Abstract
The nervous system emerges from a series of genetic programs that generate a remarkable array of neuronal cell types. Each cell type must acquire a distinct anatomical position, morphology, and function, enabling the generation of specialized circuits that drive animal behavior. How are these diverse cell types and circuits patterned along the anterior-posterior (A-P) axis of the animal body? Hox genes encode transcription factors that regulate cell fate and patterning events along the A-P axis of the nervous system. While most of our understanding of Hox-mediated control of neuronal development stems from studies in segmented animals like flies, mice, and zebrafish, important new themes are emerging from work in a non-segmented animal: the nematode Caenorhabditis elegans. Studies in C. elegans support the idea that Hox genes are needed continuously and across different life stages in the nervous system; they are not only required in dividing progenitor cells, but also in post-mitotic adult neurons. In C. elegans embryos and young larvae, Hox genes control progenitor cell specification, cell survival, and neuronal migration, consistent with their neural patterning roles in other animals. In late larvae and adults, C. elegans Hox genes control neuron type-specific identity features critical for neuronal function, thereby extending the Hox functional repertoire beyond early patterning. Here, we provide a comprehensive review of Hox studies in the C. elegans nervous system. To relate to readers outside the C. elegans community, we highlight conserved roles of Hox genes in patterning the nervous system of invertebrate and vertebrate animals. We end by calling attention to new functions in adult post-mitotic neurons for these paradigmatic regulators of cell fate.
Keywords: Hox transcription factors, C. elegans, neural progenitors, cell survival, cell migration, neuronal terminal identity
1. Introduction
Building a nervous system is a multi-step process. Following gastrulation, the ectoderm generates neural stem cells which give rise to specified neuronal and glial precursor cells [1]. These precursors then divide to produce post-mitotic neurons and glia. After exiting the cell cycle, newborn cells must differentiate and migrate to specific positions along the anterior-posterior (A-P) axis of the nervous system. Once settled into a defined location, neurons acquire region-specific identities and often undergo dramatic morphological changes to generate axons and dendrites. Finally, neurons establish specific connections (synapses) with other neurons to form neural circuits. The function of these circuits throughout life relies on the ability of post-mitotic neurons to acquire and maintain terminal identity features, such as expression of neurotransmitter (NT) biosynthesis proteins, NT receptors, neuropeptides, and ion channels [2, 3]. In this review, we collectively refer to the series of events from gastrulation to the generation of axons and dendrites as early steps of nervous system development. We refer to the subsequent processes of synapse formation and the control of neuronal terminal identity features as late steps of neuronal development.
Both early and late steps of nervous system development must be precisely controlled to generate distinct cell types and circuits at specific positions along the A-P axis. Studies in all major model organisms have provided compelling evidence that the Hox gene family plays fundamental roles during the early steps of neuronal development (reviewed in [4–6]). However, it remains largely unknown whether Hox genes are essential for the execution of later steps of neuronal development. In this review, we specifically focus on the function of Hox genes in the nervous system of the nematode Caenorhabditis elegans. Like in all other model organisms, Hox genes in C. elegans play critical roles during early patterning events of the nervous system by controlling cell proliferation, survival, and migration. However, the ease of conducting temporally controlled gene inactivation studies in C. elegans has enabled researchers to bypass early pleiotropies and discover new (non-canonical) roles for Hox genes during late developmental and adult stages. Here, we begin with a brief introduction to Hox genes. In subsequent sections, we highlight the early (section 4) and late (section 5) roles of Hox genes in the C. elegans nervous system.
2. Hox genes: spatial collinearity and homeotic transformations
Hox genes encode conserved transcription factors renowned for their roles in A-P body patterning and segmentation [7]. Hox mutations were first discovered in the fruit fly Drosophila melanogaster in the early 20th century by Calvin Bridges, a student of Thomas Hunt Morgan. Bridges began his foundational studies of Hox genes following reports by William Bateson in the late 19th century of “freak” animals, such as a moth with its legs transformed into wings. Bateson coined the term “homeosis” to describe this kind of transformation of one body part into the likeness of another, from the Greek homoios, meaning “similar” or “same”. Bridges’ discovery of fly mutants with striking homeotic transformations inspired later work on Hox genes [8]. Specifically, Bridges identified flies with duplicated thoracic segments, whose halteres (structures essential for flight) were transformed into a second pair of wings. The genes that control this homeotic transformation belong to the bithorax complex (BX-C) of genes. Genetic studies performed by Bridges and Morgan provided the foundation for Edward Lewis and Thomas Kaufman to later characterize systematically the functions of Hox genes of the BX-C complex. Lewis, along with Christiane Nusslein-Volhard and Eric Wieschaus, determined that Hox genes are required to pattern and segment the fly body, a discovery for which they shared the 1995 Nobel Prize in Physiology or Medicine.
