Abstract
In humans, mutations in calmodulin cause cardiac arrhythmia. These mutations disrupt the ability of calmodulin to sense calcium concentrations and correctly regulate two central calcium channels, together obstructing heart rhythm. This correlation is well established, but also surprising since calmodulin is expressed in all tissues and interacts with hundreds of proteins. Until now, most studies have focused on cardiac cell function and regulation of specific cardiac targets, and thus, potential other effects of these mutations have largely been unexplored. Here, we introduce the nematode Caenorhabditis elegans as an in vivo model to study effects of three human calmodulin mutations with different impairment on calcium binding. We find that arrhythmic effects of the calmodulin mutations N54I and D96V can be recapitulated in disruption of two rhythmic behaviors, pharynx pumping and defecation motor program. Interestingly, we also find that these mutations affect neuronal function, but in different ways. Whereas D96V sensitizes signaling at the neuromuscular junction, N54I has a protective effect. The mutation N98S did not affect rhythmic behavior, but impaired chemosensing. Therefore, pathogenic calmodulin mutations act through different mechanisms in rhythmic behavior and neuronal function in C. elegans, emphasizing the strength of using live multicellular models. Finally, our results support the hypothesis that human calmodulin mutations could also contribute to neurological diseases.
Graphical Abstract
Graphical Abstract.
Introduction
Across the evolutionary tree of life, calcium (Ca2+) signaling controls hundreds of processes including fertilization, apoptosis, cell migration, muscle contraction, vesicle release and memory formation. These finely regulated processes are managed by Ca2+ channels and sensor proteins that precisely relay signal information to downstream targets (1,2).
Calmodulin is a Ca2+ sensor that binds Ca2+ ions entering the cell. Calmodulin changes conformation upon Ca2+ binding and consequently affinity toward its more than 300 target proteins. In this way, calmodulin modulates the responses of its target proteins to the incoming Ca2+ signals (3,4). Calmodulin is one of the most well-conserved proteins known (5). Humans have three calmodulin genes located on separate chromosomes. They all encode the exact same protein. Before 2012, no protein altering genetic variants (mutations) within calmodulin had been reported, underscoring its extraordinary conservation. In 2012, we published the first cases of calmodulin mutations identified in patients suffering from cardiac arrhythmia (6). Since then, several reports have confirmed that, although extremely rare, mutations in calmodulin cause cardiac arrhythmias in humans, primarily catecholaminergic polymorphic ventricular tachycardia (CPVT) or long QT syndrome (LQTS) (3,7,8). In most cases, the disease is present already in childhood, in some cases even prenatally, leading to a number of tragic cardiac arrests and deaths in children.
The arrhythmogenic calmodulin missense mutations demonstrate a reduction in Ca2+ affinity and reduced interaction to or regulation of the voltage-gated Ca2+ channel isoform 1.2 (CaV1.2) and/or ryanodine receptor type 2 (RyR2). Generally, we find that mutations displaying strong impairment of Ca2+ binding and dysregulation of CaV1.2 are linked with LQTS. On the other hand, calmodulin mutations carried by individuals affected by CPVT impose dysregulation of RyR2 (9,10).
In this study, we included the mutation D96V as an example of an LQTS mutation (Table 1). This mutation was found in a child, who presented with bradycardia already during gestation and markedly prolonged QTc interval from birth. In her first years of life, she suffered from cardiac arrest and numerous cases of ventricular fibrillation (7). At the protein level, the D96V mutation strongly impairs Ca2+ binding and CaV1.2 binding and regulation. Hence, we categorize D96V as a ‘severe’ calmodulin mutation (Table 1). As an example of a CPVT mutation, we included N54I (Table 1). This mutation was found in a large family, where most carriers suffered from cardiac arrest or syncope during exercise or acute stress later in childhood (6). In contrast to D96V, the impact of N54I on Ca2+ binding and CaV1.2 binding and regulation are very low (Table 1), and we therefore consider N54I a ‘mild’ mutation. Finally, we included N98S, which has been found in different patients, who suffer from LQTS, CPVT or a combination of these (6,8). The effects of N98S on Ca2+ binding and CaV1.2 binding and regulation are intermediate of D96V and N54I (Table 1). It is important to stress that our mild, intermediate and severe classifications only relate directly to the impact on Ca2+ binding. In patients, all the mutations have a very severe impact on cardiac health.
Table 1.
Overview of arrhythmogenic calmodulin variants
| N54I | D96V | N98S | |
|---|---|---|---|
| Amino acid substitution (HGVS nomenclature)a | NP_008819.1:p.Asn54Ile | NP_001734.1:p.Asp96Val | NP_008819.1:p.Asn98Ser |
| SNP IDa | rs267607276 | rs730882254 | rs267607277 |
| Patient diagnoses | CPVT | LQTS | CPVT/LQTS |
| Mutation position | N-lobe | C-lobe | C-lobe |
| Impact on Ca2+ binding | Mild | Severe | Intermediate |
| Impact on CaV1.2 binding | Mild | Severe | Intermediate |
| Impact on CaV1.2 regulation | Mild | Severe | Intermediate |
| Impact on RyR2 binding | Mild | Severe | Intermediate |
| Impact on RyR2 regulation | Severe | Severe | Severe |
| References | 6,8,14,15,57–60 | 7,8,13,14,57,58,61 | 6,8,13–15,57,58,61–64 |
aWe here exemplify positions of specific patient calmodulin mutations in CALM1 or CALM2, but some mutations have been observed in more than one of the three genes CALM1–3. HGVS, Human Genome Variation Society, SNP, single nucleotide polymorphism.
Until now, most research on calmodulin mutations has been focused on cardiac mechanisms, primarily in cells, and in vitro studies of interactions with specific molecular targets (11–14). Given the wide span of calmodulin-target interactions, it remains a conundrum whether calmodulin mutations affect other tissues than the heart and maybe even cause other diseases. Such studies require more complex models, but few studies in animals have been conducted. Two studies found that in zebrafish, cardiac arrhythmia could be induced by calmodulin mutations N54I, N98S and D130G, but effects on other tissues were not studied (15,16). Similarly, mice carrying the mutation N98S display cardiac effects, but no other phenotypes were reported (17). Although non-cardiac phenotypes can be explored in these models, they are less suited for such studies, as they are expensive, challenging to genetically modify and limited by ethical considerations. Thus, there is a need for a system without these limitations to explore other effects of calmodulin mutations.
With this study we explore the potential of Caenorhabditis elegans as a model system to study calmodulin mutations. Central Ca2+ signaling pathways, proteins and tissues are well conserved in this nematode. C. elegans are small (approximately 1 mm), have a short life cycle (three days from egg to adult) and can easily be genetically modified by editing or genetic crossing to explore specific pathways (18). C. elegans do not have a heart, but the effects of human mutations causing CPVT and LQTS display arrhythmic pharyngeal pumping in the feeding organ (19–21). This is an important advantage, since very pathogenic mutations can be studied without causing cardiac arrest.
We hypothesized that C. elegans could serve as a model to study arrhythmogenic human calmodulin mutations and in addition provide a feasible platform to study how these mutations affect Ca2+ regulated functions in other tissues. Using CRISPR/Cas9, we designed a C. elegans strain expressing the human variant of calmodulin and found that the humanized strain was viable and healthy. In this strain, we introduced three different pathogenic mutations N54I, N98S and D96V identified in humans, representing mild, intermediate and severe defects on Ca2+ binding.
We found that the mild N54I mutation and the severe D96V mutation affected rhythmic behavior in both pharynx pumping and defecation, whereas the intermediate N98S mutation had no effects. Remarkably, N54I, D96V and N98S all affected neuronal function but in different ways, potentially affecting different targets in neurons.
Results
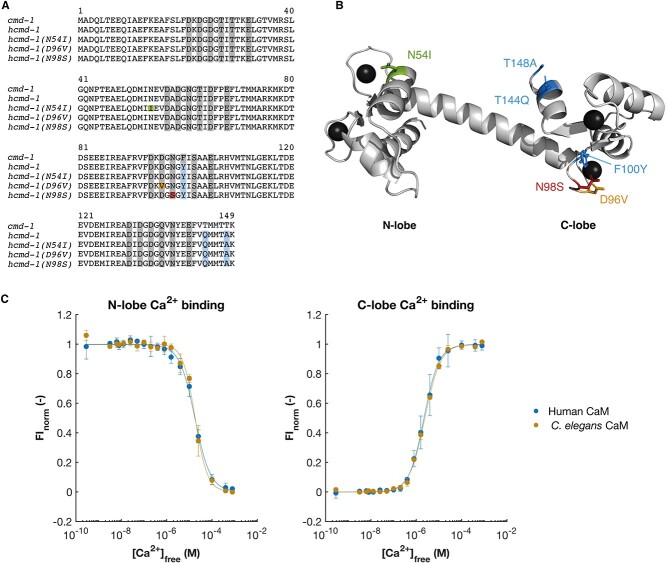
The calmodulin proteins in C. elegans and humans differ in only three amino acid positions: At position 100, there is a conservative substitution from phenylalanine in C. elegans to tyrosine in humans (F100Y). The two other substitutions are T144Q and T148A in the C-terminal end (Fig. 1A and B). As the protein is highly conserved, we first asked if the differences between calmodulin found in humans and in C. elegans were detrimental for the Ca2+ sensing properties of calmodulin. We expressed and purified the native C. elegans and human protein variants and measured their Ca2+-binding affinities. Calmodulin has two lobes, which each bind two Ca2+ ions (Fig. 1B). In both lobes, we found no difference in the Ca2+-binding properties of the human and C. elegans protein variants (Fig. 1C and Table 2). We also found no difference in the Ca2+-dependent interaction of calmodulin to the predicted calmodulin-binding domains of the two important interaction partners, egg-laying defective 19 (EGL-19) (orthologue of CaV1.2) and uncoordinated 68 (UNC-68) (orthologue of RyR2) (Supplementary Material, Fig. S1). As there were no differences in the binding properties of human and C. elegans calmodulin, a C. elegans strain expressing human calmodulin is an appropriate background to study the effects of mutations in the human calmodulin protein.
Figure 1.
