mechanisms (Ratzka et al., 2002; Okamura et al., 2022). Our endeavors to explore these mechanistic frameworks continue to build new chapters in sulfur biology research in the face of grand challenges in crop production.
Acknowledgements
We thank the Journal of Experimental Botany for supporting our research community and providing us with an opportunity to present the updates in this Special Issue.
Contributor Information
Hideki Takahashi, Department of Biochemistry and Molecular Biology, Michigan State University, East Lansing, MI, USA.
Frédéric Marsolais, Genomics and Biotechnology, London Research and Development Centre, Agriculture and Agri-Food Canada, London, Ontario, Canada; Department of Biology, Western University, London, Ontario, Canada.
Ann Cuypers, Environmental Biology, Centre for Environmental Sciences, Hasselt University, Diepenbeek, Belgium.
Stanislav Kopriva, Institute for Plant Sciences, Cluster of Excellence on Plant Sciences, University of Cologne, Cologne, Germany.
References
- Aarabi F, Kusajima M, Tohge T, et al. 2016. Sulfur deficiency-induced repressor proteins optimize glucosinolate biosynthesis in plants. Science Advances 2, e1601087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aarabi F, Rakpenthai A, Barahimipour R, et al. 2021. Sulfur deficiency-induced genes affect seed protein accumulation and composition under sulfate deprivation. Plant Physiology 187, 2419–2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apodiakou A, Hoefgen R.. 2023. New insights into the regulation of plant metabolism by O-acetyl-serine: sulfate and beyond. Journal of Experimental Botany 74, 3361–3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aroca A, Benito JM, Gotor C, Romero LC.. 2017. Persulfidation proteome reveals the regulation of protein function by hydrogen sulfide in diverse biological processes in Arabidopsis. Journal of Experimental Botany 68, 4915–4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balk J, Schaedler TA.. 2014. Iron cofactor assembly in plants. Annual Review of Plant Biology 65, 125–153. [DOI] [PubMed] [Google Scholar]
- Bonnot T, Bachelet F, Boudet J, Le Signor C, Bancel E, Vernoud V, Ravel C, Gallardo K.. 2023. Sulfur in determining seed protein composition: present understanding of its interaction with abiotic stresses and future directions. Journal of Experimental Botany 74, 3276–3285. [DOI] [PubMed] [Google Scholar]
- Bonnot T, Martre P, Hatte V, et al. 2020. Omics data reveal putative regulators of einkorn grain protein composition under sulfur deficiency. Plant Physiology 183, 501–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caubrière D, Moseler A, Rouhier N, Couturier J.. 2023. Diversity and roles of cysteine desulfurases in photosynthetic organisms. Journal of Experimental Botany 74, 3345–3360. [DOI] [PubMed] [Google Scholar]
- Cernadas RA, Doyle EL, Niño-Liu DO, et al. 2014. Code-assisted discovery of TAL effector targets in bacterial leaf streak of rice reveals contrast with bacterial blight and a novel susceptibility gene. PLoS Pathogens 10, e1003972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan KX, Phua SY, Van Breusegem F.. 2019. Secondary sulfur metabolism in cellular signalling and oxidative stress responses. Journal of Experimental Botany 70, 4237–4250. [DOI] [PubMed] [Google Scholar]
- Clemens S, Ma JF.. 2016. Toxic heavy metal and metalloid accumulation in crop plants and foods. Annual Review of Plant Biology 67, 489–512. [DOI] [PubMed] [Google Scholar]
- Dong Y, Silbermann M, Speiser A, et al. 2017. Sulfur availability regulates plant growth via glucose–TOR signaling. Nature Communications 8, 1174. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Hell R, Wirtz M.. 2011. Molecular biology, biochemistry and cellular physiology of cysteine metabolism in Arabidopsis thaliana. The Arabidopsis Book 9, e0154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriet C, Balliau T, Aimé D, Le Signor C, Kreplak J, Zivy M, Gallardo K, Vernoud V.. 2021. Proteomics of developing pea seeds reveals a complex antioxidant network underlying the response to sulfur deficiency and water stress. Journal of Experimental Botany 72, 2611–2626. [DOI] [PubMed] [Google Scholar]
