Abstract
Background
Whether non-alcoholic fatty liver disease (NAFLD) causes cardiovascular disease (CVD) and type 2 diabetes (T2D) is unclear and possible differences between ethnicities have not been thoroughly explored. We used Mendelian randomization (MR) to assess the role of NAFLD in CVD and T2D risk in Europeans and East Asians.
Methods
We conducted a MR study using genetic predictors of alanine aminotransferase (ALT), liability to NAFLD, aspartate transaminase (AST), liver magnetic resonance imaging corrected T1 and proton density fat fraction and combined them with genome-wide association studies (GWAS) summary statistics of CVD, T2D and glycaemic traits (sample size ranging from 14 400 to 977 320). Inverse-variance weighted analysis was used to assess the effect of NAFLD in these outcomes, with sensitivity analyses and replication in FinnGen. We conducted analyses in East Asians using ethnicity-specific genetic predictors of ALT and AST, and the respective outcome GWAS summary statistics.
Results
In Europeans, higher ALT was associated with higher T2D risk (odds ratio: 1.77 per standard deviation, 95% CI 1.5 to 2.08), with similar results for other exposures, across sensitivity analyses and in FinnGen. Although NAFLD proxies were related to higher coronary artery disease (CAD) and stroke risk, sensitivity analyses suggested possible bias by horizontal pleiotropy. In East Asians, higher ALT was possibly associated with higher T2D risk, and ALT and AST were inversely associated with CAD.
Conclusions
NAFLD likely increases the risk of T2D in Europeans and East Asians. Potential differential effects on CAD between Europeans and East Asians require further investigation.
Keywords: Atrial fibrillation, coronary artery disease, heart failure, NAFLD, Mendelian randomization, stroke, type 2 diabetes
Key Messages.
Non-alcoholic fatty liver disease (NAFLD) likely increases the risk of type 2 diabetes in Europeans and East Asians.
A potential protective effect of NAFLD on coronary artery disease in East Asians requires replication.
Our study highlights the importance of Mendelian randomization studies in different ethnic populations to provide contextually specific evidence.
Introduction
People with non-alcoholic fatty liver disease (NAFLD) have increased risk of cardiovascular disease (CVD) and type 2 diabetes (T2D) based on meta-analysis of large observational studies.1,2 Although studies using proxies of NAFLD [e.g. elevated aspartate transaminase [AST] and alanine aminotransferase (ALT)] also showed a positive relation with T2D risk,3,4 associations with CVDs were not always evident in earlier studies,5 with inverse associations of ALT with CVDs also reported in other studies.6,7 Given that these studies were mainly observational in nature, confounding and selection bias cannot be ruled out.
Mendelian randomization (MR) studies, a design more robust to confounding than observational studies,8 has found potential bidirectional effects suggesting higher ALT increases risk of T2D and T2D liability potentially increases ALT in Europeans.9 Earlier MR studies did not find evidence of an effect of NAFLD on coronary artery disease (CAD) in Europeans, but found a potentially protective effect in East Asians.10,11 These differences could reflect real ethnic or contextual differences or be driven by low statistical power and choice of instruments, in particular the East Asian MR study relied on only two instruments.11 A subsequent larger MR study suggested ALT likely increases CAD and ischaemic stroke risk in Europeans, but did not consider other CVDs (e.g. atrial fibrillation, heart failure).12 Genetic studies using more direct measures of NAFLD [e.g. liver magnetic resonance imaging corrected T1 (MRI cT1)] found evidence that higher cT1 resulted in more adverse cardio metabolic traits.13 In order to comprehensively evaluate the role of NAFLD in CVD and T2D in Europeans and East Asians, we conducted one of the largest MR studies to explore the potential effect of NAFLD, proxied by ALT, NAFLD liability, AST, liver MRI cT1 and proton density fat fraction (PDFF) on CVDs, T2D and glycaemic traits using summary genetic associations from relevant genome-wide association studies (GWAS) in people of European and East Asian descent (exposures were ALT and AST only in analyses of East Asians).
Methods
In this MR study, we combined summary data for genetic associations with NAFLD proxies and with cardiovascular end points and T2D risk to gain insights into the role of NAFLD in the aetiology of cardiometabolic diseases. MR studies rely on three core assumptions.8 First, the genetic instruments should be associated with the exposure. Second, there should be no confounding between genetic instruments and outcomes. Third, any associations of the genetic instruments with the outcomes should be fully mediated by the exposure. In order to estimate the magnitude of the effect of exposure on outcome, additional assumptions are required (i.e. stable unit treatment value assumption, homogeneity and monotonicity), as described in the Supplementary material (available as Supplementary data at IJE online).14,15
Exposure data sets in European-based populations
In this study, we considered ALT level (SD), liability to NAFLD defined based on elevated ALT (log odds), AST level (SD), liver MRI cT1 (SD) and PDFF (SD) as NAFLD proxies, and extracted summary results from GWAS for each of these as provided in the original GWAS investigations, and as detailed below.
Liver enzymes (ALT and AST)
We extracted strong (P-value < 5 × 10–8) and independent (r2 < 0.001) genetic predictors of ALT and AST from summary genetic associations provided by Neale Lab in ≤344 292 participants (54% women) of White British ancestry from UK Biobank. These summary data were analysed via the Integrative Epidemiology Unit (IEU) Open GWAS project database retrieved via the Two-Sample MR R package.16–18 After excluding instruments that were not bi-allelic, there were 195 ALT instruments and 228 AST instruments.
Liability to NAFLD
We extracted strong (P-value < 5 × 10–8) and independent (r2 < 0.001) autosomal genetic predictors of NAFLD from a GWAS in the Million Veteran Program (N = 164 197), restricted to participants of European descent who were predominantly men.19 NAFLD was defined as having high ALT defined as >40 U/L in men and >30 U/L in women on at least two occasions that were between 6 months and 2 years apart. In addition, cases had to have no known case of liver disease, such as alcohol-related diseases, hepatitis or hepatic cancer. The GWAS identified 55 genome-wide significant variants. After excluding single-nucleotide polymorphisms (SNPs) in high LD (r2 ≥ 0.001), there were 46 instruments for NAFLD liability.
