Abstract
Macroautophagy/autophagy is a conserved catabolic pathway that is vital for maintaining cell homeostasis and promoting cell survival under stressful conditions. Dysregulation of autophagy is associated with a variety of human diseases, such as cancer, neurodegenerative diseases, and metabolic disorders. Therefore, this pathway must be precisely regulated at multiple levels, involving epigenetic, transcriptional, post-transcriptional, translational, and post-translational mechanisms, to prevent inappropriate autophagy activity. In this review, we focus on autophagy regulation at the transcriptional level, summarizing the transcription factors that control autophagy gene expression in both yeast and mammalian cells. Because the expression and/or subcellular localization of some autophagy transcription factors are altered in certain diseases, we also discuss how changes in transcriptional regulation of autophagy are associated with human pathophysiologies.
Subject terms: Macroautophagy, Epigenetics
Facts
Autophagy is under the control of a wide range of transcription regulators, and some of them can respond to changes in the environment. Therefore, autophagy is under precise control at the transcriptional level.
Some transcription factors are altered under disease conditions, and the subsequent dysregulation of autophagy is highly associated with certain diseases.
Transcription factors regulating autophagy have the potential to be developed into therapeutic targets.
Open questions
Most studies focus on the transcriptional regulation of nonselective autophagy; but are there specific transcription factors involved in selective autophagy?
Some autophagy transcription factors have transcription-independent functions, and some can regulate other cellular processes besides autophagy; will alteration of these functions interfere with the expected upregulation or downregulation of autophagy when modifying the activity of these transcription factors?
The same transcription factor may regulate autophagy in opposite directions depending on the cell type, and different cells have distinct responses to autophagy alterations. How are these transcription factors controlled in a cell type-specific manner?
Introduction
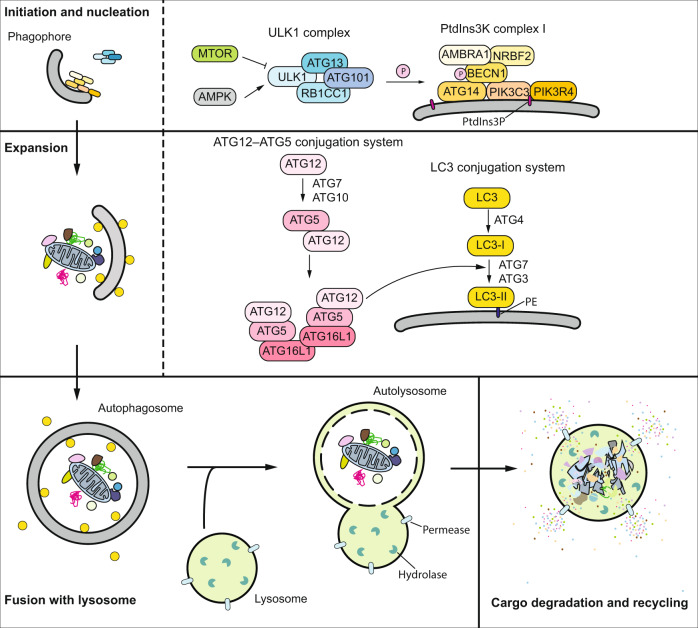
Macroautophagy (hereafter autophagy) is a conserved cellular degradation and recycling process, where a double-membrane structure termed the phagophore sequesters autophagic cargos and sends them to the lysosome (or the vacuole in plants and fungi) [1]. Depending on the specificity of the cargos, autophagy can be either nonselective or selective: nonselective autophagy cargos are relatively random portions of the cytoplasm, whereas the selective autophagic cargos are usually specific proteins or organelles, which are targeted to the phagophore through receptors and scaffold proteins [2]. The mechanism of autophagy has been well-documented, and is carried out by autophagy related (ATG) proteins. The entire autophagy process can be broken down into four sequential steps: (1) initiation and phagophore nucleation, (2) phagophore expansion and maturation into a completed autophagosome, (3) fusion between the autophagosome and an endosome and/or lysosome/vacuole, and (4) cargo degradation and recycling (Fig. 1) [3]. Because the autophagy process is conserved in yeast and mammalian cells and most mammalian ATG protein homologs can be found in yeast, here, we will only introduce the autophagy mechanism in mammalian cells due to space limitations. Yeast Atg proteins and protein complexes involved in each step of autophagy are listed in Table 1.
Fig. 1. Autophagy in mammalian cells and the essential protein complexes.
Following the induction of the ULK1 complex, which phosphorylates and activates PtdIns3K complex I, nucleation of the phagophore occurs. The expansion is facilitated by the two ubiquitin-like systems, which mediate the formation of the ATG12–ATG5-ATG16L1 complex and the conjugation of Atg8-family proteins with PE. When the expanding membrane closes and a complete autophagosome is generated, the outer membrane will fuse with the lysosome and forms an autolysosome; potential fusion with an endosome to generate an intermediate amphisome is not depicted for simplicity. The cargo within the autophagosome will subsequently be degraded and recycled.
Table 1.
Atg proteins in yeast and the primary autophagy step they are involved in [136].
| Complex | Atg components | Function | |
|---|---|---|---|
| Atg1 complex | Atg1, Atg13, Atg17, Atg29, Atg31 | Induction and nucleation | |
| PtdIns3K complex I | Atg14, Atg38, Vps15, Vps30, Vps34 | Induction and nucleation | |
| Conjugation system | Atg12–Atg5 conjugation system | Atg5, Atg7, Atg10, Atg12, Atg16 | Expansion |
| Atg8–PE conjugation system | Atg3, Atg4, Atg7, Atg8, Atg12–Atg5-Atg16 complex | Expansion | |
In mammalian cells, in response to MTORC1 (negative) and AMPK (positive) signaling, autophagy induction is regulated by the ULK1 kinase complex, which consists of the catalytic subunit ULK1, regulatory scaffold proteins ATG13 and RB1CC1, and the stabilizing protein ATG101 [4]. The activated ULK1 complex phosphorylates and activates the class III phosphatidylinositol 3-kinase (PtdIns3K) complex, which is comprised of the catalytic subunit PIK3C3, the putative tumor suppressor BECN1 and regulators including PIK3R4, ATG14, NRBF2 and AMBRA1 (Fig. 1) [5, 6]. Activated PtdIns3K produces phosphatidylinositol-3-phosphate (PtdIns3P), defining the region of phagophore initiation through recruiting PtdIns3P-effector proteins including WIPI2 and ZFYVE1/DFCP1 [7]. Following nucleation, the phagophore expands through the action of two ubiquitin-like systems. In the first system, ATG12 is conjugated with ATG5 through the E1-like enzyme ATG7 and the E2-like enzyme ATG10, and then forms a complex with ATG16L1. In the second system, Atg8-family proteins, including MAP1LC3/LC3 and GABARAP subfamilies, undergo cleavage by ATG4, followed by their conjugation to phosphatidylethanolamine (PE) via the E1-like enzyme ATG7, the E2-like enzyme ATG3 and the E3-like ATG12–ATG5-ATG16L1 complex (Fig. 1) [8]. ATG16L1 binds to WIPI2 directly, allowing the conjugation to occur at the phagophore membrane [9]. The phagophore will expand into a cup-shaped structure that sequesters cytoplasmic components and the closure of the phagophore generates the autophagosome. Ultimately, the outer membrane of the autophagosome fuses first with an endosome and subsequently, or directly, with the lysosome, where the inner membrane and engulfed contents are degraded and recycled following permease-mediated release back into the cytosol [10].
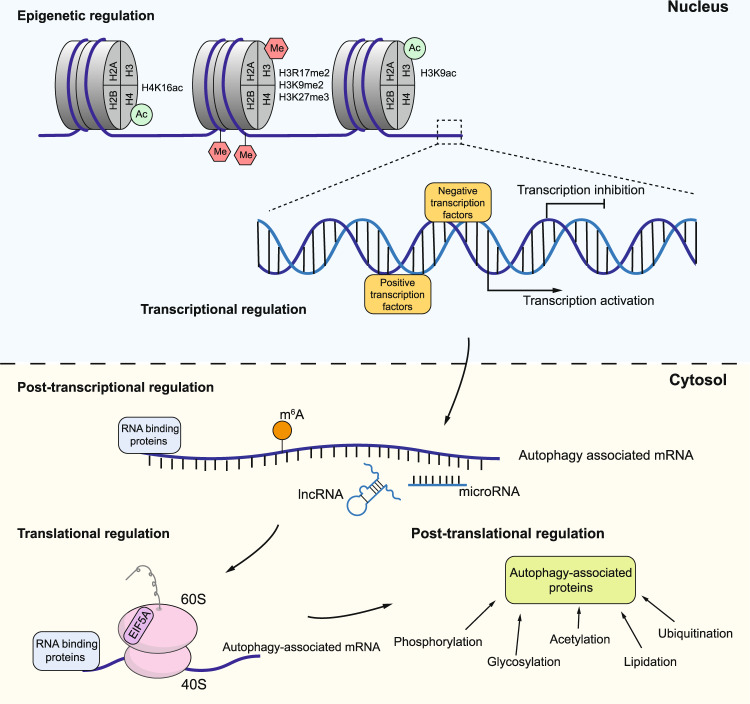
Under normal conditions, autophagy is at a basal level to maintain cellular homeostasis. When the cells are exposed to stressful conditions, such as nutrient deprivation and hypoxia, autophagy is induced to a high level and promotes the turnover of cytoplasmic material for energy replenishment and removing superfluous or damaged organelles. Therefore, autophagy must be well controlled and either too much or too little autophagy is associated with various types of diseases [11]. Autophagy regulation occurs at multiple levels, including epigenetic, transcriptional, post-transcriptional, translational, and post-translational regulation (Fig. 2). In this review, we focus on the transcriptional regulation of autophagy in both the yeast S. cerevisiae and mammalian systems, summarizing the transcription factors that control autophagy gene expression and discussing their implications in human diseases.
Fig. 2. Autophagy regulation at different levels.
In the nucleus, autophagy can be regulated at the epigenetic level, including DNA methylation and histone modifications (such as methylation and acetylation) and at the transcription level, which is basically controlled by the binding and release of transcription factors. After transcription, post-transcriptional regulation is performed by RNA binding proteins, non-coding RNAs and mRNA modification. Translation of autophagic mRNA can be regulated by some RNA binding proteins and controlled by the proteins in the core translation system, such as the requirement of EIF5A in ATG3 translation. After ATG protein synthesis, specific components can undergo post-translational modification, which affects protein activity and stability.
Transcriptional regulation of yeast autophagy
In 2015, a large-scale analysis of DNA binding proteins was published, which identified several transcription factors of ATG genes, including both positive and negative regulators (Table 2) [12]. The activities of some transcription factors can be regulated by post-translational modifications in response to nutrient change. TORC1 is the best-known kinase regulated by nutrient status. When TORC1 is inactivated by nitrogen starvation, the activated phosphatase Sit4 and PP2A (Pph21/Pph22-Tpd3-Cdc55) will induce the translocation of the two GATA type transcription factors, Gln3 and Gat1, into the nucleus [13], where they promote the transcription of ATG7, ATG8, ATG9, ATG29 and ATG32 [12]. Gcn4 is another transcription factor that is induced when TORC1 is inhibited by nutrient deprivation and gcn4 deletion leads to a decrease in ATG1 mRNA level and autophagy activity during starvation [12, 14]. A recent study reported that Gcn4 is essential for the specific binding of Rpb9, an RNA polymerase II component, to the promoter of ATG1, thus promoting the transcription of ATG1 during starvation [15]. Additionally, Gcn4 is responsible for the expression of ATG41, a gene required for efficient autophagy (Fig. 3) [16].
Table 2.
