Abstract
Background
Classically, IgA in the gut prevents the invasion of microorganisms to systemic organs through the process of neutralization and immune exclusion. Interestingly, recent reports suggest that IgA might help in biofilm formation and promote bacterial growth inside the intestine.
Methods
In this study, we used flow cytometry, ELISA, and chemical models of colitis to test whether the quality and quantity of IgA can select for bacterial persistence in the gut.
Results
We found that members of Proteobacteria, such as γ-Proteobacteria and SFB, are preferentially coated by IgA in WT mice. In the partial absence of either T-dependent or -independent IgA responses, there are no significant differences in the frequency of bacteria coated with IgA in mice. However, Rag−/− mice that lack all antibodies had a severe reduction in Proteobacteria and were resistant to DSS-induced colitis, suggesting that secretory IgA might be essential for differential retention of these taxa in the mouse gut. Rag−/− littermates in the F2 generation generated from (B6 × Rag−/−) F1 mice acquired the underrepresented bacteria taxa such as γ-Proteobacteria through vertical transmission of flora. They died soon after weaning, possibly due to the acquired flora. Additionally, continued exposure of Rag−/− mice to B6 flora by cohousing mice led to the acquisition of γ-Proteobacteria and mortality.
Conclusions
Together, our results indicate that host survival in the complete absence of an IgA response necessitates the exclusion of specific bacterial taxa from the gut microbiome.
Keywords: IgA, gut microbiota, cohousing, bacterial diversity, colonization, vertical transmission
Introduction
The intestinal tracts of mice and humans, which are essentially sterile in utero, get colonized with microbes during birth and in the early neonatal period. Although over 50 bacterial and 10 archaeal divisions or phyla have been identified,1 microbial diversity in the intestine is restricted, and dominated by just 2 divisions of bacteria (Bacteroidetes and Firmicutes) and 1 member of Archaea (Methanobrevibacter smithii)2,3 indicating active selection mechanisms.
The microbiota of the gut contains autochthonous “permanent” residents as well as allochthonous “transient” residents. Most autochthonous residents cannot proliferate in environmental reservoirs outside the host and rely on vertical transmission from parent to offspring.4 Indeed, studies performed with mice have revealed that the microbiota of littermate mice is more similar than the microbiota of genetically identical mice that differ in maternal origin.5 In this context, it is recommended that pups from heterozygous breeding should be used for metagenomics studies to normalize for microbial contributions.6 An alternative approach for normalizing the microbiota is to cohouse adult mice from different strains for extensive periods, allowing mice to acquire heterologous flora as they feed on feces or ingest feces while self-grooming. This technique is simple and can also be used to study flora acquisition in early life by cohousing dams from different strains with their new-born litters.
In addition to microbes, the neonate mice also receive IgA from the breast milk when nursed by the dams. IgA has been shown to be a major player in shaping microbial colonization of the infant gut, and in influencing innate immune cell development.7,8 After weaning, the passive IgA is replaced by active IgA that is made following interactions of the intestinal immune system with bacterial components. The secreted IgA coats luminal bacteria and restricts their access to intestinal tissue and subsequent translocation into internal organs.9,10 The IgA secreted in the gut can originate from T cell-dependent or -independent mechanisms in response to the microflora. Most commensal bacteria in the gut are coated by T-independent IgA.11 Certain bacteria like SFB induce huge amounts of nonspecific IgA that can coat other bacteria.12 Further, IgA marks the disease driving members in the intestine. Colitogenic microbes were found to be highly coated with IgA in human.13 A previous report from our lab studying immunocompetent mouse strains showed that immune exclusion by IgA in the gut is a driving factor against susceptibility to induced colitis models.14
We report here that members of the phyla Proteobacteria are IgA coated in the wild-type mice and are underrepresented in CBA/N mice that make poor T-independent type 2 immune response15 and have significantly reduced IgA-coated bacteria in the gut. This was also true for Rag−/− mice that lack all antibodies suggesting that IgA might be required for Proteobacteria to take up residence in the gut. Since IgA is essential for biofilm formation,16,17 we explored the possibility that retention of certain bacteria in the gut might require repeated exposure to the organism. We found that Proteobacteria can be obtained by horizontal and vertical transfer of microbes from wild-type mice. However, the acquisition of such flora led to mortality. Together, our data indicate that ongoing exposure to environmental bacteria, especially by the feco-oral route can alter the microbial composition of the gut and may lead to disease if the new entrants are pathogens and/or if the individual is immunocompromised.
Materials and Methods
Mice
C57BL/6ByJ (B6), Rag1−/− (Rag−/−), TCRβ−/− obtained from the Jackson laboratories were maintained in the small animal facility of the National Institute of Immunology. Male or female mice 6–8 weeks of age were used for experiments, unless otherwise indicated. B6 × Rag−/− crosses to generate F1 and F2 were set up in-house. For cohousing experiments, B6 and Rag−/− females were cohoused.
Estimation of Fecal-Bacterial Loads
To determine fecal-bacterial loads, fecal pellets were collected, weighed, and stored at −20 °C. DNA was subsequently extracted using the Qiagen DNA Stool Mini kit as per kit protocol and quantified using Nanodrop (Thermo Scientific, NanoDrop Products). The yield was calculated as DNA per gm of fecal weight.
Estimation of Immunoglobulin (Ig) Amounts
Mice were bled, and serum collected by centrifuging at 16 000g for 10 min. Serum Ig was estimated on plates coated with 2 µg/mL of goat anti-mouse Ig (Southern). Captured Ig was detected with goat anti-mouse Ig-HRP (Southern).
