Abstract
IMGT, the international ImMunoGeneTics database (http://imgt.cines.fr:8104 ), is a high-quality integrated database specialising in Immunoglobulins (Ig), T cell Receptors (TcR) and Major Histocompatibility Complex (MHC) molecules of all vertebrate species, created in 1989 by Marie-Paule Lefranc, Université Montpellier II, CNRS, Montpellier, France (lefranc@ ligm.igh.cnrs.fr ). At present, IMGT includes two databases: IMGT/LIGM-DB, a comprehensive database of Ig and TcR from human and other vertebrates, with translation for fully annotated sequences, and IMGT/HLA-DB, a database of the human MHC referred to as HLA (Human Leucocyte Antigens). The IMGT server provides a common access to expertized genomic, proteomic, structural and polymorphic data of Ig and TcR molecules of all vertebrates. By its high quality and its easy data distribution, IMGT has important implications in medical research (repertoire in autoimmune diseases, AIDS, leukemias, lymphomas), therapeutic approaches (antibody engineering), genome diversity and genome evolution studies. IMGT is freely available at http://imgt.cines.fr:8104 . The IMGT Index is provided at the IMGT Marie-Paule page (http://imgt.cines.fr:8104/textes/IMGTindex.html ).
INTRODUCTION
IMGT, the international ImMunoGeneTics database (1), created in 1989 by Marie-Paule Lefranc (Université Montpellier II, CNRS, Montpellier, France; lefranc@ligm.igh.cnrs.fr), is a high quality integrated database specialising in Immunoglobulins (Ig), T cell Receptors (TcR) and Major Histocompatibility Complex (MHC) molecules of all vertebrate species (IMGT home page at http://imgt.cines.fr:8104 ). IMGT comprises expertly annotated data and consists at present of two databases: IMGT/LIGM-DB, a comprehensive database of Ig and TcR from human and other vertebrates, with translation for fully annotated sequences, and IMGT/HLA-DB, a database of the human MHC referred to as HLA (Human Leucocyte Antigens). IMGT distributes high quality data with an important increment value added by the IMGT expert annotations. IMGT is a unique immunogenetics database for the genome, proteome, structure and polymorphism data of Ig, TcR and HLA. An ontology has been constructed by IMGT in order to provide controlled vocabulary and annotation rules which are indispensable to ensure accuracy, consistency and coherence in IMGT (2). The IMGT Index is available at the IMGT Marie-Paule page (http://imgt.cines.fr:8104/textes/IMGTindex.html ).
IMGT/LIGM-DB ORGANIZATION AND CONTENT
IMGT/LIGM-DB development is mainly based on a relational model organization. The database is maintained with SYBASE as relational DBSM (Data Base System Manager). IMGT closely follows the content of Ig and TcR sequences from generalist databases and currently contains more than 34 300 nucleic acid sequences of Ig or TcR from 81 vertebrate species, and translation for fully annotated sequences. IMGT sequences are identified by the EMBL/GenBank/DDBJ (3–5) accession number. IMGT standardized keywords for Ig and TcR have been assigned to all entries. 177 feature labels are necessary to describe all structural and functional subregions that compose Ig and TcR sequences, whereas only seven of them are available in EMBL, GenBank or DDBJ. Annotation of sequences with these labels constitutes the main part of the expertize. All IMGT/LIGM-DB information is available through search criteria described by Lefranc et al. (1).
IMGT/HLA-DB ORGANIZATION AND CONTENT
IMGT/HLA-DB is a specialist sequence database for sequences of the human major histocompatibility system. It includes the HLA sequences named by the WHO Nomenclature Commiteee for Factors of the HLA System. The database provides users with online tools and facilities for the retrieval and analysis of HLA sequences. These include allele reports, alignment tools and a detailed database of all source cells. The online IMGT/HLA-DB submission tool allows the submission of both new and confirmatory alleles to the WHO Committee for Factors of the HLA system. The latest version (Release 1.4, October 1999) contains 1015 HLA alleles derived from over component 2275 EMBL/GenBank/DDBJ sequences and since its release in December 1998 the IMGT/HLA-DB website has received over 87 000 hits. The database currently focuses on the human MHC but will be used as a model system to provide a specialist database for the MHC sequences of other species.