Yeast transcription factors regulating ATG gene expression.
| Transcription factor | Conditions | Effects (+, positive; − negative) | Target genes | Ref. |
|---|---|---|---|---|
| Rsc1 | Starvation | + | ATG8 | [137] |
| Fyv5 | Nutrient-rich and starvation | – | ATG1, 7, 8, 9,14, 29, 32 | [12] |
| Gat1 | Starvation | + | ATG7, 8, 9, 29, 32 | [12] |
| Gcn4 | Starvation | + | ATG1,41 | [12, 16] |
| Gln3 | Starvation | + | ATG7, 8, 9, 29, 32 | [12] |
| Rph1 | Nutrient-rich | – | ATG7, 8, 9, 14, 29, 32 | [19] |
| Sfl1 | Nutrient-rich | + | ATG1, 7, 8, 9, 14, 29, 32 | [12] |
| Sko1 | Nutrient-rich and starvation | – | ATG1, 7, 8, 32 | [12] |
| Spt10 | Nutrient-rich | – | ATG1, 7, 9, 14, 32 | [12] |
| Spt4-Spt5 | Nutrient-rich | – | ATG8, 41 | [21] |
| Starvation | + | ATG41 | ||
| Swi5 | Nutrient-rich | + | ATG7, 8, 9, 14, 29 | [12] |
| Ume6 | Nutrient-rich | – | ATG8 | [18] |
| Zap1 | Nutrient-rich | – | ATG1, 7, 8, 9, 14, 29, 32 | [12] |
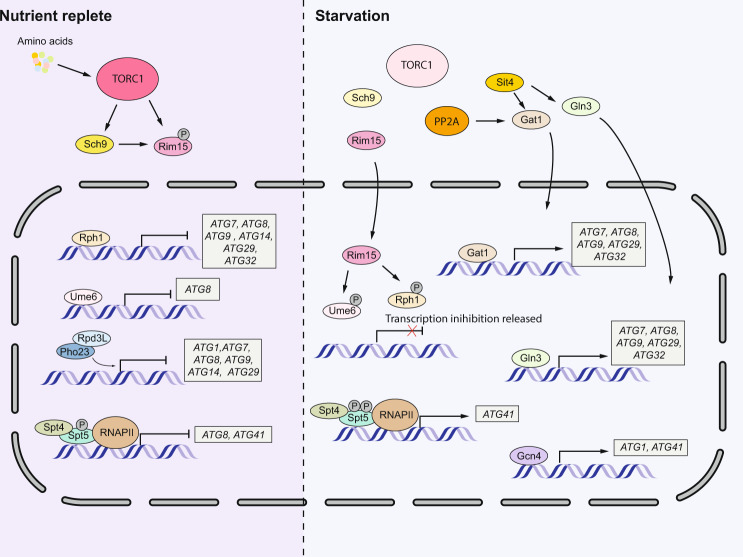
Fig. 3. Transcriptional regulation of autophagy in yeast.
Under nutrient-rich conditions, TORC1 is activated. Multiple transcription repressors can bind to the promoter region of some core ATG genes, thus inhibiting autophagy. During starvation, TORC1 is inactivated. Rim15 will translocate into the nucleus and inhibit the transcription repression due to Ume6 and Rph1. Gln3, Gat1 and Gcn4 will also translocate into the nucleus and induce the transcription of ATG genes. The Spt4-Spt5 complex plays dual roles in regulating ATG gene transcription, inhibiting transcription when nutrients are replete and promoting transcription during starvation, which depends on the phosphorylation status of Spt5.
Rim15 also plays a pivotal role in controlling the function of autophagy transcription factors in yeast. Under nutrient-rich conditions, Rim15 is phosphorylated by TORC1 and Sch9, which inhibits its nuclear translocation. During nitrogen starvation, TORC1 is inhibited and Tps2 is involved in the dephosphorylation of Rim15, promoting its nuclear localization [17]. Ume6 is a negative autophagy regulator regulated by Rim15, which binds to the ATG8 promoter and suppresses its transcription under the nutrient-rich conditions. During nitrogen starvation, Ume6 undergoes phosphorylation by Rim15, leading to the derepression of ATG8 and thus promoting ATG8 transcription (Fig. 3) [18]. Similarly, Rph1 inhibits the transcription of ATG7, ATG8, ATG9, ATG14 and ATG29 under nutrient-rich conditions, but phosphorylation by Rim15 upon nutrient deprivation releases Rph1 from these ATG genes (Fig. 3) [19].
Pho23 is another negative regulator that represses the transcription of multiple ATG genes including ATG1, ATG7, ATG8, ATG9, ATG14 and ATG29 when nutrients are replete [20]. During nitrogen starvation, the repression of most of the ATG genes is released, whereas ATG9 transcription is still inhibited by Pho23. Accordingly, pho23-deleted cells have more autophagosomes and higher autophagy activity after starvation [20]. The Spt4-Spt5 complex was recently demonstrated to play dual roles in regulating ATG gene transcription. This complex negatively regulates ATG8 and ATG41 expression under growing conditions. However, during nitrogen starvation, Spt5 is phosphorylated by the Sgv1-Bur2 complex, releasing the inhibitory effect of Spt4 and allowing the complex to function in transcriptional elongation of ATG41, thus promoting autophagy (Fig. 3) [21].
The transcription factors mentioned above are the ones regulating nonselective autophagy. Some ATG genes encoding proteins that specifically function in selective autophagy are also regulated by transcription factors. For example, Atg32 is a mitochondrial membrane protein and the receptor for yeast mitophagy, the selective autophagic degradation of mitochondria [22]. The Ume6-Sin3-Rpd3 complex represses ATG32 transcription in budding yeasts cultured with a fermentable carbon source, where the number of mitochondria and mitophagy activity remain low [23]. Paf1 is another transcription factor suppressing ATG32 transcription, which binds to the ATG32 promoter in both fermentable and non-fermentable media. Therefore, paf1-deleted cells show a higher mitophagy activity after prolonged culture in lactic acid medium [24]. In a very recent study, Dep1, a component of the Rpd3 complex, was found to be critical for ATG32 transcription and mitophagy [25]. It is of note that from an analysis of DNA binding proteins [12], multiple ATG32 regulators were found (Table 2). However, this study was conducted under growing, glucose-starvation and nitrogen-starvation conditions. It would be interesting to see whether these proteins can also regulate ATG32 transcription under a condition that is more specific to mitophagy induction.
Transcriptional regulation of mammalian autophagy
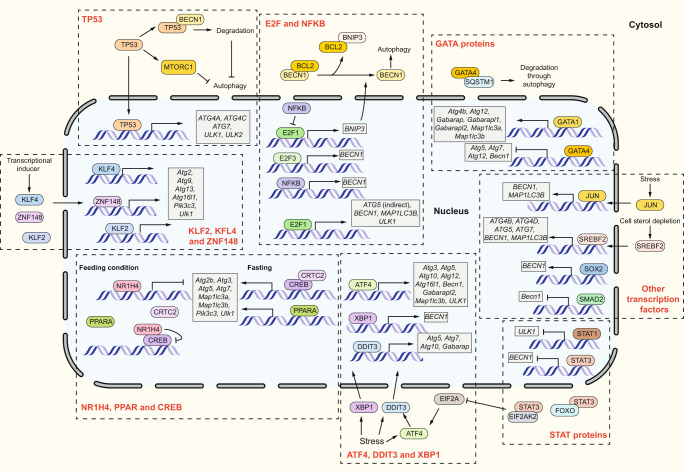
Transcription factors can play dual roles in regulating mammalian autophagy, either through transcription-dependent or transcription-independent pathways and either upregulating or downregulating autophagy. In this section, we summarize the transcription factors regulating the expression of autophagy genes (Table 3, Figs. 4 and 5) and discuss some major and complex transcription systems in autophagy regulation. Because some transcription factors cooperate and a transcription factor itself can be the target of other such factors, we also briefly discuss how transcription factors work together in autophagy regulation.
Table 3.
Mammalian transcription factors regulating ATG gene expression.
| Transcription factor | Effects (+, positive; – negative) | Target genesa | Ref. |
|---|---|---|---|
| ATF4 | + | Atg3, Atg5, Atg10, Atg12, Atg16l1, Becn1, Gabarapl2, Map1lc3b, ULK1 | [138, 139] |
| CREB | + | Atg2b, Atg3, Atg5, Atg7, Ulk1 | [71] |
| DDIT3/CHOP | + | Atg5, Atg7, Atg10, Gabarap | [139] |
| E2F1 | + | BECN1, MAP1LC3B, ULK1 | [60, 62] |
| E2F3 | + | BECN1 | [60] |
| FOXO1 | + | Atg12, Map1lc3b, Gabarapl1, Pik3c3 | [37, 38] |
| FOXO3 | + | Atg5, Atg10, Atg12, Atg14, Becn1, Map1lc3b, Pik3c3, Ulk1, Ulk2, Wipi2 | [34–36] |
| GATA1 | + | Atg4b, Atg12, Gabarap, Gabarapl1, Gabarapl2, Map1lc3a, Map1lc3b | [66] |
| GATA4 | – | Atg5, Atg7, Atg12, Becn1 | [67] |
| JUN | + | BECN1, MAP1LC3B | [140, 141] |
| KLF2 | + | Atg2, Atg9, Atg13, Atg16l1, Pik3c3, Ulk1 | [142] |
| KLF4 | + | Atg2, Atg9, Atg13, Atg16l1, Pik3c3, Ulk1 | [142] |
| MITF1 | + | ATG4A, ATG9B, ATG16L1, MAP1LC3B, WIPI1 | [29, 30] |
| NFKB | + | BECN1 | [61] |
| NR1H4/FXR | – | Atg2a, Atg2b, Atg3, Atg5, Atg7, Atg10, Atg16l1, Gabarap, Map1lc3a, Map1lc3b, Pik3c3, Ulk1, Wipi1, Wipi2 | [71, 72] |
| PPARA | + | Atg7, Map1lc3a, Map1lc3b, Pik3c3 | [72] |
| SMAD2 | – | Becn1 | [143] |
| SOX2 | + | BECN1 | [144] |
| SREBF2/SREBP2 | + | ATG4B, ATG4D, ATG5, ATG7, BECN1, MAP1LC3B | [145, 146] |
| STAT1 | – | ULK1 | [147] |
| STAT3 | – | BECN1 | [148] |
| TFE3 | + | ATG3, ATG4A, ATG5, ATG9B, ATG16L1, GABARAPL1, GABARAPL2, MAP1LC3B, WIPI1 | [30] |
| TFEB | + | ATG9B, ATG16L1, GABARAPL1, MAP1LC3B, WIPI1 | [27, 30] |
| TP53 | + (nuclear TP53) | ATG4A, ATG4C, ATG7, ULK1, ULK2 | [47, 48] |
| XBP1 | + | BECN1 | [78] |
| ZKSCAN3 | – | MAP1LC3B, ZFYVE1/DFCP1, ULK1, WIPI2 | [32] |
| ZNF148 | + | Atg2, Atg9, Atg13, Atg16l1, Pik3c3, Ulk1 | [142] |
aBased on convention, human genes are indicated with all uppercase letters and mouse genes with only the first letter capitalized.
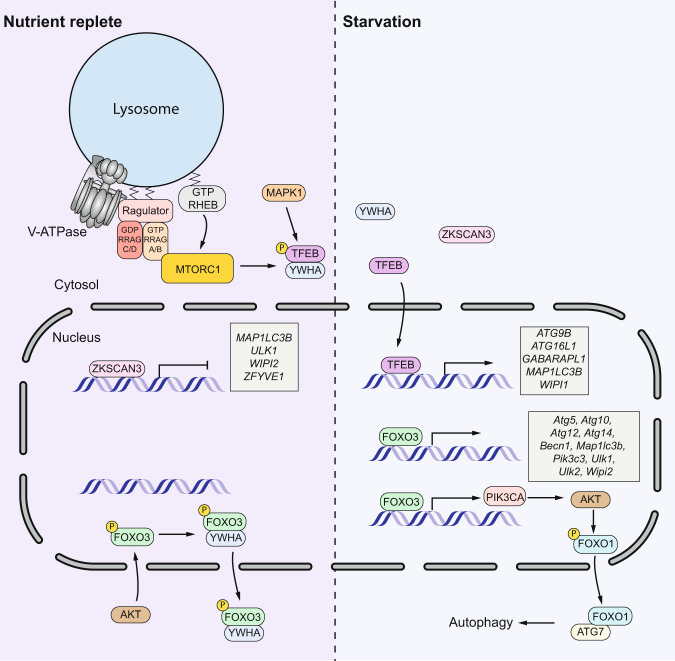
Fig. 4. The regulation of autophagy in mammalian cells through TFEB, ZKSCAN3 and FOXO proteins.