DSS Colitis
Colitis was induced in mice by adding 2.5% DSS (M.W. 36 000–50 000 Da, MP Biomedicals) to autoclaved drinking water. The DSS was replaced every third day. Mortality, weight loss, and disease activity were recorded over time. For scoring of disease index, 0 = normal fecal pellet, 1 = few formed pellets to semisolid stool, 2 = semisolid to fluid stool with or without blood, 3 = bloody stool, 4 = bloody fluid, 5 = dead on arrival.
IgA Coating and Separation of IgA-Coated Fecal Bacteria
To estimate frequency of IgA-coated bacteria, fecal pellets were collected, homogenized in sterile PBS and centrifuged at 800g for 5 min to remove debris. The supernatant was collected and centrifuged at 9200g for 10 min to pellet fecal bacteria and the pelleted bacteria were stained with biotinylated goat anti-mouse IgA (Southern Biotech) for 30 min at 4 °C, washed twice with PBS 0.05 M EDTA and counterstained with streptavidin-APC Cy7 (BD) and nucleic acid stain Syto-13 (Invitrogen) in saline. IgA-coated and -uncoated bacteria were separated on streptavidin-MACS columns (Miltenyi Biotech) after staining with biotinylated goat anti-mouse IgA (Southern). Preparations were routinely >85% pure.
Staining of Fecal Samples With Anti-mouse IgA
Fecal samples will be stained with biotinylated anti-mouse IgA for 30 min, washed with Versene (Invitrogen), counterstained with streptavidin-APC-Cy7, washed, stained with Syto-13 and analyzed by flow cytometry.
Real-Time PCR
25 ng of fecal genomic DNA along with bacteria specific primers (500 nM) and Power SYBR Green PCR master mix (Applied Biosystems) were used for real-time PCR in an ABI Prism 7000 cycler. Amplification conditions were 95 °C for 5 min was followed by 40 cycles of 95 °C for 60 s, 50–67 °C for 45 s, and 72 °C for 60 s. Results were calculated using ΔΔCt method, to determine the relative change in expression. Eubacteria universal primer were used to normalize for difference if any in the input DNA.
For bacterial translocation into mesenteric lymph nodes, genomic DNA from the tissue was extracted with HiYieldGenomic DNA Mini Kit (Real Biotech), as recommended. Bacterial loads in tissue were determined by PCR with primers detecting all bacteria and expressed relative to GAPDH. Primers used in the study are listed in Table 1.
Table 1.
Sequence of primers used in the study.
| Bacteria | Primers |
|---|---|
| Eubacteria (all groups) | 5ʹ-ACTCCTACGGGAGGCAGCAG-3ʹ 5ʹ-ATTACCGCGGCTGCTGG-3ʹ |
| Actinobacteria | 5ʹ-CGCGGCCTATCAGCTTGTTG-3ʹ 5ʹ-ATTACCGCGGCTGCTGG-3ʹ |
| Bacteroidetes | 5ʹ-GGARCATGTGGTTTAATTCGATGAT-3ʹ 5ʹ-AGCTGACGACAACCATGCAG-3ʹ |
| Firmicutes | 5ʹ-GGAGYATGTGGTTTAATTCGAAGCA-3ʹ 5ʹ-AGCTGACGACAACCATGCAC-3ʹ |
| Bifidobacterium | 5ʹ-TCGCGTC(C/T)GGTGTGAAAG-3ʹ 5ʹ-CCACATCCAGC(A/G)TCCAC-3ʹ |
| Lactobacillus | 5ʹ-AGCAGTAGGGAATCTTCCA-3ʹ 5ʹ-CACCGCTACACATGGAG-3ʹ |
| Bacillus | 5ʹ-GCGGCGTGCCTAATACATGC-3ʹ 5ʹ-CTTCATCACTCACGCGGCGT-3ʹ |
| Bacteroides–Prevotella–Porphyromonas (BPP) | 5ʹ-GGTGTCGGCTTAAGTGCCAT-3ʹ 5ʹ-CGGA(C/T)GTAAGGGCCGTGC-3ʹ |
| SFB | 5ʹ-GACGCTGAGGCATGAGAGCAT-3ʹ 5ʹ-GACGGCACGGATTGTTATTCA-3ʹ |
| Helicobacter | 5ʹ-CTTAACCATAGAACTGCATTTGAAACTAC-3ʹ 5ʹ-GGTCGCCTTCGCAATGAGTA-3ʹ |
| Enterobacteraiceace | 5ʹ-GTGCCAGCMGCCGCGGTAA-3ʹ 5ʹ-GCCTCAAGGGCACAACCTCCAAG-3ʹ |
| Alphaproteobacteria | 5ʹ-CIAGTGTAGAGGTGAAATT-3ʹ 5ʹ-CCCCGTCAATTCCTTTGAGTT-3ʹ |
| Betaproteobacteria | 5ʹ-TCACTGCTACACGYG-3ʹ 5ʹ-ACTCCTACGGGAGGCAGCAG-3ʹ |
| Gammaproteobacteria | 5ʹ-TCGTCAGCTCGTGTYGTGA-3ʹ 5ʹ-CGTAAGGGCCATGATG-3ʹ |
| Faecalibacterium prausnitzii | 5ʹ-AGATGGCCTCGCGTCCGA-3ʹ 5ʹ-CCGAAGACCTTCTTCCTCC-3ʹ |
| Peptostreptococcus productus | 5ʹ-AACTCCGGTGGTATCAGATG-3ʹ 5ʹ-GGGGCTTCTGAGTCAGGTA-3ʹ |
| Clostridium clostridiiforme | 5ʹ-CCGCATGGCAGTGTGTGAAA-3ʹ 5ʹ-CTGCTGATAGAGCTTTACATA-3ʹ |
Statistical Analyses
Data were analyzed by Student’s t-test. Error bars indicate standard error of mean.