Liver MRI cT1 and PDFF
We extracted strong (P-value < 5 × 10–8) and independent (r2 < 0.001) genetic predictors of liver MRI-based cT1 and PDFF from a published GWAS using a subset of UK Biobank (n = 14 440).13 cT1 measured T1 relaxation time, which is related to fibrosis and inflammation, but corrected for the presence of iron. PDFF measured liver fat concentration. After screening, we selected five cT1 variants and four PDFF variants.
Details of these studies can be found in Supplementary Table S1 and the Supplementary material (available as Supplementary data at IJE online).
Outcome data sets for the study in European-based populations
We extracted genetic associations for NAFLD proxies-associated SNPs with outcomes from large GWAS of CVDs, T2D and glycaemic traits, mainly comprising European participants. Outcomes included CAD (CARDIoGRAM),20 overall stroke, ischaemic stroke and its subtypes (cardioembolic, large artery and small vessel stroke) (MEGASTROKE),21 atrial fibrillation,22 heart failure (HERMES consortium),23 T2D (DIAMANTE consortium),24 glucose, glycated haemoglobin and insulin (MAGIC).25 Details of these GWAS, such as sample size and case/control definitions, can be found in Supplementary Table S1 (available as Supplementary data at IJE online). The outcome GWAS were harmonized with the exposure GWAS to the same effect alleles. Effect allele frequencies (minor allele frequency ≤ 0.42) were used to harmonize palindromic SNPs. Proxy SNPs (r2 ≥ 0.8) were identified for outcomes available in MR Base.
Statistical analyses
We calculated the variance of each exposure explained by the instruments (R2) and approximated F-statistics to quantify the strength of the instruments using established formula used in previous studies.26,27 In our main analyses, we obtained effect estimates using the inverse-variance weighted (IVW) method with multiplicative random effects, which assumes balanced pleiotropy.28 We assessed between SNP heterogeneity using Cochran’s Q test. Between SNP heterogeneity may be caused by some SNPs having horizontal pleiotropic effects. MR–Egger, weighted median and MR–RAPS (robust adjusted profile score) estimates were computed as sensitivity analyses. These methods relax the no horizontal pleiotropy assumption but have additional assumptions (see Supplementary material, available as Supplementary data at IJE online for details).29–31 MR–Egger intercept P-values of <0.05 were used as statistical tests of the potential presence of unbalanced horizontal pleiotropy.29 Consistent findings across methods strengthen the evidence for causation.32 Given that MR–Egger is particular sensitive to outliers and may result in reversed estimates, we additionally used radial MR–IVW and radial MR–Egger to identify possible outliers and repeated the analyses without these SNPs in radial MR–IVW/MR–Egger as sensitivity analyses.33
Exploring possible horizontal pleiotropic effects via established risk factors for CVD/T2D
To complement the sensitivity analyses exploring evidence of any directional horizontal pleiotropy, we explored potential specific horizontal pleiotropic paths by examining the associations of the genetic instruments for each exposure with established risk factors for CVD/T2D [educational attainment,34 body mass index (BMI),35 alcohol,36 tobacco use36 and moderate to vigorous physical activity]37 using respective summary GWAS results (Supplementary Table S1, available as Supplementary data at IJE online). Specifically, we used IVW MR (i.e. exactly the same as for the main MR analyses) to test potential effects of the NAFLD exposures on these risk factors. We used a Bonferroni multiple testing P-value threshold accounting for the number of exposures and risk factors to identify specific pleiotropic paths (0.05/25 = 0.002). Any evidence of potential effects of NAFLD exposures on these cardiometabolic risk factors could indicate horizontal pleiotropy (i.e. an independent path from the genetic instruments to one or more of our outcomes) or vertical pleiotropy (i.e. the risk factors mediate the potential effect of NAFLD on outcome).
Replication in European-based populations using FinnGen
We sought replication of our main MR results of NAFLD proxies on CVDs and T2D using the FinnGen study—a combination of biobanks of 218 792 participants of European descent (Release 5). The distribution of common alleles is similar between Finnish and other European populations although the former had a higher frequency of deleterious and rare variants.38 Details of the FinnGen study can be found in Supplementary Table S2 (available as Supplementary data at IJE online).
Exposure and outcome data sets in East Asian-based populations
We extracted strong (P-value < 5 × 10–8) and independent (r2 < 0.001) instruments for ALT and AST from Biobank Japan GWAS (up to n = 134 182), which gave 27 and 25 instruments for ALT and AST. We then assessed the role of ALT and AST in CVDs, derived from a patient cohort of 179 660 participants of East Asian descent who were diagnosed with 1 or more of 47 diseases (e.g. common conditions such as CVDs, T2D, cancer, dyslipidemia, cataract, allergies and chronic respiratory diseases in Biobank Japan, as described previously) and 32 793 population-based controls,39,40 GWAS of T2D using a large GWAS in East Asians41 and glycaemic traits based on summary statistics of MAGIC, restricted to East Asians (up to n = 33 741).25 Details of Biobank Japan, the GWAS of T2D and glycaemic traits can be found in Supplementary Table S3 (available as Supplementary data at IJE online).
