Abstract
Proteins from the BCL-2 family control cell survival and apoptosis in health and disease, and regulate apoptosis-unrelated cellular processes. BCL-Gonad (BCL-G, also known as BCL2-like 14) is a non-typical protein of the family as its long isoform (BCL-GL) consists of BH2 and BH3 domains without the BH1 motif. BCL-G is predominantly expressed in normal testes and different organs of the gastrointestinal tract. The complexity of regulatory mechanisms of BCL-G expression and post-translational modifications suggests that BCL-G may play distinct roles in different types of cells and disorders. While several genetic alterations of BCL2L14 have been reported, gene deletions and amplifications prevail, which is also confirmed by the analysis of sequencing data for different types of cancer. Although the studies validating the phenotypic consequences of genetic manipulations of BCL-G are limited, the role of BCL-G in apoptosis has been undermined. Recent studies using gene-perturbation approaches have revealed apoptosis-unrelated functions of BCL-G in intracellular trafficking, immunomodulation, and regulation of the mucin scaffolding network. These studies were, however, limited mainly to the role of BCL-G in the gastrointestinal tract. Therefore, further efforts using state-of-the-art methods and various types of cells are required to find out more about BCL-G activities. Deciphering the isoform-specific functions of BCL-G and the BCL-G interactome may result in the designing of novel therapeutic approaches, in which BCL-G activity will be either imitated using small-molecule BH3 mimetics or inhibited to counteract BCL-G upregulation. This review summarizes two decades of research on BCL-G.
Subject terms: Prognostic markers, Tumour-suppressor proteins, Cancer genetics, Gene expression, Gene regulation
Facts
Two validated protein isoforms of BCL-G: BCL-GL and BCL-GS are generated in humans as a result of alternative splicing.
BCL-G is predominantly expressed in normal testes, and organs of the gastrointestinal tract already at the early stages of fetal development.
BCL-G level and activity are under the control of multiple proteins, including transcriptional regulators such as p53, PAR bZIP, IRF-1, STAT1, NF-κB and G9a, and a post-translational modifier FAU.
The involvement of BCL-G in pro-apoptotic activity has been undermined by recent studies in mouse models using genetic manipulation approaches and complementary methods.
Open questions
Do validated human isoforms of BCL-G differ in cellular functions?
Is BCL-G evolutionary conserved and plays similar roles across different species?
Is the regulation of BCL2L14 expression and BCL-G activity cell type-specific, and does BCL-G play a considerable role beyond the gastrointestinal tract?
Could mimicking activity of BCL-G be a therapeutic approach in inflammation-associated colon diseases considering its role in the gastrointestinal tract?
Introduction
Members of the B-cell lymphoma-2 (BCL-2) family of proteins are involved in the control of cell survival and death in specific physiological and pathological contexts [1]. It has been widely demonstrated and discussed that different BCL-2-like proteins can exert apoptotic and non-apoptotic functions, while they exhibit redundancy in selected cellular processes and cell types [2–9]. The BCL-2 family comprises at least 20 proteins, which can be classified based on apoptosis-related activity and the presence of the structural regions named BCL-2 homology (BH) domains. In general, four BH domains are found in pro-survival proteins, while pro-apoptotic proteins either lack the BH4 domain as shown for BAX, BAK, and BOK, or possess exclusively BH3 domain (BH3-only proteins). Cell fate is determined by complex protein-protein interactions between different members of the family [10–12]. Clinical implications of these interactions have been substantiated with the advent of a novel group of small-molecule agents called BH3 mimetics. BH3 mimetics resemble the activity of specific BH3-only proteins, therefore they induce apoptosis via the inhibition of pro-survival proteins [13–16]. Venetoclax, a first-in-class selective inhibitor of BCL-2 was primarily approved in 2016 by FDA for the treatment of patients with chronic lymphocytic leukemia [17], and then received approval as a therapeutic regimen in other hematological malignancies [18]. In addition, several other selective and dual-targeting BH3 mimetics have demonstrated promising activity in disease, and they undergo extensive investigation. However, the activity of these drugs is (1) restricted to cell types that exhibit dependence on particular BCL-2-like proteins, and (2) limited by adaptive resistance resulting from the application of BH3 mimetics [19–23]. In this respect, unraveling cell dependence on particular pro-survival proteins and deciphering pro-apoptotic BCL-2-like proteins that fuel cell death response, in addition to delineating mechanisms of apoptosis-unrelated functions of BCL-2-like proteins can provide basis for efficient targeting of cancers and non-cancerous disorders. This review summarizes current knowledge on the structure and function of BCL-Gonad (BCL-G), which is an unusual BCL-2-like protein as (1) its long isoform (BCL-GL) consists of BH2 and BH3 domains without the BH1 motif, and (2) its role in apoptosis has been undermined by recent studies using genetic manipulation approaches and complementary methods.
BCL-G: characteristics of gene and corresponding protein
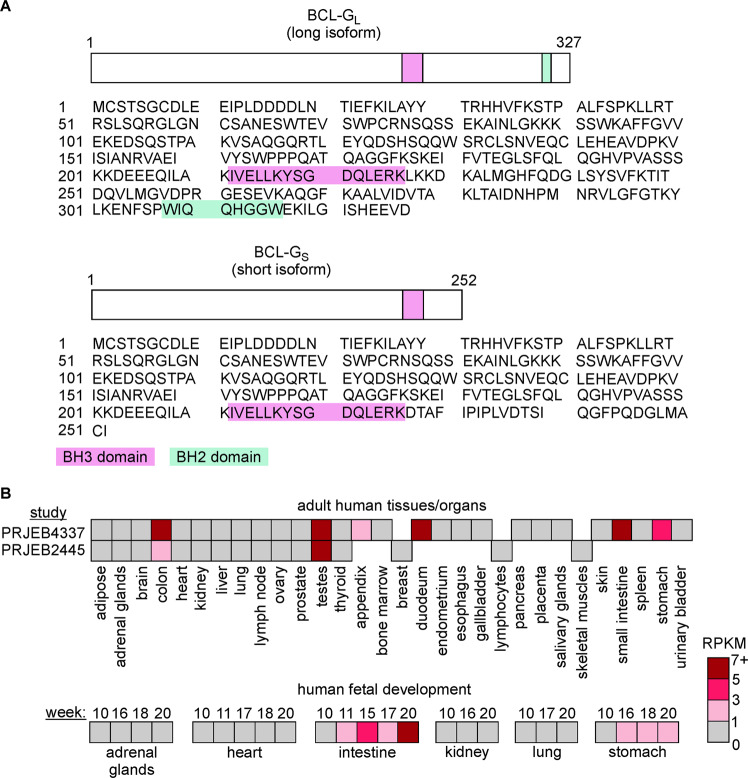
cDNA of BCL-G (also known as BCL2-like 14) was cloned and initially characterized in Reed’s lab in 2001 [24]. BCL2L14 encoding BCL-G is located on chromosome 12. As a result of alternative splicing, BCL-G proteins of different lengths can be generated in humans, including short (BCL-GS), median (BCL-GM), and long (BCL-GL) isoforms consisting of 252, 276, and 327 amino acid residues, respectively [24, 25]. It was shown that BCL-G isoforms differed within their C-terminal region. The BH3 domain, which is typical of the vast majority of proteins from the BCL-2 family, was found in all three isoforms, while the presence of the BH2 domain was restricted to BCL-GL [24, 25] (Fig. 1A). Currently available data on Ensembl genome browser indicate that human BCL2L14 gives rise to 14 transcripts, from which only eight contain an open reading frame (www.ensembl.org). Notably, a splice variant potentially translated into the 276-amino acid isoform of BCL-G is labeled as nonsense-mediated decay (www.ensembl.org), suggesting that BCL-GM protein might not have a biological function. The open reading frame of porcine Bcl-G contains five exons and encodes 329-amino acid protein that demonstrated 71% identity with human BCL-G [26], while in mice, only one isoform resembling human BCL-GL was found [27]. The similarity between murine Bcl-G and human BCL-GL was substantiated by the demonstration that BCL-GL exhibited a very low affinity to BCL-XL, while BCL-GL binding to BCL-XL could be enhanced upon deletion of the BH2 domain both in humans and mice [24, 28].
Fig. 1. Domain structure of BCL-G and expression of BCL2L14 in human tissues and organs.
A Amino acid sequences of human long and short isoforms of BCL-G: BCL-GL and BCL-GS, respectively were obtained from uniprot.org. The BH3 domain (amino acids from 212 to 226) and BH2 domain (amino acids from 308 to 315) are marked. B Upper panel: Transcript levels of BCL2L14 were assessed by RNA sequencing in different human tissues and organs (BioProject dataset, study PRJEB4337 including samples from 95 individuals representing 27 different tissues, also published [117]; and PRJEB2445 involving 16 tissue samples). Lower panel: Transcript levels of BCL2L14 were assessed by RNA sequencing in different human fetal tissues between the 10th and 20th weeks of fetal development (BioProject dataset, study PRJNA270632 including 35 human fetal samples from 6 tissues, also published [118]). RPKM reads per kilobase of transcript per million reads mapped.