Given this rich history, it is not surprising that Drosophila has been the premier model to investigate the role of Hox genes in body patterning and segmentation along the A-P axis [9]. Much of our understanding of Hox-mediated control of neuronal development has emerged from studies in the ventral nerve cord of Drosophila, including the discovery of body region-specific neuromuscular networks (Reviewed in [10, 11]). In vertebrates, studies of the spinal cord, hindbrain, and limb systems have also provided key insights into the function of Hox genes in body patterning, segmentation, and nervous system development [5, 6, 12]. Three highly conserved themes have emerged from all these studies of bilaterian animals: (1) Hox genes are organized into chromosomal clusters, (2) the sequence of expression domains of each Hox gene along the A-P body axis matches the order of the genes in the Hox cluster (spatial collinearity), and (3) mutations in Hox genes can have dramatic (e.g., homeotic transformations) as well as subtle effects in animal development.
This review specifically focuses on the function of Hox genes in the C. elegans nervous system for two reasons. First, we compare and contrast C. elegans studies with observations in Drosophila and vertebrates, aiming to highlight the remarkable conservation of Hox gene functions in early patterning of the nervous system. Second, we call attention to novel functions of Hox genes recently discovered in adult C. elegans neurons, i.e., a continuous requirement to maintain cellular identity. Such new functions may be conserved in other cell types and organisms.
Hox genes in C. elegans
Although Hox gene functions have been traditionally studied in animals with segmented structures within the nervous system (e.g., rhombomeres in the vertebrate hindbrain [13–15]), the non-segmented nervous system of the nematode C. elegans has also been an invaluable platform. The nematode nervous system is simple and well-characterized, offering single-cell resolution. The adult hermaphrodite and male contain 302 and 387 neurons, respectively. All neurons in both sexes have been described and named, and their complete lineages are known [16–21]. Furthermore, all neuronal connections (synapses) have been mapped for both nematode sexes [22] along with the transcriptional profiles of all neurons in the mature hermaphrodite [23]. Finally, the nematode has (a) a short lifespan of about three weeks, (b) is amenable to powerful genetic approaches, and (c) only six Hox genes (compared to eight in Drosophila and thirty-nine in mice) are embedded in its genome.
Hox genes encode homeodomain proteins – transcription factors defined by the presence of a 60 amino acid-long homeodomain essential for DNA contact [24]. In total, there are 102 homeodomain transcription factors in C. elegans. Only 6 of these 102 are Hox transcription factors, and these 6 Hox proteins are the focus of this review. We refer readers interested in the remaining 96 homeodomain transcription factors to another comprehensive review [25].
The six genes in the C. elegans Hox cluster span 5 Mb of chromosome III (Fig. 1 A). Unlike other bilaterians, C. elegans only contains 4 Hox ortholog groups: Hox1, Hox5, Hox6-8, and Hox9-13 [26]. There is a single anterior Hox gene ceh-13 (Lab/Hox1), two midbody Hox genes lin-39 (Scr/Dfd/Hox3-5) and mab-5 (Antp/Hox6-8), and three posterior Hox genes egl-5 (Abd-A/Abd-B/Hox9-13), nob-1 (Abd-B/Hox9-13) and php-3 (Abd-B/Hox9-13). Except for ceh-13, which is positioned between lin-39 and mab-5 due to a genomic inversion event, spatial collinearity is observed for all C. elegans Hox genes both in embryonic (Fig. 1 B) and postembryonic tissues (Fig. 1 C) [26–28]. Over the past three decades, the nematode C. elegans has been a prime model to systematically investigate the function of Hox genes in neurodevelopment.
Figure 1. Spatial collinearity is conserved in the C. elegans Hox cluster.

(A) Depiction of Hox clusters in C. elegans (top) and Drosophila (bottom). Colors are used to depict the closely related orthologs. (B) Expression of Hox genes in C. elegans 1.5-fold embryo. (C) Expression of Hox genes in larval and adult stage C. elegans. A, Anterior; P, posterior; D, dorsal; V, ventral.