All analyses were performed using R Version 4.0.4 [R Core Team (2021). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/] and the R package (‘TwoSampleMR’).17
Results
MR in European-based populations
A full list of instruments for ALT, liability to NAFLD, AST, cT1 and PDFF is shown in Supplementary Table S4 (available as Supplementary data at IJE online). Variance explained by the instruments was 4.8% (ALT), 6.5% (AST), 5.1% (cT1) and 3.7% (PDFF). After data harmonization, 155–188 ALT instruments, 36–44 instruments for liability to NAFLD, 185–215 AST instruments and 4–5 instruments for cT1 and 3–4 instruments for PDFF were used. Weak instrument bias was unlikely based on the F-statistics (Mean FALT: 75–83; Mean FNAFLD: 86–104; Mean FAST: 79–100; Mean FcT1: 152–180; Mean FPDFF: 79–134). There was no strong statistical evidence of associations between genetic instruments of NAFLD proxies and most cardiometabolic risk factors (Supplementary Table S5, available as Supplementary data at IJE online), except for PDFF instruments with smoking heaviness (P-value: 0.0017).
Potential effects of ALT and liability to NAFLD on cardiometabolic outcomes in European-based populations
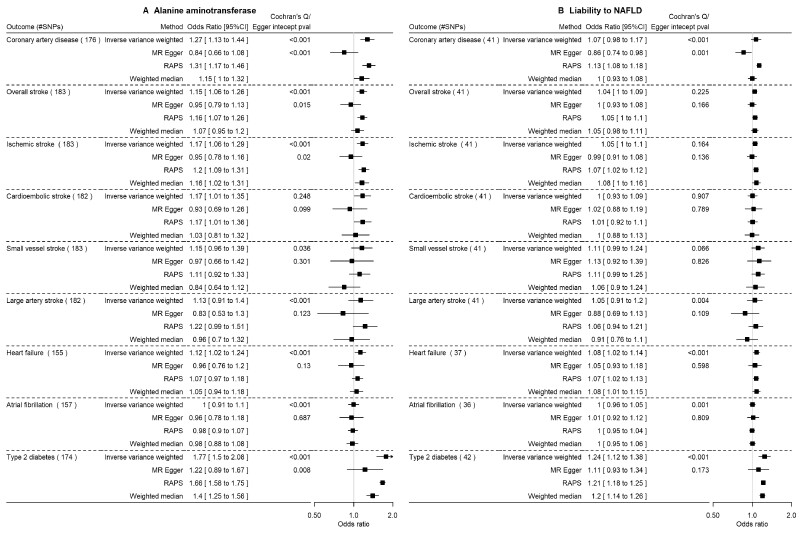
Figure 1 and Supplementary Table S6 (available as Supplementary data at IJE online) show the main (IVW) and sensitivity MR estimates of the effects of ALT and liability to NAFLD on each outcome. In these IVW analyses, there was evidence that higher ALT levels resulted in higher risk of CAD [odds ratio (OR) per SD, 1.27 95% CI 1.13 to 1.44], overall stroke (OR 1.15, 95% CI 1.06 to 1.26), likely driven by a potential effect on ischaemic stroke and its subtypes, heart failure (OR 1.12, 95% CI 1.02 to 1.24) and T2D (OR 1.77, 95% CI 1.5 to 2.08). Most sensitivity analyses were consistent with the main IVW results although there was statistical evidence of between SNP heterogeneity and some statistical support for unbalanced horizontal pleiotropy based on the MR–Egger test, with corresponding directionally different estimates from MR–Egger (except T2D). These associations were largely replicated using liability to NAFLD as the exposure.
Figure 1.
The impact of alanine aminotransferase level (SD) and liability to NAFLDa (per log odds) on cardiovascular diseases and type 2 diabetes using Mendelian randomization in predominantly European populations. SD, standard deviation; NAFLD, non-alcoholic fatty liver disease; RAPS, robust adjusted profile score. SDs and odds as provided in the original genome-wide association studies. aDefined as a high alanine aminotransferase level using sex-specific thresholds
Potential effects of AST and MRI-based measures (cT1 and PDFF) on cardiometabolic outcomes in European-based populations
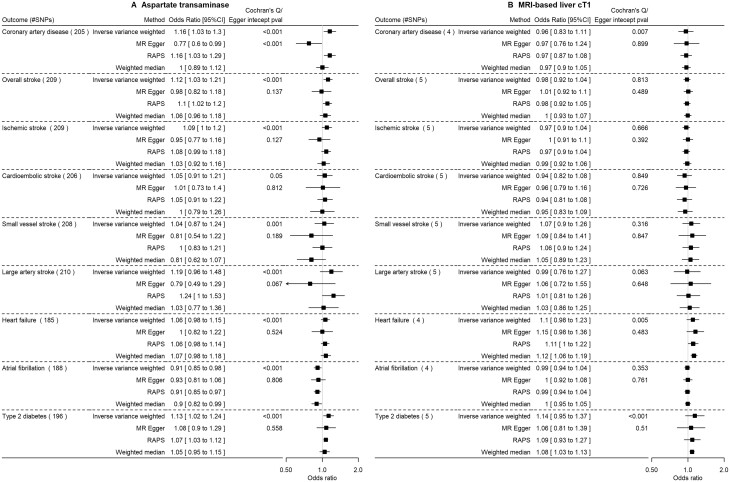
Figure 2, Supplementary Figure S1 and Supplementary Table S6 (available as Supplementary data at IJE online) show the main and sensitivity analyses. AST resulted in higher risk of CAD, overall stroke and T2D, and potentially lower risk of atrial fibrillation. Higher cT1 was possibly associated with higher heart failure and T2D risk, but these results were imprecise with wide CIs. PDFF did not appear to be a potential risk factor for any of the outcomes (Supplementary Figure S1, available as Supplementary data at IJE online). However, most of these estimates had wider confidence interval due to lower statistical power compared with ALT, liability to NAFLD and AST.
Figure 2.