Expression of Bcl2l14 in normal tissues
BCL-GL transcript was initially detected in several normal human organs, including the testes, prostate, lung, bone marrow, colon, and pancreas, whereas BCL-GS and BCL-GM were found exclusively in testes [24, 25]. In a more recent study, high levels of both short and long isoforms of BCL-G were found in the human stomach, small intestine, colon, testes, and lymph nodes, but not in the heart and brain, while in the spleen, only BCL-GL was expressed [29]. Therefore, considering BCL-G isoforms separately could be useful to fully delineate their biological functions. In two other BioProject datasets (www.ncbi.nlm.nih.gov/bioproject) involving RNA-seq, a high level of BCL-G mRNA was assessed in a few adult human organs, including the colon, duodenum, small intestine, stomach, appendix, and testes (Fig. 1B, upper panel). Notably, high expression of BCL2L14 in the gastrointestinal tract was found early during human fetal development, while BCL-G transcript level was low or undetectable in fetal adrenal glands, heart, kidney, and lungs up to 20th week (Fig. 1B, lower panel). Using highly specific monoclonal antibodies, murine Bcl-G was predominantly detected in the small intestine and colon, ciliated epithelial cells in the trachea, bronchi and lungs, CD8+ dendritic cells, bladder, uterus, stratified squamous epithelia of the tongue, salivary and lacrimal glands, and late-stage spermatids of the germinal epithelium [30]. In turn, Bcl2l14 was not expressed in the interstitial Leydig cells, in cells undergoing earlier stages of spermatogenesis, kidneys, liver, and brain [30]. The transcript level of porcine Bcl-G was assessed at a high level in the heart, lymph nodes, spleen, tonsil, lung, liver, and thymus, while the lowest level was found in the kidney [31].
Regulation of BCL-G level and activity
Transcriptional regulation
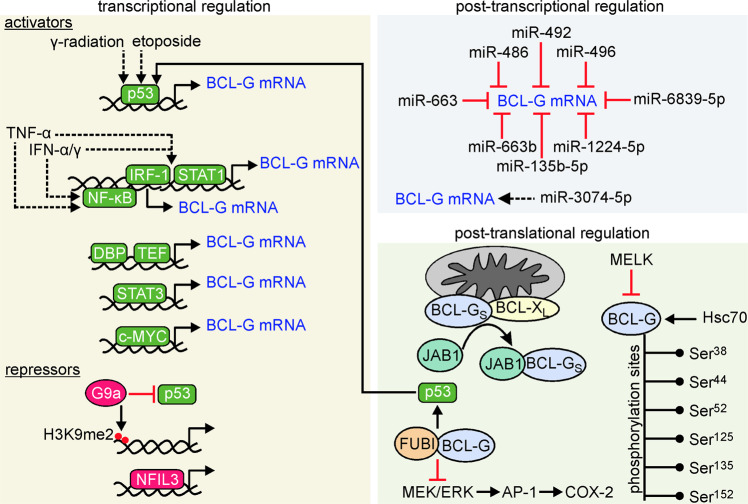
Several transcriptional regulators of BCL2L14 expression were identified (Fig. 2). Proline- and acid-rich basic region leucine zipper (PAR bZIP) proteins were shown to control BCL2L14 expression, particularly the BCL-GS isoform in human embryonic kidney [32], but not in mice [33]. The human BCL2L14 promoter was especially responsive to activation by the thyrotroph embryonic factor (TEF) in a p53-independent manner, and sensitive to suppression by the nuclear factor interleukin-3-regulated (NFIL3) [32]. In addition, although the BCL2L14 promoter was also activated by the D-site binding protein (DBP), it was demonstrated that the DBP variant lacking the transactivation domain reduced the formation of active TEF dimers and affected BCL-GS expression [32]. A functional intronic p53-binding site was uncovered in BCL2L14, and the BCL-G transcript level increased after tetracycline-inducible activation of p53 [34]. Pharmacological inhibition of G9a, a transcriptional repressor that di-methylates histone 3 lysine 9 (H3K9me2), resulted in striking Bcl-G upregulation via recruitment of p53 to Bcl2l14. Notably, this mechanism was not universal for the regulation of all p53-dependent genes [35]. Accordingly, Bcl-G was substantially upregulated in hepatocyte-specific G9a-deficient (G9aΔHep) mice, and expression of BCL-G and G9a were negatively correlated in the human liver [35]. Knockout of Wdr5, encoding a protein involved in chromatin modifications and regulation of mouse embryonic stem cell differentiation, was associated with the downregulation of Bcl2l14 expression in a p53-dependent manner [36]. p53 might contribute to elevated Bcl2l14 expression in mice exposed to γ-radiation, whereas high levels of Bcl-G were found in the splenic white pulp, predominantly in cells with fragmented DNA [37]. Other transcriptional regulators of Bcl2l14 expression such as c-Myc and Stat3 were also considered [37]. On the other hand, no changes in the BCL-GS transcript level and a mild decrease in BCL-GL mRNA abundance were found in normal human colon cell line exposed to nutlin-3, an activator of p53 [38], suggesting cell type-dependent regulation of BCL2L14 expression by p53.
Fig. 2. Positive and negative regulators of BCL-G level and activity.
In transcriptional regulation, activators are marked in green, while transcriptional repressors are shown in red. For interaction with JAB1, BCL-GS was shown to be relocated from the association with either BCL-XL or BCL-2 [46]. Dotted lines are used to show indirect influence. The phosphorylation sites of human BCL-G were retrieved from www.phosphosite.org.
Interferon regulatory protein 1 (IRF-1)- and signal transducer and activator of transcription 1 (STAT1)-binding sites were also identified in the promoter of BCL2L14. In silico analysis of the promoter region revealed interferon regulatory factor element (IRF-E) and interferon-γ (IFN-γ)-activated site (GAS) between −204 and −139, in addition to cAMP responsive element (CRE) that was found downstream [39]. It was demonstrated that co-treatment with IFN-α and IFN-γ substantially increased the mRNA level of BCL-G in human hepatoma cells in both IRF-1- and STAT1-dependent manner [39]. IFN-γ and tumor necrosis factor α (TNF-α) synergistically upregulated short and long isoforms of BCL-G in colonic epithelial cells, while this required STAT1, p65/NF-κB, as well as Brahma (BRM) and Brahma-related gene 1 (BRG1), which are SWI/SNF-associated chromatin proteins [29]. IFN-α2b was also shown to upregulate different BCL-G splice variants in vivo [40].
Post-transcriptional regulation
Post-transcriptional regulation of BCL-G transcript by different microRNAs (miRs) was demonstrated in different cell types (Fig. 2). BCL-G mRNA was identified as a direct target for miR-663b as miR-663b could complementarily bind within the 3’UTR of BCL-G mRNA in human endometrial cancer cells [41]. BCL-G transcript was targeted by miR-496 in neuroblastoma cells [42], and by miR-486 and miR-663 in fibroblasts and keratinocytes [43]. In turn, it was demonstrated that overexpression of miR-3074-5p was accompanied by a significant increase in BCL-G protein level in human extravillous trophoblast cells [44], although it was not investigated whether this miR was directly associated with BCL-G mRNA, or this was an effect of indirect regulation. miR-135b-5p, miR-492, miR-1224-5p, and miR-6839-5p were found as potentially involved in BCL-G regulation in chondrosarcoma cells [45].
Post-translational regulation of BCL-G
BCL-GS was shown to interact with JUN activation domain-binding protein 1 (JAB1) both in vitro and in vivo, and JAB1 and BCL-GS co-localized in the cytoplasm [46] (Fig. 2). This interaction could, therefore, affect the intracellular distribution of BCL-G as it was demonstrated that BCL-GS was confined to intracellular organelles [24, 47, 48], whereas BCL-GL was diffused in the cytosol [24]. In addition, BCL-GS bound JAB1 preferentially over BCL-XL/BCL-2 when all these proteins were co-expressed [46]. Similar observations were made for porcine BCL-G and JAB1 [31], which is identical to human JAB1 [49].
BCL-G was also identified as a substrate for Ser/Thr maternal embryonic leucine-zipper kinase (MELK), and MELK interacted with BCL-G via the N-terminal moiety of BCL-G [50]. The regulation of Bcl-G by porcine Melk, which exhibits 91% similarity to human MELK, was also shown in swine umbilical vein endothelial cells (SUVECs) [51]. In addition, several putative phosphorylation sites were identified in human BCL-G (www.phosphosite.org) (Fig. 2). The consequences of these modifications and the kinases responsible for them remain, however, to be determined.
Ubiquitin-like modification of BCL-G by monoclonal non-specific suppressor factor β (MNSFβ), known as Finkel-Biskis-Reilly murine sarcoma virus-associated ubiquitously expressed (FAU), was also investigated. FAU encodes a fusion protein that consists of the N-terminal ubiquitin-like FUBI (Ubi-L) domain and C-terminal protein S30, whereas the activity of FAU protein is associated with post-translational modification of target proteins by transferring FUBI (Ubi-L) moiety [52]. FUBI (Ubi-L) was shown to covalently bind to Bcl-G through an isopeptide bond between the C-terminal Gly74 residue in FUBI (Ubi-L) and Lys110 in Bcl-G [27]. Bcl-G-Ubi-L adduct was detected in the spleen, thymus, and brain, but not in the testes [27], indicating that its presence did not overlap with a tissue-specific abundance of Bcl-G. A similar association was demonstrated for porcine Mnsfβ and Bcl-G [53]. The significance of BCL-G modification by FUBI is unlikely to be associated with the formation of polyubiquitin-like chains to promote proteasomal degradation as Lys48 is not conserved in ubiquitin-like proteins [52]. Accordingly, either ectopic overexpression or downregulation of FAU did not affect the BCL-G protein level [54]. The association between BCL-G and MNSFβ might have, however, an influence on intracellular signaling. It was demonstrated that Bcl-G-Mnsfβ inhibited mitogen-activated protein kinase (MAPK) pathway in both unstimulated and lipopolysaccharide (LPS)-exposed murine macrophages [55]. It was also shown that co-transfection with Mnsfβ and Bcl-G reduced S-nitrosoglutathione-induced extracellular signal-regulated kinase 1/2 (Erk-1/2) phosphorylation in macrophages [56]. This was accompanied by elevated expression of p53, decreased Cox-2 activity as a result of the downregulation of the activator protein 1 (AP-1) signaling cascade, and apoptosis [56]. In this respect, it can be speculated that a positive feedback loop may exist in the p53-dependent regulation of Bcl-G (Fig. 2). In addition, the contribution of heat shock proteins to the regulation of Bcl-G stability should be considered as it was demonstrated that siRNA-mediated downregulation of Hsc70 was associated with a substantial decrease in the level of Bcl-G protein in mouse macrophages [57].