Generation of C. elegans strains expressing the human calmodulin protein and three cardiac mutations. (A) Protein sequence alignment of the calmodulin protein expressed in the C. elegans strains in this study: wild-type calmodulin in N2 (expressed from cmd-1), humanized calmodulin expressed in hcmd-1 and three strains expressing the human calmodulin with additional arrhythmogenic mutations (hcmd-1(N54I), hcmd-1(D96V) and hcmd-1(N98S)). Note that the immature numbering of calmodulin residues is used. The initial methionine is cleaved off the mature protein, in which the numbering would be N53I, D95V and N97S. Residues involved in Ca2+ binding are indicated in gray. Residues that differ between human and worm are indicated in blue, and positions of arrhythmia mutations are indicated in green (N54I), orange (D96V) and red (N98S). (B) Protein structure of human calmodulin (PDB: 1cll) in gray with four Ca2+ ions bound (black). Colors as in A. (C) Ca2+ binding to purified human (blue) or C. elegans (orange) calmodulin protein. Binding of Ca2+ induces a structural change in the protein that can be measured as a change in intrinsic fluorescence. P-values from statistical tests are given in Supplementary Material, Table S1. CaM, calmodulin, Flnorm, normalized fluorescence intensity.
Table 2.
Ca2+-binding properties of human and C. elegans calmodulin protein
| N-domain Dissociation Constant, KD,app (mean ± SD, μm) | N-domain Hill Coefficient, n (mean ± SD) | C-domain Dissociation Constant, KD,app (mean ± SD, μm) | C-domain Hill Coefficient, n (mean ± SD) | |
|---|---|---|---|---|
| Human CaM | 17.6 ± 2.9 | 1.30 ± 0.33 | 2.37 ± 0.93 | 1.36 ± 0.21 |
| C. elegans CaM | 18.52 ± 3.4 | 1.63 ± 0.20 | 2.53 ± 0.24 | 1.16 ± 0.15 |
| P-value | 0.74 | 0.23 | 0.79 | 0.26 |
The Ca2+ affinity measured as apparent dissociation constants, KD,app, and Hill coefficients, n, for the two lobes were extracted from a Hill fit of the CaM Ca2+ titration data. The data were compared using Student’s t-test.
Whereas humans have three calmodulin-encoding genes (CALM1-3), C. elegans only expresses calmodulin from one gene (cmd-1). As discussed above, there are only three amino acid differences (F100Y, T144Q, T148A) in the protein produced from the C. elegans and the human genes. We genetically engineered C. elegans with CRISPR/Cas9 to create a ‘humanized’ mutant hcmd-1(F100Y, T144Q, T148A) (from here referred to as hcmd-1). This ‘humanized’ C. elegans strain expresses the human calmodulin protein from a modified version of the endogenous cmd-1 gene (Figs 1A, B and 7 in Materials and Methods). To examine the effect of the pathogenic mutations N54I, D96V and N98S, we used CRISPR/Cas9 to introduce them in the hcmd-1 mutant background. These new strains are referred to as hcmd-1(N54I), hcmd-1(D96V) and hcmd-1(N98S), respectively (Fig. 1A, B and Table 1).
Figure 7.
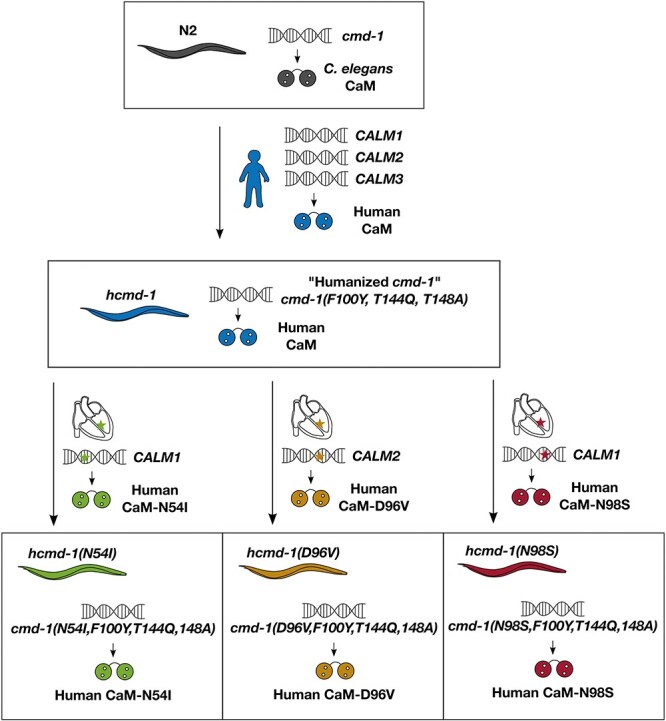
The calmodulin gene cmd-1 in wild-type N2 was modified to express the human form of calmodulin, before introduction of an additional three arrhythmogenic mutations. There are three amino acid differences between human and C. elegans calmodulin protein. In hcmd-1, the endogenous cmd-1 calmodulin gene was modified using CRISPR/Cas9 to express the human form of the protein. Afterwards, three different edits were further introduced to generate three different strains expressing calmodulin with amino acid substitutions identical to those found in arrhythmia patients.
The D96V mutation dramatically affects C. elegans size and fertility
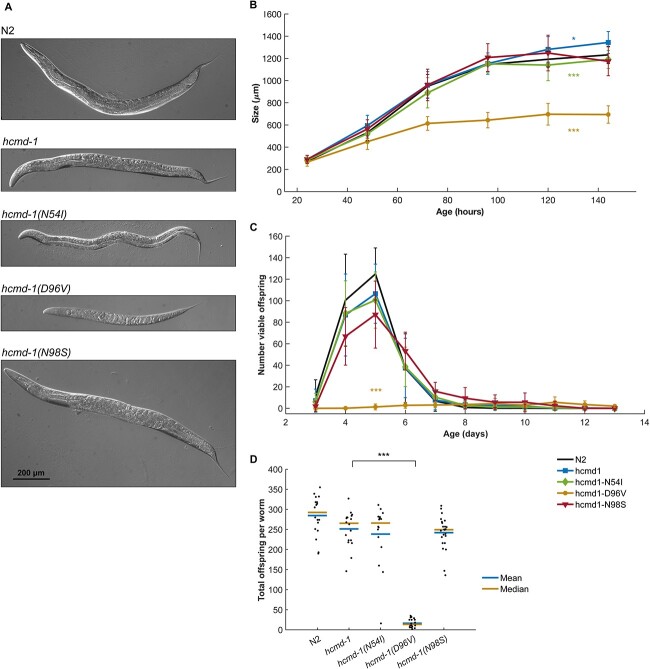
Since calmodulin is widely involved in Ca2+ signaling pathways in the cell, we first asked how these hcmd-1 mutations affected general worm health. We addressed this question by quantifying the size and fertility of the worms (Fig. 2).
Figure 2.
C. elegans carrying the D96V mutation have reduced growth and fertility. (A) Representative examples of four-day-old (4d) C. elegans worms (except hcmd-1(D96V), 6d) expressing the calmodulin protein variants indicated in Figure 1. (B) Length of worms as a function of age. Each data point corresponds to 7–69 worms, pooled from six independent experiments. Error bars show standard deviation. (C) Number of viable offspring per worm per day. n = 27–29 for each strain, pooled from three independent experiments. Error bars show standard deviation. (D) Total number of viable offspring per worm. n = 16–24 for each strain, pooled from three independent experiments. Statistical tests: B, two-way analysis of variance (ANOVA) and Tukey–Kramer post-hoc test. C, repeated measures ANOVA and Tukey–Kramer post-hoc test. D, Student’s t-test with Bonferroni correction. P-values from statistical tests are given in Supplementary Material, Table S1.
The hcmd-1 mutants were slightly, but significantly, larger in size after reaching adulthood compared to wild-type N2 worms, but had otherwise overall normal morphology (Fig. 2A and B). When comparing hcmd-1 to the three strains carrying arrhythmogenic mutations, the D96V mutation, which has the most severe impact on Ca2+ binding, gave the most dramatic effects. The hcmd-1(D96V) worms were small and carried few eggs in utero (Supplementary Material, Figs S2 and S3). Introduction of the mild N54I mutation conferred a small but significant reduction in size after reaching adulthood, but no other effects on morphology were observed. The hcmd-1(N98S) mutants had a very high number of eggs in utero suggesting an Egl phenotype, but in terms of size hcmd-1(N98S) mutants were not different from hcmd-1 worms (Supplementary Material, Figs S2 and S3).
Humanizing calmodulin in hcmd-1 mutants caused an insignificant reduction in viable offspring compared to the wild-type strain N2 (Fig. 2C and D). The introduction of N54I and N98S did not further affect fertility. Furthermore, the N98S mutation caused a slight but insignificant delay in egg laying (Fig. 2C). On the contrary, the D96V mutation caused a significant reduction in fertility. On average, the D96V mutants produced 93.5% fewer viable offspring than the hcmd-1 strain. Further, most hcmd-1(D96V) worms had their first offspring at day 5 or 6, as opposed to day 3 or 4 for the other strains. Therefore, throughout this study we used six-day-old worms of hcmd-1(D96V), whereas all other strains were used at four-day-old to match their stage of development. From differential interference contrast (DIC) microscopy (Supplementary Material, Fig. S2) and DAPI staining (Supplementary Material, Fig. S3), it was clear that hcmd-1(D96V) mutants had under-proliferated germlines with very few germ cells which is consistent with the very reduced fertility.
Together, these experiments showed that the humanizing genetic edits and the two arrhythmogenic mutations N54I and N98S had no or minor effects on worm fertility and growth. On the other hand, the arrhythmogenic mutation D96V had severe effects on both fertility and growth. Interestingly, this suggests that the severity of Ca2+-binding defects in human calmodulin variants is reflected in phenotypic strength in C. elegans.
N54I and D96V mutations disrupt pharynx pumping
The pharynx is the feeding organ of C. elegans, and it shows many physiological and mechanical properties similar to the human heart (22,23). The pharynx is composed of 20 muscle cells, which are connected by gap junctions and controlled by a mostly autonomous nervous system. Rhythmic contractions (pumping) of the pharynx allow C. elegans to eat bacteria. In the presence of food, the pharynx pumps at a high frequency, and both human CPVT and LQTS mutations in other genes can disrupt its rhythmicity (19–21).