The impact of aspartate transaminase level (SD) and MRI-based liver cT1 (SD) on cardiovascular diseases and type 2 diabetes using Mendelian randomization in predominantly European populations. SD, standard deviation (as provided in the original genome-wide association studies); MRI-based liver cT1, magnetic resonance imaging-based liver corrected T1; RAPS, robust adjusted profile score
Potential effects of NAFLD proxies on glycaemic traits in European-based populations and replication in FinnGen
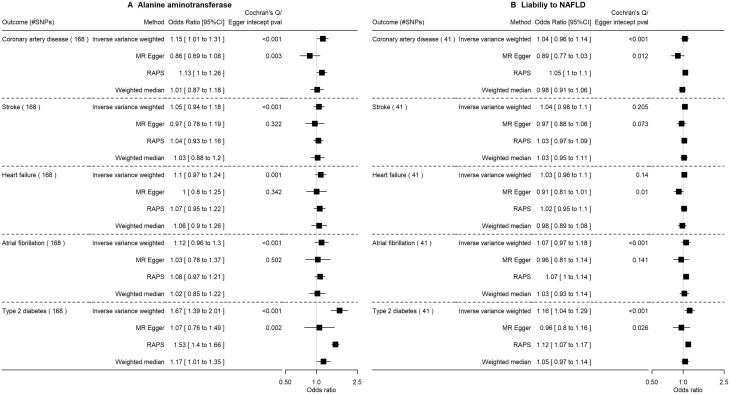
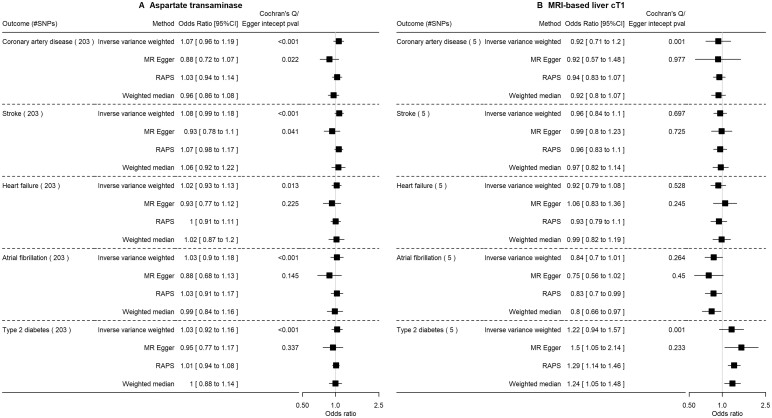
Only ALT, liability to NAFLD and AST were associated with higher insulin (Supplementary Figures S2–S4 and Supplementary Table S6, available as Supplementary data at IJE online). The findings from FinnGen suggested NAFLD proxies likely increase the risk of T2D, in particular cT1, although MR–Egger analyses sometimes gave directionally opposite estimates (Figures 3 and 4, and Supplementary Figure S1, available as Supplementary data at IJE online). ALT but not other measures of NAFLD potentially increased risk of CAD.
Figure 3.
The impact of alanine aminotransferase level (SD) and liability to NAFLDa (per log odds) on cardiovascular diseases and type 2 diabetes in FinnGen using Mendelian randomization. SD, standard deviation; NAFLD, non-alcoholic fatty liver disease; RAPS, robust adjusted profile score. SDs and odds as provided in the original genome-wide association studies. aDefined as a high alanine aminotransferase level using sex-specific thresholds
Figure 4.
The impact of aspartate transaminase level (SD) and MRI-based liver cT1 (SD) on cardiovascular diseases and type 2 diabetes in FinnGen using Mendelian randomization. SD, standard deviation (as provided in the original genome-wide association studies); MRI-based liver cT1, magnetic resonance imaging-based liver corrected T1; RAPS, robust adjusted profile score
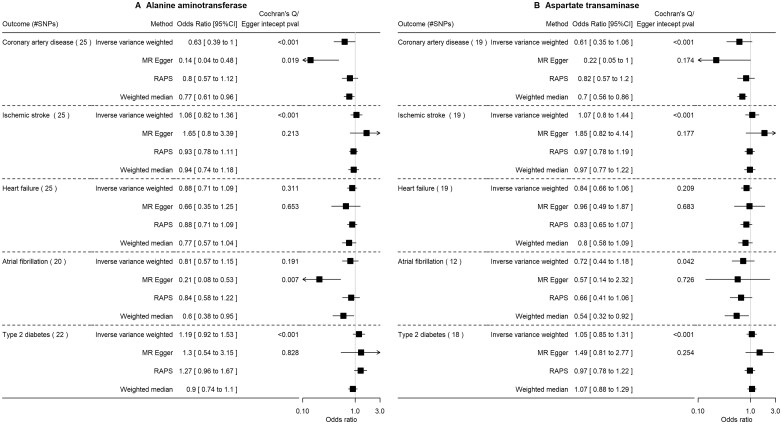
Potential effects of ALT and AST on cardiometabolic outcomes in East Asian-based populations
A full list of SNPs selected as instruments for ALT and AST in East Asians are shown in Supplementary Table S7 (available as Supplementary data at IJE online). Variance explained by the instruments was 1.3% (ALT) and 1.2% (AST). After data harmonization, 20–25 ALT instruments and 12–19 AST were used, with numbers varying by outcome (Supplementary Table S7, available as Supplementary data at IJE online). Weak instrument bias was unlikely based on F-statistics (Mean FALT: 52–61; Mean FAST: 72–76).
Figure 5 and Supplementary Table S8 (available as Supplementary data at IJE online) show the main and sensitivity analyses. Higher ALT levels potentially resulted in lower risk of CAD (OR: 0.63 per SD of log, 95% CI 0.39 to 1.00), with consistent findings in sensitivity analyses, and a potential effect of increasing T2D risk (OR: 1.19, 95% CI 0.92 to 1.53). Whilst there was evidence for heterogeneity across SNPs, overall horizontal pleiotropy was not evident based on the MR–Egger intercept except for the CAD and atrial fibrillation analyses. The relation of AST for these outcomes was consistent with ALT, apart from the null findings for T2D. ALT was associated with lower insulin, but not other glycaemic traits (Supplementary Figure S5 and Supplementary Table S8, available as Supplementary data at IJE online). These East Asian results had lower power and hence less precision than those for people of European ancestry.