Disease-associated genetic alterations of BCL2L14
A few types of genetic alterations of BCL2L14 with specific biological consequences were reported. BCL2L14-containing region of chromosome 12 was found commonly deleted in pre-B acute lymphoblastic leukemia (ALL) [58]. In addition, BCL2L14 was found within the minimal deleted region in acute myeloid leukemia (AML), however, its expression did not differ between AML with and without 12p13 deletion [59]. Allelic losses within chromosome 12 were also reported in patients with prostate cancer [60, 61], oligodontia, and thrombocytopenia [62]. An in-frame insertion of the 33-nucleotide fragment derived from exon 4 of BCL2L14 corresponding to the median isoform was detected between exon 5 of TEL and exon 2 of AML1, however, the functional consequences were not determined [63]. BCL2L14 was identified in a deleted region accompanying translocation leading to ETS variant transcription factor 6 (ETV6)-Runt-related transcription factor 1 (RUNX1) fusion protein in ALL cells [64]. In addition, BCL2L14-ETV6 fusion gene was mainly present in aggressive triple-negative breast cancer (TNBC) characterized by extensive necrosis, high tumor grade, and mesenchymal phenotype [65]. The frequency of this genetic alteration ranged from 4.4 to 12.2% of TNBC cases, while a fusion between exon 2 of ETV6 and exon 4 of BCL2L14 was the most commonly detected [65]. It was also demonstrated that ectopically expressed BCL2L14-ETV6 exerted cytosolic location, and increased cell motility and invasive potential in both TNBC and benign breast epithelial cells. Notably, a product of BCL2L14-ETV6 rearrangement promoted epithelial-mesenchymal transition and was associated with cell resistance to paclitaxel [65]. As ETV6 is a ubiquitously expressed transcriptional repressor, which was shown to form fusion genes with oncogenic consequences in a number of hematological malignancies and solid tumors [66, 67], it would be crucial to determine whether BCL2L14-ETV6 fusion occurs in other types of cancer. In addition, a single nucleotide polymorphism variant of BCL2L14 (rs1544669) in former smokers was associated with a 36% increase in risk for lung cancer [68]. As genetic aberrations of BCL2L14 rarely involved point mutations [69, 70], publicly available datasets of 310 cancer studies (www.cbioportal.org/) were analyzed with regard to BCL2L14 genetic alterations (Table 1). The overall frequency of alterations did not exceed 8% in any type of cancer (Table 1). It was accordingly demonstrated that gene amplifications and deep deletions were the most frequent alterations except for non-melanoma skin cancer (Table 1). Therefore, an in-depth investigation of the significance of genetic deletions and amplifications of the part of chromosome 12 containing BCL2L14 might answer the question about their potential roles in various diseases.
Table 1.
Ten human cancers with the highest frequency of genetic alterations of BCL2L14 based on 310 cancer studies available on cBioPortal (www.cbioportal.org/).
| Alteration frequency | |||||
|---|---|---|---|---|---|
| Cancer type | Number of cases | Amplification | Deep deletion | Mutation | Structural variant |
| Ovarian cancer | 1106 | 7.23% | 0.54% | – | – |
| Germ cell tumor | 388 | 7.47% | – | – | – |
| B-lymphoblastic leukemia/lymphoma | 1026 | 0.39% | 7.02% | – | – |
| Hodgkin lymphoma | 28 | 7.14% | – | – | – |
| Lung cancer | 39 | 2.56% | 2.56% | – | – |
| Invasive breast carcinoma | 950 | 2.84% | 0.74% | 0.21% | – |
| T-lymphoblastic leukemia/lymphoma | 92 | – | 3.26% | – | – |
| Breast cancer | 5328 | 1.93% | 0.88% | 0.02% | 0.08% |
| Skin cancer, non-melanoma | 364 | – | – | 2.75% | – |
| Pancreatic cancer | 2508 | 1.79% | 0.16% | 0.16% | – |
Only studies reporting combined data on structural variants, mutation and copy number alterations were selected for table preparation.
Role of BCL-G in normal and diseased cells—is BCL-G pro-apoptotic, pro-survival, or apoptosis-unrelated protein?
Over 20 years of research on BCL-G, a number of studies have reported changes in the levels of BCL-G transcript and/or protein in different biological systems. It was shown that CD3/CD28 stimulation promoted the formation of the Bcl-G-Mnsfβ complex that was accompanied by inhibition of Erk-1/2 and reduced secretion of interleukin-4, as well as apoptosis in Th cells and purified splenic T cells [71]. BCL2L14 was upregulated in CD4+ T cells in patients with systemic lupus erythematosus [72], in the bone marrow of myelodysplasia patients treated with arsenic trioxide and ascorbic acid [73], under hyperbaric air conditions in human diploid embryonic lung fibroblasts [74], and in tongue squamous cell carcinoma cells as a result of hypomethylation of BCL2L14 [75]. Alterations of DNA methylation patterns to regulate BCL-G levels were also found in human colon adenocarcinoma [76], and lupus [77]. BCL2L14 was upregulated by nanoparticulate tetraiodothyroacetic acid (tetrac), which reduced viable cell numbers more efficiently than unmodified tetrac in estrogen receptor-negative human breast cancer cells [78, 79]. Upregulation of BCL2L14 was demonstrated in human osteosarcoma cells exposed to either bilirubin or lithocholic acid, which were shown to have deleterious consequences on osteoblasts, while ursodeoxycholic acid attenuated this effect, suggesting that BCL-G might be involved in osteoporosis in patients with liver diseases [80]. BCL-G mRNA levels also increased in dexamethasone-induced glycogen synthase kinase-3 beta (GSK-3β)-mediated apoptosis in osteoblasts [81]. In a more recent study, it has been demonstrated that BCL-GL was overexpressed in medullary breast carcinoma compared with other subtypes of breast cancer [82]. Thus, the tumor subtype might explain inconsistent observations on either higher or lower levels of BCL-G in breast cancer cells in comparison to normal cells [50, 83]. In turn, lower levels of BCL-G were detected in patient biopsies and cell lines of prostate cancer compared with normal prostate [60, 84], and the transcript level of BCL-G was significantly decreased in human chondrosarcoma cells after iodine-125 (125I) seed irradiation [45]. miR-486/miR-663-dependent reduction of BCL-G level accompanied the healing of the thermal injury in the skin [43]. Also, the role of BCL-G in male fertility [85], pregnancy [86], and the correlation between BCL-G level and survival of cancer patients [83, 87] might be considered but require further investigation.
In several studies, RNA interference techniques or overexpression assays were used to determine the phenotypic consequences of direct manipulations of BCL-G level. It was shown that attenuation of BCL2L14 expression accompanied the development of resistance to neratinib in breast cancer cells [88]. Antisense oligonucleotide-mediated downregulation of Bcl-G was followed by reduced proliferative potential of mitogen-stimulated T cells [27]. In addition, it was shown that suppression of BCL2L14 protected kidney epithelial cells from detrimental effects of glucose and oxygen deprivation, and limited nephrotoxicity of cisplatin [89], attenuated UV-induced apoptosis in prostate carcinoma and breast cancer cells [83, 84], and prevented apoptosis induced by ectopic FAU overexpression in human T-lymphoblastic leukemia cells and embryonic kidney-derived cells [54]. Either downregulation of BCL-G or overexpression of miR-663b, which was involved in suppression of BCL-G, counteracted the influence of pterostilbene, a phenolic compound extracted from the Vitis sp., on endometrial cancer cells [41]. More recently, miR-496 was speculated to protect from cerebral ischemia-reperfusion injury via downregulating Bcl-G, while restoration of Bcl-G exhibited more detrimental effects [42]. In addition, BCL-G has been identified during an RNA-seq search for human immunodeficiency virus (HIV) restriction genes involved in response to IFN-α2b [40]. In this respect, BCL-G was markedly upregulated in activated CD4+ T cells, while high levels of BCL-G were associated with a decline of HIV RNA in plasma. The capability of BCL-G to diminish the replication of HIV was also confirmed in vitro [40]. Bcl-G was also shown to prevent hepatocarcinogenesis induced by diethylnitrosamine in mice [35]. Mechanistically, Bcl-G was involved in DNA damage-induced apoptosis in hepatocytes following G9a inhibition [35].