Under nutrient-replete conditions, TFEB is phosphorylated by both MTORC1 and MAPK1, and phosphorylated TFEB will be retained int the cytosol. At the same time, ZKSCAN3 suppresses the transcription of autophagy and lysosome genes. FOXO3 is phosphorylated by AKT and binds to YWHA/14-3-3 proteins, which will exit from the nucleus. Upon starvation, dephosphorylated TFEB is transported into the nucleus and transactivates a series of genes involved in autophagy and lysosome function. In contrast, ZKSCAN3 is pushed outside the nucleus. FOXO3 maintains its nuclear localization and induces the transcription of multiple ATG genes. FOXO3 also promotes the expression of PIK3CA which induces the subsequent phosphorylation of FOXO1. Phosphorylated FOXO1 will move into the cytosol and binds to ATG7 to induce autophagy through a transcription-independent pathway.
Fig. 5. Transcription regulation of autophagy in mammalian cells.
The transcription factors that regulate autophagy (in addition to TFEB, ZKSCAN3 and FOXO proteins) are summarized. The transcription-independent functions of some proteins are also included.
TFEB and ZKSCAN3
TFEB (transcription factor EB) is a one of the microphthalmia (MiT/TFE) transcription factors belonging to the basic helix-loop-helix leucine-zipper family, and is considered as the master transcriptional regulator of autophagy through promoting the transcription of genes involved in lysosomal biogenesis and autophagy [26]. The subcellular localization and transcription activity of TFEB is mainly determined by its phosphorylation status, with MTORC1 as the one of the major kinases. When nutrients are replete, TFEB is phosphorylated by MTORC1, then interacts with YWHA/14-3-3 and remains in the cytosol, whereas starvation leads to the inactivation of MTORC1 and a rapid translocation of dephosphorylated TFEB from the cytosol to the nucleus where it transactivates multiple genes in lysosome biogenesis and the autophagy pathway including MAP1LC3B, ATG9B, ATG16L1, UVRAG and WIPI1 (Fig. 4) [27, 28]. Besides MTORC1, MAPK1 is another kinase that leads to the inhibitory phosphorylation of TFEB under nutrient-replete conditions [27]. The other two transcription factors in the MiT/TFE family, TFE3 and MITF, are also important to drive the expression of genes involved in lysosome biogenesis and autophagy (Table 3) and the regulation of their subcellular localizations is similar to TFEB [29–31]. ZKSCAN3 belongs to a family of zinc finger transcription factors and represents the counterpart of TFEB. When nutrients are replete, the transcription of some core ATG genes (Table 3) and genes encoding proteins involved in autophagosome-lysosome fusion (such as STX5 and SEC22B) and lysosome functions (such as ATP6V1A and CTSA) are suppressed by ZKSCAN3. In contrast, the transcription of genes encoding negative autophagy regulators such as RPTOR and AKT, are promoted by ZKSCAN3. Starvation leads to the translocation of ZKSCAN3 to the cytoplasm, which releases the transcription suppression of autophagy genes (Fig. 4) [32].
FOXO
FOXO (forkhead box O) family proteins are transcription factors with important roles in metabolism, cellular proliferation, and stress tolerance [33]. FOXO1 and FOXO3 are the two major FOXO transcription factors regulating autophagy. In muscle cells, FOXO3 promotes the transcription of a series of Atg genes (Table 3) [34], and genes coding for positive autophagy regulators such as Bnip3 [35]. Transactivation of ATG genes by FOXO3 is also reported in neural stem cells, and FOXO3 is required to maintain autophagy [36]. Similarly, FOXO1 also promotes autophagy through transactivating multiple Atg genes (Table 3) [37, 38]. Additionally, acetylated FOXO1 binds to ATG7 in the cytosol, which is required for autophagy induction, indicating a transcription-independent role of FOXO1 in autophagy regulation (Fig. 4) [39].
Similar to TFEB, the subcellular localization and activity of FOXO family proteins is determined by their post-translational modifications, but it is more complicated [40]. In response to growth factors, FOXO3 is phosphorylated by AKT, which induces binding with YWHA/14-3-3 proteins and exclusion from the nucleus, thus inhibiting its transcriptional activity (Fig. 4) [41, 42]. In addition, FOXO3 can induce AKT activity through increasing the transcription of PIK3CA, which promotes the phosphorylation of FOXO1 and its cytosolic localization, the critical step of FOXO1-induced autophagy via the transcription-independent pathway (Fig. 4) [43]. However, it is still not very clear how the FOXO1-ATG7 interaction compensates for the impaired expression of some autophagy genes. Under nutrient stress, activated AMPK phosphorylates FOXO3 and activates its transcriptional activity, thus promoting the expression of some autophagy-related genes [44, 45]. Besides the two kinases mentioned above, phosphorylation by other kinases and other post-translational modifications also control FOXO protein activity and together contribute to autophagy regulation [46].
TP53
TP53 is a transcription factor regulating the cell cycle and, in addition to its well-known function in tumor suppression, other aspects of TP53 function have been defined, among which is its effect on autophagy. TP53 plays dual roles in autophagy regulation, depending on its subcellular localization. In the nucleus, TP53 transactivates various ATG genes (Table 3) [47, 48] and genes encoding MTOR inhibitors, such as PRKAB1/AMPKβ1, TSC2 and DEPTOR [49, 50]. DRAM1, encoding a lysosomal protein that is critical for the acidification of lysosomes, is also activated by nuclear TP53, and TP53-induced autophagy is dependent on DRAM1 [51]. In contrast, cytosolic TP53 inhibits autophagy, mainly through a transcription-independent pathway such as inhibiting AMPK, activating MTOR and promoting BECN1 ubiquitination and degradation (Fig. 5). Physiological autophagy inducers cause destruction of the cytosolic pool of TP53 leading to autophagy induction [52–54]. TP53 also regulates mitophagy. Besides the inhibition by the cytosolic TP53 via binding to PRKN [55], nuclear TP53 also suppresses mitophagy through transcriptional repression of PINK1, another important protein in mitophagy [56].
E2F1 and NFKB
E2F1 and NFKB are two well-defined transcription factors for their roles in regulating the cell cycle and immune responses, respectively. E2F1 and NFKB oppositely regulate the expression of Bnip3, a hypoxia-induced activator that is required for hypoxia-induced autophagy [57–59]. Under normal conditions, NFKB represses Bnip3 gene transcription via inhibiting E2F1 binding to the Bnip3 promoter region, whereas hypoxia reduces NFKB expression, thus releasing the repression and promoting E2F1-dependent Bnip3 transcription (Fig. 5) [58, 59]. BECN1 is another common target of E2F1 and NFKB, but, unlike BNIP3, both E2F1 and NFKB induce the transcription of BECN1 [60, 61]. In addition, E2F1 induces the transcription of several other ATG genes (Table 3) as well as DRAM1 [62]. Of note, the fact that E2F1 lacking the transactivation domain is still able to activate autophagy, associated with increasing ATG5 expression, indicates that E2F1 may have an alternative or indirect way to upregulate autophagy [63]. In a recent paper, E2F1 is reported to suppress mitophagy via upregulating the transcription of MFN2, which is involved in mitochondria fusion [64].
GATA family
GATA transcription factors are a conserved family of zinc-finger transcription factors that fulfill multiple regulatory roles including those involved in development and cell fate decisions [65]. Regarding autophagy, GATA1 activates the transcription of Atg8-family genes, Atg4b, Atg12 and genes involved in lysosome biogenesis and function such as Lamp1 and Atp6v0e [66]. In rat cardiomyocytes, GATA4 inhibits doxorubicin (DOX)-induced autophagy partially through upregulating the transcription of Bcl2; the BCL2 protein binds and sequesters BECN1 [67, 68]. In addition, overexpressing GATA4 suppresses the DOX-induced expression of Atg5, Atg7, Atg12 and Becn1, which may also contribute to autophagy inhibition [67]. Interestingly, this study also found that DOX-induced autophagy is accompanied by decreasing GATA4 protein level [67]. In a later study, GATA4 was found as a selective autophagy cargo through binding to SQSTM1. When a senescence signal is activated, the interaction between GATA4 and SQSTM1 is reduced and accumulated GATA4 initiates a senescence-associated secretory phenotype, indicating a role of selective autophagic degradation of GATA4 in protecting cells from senescence [69].
NR1H4, PPAR and CREB
NR1H4/FXR (nuclear receptor subfamily 1 group H member 4) is an energy sensor that is activated by bile acids in the fed liver [70]. Two studies confirm the role of NR1H4 in suppressing the expression of a wide range of autophagy genes in the liver under fed conditions (Table 3), but the regulatory mechanisms reported are different [71, 72]. In the paper published by Seok et al., activated NR1H4 inhibits the transcription of autophagy genes through disrupting the interaction between CREB and its coactivator CRTC2, which promotes autophagy gene transcription during starvation. Most CREB-binding autophagy genes have shared binding sites with NR1H4, and the downregulation of CREB inhibits lipophagy (autophagic degradation of lipids) during starvation (Fig. 5) [71]. However, with the discovery that autophagy and the expression of autophagy genes are induced under fed conditions by PPARA (peroxisome proliferator activated receptor alpha) agonist and suppressed during starvation by NR1H4 agonist, Lee et al. suggested that PPARA, which is activated in fasted liver, competes with NR1H4 in binding to the shared sites in autophagy gene promoters with opposite transcription outputs (Fig. 5) [72].
In addition to PPARA, PPARG also regulates autophagy as a transcription factor, but how this regulation occurs is more complex and remains controversial [73]. PPARG induces autophagy in breast cancer cells through activating the transcription of HIF1A and BNIP3 [74]. In hepatocytes, PPARG activation induces autophagy through transactivating NEDD4, an E3 ubiquitin ligase acting as a positive autophagy regulator [75]. However, autophagy inhibition by PPARG agonists is found in neurons after injury [76, 77], indicating a complicated autophagy regulation by PPARG possibly depending on cell types.
Interaction between autophagy transcription factors
As mentioned previously, some autophagy transcription factors can function independently from their transcription activity. This includes the regulation of other transcription factors through direct binding. For instance, FOXO family proteins are regulated by XBP1 and STAT3 independently from their roles as transcription factors. XBP1 is a transcription factor and a crucial signal transducer in the endoplasmic reticulum (ER) stress response. In response to ER stress, XBP1 transcripts undergo splicing by ERN1/IRE1 to generate XBP1s, which can trigger an autophagic response through promoting BECN1 transcription [78]. However, XBP1s also binds to FOXO1 and leads to its degradation through the proteasome [79]. Additionally, the unspliced isoform of XBP1 can also bind and induce FOXO1 degradation [80]. STAT3 can function as a transcription factor to suppress the transcription of BECN1 and transactivate some negative autophagy regulatory genes (such as PIK3R1 and BCL2) [81]; unphosphorylated cytosolic STAT3 interacts with FOXO1 and FOXO3, sequestering them in the cytosol and inhibiting their function in promoting transcription of autophagy genes [82]. In addition, cytosolic STAT3 also suppresses autophagy through interacting with and inhibiting EIF2AK2 and the subsequent phosphorylation of EIF2A, which promotes ATF4 expression, a transcription factor that transactivates multiple autophagy genes (Table 3) [83].