3. Hox gene functions in early patterning of the C. elegans nervous system
Unlike other model organisms, single (null) mutant analysis for five of the six C. elegans Hox genes (lin-39, mab-5, egl-5, nob-1, php-3) indicates that they are not required for organismal survival [29–34]; only the anterior Hox gene ceh-13 (“C. elegans homeobox”-13) is required for survival to adulthood [35]. Although double nob-1; php-3 mutants are lethal [36], double and triple null mutants for midbody Hox genes lin-39 and mab-5 and the posterior Hox gene egl-5 are viable. Hence, C. elegans provides the opportunity to investigate Hox gene function both in early and late stages of nervous system development. To date, much of what we know about Hox genes in the C. elegans nervous system is derived from single and double mutant analysis. In the next three sections (3.1 – 3.3), we highlight key roles for Hox in early patterning of the C. elegans nervous system.
3.1. Control of neuroblast cell divisions
Hox genes influence the timing and number of cell divisions in Drosophila and vertebrate neural stem cells [37–40]. Because the timing and location of all cell divisions are well-defined in C. elegans, it has been possible to perform in-depth investigations into the functions of Hox genes in neuroblast divisions.
A null mutation in ceh-13 or a mutation that eliminates expression of both nob-1 and php-3 cause severe defects in the organization of the nervous system, intestine, body wall muscle, and epidermis, leading to early lethality [35, 36, 41, 42]. The nervous system is especially affected in these Hox mutants relative to other tissues. For example, the most severe organizational defects in ceh-13 mutants occur in the DA and SAB motor neuron classes, which become anteriorly displaced by several cell diameters [42]. Morphological defects in both ceh-13 and nob-1;php-3 mutants were initially attributed to abnormal cell migration [35, 36], but recent work has also revealed a role for these Hox genes in the control of embryonic cell divisions, especially in the nervous system [42]. For example, the precursor cells that normally differentiate to generate the sheath (glial) cells that wrap around the ADE sensory neuron fail to exit the cell cycle in ceh-13 mutants [42]. These animals also have delayed cell divisions in the neuroblasts that generate the DA, DD, and SAB motor neurons. nob-1; php-3 mutants also have delayed cell divisions; these occur in posterior neuroblasts that generate interneurons (PVQ class), sensory neurons (PHB class), and motor neurons (HSN class) [42]. Additionally, the neuroblast that normally generates the PLM and ALN sensory neurons in the posterior fails to divide entirely [42].
3.2. Control of neuronal lineage and cell survival
A distinguishing feature of C. elegans neurogenesis is that neurons emerge non-clonally from independent lineages or sub-lineages spanning the A-P axis. While some sub-lineages exclusively generate neurons, others also generate non-neuronal cells like muscle and hypodermis. Within a sub-lineage, neurons often do not share common features, i.e., they do not use the same neurotransmitter (NT) nor perform the same functions [43]. In C. elegans, studies on ceh-13 and mab-5 suggest that Hox gene expression is controlled, at least in part, by a lineage-based molecular mechanism; specific cell lineages and sub-lineages autonomously activate ceh-13 and mab-5 genes independently of cell position along the A-P axis [41, 44]. In addition, Wnt signaling is necessary for Hox gene expression in C. elegans lineages [45–47]. Wnt signaling controls Hox expression in Drosophila and vertebrate embryos as well [48, 49], constituting a conserved control mechanism of Hox gene expression. Further, neuronal lineages along the A-P axis of the nematode are controlled in part by Hox gene activity. This has been most evident from extensive studies on the patterning of neuroectodermal blast cell lineages in both C. elegans sexes (section 3.2.1) and sensory neurons of the male tail (section 3.2.2).
3.2.1. Lineage transformations in the neuroectoderm
The twelve ventral neuroectodermal blast cells called P1-12 that neatly align along the A-P axis of the C. elegans body constitute one of the most well-studied examples of neuronal lineages controlled by Hox gene activity (Fig. 2 A). Early in L1, P cells divide to produce two cells: the anterior daughter cell (Pn.a) is a neuroblast, whereas the posterior daughter (Pn.p) is an epidermal cell. Each P cell will ultimately produce one epidermal and several distinct neuronal descendants (Fig. 2 B) [50]. Anterior descendants of P (Pn.a) become motor neurons (AS, VA, VB, VC, and VD classes) of the ventral nerve cord [50], an analogous structure to the vertebrate spinal cord. The motor neurons of the ventral nerve cord provide a straightforward context within which to interpret lineage transformations in Hox mutants for several reasons. First, the expression of each Hox gene is generally uniform in P descendants within each Hox expression domain [51, 52]. Second, Hox expression domains establish variations in the patterning of fates along the ventral nerve cord motor neurons [51, 53, 54]. Third, ventral nerve cord motor neurons are linearly arrayed along the A-P axis and have region-specific characteristics and identities.
Figure 2. Hox genes pattern the neuroectoderm in C. elegans.