All the above-mentioned studies predominantly reported observations accompanying the activity of diverse perturbants and their effects on phenotypic changes rather than provided robust results to exhaustively confirm context-dependent genetic dependence on BCL-G. In this respect, a number of technical and experimental limitations could affect the conclusions [90, 91], and these papers should be mainly treated as a source for additional hypotheses that need to be verified (Table 2). This can be exemplified by the significance of MELK, which was indicated as a potential drug target, but it was shown dispensable for cancer cell viability when CRISPR-mediated MELK knockout experimental models were applied [92–96]. For this reason, the biological consequences of the interaction between MELK and BCL-G [50, 51] remain to be determined as it has been demonstrated that MELK expression correlated with tumor mitotic activity in one of the studies using MELK knockout [95]. Since CRISPR-Cas9 is currently one of the most efficient techniques to generate a complete loss-of-function allele in a gene of interest, is less prone to off-target effects and less affected by gene expression than RNA interference approaches [91], only a few studies have used state-of-the-art genetic manipulations followed by complementary methods to validate the consequences of BCL2A14 knockout [28–30, 38] (Table 2). Consequently, these studies have consistently excluded the contribution of BCL-G to the regulation of apoptosis [28–30, 38], while a pro-apoptotic function of BCL-G was questioned earlier (reviewed in [97]) despite the presence of exclusively BH3 domain in BCL-GS. It was demonstrated that murine Bcl-G might not act as a typical BH3-only protein. Bcl-G did not interact with either Bax or Bak, and could weakly associate with pro-survival proteins from the Bcl-2 family, including Bcl-2, Bcl-XL, Mcl-1, Bcl-w, and Bfl-1 [28]. Notably, the deletion of the BH3 domain in Bcl-G did not affect the interactions between Bcl-G and all five pro-survival proteins, which was further evidenced by the replacement of the Bim BH3 domain with Bcl-G-derived BH3 moiety [28]. In addition, Bcl2l14-deleted mice had intact gastrointestinal tract [28], including normal architecture and lengths of colon crypts, which was associated with unaffected proliferation and survival of epithelial cells [38]. Importantly, when Bcl-g−/− mice were grown beyond 1 year, no spontaneous tumor was formed in the entire gastrointestinal tract [38]. Bcl-G was also dispensable for apoptosis in splenic dendritic cells, either in the presence or absence of granulocyte-macrophage colony-stimulating factor [28]. This could suggest that a functional redundancy for Bcl-G could exist as demonstrated for other proteins of the Bcl-2 family [5–7]. However, no changes in the expression of Bcl-2-like proteins were found in the study by Nguyen et al. [38], suggesting that the activity of BCL-G is unlikely to be shared by other proteins from the BCL-2 family at least in adult colonic crypts. Moreover, although an increase in the transcript levels of both BCL-GL and BCL-GS were found during IFN-γ/TNF-α-induced apoptosis in human colorectal cancer cells, downregulation and isoform-specific overexpression of BCL-G revealed that cell death was not dependent on any isoform of BCL-G [29]. In turn, a few apoptosis-unrelated functions of BCL-G were demonstrated. It was shown that murine Bcl-G could be involved in vesicle trafficking and protein transport by interacting with the TRAPP complex, specifically Trappc3, Trappc4, Trappc5, and Trappc6b proteins in intestinal epithelial cells [28]. Any protein of the Bcl-2 family was not found in association with Bcl-G in this experiment employing co-immunoprecipitation [28] that supported the biological function of BCL-G apart from the BCL-2-like proteins. In addition, the biological relevance of the association between the TRAPP complex and BCL-G has been reinforced by a more recent study showing a similar interactome in human cells [98]. It was also demonstrated that Bcl-G regulated the stability of the mucin scaffolding network, and accelerated progression of colitis-associated cancer upon loss of Bcl-G was demonstrated in mice [38]. In this respect, disruption of Bcl-G activity might be linked with colon tumorigenesis [38] as confirmed by a significantly reduced expression of BCL2L14 in human late-stage colorectal tumors [29, 38]. Also, the depletion of BCL-G affected IFN-γ/TNF-α-induced secretion of inflammatory chemokines CCL5 and CCL20 [29], indicating that BCL-G regulated human gut homeostasis through immunomodulatory activity rather than promoted apoptosis.
Table 2.
The biological activity of BCL-G demonstrated with distinct strength of evidence in diverse biological systems.
| Major observations and findings | Experimental model | Activity of BCL-G | Strength of evidencea | Ref. |
|---|---|---|---|---|
| BCL-G is involved in vesicle trafficking and protein transport via interaction with the TRAPP complex | Bcl-g−/− mice; murine intestinal epithelial cells | Apoptosis-unrelatedb | High (in vivo and ex vivo; Bcl-g KO mice) | [28] |
| BCL-G prevents the progression of colitis-associated cancer via regulation of the mucin scaffolding network | Bcl-g−/− mice; murine small intestinal crypts | Apoptosis-unrelatedb | High (in vivo and ex vivo; Bcl-g KO mice) | [38] |
| BCL-G exerts an immunomodulatory activity via the regulation of secretion of chemokines: CCL5 and CCL20 | Intestinal epithelial cells | Apoptosis-unrelatedb | High (BCL2L14 KO; induced overexpression of BCL-G; KD—siRNA) | [29] |
| BCL-G is involved in response to IFN-α2b by diminishing HIV replication | AIDS | Apoptosis-unrelated | Moderate (ex vivo; induced overexpression of BCL-G) | [40] |
| BCL-G contributes to DNA damage-induced apoptosis after G9a inhibition, and is involved in hepatocarcinogenesis | G9aΔHep mice; hepatocytes | Pro-apoptotic | Moderate (in vivo and ex vivo—G9aΔHep mice; induced overexpression of BCL-G; KD—shRNA) | [35] |
| BCL-GL promotes apoptosis in CD4+ T cells isolated from patients with systemic lupus erythematosus (SLE) | SLE | Pro-apoptotic | Moderate (ex vivo; induced overexpression of BCL-G; KD—shRNA) | [72] |
| BCL-G promotes apoptosis accompanying cerebral ischemia-reperfusion (I/R) injury | Neuroblastoma cells | Pro-apoptotic | Moderate (induced overexpression of BCL-G; KD—siRNA) | [42] |
| BCL-GL enhances basal apoptosis in COS7 cells | Monkey kidney fibroblast-like cells | Pro-apoptotic | Moderate (induced overexpression of BCL-G) | [50] |
| BCL-G contributes to ultraviolet-induced apoptosis | Breast/prostate cancer and embryonic kidney cells | Pro-apoptotic | Moderate (KD—siRNA) | [54, 83, 84] |
| BCL-G contributes to detrimental effects of glucose/oxygen deprivation, and nephrotoxicity of cisplatin | Kidney epithelial cells | Pro-apoptotic | Moderate (KD—shRNA) | [89] |
| BCL-G contributes to pterostilbene-induced apoptosis | Endometrial cancer | Pro-apoptotic | Moderate (KD—siRNA) | [41] |
| BCL-G downregulation accompanies acquisition of resistance to neratinib | Breast cancer | Pro-apoptoticc | Moderate (KD—shRNA) | [88] |
| BCL-G is upregulated by nano-particulate tetraiodothyroacetic acid | Breast cancer | Pro-apoptoticc | Low | [78] |
| BCL-G is upregulated after exposure to bilirubin or lithocholic acid | Osteosarcoma | Pro-apoptoticc | Low | [80] |
| BCL-G is upregulated during dexamethasone-induced apoptosis | Murine osteoblasts | Pro-apoptoticc | Low | [81] |
| BCL-G downregulation accompanies the healing of the skin | Thermal injury | Not yet determined | Low (ex vivo) | [43] |
| BCL-G is upregulated in bone marrow of patients treated with arsenic trioxide and ascorbic acid | Myelodysplasia | Not yet determined | Low (ex vivo) | [73] |
| BCL-G downregulation accompanies reduced activation of T cells | Murine T cells | Not yet determined | Low | [27] |
| BCL-G downregulation accompanies iodine-125 seed irradiation | Chondrosarcoma | Not yet determined | Low | [45] |
| BCL-G is upregulated under hyperbaric air conditions | Embryonic lung fibroblasts | Not yet determined | Low | [74] |
Unless stated otherwise, the observations and findings were made in vitro and using human cells.
KD knockdown, KO knockout.
a“Low” (observations), “moderate” (observations validated using a single method, or methods with high susceptibility to off-target effects [91]), “high” (solid results obtained using state-of-the-art genetic manipulations and numerous complementary methods [91]).
bPro-apoptotic activity of BCL-G was questioned in experiments performed in parallel.
cExpected pro-apoptotic activity of BCL-G requiring extensive experimental confirmation.