We quantified how the calmodulin mutations affected pharynx pumping in C. elegans (Fig. 3A). As positive control, the egl-19(n2368) mutant was included. This mutant carries a point mutation in the voltage-gated Ca2+ channel EGL-19. The n2368 allele is a known LQTS mutation in the orthologous human protein CaV1.2 (21,24,25).
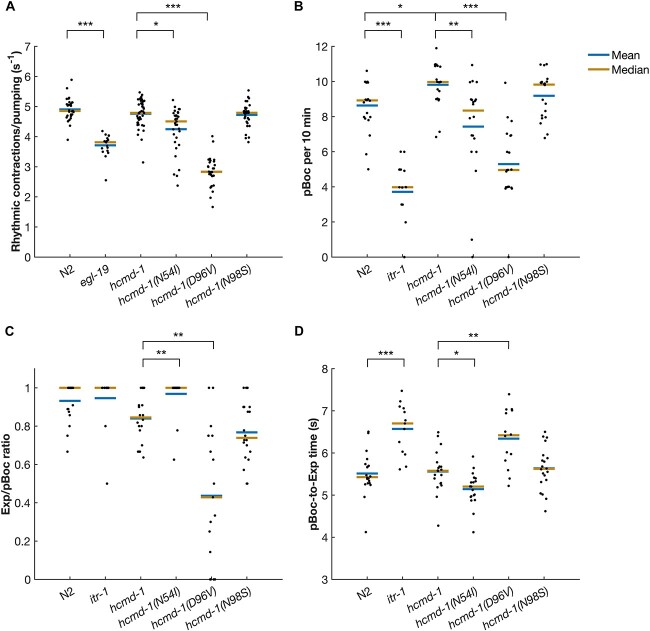
Figure 3.
N54I and D96V mutations impair rhythmic behavior in pharynx pumping and DMP. (A) The number of pharynx pumps was manually counted from movies of worms feeding on OP50. n = 18–43, pooled from four independent experiments. (B–D) The two muscle contractions pBoc (P) and Exp (X) in DMP were recorded by observing actively feeding worms. n = 15–20, pooled from 10 independent experiments. Example ethograms from DMP experiments are shown in Supplementary Material, Figure S4. (B) Number of pBoc contractions per 10 min. (C) Calculated number of Exp contractions per pBoc per worm. (D) Average time from pBoc to Exp. Statistical tests: A–C, Wilcoxon rank-sum test with Bonferroni correction. D, Student’s t-test with Bonferroni correction. P-values from statistical tests are given in Supplementary Material, Table S1.
The humanizing genetic edits in calmodulin did not affect pharynx pumping rate, and neither did the N98S mutation (Fig. 3A). On the other hand, the mild N54I mutation caused a significant 10.8% reduction in pumping rate compared to hcmd-1. The severe D96V mutation reduced the pumping rate by 40.5%, which was more than the egl-19(n2368) LQTS positive control. Thus, N54I and D96V both affected pharynx pumping rate in C. elegans, despite having different Ca2+-binding properties and being located in the N- and C-terminal lobes, respectively.
N54I and D96V mutations disrupt rhythmical behavior in the defecation motor program
Motivated by the effects on pharynx pumping and the difference in impact from the N54I and D96V mutations, we turned our attention to another rhythmic behavior in C. elegans, namely the defecation motor program (DMP), which is regulated by Ca2+ signaling (26). This autonomous motor program is activated approximately once every minute in actively feeding worms and consists of a distinct set of muscle contractions (27). The DMP cycle is initiated by an easily recognizable contraction in the posterior body-wall muscles (pBoc, P). This is followed by a smaller contraction in the anterior body-wall muscles (aBoc). Finally, the gut content is expelled by a characteristic contraction of enteric muscles—referred to as Expulsion (Exp, X). Knock-down studies using RNAi against calmodulin in the intestine have shown titratable effects on the DMP cycle regulation, accompanied by disrupted Ca2+ signals (28).
Several genes are known to regulate specific steps of the DMP cycle. Here, we included the itr-1(sa73) mutant strain as a positive control. These mutants carry a mutation in the nematode IP3 receptor, which is a Ca2+ channel and a main initiator of the DMP cycle. We find that itr-1(sa73) mutants initiate fewer DMP cycles (Fig. 3B) as previously reported (29).
The humanizing mutations caused a small but significant increase in cycle frequency compared to N2 (Fig. 3B and Supplementary Material, Fig. S4). Like for the rate of pharynx pumping, we found a reduction in DMP cycle frequency for hcmd-1(N54I) and hcmd-1(D96V) mutants compared to hcmd-1, measured as the number of pBoc contractions per 10 min (Fig. 3B and Supplementary Material, Fig. S4). The N98S mutation did not have any effects on DMP cycle frequency.
We then asked whether these mutations affected the ability of the worms to complete the full motor program by quantifying the number of finalized Exp events relative to initiated pBoc events (Fig. 3C). The hcmd-1(D96V) mutant frequently failed to complete the DMP cycle (43.7% complete cycles). Conversely, hcmd-1(N54I) mutants displayed significantly more complete DMP cycles (96.9%) compared to the hcmd-1 controls (84.0%). Finally, we quantified the time interval between pBoc and Exp contractions (Fig. 3D). The D96V mutation caused a 14.0% increase in pBoc-to-Exp time, whereas the N54I mutants had reduced time interval by 7.5% compared to hcmd-1 controls.
In summary, we found that the humanizing edits and the N98S mutation had no or small effects on DMP regulation. On the other hand, regulation of DMP cycle initiation and termination was impaired in hcmd-1(D96V) mutants. The N54I mutation caused a reduction in the initiation frequency of the DMP cycle, but a small increase in termination efficiency.
Rhythmic behaviors are not altered by calmodulin mutations due to a systemic disruption of muscle function
Together, the data presented above suggest that the arrhythmogenic calmodulin mutations N54I and D96V, but not N98S, affect rhythmicity in C. elegans. Since calmodulin is involved in many Ca2+ signaling pathways and thus is key in regulating muscle function, we speculated whether these effects could result from a general impairment of muscle function.
To answer this question, we turned our attention to motility as a general measure of muscle function. We measured the body bend frequency (thrashing) during swimming (Fig. 4A). As positive control, we included the slowly moving unc-68(r1162) strain, which expresses a loss-of-function version of the Ca2+ channel UNC-68, the nematode ryanodine receptor. We did not see any effects on thrashing frequency by the humanizing edits or the N98S mutation. The N54I mutation decreased thrashing frequency by 31.4%, while the D96V mutation resulted in a 20.3% increase, compared to hcmd-1 (Fig. 4A). It is intriguing that two arrhythmogenic mutations had opposite effects and that mutants harboring the severe D96V mutation displayed increased thrashing frequency. Therefore, we quantified maximum speed of worms moving on nematode growth medium (NGM) plates with food, and we found the same trends as for thrashing, however not statistically significant (Fig. 4B). When assessing the average speed measured as the track length moved in 30 s, we did not find any significant effects of the genetic modifications (Fig. 4C).
Figure 4.
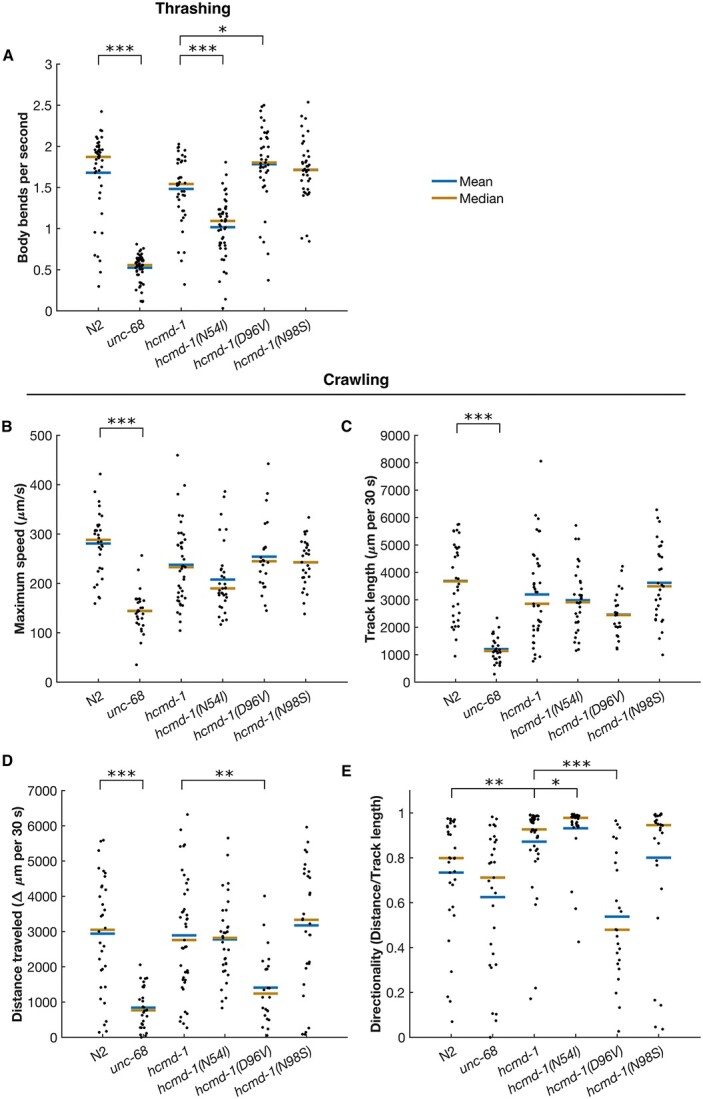
N54I and D96V have opposite effects on worm motility. (A) Average speed of body bends during swimming/thrashing. n = 39–53, pooled from five independent experiments. (B–E) Motility of worms on plates. n = 24–41, pooled from four individual experiments. (B) Maximum speed. (C) Total length of track in 30 s (speed). (D) Net distance traveled from start to end in 30 s. (E) Directionality measured as the net distance traveled relative to the total track length. Statistical tests: A–B, Student’s t-test with Bonferroni correction. C–E, Wilcoxon rank-sum test with Bonferroni correction. P-values from statistical tests are given in Supplementary Material, Table S1.