Furthermore, some transcription factors are the target of other such factors. For instance, it is reported that FOXO1 induces Tfeb expression through directly binding to its promoter region and FOXO1 inhibition reduces Tfeb mRNA and protein level, accompanied with decreased autophagy [84]. Besides, Tfeb transcription is also controlled by NR1H4, CREB and PPARA, which are transcription factors that can independently regulate autophagy gene expression [71, 85]. Additionally, GATA1 promotes the generation of Foxo3 transcripts, and Foxo3 knockdown reduces GATA1-induced Map1lc3b expression by approximately 50%, suggesting that GATA1 either functions directly through binding to the autophagy gene loci or indirectly through transactivating Foxo3 [66]. The independent and cooperative functions of these transcription factors establish a complicated but well controlled system to regulate autophagy at the transcription level.
Application of autophagy transcription regulation: from the perspective of diseases
Because autophagy is pivotal for cell survival under stress conditions and the clearance of protein aggregates and superfluous or damaged organelles, it is critical for maintaining cell homeostasis, and the dysregulation of autophagy is correlated with multiple human diseases, including cancer, neurodegenerative diseases, and metabolic disorders [11]. With many studies revealing an increasing number of autophagy transcription factors in the past several decades, recent studies have started to investigate how alterations of the activities of these transcription factors contribute to human diseases and whether they can be developed into therapeutic targets. In this section, we briefly summarize studies indicating the association between transcriptional regulation of autophagy and diseases (Table 4).
Table 4.
Studies showing the implication of autophagy transcription factors in human diseases.
| Transcription factor (TF) | Type of disease | Changes in TF (pathological or artificial) | Results and/or applications | Ref. |
|---|---|---|---|---|
| Cancers | ||||
| TP53 | Colorectal Cancer | Expressing cancer-associated R175H, R248W and R273H mutants | Decreased TSC2 expression and hyperactivity of MTORC1, which may result in autophagy suppression. | [87] |
| Non-small cell lung carcinoma | ||||
| Pancreatic ductal adenocarcinoma | Expressing cancer-associated R273H and R175H mutants in TP53 null cancer cells | Inhibition of autophagy gene transcription and lower autophagy activity, which promotes cancer cell proliferation. | [88] | |
| Non-small cell lung carcinoma | ||||
| Pancreatic ductal adenocarcinoma | Knocking down TP53R273H or TP53R175H mutants carried by cancer cells | Elevated autophagy gene transcription and autophagy activity. | [88] | |
| Breast cancer | ||||
| Head and neck cancer | Overexpressing the TP53R175H mutant | Inhibition of PRKAA2/AMPK, which may result in autophagy inhibition. | [89] | |
| TFEB | Prostate cancer | Upregulation by AR (androgen receptor) activation | Required for maximal androgen-mediated autophagy and cell growth. | [93] |
| Non-small cell lung cancer | Upregulation in some tumors from patients | Silencing TFEB reduces the migration ability of lung cancer cells. | [94] | |
| Colorectal cancer | Downregulation in cancer tissue | TFEB expression is positively correlated with malignant progression. Silencing TFEB reduces BECN1 expression and inhibits cell growth and migration. | [95] | |
| Nuclear translocation after doxorubicin treatment | Silencing TFEB inhibits doxorubicin-induced autophagy and promotes cell sensitivity to chemotherapy. | [96] | ||
| Breast cancer | Upregulation in some tumor samples | TFEB expression is positively correlated with poor prognosis. | [97] | |
| Pancreatic ductal adenocarcinoma | Upregulation in PDA | Critical for autophagy activation and metabolic reprogramming. | [98] | |
| Upregulation after TGFB1 treatment | Mediating autophagy upregulation, which is critical for TGFB1-induced cell metastasis. | [99] | ||
| FOXO3 | Hepatocellular carcinoma | Nuclear localization during hypoxia | Mediating hypoxia-induced autophagy and resistance to chemosensitivity. | [103] |
| Neurodegenerative diseases | ||||
| TFEB | Alzheimer disease | Downregulation in nuclear fractions in AD brain | — | [105] |
| Exogenous expression of TFEB | Reduction of Aβ level and amyloid plaques. | [106] | ||
| Optogenetic TFEB induction | Promoted clearance of MAPT/tau. | [107] | ||
| Parkinson disease | Downregulation in nuclear fractions in PD midbrains | Overexpressing TFEB stimulates autophagy and protects neurons from SNCA toxicity. | [111] | |
| Overexpression or pharmacological activation | Induced clearance of SNCA aggregates. | [112] | ||
| Pharmacological activation | Activated autophagy and promoted clearance of protein aggregates. | [114] | ||
| Huntington disease | Pharmacological activation | Activated autophagy and promoted clearance of protein aggregates. | [114] | |
| Exogenous expression of TFEB | Activated autophagy and induced clearance of mHTT. | [113] | ||
| Spinal and bulbar muscular atrophy | Inactivation in motor neurons | Impaired autophagy activity and promoted pathogenesis. | [115] | |
| Upregulation in skeletal muscle | Enhanced autophagy activity. | [116] | ||
| Amyotrophic lateral sclerosis | Downregulation in nuclear fractions | — |
[105] [118] |
|
| Decreased expression in spinal cords and cell model | TFEB overexpression increases BECN1 expression and cell survival. | [117] | ||
| PPARA | Alzheimer disease | Pharmacological activation | Induced autophagy activities and reduced amyloid pathology. | [119] |
| Pharmacological activation | Induced TFEB expression, autophagy activities and enhanced Aβ degradation. | [120] | ||
| Parkinson disease | Activation via treadmill exercise | Higher TFEB and autophagy-lysosome gene expression and SNCA accumulation prevention. | [121] | |
| Metabolic disorders | ||||
| TFEB | Obesity | Downregulation in diabetic heart | Inhibited autophagy and higher risk of cardiac injury. | [129] |
| Pharmacological activation | Induced autophagy activity and decreased intracellular lipid. | [130] | ||
| E2F1 | Obesity | Upregulated in omental adipose tissue in obesity | Increased autophagy gene expression and cell sensitivity to stress-induced autophagy. | [125] |
| Upregulated in omental adipose tissue in obesity | Induced autophagy activity, which is correlated with obesity-related phenotypes. | [126] | ||
| Knockout in vivo or in adipocytes | Autophagy suppression and white adipose tissue browning promotion. | [127] | ||
| FOXO3 | Obesity | Upregulated in visceral adipose tissue in obesity | Downregulating FOXO3 inhibits autophagy and lipid accumulation. | [128] |
Cancer
The dual roles of autophagy in cancer have been demonstrated by a wide range of research; defective autophagy contributes to cancer initiation due to the deficiency in the clearance of intracellular waste and subsequently induced genome instability, while elevated autophagy would help cancer cell survival under stress conditions, such as nutrient deprivation, hypoxia and damage from anti-cancer drugs [3]. The autophagy transcription factor TP53 is one of the most commonly mutated genes in various cancer types. In 2008, via expressing 22 TP53 single amino acid mutants occurring in colon cancer cells in TP53 null cells, researchers found that one third of these mutations inhibit autophagy. A significant correlation between the TP53 mutants in the cytoplasm and their ability to repress autophagy was also discovered, which suggests the cytosolic mutant TP53 may lose the ability to promote the transcription of autophagic genes or inhibit autophagy through the transcription-independent pathway [86]. Cancer-associated TP53 mutations, including R175H, R248W, and R273H, reduce the transcription of TSC2, an inhibitor of MTOR, subsequently leading to higher MTOR activity, which may contribute to autophagy suppression [87]. Expressing TP53R273H or TP53R175H in TP53 null cells also reduces autophagy gene transcription including ATG12, BECN1 and DRAM1, as well as autophagy activity [88]. It is also reported that TP53R175H preferably binds to PRKAA2/AMPK and inhibits its activation even under conditions of energy stress, indicating a transcription-independent role of these cancer-associated mutations in inhibiting autophagy [89]. Because of the high correlation between TP53 mutation and cancer, drugs targeting TP53 have been one focus of research [90]. For example, in tamoxifen-resistant breast cancer cells, combined treatment of Cl-amidine and docetaxel enhances TP53 nuclear accumulation, inhibits MTOR signaling, elevates autophagy and induces cell apoptosis [91]. Additionally, in cervical cancer cells, SNX-2112 increases TP53 expression and induces autophagy, which is necessary for cell apoptosis [92].
TFEB is important in tumor growth and metastasis and correlates with tumor malignancy, worse prognosis, and resistance to therapy [93–97]. Elevated TFEB expression and nuclear localization have been detected in pancreatic cancer cells, and the subsequent activation of autophagy and lysosome functions are critical to meet the metabolic requirement of the cancer cells [98]. TFEB-driven autophagy is also required for TGFB1-induced pancreatic cancer cell migration and metastasis [99]. In prostate cancer, androgen-induced TFEB expression and activity promotes autophagy and prostate cancer growth [93]. Additionally, in colorectal cancer, TFEB expression is positively correlated with infiltration and metastasis rate [95] and TFEB is involved in the autophagy-mediated resistance to chemotherapy [96]. Therefore, a variety of studies have suggested the potential of TFEB inhibition as a therapeutic approach for cancer treatment. For instance, silencing TFEB sensitizes glioblastoma to radiotherapy [100]. Alantolactone treatment reduces TFEB expression in pancreatic cancer cells, which contributes to the lysosome dysfunction, autophagosome accumulation and the consequent induction of cell apoptosis [101]. Knocking down TFEB can also sensitize pancreatic cancer cells with KRAS mutations to MAP2K/MEK inhibitors [102]. In addition to TFEB, FOXO3-induced autophagy is reported to mediate sorafenib resistance in hepatocellular carcinoma and FOXO3-targeted therapy is proposed to be a promising approach to improve clinical prognosis [103].
Neurodegenerative disorders
Deficient autophagy is highly correlated with neurodegenerative diseases due to the impaired clearance of aggregate-prone proteins such as the amyloid β-protein (Aβ) and MAPT/tau in Alzheimer disease (AD), SNCA/α-synuclein in Parkinson disease (PD) and mutant HTT (mHTT) in Huntington disease (HD) [104]. Therefore, modulating autophagy transcription factors to activate autophagy has the potential to alleviate neurodegenerative diseases. In AD brains, nuclear TFEB is reduced [105] and the fact that overexpressing TFEB and inducing TFEB nuclear localization promote the clearance of Aβ and MAPT/tau supports the close association between TFEB dysregulation and AD [106, 107]. Therefore, some small molecules that activate TFEB, such as celastrol, an MTORC1 inhibitor, and trametinib, a MAP2K1 and MAP2K2 inhibitor, may have the potential to treat AD [108–110]. As with AD, reduced nuclear TFEB is also found in PD brains [111]. Autophagy induced by overexpression or pharmacological activation of TFEB promotes the clearance of SNCA and protects neurons from SNCA toxicity [111, 112]. This is also true for HD, where overexpressing TFEB reduces the mHTT level in the HD mouse model [113]. In a more recent study, SMK-17 is found to upregulate autophagy through inducing TFEB nuclear localization and to reduce protein aggregates in both PD and HD cell models [114]. Besides these three major neurodegenerative diseases, in spinal and bulbar muscular atrophy (SBMA), polyglutamine-expanded AR (androgen receptor), the major cause of this disease, interferes with TFEB transactivation function, leading to decreased autophagy in motor neurons and promoting SBMA pathogenesis. More importantly, deficient autophagy flux can be rescued by overexpressing TFEB, highlighting the potential of TFEB as a therapeutic target [115]. However, in the skeletal muscle cells from SBMA mice, higher TFEB activity and autophagy are reported, suggesting the distinct roles of autophagy in neurons and muscles in neuromuscular diseases [116]. Additionally, in amyotrophic lateral sclerosis/ALS brains from mouse model or patients, reduced TFEB expression and nuclear fraction are discovered and overexpressing TFEB improves the survival and proliferation of amyotrophic lateral sclerosis neurons [105, 117, 118].