Figure 5.
The impact of alanine aminotransferase level (per SD of log) and aspartate transaminase (per SD of log) on cardiovascular diseases and type 2 diabetes using Mendelian randomization in East Asians. SD, standard deviation (as provided in the original genome-wide association studies); RAPS, robust adjusted profile score
Discussion
This is one of the largest MR studies to explore the effects of NAFLD proxies on CVDs and T2D in Europeans and East Asians. In larger samples, we confirmed potential effects of NAFLD on T2D.9,10 We also found some evidence of a potential effect on increasing risk of CAD, overall stroke (largely driven by ischaemic stroke) and heart failure, and a potential protective effect of AST on atrial fibrillation. For CAD, the effect was likely biased by unbalanced horizontal pleiotropy whereas for other outcomes sensitivity analyses were consistent with the main analyses, though some MR–Egger estimates were too imprecise to be interpreted. Whilst ours is the largest study in East Asians to date, we had less statistical power than in analyses of Europeans. Nonetheless there was consistent evidence of NAFLD potentially increasing risk of T2D. We also found a potentially protective effect of NAFLD on CAD in East Asians, which is consistent with a previous smaller MR study in Chinese adults (N = 10 623, two instruments).11
Previous studies have shown overlap in risk profile between NAFLD and metabolic syndrome and subsequent risk of T2D and CVDs.42 Correspondingly, potential mechanisms for an effect of NAFLD on T2D include exacerbation of hepatic and peripheral insulin resistance, which is also supported by the effects we found on insulin, and inflammation.43 Possible mechanisms underlying potential effects on CAD and (ischaemic) stroke also include inflammation and a possible predisposition to atherogenic dyslipidemia.43 Given that sex hormones such as testosterone7 are produced in the liver and have a well-established role in protecting against T2D but may cause CVD, this may partly explain the diverging effect of NAFLD in CVD and T2D risk, although it remains unclear why the opposite effects were not evident in Europeans.
Differences in association by ethnic background have also been reported in some observational studies.6,44 In a previous review, ALT was associated with higher risk of CVDs in studies amongst people of Asian origin but not Europeans.6 However, there was substantial heterogeneity between studies, with ethnic differences possibly due to differences in residual confounding and selection bias.44 Selection bias could also influence our results, including the notable difference in direction of potential effect of NAFLD on CAD. Liver disease is more prevalent in East Asians and a more common cause of death than in Europeans.45 By definition, to be included, the GWAS identifying instruments for NAFLD participants had to be alive, meaning that in the East Asian analyses, it is possible that there is greater selection on factors that prevent death due to liver disease and that could introduce selection bias. As such, the biases introduced by competing risk of death from liver disease could lead to the observed inverse association of ALT with CAD in East Asians.46 Alternatively, potential drivers or consequences of ALT, such as chronic hepatitis and sex hormones, could differ by ethnicity and thereby result in instruments representing different exposures by ethnicity.47 For instance, elevated ALT has been shown to associate with lower triglycerides in East Asians11 but not in Europeans.13 Our findings further highlight the need for genetic studies, including MR, to be conducted in large samples of different ancestral origins given possible limitations in external validity across ancestries.
There were some limitations in this study. The robustness of MR studies relies on the three key assumptions described above.8 We chose strong instruments from large studies to reduce the risk of weak instrument bias and the high F-statistics support that. We used multiple sensitivity analyses to rule out possible bias due to unbalanced horizontal pleiotropy, as well as testing the possible association between instruments and factors that could constitute specific horizontal pleiotropic paths. However, some of the NAFLD risk alleles, such as ones in PNPLA3 (rs738409) and TM6SF2 (rs58542926), were associated with lower very-low-density lipoprotein cholesterol (vLDL) secretion and have neutral or even protective effects against CAD.48–50 These may partly contribute to horizontal pleiotropy and hence distorted MR–Egger estimates, such as the analyses for CAD and stroke. However, the positive association of NAFLD in T2D risk seen in our analyses partly suggests that the association of NAFLD with CVD cannot be completely explained by horizontal pleiotropy given that NAFLD might impact CVD via its detrimental effect on T2D risk, a known cause of CVD, and thus provides a potential causal pathway.51 Other sensitivity analyses relying on other assumptions generally gave consistent findings although we could not rule out the possibility of systematic biases such as selection bias, which may explain the inverse association of AST with atrial fibrillation in Europeans. Lastly, we were unable to explore possible non-linearity given that we relied on summary statistics.
Our study implies that NAFLD contributes to increased risk of T2D in both Europeans and East Asians, and with potential differential effects on CAD between Europeans and East Asians. Factors contributing to increased risk of NAFLD, such as obesity arising from overnutrition and sedentary behaviour, should be targeted to reduce the risk of NAFLD and subsequent elevated risk of CVDs and T2D.52 Given that some medications for CVDs and T2D, such as statins, metformin and SGLT2 inhibitors, could also impact AST, ALT and liver dysfunction risk,53–55 these may be unexplored pathways by which these medications influence the risk of these diseases and can be further examined using MR studies.56,57
Ethics approval
This study only used publicly available data and hence no ethics approval was required. Details of ethical approval and participant consent for each of the studies that contributed to the GWAS can be found in the original publications.