Conclusions and future perspectives
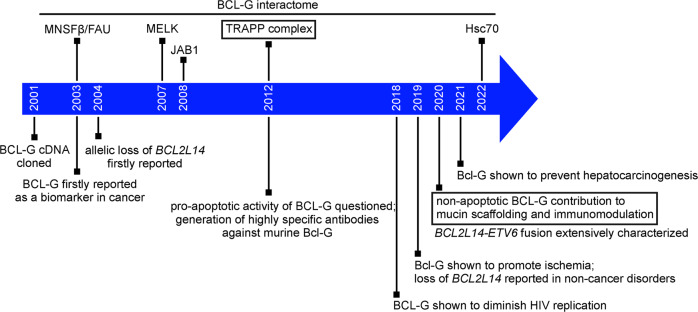
In spite of more than two decades of investigation on BCL-G, several crucial milestones (Fig. 3), and recent studies using state-of-the-art genetic techniques that have implied apoptosis-unrelated functions of BCL-G in the gastrointestinal tract (Table 2), the biological role of BCL-G remains to be fully defined. Further efforts focusing on various types of cells are required to find out more about the cell type- and context-specific activities of BCL-G. It also needs to be determined how conserved BCL-G is and whether it plays similar or different functions across species. Although the phylogenetic tree of BCL-2 homologous proteins, rooted with CED-9 sequences from Caenorhabditis and including BCL-G, has been demonstrated [99], BCL-G remains understudied in this respect. In general, the phylogenetic analyses have revealed quite similar structures and mechanisms of interactions between BCL-2-like proteins despite their either convergent or divergent history of evolution [100–107]. In addition, as alternatively spliced variants lacking certain motifs are found in the family as exemplified by BCL-XS and BCL-GS, it has been speculated that some BCL-2-like proteins derive from a common ancestry with multidomain members through gene duplication and exon loss [108–110]. Therefore, it will be crucial to delineate whether BCL-GS and BCL-GL resemble two isoforms of BCL-X, BCL-XS and BCL-XL exerting diverse activity [111]. In contrast to BCL-XL, however, the BCL-GL isoform contains BH2 and BH3 domains without the BH1 domain, which is not typical for proteins of the BCL-2 family. In this respect, BCL-2 family Kin (BFK) is another example of a BCL-2-like protein, which is structurally similar to BCL-GL [112]. A recently published report on BFK might shed light on the directions of further research on BCL-G [113]. Biophysical interaction analysis revealed that full-length BFK did not interact with other BCL-2-like proteins, but instead it was functionally reminiscent of BID, in which the BH3 domain was released upon caspase-mediated cleavage and truncated form (tBID) underwent a conformational alteration [113]. In addition, as protein modifications with ubiquitin-like moieties were shown to regulate the activity of target proteins, their intracellular localization and interactions with other proteins [114], the activity of FAU via BCL-G should be taken into account when the role of BCL-G is identified. The interaction of BCL-G with FAU is also likely to be of clinical significance as decreased FAU levels related to unfavorable outcomes were found in a number of tumors, including prostate, breast, and ovarian cancer [83, 84, 115]. The FAU polymorphic variant (rs769440) has been recently associated with recurrent pregnancy loss [116]. Therefore, understanding the BCL-G interactome may facilitate the design of novel therapies, in which the activity of BCL-G will be either imitated using small-molecule BH3 mimetics or inhibited to the attenuate effects of BCL-G upregulation. It will be also valuable to assess BCL-G levels under clinically-relevant scenarios. In this respect, G9a expression, and the correlation between G9a and clinical outcomes will be evaluated in patients with vulvar cancer (recruiting study NCT03695809). For now, BCL-G remains a protein with an apoptosis-unrelated role in intracellular trafficking, immunomodulatory functions and regulation of the mucin scaffolding network within the gastrointestinal tract.
Fig. 3. A timeline of major milestones in BCL-G investigation.
Discoveries made using state-of-the-art genetic manipulation approaches and complementary methods to validate the role of BCL-G are framed.
Author contributions
MLH performed review concept, drafted the manuscript and prepared figures; MC revised the manuscript. Both authors read and approved the final manuscript.
Funding
This work was supported by the research funding of Medical University of Lodz (503/1-156-01/503-11-001).
Data availability
This review article does not present any new primary data. References for publicly available datasets concerning gene expression, genetic alterations and protein structure are given in figure and table legends.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Singh R, Letai A, Sarosiek K. Regulation of apoptosis in health and disease: the balancing act of BCL-2 family proteins. Nat Rev Mol Cell Biol. 2019;20:175–93. doi: 10.1038/s41580-018-0089-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hartman ML. Non-apoptotic cell death signaling pathways in melanoma. Int J Mol Sci. 2020;21:2980. doi: 10.3390/ijms21082980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hartman ML, Czyz M. BCL-w: apoptotic and non-apoptotic role in health and disease. Cell Death Dis. 2020;11:260. doi: 10.1038/s41419-020-2417-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kurschat C, Metz A, Kirschnek S, Häcker G. Importance of Bcl-2-family proteins in murine hematopoietic progenitor and early B cells. Cell Death Dis. 2021;12:784. doi: 10.1038/s41419-021-04079-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eichhorn JM, Alford SE, Sakurikar N, Chambers TC. Molecular analysis of functional redundancy among anti-apoptotic Bcl-2 proteins and its role in cancer cell survival. Exp Cell Res. 2014;322:415–24. doi: 10.1016/j.yexcr.2014.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Delbridge AR, Aubrey BJ, Hyland C, Bernardini JP, Di Rago L, Garnier JM, et al. The BH3-only proteins BIM and PUMA are not critical for the reticulocyte apoptosis caused by loss of the pro-survival protein BCL-XL. Cell Death Dis. 2017;8:e2914. doi: 10.1038/cddis.2017.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schenk RL, Tuzlak S, Carrington EM, Zhan Y, Heinzel S, Teh CE, et al. Characterisation of mice lacking all functional isoforms of the pro-survival BCL-2 family member A1 reveals minor defects in the haematopoietic compartment. Cell Death Differ. 2017;24:534–45. doi: 10.1038/cdd.2016.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gross A, Katz SG. Non-apoptotic functions of BCL-2 family proteins. Cell Death Differ. 2017;24:1348–58. doi: 10.1038/cdd.2017.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gabellini C, Trisciuoglio D, Del Bufalo D. Non-canonical roles of Bcl-2 and Bcl-xL proteins: relevance of BH4 domain. Carcinogenesis. 2017;38:579–87. doi: 10.1093/carcin/bgx016. [DOI] [PubMed] [Google Scholar]
- 10.Czabotar PE, Lessene G, Strasser A, Adams JM. Control of apoptosis by the BCL-2 protein family: implications for physiology and therapy. Nat Rev Mol Cell Biol. 2014;15:49–63. doi: 10.1038/nrm3722. [DOI] [PubMed] [Google Scholar]
- 11.Dadsena S, King LE, García-Sáez AJ. Apoptosis regulation at the mitochondria membrane level. Biochim Biophys Acta Biomembr. 2021;1863:183716. doi: 10.1016/j.bbamem.2021.183716. [DOI] [PubMed] [Google Scholar]
- 12.Kale J, Osterlund EJ, Andrews DW. BCL-2 family proteins: changing partners in the dance towards death. Cell Death Differ. 2018;25:65–80. doi: 10.1038/cdd.2017.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Strasser A, Vaux DL. Cell death in the origin and treatment of cancer. Mol Cell. 2020;78:1045–54. doi: 10.1016/j.molcel.2020.05.014. [DOI] [PubMed] [Google Scholar]
- 14.Hartman ML, Czyz M. Pro-apoptotic activity of BH3-only proteins and BH3 mimetics: from theory to potential cancer therapy. Anticancer Agents Med Chem. 2012;12:966–81. doi: 10.2174/187152012802650084. [DOI] [PubMed] [Google Scholar]
- 15.Campbell KJ, Tait SWG. Targeting BCL-2 regulated apoptosis in cancer. Open Biol. 2018;8:180002. doi: 10.1098/rsob.180002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Merino D, Kelly GL, Lessene G, Wei AH, Roberts AW, Strasser A. BH3-mimetic drugs: blazing the trail for new cancer medicines. Cancer Cell. 2018;34:879–91. doi: 10.1016/j.ccell.2018.11.004. [DOI] [PubMed] [Google Scholar]
- 17.Deeks ED. Venetoclax: first global approval. Drugs. 2016;76:979–87. doi: 10.1007/s40265-016-0596-x. [DOI] [PubMed] [Google Scholar]
- 18.Daver N, Wei AH, Pollyea DA, Fathi AT, Vyas P, DiNardo CD. New directions for emerging therapies in acute myeloid leukemia: the next chapter. Blood Cancer J. 2020;10:107. doi: 10.1038/s41408-020-00376-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Diepstraten ST, Anderson MA, Czabotar PE, Lessene G, Strasser A, Kelly GL. The manipulation of apoptosis for cancer therapy using BH3-mimetic drugs. Nat Rev Cancer. 2022;22:45–64. doi: 10.1038/s41568-021-00407-4. [DOI] [PubMed] [Google Scholar]