Because PPARA can promote the transcription of TFEB, PPARA activation can also alleviate neurodegenerative pathogenesis. It is reported that the activation of autophagy by PPARA agonist improves the clearance of Aβ and mitigates AD pathology and cognitive decline in the murine model [119]. A later study demonstrated that this pathway is mediated by TFEB because Tfeb knockdown inhibits the degradation of Aβ induced by the activation of PPARA in mouse astrocytes [120]. Additionally, PPARA activation by treadmill exercise induces Tfeb transcription and the expression of autophagy-lysosome genes, which is essential to prevent SNCA accumulation [121]. In the mouse model of HD, the induction of PPARGC1A/PGC1A, the coactivator of PPARG, activates the transcription of Tfeb, which contributes to the mHTT aggregate reduction [122].
Here, we only summarize the implication of these autophagic transcription factors in neurodegenerative disorders depending on their transcriptional activity, focusing on TFEB. It should be kept in mind that these proteins may have functions independent from being a transcription factor. Therefore, even though gene overexpression or pharmacological activation are potential therapeutic approaches to neurodegenerative diseases, there are some limitations. For instance, a study from Brattas et al. reported that overexpressing TFEB does not reduce the mHTT aggregates, whereas overexpressing BECN1 can partially clear the aggregates at the early stage of the disease [123]. A later study identified a prion-like domain at the N terminus of TFEB that mediates the coaggregation of TFEB and mHTT and this may explain the failure of overexpressed TFEB in the clearance of mHTT aggregates [124]. In addition, BECN1 overexpression cannot rescue the mHTT-associated phenotype at a later stage of HD when prominent mHTT accumulation occurs, suggesting that activating autophagy at a particular stage of the disease is important in the outcome [123].
Metabolic disorders
Because autophagy is a critical process to maintain the balance of metabolites, dysregulation of autophagy has been seen in metabolic disorders such as obesity and diabetes [11]. The association between autophagy and obesity is complicated: autophagy may both contribute to or inhibit obesity, and autophagy may have different effects in different tissues in an obese model. In omental adipose tissue, elevated E2F1 and autophagy gene expression are reported, and these are correlated with obesity-associated cardio-metabolic risk signature [125, 126]. In another study, e2f1 knockout promotes white adipose tissue browning through inhibiting autophagy, and therefore, it could become a potential therapeutic treatment against obesity through reducing excessive energy storage in white adipose tissue [127]. Additionally, in the adipose tissue of high fat-diet induced obese mouse, the FOXO3 protein level and some Atg gene transcripts are upregulated and Foxo3 knockdown inhibits lipid accumulation [128]. However, in the heart from a diet-induced obesity mouse model, TFEB with inhibitory phosphorylation increases and autophagosome turnover is inhibited [129]. In line with this, another study reported that the chemical MSL enhances autophagy through activating PPP3/calcineurin to induce TFEB dephosphorylation and nuclear translocation. More importantly, this molecule can induce the clearance of intracellular lipids and improve the metabolic profile of obese mice [130].
In this section, we briefly discussed how autophagy transcription factors are altered in cancer, neurodegenerative diseases and metabolic disorders and their potential to become therapeutic targets, which are summarized in Table 4. However, diseases associated with altered autophagy transcription factors are not limited to the three mentioned here. For example, TFEB and autophagy are important in the immune response, suggesting a close correlation with infectious diseases [131]. Additionally, TFEB and FOXO also control the expression of many autophagy genes in the context of aging [132]. These findings further highlight the importance of fine-tuning of autophagy in maintaining cell homeostasis.
Discussion
In the past decades, a large number of transcriptional regulators of autophagy have been discovered in both yeast and mammalian systems. While one transcription factor can target multiple autophagy genes, the same autophagy gene can be regulated by multiple transcription factors, which allows the precise control of autophagy under different conditions. Some core ATG genes, including BECN1, ULK1 and MAP1LC3B are common targets of these transcription factors (Table 3), indicating their key roles in modulating autophagy. The importance of transcriptional regulation of autophagy is also reflected by the alteration of some transcription factors under pathological status, such as the mutations in TP53 in tumors and the change of TFEB expression and subcellular localization in neurodegenerative diseases, and the concomitant abnormal autophagy. Therefore, these transcription factors have the potential to be developed into biomarkers for disease diagnosis and targets in therapy, but their transcription-independent roles may lead to some limitations and should always be taken into consideration. Additionally, as different cells have distinct responses to the change in autophagy, the assessment of autophagy transcription factor modulation should also be cell specific.
In this review, we mainly focused on nonselective autophagy and briefly discussed how mitophagy is regulated at the transcriptional level [25, 55, 56]. Because few studies focus on how selective autophagy is controlled by transcription factors, it will be important and worthwhile to understand whether and how different transcription factors regulate selective types of autophagy and if their modulation can be exploited for therapeutic purposes.
As mentioned previously, autophagy regulation can occur at multiple levels, which together controls the fine-tuning of this process (Fig. 2). In the nucleus, transcription regulation cooperates closely with epigenetic regulation. First, modifications on DNA and histones influence chromatin structure, thus affecting the accessibility to transcription factors [133]. Second, histone modifiers can work together with transcription factors and repressors. For instance, CARM1 not only causes H3R17 dimethylation during glucose starvation, but also functions as the coactivator of TFEB [134]. Additionally, EHMT2/G9a induces H3K9 methylation and recruits the transcription repressor BRD4, both inhibiting the transcription of autophagy genes [135]. More studies analyzing the integration of these factors would deepen our knowledge about the complicated network of autophagy regulation in the nucleus.
Author contributions
YL wrote and DJK edited the manuscript.
Funding
This work was supported by NIH grant GM131919.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Klionsky DJ, Emr SD. Autophagy as a regulated pathway of cellular degradation. Science. 2000;290:1717–21. doi: 10.1126/science.290.5497.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gatica D, Lahiri V, Klionsky DJ. Cargo recognition and degradation by selective autophagy. Nat Cell Biol. 2018;20:233–42. doi: 10.1038/s41556-018-0037-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ariosa AR, Lahiri V, Lei Y, Yang Y, Yin Z, Zhang Z, et al. A perspective on the role of autophagy in cancer. Biochim Biophys Acta Mol Basis Dis. 2021;1867:166262. doi: 10.1016/j.bbadis.2021.166262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wong PM, Puente C, Ganley IG, Jiang X. The ULK1 complex: sensing nutrient signals for autophagy activation. Autophagy. 2013;9:124–37. doi: 10.4161/auto.23323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lu J, He L, Behrends C, Araki M, Araki K, Jun Wang Q, et al. NRBF2 regulates autophagy and prevents liver injury by modulating Atg14L-linked phosphatidylinositol-3 kinase III activity. Nat Commun. 2014;5:3920. doi: 10.1038/ncomms4920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fimia GM, Stoykova A, Romagnoli A, Giunta L, Di Bartolomeo S, Nardacci R, et al. Ambra1 regulates autophagy and development of the nervous system. Nature. 2007;447:1121–5. doi: 10.1038/nature05925. [DOI] [PubMed] [Google Scholar]
- 7.Nascimbeni AC, Codogno P, Morel E. Phosphatidylinositol-3-phosphate in the regulation of autophagy membrane dynamics. FEBS J. 2017;284:1267–78. doi: 10.1111/febs.13987. [DOI] [PubMed] [Google Scholar]
- 8.Yin Z, Popelka H, Lei Y, Yang Y, Klionsky DJ. The roles of ubiquitin in mediating autophagy. Cells. 2020;9:2025. doi: 10.3390/cells9092025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dooley HC, Razi M, Polson HE, Girardin SE, Wilson MI, Tooze SA. WIPI2 links LC3 conjugation with PI3P, autophagosome formation, and pathogen clearance by recruiting Atg12-5-16L1. Mol Cell. 2014;55:238–52. doi: 10.1016/j.molcel.2014.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Diao J, Liu R, Rong Y, Zhao M, Zhang J, Lai Y, et al. ATG14 promotes membrane tethering and fusion of autophagosomes to endolysosomes. Nature. 2015;520:563–6. doi: 10.1038/nature14147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lei Y, Klionsky DJ. The emerging roles of autophagy in human diseases. Biomedicines. 2007;9:1651. doi: 10.3390/biomedicines9111651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bernard A, Jin M, Xu Z, Klionsky DJ. A large-scale analysis of autophagy-related gene expression identifies new regulators of autophagy. Autophagy. 2015;11:2114–22. doi: 10.1080/15548627.2015.1099796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tate JJ, Georis I, Dubois E, Cooper TG. Distinct phosphatase requirements and GATA factor responses to nitrogen catabolite repression and rapamycin treatment in Saccharomyces cerevisiae. J Biol Chem. 2010;285:17880–95. doi: 10.1074/jbc.M109.085712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Valenzuela L, Aranda C, Gonzalez A. TOR modulates GCN4-dependent expression of genes turned on by nitrogen limitation. J Bacteriol. 2001;183:2331–4. doi: 10.1128/JB.183.7.2331-2334.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang T, Jiang G, Zhang Y, Lei Y, Liu S, Li H, et al. The RNA polymerase II subunit Rpb9 activates ATG1 transcription and autophagy. EMBO Rep. 2022;23:e54993. doi: 10.15252/embr.202254993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yao Z, Delorme-Axford E, Backues SK, Klionsky DJ. Atg41/Icy2 regulates autophagosome formation. Autophagy. 2015;11:2288–99. doi: 10.1080/15548627.2015.1107692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim B, Lee Y, Choi H, Huh WK. The trehalose-6-phosphate phosphatase Tps2 regulates ATG8 transcription and autophagy in Saccharomyces cerevisiae. Autophagy. 2021;17:1013–27. doi: 10.1080/15548627.2020.1746592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bartholomew CR, Suzuki T, Du Z, Backues SK, Jin M, Lynch-Day MA, et al. Ume6 transcription factor is part of a signaling cascade that regulates autophagy. Proc Natl Acad Sci USA. 2012;109:11206–10. doi: 10.1073/pnas.1200313109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bernard A, Jin M, Gonzalez-Rodriguez P, Fullgrabe J, Delorme-Axford E, Backues SK, et al. Rph1/KDM4 mediates nutrient-limitation signaling that leads to the transcriptional induction of autophagy. Curr Biol. 2015;25:546–55. doi: 10.1016/j.cub.2014.12.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jin M, He D, Backues SK, Freeberg MA, Liu X, Kim JK, et al. Transcriptional regulation by Pho23 modulates the frequency of autophagosome formation. Curr Biol. 2014;24:1314–22. doi: 10.1016/j.cub.2014.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wen X, Gatica D, Yin Z, Hu Z, Dengjel J, Klionsky DJ. The transcription factor Spt4-Spt5 complex regulates the expression of ATG8 and ATG41. Autophagy. 2020;16:1172–85. doi: 10.1080/15548627.2019.1659573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kanki T, Wang K, Baba M, Bartholomew CR, Lynch-Day MA, Du Z, et al. A genomic screen for yeast mutants defective in selective mitochondria autophagy. Mol Biol Cell. 2009;20:4730–8. doi: 10.1091/mbc.e09-03-0225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aihara M, Jin X, Kurihara Y, Yoshida Y, Matsushima Y, Oku M, et al. Tor and the Sin3-Rpd3 complex regulate expression of the mitophagy receptor protein Atg32 in yeast. J Cell Sci. 2014;127:3184–96. doi: 10.1242/jcs.153254. [DOI] [PubMed] [Google Scholar]