Results
Bacterial Diversity and Variable Coating of Fecal Bacteria With IgA in B6 Mice
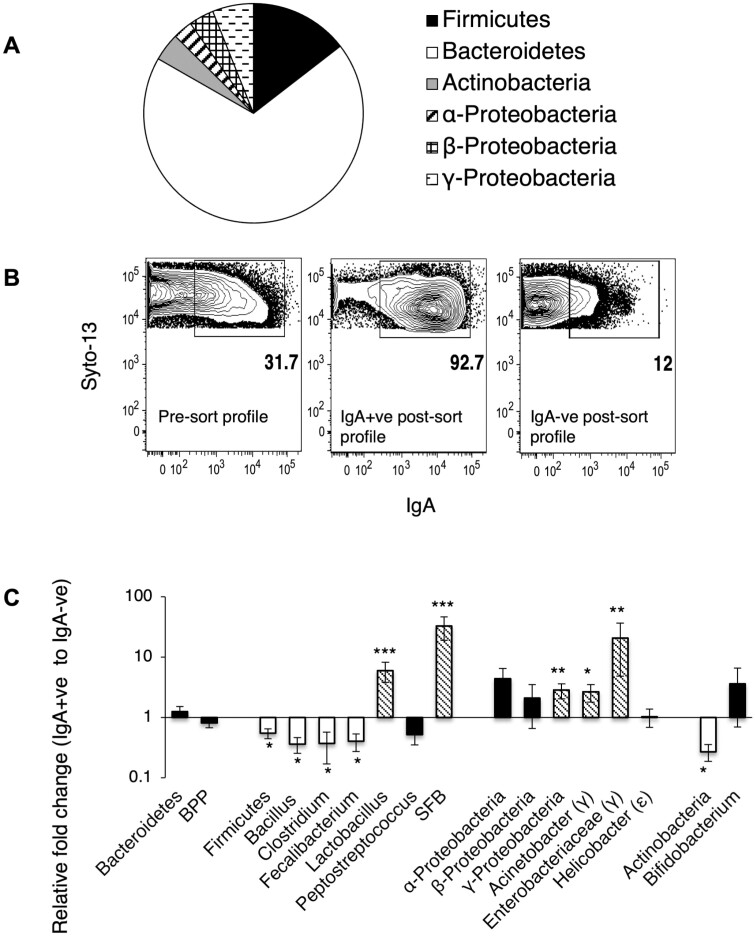
Mouse fecal bacteria belong predominantly to 4 phyla-Bacteroidetes, Firmicutes, Actinobacteria, and Proteobacteria. In our B6 colony we find phylum Bacteroidetes constitute the bulk of the bacterial diversity followed by phyla Proteobacteria, Firmicutes, and Actinobacteria (Figure 1A). Previous reports have indicated that 10%–50% of fecal bacteria are coated with IgA in B6 mice in various animal facilities.11,13,18 We found that in our colony of B6 mice, around 30% of fecal bacteria are coated with IgA (Figure 1B). To determine whether specific members of the bacterial community are targeted with IgA in B6 mice, we separated IgA-bound and -unbound bacteria by magnetic cell sorting (Figure 1B) and amplified bacterial 16S rDNA with phylum-, family-, or taxon-specific primers to determine whether specific members were enriched in the IgA-bound and -unbound fractions. We used equal input DNA to measure the abundance of specific groups relative to total bacterial DNA (amplified with Eubacteria) in the IgA-bound and -unbound fractions. We found that there was no clear separation of taxa into the 2 fractions. However, that Phylum Actinobacteria and Phylum Firmicutes (and members Bacillus, Clostridium clostridioforme, and Faecalibacterium prausnitzii) were enriched in the IgA-unbound fraction, and subphylum γ-Proteobacteria (and members Acinetobacter and Enterobacteriaceae) and members of phylum Firmicutes (Lactobacillus and SFB) were enriched in the IgA-coated fraction (Figure 1C). Many bacteria were equally distributed in both IgA-coated and -uncoated fractions. These include phylum Bacteroidetes and its members Bacteroides–Prevotella–Porphyromonas (BPP), subphyla α-Proteobacteria, and β-Proteobacteria, and other taxa like Bifidobacterium (phylum Actinobacteria), Helicobacter (ε-Proteobacteria), and Peptostreptococcus (phylum Firmicutes). It is intriguing that a fraction of these taxa remain uncoated despite the presence of secreted IgA that can coat it. A possible explanation is that they represent new entrants in the gut, and that mice are being continuously colonized.
Figure 1.
Bacterial diversity and variable coating of fecal bacteria with IgA in B6 mice. (A) Distribution of various bacterial phyla in fecal pellets from B6 mice. Pie chart is plotted by using 1/ΔCt × 100 normalized to Eubacteria. Data represent distribution in 12 B6 over 3 independent experiments. (B) Representative MACS sort profile of IgA-bound and -unbound fraction from B6 fecal pellets. (C) Relative abundance of bacteria in IgA+ve (ie, fraction bound by IgA) versus bacteria in IgA-ve fraction (ie, IgA-unbound fraction) in B6 fecal pellet. Results represent pooled data from across 6 independent experiments (*P < .05, **P < .01, ***P < .001).
Effect of Partial Absence of IgA on Bacterial Diversity in Fecal Pellets
IgA in the gut is generated following stimulation of B cells in Peyer’s patches or intestine through T-dependent or -independent mechanisms. To determine whether IgA coating was dominated by T-dependent or -independent IgA, we assessed IgA coating of bacteria in fecal pellets of T cell-deficient (TCRβ−/−) mice that cannot mount T-dependent antibody responses and of xid (CBA/N) mice that make poor T-independent antibody responses. We found that IgA coating was unaffected in both these strains (Figure 2A).