Finally, we asked if the crawling worms moved directly forward, or if they turned or reversed during movement. Thus, we quantified the net distance traveled from start to end of the 30 s migration. As a measure of directionality, we calculated the ratio between the net distance moved and the track length for each worm (Fig. 4D and E) (30). We found that the humanizing genetic edits caused a significant 10.6% increase in directionality, which was further increased by the N54I mutation by 6.7%. N98S gave an insignificant 8.4% decrease, whereas the D96V mutation had the strongest effect on directionality, showing a 38.4% reduction compared to hcmd-1.
In summary, we found that both the humanizing genetic edits and that the N54I and D96V mutations affected motility, the latter two in opposite directions. We however note that all strains were motile, implying that disruption of rhythmicity cannot alone be explained by a general impairment of muscle function.
Arrhythmogenic mutations influence neuronal function
We found that N54I and D96V had opposite effects on DMP termination and directionality during movement. These functions are under neuronal regulation (27,31,32). Calmodulin is expressed in neurons (33), and Ca2+ signaling plays an important role in the release of neurotransmitters and proper neurotransmission (34). Therefore, we hypothesized that the calmodulin mutations could affect the function of neurons innervating muscles involved in rhythmic behavior.
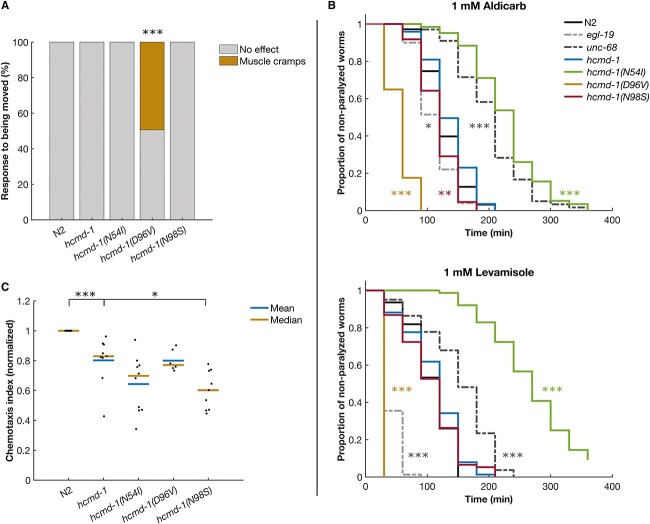
Interestingly, during maintenance of the worms, we observed that hcmd-1(D96V) exhibited systemic muscle cramps or full body convulsions (Fig. 5A, Supplementary Material, Movies S1 and S2). These convulsions were induced by moving the worms with a platinum picker between NGM plates during routine maintenance. Furthermore, we observed that when moving the same worm repeatedly, the convulsions increased in frequency and duration (data not shown). Since similar convulsions have been reported as a result of defective neurotransmission (35), we speculated that the calmodulin mutations negatively affect neuromuscular signaling.
Figure 5.
C. elegans with the D96V calmodulin mutation are hypersensitive to aldicarb and levamisole, whereas N54I carriers are resistant. (A) Systemic full body convulsions when transferring worms with platinum picker. n = 75 for each strain. See Supplementary Material, Movies S1 and S2 for an example. (B) Worms were placed on NGM plates with 1 mm aldicarb (top) or levamisole (bottom). Every 30 min, paralyzed worms were counted and removed. n = 57–79, pooled from three independent experiments. Asterisks in gray indicate differences from N2. Asterisks in color indicate differences from hcmd-1. (C) Worms were placed on plates with drops of the chemoattractant benzaldehyde or a control buffer. A chemotaxis index of 1 indicates that all worms moved to the chemoattractant. A chemotaxis index of 0 indicates random movement. n = 6–11 experiments combining a total of 211–1463 worms. Statistical tests: A, Fisher’s Exact test. B, Bonferroni-corrected log rank test. C, Bonferroni-corrected Wilcoxon rank-sum test. P-values from statistical tests are given in Supplementary Material, Table S1.
We addressed this using a well-established paralysis assay with aldicarb and levamisole to identify presynaptic (neuronal) and postsynaptic (muscular) effects, respectively. These drugs target cholinergic signaling at the neuromuscular junction (36,37). As controls, we again included egl-19(n2368) and unc-68(r1162), as egl-19 and unc-68 both affect cholinergic signaling (24,38,39). Aldicarb (a cholinesterase inhibitor) causes accumulation of acetylcholine in the synapse. If a mutation affects the efficiency of neurotransmitter release, paralysis will happen faster or slower. Consistent with previous studies, the unc-68 null allele was hyposensitive to aldicarb (38), and the LQTS egl-19 allele was slightly, but significantly, hypersensitive (Fig. 5B, upper panel). Interestingly, we found that hcmd-1(N54I) were hyposensitive to aldicarb, whereas hcmd-1(D96V) were hypersensitive. There was a small although significant increase in sensitivity for hcmd-1(N98S) as well (Fig. 5B, upper panel).
Levamisole (a nicotinic acetylcholine receptor agonist) directly activates the acetylcholine receptor causing continued muscle stimulation and therefore identifies postsynaptic effects (40). We found a protective effect of N54I and increased sensitivity in the hcmd-1(D96V) mutant, indicating that these mutations also affect acetylcholine reception and/or its downstream targets, again in opposite directions (Fig. 5B, lower panel). This is consistent with calmodulin having both pre- and postsynaptic effects and importantly shows that these functions can be separated by specific mutations in calmodulin.
To determine if the presynaptic effects of calmodulin mutations would manifest in other neuronal defects, we examined their effect on chemotaxis. Here, worms are allowed to move freely on a plate prepared with two drops of benzaldehyde and two drops of a control buffer in opposite quadrants and with equal distance to the center of the plate. Benzaldehyde is a chemoattractant sensed by the AWC neurons in C. elegans (41). Azide was applied to each spot to paralyze the worms upon arrival at the test or control spots. After 60 min, we found that almost all N2 controls had found their way to the chemoattractant. The hcmd-1 worms were 19.8% less efficient in sensing the chemoattractant compared to N2 (Fig. 5C). Chemotaxis was further reduced in the hcmd-1(N54I) and hcmd-1(N98S) mutants by 19.8% and 25.0%, respectively. Remarkably, despite that D96V has the most severe Ca2+-binding defect, the chemotaxis index of the hcmd-1(D96V) mutants was not different compared to hcmd-1.
Together, we find neuronal effects of arrhythmogenic calmodulin mutations in C. elegans. Briefly, D96V strongly attenuates signaling at the neuromuscular junction. N54I has opposite effects to D96V at the neuromuscular junction. Although hcmd-1(N98S) has shown no or weak effects in rhythmic behavior and motility, we find that thus mutant shows the strongest chemotaxis defect.
Discussion
Calmodulin is extremely conserved across species but not trivial to study in humans where carriers of calmodulin mutations are exceedingly rare and deadly. Furthermore, calmodulin is highly pleiotropic which adds to the complexity of studying calmodulin variants in vivo, but at the same time stresses the need for using model organisms. To overcome these obstacles, we have established C. elegans as a model to study human calmodulin mutations. In C. elegans, the calmodulin gene cmd-1 is essential and RNAi results in embryonic lethality (42). Therefore, the role of calmodulin in C. elegans is largely underexplored, and no mutation has yet been characterized. The C. elegans calmodulin protein differs from human calmodulin only in three amino acids, and we find that the two proteins have similar Ca2+ and target-binding properties in vitro. Consistent with this, humanizing the native C. elegans calmodulin in vivo does not cause any strong phenotypes across the investigated traits. This is in stark contrast to the introduction of calmodulin variants known to affect Ca2+ binding and to cause cardiac arrhythmia in humans, which cause a range of phenotypes in the worms.
C. elegans as a model of arrhythmia induced by human calmodulin mutations
Together, we find that the arrhythmogenic effect of the mutations N54I and D96V, but not N98S, can be recapitulated in assays of pharynx pumping and DMP in C. elegans (Fig. 6).
Figure 6.
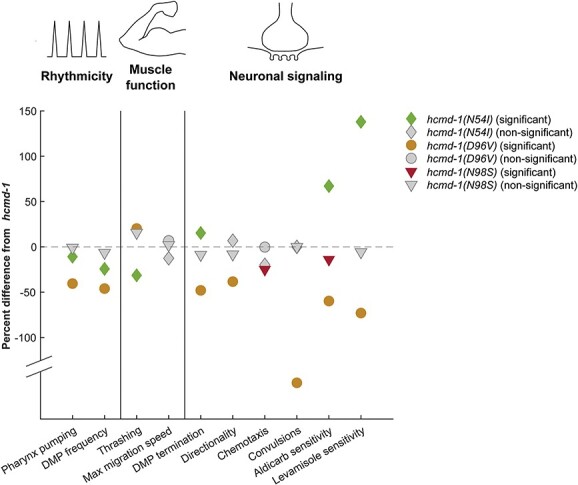
Data summary. Mean values for indicated experiments were extracted, and the difference from hcmd-1 was calculated. For convulsions, the results are shown as ‘lack of convulsions’ to reflect the negative effects of calmodulin mutations. For the paralysis assays, the mean paralysis time was used to calculate the difference. Color-filled symbols indicate statistical significance in the figures presented.
The CPVT-mutation N54I has mild or no effects on Ca2+ binding, and it is one of the only mutations found in the N-terminal lobe of the protein (3,6,8). N54I can induce arrhythmia in zebrafish (15). In this study, we found significant effects on rhythmical behavior in C. elegans, but the animals were otherwise healthy with normal morphology, fertility and development.
On the contrary, the LQTS-mutation D96V is one of the most severe calmodulin mutations, manifesting already prenatally (7). In C. elegans, D96V also manifested severely, resulting in animals that could only just reproduce. The mutation had strong effects on rhythmical behavior, correlating well with the phenotype found in humans. hcmd-1(D96V) worms were fully motile with a slight increase in speed compared to controls. This intact muscle function is important to note as it demonstrates that D96V does not systemically disrupt all Ca2+ signaling in C. elegans.