- 24.Zheng L, Shu WJ, Li YM, Mari M, Yan C, Wang D, et al. The Paf1 complex transcriptionally regulates the mitochondrial-anchored protein Atg32 leading to activation of mitophagy. Autophagy. 2020;16:1366–79. doi: 10.1080/15548627.2019.1668228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Camougrand N, Vigie P, Dompierre J, Massoni-Laporte A, Lasserre JP, Bhatia-Kissova I. The Dep1 protein: A new regulator of mitophagy in yeast. Biochem Biophys Res Commun. 2022;635:218–26. doi: 10.1016/j.bbrc.2022.10.052. [DOI] [PubMed] [Google Scholar]
- 26.Napolitano G, Ballabio A. TFEB at a glance. J Cell Sci. 2016;129:2475–81. doi: 10.1242/jcs.146365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Settembre C, Di Malta C, Polito VA, Garcia Arencibia M, Vetrini F, Erdin S, et al. TFEB links autophagy to lysosomal biogenesis. Science. 2011;332:1429–33. doi: 10.1126/science.1204592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roczniak-Ferguson A, Petit CS, Froehlich F, Qian S, Ky J, Angarola B, et al. The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Sci Signal. 2012;5:ra42. doi: 10.1126/scisignal.2002790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moller K, Sigurbjornsdottir S, Arnthorsson AO, Pogenberg V, Dilshat R, Fock V, et al. MITF has a central role in regulating starvation-induced autophagy in melanoma. Sci Rep. 2019;9:1055. doi: 10.1038/s41598-018-37522-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martina JA, Diab HI, Lishu L, Jeong AL, Patange S, Raben N, et al. The nutrient-responsive transcription factor TFE3 promotes autophagy, lysosomal biogenesis, and clearance of cellular debris. Sci Signal. 2014;7:ra9. doi: 10.1126/scisignal.2004754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martina JA, Puertollano R. Rag GTPases mediate amino acid-dependent recruitment of TFEB and MITF to lysosomes. J Cell Biol. 2013;200:475–91. doi: 10.1083/jcb.201209135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chauhan S, Goodwin JG, Chauhan S, Manyam G, Wang J, Kamat AM, et al. ZKSCAN3 is a master transcriptional repressor of autophagy. Mol Cell. 2013;50:16–28. doi: 10.1016/j.molcel.2013.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eijkelenboom A, Burgering BM. FOXOs: signalling integrators for homeostasis maintenance. Nat Rev Mol Cell Biol. 2013;14:83–97. doi: 10.1038/nrm3507. [DOI] [PubMed] [Google Scholar]
- 34.Zhao J, Brault JJ, Schild A, Cao P, Sandri M, Schiaffino S, et al. FoxO3 coordinately activates protein degradation by the autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell Metab. 2007;6:472–83. doi: 10.1016/j.cmet.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 35.Mammucari C, Milan G, Romanello V, Masiero E, Rudolf R, Del Piccolo P, et al. FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab. 2007;6:458–71. doi: 10.1016/j.cmet.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 36.Audesse AJ, Dhakal S, Hassell LA, Gardell Z, Nemtsova Y, Webb AE. FOXO3 directly regulates an autophagy network to functionally regulate proteostasis in adult neural stem cells. PLoS Genet. 2019;15:e1008097. doi: 10.1371/journal.pgen.1008097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sengupta A, Molkentin JD, Yutzey KE. FoxO transcription factors promote autophagy in cardiomyocytes. J Biol Chem. 2009;284:28319–31. doi: 10.1074/jbc.M109.024406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu HY, Han J, Cao SY, Hong T, Zhuo D, Shi J, et al. Hepatic autophagy is suppressed in the presence of insulin resistance and hyperinsulinemia: inhibition of FoxO1-dependent expression of key autophagy genes by insulin. J Biol Chem. 2009;284:31484–92. doi: 10.1074/jbc.M109.033936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao Y, Yang J, Liao W, Liu X, Zhang H, Wang S, et al. Cytosolic FoxO1 is essential for the induction of autophagy and tumour suppressor activity. Nat Cell Biol. 2010;12:665–75. doi: 10.1038/ncb2069. [DOI] [PubMed] [Google Scholar]
- 40.Wang X, Hu S, Liu L. Phosphorylation and acetylation modifications of FOXO3a: Independently or synergistically? Oncol Lett. 2017;13:2867–72. doi: 10.3892/ol.2017.5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brunet A, Bonni A, Zigmond MJ, Lin MZ, Juo P, Hu LS, et al. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell. 1999;96:857–68. doi: 10.1016/S0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- 42.Tzivion G, Dobson M, Ramakrishnan G. FoxO transcription factors; Regulation by AKT and 14-3-3 proteins. Biochim Biophys Acta. 2011;1813:1938–45. doi: 10.1016/j.bbamcr.2011.06.002. [DOI] [PubMed] [Google Scholar]
- 43.Zhou J, Liao W, Yang J, Ma K, Li X, Wang Y, et al. FOXO3 induces FOXO1-dependent autophagy by activating the AKT1 signaling pathway. Autophagy. 2012;8:1712–23. doi: 10.4161/auto.21830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Greer EL, Oskoui PR, Banko MR, Maniar JM, Gygi MP, Gygi SP, et al. The energy sensor AMP-activated protein kinase directly regulates the mammalian FOXO3 transcription factor. J Biol Chem. 2007;282:30107–19. doi: 10.1074/jbc.M705325200. [DOI] [PubMed] [Google Scholar]
- 45.Sanchez AM, Csibi A, Raibon A, Cornille K, Gay S, Bernardi H, et al. AMPK promotes skeletal muscle autophagy through activation of forkhead FoxO3a and interaction with Ulk1. J Cell Biochem. 2012;113:695–710. doi: 10.1002/jcb.23399. [DOI] [PubMed] [Google Scholar]
- 46.Cheng Z. The FoxO-autophagy axis in health and disease. Trends Endocrinol Metab. 2019;30:658–71. doi: 10.1016/j.tem.2019.07.009. [DOI] [PubMed] [Google Scholar]
- 47.Gao W, Shen Z, Shang L, Wang X. Upregulation of human autophagy-initiation kinase ULK1 by tumor suppressor p53 contributes to DNA-damage-induced cell death. Cell Death Differ. 2011;18:1598–607. doi: 10.1038/cdd.2011.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kenzelmann Broz D, Spano Mello S, Bieging KT, Jiang D, Dusek RL, Brady CA, et al. Global genomic profiling reveals an extensive p53-regulated autophagy program contributing to key p53 responses. Genes Dev. 2013;27:1016–31. doi: 10.1101/gad.212282.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Feng Z, Hu W, de Stanchina E, Teresky AK, Jin S, Lowe S, et al. The regulation of AMPK beta1, TSC2, and PTEN expression by p53: stress, cell and tissue specificity, and the role of these gene products in modulating the IGF-1-AKT-mTOR pathways. Cancer Res. 2007;67:3043–53. doi: 10.1158/0008-5472.CAN-06-4149. [DOI] [PubMed] [Google Scholar]
- 50.Cui D, Dai X, Gong L, Chen X, Wang L, Xiong X, et al. DEPTOR is a direct p53 target that suppresses cell growth and chemosensitivity. Cell Death Dis. 2020;11:976. doi: 10.1038/s41419-020-03185-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Crighton D, Wilkinson S, O’Prey J, Syed N, Smith P, Harrison PR, et al. DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell. 2006;126:121–34. doi: 10.1016/j.cell.2006.05.034. [DOI] [PubMed] [Google Scholar]
- 52.Tasdemir E, Maiuri MC, Galluzzi L, Vitale I, Djavaheri-Mergny M, D’Amelio M, et al. Regulation of autophagy by cytoplasmic p53. Nat Cell Biol. 2008;10:676–87. doi: 10.1038/ncb1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Green DR, Kroemer G. Cytoplasmic functions of the tumour suppressor p53. Nature. 2009;458:1127–30. doi: 10.1038/nature07986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tripathi R, Ash D, Shaha C. Beclin-1-p53 interaction is crucial for cell fate determination in embryonal carcinoma cells. J Cell Mol Med. 2014;18:2275–86. doi: 10.1111/jcmm.12386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hoshino A, Mita Y, Okawa Y, Ariyoshi M, Iwai-Kanai E, Ueyama T, et al. Cytosolic p53 inhibits Parkin-mediated mitophagy and promotes mitochondrial dysfunction in the mouse heart. Nat Commun. 2013;4:2308. doi: 10.1038/ncomms3308. [DOI] [PubMed] [Google Scholar]
- 56.Goiran T, Duplan E, Rouland L, El Manaa W, Lauritzen I, Dunys J, et al. Nuclear p53-mediated repression of autophagy involves PINK1 transcriptional down-regulation. Cell Death Differ. 2018;25:873–84. doi: 10.1038/s41418-017-0016-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ney PA. Mitochondrial autophagy: Origins, significance, and role of BNIP3 and NIX. Biochim Biophys Acta. 2015;1853:2775–83. doi: 10.1016/j.bbamcr.2015.02.022. [DOI] [PubMed] [Google Scholar]
- 58.Tracy K, Dibling BC, Spike BT, Knabb JR, Schumacker P, Macleod KF. BNIP3 is an RB/E2F target gene required for hypoxia-induced autophagy. Mol Cell Biol. 2007;27:6229–42. doi: 10.1128/MCB.02246-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shaw J, Yurkova N, Zhang T, Gang H, Aguilar F, Weidman D, et al. Antagonism of E2F-1 regulated Bnip3 transcription by NF-kappaB is essential for basal cell survival. Proc Natl Acad Sci USA. 2008;105:20734–9. doi: 10.1073/pnas.0807735105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang B, Ling S, Lin WC. 14-3-3Tau regulates Beclin 1 and is required for autophagy. PLoS One. 2010;5:e10409. doi: 10.1371/journal.pone.0010409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Copetti T, Bertoli C, Dalla E, Demarchi F, Schneider C. p65/RelA modulates BECN1 transcription and autophagy. Mol Cell Biol. 2009;29:2594–608. doi: 10.1128/MCB.01396-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Polager S, Ofir M, Ginsberg D. E2F1 regulates autophagy and the transcription of autophagy genes. Oncogene. 2008;27:4860–4. doi: 10.1038/onc.2008.117. [DOI] [PubMed] [Google Scholar]
- 63.Garcia-Garcia A, Rodriguez-Rocha H, Tseng MT, Montes de Oca-Luna R, Zhou HS, McMasters KM, et al. E2F-1 lacking the transcriptional activity domain induces autophagy. Cancer Biol Ther. 2012;13:1091–101. doi: 10.4161/cbt.21143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bucha S, Mukhopadhyay D, Bhattacharyya NP. E2F1 activates MFN2 expression by binding to the promoter and decreases mitochondrial fission and mitophagy in HeLa cells. FEBS J. 2019;286:4525–41. doi: 10.1111/febs.14980. [DOI] [PubMed] [Google Scholar]