- Ito T, Kitaiwa T, Nishizono K, et al. 2022. Glutathione degradation activity of γ-glutamyl peptidase 1 manifests its dual roles in primary and secondary sulfur metabolism in Arabidopsis. The Plant Journal 111, 1626–1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Ohkama-Ohtsu N.. 2023. Degradation of glutathione and glutathione conjugates in plants. Journal of Experimental Botany 74, 3313–3327. [DOI] [PubMed] [Google Scholar]
- Iven V, Vanbuel I, Hendrix S, Cuypers A.. 2023. The glutathione-dependent alarm triggers signalling responses involved in plant acclimation to cadmium. Journal of Experimental Botany 74, 3300–3312. [DOI] [PubMed] [Google Scholar]
- Jacoby RP, Koprivova A, Kopriva S.. 2021. Pinpointing secondary metabolites that shape the composition and function of the plant microbiome. Journal of Experimental Botany 72, 57–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama-Nakashita A, Nakamura Y, Tohge T, Saito K, Takahashi H.. 2006. Arabidopsis SLIM1 is a central transcriptional regulator of plant sulfur response and metabolism. The Plant Cell 18, 3235–3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller SM, Wang S, Telman W, et al. 2017. The redox-sensitive module of cyclophilin 20-3, 2-cysteine peroxiredoxin and cysteine synthase integrates sulfur metabolism and oxylipin signaling in the high light acclimation response. The Plant Journal 91, 995–1014. [DOI] [PubMed] [Google Scholar]
- Okamura Y, Dort H, Reichelt M, Tunström K, Wheat CW, Vogel H.. 2022. Testing hypotheses of a coevolutionary key innovation reveals a complex suite of traits involved in defusing the mustard oil bomb. Proceedings of the National Academy of Sciences USA 119, e2208447119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedroletti L, Moseler A, Meyer AJ.. 2023. Assembly, transfer, and fate of mitochondrial iron–sulfur clusters. Journal of Experimental Botany 74, 3328–3344. [DOI] [PubMed] [Google Scholar]
- Rakpenthai A, Apodiakou A, Whitcomb SJ, Hoefgen R.. 2022. In silico analysis of cis-elements and identification of transcription factors putatively involved in the regulation of the OAS cluster genes SDI1 and SDI2. The Plant Journal 110, 1286–1304. [DOI] [PubMed] [Google Scholar]
- Ratzka A, Vogel H, Kliebenstein DJ, Mitchell-Olds T, Kroymann J.. 2002. Disarming the mustard oil bomb. Proceedings of the National Academy of Sciences USA 99, 11223–11228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama R, Li R, Kuwahara A, et al. 2021. Retrograde sulfur flow from glucosinolates to cysteine in Arabidopsis thaliana. Proceedings of the National Academy of Sciences USA 118, e2017890118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S-K, Chen J, Zhao F-J.. 2023. Regulatory mechanisms of sulfur metabolism affecting tolerance and accumulation of toxic trace metals and metalloids in plants. Journal of Experimental Botany 74, 3286–3299. [DOI] [PubMed] [Google Scholar]
- Sun S-K, Xu X, Tang Z, Tang Z, Huang X-Y, Wirtz M, Hell R, Zhao F-J.. 2021. A molecular switch in sulfur metabolism to reduce arsenic and enrich selenium in rice grain. Nature Communications 12, 1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, Kopriva S, Giordano M, Saito K, Hell R.. 2011. Sulfur assimilation in photosynthetic organisms: molecular functions and regulations of transporters and assimilatory enzymes. Annual Review of Plant Biology 62, 157–184. [DOI] [PubMed] [Google Scholar]
- Wang W, Liu J, Mishra B, Mukhtar MS, McDowell JM.. 2022. Sparking a sulfur war between plants and pathogens. Trends in Plant Science 27, 1253–1265. [DOI] [PubMed] [Google Scholar]
- Watanabe M, Mochida K, Kato T, Tabata S, Yoshimoto N, Noji M, Saito K.. 2008. Comparative genomics and reverse genetics analysis reveal indispensable functions of the serine acetyltransferase gene family in Arabidopsis. The Plant Cell 20, 2484–2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirtz M, Leemhuis W, Hell R.. 2023. Dynamic association of the plastid localized cysteine synthase complex is vital for efficient cysteine production, photosynthesis, and granal thylakoid formation in transgenic tobacco. Journal of Experimental Botany 74, 3379–3394. [DOI] [PubMed] [Google Scholar]