Supplementary Material
Acknowledgements
Data on CAD have been contributed by CARDIoGRAMplusC4D investigators and have been downloaded from www.CARDIORAMPLUSC4D.ORG. We thank MEGASTROKE for providing the summary statistics for stroke, which have been downloaded from http://www.megastroke.org/index.html. The MEGASTROKE project received funding from sources specified at http://www.megastroke.org/acknowledgments.html. The list of authors in the MEGASTROKE Consortium can be found in the Supplementary Materials (available as Supplementary data at IJE online). We thank HERMES Consortium and the atrial fibrillation GWAS investigators for providing the summary statistics for heart failure and atrial fibrillation, respectively, which have been accessed via IEU GWAS database (https://gwas.mrcieu.ac.uk/). We thank MAGIC for providing summary statistics for glucose, HbA1c and insulin, which have been downloaded from https://www.magicinvestigators.org. We thank DIAMANTE consortium for providing the summary statistics for type 2 diabetes, which have been accessed via https://diagram-consortium.org/. We thank SSGAC for providing the summary statistics for years of education attainment, which have been downloaded from https://www.thessgac.org/data. We thank GSCAN for providing the summary statistics for alcohol and cigarette use phenotypes, which have been downloaded from https://conservancy.umn.edu/handle/11299/201564. We thank the physical activity GWAS investigators for depositing the summary statistics for physical activity, which have been accessed via IEU GWAS database (https://gwas.mrcieu.ac.uk/). Data on ALT and AST (Neale Lab) were extracted from summary genome-wide association study results in UK Biobank, available in the IEU GWAS database (https://gwas.mrcieu.ac.uk/). We want to acknowledge the participants and investigators of FinnGen study (https://www.finngen.fi/en). We thank Biobank Japan Project and AGEN consortium (type 2 diabetes) for providing the summary statistics of phenotypes used in the analyses related to East Asians (http://jenger.riken.jp/en/ and https://blog.nus.edu.sg/agen/summary-statistics/t2d-2020/).
Conflict of interest
D.A.L. receives support from several national and international government and charitable research funders, as well as from Medtronic Ltd and Roche Diagnostics for research unrelated to that presented here. All other authors declare they have no conflict of interest, financial or otherwise.
Contributor Information
Shiu Lun Au Yeung, School of Public Health, LKS Faculty of Medicine, The University of Hong Kong, Hong Kong SAR, China.
Maria Carolina Borges, MRC Integrative Epidemiology Unit at the University of Bristol, Bristol, UK; Population Health Sciences, Bristol Medical School, University of Bristol, Bristol, UK.
Tommy Hon Ting Wong, School of Public Health, LKS Faculty of Medicine, The University of Hong Kong, Hong Kong SAR, China.
Deborah A Lawlor, MRC Integrative Epidemiology Unit at the University of Bristol, Bristol, UK; Population Health Sciences, Bristol Medical School, University of Bristol, Bristol, UK; National Institute for Health Research Bristol Biomedical Research Centre, University Hospitals Bristol NHS Foundation Trust and University of Bristol, Bristol, UK.
C Mary Schooling, School of Public Health, LKS Faculty of Medicine, The University of Hong Kong, Hong Kong SAR, China; School of Public Health and Health Policy, City University of New York, New York, USA.
Data availability
All data used in this study can be found in the cited references, URLs in the Acknowledgements and Supplementary material (available as Supplementary data at IJE online).
Supplementary data
Supplementary data are available at IJE online.
Author contributions
S.L.A.Y. designed the study, wrote the analysis plan and interpreted the results. S.L.A.Y. undertook analyses with feedback from M.C.B., D.A.L. and C.M.S. M.C.B. and D.A.L. provided additional feedback on the interpretation of the results. T.H.T.W. conducted the PDFF and Radial MR-related analyses. S.L.A.Y. wrote the first draft of the manuscript with critical feedback and revisions from M.C.B., D.A.L., T.H.T.W. and C.M.S. All authors gave final approval of the version to be published. S.L.A.Y. had primary responsibility for final content.
Funding
This study was supported by the Pre-emptive retention/Start up fund, Li Ka Shing Faculty of Medicine, The University of Hong Kong (S.L.A.Y.). M.C.B. was supported by Medical Research Council (MRC) Skills Development Fellowship (MR/P014054/1). M.C.B. and D.A.L.’s contribution to this study is supported by the British Heart Foundation (AA/18/7/34219 and CH/F/20/90003) and M.C.B. and D.A.L. work in a unit that receives funding from the University of Bristol and United Kingdom MRC (MC_UU_00011/6). The funders had no role in the design, analyses, interpretation of results or writing of the paper. The views expressed in this paper are those of the authors and not necessarily those of any of the funders.