- 65.Tremblay M, Sanchez-Ferras O, Bouchard M. GATA transcription factors in development and disease. Development. 2007;145:dev164384. doi: 10.1242/dev.164384. [DOI] [PubMed] [Google Scholar]
- 66.Kang YA, Sanalkumar R, O’Geen H, Linnemann AK, Chang CJ, Bouhassira EE, et al. Autophagy driven by a master regulator of hematopoiesis. Mol Cell Biol. 2012;32:226–39. doi: 10.1128/MCB.06166-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kobayashi S, Volden P, Timm D, Mao K, Xu X, Liang Q. Transcription factor GATA4 inhibits doxorubicin-induced autophagy and cardiomyocyte death. J Biol Chem. 2010;285:793–804. doi: 10.1074/jbc.M109.070037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kobayashi S, Lackey T, Huang Y, Bisping E, Pu WT, Boxer LM, et al. Transcription factor gata4 regulates cardiac BCL2 gene expression in vitro and in vivo. FASEB J. 2006;20:800–2. doi: 10.1096/fj.05-5426fje. [DOI] [PubMed] [Google Scholar]
- 69.Kang C, Xu Q, Martin TD, Li MZ, Demaria M, Aron L, et al. The DNA damage response induces inflammation and senescence by inhibiting autophagy of GATA4. Science. 2015;349:aaa5612. doi: 10.1126/science.aaa5612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ma K, Saha PK, Chan L, Moore DD. Farnesoid X receptor is essential for normal glucose homeostasis. J Clin Invest. 2006;116:1102–9. doi: 10.1172/JCI25604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Seok S, Fu T, Choi SE, Li Y, Zhu R, Kumar S, et al. Transcriptional regulation of autophagy by an FXR-CREB axis. Nature. 2014;516:108–11. doi: 10.1038/nature13949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lee JM, Wagner M, Xiao R, Kim KH, Feng D, Lazar MA, et al. Nutrient-sensing nuclear receptors coordinate autophagy. Nature. 2014;516:112–5. doi: 10.1038/nature13961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Faghfouri AH, Khajebishak Y, Payahoo L, Faghfuri E, Alivand M. PPAR-gamma agonists: Potential modulators of autophagy in obesity. Eur J Pharm. 2021;912:174562. doi: 10.1016/j.ejphar.2021.174562. [DOI] [PubMed] [Google Scholar]
- 74.Zhou J, Zhang W, Liang B, Casimiro MC, Whitaker-Menezes D, Wang M, et al. PPARgamma activation induces autophagy in breast cancer cells. Int J Biochem Cell Biol. 2009;41:2334–42. doi: 10.1016/j.biocel.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Liu J, Yao Q, Xiao L, Ma W, Li F, Lai B, et al. PPARgamma induces NEDD4 gene expression to promote autophagy and insulin action. FEBS J. 2020;287:529–45. doi: 10.1111/febs.15042. [DOI] [PubMed] [Google Scholar]
- 76.Qin H, Tan W, Zhang Z, Bao L, Shen H, Wang F, et al. 15d-prostaglandin J2 protects cortical neurons against oxygen-glucose deprivation/reoxygenation injury: involvement of inhibiting autophagy through upregulation of Bcl-2. Cell Mol Neurobiol. 2015;35:303–12. doi: 10.1007/s10571-014-0125-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yao J, Zheng K, Zhang X. Rosiglitazone exerts neuroprotective effects via the suppression of neuronal autophagy and apoptosis in the cortex following traumatic brain injury. Mol Med Rep. 2015;12:6591–7. doi: 10.3892/mmr.2015.4292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Margariti A, Li H, Chen T, Martin D, Vizcay-Barrena G, Alam S, et al. XBP1 mRNA splicing triggers an autophagic response in endothelial cells through BECLIN-1 transcriptional activation. J Biol Chem. 2013;288:859–72. doi: 10.1074/jbc.M112.412783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhou Y, Lee J, Reno CM, Sun C, Park SW, Chung J, et al. Regulation of glucose homeostasis through a XBP-1-FoxO1 interaction. Nat Med. 2011;17:356–65. doi: 10.1038/nm.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhao Y, Li X, Cai MY, Ma K, Yang J, Zhou J, et al. XBP-1u suppresses autophagy by promoting the degradation of FoxO1 in cancer cells. Cell Res. 2013;23:491–507. doi: 10.1038/cr.2013.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.You L, Wang Z, Li H, Shou J, Jing Z, Xie J, et al. The role of STAT3 in autophagy. Autophagy. 2015;11:729–39. doi: 10.1080/15548627.2015.1017192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Oh HM, Yu CR, Dambuza I, Marrero B, Egwuagu CE. STAT3 protein interacts with Class O Forkhead transcription factors in the cytoplasm and regulates nuclear/cytoplasmic localization of FoxO1 and FoxO3a proteins in CD4(+) T cells. J Biol Chem. 2012;287:30436–43. doi: 10.1074/jbc.M112.359661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shen S, Niso-Santano M, Adjemian S, Takehara T, Malik SA, Minoux H, et al. Cytoplasmic STAT3 represses autophagy by inhibiting PKR activity. Mol Cell. 2012;48:667–80. doi: 10.1016/j.molcel.2012.09.013. [DOI] [PubMed] [Google Scholar]
- 84.Liu L, Tao Z, Zheng LD, Brooke JP, Smith CM, Liu D, et al. FoxO1 interacts with transcription factor EB and differentially regulates mitochondrial uncoupling proteins via autophagy in adipocytes. Cell Death Disco. 2016;2:16066. doi: 10.1038/cddiscovery.2016.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ghosh A, Jana M, Modi K, Gonzalez FJ, Sims KB, Berry-Kravis E, et al. Activation of peroxisome proliferator-activated receptor alpha induces lysosomal biogenesis in brain cells: implications for lysosomal storage disorders. J Biol Chem. 2015;290:10309–24. doi: 10.1074/jbc.M114.610659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Morselli E, Tasdemir E, Maiuri MC, Galluzzi L, Kepp O, Criollo A, et al. Mutant p53 protein localized in the cytoplasm inhibits autophagy. Cell Cycle. 2008;7:3056–61. doi: 10.4161/cc.7.19.6751. [DOI] [PubMed] [Google Scholar]
- 87.Agarwal S, Bell CM, Taylor SM, Moran RG. p53 deletion or hotspot mutations enhance mTORC1 activity by altering lysosomal dynamics of TSC2 and Rheb. Mol Cancer Res. 2016;14:66–77. doi: 10.1158/1541-7786.MCR-15-0159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cordani M, Oppici E, Dando I, Butturini E, Dalla Pozza E, Nadal-Serrano M, et al. Mutant p53 proteins counteract autophagic mechanism sensitizing cancer cells to mTOR inhibition. Mol Oncol. 2016;10:1008–29. doi: 10.1016/j.molonc.2016.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhou G, Wang J, Zhao M, Xie TX, Tanaka N, Sano D, et al. Gain-of-function mutant p53 promotes cell growth and cancer cell metabolism via inhibition of AMPK activation. Mol Cell. 2014;54:960–74. doi: 10.1016/j.molcel.2014.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rahman MA, Park MN, Rahman MH, Rashid MM, Islam R, Uddin MJ, et al. p53 modulation of autophagy signaling in cancer therapies: perspectives mechanism and therapeutic targets. Front Cell Dev Biol. 2022;10:761080. doi: 10.3389/fcell.2022.761080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Li F, Miao L, Xue T, Qin H, Mondal S, Thompson PR, et al. Inhibiting PAD2 enhances the anti-tumor effect of docetaxel in tamoxifen-resistant breast cancer cells. J Exp Clin Cancer Res. 2019;38:414. doi: 10.1186/s13046-019-1404-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hu L, Wang Y, Chen Z, Fu L, Wang S, Zhang X, et al. Hsp90 Inhibitor SNX-2112 enhances TRAIL-induced apoptosis of human cervical cancer cells via the ROS-mediated JNK-p53-autophagy-DR5 pathway. Oxid Med Cell Longev. 2019;2019:9675450. doi: 10.1155/2019/9675450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Blessing AM, Rajapakshe K, Reddy Bollu L, Shi Y, White MA, Pham AH, et al. Transcriptional regulation of core autophagy and lysosomal genes by the androgen receptor promotes prostate cancer progression. Autophagy. 2017;13:506–21. doi: 10.1080/15548627.2016.1268300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Giatromanolaki A, Kalamida D, Sivridis E, Karagounis IV, Gatter KC, Harris AL, et al. Increased expression of transcription factor EB (TFEB) is associated with autophagy, migratory phenotype and poor prognosis in non-small cell lung cancer. Lung Cancer. 2015;90:98–105. doi: 10.1016/j.lungcan.2015.07.008. [DOI] [PubMed] [Google Scholar]
- 95.Liang J, Jia X, Wang K, Zhao N. High expression of TFEB is associated with aggressive clinical features in colorectal cancer. Onco Targets Ther. 2018;11:8089–98. doi: 10.2147/OTT.S180112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Fang LM, Li B, Guan JJ, Xu HD, Shen GH, Gao QG, et al. Transcription factor EB is involved in autophagy-mediated chemoresistance to doxorubicin in human cancer cells. Acta Pharm Sin. 2017;38:1305–16. doi: 10.1038/aps.2017.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Giatromanolaki A, Sivridis E, Kalamida D, Koukourakis MI. Transcription factor EB expression in early breast cancer relates to lysosomal/autophagosomal markers and prognosis. Clin Breast Cancer. 2017;17:e119–e125. doi: 10.1016/j.clbc.2016.11.006. [DOI] [PubMed] [Google Scholar]
- 98.Perera RM, Stoykova S, Nicolay BN, Ross KN, Fitamant J, Boukhali M, et al. Transcriptional control of autophagy-lysosome function drives pancreatic cancer metabolism. Nature. 2015;524:361–5. doi: 10.1038/nature14587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.He R, Wang M, Zhao C, Shen M, Yu Y, He L, et al. TFEB-driven autophagy potentiates TGF-beta induced migration in pancreatic cancer cells. J Exp Clin Cancer Res. 2019;38:340. doi: 10.1186/s13046-019-1343-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mitrakas AG, Kalamida D, Giatromanolaki A, Pouliliou S, Tsolou A, Kyranas R, et al. Autophagic flux response and glioblastoma sensitivity to radiation. Cancer Biol Med. 2018;15:260–74. doi: 10.20892/j.issn.2095-3941.2017.0173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.He R, Shi X, Zhou M, Zhao Y, Pan S, Zhao C, et al. Alantolactone induces apoptosis and improves chemosensitivity of pancreatic cancer cells by impairment of autophagy-lysosome pathway via targeting TFEB. Toxicol Appl Pharm. 2018;356:159–71. doi: 10.1016/j.taap.2018.08.003. [DOI] [PubMed] [Google Scholar]
- 102.Zhao B, Dierichs L, Gu JN, Trajkovic-Arsic M, Axel Hilger R, Savvatakis K, et al. TFEB-mediated lysosomal biogenesis and lysosomal drug sequestration confer resistance to MEK inhibition in pancreatic cancer. Cell Death Disco. 2020;6:12. doi: 10.1038/s41420-020-0246-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Liang C, Dong Z, Cai X, Shen J, Xu Y, Zhang M, et al. Hypoxia induces sorafenib resistance mediated by autophagy via activating FOXO3a in hepatocellular carcinoma. Cell Death Dis. 2020;11:1017. doi: 10.1038/s41419-020-03233-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Menzies FM, Fleming A, Caricasole A, Bento CF, Andrews SP, Ashkenazi A, et al. Autophagy and Neurodegeneration: Pathogenic Mechanisms and Therapeutic Opportunities. Neuron. 2017;93:1015–34. doi: 10.1016/j.neuron.2017.01.022. [DOI] [PubMed] [Google Scholar]