- 20.Montero J, Haq R. Adapted to survive: targeting cancer cells with BH3 mimetics. Cancer Discov. 2022;12:1217–32. doi: 10.1158/2159-8290.CD-21-1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Townsend PA, Kozhevnikova MV, Cexus ONF, Zamyatnin AA, Jr, Soond SM. BH3-mimetics: recent developments in cancer therapy. J Exp Clin Cancer Res. 2021;40:355. doi: 10.1186/s13046-021-02157-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fairlie WD, Lee EF. Targeting the BCL-2-regulated apoptotic pathway for the treatment of solid cancers. Biochem Soc Trans. 2021;49:2397–410. doi: 10.1042/BST20210750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Z, Bai L, Hou L, Deng H, Luan S, Liu D, et al. Trends in targeting Bcl-2 anti-apoptotic proteins for cancer treatment. Eur J Med Chem. 2022;232:114184. doi: 10.1016/j.ejmech.2022.114184. [DOI] [PubMed] [Google Scholar]
- 24.Guo B, Godzik A, Reed JC. Bcl-G, a novel pro-apoptotic member of the Bcl-2 family. J Biol Chem. 2001;276:2780–5. doi: 10.1074/jbc.M005889200. [DOI] [PubMed] [Google Scholar]
- 25.Montpetit A, Boily G, Sinnett D. A detailed transcriptional map of the chromosome 12p12 tumour suppressor locus. Eur J Hum Genet. 2002;10:62–71. doi: 10.1038/sj.ejhg.5200766. [DOI] [PubMed] [Google Scholar]
- 26.Jiang P, Li D, Bi L, Zhang D. BCL-G as a new candidate gene for immune responses in pigs: bioinformatic analysis and functional characterization. Vet Immunol Immunopathol. 2012;150:112–7. doi: 10.1016/j.vetimm.2012.08.001. [DOI] [PubMed] [Google Scholar]
- 27.Nakamura M, Tanigawa Y. Characterization of ubiquitin-like polypeptide acceptor protein, a novel pro-apoptotic member of the Bcl2 family. Eur J Biochem. 2003;270:4052–8. doi: 10.1046/j.1432-1033.2003.03790.x. [DOI] [PubMed] [Google Scholar]
- 28.Giam M, Okamoto T, Mintern JD, Strasser A, Bouillet P. Bcl-2 family member Bcl-G is not a proapoptotic protein. Cell Death Dis. 2012;3:e404. doi: 10.1038/cddis.2012.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Woznicki JA, Flood P, Bustamante-Garrido M, Stamou P, Moloney G, Fanning A, et al. Human BCL-G regulates secretion of inflammatory chemokines but is dispensable for induction of apoptosis by IFN-γ and TNF-α in intestinal epithelial cells. Cell Death Dis. 2020;11:68. doi: 10.1038/s41419-020-2263-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Giam M, Mintern JD, Rautureau GJ, Hinds MG, Strasser A, Bouillet P. Detection of Bcl-2 family member Bcl-G in mouse tissues using new monoclonal antibodies. Cell Death Dis. 2012;3:e378. doi: 10.1038/cddis.2012.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiang P, Wang X, Chen X, Wang Y, Kang Z, Wang J, et al. A potential molecular model for studying apoptosis enhanced by the interaction of BCL-G with JAB1 in swine. Oncotarget. 2016;7:62912–24. doi: 10.18632/oncotarget.11230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Benito A, Gutierrez O, Pipaon C, Real PJ, Gachon F, Ritchie AE, et al. A novel role for proline- and acid-rich basic region leucine zipper (PAR bZIP) proteins in the transcriptional regulation of a BH3-only proapoptotic gene. J Biol Chem. 2006;281:38351–7. doi: 10.1074/jbc.M607004200. [DOI] [PubMed] [Google Scholar]
- 33.Ritchie A, Gutierrez O, Fernandez-Luna JL. PAR bZIP-bik is a novel transcriptional pathway that mediates oxidative stress-induced apoptosis in fibroblasts. Cell Death Differ. 2009;16:838–46. doi: 10.1038/cdd.2009.13. [DOI] [PubMed] [Google Scholar]
- 34.Miled C, Pontoglio M, Garbay S, Yaniv M, Weitzman JB. A genomic map of p53 binding sites identifies novel p53 targets involved in an apoptotic network. Cancer Res. 2005;65:5096–104. doi: 10.1158/0008-5472.CAN-04-4232. [DOI] [PubMed] [Google Scholar]
- 35.Nakatsuka T, Tateishi K, Kato H, Fujiwara H, Yamamoto K, Kudo Y, et al. Inhibition of histone methyltransferase G9a attenuates liver cancer initiation by sensitizing DNA-damaged hepatocytes to p53-induced apoptosis. Cell Death Dis. 2021;12:99. doi: 10.1038/s41419-020-03381-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li Q, Mao F, Zhou B, Huang Y, Zou Z, denDekker AD, et al. p53 integrates temporal WDR5 inputs during neuroectoderm and mesoderm differentiation of mouse embryonic stem cells. Cell Rep. 2020;30:465–80.e6. doi: 10.1016/j.celrep.2019.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Finnberg N, Wambi C, Ware JH, Kennedy AR, El-Deiry WS. Gamma-radiation (GR) triggers a unique gene expression profile associated with cell death compared to proton radiation (PR) in mice in vivo. Cancer Biol Ther. 2008;7:2023–33. doi: 10.4161/cbt.7.12.7417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nguyen PM, Dagley LF, Preaudet A, Lam N, Giam M, Fung KY, et al. Loss of Bcl-G, a Bcl-2 family member, augments the development of inflammation-associated colorectal cancer. Cell Death Differ. 2020;27:742–57. doi: 10.1038/s41418-019-0383-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang XN, Liu JX, Hu YW, Chen H, Yuan ZH. Hyper-activated IRF-1 and STAT1 contribute to enhanced interferon stimulated gene (ISG) expression by interferon alpha and gamma co-treatment in human hepatoma cells. Biochim Biophys Acta. 2006;1759:417–25. doi: 10.1016/j.bbaexp.2006.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.El-Diwany R, Soliman M, Sugawara S, Breitwieser F, Skaist A, Coggiano C, et al. CMPK2 and BCL-G are associated with type 1 interferon-induced HIV restriction in humans. Sci Adv. 2018;4:eaat0843. doi: 10.1126/sciadv.aat0843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang YL, Shen Y, Xu JP, Han K, Zhou Y, Yang S, et al. Pterostilbene suppresses human endometrial cancer cells in vitro by down-regulating miR-663b. Acta Pharm Sin. 2017;38:1394–400. doi: 10.1038/aps.2017.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yao X, Yao R, Yi J, Huang F. Upregulation of miR-496 decreases cerebral ischemia/reperfusion injury by negatively regulating BCL2L14. Neurosci Lett. 2019;696:197–205. doi: 10.1016/j.neulet.2018.12.039. [DOI] [PubMed] [Google Scholar]
- 43.Wu SG, Li HT, Wang LL, Yan L. Lidocaine promotes fibroblast proliferation after thermal injury via up-regulating the expression of miR-663 and miR-486. Kaohsiung J Med Sci. 2020;36:274–80. doi: 10.1002/kjm2.12166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gu Y, Shi Y, Yang Q, Gu WW, He YP, Zheng HJ, et al. miR-3074-5p promotes the apoptosis but inhibits the invasiveness of human extravillous trophoblast-derived HTR8/SVneo cells in vitro. Reprod Sci. 2018;25:690–9. doi: 10.1177/1933719117725823. [DOI] [PubMed] [Google Scholar]
- 45.Li F, Xu J, Zhu Y, Sun L, Zhou R. Analysis of cells proliferation and microRNAs expression profile in human chondrosarcoma SW1353 cells exposed to iodine-125 seeds irradiation. Dose Response. 2020;18:1559325820920525. doi: 10.1177/1559325820920525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu X, Pan Z, Zhang L, Sun Q, Wan J, Tian C, et al. JAB1 accelerates mitochondrial apoptosis by interaction with proapoptotic BclGs. Cell Signal. 2008;20:230–40. doi: 10.1016/j.cellsig.2007.10.012. [DOI] [PubMed] [Google Scholar]
- 47.Oltersdorf T, Elmore SW, Shoemaker AR, Armstrong RC, Augeri DJ, Belli BA, et al. An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature. 2005;435:677–81. doi: 10.1038/nature03579. [DOI] [PubMed] [Google Scholar]
- 48.Becattini B, Kitada S, Leone M, Monosov E, Chandler S, Zhai D, et al. Rational design and real time, in-cell detection of the proapoptotic activity of a novel compound targeting Bcl-X(L) Chem Biol. 2004;11:389–95. doi: 10.1016/j.chembiol.2004.02.020. [DOI] [PubMed] [Google Scholar]
- 49.Jiang P, Wang J, Kang Z, Li D, Zhang D. Porcine JAB1 significantly enhances apoptosis induced by staurosporine. Cell Death Dis. 2013;4:e823. doi: 10.1038/cddis.2013.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lin ML, Park JH, Nishidate T, Nakamura Y, Katagiri T. Involvement of maternal embryonic leucine zipper kinase (MELK) in mammary carcinogenesis through interaction with Bcl-G, a pro-apoptotic member of the Bcl-2 family. Breast Cancer Res. 2007;9:R17. doi: 10.1186/bcr1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen P, Wang J, Wang X, Chen X, Li C, Tan T. Cloning, tissue distribution, expression pattern, and function of porcine maternal embryonic leucine zipper kinase. Ann Transl Med. 2020;8:239. doi: 10.21037/atm.2020.03.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Martín-Villanueva S, Gutiérrez G, Kressler D, de la Cruz J. Ubiquitin and ubiquitin-like proteins and domains in ribosome production and function: chance or necessity? Int J Mol Sci. 2021;22:4359. doi: 10.3390/ijms22094359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen P, Wang J, Wang X, Wang Y, Xu C, Ji C. Porcine ubiquitin-like protein MNSFβ promotes cell apoptosis and covalently binds to BCL-G to enhance staurosporine-induced apoptosis. Ann Transl Med. 2020;8:1306. doi: 10.21037/atm-20-6348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pickard MR, Mourtada-Maarabouni M, Williams GT. Candidate tumour suppressor Fau regulates apoptosis in human cells: an essential role for Bcl-G. Biochim Biophys Acta. 2011;1812:1146–53. doi: 10.1016/j.bbadis.2011.04.009. [DOI] [PubMed] [Google Scholar]