IMGT, AN INTEGRATED DATABASE
Genome
IMGT reference sequences. IMGT reference sequences for Ig and TcR have been defined. They are listed in the germline gene tables of the IMGT Repertoire (6–12). The IMGT reference directory is crucial for the high quality of IMGT/DNAPLOT results (13,14).
IMGT gene name nomenclature. The objective is to provide immunologists and geneticists with a unique nomenclature per locus which will allow extraction and comparison of data for the complex B and T cell antigen receptor molecules, and the MHC, whatever the species.
Proteome
IMGT unique numbering. A uniform numbering system for Ig and TcR sequences of all species has been established by Marie-Paule Lefranc to facilitate sequence comparison and cross-referencing between experiments from different laboratories whatever the antigen receptor (Ig or TcR), the chain type or the species (1,13,15,16). The IMGT unique numbering has allowed to redefine the limits of the framework (FR) and complementarity determining regions (CDR) of the Ig and TcR variable domain. Tables of FR-IMGT and CDR-IMGT lengths are available from the IMGT Repertoire.
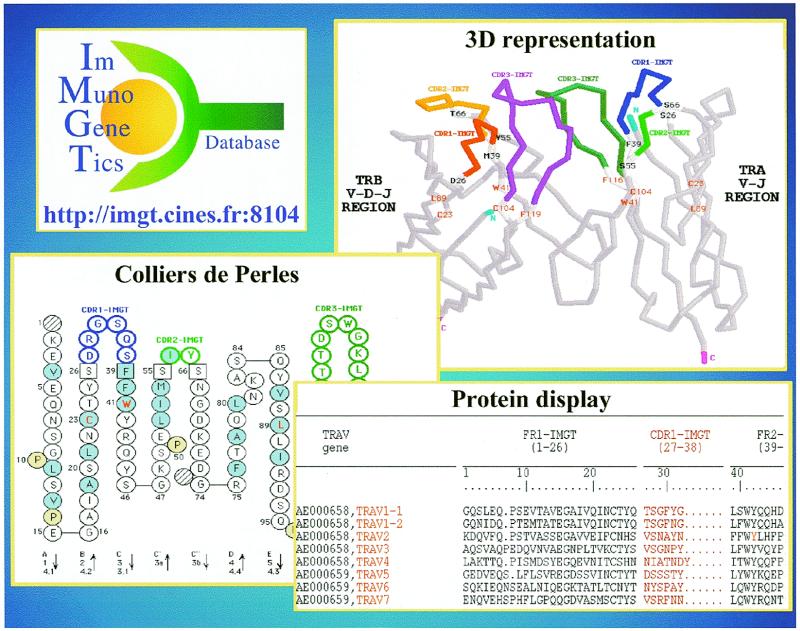
Protein displays. Protein displays are provided, in the IMGT Repertoire, for all the human germline variable regions of Ig and TcR (12) (Fig. 1), and in IMGT/HLA-DB for the human MHC.
Figure 1.
Examples of TcR V-REGION data in the IMGT Repertoire: protein display, 2D representation (Colliers de Perles) and 3D representation.
Polymorphic data
IMGT mutation and allele polymorphism description. The IMGT unique numbering has allowed a standardized IMGT description of mutations and the description of allele polymorphisms and somatic hypermutations for the Ig and TcR. Alignments of alleles (13) and tables of alleles (7–11) have been set up for the coding region of the Ig and TcR germline genes (IMGT Repertoire) and for the human MHC genes (IMGT/HLA-DB).
Probes and RFLP. A new section of the IMGT Repertoire, currently developed, contains data on probes used for the analysis of Ig and TcR gene rearrangements and expressions, and RFLP (Restriction Fragment Length Polymorphism) studies.
Structural data
2D representations (Colliers de Perles) and the IMGT/Colliers de Perles tool. 2D graphical representations designated as Colliers de Perles (1,13) are provided for all the human germline variable regions of Ig and TcR (Fig. 1). The most recent 2D representations were generated with the IMGT/Colliers de Perles tool, developed by Gérard Mennessier (LPM, Montpellier, France).