Figure 2.
Effect on bacterial diversity in CBA/N and TCRβ−/− mice. (A) Frequency of IgA-coated bacteria in fecal pellets of B6, TCRβ−/−, and CBA/N mice. Data from n = 6, over 2 independent experiments. (B, C) Relative abundance of various bacterial phyla, subphyla, and species in fecal pellets of CBA/N and TCRβ−/− mice with respect to B6 mice. Data pooled from n = 6 mice over 2 independent experiments (*P < .05, **P < .01, ***P < .001).
To determine whether microbial diversity was affected by the presence or absence of T-dependent and -independent IgA, we assessed bacterial composition by amplification of 16S rDNA with primers that either amplify all bacteria or are phylum/genus/family/species-specific in fecal pellets of these mice. We found that the fecal-bacterial diversity in the 2 strains was largely similar to that of B6 mice with few differences such as lower representation of Actinobacteria and higher representation of β-Proteobacteria in the CBA/N strain and higher representation of both Actinobacteria and γ-Proteobacteria in the TCRβ−/− strain (Figure 2B and C). Since both TCRβ−/− and CBA/N mice still had secretory IgA in the gut, we hypothesized that a more drastic change in colonization of the microbe might be observed in mice completely deficient in gut IgA.
IgA-Coating Frequencies and Bacterial Diversity in Mice With That Lack Antibody Responses
In order to understand how the colonization and retention of microbiota is affected in the complete absence of secretory IgA in the gut, we assessed the microbial composition in fecal pellets of B6 and Rag−/− mice. As expected, the fecal bacteria in the Rag−/− mice were not coated with IgA (Figure 3A). Like B6 mice, the microbiota in Rag−/− mice were dominated by Firmicutes and Bacteroidetes (Figure 3B). We found no difference in the overall representation of 3 major phyla—Bacteroidetes, Firmicutes, and Actinobacteria. However, representation of Faecalibacterium was higher and that of SFB was lower (both members of phylum Firmicutes) in Rag−/− mice (Figure 3C). A severe reduction was seen in the proportions of α-Proteobacteria and γ-Proteobacteria in Rag−/− mice, and proportions of Enterobacteriaceae and Acinetobacter (other members of this phylum) were also reduced (Figure 3C). The fecal-bacterial load, represented by the amount of DNA obtained per gram of fecal weight, was lower in Rag−/− mice although the difference was not statistically significant (Figure 3D). Since Rag−/− mice cannot produce IgA, the primary antibody required to limit bacterial translocation to systemic organs, we reasoned that these mice might have higher bacterial loads in the peripheral organs like MLN and spleen. Total bacterial loads in MLN and spleen, as measured with primers for Eubacteria and normalized to the housekeeping gene GAPDH, was not different between B6 mice and Rag−/− mice (Figure 3E). These data indicate that in Rag−/− mice components of the innate immunity like dendritic cells, macrophages, and anti-microbial peptides at the epithelial barrier are enough to restrain bacteria to the lumen.
Figure 3.
Effect on bacterial diversity in the absence of IgA. (A) Flow cytometry profile of IgA-coated fecal bacteria in B6 and Rag−/− mice. (B) Distribution of microbiota in Rag−/− mice. Pie chart is plotted by using 1/ΔCt × 100 normalized to Eubacteria. n = 9. (C) Relative abundance in Rag−/− mice with respect to B6 mice of various bacterial phyla, subphyla, and species. Data pooled from 12 to 13 mice over 3 experiments. P values indicated wherever significant. (D) Fecal-bacterial loads in untreated B6 and Rag−/− mice. Data pooled from 8 mice over 2 experiments. (E) Bacterial loads in mesenteric lymph nodes and spleen expressed as a ratio of Eubacteria Ct to GAPDH Ct. Data pooled from 3 independent experiments (*P < .05).
Effect of Vertical Transmission of B6 Flora to Rag−/− Mice
Neonates are stably colonized soon after birth with maternally transmitted microbes and thus, the differences in microbial composition between Rag−/− and B6 adults may reflect differential familial transmission of specific taxa. Hence, we determined whether vertical transmission of bacteria from B6 dams to Rag−/− pups would lead to stable equilibration of flora in the 2 strains despite the absence of IgA. To do this, we crossed B6 and Rag−/− mice, and the pups from the F1 generation were then intercrossed and Rag−/− and B6 littermates in the F2 generation were identified by the presence or absence of serum immunoglobulins (Figure 4A). When fecal samples were analyzed, most taxa were present in similar proportions indicating equilibration of flora between both strains (Figure 4B). Representation of γ-Proteobacteria was found to be higher in Rag−/− littermates (Figure 4B). These data indicate that Rag−/− pups born to and nursed by F1 dams and exposed to B6 flora early in life, can acquire and retain the otherwise underrepresented bacterial taxa as adults. The Rag−/− littermates that acquired the “pro-inflammatory” members started dying soon after weaning (data not shown) indicating that the acquired taxa have pathogenic potential, and that an adaptive immune system is needed to contain them.
Figure 4.
Vertical transmission leads to equilibration of flora between littermates. (A) Serum ELISA for the identification of Rag−/− and B6 pups obtained from F1 × F1 crosses. (B) Fold expression of various bacterial taxa in the fecal sample of Rag−/− mice as compared with Rag+/+ mice. Data pooled from 8 to 12 mice over 2 experiments (*P < .05, **P < .01).