This study did not find arrhythmia effects of N98S in humanized C. elegans. Interestingly, this mutation has been found in several patients, who have been diagnosed with CPVT, LQTS or a combination of the two (6,8). Due to this incongruity, the N98S mutation has been modeled in mice, which did demonstrate arrhythmic effects, however not very dramatic (17). Whether our results are a consequence of the N98S mutation having a special interaction pattern generally or because it does not affect rhythmical behavior in nematodes specifically, remains to be shown. Other studies also find that not all human arrhythmia mutations cause arrhythmia in C. elegans (19,20).
Although biochemically very different, both N54I and D96V mutations affect worm rhythmicity. N54I is located in the N-terminal lobe and is not involved in Ca2+ binding. In contrast, D96V strongly affects a central Ca2+-binding residue in the C-terminal lobe. These observations emphasize the importance of using both in vitro and in vivo approaches to study human disease, as they contribute complementing observations. Moreover, it displays the strength of using simple model organisms as the phenotypic differences observed could not have been predicted solely based upon Ca2+ binding.
Calmodulin mutations impair neuronal function in C. elegans
Several previous studies have found that calmodulin is important for neuronal development and function in both C. elegans, Drosophila and mice (33,43,44). A robust association between neuronal effects and arrhythmogenic calmodulin mutations has, however, never been demonstrated. We find different neuronal effects for N54I, D96V and N98S. The mutation D96V increases aldicarb/levamisole sensitivity and decreases directional movement and DMP termination efficiency, which are also regulated by neurons (Fig. 6). Interestingly, the N54I mutation displays an opposite pattern, as it gives resistance to aldicarb and levamisole and increases DMP termination efficiency. We propose that these two mutations cause similar effects on rhythmic behavior but disrupt neuronal function through different downstream mechanisms. For example, D96V may affect a range of targets through disrupted Ca2+ sensing, while N54I may affect specific targets that depend on biochemical interactions with this specific residue.
Supporting this notion, only the hcmd-1(D96V) mutants display whole body convulsions after being transferred between plates. Similar convulsions have previously been reported following treatment of uncoordinated-43 (unc-43) null mutants with the γ-aminobutyric acid (GABA) receptor antagonist pentylenetetrazole (35). unc-43 encodes a homolog of the Ca2+/calmodulin-dependent protein kinase II (CaMKII) which is considered a general regulator of synaptic plasticity (35,45). There are few studies available on the effects of arrhythmogenic calmodulin mutations on CaMKII, and the results are inconsistent (16,46,47). Therefore, further studies are required to elucidate whether hcmd-1(D96V) mutants experience whole body convulsions due to defective signaling via unc-43 or GABAergic signaling.
The difference between D96V and N54I in aldicarb and levamisole resistance is interesting. We speculate that N54I causes resistance toward chronic excitation of neurons. Such a protective effect is supported by a recent study showing that the decline of neuronal plasticity of O2-sensing neurons (URX, AQR and PQR) with age is mediated via calmodulin in C. elegans (48). This study finds that knock down of calmodulin using cell-specific RNAi can counteract plasticity decline observed with age. We propose that a similar mechanism could be at play for the N54I mutation which may not interact correctly with specific targets and in this regard mimics a calmodulin knock down. The aldicarb/levamisole sensitivity observed for the severe D96V mutation could be due to a near complete and systemic disruption of Ca2+/calmodulin signaling. Alternatively, this mutation may affect other targets than those affected by the N54I mutation, such as isoforms of voltage-gated Ca2+ channels, which are also affected in the heart (14) or unc-43/CaMKII and/or the GABA signaling pathway as discussed above.
That different downstream molecular mechanisms are influenced by the calmodulin mutations is further supported by the chemotaxis analysis. Surprisingly, the D96V mutation with strongest impact on Ca2+ did not affect chemosensing. The neurological effects of D96V are therefore not systemic. On the contrary, the N98S mutation significantly impaired chemotaxis and aldicarb sensitivity but had no or minor effects on rhythmicity and motility. In chemotaxis, we also found that the humanizing edits in themselves significantly impacted benzaldehyde sensing. Benzaldehyde is one of several chemoattractants used in C. elegans (49). Whether the effects of the calmodulin mutations on chemotaxis could be chemoattractant specific remains to be shown. Together these observations support the hypothesis that calmodulin mutations can impact neuronal function, and that different calmodulin mutations can affect different downstream pathways in neurons.
There are several other assays to study neuronal regulated behavior in worms. C. elegans display an interesting phenotype called social feeding where members of a population aggregate on the bacterial lawn (50). Social feeding is considered a model of psychiatric illness, and many antipsychotic drugs inhibit social feeding (51). Interestingly, RNAi mediated knock down of calmodulin also inhibits social feeding, further supporting an important neuronal role of calmodulin.
Implications for understanding human calmodulin mutations
Human calmodulin mutations are extremely rare. New patient carriers are collected in the International Calmodulinopathy Registry (8), which in 2019 reported 74 subjects, of whom 86.5% suffered from cardiac arrhythmia. Interestingly, 13 of these subjects also showed mild-to-severe neurological impairment. Whether these neurological effects were consequences of life-threatening cardiac events early in life is unknown. With the data presented here in mind, the hypothesis that calmodulin mutations could have neurological consequences in humans seems very likely.
In this study, we establish C. elegans as a model of cardiac arrhythmia induced by human calmodulin mutations. We demonstrate that the three arrhythmogenic mutations N54I, D96V and N98S have neurological effects, likely through different mechanisms. We thus suggest that neurological consequences of calmodulin mutations will originate from different molecular pathways, and their severity may not be predictable from what we know about calmodulin mutations today. These results are new to the domain of calmodulin mutations and underscore how animal models can contribute new and more complex information. Calmodulin is highly pleiotropic, but the fact that the studied human calmodulin mutations display separable phenotypes is promising from a drug development point of view. Future studies into underlying mechanisms have potential to form the basis for designing highly specific drug interventions impacting specific downstream pathways.
Materials and Methods
C. elegans strains and maintenance
The wild-type N2 strain was used as basis for generation of the hcmd-1 strains (Fig. 7): OLS160 hcmd-1(aar160[F100Y, T144Q, T148A]) referred to as hcmd-1, OLS178 hcmd-1(aar178[N54I]) referred to as hcmd-1(N541), OLS183 hcmd-1(aar183[(D96V]) referred to as hcmd-1(D96V) and OLS171 hcmd-1(aar171[(N98S]) referred to as hcmd-1(N98S). The strains JT73 itr-1(sa73), MT6129 egl-19(n2368) and TR2171 unc-68(r1162) were purchased from the Caenorhabditis Genetic Center (CGC, Minnesota, USA).
All strains were maintained on NGM plates at 20°C. NGM plates were prepared from 3 g/L NaCl, 2.5 g/L soy peptone, 17 g/L agar, supplemented with 5 mg/mL cholesterol, 1 mm CaCl2, 1 mm MgSO4 and 25 mm potassium phosphate buffer (stock is 132 mm K2HPO4, 868 mm KH2PO4, (pH 6)) and spotted with OP50 Escherichia coli grown over-night in LB medium (10 g/L NaCl, 10 g/L soy peptone, 5 g/L agar, pH 7.5). For maintenance, eggs were transferred to fresh plates 1–2 times per week. Unless otherwise stated, worms were used in experiments as four-day-old adults, except hcmd-1(D96V), which were six days old.
CRISPR/Cas9 Oligos
crRNAs and single-stranded oligodeoxynucleotides (ssODNs) were designed in the online software Benchling (https://benchling.com) and ordered with PAGE purification from Sigma-Aldrich. crRNAs cleaving >10 bases away from the edit site were chosen when possible. ssODNs were made with 35–40 bp homology arms, and for silent mutations codon usage was accounted for (52).
Trans-activating CRISPR (tracr)RNA, guide CRISPR (cr)RNAs and ssODN repair templates were dissolved in TRIS buffer and kept at −20°C. Sequences and stock concentrations can be found in Supplementary Material, Tables S2 and S3.
CRISPR/Cas9 injection protocol
The humanized calmodulin worm strain hcmd-1 and the following disease mutants were made using the dpy-10 co-CRISPR strategy described by Arribere and colleagues (53) and the direct delivery of in vitro-synthesized and -assembled Cas9-crRNA-tracrRNA complexes described by Paix and colleagues (54). For all injections, the 10 μL injection mixture was prepared as described (54), with modified concentrations. 15 μm tracrRNA, 13 μm crRNA-X and 2 μm dpy-10(cn64) were mixed and placed in a thermocycler (5 min at 95°C, 5 min at 21°C). 15 μm Cas9 (produced in-house) was added and left at room temperature for 5 min. Last, ssODN-X, ssODN dpy-10(cn64) and nuclease-free H2O up to 10 μL was added. The injection mix was spun down at 13200 rpm for 4 min at 4°C, incubated at 37°C for 30 min and spun down again for 10 min. N2 worms were injected to make the hcmd-1 strain which was backcrossed against N2 before introducing the disease mutations (N54I, N98S and D96V). Disease mutants were backcrossed three times to hcmd-1.
Microinjection needles
Needles for microinjection were made by pulling Borosilicate Glass Capillaries (1B110F-4) (World precision instruments) using Narishige’s PC-100 micropipette puller with heater 1 set at 58 and no setting for heater 2. Injections were made using a FemtoJet® 4× (Eppendorf) with an injection pressure of 200 hPa.
Mounting C. elegans for injections
Young adult worms were transferred to a droplet of Halocarbon oil on a glass slide with a 2% dried agarose pad. DIC on a Leica DNI300B microscope was applied for visualization of animal gonads. Animals were recovered in S-basal buffer (100 mm NaCl, 50 mm KH2PO4) and one to three animals were placed on each NGM plate to lay eggs for four days at 16°C.
Screening for transgenes and mutants
F1 animals on jackpot brood plates (54) were screened by identifying individuals with a roller phenotype (53). Briefly, rollers were singled out and tested by polymerase chain reaction (PCR) after egg lay to identify individuals with the desired edit, and F2 wild-type worms from positive mothers were then singled out and tested by PCR to identify homozygote animals of the desired edit. Finally, the CRISPR edits were verified by Illumina sequencing.