- 105.Wang H, Wang R, Xu S, Lakshmana MK. Transcription Factor EB Is Selectively Reduced in the Nuclear Fractions of Alzheimer’s and Amyotrophic Lateral Sclerosis Brains. Neurosci J. 2016;2016:4732837. doi: 10.1155/2016/4732837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Xiao Q, Yan P, Ma X, Liu H, Perez R, Zhu A, et al. Neuronal-Targeted TFEB Accelerates Lysosomal Degradation of APP, Reducing Abeta Generation and Amyloid Plaque Pathogenesis. J Neurosci. 2015;35:12137–51. doi: 10.1523/JNEUROSCI.0705-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Binder JL, Chander P, Deretic V, Weick JP, Bhaskar K. Optical induction of autophagy via Transcription factor EB (TFEB) reduces pathological tau in neurons. PLoS One. 2020;15:e0230026. doi: 10.1371/journal.pone.0230026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Yang C, Su C, Iyaswamy A, Krishnamoorthi SK, Zhu Z, Yang S, et al. Celastrol enhances transcription factor EB (TFEB)-mediated autophagy and mitigates Tau pathology: Implications for Alzheimer’s disease therapy. Acta Pharm Sin B. 2022;12:1707–22. doi: 10.1016/j.apsb.2022.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Chun YS, Kim MY, Lee SY, Kim MJ, Hong TJ, Jeon JK, et al. MEK1/2 inhibition rescues neurodegeneration by TFEB-mediated activation of autophagic lysosomal function in a model of Alzheimer’s Disease. Mol Psychiatry. 2022;27:4770–80. doi: 10.1038/s41380-022-01713-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Gu Z, Cao H, Zuo C, Huang Y, Miao J, Song Y, et al. TFEB in Alzheimer’s disease: From molecular mechanisms to therapeutic implications. Neurobiol Dis. 2022;173:105855. doi: 10.1016/j.nbd.2022.105855. [DOI] [PubMed] [Google Scholar]
- 111.Decressac M, Mattsson B, Weikop P, Lundblad M, Jakobsson J, Bjorklund A. TFEB-mediated autophagy rescues midbrain dopamine neurons from alpha-synuclein toxicity. Proc Natl Acad Sci USA. 2013;110:E1817–1826. doi: 10.1073/pnas.1305623110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kilpatrick K, Zeng Y, Hancock T, Segatori L. Genetic and chemical activation of TFEB mediates clearance of aggregated alpha-synuclein. PLoS One. 2015;10:e0120819. doi: 10.1371/journal.pone.0120819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Vodicka P, Chase K, Iuliano M, Tousley A, Valentine DT, Sapp E, et al. Autophagy Activation by Transcription Factor EB (TFEB) in Striatum of HDQ175/Q7 Mice. J Huntingt Dis. 2016;5:249–60. doi: 10.3233/JHD-160211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kataura T, Tashiro E, Nishikawa S, Shibahara K, Muraoka Y, Miura M, et al. A chemical genomics-aggrephagy integrated method studying functional analysis of autophagy inducers. Autophagy. 2021;17:1856–72. doi: 10.1080/15548627.2020.1794590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Cortes CJ, Miranda HC, Frankowski H, Batlevi Y, Young JE, Le A, et al. Polyglutamine-expanded androgen receptor interferes with TFEB to elicit autophagy defects in SBMA. Nat Neurosci. 2014;17:1180–9. doi: 10.1038/nn.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Chua JP, Reddy SL, Merry DE, Adachi H, Katsuno M, Sobue G, et al. Transcriptional activation of TFEB/ZKSCAN3 target genes underlies enhanced autophagy in spinobulbar muscular atrophy. Hum Mol Genet. 2014;23:1376–86. doi: 10.1093/hmg/ddt527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Chen Y, Liu H, Guan Y, Wang Q, Zhou F, Jie L, et al. The altered autophagy mediated by TFEB in animal and cell models of amyotrophic lateral sclerosis. Am J Transl Res. 2015;7:1574–87. [PMC free article] [PubMed] [Google Scholar]
- 118.Cunningham KM, Maulding K, Ruan K, Senturk M, Grima JC, Sung H, et al. TFEB/Mitf links impaired nuclear import to autophagolysosomal dysfunction in C9-ALS. Elife. 2020;9:e59419. doi: 10.7554/eLife.59419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Luo R, Su LY, Li G, Yang J, Liu Q, Yang LX, et al. Activation of PPARA-mediated autophagy reduces Alzheimer disease-like pathology and cognitive decline in a murine model. Autophagy. 2020;16:52–69. doi: 10.1080/15548627.2019.1596488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Raha S, Ghosh A, Dutta D, Patel DR, Pahan K. Activation of PPARalpha enhances astroglial uptake and degradation of beta-amyloid. Sci Signal. 2021;14:eabg4747. doi: 10.1126/scisignal.abg4747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Dutta D, Paidi RK, Raha S, Roy A, Chandra S, Pahan K. Treadmill exercise reduces alpha-synuclein spreading via PPARalpha. Cell Rep. 2022;40:111058. doi: 10.1016/j.celrep.2022.111058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Tsunemi T, Ashe TD, Morrison BE, Soriano KR, Au J, Roque RA, et al. PGC-1alpha rescues Huntington’s disease proteotoxicity by preventing oxidative stress and promoting TFEB function. Sci Transl Med. 2012;4:142ra197. doi: 10.1126/scitranslmed.3003799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Brattas PL, Hersbach BA, Madsen S, Petri R, Jakobsson J, Pircs K. Impact of differential and time-dependent autophagy activation on therapeutic efficacy in a model of Huntington disease. Autophagy. 2021;17:1316–29. doi: 10.1080/15548627.2020.1760014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Yang J, Xu H, Zhang C, Yang X, Cai W, Chen X. A prion-like domain of TFEB mediates the co-aggregation of TFEB and mHTT. Autophagy. 2023;19:544–50. doi: 10.1080/15548627.2022.2083857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Haim Y, Bluher M, Slutsky N, Goldstein N, Kloting N, Harman-Boehm I, et al. Elevated autophagy gene expression in adipose tissue of obese humans: A potential non-cell-cycle-dependent function of E2F1. Autophagy. 2015;11:2074–88. doi: 10.1080/15548627.2015.1094597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kovsan J, Bluher M, Tarnovscki T, Kloting N, Kirshtein B, Madar L, et al. Altered autophagy in human adipose tissues in obesity. J Clin Endocrinol Metab. 2011;96:E268–277. doi: 10.1210/jc.2010-1681. [DOI] [PubMed] [Google Scholar]
- 127.Xiong M, Hu W, Tan Y, Yu H, Zhang Q, Zhao C, et al. Transcription factor E2F1 knockout promotes mice white adipose tissue browning through autophagy inhibition. Front Physiol. 2021;12:748040. doi: 10.3389/fphys.2021.748040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Zhang X, Liu Q, Zhang X, Guo K, Zhang X, Zhou Z. FOXO3a regulates lipid accumulation and adipocyte inflammation in adipocytes through autophagy: Role of FOXO3a in obesity. Inflamm Res. 2021;70:591–603. doi: 10.1007/s00011-021-01463-0. [DOI] [PubMed] [Google Scholar]
- 129.Trivedi PC, Bartlett JJ, Perez LJ, Brunt KR, Legare JF, Hassan A, et al. Glucolipotoxicity diminishes cardiomyocyte TFEB and inhibits lysosomal autophagy during obesity and diabetes. Biochim Biophys Acta. 2016;1861:1893–910. doi: 10.1016/j.bbalip.2016.09.004. [DOI] [PubMed] [Google Scholar]
- 130.Lim H, Lim YM, Kim KH, Jeon YE, Park K, Kim J, et al. A novel autophagy enhancer as a therapeutic agent against metabolic syndrome and diabetes. Nat Commun. 2018;9:1438. doi: 10.1038/s41467-018-03939-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Brady OA, Martina JA, Puertollano R. Emerging roles for TFEB in the immune response and inflammation. Autophagy. 2018;14:181–9. doi: 10.1080/15548627.2017.1313943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Lapierre LR, Kumsta C, Sandri M, Ballabio A, Hansen M. Transcriptional and epigenetic regulation of autophagy in aging. Autophagy. 2015;11:867–80. doi: 10.1080/15548627.2015.1034410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Morgan MAJ, Shilatifard A. Reevaluating the roles of histone-modifying enzymes and their associated chromatin modifications in transcriptional regulation. Nat Genet. 2020;52:1271–81. doi: 10.1038/s41588-020-00736-4. [DOI] [PubMed] [Google Scholar]
- 134.Shin HJ, Kim H, Oh S, Lee JG, Kee M, Ko HJ, et al. AMPK-SKP2-CARM1 signalling cascade in transcriptional regulation of autophagy. Nature. 2016;534:553–7. doi: 10.1038/nature18014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Sakamaki JI, Wilkinson S, Hahn M, Tasdemir N, O’Prey J, Clark W, et al. Bromodomain Protein BRD4 Is a Transcriptional Repressor of Autophagy and Lysosomal Function. Mol Cell. 2017;66:517–532.e519. doi: 10.1016/j.molcel.2017.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Lei Y, Huang Y, Wen X, Yin Z, Zhang Z, Klionsky DJ. How cells deal with the fluctuating environment: autophagy regulation under stress in yeast and Mammalian systems. Antioxidants (Basel) 2022;11:304. doi: 10.3390/antiox11020304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Yu F, Imamura Y, Ueno M, Suzuki SW, Ohsumi Y, Yukawa M, et al. The yeast chromatin remodeler Rsc1-RSC complex is required for transcriptional activation of autophagy-related genes and inhibition of the TORC1 pathway in response to nitrogen starvation. Biochem Biophys Res Commun. 2015;464:1248–53. doi: 10.1016/j.bbrc.2015.07.114. [DOI] [PubMed] [Google Scholar]
- 138.Pike LR, Singleton DC, Buffa F, Abramczyk O, Phadwal K, Li JL, et al. Transcriptional up-regulation of ULK1 by ATF4 contributes to cancer cell survival. Biochem J. 2013;449:389–400. doi: 10.1042/BJ20120972. [DOI] [PubMed] [Google Scholar]
- 139.B’Chir W, Maurin AC, Carraro V, Averous J, Jousse C, Muranishi Y, et al. The eIF2alpha/ATF4 pathway is essential for stress-induced autophagy gene expression. Nucleic Acids Res. 2013;41:7683–99. doi: 10.1093/nar/gkt563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Li DD, Wang LL, Deng R, Tang J, Shen Y, Guo JF, et al. The pivotal role of c-Jun NH2-terminal kinase-mediated Beclin 1 expression during anticancer agents-induced autophagy in cancer cells. Oncogene. 2009;28:886–98. doi: 10.1038/onc.2008.441. [DOI] [PubMed] [Google Scholar]
- 141.Sun T, Li D, Wang L, Xia L, Ma J, Guan Z, et al. c-Jun NH2-terminal kinase activation is essential for up-regulation of LC3 during ceramide-induced autophagy in human nasopharyngeal carcinoma cells. J Transl Med. 2011;9:161. doi: 10.1186/1479-5876-9-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Salmon M, Spinosa M, Zehner ZE, Upchurch GR, Ailawadi G. Klf4, Klf2, and Zfp148 activate autophagy-related genes in smooth muscle cells during aortic aneurysm formation. Physiol Rep. 2019;7:e14058. doi: 10.14814/phy2.14058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Pan CC, Kumar S, Shah N, Bloodworth JC, Hawinkels LJ, Mythreye K, et al. Endoglin regulation of Smad2 function mediates Beclin1 expression and endothelial autophagy. J Biol Chem. 2015;290:14884–92. doi: 10.1074/jbc.M114.630178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Zhu Y, Huang S, Chen S, Chen J, Wang Z, Wang Y, et al. SOX2 promotes chemoresistance, cancer stem cells properties, and epithelial-mesenchymal transition by beta-catenin and Beclin1/autophagy signaling in colorectal cancer. Cell Death Dis. 2021;12:449. doi: 10.1038/s41419-021-03733-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Seo YK, Jeon TI, Chong HK, Biesinger J, Xie X, Osborne TF. Genome-wide localization of SREBP-2 in hepatic chromatin predicts a role in autophagy. Cell Metab. 2011;13:367–75. doi: 10.1016/j.cmet.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Cheng C, Deng X, Xu K. Increased expression of sterol regulatory element binding protein‑2 alleviates autophagic dysfunction in NAFLD. Int J Mol Med. 2018;41:1877–86. doi: 10.3892/ijmm.2018.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Goldberg AA, Nkengfac B, Sanchez AMJ, Moroz N, Qureshi ST, Koromilas AE, et al. Regulation of ULK1 expression and autophagy by STAT1. J Biol Chem. 2017;292:1899–909. doi: 10.1074/jbc.M116.771584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Miao LJ, Huang FX, Sun ZT, Zhang RX, Huang SF, Wang J. Stat3 inhibits Beclin 1 expression through recruitment of HDAC3 in nonsmall cell lung cancer cells. Tumour Biol. 2014;35:7097–103. doi: 10.1007/s13277-014-1961-6. [DOI] [PubMed] [Google Scholar]