3D representations. In order to establish the first common data access to all structural data concerning the Ig and TcR, structural data are retrieved from PDB (17,18) and from the literature. The IMGT unique numbering of the amino acids and the IMGT standardization rules have been applied to the PDB files. 3D representations of Ig and TcR V-REGIONs are available from the IMGT Repertoire. This visualization permits rapid correlation between protein sequences and 3D structure (Fig. 1).
IMGT, A HIGH QUALITY DATABASE BASED ON THE IMGT-ONTOLOGY
Ontology
IMGT has developed a formal specification of the terms to be used in the domain of immunogenetics and bioinformatics. This has been the basis of the IMGT-ONTOLOGY (2), the first ontology in the domain, which allows the management of the immunogenetics knowledge for all vertebrate species.
IMGT data coherence
Control of coherence in IMGT combines data integrity control and biological data evaluation (19).
IMGT INTEROPERABILITY AND DATA DISTRIBUTION
Since July 1995, IMGT/LIGM-DB has been available on the web at the IMGT home page. IMGT/HLA-DB is available since December 1998. IMGT provides the immunologists with an easy to use and friendly interface. IMGT has an exceptional response with more than 12 000 requests a week. From January 1996 to October 1999, IMGT WWW server at Montpellier was accessed by more than 60 000 sites. IMGT data are also distributed by EBI (distribution of CD-ROM, network fileserver: netserv@ebi.ac.uk and anonymous FTP server), by the CINES anonymous FTP server. IMGT is available from many SRS sites. To facilitate the integration of IMGT data into applications developed by other laboratories, we have built an Application Programming Interface to access the database and its software tools (1,19).
ELECTRONIC AND MAILING ADDRESSES
IMGT home page: http://imgt.cines.fr:8104 (IMGT contact lefranc@ligm.igh.cnrs.fr ).
IMGT page at EBI (flat file release and sequence submission information): http://www.ebi.ac.uk/imgt/
IMGT/LIGM-DB: http://imgt.cines.fr:8104 (contacts lefranc@ ligm.igh.cnrs.fr , giudi@ligm.igh.cnrs.fr ).
IMGT/HLA-DB: http://www.ebi.ac.uk/imgt/hla/ (contacts jrobinso@ebi.ac.uk , julia@icrf.icnet.uk , marsh@icrf.icnet.uk ).
Anonymous FTP servers: ftp.ebi.ac.uk, ftp://imgt.cines.fr/pub/IMGT (contact denys.chaume@igh.cnrs.fr ).
IMGT Initiator and Coordinator: Marie-Paule Lefranc, IMGT, the International ImMunoGeneTics database, Laboratoire d’ImmunoGénétique Moléculaire, LIGM, UPR CNRS 1142, IGH, rue de la Cardonille, 34396 Montpellier Cedex 5, France. Tel: +33 (0)4 99 61 99 65; Fax: +33 (0)4 99 61 99 01; Email: lefranc@ligm.igh.cnrs.fr
CITING IMGT
Authors who make use of the information provided by IMGT should cite this article as a general reference for the access to and content of IMGT, and quote the IMGT home page URL, http://imgt.cines.fr:8104
Acknowledgments
ACKNOWLEDGEMENTS
We are deeply grateful to Gérard Mennessier (LIGMotif and IGMT/Colliers de Perles tools), Hans-Helmar Althaus and Werner Müller (DNAPLOT program), Johanne Abad, Sylvaine Artero, Valerie Barbié, Nathalie Bosc, Valérie Contet, Géraldine Folch, Oksana Kravchuk, Christèle Martinez, Violaine Moreau, Olga Posukh and Dominique Scaviner (IMGT/LIGM-DB), Natasja de Groot (IMGT/MHC). IMGT is funded by the European Union’s BIOTECH programme BIO4CT96-0037, the CNRS (Centre National de la Recherche Scientifique), and the MENRT (Ministère de l’Education Nationale, de la Recherche et de la Technologie). Subventions have been received from the Imperial Cancer Research Fund, the National Institute of Health, NMDP (National Marrow Donor Program), the Anthony Nolan Bone Marrow Trust, ARC (Association pour la Recherche sur le Cancer), ARP (Association de Recherche sur la Polyarthrite), FRM (Fondation pour la Recherche Médicale), Ligue Nationale contre le Cancer, and the Région Languedoc-Roussillon.