Horizontal Transmission of B6 Flora to Rag−/− Mice by Cohousing
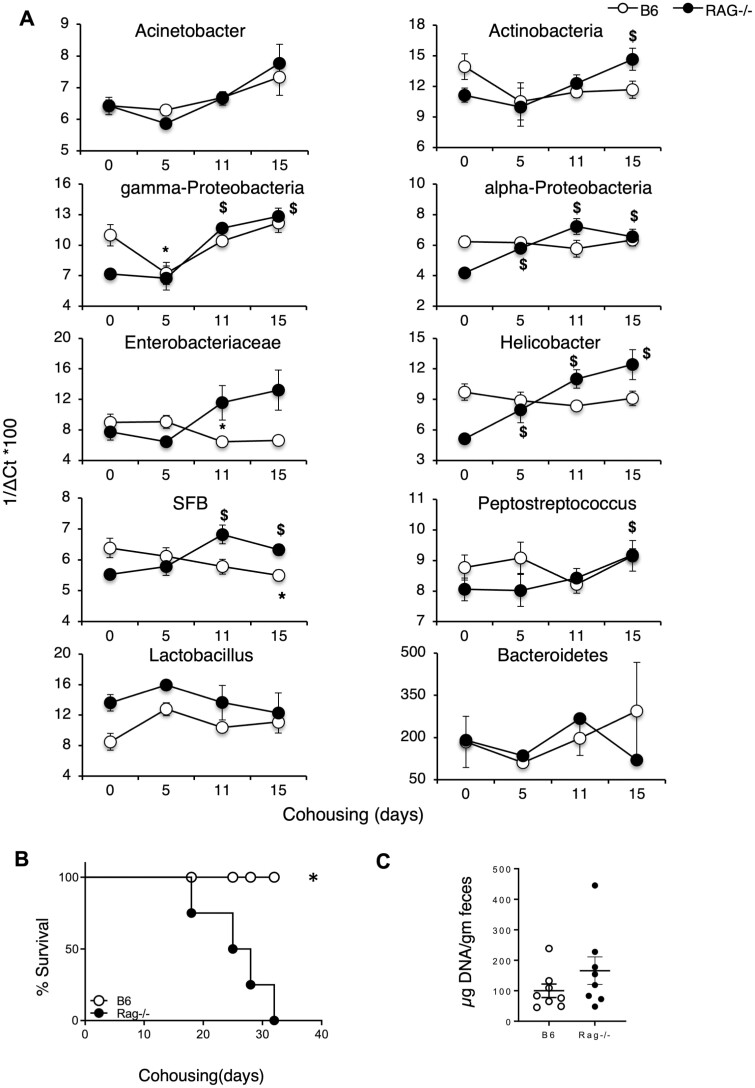
It has been reported that the intestinal microbiome of adult immunocompetent mice is quite resilient and can resist colonization by non-autochthonous bacteria in cohousing experiments.19 We tested whether Rag−/− mice could be colonized by B6 flora in such horizontal transfer experiments. Adult female Rag−/− mice were cohoused with adult female B6 mice to provide continuous exposure of B6 flora to Rag−/− mice. Fecal samples were collected at various time points post-cohousing and bacterial composition was compared with the flora at the start of the experiment (Figure 5A). We found that cohoused Rag−/− mice started dying by day 15 (Figure 5B) and this correlated with the acquisition of γ-Proteobacteria and SFB by day 11, and high abundance of Actinobacteria, α-Proteobacteria, Helicobacter, and Enterobacteriaceae (Figure 5A) in the Rag−/− mice. Fecal microbial loads were also slightly higher in Rag−/− mice at this time as compared with the loads in cohoused B6 controls (Figure 5C) indicating that the acquired flora can expand in the absence of IgA.
Figure 5.
Cohabitation leads to changes in flora distribution. (A) Distribution of various bacterial taxa in fecal samples collected from Rag−/− mice cohoused with B6 mice over time. Panel includes bacterial phyla and subphyla assessed. Data represent distribution in 8 mice pooled over 2 experiments. Results have been calculated as 1/ΔCt ×100 after normalizing for Eubacteria. P values indicated as *P < .05 for B6 mice compared with its day0; $P < .05 for Rag−/− mice compared with its day 0. (B) Survival post-cohousing of Rag−/− mice is plotted over time. (C) Bacterial loads in feces of B6 versus Rag−/− mice. P values indicated as *P < .05. Data pooled from 4 independent experiments.
Physiological Relevance of the Presence of Pro-inflammatory Bacteria in Mice
The taxa that are underrepresented taxa in Rag−/− mice, namely α- and γ-Proteobacteria, have been deemed as “pro-inflammatory” in experimental colitis models.18,20 Indeed, we found that when colitis was induced with 2.5% DSS in drinking water, Rag−/− mice survived and showed minimal weight loss and disease activity as compared with B6 mice (Figure 6A–C). These findings support an earlier report showing that the severity of DSS-induced colitis was markedly reduced in Rag−/− mice21 and resistance to DSS colitis. These data further indicate that colitogenic members have been selected against in mice lacking IgA and raise the possibility that the retention of such colitogenic members may be facilitated by IgA coating.
Figure 6.
Physiological outcome of reduced bacterial diversity in Rag−/− mice. (A) Survival, (B) weight change, and (C) disease index over time in B6 and Rag−/− mice given 2.5% DSS in drinking water. Data are mean of 6 mice per group. P values indicated wherever significant (*P < .05, $P < .01, #P < .001).