References
- 1. Mantovani A, Csermely A, Petracca G. et al. Non-alcoholic fatty liver disease and risk of fatal and non-fatal cardiovascular events: an updated systematic review and meta-analysis. Lancet Gastroenterol Hepatol 2021;6:903–13. [DOI] [PubMed] [Google Scholar]
- 2. Mantovani A, Byrne CD, Bonora E, Targher G.. Nonalcoholic fatty liver disease and risk of incident type 2 diabetes: a meta-analysis. Diabetes Care 2018;41:372–82. [DOI] [PubMed] [Google Scholar]
- 3. Fraser A, Harris R, Sattar N, Ebrahim S, Davey Smith G, Lawlor DA.. Alanine aminotransferase, gamma-glutamyltransferase, and incident diabetes: the British Women's Heart and Health Study and meta-analysis. Diabetes Care 2009;32:741–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chen SC, Tsai SP, Jhao JY, Jiang WK, Tsao CK, Chang LY.. Liver fat, hepatic enzymes, alkaline phosphatase and the risk of incident type 2 diabetes: a prospective study of 132,377 adults. Sci Rep 2017;7:4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ghouri N, Preiss D, Sattar N.. Liver enzymes, nonalcoholic fatty liver disease, and incident cardiovascular disease: a narrative review and clinical perspective of prospective data. Hepatology 2010;52:1156–61. [DOI] [PubMed] [Google Scholar]
- 6. Kunutsor SK, Apekey TA, Khan H.. Liver enzymes and risk of cardiovascular disease in the general population: a meta-analysis of prospective cohort studies. Atherosclerosis 2014;236:7–17. [DOI] [PubMed] [Google Scholar]
- 7. Schooling CM, Kelvin EA, Jones HE.. Alanine transaminase has opposite associations with death from diabetes and ischemic heart disease in NHANES III. Ann Epidemiol 2012;22:789–98. [DOI] [PubMed] [Google Scholar]
- 8. Davies NM, Holmes MV, Davey Smith G.. Reading Mendelian randomisation studies: a guide, glossary, and checklist for clinicians. BMJ 2018;362:k601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. De Silva NMG, Borges MC, Hingorani AD. et al. ; UCLEB consortium. Liver function and risk of type 2 diabetes: bidirectional Mendelian randomization study. Diabetes 2019;68:1681–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Liu J, Au Yeung SL, Lin SL, Leung GM, Schooling CM.. Liver enzymes and risk of ischemic heart disease and type 2 diabetes mellitus: a Mendelian randomization study. Sci Rep 2016;6:38813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Xu L, Jiang CQ, Lam TH. et al. Mendelian randomization estimates of alanine aminotransferase with cardiovascular disease: Guangzhou Biobank Cohort study. Hum Mol Genet 2017;26:430–37. [DOI] [PubMed] [Google Scholar]
- 12. Pazoki R, Vujkovic M, Elliott J, VA Million Veteran Program et al Genetic analysis in European ancestry individuals identifies 517 loci associated with liver enzymes. Nat Commun 2021;12:2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Parisinos CA, Wilman HR, Thomas EL. et al. Genome-wide and Mendelian randomisation studies of liver MRI yield insights into the pathogenesis of steatohepatitis. J Hepatol 2020;73:241–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Labrecque J, Swanson SA.. Understanding the assumptions underlying instrumental variable analyses: a brief review of falsification strategies and related tools. Curr Epidemiol Rep 2018;5:214–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Burgess S, Small DS.. Predicting the direction of causal effect based on an instrumental variable analysis: a cautionary tale. J Causal Inference 2016;4:49–59. [Google Scholar]
- 16.Neale Lab—UK Biobank GWAS. http://www.nealelab.is/uk-biobank (23 October 2021, date last acccessed).
- 17. Hemani G, Zheng J, Elsworth B. et al. The MR-Base platform supports systematic causal inference across the human phenome. Elife 2018;7:e34408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Elsworth B, Lyon M, Alexander T. et al. The MRC IEU OpenGWAS data infrastructure. bioRxiv2020; 10.1101/2020.08.10.244293. [DOI]
- 19. Vujkovic M, Ramdas S, Lorenz KM. et al. A genome-wide association study for nonalcoholic fatty liver disease identifies novel genetic loci and trait-relevant candidate genes in the Million Veteran Program. medRxiv2021; 10.1101/2020.12.26.20248491. [DOI]
- 20. Nikpay M, Goel A, Won HH. et al. A comprehensive 1,000 genomes-based genome-wide association meta-analysis of coronary artery disease. Nat Genet 2015;47:1121–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Malik R, Chauhan G, Traylor M. et al. ; MEGASTROKE Consortium. Multiancestry genome-wide association study of 520,000 subjects identifies 32 loci associated with stroke and stroke subtypes. Nat Genet 2018;50:524–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Roselli C, Chaffin MD, Weng LC. et al. Multi-ethnic genome-wide association study for atrial fibrillation. Nat Genet 2018;50:1225–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shah S, Henry A, Roselli C. et al. ; Regeneron Genetics Center. Genome-wide association and Mendelian randomisation analysis provide insights into the pathogenesis of heart failure. Nat Commun 2020;11:163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mahajan A, Taliun D, Thurner M. et al. Fine-mapping type 2 diabetes loci to single-variant resolution using high-density imputation and islet-specific epigenome maps. Nat Genet 2018;50:1505–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chen J, Spracklen CN, Marenne G. et al. ; Meta-Analysis of Glucose and Insulin-related Traits Consortium (MAGIC). The trans-ancestral genomic architecture of glycemic traits. Nat Genet 2021;53:840–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yarmolinsky J, Bonilla C, Haycock PC. et al. ; PRACTICAL Consortium. Circulating selenium and prostate cancer risk: a Mendelian randomization analysis. J Natl Cancer Inst 2018;110:1035–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Au Yeung SL, Zhao JV, Schooling CM.. Evaluation of glycemic traits in susceptibility to COVID-19 risk: a Mendelian randomization study. BMC Med 2021;19:72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bowden J, Del Greco MF, Minelli C, Davey Smith G, Sheehan N, Thompson J.. A framework for the investigation of pleiotropy in two-sample summary data Mendelian randomization. Stat Med 2017;36:1783–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bowden J, Davey Smith G, Burgess S.. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol 2015;44:512–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bowden J, Davey Smith G, Haycock PC, Burgess S.. Consistent estimation in Mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol 2016;40:304–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhao QY, Wang JS, Hemani G, Bowden J, Small DS.. Statistical inference in two-sample summary-data Mendelian randomization using robust adjusted profile score. Ann Stat 2020;48:1742–69. [Google Scholar]