REFERENCES
- 1.Lefranc M.-P., Giudicelli,V., Ginestoux,C., Bodmer,J., Müller,W., Bontrop,R., Lemaitre,M., Malik,A., Barbié,V. and Chaume,D. (1999) Nucleic Acids Res., 27, 209–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Giudicelli V. and Lefranc,M.-P. (1999) Bioinformatics, in press. [DOI] [PubMed] [Google Scholar]
- 3.Stoesser G., Tuli,M.A., Lopez,R. and Sterk,P. (1999) Nucleic Acids Res., 27, 18–24. Updated article in this issue: Nucleic Acids Res. (2000), 28, 19–23.9847133 [Google Scholar]
- 4.Benson D.A., Boguski,M.S., Lipman,D.J., Ostell,J., Ouellette,B.F.F., Rapp,B.A. and Wheeler,D.L. (1999) Nucleic Acids Res., 27, 12–17. Updated article in this issue: Nucleic Acids Res. (2000), 28, 15–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sugawara H., Miyazaki,S., Gojobori,T. and Tateno,Y. (1999) Nucleic Acids Res., 27, 25–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lefranc M.-P. (1998) Exp. Clin. Immunogenet., 15, 1–7. [DOI] [PubMed] [Google Scholar]
- 7.Pallarès N., Frippiat,J.-P., Giudicelli,V. and Lefranc,M.-P. (1998) Exp. Clin. Immunogenet., 15, 8–18. [DOI] [PubMed] [Google Scholar]
- 8.Barbié V. and Lefranc,M.-P. (1998) Exp. Clin. Immunogenet., 15, 171–183. [DOI] [PubMed] [Google Scholar]
- 9.Martinez C. and Lefranc,M.-P. (1998) Exp. Clin. Immunogenet., 15, 184–193. [DOI] [PubMed] [Google Scholar]
- 10.Pallarès N., Lefebvre,S., Contet,V., Matsuda,F. and Lefranc,M.-P. (1999) Exp. Clin. Immunogenet., 16, 36–60. [DOI] [PubMed] [Google Scholar]
- 11.Ruiz M., Pallarès,N., Contet,V., Barbié,V. and Lefranc,M.-P. (1999) Exp. Clin. Immunogenet., 16, 173–184. [DOI] [PubMed] [Google Scholar]
- 12.Scaviner D., Barbié,V., Ruiz,M. and Lefranc,M.-P. (1999) Exp. Clin. Immunogenet., 16, in press. [DOI] [PubMed] [Google Scholar]
- 13.Lefranc M.-P., Giudicelli,V., Busin,C., Bodmer,J., Müller,W., Bontrop,R., Lemaitre,M., Malik,A. and Chaume,D. (1998) Nucleic Acids Res., 26, 297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Giudicelli V., Chaume,D., Mennessier,G., Althaus,H.H., Müller,W., Bodmer,J., Malik,A. and Lefranc,M.-P. (1998) Proceedings of the Ninth World Congress on Medical Informatics, MEDINFO’ 98, 351–355. [PubMed]
- 15.Lefranc M.-P. (1997) Immunol. Today, 18, 509. [DOI] [PubMed] [Google Scholar]
- 16.Lefranc M.-P. (1999) The Immunologist, 7, 132–136. [Google Scholar]
- 17.Bernstein F.C., Koetzle,T.F., Williams,G.J., Meyer,E.E.,Jr, Brice,M.D., Rodgers,J.R., Kennard,O., Shimanouchi,T. and Tasumi,M. (1977) J. Mol. Biol., 112, 535–542. [DOI] [PubMed] [Google Scholar]
- 18.Abola E.E., Sussman,J.L., Prilusky,J. and Manning,N.O. (1997) Methods Enzymol., 277, 556–571. [DOI] [PubMed] [Google Scholar]
- 19.Giudicelli V., Chaume,D. and Lefranc,M.-P. (1998) Proceedings of the Sixth International Conference on Intelligent Systems for Molecular Biology, ISBM-98, 59–68. [PubMed]