Discussion
A layer of simple epithelium that aids absorption of nutrients lines the intestine and forms a physical barrier separating luminal microbes from the underlying lamina propria and systemic organs. Immune protection against infection by intestinal pathogens, containment of bacteria to the lumen, and maintenance of gut homeostasis in individuals is mediated by a variety of immune cells including CD8 cells in the epithelial layer (IELs), and neutrophils, basophils, macrophages, and dendritic cells and IgA-secreting plasma cells in the subepithelial lamina propria.22–25 In mice harboring conventional flora, the gut-associated lymphoid tissue, unlike other systemic lymphoid organs, is in a state of chronic stimulation with continuous sampling of luminal antigens. Occasional intrusion by luminal bacteria is taken care of by innate immune cells and effector T cells. This homeostasis can break down due to a combination of factors including barrier dysfunction, pathogen entry, dysbiosis, and intestinal injury, resulting in greater bacterial entry and recruitment of cells to the site of the breach. Alterations in the composition of the gut microbiota, known as dysbiosis, have been associated with several chronic metabolic and intestinal diseases.
IgA is the predominant immunoglobulin isotype found in the gut and is produced by T-dependent and -independent immune responses. Earlier studies have shown that the steady-state intestinal flora is predominantly coated by IgA generated from T-independent immune responses.11 To determine how microbial composition in the gut is affected in the absence of T-independent (TI) IgA, we used CBA/N mice with a mutation in Btk gene. These mice have a defect in B cells involved in secreting TI IgA.26 Furthermore, humans with mutation in Btk have been shown to be highly susceptible to IBD which was corroborated with mouse models of colitis using Btk-deficient mice.27 We did not find any significant differences between wild-type and CBA/N mice in IgA-coated frequency of fecal bacteria (Figure 2A) or any of the major Phylum we assessed (Figure 2B). To determine how microbial composition was altered in the absence of T-dependent IgA, we studied the TCRβ−/− mice (deficient in T-dependent immune responses). Previous studies have shown TCRβ−/− mice to be more susceptible to induced-colitis models, however, the susceptibility is largely dependent on the nature of resident microflora in the gut.28–30 We found higher proportions of γ-Proteobacteria and Actinobacteria in TCRβ−/− mice when compared with WT mice (Figure 2C). The relative abundance of other members of the gut microbiota was largely unaltered.
We hypothesized that to see significant differences in microbiota we need to study microbiota retention and colonization in the complete absence of immunoglobulins. We examined the microbial composition in Rag−/− mice versus WT mice. Rag−/− mice lacks adaptive immune responses and hence would lack any secretory antibodies in the gut. We observed a statistically significant reduction in the abundance of Proteobacteria in feces obtained from Rag−/− mice. Specifically, we observed a reduction in α-Proteobacteria and γ-Proteobacteria (Figure 3A–C). Our data agree with previous reports that show reduced bacterial diversity in Rag−/− mice.18
Next, we wanted to know whether the potentially colitogenic flora (members of Proteobacteria) could be transferred to the Rag−/− mice by crossing B6 × Rag−/− mice. The pups from F1 progeny of B6 × Rag−/− mice were further bred and the resultant F2 progeny of the B6 and Rag−/−cross were compared for microbial diversity. The taxa that were originally less represented in the Rag−/− mice, were found to be equally represented in Rag−/− and B6 littermates of F2 progeny. Thus, Rag−/− mice can acquire the missing bacterial members upon exposure to B6 littermates (Figure 4A and B). However, Rag−/− pups from the F2 progeny started dying shortly after weaning indicating the need for an adaptive immune system especially IgA to allow for the persistence of certain bacteria. While we were unable to record the exact days when the Rag−/− pups died, their death post-weaning suggests that once the maternally derived immunoglobulins are lost, the potentially pathogenic microbes obtained from the dams can be fatal to the immune-deficient host.
Subsequently, we wanted to test whether we can achieve horizontal transmission of microbiota from B6 to Rag−/− mice by cohousing the 2 strains and whether that also led to the mortality in the Rag−/− mice. This method takes advantage of the coprophagic nature of mice to enable transfer of microbes across co-caged mice. Earlier reports have shown that potential colitogenic flora can be transferred between 2 mouse strains by simply cohousing them.31,32 In fact, a study has previously demonstrated equilibration of microbial communities between B6 and Rag−/− mice post-cohousing for 3 weeks.11 However, it is also important to note that previous data from our lab,14 and other groups,33 have shown that microbiota cannot be transferred by cohousing immune-competent mouse strains with conventional flora. We followed the transfer kinetics of individual microbes after cohousing B6 and Rag−/− mice. We report that Rag−/− mice cohoused with B6 mice gained α-Proteobacteria, gamma-Proteobacteria by day 15 (Figure 5A). Also, total microbial loads were slightly higher in Rag−/− mice on day 15 post-cohousing compared with B6 mice (Figure 5C). Cohoused Rag−/− mice also started dying at this time point supporting our hypothesis that acquiring the flora from B6 mice is causing morality due to the absence of an adaptive immune response. Other studies where specific-pathogen-free mice were cohoused with pet store mice, a mortality rate of approximately 20% was reported, suggesting that introduction of unfamiliar microbiota can lead to morality in mice.34,35 Interestingly, adult Rag−/− mice died within 2 weeks of cohousing exposure but the Rag−/− pups obtained from B6 × Rag−/− breeding only die after weaning (~4–5 weeks), showing that maternal milk IgA can provide protection, and adult Rag−/− mice cohoused with B6 mice die only when they are exposed via continued cohousing coprophagy from B6 littermates post-weaning. Interestingly, it has been shown that exposure to pathobionts in Rag−/− mice versus WT mice causes an expansion of the pathobionts which is in sync with our findings. This study, however, has no data for mortality, a gap filled by our manuscript.36
We also observed that certain bacterial members which are in lower abundance in non-cohoused Rag−/− mice have been associated with causing enhanced intestinal inflammation in other mouse models. For example, members of the phylum Proteobacteria such as Enterobacteriaceae have been associated with enhanced intestinal inflammation.20 Enterobacteriaceae have been reported to reside in the healthy gut at low levels and are amongst the most commonly overgrown symbionts in many conditions involving inflammation, such as IBD, colorectal cancer, and antibiotic treatment.37 Previous study from our lab also found association of members of Proteobacteria with enhanced susceptibility to DSS-induced colitis.14 However, it is important to note that in the previous report, the potential colitogenic bacteria were found to be enriched in IgA-uncoated fraction. The differences seen in this study might be attributed to chronologically separated lines of wild-type mice and the fact that microbiota can be readily influenced by changes in environment and/or diet.38 The possible factors which lead to such differences were outside the scope of the current study. Both the studies corroborate the role of IgA in shaping the murine gut microbiota and providing protection in induced-colitis models. We hypothesized that decreased abundance of potentially colitogenic bacteria in the Rag−/− mice would mean lower susceptibility to colitis inducing agents. Indeed, we found Rag−/− mice to be less susceptible to DSS-induced colitis compared with B6 mice indicating that non-IgA-coated taxa still found in Rag−/− mice do not cause problems even after DSS because that is not their colonization strategy (Figure 6A–C).