Genotyping of C. elegans stains
During gene editing work and before every experiment, correct identity of worms was ensured by PCR (N2, TR2171 and hcmd-1 strains) or by visual inspection/quantification of characteristic traits. For PCR, 1–10 worms were frozen in lysis buffer (50 mm KCl, 10 mm Tris (pH 8.3), 2.5 mm MgCl2, 0.45% NP-40, 0.45% Tween-20, 0.01% (w/v) gelatin) at -80°C for at least 30 min. The lysates were then heated to 60°C for 60 min, followed by 95°C for 15 min. PCR reactions were prepared using primers indicated in Supplementary Material, Table S4 in a DreamTaq polymerase master mix (Thermo Scientific, K9022), following the manufacturer’s instructions. The PCR reactions were then analyzed in 1% or 2% agarose gels stained with Sybr Safe (Invitrogen, S33102) and developed on a ChemiDoc MP imager (Bio-Rad).
Fertility assay
To measure the number of viable offspring for each worm, eggs of each strain were placed on separate NGM plates. After 3 days and subsequently every 24 h, the now adult worms were moved to new plates. The plates that worms were removed from were returned to the incubator for another two days to allow the offspring to develop to large larvae, easing subsequent quantification. Plates were then removed from the incubator, and the offspring was counted. In some cases, plates were stored at 4°C before counting the offspring.
C. elegans size measurements
Adult hermaphrodites were placed on fresh NGM plates and allowed to lay eggs for 2 h. The adults were then removed. The offspring was imaged at 24 h intervals on an Olympus SZ61 stereomicroscope with an ANDOR Zyla 4.2 sCMOS camera and KL 300 LED lamp, managed by NIS Elements software. The length of each worm was analyzed using the Segmented Line tool in Fiji ImageJ.
Muscle convulsion assay
Adult worms were transferred to fresh NGM plates. Worms from each strain were then moved one at a time to another place on the plate using a platinum picker in the same way as during routine maintenance. After the worm had been placed on the plate, it was observed whether it began normal movement or convulsed. The experiment was blinded and performed by two different individuals.
DMP assay
Adult worms were moved to a fresh NGM plate. Each worm was followed for 10 min while feeding on OP50 and then sacrificed. If the worm moved away from the bacterial lawn, the experiment was stopped. pBoc and Exp muscle contractions were recorded using custom-written software. aBoc contractions were not recorded as they can easily be overlooked. The experiment was blinded.
Pharynx pump assay
Adult worms were imaged for 15–20 s on an Olympus SZ61 stereomicroscope equipped with an ANDOR Zyla 4.2 sCMOS camera and KL 300 LED lamp, managed by NIS Elements software. The number of pumps was manually counted using the Cell counter tool in Fiji ImageJ.
Thrashing
Adult worms were transferred to droplets of 100 μL S-basal (100 mm NaCl, 50 mm KH2PO4 (pH 6)), avoiding clumps of bacteria in the suspension. The worms were imaged for 30 s at 15 fps on an Olympus SZX16 stereomicroscope equipped with a DP72 camera and managed with Olympus CellB software.
Motility on plates
NGM plates with thin, homogeneous OP50 lawns were generated by evenly distributing four to five drops of an OP50 over-night culture on NGM plates 12–16 h before the experiment. Adult worms were placed in a corner of the plate and allowed to move away from this region for at least 15 min. The worms were imaged for at 15 fps on an Olympus SZX16 stereomicroscope equipped with a DP72 camera and managed by Olympus CellB software.
Imaging of C. elegans morphology
Adult worms were placed on agarose pads with a 10 μL drop of S-basal and covered with a coverslip. The worms were imaged on an Olympus IX83 inverted microscope equipped with a Hamamatsu Orca-Flash 4.0 camera, using a 10× UPLSAPO objective and managed by Olympus CellSens software. For DAPI staining, worms were fixed in ice-cold methanol for 5 min before being mounted in EverBrite™ Hardset Mounting medium (Biotium 23004).
Paralysis assay
NGM plates containing either aldicarb (Sigma-Aldrich, 33386) or levamisole (MP Biomedicals, 155228) were prepared by first making 100 mm stock solutions in 70% ethanol (aldicarb) or H2O (levamisole). The stock solutions were added to NGM medium directly before casting plates to reach a final concentration of 1 mm. 30 μL over-night OP50 bacterial culture was added 30 min prior to the start of the assay. 20–25 age-matched worms were transferred to the drug-containing plates. Every 30 min the worms were poked up to three times with a platinum picker. If this did not provoke movement and there was no visible pharynx pumping, the worms were scored as paralyzed and removed.
Chemotaxis assay
5 cm NGM plates were marked into four equal quadrants with a 1 cm radius circle made with origo at the center of the plate. Each pair of opposite quadrants was labelled C (control) or T (test) and a test-spot was marked in the center of the quadrant, at least 2 cm from the center circle. To this, thrice washed adult worms were applied to the center. Simultaneously, 1 μL 1 M NaN3 was applied to the test-spot in all quadrants along with 1 μL 1:200 benzaldehyde in EtOH (B1334, Sigma-Aldrich) in the T quadrants and 1 μL EtOH in C quadrants. After 60 min, the number of worms in each quadrant was manually counted.
Calmodulin protein expression and purification
Full-length human and C. elegans calmodulin were expressed in E. coli Rosetta (DE3), both from a modified pET vector containing an N-terminal maltose-binding protein (MBP) and a tobacco etch virus (TEV) cleavage site. The MBP–TEV–calmodulin fusion protein was purified on an amylose affinity column (New England Biolabs) and subsequently cleaved by a TEV protease. Following cleavage, calmodulin, TEV and MBP were separated by anion ion exchange chromatography using a Q-sepharose column (GE Healthcare). The calmodulin-containing fractions were pooled and concentrated before removing residual Ca2+ by adding 20 mm ethylenediaminetetraacetic acid (EDTA) to the sample prior to applying it to a Superdex 75 size exclusion column (GE Healthcare). The identity and integrity of each protein preparation was confirmed by matrix-assisted laser desorption/ionization-time of flight mass spectrometry. Protein concentrations were determined by absorption at 280 nm.
Calmodulin Ca 2+-binding affinity
Ca2+ titrations were done as previously described (9). Briefly, this assay was based on measuring intrinsic calmodulin fluorescence using a spectrofluorometer (HORIBA Jobin Yvon, FluoroMax-4P). In preparation for this, discontinuous titration points of different free Ca2+ concentrations, [Ca2+]free, were prepared and added 30 μm calmodulin before sequentially transferring each solution to a 3 by 3 mm cuvette for fluorescence emission spectra recording. Measurements were done in triplicate.
Ca2+-binding curves for the N- and C-lobes of calmodulin were extracted from the fluorescence intensity (FI) signals from Phe and Tyr emission at 280 and 310 nm, respectively, and the data were fitted according to the generic Hill equation:
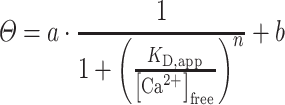 |
where b and a indicate the initial FI-plateau and the span in FI from low to high [Ca2+]free, respectively. n is the Hill coefficient and KD,app is the apparent Ca2+ dissociation constant. Curve fitting was done to the raw FI data using GraphPad Prism 8.4.2, whereas data normalization was only done for visualization purposes.
Calmodulin-binding affinity to EGL-19 and UNC-68 peptides
A two-dimensional titration assay was used to determine the affinity of human and C. elegans calmodulin for binding to EGL-19 and UNC-68 peptides at 4 discrete [Ca2+]. Peptides with an N-terminal 5-TAMRA (5-carboxytetramethylrhodamine) label were obtained from Proteogenix. The separate [Ca2+] conditions were mixed by an automated liquid handling robot (Hamilton MicroLab STARlet). 3 μL CaM (∼600 μm) was added to the first column of a 384-well plate (Corning) allowing for 24 dilutions of CaM (column-wise) at each of the four different free Ca2+ concentrations (row-wise). Meanwhile, the peptide concentration was kept constant (∼30 nm) in all wells. Calmodulin binding to either peptide was then monitored by measuring the fluorescence anisotropy (FA) signal of the 5-TAMRA-labeled peptide in a fluorescence plate reader (Spark Tecan, Zurich, Switzerland). For further details and graphical explanations of the assay, please refer to our previous work (55). The data from the resulting Ca2+-dependent calmodulin/EGL-19 or calmodulin/UNC-68-binding curves were fitted according to a stoichiometric equation:
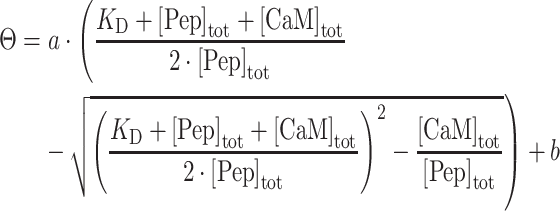 |
where b and a indicate the initial FA-plateau and the span in FA from low to high [CaM], respectively. [Pep]tot and [CaM]tot are the total concentrations of peptide and calmodulin, respectively, and KD is the apparent calmodulin dissociation constant. Measurements were done in triplicate and curve fitting was done to the raw FA data using GraphPad Prism 8.4.2.
Image analysis
All image analysis was performed in Fiji ImageJ (freely available from NIH). In images of morphology, brightness/contrast settings were gently adjusted without compromising the image data. Time-lapse acquisitions of motility were analyzed using the wrMTRCK plugin (56).
Data analysis and statistics
Numeric data were collected in Microsoft Excel. Figure graphs and statistical tests were prepared using MathWorks MATLAB. All data were evaluated for normal distribution before selecting statistical analyses. Statistical significance is indicated as *P < 0.05, **P < 0.01, ***P < 0.001. Where indicated these levels are Bonferroni-corrected. All statistical tests and P-values are given in Supplementary Material, Table S1. Figures were prepared in Inkscape.
Supplementary Material
Acknowledgements
We thank students and members of the laboratory for contributing to the optimization of assays and establishment of the worm strains. In particular, we thank Nikolaj Døssing Bak for his work on developing protocols for motility assays. We also owe big thanks to Jesper Halkjær Jensen for his help with coding and statistics. Some strains were provided by the CGC, which is funded by the National Institute of Health Office of Research Infrastructure Programs (P40 OD010440).
Conflict of Interest statement. All authors declare no conflict of interest.