- 55.Nakamura M, Yamaguchi S. The ubiquitin-like protein MNSFbeta regulates ERK-MAPK cascade. J Biol Chem. 2006;281:16861–9. doi: 10.1074/jbc.M509907200. [DOI] [PubMed] [Google Scholar]
- 56.Watanabe J, Nakagawa M, Watanabe N, Nakamura M. Ubiquitin-like protein MNSFβ covalently binds to Bcl-G and enhances lipopolysaccharide/interferon γ-induced apoptosis in macrophages. FEBS J. 2013;280:1281–93. doi: 10.1111/febs.12120. [DOI] [PubMed] [Google Scholar]
- 57.Nakamura M, Fukuma Y, Notsu K, Kono M. Quercetin and HSC70 coregulate the anti-inflammatory action of the ubiquitin-like protein MNSFβ. Mol Biol Rep. 2022;49:1213–22. doi: 10.1007/s11033-021-06949-y. [DOI] [PubMed] [Google Scholar]
- 58.Montpetit A, Larose J, Boily G, Langlois S, Trudel N, Sinnett D. Mutational and expression analysis of the chromosome 12p candidate tumor suppressor genes in pre-B acute lymphoblastic leukemia. Leukemia. 2004;18:1499–504. doi: 10.1038/sj.leu.2403441. [DOI] [PubMed] [Google Scholar]
- 59.Feurstein S, Rücker FG, Bullinger L, Hofmann W, Manukjan G, Göhring G, et al. Haploinsufficiency of ETV6 and CDKN1B in patients with acute myeloid leukemia and complex karyotype. BMC Genomics. 2014;15:784. doi: 10.1186/1471-2164-15-784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kibel AS, Huagen J, Guo C, Isaacs WB, Yan Y, Pienta KJ, et al. Expression mapping at 12p12-13 in advanced prostate carcinoma. Int J Cancer. 2004;109:668–72. doi: 10.1002/ijc.20060. [DOI] [PubMed] [Google Scholar]
- 61.Kluth M, Ahrary R, Hube-Magg C, Ahmed M, Volta H, Schwemin C, et al. Genomic deletion of chromosome 12p is an independent prognostic marker in prostate cancer. Oncotarget. 2017;6:27966–79. doi: 10.18632/oncotarget.4626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ross J, Fennis W, de Leeuw N, Cune M, Willemze A, Rosenberg A, et al. Concurrent manifestation of oligodontia and thrombocytopenia caused by a contiguous gene deletion in 12p13.2: a three-generation clinical report. Mol Genet Genom Med. 2019;7:e679. doi: 10.1002/mgg3.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Abdelhaleem M, Yi Q, Beimnet K, Hitzler J. A novel TEL-AML1 fusion transcript involving the pro-apoptotic gene BCL-G in pediatric precursor B acute lymphoblastic leukemia. Leukemia. 2006;20:1294. doi: 10.1038/sj.leu.2404249. [DOI] [PubMed] [Google Scholar]
- 64.Al-Shehhi H, Konn ZJ, Schwab CJ, Erhorn A, Barber KE, Wright SL, et al. Abnormalities of the der(12)t(12;21) in ETV6-RUNX1 acute lymphoblastic leukemia. Genes Chromosomes Cancer. 2013;52:202–13. doi: 10.1002/gcc.22021. [DOI] [PubMed] [Google Scholar]
- 65.Lee S, Hu Y, Loo SK, Tan Y, Bhargava R, Lewis MT, et al. Landscape analysis of adjacent gene rearrangements reveals BCL2L14-ETV6 gene fusions in more aggressive triple-negative breast cancer. Proc Natl Acad Sci USA. 2020;117:9912–21. doi: 10.1073/pnas.1921333117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Biswas A, Rajesh Y, Mitra P, Mandal M. ETV6 gene aberrations in non-haematological malignancies: a review highlighting ETV6 associated fusion genes in solid tumors. Biochim Biophys Acta Rev Cancer. 2020;1874:188389. doi: 10.1016/j.bbcan.2020.188389. [DOI] [PubMed] [Google Scholar]
- 67.Loo SK, Yates ME, Yang S, Oesterreich S, Lee AV, Wang XS. Fusion-associated carcinomas of the breast: diagnostic, prognostic, and therapeutic significance. Genes Chromosomes Cancer. 2022;61:261–73. doi: 10.1002/gcc.23029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Spitz MR, Gorlov IP, Dong Q, Wu X, Chen W, Chang DW, et al. Multistage analysis of variants in the inflammation pathway and lung cancer risk in smokers. Cancer Epidemiol Biomark Prev. 2012;21:1213–21. doi: 10.1158/1055-9965.EPI-12-0352-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Soung YH, Lee JW, Park WS, Nam SW, Lee JY, Yoo NJ, et al. BH3 domain mutation of proapoptotic genes Bad, Bmf and Bcl-G is rare in transitional cell carcinomas of the urinary bladder. Pathology. 2006;38:33–4. doi: 10.1080/00313020500455811. [DOI] [PubMed] [Google Scholar]
- 70.Yoo NJ, Soung YH, Lee SH, Jeong EG, Lee SH. Mutational analysis of the BH3 domains of proapoptotic Bcl-2 family genes Bad, Bmf and Bcl-G in laryngeal squamous cell carcinomas. Tumori. 2007;93:195–7. doi: 10.1177/030089160709300214. [DOI] [PubMed] [Google Scholar]
- 71.Nakamura M, Nakagawa M, Watanabe J. Ubiquitin-like protein MNSFβ negatively regulates T cell function and survival. Immunol Invest. 2015;44:1–12. doi: 10.3109/08820139.2014.909454. [DOI] [PubMed] [Google Scholar]
- 72.Luo N, Wu Y, Chen Y, Yang Z, Guo S, Fei L, et al. Upregulated BclG(L) expression enhances apoptosis of peripheral blood CD4+ T lymphocytes in patients with systemic lupus erythematosus. Clin Immunol. 2009;132:349–61. doi: 10.1016/j.clim.2009.05.010. [DOI] [PubMed] [Google Scholar]
- 73.Galimberti S, Guerrini F, Salvi F, Petrini I, Gioia D, Messa E, et al. Arsenic trioxide and ascorbic acid interfere with the BCL2 family genes in patients with myelodysplastic syndromes: an ex-vivo study. J Hematol Oncol. 2012;5:53. doi: 10.1186/1756-8722-5-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Oh S, Lee E, Lee J, Lim Y, Kim J, Woo S. Comparison of the effects of 40% oxygen and two atmospheric absolute air pressure conditions on stress-induced premature senescence of normal human diploid fibroblasts. Cell Stress Chaperones. 2008;13:447–58. doi: 10.1007/s12192-008-0041-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang S, Feng XL, Shi L, Gong CJ, He ZJ, Wu HJ, et al. Genome-wide analysis of DNA methylation in tongue squamous cell carcinoma. Oncol Rep. 2013;29:1819–26. doi: 10.3892/or.2013.2309. [DOI] [PubMed] [Google Scholar]
- 76.Balasubramanian D, Akhtar-Zaidi B, Song L, Bartels CF, Veigl M, Beard L, et al. H3K4me3 inversely correlates with DNA methylation at a large class of non-CpG-island-containing start sites. Genome Med. 2012;28:47. doi: 10.1186/gm346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Coit P, Jeffries M, Altorok N, Dozmorov MG, Koelsch KA, Wren JD, et al. Genome-wide DNA methylation study suggests epigenetic accessibility and transcriptional poising of interferon-regulated genes in naïve CD4+ T cells from lupus patients. J Autoimmun. 2013;43:78–84. doi: 10.1016/j.jaut.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Glinskii AB, Glinsky GV, Lin HY, Tang HY, Sun M, Davis FB, et al. Modification of survival pathway gene expression in human breast cancer cells by tetraiodothyroacetic acid (tetrac) Cell Cycle. 2009;8:3562–70. doi: 10.4161/cc.8.21.9963. [DOI] [PubMed] [Google Scholar]
- 79.Davis PJ, Glinsky GV, Lin HY, Leith JT, Hercbergs A, Tang HY, et al. Cancer cell gene expression modulated from plasma membrane integrin αvβ3 by thyroid hormone and nanoparticulate tetrac. Front Endocrinol. 2015;5:240. doi: 10.3389/fendo.2014.00240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ruiz-Gaspà S, Guañabens N, Jurado S, Dubreuil M, Combalia A, Peris P, et al. Bile acids and bilirubin effects on osteoblastic gene profile. Implications in the pathogenesis of osteoporosis in liver diseases. Gene. 2020;725:144167. doi: 10.1016/j.gene.2019.144167. [DOI] [PubMed] [Google Scholar]
- 81.Nie Z, Chen S, Deng S, Long L, Peng P, Gao M, et al. Gene expression profiling of osteoblasts subjected to dexamethasone-induced apoptosis with/without GSK3β-shRNA. Biochem Biophys Res Commun. 2018;506:41–47. doi: 10.1016/j.bbrc.2018.10.043. [DOI] [PubMed] [Google Scholar]
- 82.Romero P, Benhamo V, Deniziaut G, Fuhrmann L, Berger F, Manié E, et al. Medullary breast carcinoma, a triple-negative breast cancer associated with BCLG overexpression. Am J Pathol. 2018;188:2378–91. doi: 10.1016/j.ajpath.2018.06.021. [DOI] [PubMed] [Google Scholar]
- 83.Pickard MR, Green AR, Ellis IO, Caldas C, Hedge VL, Mourtada-Maarabouni M, et al. Dysregulated expression of Fau and MELK is associated with poor prognosis in breast cancer. Breast Cancer Res. 2009;11:R60. doi: 10.1186/bcr2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pickard MR, Edwards SE, Cooper CS, Williams GT. Apoptosis regulators Fau and Bcl-G are down-regulated in prostate cancer. Prostate. 2010;70:1513–23. doi: 10.1002/pros.21186. [DOI] [PubMed] [Google Scholar]