Collectively, our data would suggest that mice are being colonized with microbiota continuously and that in the absence of enough immunoglobulins acquiring “inflammatory” bacteria is harmful for an immunocompromised host.
Acknowledgments
We would like to thank Dr. Nagarajan for breeding and maintenance of mouse strains.
Contributor Information
Suman Gupta, Mucosa Immunology Laboratory, National Institute of Immunology, New Delhi, India; Department of Medicine, Cedars-Sinai Medical Center, Los Angeles, California, USA.
Sneh Lata Gupta, Mucosa Immunology Laboratory, National Institute of Immunology, New Delhi, India; Department of Pediatrics, Division of Infectious Disease, School of Medicine, Emory University, Atlanta, Georgia, USA.
Aashima Singh, Mucosa Immunology Laboratory, National Institute of Immunology, New Delhi, India.
Neelam Oswal, Mucosa Immunology Laboratory, National Institute of Immunology, New Delhi, India; Center for Discovery and Innovation, Hackensack University Medical Center, Nutley, New Jersey, USA.
Vineeta Bal, Mucosa Immunology Laboratory, National Institute of Immunology, New Delhi, India; Indian Institute of Science Education and Research, Pune, India.
Satyajit Rath, Mucosa Immunology Laboratory, National Institute of Immunology, New Delhi, India; Translational Health Science and Technology Institute, Faridabad, India.
Anna George, Mucosa Immunology Laboratory, National Institute of Immunology, New Delhi, India.
Srijani Basu, Mucosa Immunology Laboratory, National Institute of Immunology, New Delhi, India; Department of Medicine, Weill Cornell Medicine, New York, New York, USA.
Funding
This work was supported in part by grants from the Department of Biotechnology, Government of India (BT/PR-14592/BRB/10/858/2010 to S.R. and BT/PR13699/BRB/10/1514/2016 to A.G.) and the Science and Engineering Research Board, Government of India (to A.G. and V.B.). The National Institute of Immunology is supported by the Department of Biotechnology, Government of India.
Conflicts of Interest
None declared.
Ethical Approval
Experimentation on animals was carried out by protocols approved by the Institutional Animal Ethics Committee of the National Institute of Immunology.
Data Availability
No new data were created or analyzed.
References
- 1. DeLong EF, Pace NR.. Environmental diversity of bacteria and archaea. Syst Biol. 2001;50(4):470–478. [PubMed] [Google Scholar]
- 2. Eckburg PB, Bik EM, Bernstein CN, et al. Diversity of the human intestinal microbial flora. Science. 2005;308(5728):1635–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bäckhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI.. Host-bacterial mutualism in the human intestine. Science. 2005;307(5717):1915–1920. [DOI] [PubMed] [Google Scholar]
- 4. Ley RE, Peterson DA, Gordon JI.. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006;124(4):837–848. [DOI] [PubMed] [Google Scholar]
- 5. Ley RE, Bäckhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI.. Obesity alters gut microbial ecology. Proc Natl Acad Sci U S A. 2005;102(31):11070–11075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Stappenbeck TS, Virgin HW.. Accounting for reciprocal host-microbiome interactions in experimental science. Nature. 2016;534(7606):191–199. [DOI] [PubMed] [Google Scholar]
- 7. Rogier EW, Frantz AL, Bruno MEC, et al. Secretory antibodies in breast milk promote long-term intestinal homeostasis by regulating the gut microbiota and host gene expression. Proc Natl Acad Sci U S A. 2014;111(8):3074–3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gomez de Agüero M, Ganal-Vonarburg SC, Fuhrer T, et al. The maternal microbiota drives early postnatal innate immune development. Science. 2016;351(6279):1296–1302. [DOI] [PubMed] [Google Scholar]
- 9. Cebra JJ. Influences of microbiota on intestinal immune system development. Am J Clin Nutr. 1999;69(5):1046S-1051S. [DOI] [PubMed] [Google Scholar]
- 10. Logan AC, Chow KP, George A, Weinstein PD, Cebra JJ.. Use of Peyer’s patch and lymph node fragment cultures to compare local immune responses to Morganella morganii. Infect Immun. 1991;59(3):1024–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bunker JJ, Flynn TM, Koval JC, et al. Innate and adaptive humoral responses coat distinct commensal bacteria with immunoglobulin A. Immunity. 2015;43(3):541–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Talham GL, Jiang HQ, Bos NA, Cebra JJ.. Segmented filamentous bacteria are potent stimuli of a physiologically normal state of the murine gut mucosal immune system. Infect Immun. 1999;67(4):1992–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Palm NW, de Zoete MR, Cullen TW, et al. Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell. 2014;158(5):1000–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gupta S, Basu S, Bal V, Rath S, George A.. Gut IgA abundance in adult life is a major determinant of resistance to dextran sodium sulfate-colitis and can compensate for the effects of inadequate maternal IgA received by neonates. Immunology. 2019;158(1):19–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Selinka HC, Bösing-Schneider R.. xid mice fail to express an anti-dextran immune response but carry alpha(1–3)dextran-specific lymphocytes in their potential repertoire. Eur J Immunol. 1988;18(11):1727–1732. [DOI] [PubMed] [Google Scholar]