Contributor Information
Helene H Jensen, Department of Chemistry and Bioscience, Aalborg University, Aalborg Ø 9220, Denmark.
Magnus T Frantzen, Department of Chemistry and Bioscience, Aalborg University, Aalborg Ø 9220, Denmark.
Jonas L Wesseltoft, Department of Chemistry and Bioscience, Aalborg University, Aalborg Ø 9220, Denmark.
Ana-Octavia Busuioc, Department of Chemistry and Bioscience, Aalborg University, Aalborg Ø 9220, Denmark.
Katrine V Møller, Department of Chemistry and Bioscience, Aalborg University, Aalborg Ø 9220, Denmark.
Malene Brohus, Department of Chemistry and Bioscience, Aalborg University, Aalborg Ø 9220, Denmark.
Palle R Duun, Department of Health Science and Technology, Aalborg University, Aalborg Ø 9220, Denmark.
Mette Nyegaard, Department of Health Science and Technology, Aalborg University, Aalborg Ø 9220, Denmark.
Michael T Overgaard, Department of Chemistry and Bioscience, Aalborg University, Aalborg Ø 9220, Denmark.
Anders Olsen, Department of Chemistry and Bioscience, Aalborg University, Aalborg Ø 9220, Denmark.
Funding
Lundbeckfonden (R250-2017-134 to H.H.J., R324-2019-1933 to M.T.O. and R324-2019-2035 to A.O.), the Independent Research Fund Denmark (2032-00333B to M.T.O.), Carlsberg Foundation (CF19-0668 to A.O.) and Obelske Familiefond.
Data availability
All data is available upon request.
References
- 1. Clapham, D.E. (2007) Calcium signaling. Cell, 131, 1047–1058. [DOI] [PubMed] [Google Scholar]
- 2. Berridge, M.J., Bootman, M.D. and Roderick, H.L. (2003) Calcium signalling: dynamics, homeostasis and remodelling. Nat. Rev. Mol. Cell Biol., 4, 517–529. [DOI] [PubMed] [Google Scholar]
- 3. Jensen, H.H., Brohus, M., Nyegaard, M. and Overgaard, M.T. (2018) Human Calmodulin mutations. Front. Mol. Neurosci., 11. 10.3389/fnmol.2018.00396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sorensen, A.B., Søndergaard, M.T. and Overgaard, M.T. (2013) Calmodulin in a heartbeat. FEBS J., 280, 5511–5532. [DOI] [PubMed] [Google Scholar]
- 5. Halling, D.B., Liebeskind, B.J., Hall, A.W. and Aldrich, R.W. (2016) Conserved properties of individual Ca2+-binding sites in calmodulin. Proc. Natl. Acad. Sci., 113, E1216–E1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nyegaard, M., Overgaard, M.T., Søndergaard, M.T., Vranas, M., Behr, E.R., Hildebrandt, L.L., Lund, J., Hedley, P.L., Camm, A.J., Wettrell, G. et al. (2012) Mutations in Calmodulin cause ventricular tachycardia and sudden cardiac death. Am. J. Hum. Genet., 91, 703–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Crotti, L., Johnson, C.N., Graf, E., De Ferrari, G.M., Cuneo, B.F., Ovadia, M., Papagiannis, J., Feldkamp, M.D., Rathi, S.G., Kunic, J.D. et al. (2013) Calmodulin mutations associated with recurrent cardiac arrest in infants. Circulation, 127, 1009–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Crotti, L., Spazzolini, C., Tester, D.J., Ghidoni, A., Baruteau, A., Beckmann, B., Behr, E.R., Bennett, J.S., Bezzina, C.R., Bhuiyan, Z.A. et al. (2019) Calmodulin mutations and life-threatening cardiac arrhythmias: insights from the international Calmodulinopathy registry. Eur. Heart J., 40, 2964–2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Brohus, M., Arsov, T., Wallace, D.A., Jensen, H.H., Nyegaard, M., Crotti, L., Adamski, M., Zhang, Y., Field, M.A., Athanasopoulos, V. et al. (2020) Infanticide vs. inherited cardiac arrhythmias. EP Eur., 23, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nyegaard, M. and Overgaard, M.T. (2019) The international Calmodulinopathy registry: recording the diverse phenotypic spectrum of un-CALM hearts. Eur. Heart J., 40, 2976–2978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yamamoto, Y., Makiyama, T., Harita, T., Sasaki, K., Wuriyanghai, Y., Hayano, M., Nishiuchi, S., Kohjitani, H., Hirose, S., Chen, J. et al. (2017) Allele-specific ablation rescues electrophysiological abnormalities in a human iPS cell model of long-QT syndrome with a CALM2 mutation. Hum. Mol. Genet., 26, 1670–1677. [DOI] [PubMed] [Google Scholar]
- 12. Limpitikul, W.B., Dick, I.E., Tester, D.J., Boczek, N.J., Limphong, P., Yang, W., Choi, M.H., Babich, J., DiSilvestre, D., Kanter, R.J. et al. (2017) A precision medicine approach to the rescue of function on malignant Calmodulinopathic long-QT syndrome. Circ. Res., 120, 39–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Søndergaard, M.T., Liu, Y., Brohus, M., Guo, W., Nani, A., Carvajal, C., Fill, M., Overgaard, M.T. and Chen, S.R.W. (2019) Diminished inhibition and facilitated activation of RyR2-mediated Ca2+ release is a common defect of arrhythmogenic calmodulin mutations. FEBS J., 286, 4554–4578. [DOI] [PubMed] [Google Scholar]
- 14. Limpitikul, W.B., Dick, I.E., Joshi-Mukherjee, R., Overgaard, M.T., George, A.L. and Yue, D.T. (2014) Calmodulin mutations associated with long QT syndrome prevent inactivation of cardiac L-type Ca2+ currents and promote proarrhythmic behavior in ventricular myocytes. J. Mol. Cell. Cardiol., 74, 115–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Søndergaard, M.T., Sorensen, A.B., Skov, L.L., Kjaer-Sorensen, K., Bauer, M.C., Nyegaard, M., Linse, S., Oxvig, C. and Overgaard, M.T. (2015) Calmodulin mutations causing catecholaminergic polymorphic ventricular tachycardia confer opposing functional and biophysical molecular changes. FEBS J., 282, 803–816. [DOI] [PubMed] [Google Scholar]
- 16. Berchtold, M.W., Zacharias, T., Kulej, K., Wang, K., Torggler, R., Jespersen, T., Chen, J.N., Larsen, M.R. and La Cour, J.M. (2016) The arrhythmogenic calmodulin mutation D129G dysregulates cell growth, calmodulin-dependent kinase II activity, and cardiac function in zebrafish. J. Biol. Chem., 291, 26636–26646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tsai, W.-C., Guo, S., Olaopa, M.A., Field, L.J., Yang, J., Shen, C., Chang, C.-P., Chen, P.-S. and Rubart, M. (2020) Complex arrhythmia syndrome in a knock-in mouse model carrier of the N98S Calm1 mutation. Circulation, 142, 1937–1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Corsi, A.K., Wightman, B. and Chalfie, M. (2015) A transparent window into biology: a primer on Caenorhabditis elegans. Genetics, 200, 387–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Engel, M.A., Wörmann, Y.R., Kaestner, H. and Schüler, C. (2022) An Optogenetic arrhythmia model—insertion of several Catecholaminergic polymorphic ventricular tachycardia mutations into Caenorhabditis elegans UNC-68 disturbs Calstabin-mediated stabilization of the ryanodine receptor homolog. Front. Physiol., 13. 10.3389/fphys.2022.691829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fischer, E., Gottschalk, A. and Schüler, C. (2017) An optogenetic arrhythmia model to study catecholaminergic polymorphic ventricular tachycardia mutations. Sci. Rep., 7, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schüler, C., Fischer, E., Shaltiel, L., Steuer Costa, W. and Gottschalk, A. (2015) Arrhythmogenic effects of mutated L-type Ca2+-channels on an optogenetically paced muscular pump in Caenorhabditis elegans. Sci. Rep., 5, 14427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shtonda, B. and Avery, L. (2005) CCA-1, EGL-19 and EXP-2 currents shape action potentials in the Caenorhabditis elegans pharynx. J. Exp. Biol., 208, 2177–2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Avery, L. and You, Y. (2012) C. elegans feeding. WormBook, 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Laine, V., Segor, J.R., Zhan, H., Bessereau, J.-L. and Jospin, M. (2014) Hyperactivation of L-type voltage-gated Ca2+ channels in Caenorhabditis elegans striated muscle can result from point mutations in the IS6 or the IIIS4 segment of the 1 subunit. J. Exp. Biol., 217, 3805–3814. [DOI] [PubMed] [Google Scholar]
- 25. Kerr, R., Lev-Ram, V., Baird, G., Vincent, P., Tsien, R.Y. and Schafer, W.R. (2000) Optical imaging of calcium transients in neurons and pharyngeal muscle of C. elegans. Neuron, 26, 583–594. [DOI] [PubMed] [Google Scholar]
- 26. Peters, M.A., Teramoto, T., White, J.Q., Iwasaki, K. and Jorgensen, E.M. (2007) A calcium wave mediated by gap junctions coordinates a rhythmic behavior in C. elegans. Curr. Biol., 17, 1601–1608. [DOI] [PubMed] [Google Scholar]
- 27. Branicky, R. and Hekimi, S. (2006) What keeps C. elegans regular: the genetics of defecation. Trends Genet., 22, 571–579. [DOI] [PubMed] [Google Scholar]
- 28. Allman, E., Waters, K., Ackroyd, S. and Nehrke, K. (2013) Analysis of Ca2+ Signaling motifs that regulate proton Signaling through the Na+/H+ exchanger NHX-7 during a rhythmic behavior in Caenorhabditis elegans. J. Biol. Chem., 288, 5886–5895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dal Santo, P., Logan, M.A., Chisholm, A.D. and Jorgensen, E.M. (1999) The inositol trisphosphate receptor regulates a 50-second behavioral rhythm in C. elegans. Cell, 98, 757–767. [DOI] [PubMed] [Google Scholar]