- 85.Omolaoye TS, Omolaoye VA, Kandasamy RK, Hachim MY, Du Plessis SS. Omics and male infertility: highlighting the application of transcriptomic data. Life. 2022;12:280. doi: 10.3390/life12020280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gu Y, He Y, Zhang X, Shi Y, Yang Q, Yu L, et al. Deficiency of monoclonal non-specific suppressor factor beta (MNSFB) promotes pregnancy loss in mice. Mol Reprod Dev. 2015;82:475–88. doi: 10.1002/mrd.22495. [DOI] [PubMed] [Google Scholar]
- 87.Latil A, Bièche I, Chêne L, Laurendeau I, Berthon P, Cussenot O, et al. Gene expression profiling in clinically localized prostate cancer: a four-gene expression model predicts clinical behavior. Clin Cancer Res. 2003;9:5477–85. [PubMed] [Google Scholar]
- 88.Seyhan AA, Varadarajan U, Choe S, Liu W, Ryan TE. A genome-wide RNAi screen identifies novel targets of neratinib resistance leading to identification of potential drug resistant genetic markers. Mol Biosyst. 2012;8:1553–70. doi: 10.1039/c2mb05512k. [DOI] [PubMed] [Google Scholar]
- 89.Zynda ER, Schott B, Babagana M, Gruener S, Wernher E, Nguyen GD, et al. An RNA interference screen identifies new avenues for nephroprotection. Cell Death Differ. 2016;23:608–15. doi: 10.1038/cdd.2015.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kaelin WG., Jr Common pitfalls in preclinical cancer target validation. Nat Rev Cancer. 2017;17:425–40. doi: 10.1038/nrc.2017.32. [DOI] [PubMed] [Google Scholar]
- 91.Lin A, Sheltzer JM. Discovering and validating cancer genetic dependencies: approaches and pitfalls. Nat Rev Genet. 2020;21:671–82. doi: 10.1038/s41576-020-0247-7. [DOI] [PubMed] [Google Scholar]
- 92.Huang HT, Seo HS, Zhang T, Wang Y, Jiang B, Li Q, et al. MELK is not necessary for the proliferation of basal-like breast cancer cells. Elife. 2017;6:e26693. doi: 10.7554/eLife.26693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lin A, Giuliano CJ, Sayles NM, Sheltzer JM. CRISPR/Cas9 mutagenesis invalidates a putative cancer dependency targeted in on-going clinical trials. Elife. 2017;6:e24179. doi: 10.7554/eLife.24179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Settleman J, Sawyers CL, Hunter T. Challenges in validating candidate therapeutic targets in cancer. Elife. 2018;7:e32402. doi: 10.7554/eLife.32402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Giuliano CJ, Lin A, Smith JC, Palladino AC, Sheltzer JM. MELK expression correlates with tumor mitotic activity but is not required for cancer growth. Elife. 2018;7:e32838. doi: 10.7554/eLife.32838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.McDonald IM, Graves LM. Enigmatic MELK: The controversy surrounding its complex role in cancer. J Biol Chem. 2020;295:8195–203. doi: 10.1074/jbc.REV120.013433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tischner D, Villunger A. Bcl-G acquitted of murder! Cell Death Dis. 2012;3:e405. doi: 10.1038/cddis.2012.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hubel P, Urban C, Bergant V, Schneider WM, Knauer B, Stukalov A, et al. A protein-interaction network of interferon-stimulated genes extends the innate immune system landscape. Nat Immunol. 2019;20:493–502. doi: 10.1038/s41590-019-0323-3. [DOI] [PubMed] [Google Scholar]
- 99.Aouacheria A, Rech de Laval V, Combet C, Hardwick JM. Evolution of Bcl-2 homology motifs: homology versus homoplasy. Trends Cell Biol. 2013;23:103–11. doi: 10.1016/j.tcb.2012.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Moya A, Sakamaki K, Mason BM, Huisman L, Forêt S, Weiss Y, et al. Functional conservation of the apoptotic machinery from coral to man: the diverse and complex Bcl-2 and caspase repertoires of Acropora millepora. BMC Genomics. 2016;17:62. doi: 10.1186/s12864-015-2355-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Banjara S, Suraweera CD, Hinds MG, Kvansakul M. The Bcl-2 family: ancient origins, conserved structures, and divergent mechanisms. Biomolecules. 2020;10:128. doi: 10.3390/biom10010128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lanave C, Santamaria M, Saccone C. Comparative genomics: the evolutionary history of the Bcl-2 family. Gene. 2004;333:71–79. doi: 10.1016/j.gene.2004.02.017. [DOI] [PubMed] [Google Scholar]
- 103.Choudhury S. A comparative analysis of BCL-2 family. Bioinformation. 2019;15:299–306. doi: 10.6026/97320630015299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Suraweera CD, Banjara S, Hinds MG, Kvansakul M. Metazoans and intrinsic apoptosis: an evolutionary analysis of the Bcl-2 family. Int J Mol Sci. 2022;23:3691. doi: 10.3390/ijms23073691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Jette CA, Flanagan AM, Ryan J, Pyati UJ, Carbonneau S, Stewart RA, et al. BIM and other BCL-2 family proteins exhibit cross-species conservation of function between zebrafish and mammals. Cell Death Differ. 2008;15:1063–72. doi: 10.1038/cdd.2008.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Xie VC, Pu J, Metzger BP, Thornton JW, Dickinson BC. Contingency and chance erase necessity in the experimental evolution of ancestral proteins. Elife. 2021;10:e67336. doi: 10.7554/eLife.67336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zmasek CM, Zhang Q, Ye Y, Godzik A. Surprising complexity of the ancestral apoptosis network. Genome Biol. 2007;8:R226. doi: 10.1186/gb-2007-8-10-r226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Aouacheria A, Brunet F, Gouy M. Phylogenomics of life-or-death switches in multicellular animals: Bcl-2, BH3-Only, and BNip families of apoptotic regulators. Mol Biol Evol. 2005;22:2395–416. doi: 10.1093/molbev/msi234. [DOI] [PubMed] [Google Scholar]
- 109.Popgeorgiev N, Sa JD, Jabbour L, Banjara S, Nguyen TTM, Akhavan-E-Sabet A, et al. Ancient and conserved functional interplay between Bcl-2 family proteins in the mitochondrial pathway of apoptosis. Sci Adv. 2020;6:eabc4149. doi: 10.1126/sciadv.abc4149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Strasser A, Vaux DL. Viewing BCL2 and cell death control from an evolutionary perspective. Cell Death Differ. 2018;25:13–20. doi: 10.1038/cdd.2017.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Dou Z, Zhao D, Chen X, Xu C, Jin X, Zhang X, et al. Aberrant Bcl-x splicing in cancer: from molecular mechanism to therapeutic modulation. J Exp Clin Cancer Res. 2021;40:194. doi: 10.1186/s13046-021-02001-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Coultas L, Pellegrini M, Visvader JE, Lindeman GJ, Chen L, Adams JM, et al. Bfk: a novel weakly proapoptotic member of the Bcl-2 protein family with a BH3 and a BH2 region. Cell Death Differ. 2003;10:185–92. doi: 10.1038/sj.cdd.4401204. [DOI] [PubMed] [Google Scholar]
- 113.Jang DM, Oh EK, Hahn H, Kim HS, Han BW. Structural insights into apoptotic regulation of human Bfk as a novel Bcl-2 family member. Comput Struct Biotechnol J. 2022;20:745–56. doi: 10.1016/j.csbj.2022.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Salas-Lloret D, González-Prieto R. Insights in post-translational modifications: ubiquitin and SUMO. Int J Mol Sci. 2022;23:3281. doi: 10.3390/ijms23063281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Moss EL, Mourtada-Maarabouni M, Pickard MR, Redman CW, Williams GT. FAU regulates carboplatin resistance in ovarian cancer. Genes Chromosomes Cancer. 2010;49:70–77. doi: 10.1002/gcc.20721. [DOI] [PubMed] [Google Scholar]
- 116.Gu Y, Wang JM, Zhang ZF, Wang J, Cao YL, Pan CJ, et al. The association between polymorphisms of genes related to inflammation and recurrent pregnancy loss. Gynecol Endocrinol. 2018;34:349–52. doi: 10.1080/09513590.2017.1395837. [DOI] [PubMed] [Google Scholar]
- 117.Fagerberg L, Hallström BM, Oksvold P, Kampf C, Djureinovic D, Odeberg J, et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol Cell Proteom. 2014;13:397–406. doi: 10.1074/mcp.M113.035600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Szabo L, Morey R, Palpant NJ, Wang PL, Afari N, Jiang C, et al. Statistically based splicing detection reveals neural enrichment and tissue-specific induction of circular RNA during human fetal development. Genome Biol. 2015;16:126. doi: 10.1186/s13059-015-0690-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This review article does not present any new primary data. References for publicly available datasets concerning gene expression, genetic alterations and protein structure are given in figure and table legends.