- 32. Lawlor DA, Tilling K, Davey Smith G.. Triangulation in aetiological epidemiology. Int J Epidemiol 2016;45:1866–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bowden J, Spiller W, Del Greco MF. et al. Improving the visualization, interpretation and analysis of two-sample summary data Mendelian randomization via the Radial plot and Radial regression. Int J Epidemiol 2018;47:1264–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Okbay A, Beauchamp JP, Fontana MA. et al. ; LifeLines Cohort Study. Genome-wide association study identifies 74 loci associated with educational attainment. Nature 2016;533:539–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yengo L, Sidorenko J, Kemper KE. et al. ; GIANT Consortium. Meta-analysis of genome-wide association studies for height and body mass index in approximately 700000 individuals of European ancestry. Hum Mol Genet 2018;27:3641–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Liu M, Jiang Y, Wedow R. et al. ; HUNT All-In Psychiatry. Association studies of up to 1.2 million individuals yield new insights into the genetic etiology of tobacco and alcohol use. Nat Genet 2019;51:237–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Klimentidis YC, Raichlen DA, Bea J. et al. Genome-wide association study of habitual physical activity in over 377,000 UK Biobank participants identifies multiple variants including CADM2 and APOE. Int J Obes (Lond) 2018;42:1161–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lim ET, Wurtz P, Havulinna AS. et al. ; Sequencing Initiative Suomi (SISu) Project. Distribution and medical impact of loss-of-function variants in the Finnish founder population. PLoS Genet 2014;10:e1004494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ishigaki K, Akiyama M, Kanai M. et al. Large-scale genome-wide association study in a Japanese population identifies novel susceptibility loci across different diseases. Nat Genet 2020;52:669–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Low SK, Takahashi A, Ebana Y. et al. ; AFGen Consortium. Identification of six new genetic loci associated with atrial fibrillation in the Japanese population. Nat Genet 2017;49:953–58. [DOI] [PubMed] [Google Scholar]
- 41. Spracklen CN, Horikoshi M, Kim YJ. et al. Identification of type 2 diabetes loci in 433,540 East Asian individuals. Nature 2020;582:240–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yki-Jarvinen H. Non-alcoholic fatty liver disease as a cause and a consequence of metabolic syndrome. Lancet Diabetes Endocrinol 2014;2:901–10. [DOI] [PubMed] [Google Scholar]
- 43. Targher G, Corey KE, Byrne CD, Roden M.. The complex link between NAFLD and type 2 diabetes mellitus: mechanisms and treatments. Nat Rev Gastroenterol Hepatol 2021;18:599–612. [DOI] [PubMed] [Google Scholar]
- 44. Ford I, Mooijaart SP, Lloyd S. et al. The inverse relationship between alanine aminotransferase in the normal range and adverse cardiovascular and non-cardiovascular outcomes. Int J Epidemiol 2011;40:1530–38. [DOI] [PubMed] [Google Scholar]
- 45. Sarin SK, Kumar M, Eslam M. et al. Liver diseases in the Asia-Pacific region: a Lancet Gastroenterology & Hepatology Commission. Lancet Gastroenterol Hepatol 2020;5:167–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Schooling CM, Lopez PM, Yang Z, Zhao JV, Au Yeung SL, Huang JV.. Use of multivariable mendelian randomization to address biases due to competing risk before recruitment. Front Genet 2020;11:610852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lopez PM, Subramanian SV, Schooling CM.. Effect measure modification conceptualized using selection diagrams as mediation by mechanisms of varying population-level relevance. J Clin Epidemiol 2019;113:123–28. [DOI] [PubMed] [Google Scholar]
- 48. Pirazzi C, Adiels M, Burza MA. et al. Patatin-like phospholipase domain-containing 3 (PNPLA3) I148M (rs738409) affects hepatic VLDL secretion in humans and in vitro. J Hepatol 2012;57:1276–82. [DOI] [PubMed] [Google Scholar]
- 49. Kozlitina J, Smagris E, Stender S. et al. Exome-wide association study identifies a TM6SF2 variant that confers susceptibility to nonalcoholic fatty liver disease. Nat Genet 2014;46:352–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Simons N, Isaacs A, Koek GH, Kuč S, Schaper NC, Brouwers MCGJ.. PNPLA3, TM6SF2, and MBOAT7 genotypes and coronary artery disease. Gastroenterology 2017;152:912–13. [DOI] [PubMed] [Google Scholar]
- 51. Luo S, Au Yeung SL, Schooling CM.. Assessing the linear and non-linear association of HbA1c with cardiovascular disease: a Mendelian randomisation study. Diabetologia 2021;64:2502–10. [DOI] [PubMed] [Google Scholar]
- 52. Stefan N, Haring HU, Cusi K.. Non-alcoholic fatty liver disease: causes, diagnosis, cardiometabolic consequences, and treatment strategies. Lancet Diabetes Endocrinol 2019;7:313–24. [DOI] [PubMed] [Google Scholar]
- 53. Jalali M, Rahimlou M, Mahmoodi M. et al. The effects of metformin administration on liver enzymes and body composition in non-diabetic patients with non-alcoholic fatty liver disease and/or non-alcoholic steatohepatitis: an up-to date systematic review and meta-analysis of randomized controlled trials. Pharmacol Res 2020;159:104799. [DOI] [PubMed] [Google Scholar]
- 54. Wong C, Yaow CYL, Ng CH. et al. Sodium-glucose co-transporter 2 inhibitors for non-alcoholic fatty liver disease in Asian patients with type 2 diabetes: a meta-analysis. Front Endocrinol (Lausanne) 2020;11:609135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Cai T, Abel L, Langford O. et al. Associations between statins and adverse events in primary prevention of cardiovascular disease: systematic review with pairwise, network, and dose-response meta-analyses. BMJ 2021;374:n1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Luo S, Schooling CM, Wong ICK, Au Yeung SL.. Evaluating the impact of AMPK activation, a target of metformin, on risk of cardiovascular diseases and cancer in the UK Biobank: a Mendelian randomisation study. Diabetologia 2020;63:2349–58. [DOI] [PubMed] [Google Scholar]
- 57. Ference BA, Robinson JG, Brook RD. et al. Variation in PCSK9 and HMGCR and Risk of Cardiovascular Disease and Diabetes. N Engl J Med 2016;375:2144–53. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data used in this study can be found in the cited references, URLs in the Acknowledgements and Supplementary material (available as Supplementary data at IJE online).