- 30. Jensen, H.H., Pedersen, G.A., Morgen, J.J., Parsons, M., Pedersen, S.F. and Nejsum, L.N. (2019) The Na+/H+ exchanger NHE1 localizes as clusters to cryptic lamellipodia and accelerates collective epithelial cell migration. J. Physiol., 597, 849–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hums, I., Riedl, J., Mende, F., Kato, S., Kaplan, H.S., Latham, R., Sonntag, M., Traunmüller, L. and Zimmer, M. (2016) Regulation of two motor patterns enables the gradual adjustment of locomotion strategy in Caenorhabditis elegans. Elife, 5, 1–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Peliti, M., Chuang, J.S. and Shaham, S. (2013) Directional locomotion of C. elegans in the absence of external stimuli. PLoS One, 8, e78535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Vuong-Brender, T.T., Flynn, S., Vallis, Y. and de Bono, M. (2021) Neuronal calmodulin levels are controlled by CAMTA transcription factors. Elife, 10, 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Brini, M., Calì, T., Ottolini, D. and Carafoli, E. (2014) Neuronal calcium signaling: function and dysfunction. Cell. Mol. Life Sci., 71, 2787–2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Williams, S.N., Locke, C.J., Braden, A.L., Caldwell, K.A. and Caldwell, G.A. (2004) Epileptic-like convulsions associated with LIS-1 in the cytoskeletal control of neurotransmitter signaling in Caenorhabditis elegans. Hum. Mol. Genet., 13, 2043–2059. [DOI] [PubMed] [Google Scholar]
- 36. Mahoney, T.R., Luo, S. and Nonet, M.L. (2006) Analysis of synaptic transmission in Caenorhabditis elegans using an aldicarb-sensitivity assay. Nat. Protoc., 1, 1772–1777. [DOI] [PubMed] [Google Scholar]
- 37. Oh, K. and Kim, H. (2017) Aldicarb-induced paralysis assay to determine defects in synaptic transmission in Caenorhabditis elegans. BIO-PROTOCOL, 7. 10.21769/BioProtoc.2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Graham, B., Shaw, M.-A. and Hope, I.A. (2020) Single amino acid changes in the ryanodine receptor in the human population have effects in vivo on Caenorhabditis elegans neuro-muscular function. Front. Genet., 11, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yan, Z., Cheng, X., Li, Y., Su, Z., Zhou, Y. and Liu, J. (2022) Sexually dimorphic neurotransmitter release at the neuromuscular junction in adult Caenorhabditis elegans. Front. Mol. Neurosci., 14, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gottschalk, A., Almedom, R.B., Schedletzky, T., Anderson, S.D., Yates, J.R. and Schafer, W.R. (2005) Identification and characterization of novel nicotinic receptor-associated proteins in Caenorhabditis elegans. EMBO J., 24, 2566–2578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Torayama, I., Ishihara, T. and Katsura, I. (2007) Caenorhabditis elegans integrates the signals of butanone and food to enhance chemotaxis to butanone. J. Neurosci., 27, 741–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Karabinos, A., Büssing, I., Schulze, E., Wang, J., Weber, K. and Schnabel, R. (2003) Functional analysis of the single calmodulin gene in the nematode Caenorhabditis elegans by RNA interference and 4-D microscopy. Eur. J. Cell Biol., 82, 557–563. [DOI] [PubMed] [Google Scholar]
- 43. VanBerkum, M.F.A. and Goodman, C.S. (1995) Targeted disruption of Ca2+-calmodulin signaling in drosophila growth cones leads to stalls in axon extension and errors in axon guidance. Neuron, 14, 43–56. [DOI] [PubMed] [Google Scholar]
- 44. Bae, B., Gruner, H.N., Lynch, M., Feng, T., So, K., Oliver, D., Mastick, G.S., Yan, W., Pieraut, S. and Miura, P. (2020) Elimination of Calm1 long 3′-UTR mRNA isoform by CRISPR–Cas9 gene editing impairs dorsal root ganglion development and hippocampal neuron activation in mice. RNA, 26, 1414–1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yasuda, R., Hayashi, Y. and Hell, J.W. (2022) CaMKII: a central molecular organizer of synaptic plasticity, learning and memory. Nat. Rev. Neurosci., 23, 666–682. [DOI] [PubMed] [Google Scholar]
- 46. Hwang, H.S., Nitu, F.R., Yang, Y., Walweel, K., Pereira, L., Johnson, C.N., Faggioni, M., Chazin, W.J., Laver, D., George, A.L. et al. (2014) Divergent regulation of ryanodine receptor 2 calcium release channels by arrhythmogenic human calmodulin missense mutants. Circ. Res., 114, 1114–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Prakash, O., Gupta, N., Milburn, A., McCormick, L., Deugi, V., Fisch, P., Wyles, J., Thomas, N.L., Antonyuk, S., Dart, C. et al. (2022) Calmodulin variant E140G associated with long QT syndrome impairs CaMKIIδ autophosphorylation and L-type calcium channel (Cav1.2) inactivation. J. Biol. Chem., 307, 102777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Li, Q., Marcu, D.-C., Palazzo, O., Turner, F., King, D., Spires-Jones, T.L., Stefan, M.I. and Busch, K.E. (2020) High neural activity accelerates the decline of cognitive plasticity with age in Caenorhabditis elegans. Elife, 9, 1–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wes, P.D. and Bargmann, C.I. (2001) C. elegans odour discrimination requires asymmetric diversity in olfactory neurons. Nature, 410, 698–701. [DOI] [PubMed] [Google Scholar]
- 50. de Bono, M., Tobin, D.M., Davis, M.W., Avery, L. and Bargmann, C.I. (2002) Social feeding in Caenorhabditis elegans is induced by neurons that detect aversive stimuli. Nature, 419, 899–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Dwyer, D.S., Awatramani, P., Thakur, R., Seeni, R. and Aamodt, E.J. (2015) Social feeding in Caenorhabditis elegans is modulated by antipsychotic drugs and calmodulin and may serve as a protophenotype for asociality. Neuropharmacology, 92, 56–62. [DOI] [PubMed] [Google Scholar]
- 52. Sharp, P.M. and Bradnam, K.R. (1997) In Riddle, D.L., Blumenthal, T., Meyer, B.J. and Priess, J.R. (eds), C. elegans II: Appendix 3 Codon Usage in C. elegans, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 53. Arribere, J.A., Bell, R.T., Fu, B.X.H., Artiles, K.L., Hartman, P.S. and Fire, A.Z. (2014) Efficient marker-free recovery of custom genetic modifications with CRISPR/Cas9 in Caenorhabditis elegans. Genetics, 198, 837–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Paix, A., Folkmann, A., Rasoloson, D. and Seydoux, G. (2015) High efficiency, homology-directed genome editing in Caenorhabditis elegans using CRISPR-Cas9ribonucleoprotein complexes. Genetics, 201, 47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Brohus, M., Søndergaard, M.T., Wayne Chen, S.R., van Petegem, F. and Overgaard, M.T. (2019) Ca2+-dependent calmodulin binding to cardiac ryanodine receptor (RyR2) calmodulin-binding domains. Biochem. J., 476, 193–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Nussbaum-Krammer, C.I., Neto, M.F., Brielmann, R.M., Pedersen, J.S. and Morimoto, R.I. (2015) Investigating the spreading and toxicity of prion-like proteins using the metazoan model Organism C. elegans. J. Vis. Exp., 95, e52321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Vassilakopoulou, V., Calver, B.L., Thanassoulas, A., Beck, K., Hu, H., Buntwal, L., Smith, A., Theodoridou, M., Kashir, J., Blayney, L. et al. (2015) Distinctive malfunctions of calmodulin mutations associated with heart RyR2-mediated arrhythmic disease. Biochim. Biophys. Acta Gen. Subj., 1850, 2168–2176. [DOI] [PubMed] [Google Scholar]
- 58. Søndergaard, M.T., Tian, X., Liu, Y., Wang, R., Chazin, W.J., Chen, S.R.W. and Overgaard, M.T. (2015) Arrhythmogenic calmodulin mutations affect the activation and termination of cardiac ryanodine receptor-mediated Ca2+ release. J. Biol. Chem., 290, 26151–26162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wang, K., Brohus, M., Holt, C., Overgaard, M.T., Wimmer, R. and Van Petegem, F. (2020) Arrhythmia mutations in calmodulin can disrupt cooperativity of Ca2+ binding and cause misfolding. J. Physiol., 598, 1169–1186. [DOI] [PubMed] [Google Scholar]
- 60. Søndergaard, M.T., Liu, Y., Guo, W., Wei, J., Wang, R., Brohus, M., Overgaard, M.T. and Chen, S.R.W. (2020) Role of cardiac ryanodine receptor calmodulin-binding domains in mediating the action of arrhythmogenic calmodulin N-domain mutation N54I. FEBS J., 287, 2256–2280. [DOI] [PubMed] [Google Scholar]
- 61. Wang, K., Holt, C., Lu, J., Brohus, M., Larsen, K.T., Overgaard, M.T., Wimmer, R. and Van Petegem, F. (2018) Arrhythmia mutations in calmodulin cause conformational changes that affect interactions with the cardiac voltage-gated calcium channel. Proc. Natl. Acad. Sci., 115, E10556–E10565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Jiménez-Jáimez, J., Doza, J.P., Ortega, Á., Macías-Ruiz, R., Perin, F., Rodríguez-Vázquez Del Rey, M.M., Ortiz-Genga, M., Monserrat, L., Barriales-Villa, R., Blanca, E. et al. (2016) Calmodulin 2 mutation N98S is associated with unexplained cardiac arrest in infants due to low clinical penetrance electrical disorders. PLoS One, 11, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Makita, N., Yagihara, N., Crotti, L., Johnson, C.N., Beckmann, B.M., Roh, M.S., Shigemizu, D., Lichtner, P., Ishikawa, T., Aiba, T. et al. (2014) Novel calmodulin mutations associated with congenital arrhythmia susceptibility. Circ. Cardiovasc. Genet., 7, 466–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Fujita, S., Nakagawa, R., Futatani, T., Igarashi, N., Fuchigami, T., Saito, S., Ohno, S., Horie, M. and Hatasaki, K. (2019) Long QT syndrome with a de novo CALM2 mutation in a 4-year-old boy. Pediatr. Int., 61, 852–858. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data is available upon request.