- 16. Bollinger RR, Everett ML, Palestrant D, Love SD, Lin SS, Parker W.. Human secretory immunoglobulin A may contribute to biofilm formation in the gut. Immunology. 2003;109(4):580–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bollinger RR, Everett ML, Wahl SD, Lee Y-H, Orndorff PE, Parker W.. Secretory IgA and mucin-mediated biofilm formation by environmental strains of Escherichia coli: role of type 1 pili. Mol Immunol. 2006;43(4):378–387. [DOI] [PubMed] [Google Scholar]
- 18. Kawamoto S, Maruya M, Kato LM, et al. Foxp3(+) T cells regulate immunoglobulin a selection and facilitate diversification of bacterial species responsible for immune homeostasis. Immunity. 2014;41(1):152–165. [DOI] [PubMed] [Google Scholar]
- 19. Huggins MA, Jameson SC, Hamilton SE.. Embracing microbial exposure in mouse research. J Leukoc Biol. 2019;105(1):73–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lee KW, Kim M, Lee CH.. Treatment of dextran sulfate sodium-induced colitis with mucosa-associated lymphoid tissue lymphoma translocation 1 inhibitor MI-2 is associated with restoration of gut immune function and the microbiota. Infect Immun. 2018;86(12): 10.1128/iai.00091-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kim TW, Seo JN, Suh YH, et al. Involvement of lymphocytes in dextran sulfate sodium-induced experimental colitis. World J Gastroenterol. 2006;12(2):302–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Olivares-Villagómez D, Van Kaer L.. Intestinal intraepithelial lymphocytes: sentinels of the mucosal barrier. Trends Immunol. 2018;39(4):264–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sheridan BS, Lefrançois L.. Intraepithelial lymphocytes: to serve and protect. Curr Gastroenterol Rep. 2010;12(6):513–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Peterson LW, Artis D.. Intestinal epithelial cells: regulators of barrier function and immune homeostasis. Nat Rev Immunol. 2014;14(3):141–153. [DOI] [PubMed] [Google Scholar]
- 25. Cerutti A, Rescigno M.. The biology of intestinal immunoglobulin A responses. Immunity. 2008;28(6):740–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hasbold J, Klaus GG.. B cells from CBA/N mice do not proliferate following ligation of CD40. Eur J Immunol. 1994;24(1):152–157. [DOI] [PubMed] [Google Scholar]
- 27. Guan D, Wang Z, Huo J, Xu S, Lam K-P.. Bruton’s tyrosine kinase regulates gut immune homeostasis through attenuating Th1 response. Cell Death Dis. 2021;12(5):431–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tsuchiya T, Fukuda S, Hamada H, et al. Role of gamma delta T cells in the inflammatory response of experimental colitis mice. J Immunol. 2003;171(10):5507–5513. [DOI] [PubMed] [Google Scholar]
- 29. Mizoguchi E, Sadanaga T, Okada T.. Biological analyses-derived translational findings in the T cell receptor alpha chain knockout mouse as an experimental model for ulcerative colitis. Int J Transl Med. 2021;1(3):187–204. [Google Scholar]
- 30. Dianda L, Hanby AM, Wright NA, Sebesteny A, Hayday AC, Owen MJ.. T cell receptor-alpha beta-deficient mice fail to develop colitis in the absence of a microbial environment. Am J Pathol. 1997;150(1):91–97. [PMC free article] [PubMed] [Google Scholar]
- 31. Elinav E, Strowig T, Kau AL, et al. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell. 2011;145(5):745–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lipinski JH, Zhou X, Gurczynski SJ, et al. Cage environment regulates gut microbiota independent of Toll-like receptors. Infect Immun. 2021;89(9):e0018721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fransen F, Zagato E, Mazzini E, et al. BALB/c and C57BL/6 mice differ in polyreactive IgA abundance, which impacts the generation of antigen-specific IgA and microbiota diversity. Immunity. 2015;43(3):527–540. [DOI] [PubMed] [Google Scholar]
- 34. Beura LK, Hamilton SE, Bi K, et al. Normalizing the environment recapitulates adult human immune traits in laboratory mice. Nature. 2016;532(7600):512–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hamilton SE, Badovinac VP, Beura LK, et al. New insights into the immune system using dirty mice. J Immunol. 2020;205(1):3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gálvez EJC, Iljazovic A, Gronow A, Flavell R, Strowig T.. Shaping of intestinal microbiota in Nlrp6- and Rag2-deficient mice depends on community structure. Cell Rep. 2017;21(13):3914–3926. [DOI] [PubMed] [Google Scholar]
- 37. Zeng MY, Inohara N, Nuñez G.. Mechanisms of inflammation-driven bacterial dysbiosis in the gut. Mucosal Immunol. 2017;10(1):18–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ericsson AC, Franklin CL.. The gut microbiome of laboratory mice: considerations and best practices for translational research. Mamm Genome. 2021;32(4):239–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No new data were created or analyzed.