(A) Expression of Hox genes in the P cell lineage at the L1 stage. (B) Lineage diagrams indicating the descendants from each P-derived neuroblast along the A-P axis of hermaphrodites (left) and males (right). Posterior P lineage (Pn.p) generates cells of the epidermis and is excluded. Red X indicates programmed cell death. Blue text indicates neurons discussed in C-F. (C) LIN-39 patterns sex-specific VC neurons in the hermaphrodite midbody P lineage. (D) LIN-39 patterns sex-specific CA/CP neurons in the male anterior and midbody P lineage. (E) MAB-5 patterns the VB motor neurons in both sexes and the posterior P12.aap lineage in males (F).
Here, we discuss key roles for midbody (lin-39, mab-5) and posterior (egl-5) Hox genes in the establishment of cell fate in P descendants. Mutations in lin-39 were initially recovered in genetic screens for genes involved in cell survival [29, 30]. In the C. elegans hermaphrodite, lack of lin-39 gene activity leads to cell death of the sex-specific VC neurons that are used for egg-laying. Specifically, the midbody P descendants P3-P8.aap that normally generate VC neurons adopt instead the anterior or posterior Pn.aap fate of programmed cell death (Fig. 2 C) [29, 50, 54–56]. Importantly, the same phenotype is observed in animals lacking ceh-20 [57], a Hox cofactor of the extradenticle/Pbx class of homeodomain proteins [58]. Mechanistically, LIN-39 and CEH-20 form a complex that directly represses transcription of the proapoptotic gene egl-1 (Bcl-2 family member), thereby ensuring survival of the VC neurons [59]. Cooperative DNA-binding of Hox proteins with their cofactors is a conserved strategy to increase Hox binding affinity and specificity [60].
Like in hermaphrodites, lin-39 and ceh-20 are necessary for survival of midbody P descendants in the C. elegans male [55, 61]. That is, lin-39 mutants exhibit fate transformations in the midbody P lineage that generates the male-specific CA and CP neurons: P3-P6.aap cells adopt the anterior P1.aap fate of programmed cell death, while P7-P8.aap survive but produce CAs and CPs that resemble the posterior P9-P11.aap fate, and thereby fail to produce serotonin – the normal NT of CA and CP neurons (Fig. 2 D) [54, 55, 61]. Thus, lin-39 is required, in both C. elegans sexes, to distinguish motor neuron lineages in the midbody from lineages of motor neurons anterior or posterior to P3-P8 (Fig. 2 A).
Lineage transformations have also been observed in animals lacking mab-5, another midbody Hox gene. Posterior to the lin-39 expression domain (Fig. 2 A), mab-5 has a role in determining the cell fate and survival of P descendants. MAB-5 protein is expressed in P7-P12 blast cells (Fig. 2 A) [54]. In both hermaphrodite and male C. elegans, the anterior P1-10.aaap cells become the ventral B-type (VB) motor neurons, while the posterior P11-12.aaap cells undergo programmed cell death (Fig. 2 E). In mab-5 mutants, all Pn.aaap cells survive and at least one of the P descendants that normally dies, P11.aaap, generates a putative VB neuron [62]. Hence, mab-5 promotes cell death in posterior P11-12.aaap cells. Mechanistically, MAB-5 induces cell death by forming a complex with CEH-20 (extradenticle/Pbx) to activate transcription of the proapoptotic gene egl-1 [57].
In addition to promoting cell death in P11.aaap cells of both C. elegans sexes, mab-5 also controls patterning of neuronal cell lineages in Pn.aap cells in the C. elegans male. In wildtype males, the Pn.aap cells have definite fates based on position along the A-P axis: P1.aap undergoes cell death, P2.aap and P12.aap differentiate, and P(3-11).aap divide before differentiation. In mab-5 mutants, the anterior cells P(1-8).aap are unaffected, while the posterior P(9-11).aap differentiate without division and morphologically resemble P2.aap cells (Fig. 2 F) [62].
The posterior Hox gene egl-5 (Abd-A/Abd-B/Hox9-13) is expressed just posterior to the expression domain of mab-5 (Fig. 2 A). egl-5 regulates lineage patterning in the most posterior P cell P12; In egl-5 mutants of both sexes, P12 adopts a P11.p-like lineage [32]. Such lineage transformation is not observed in ceh-20 mutants, suggesting CEH-20 (extradenticle/Pbx) may not act as cofactor for the posterior Hox protein EGL-5 [57]. Interestingly, work in Drosophila and vertebrates also suggested that Abd-B proteins, like EGL-5, can function independently of Hox cofactors [63, 64].
The neuroectodermal blast cells P11 and P12 begin as developmentally equivalent left-right homologs at the L1 stage [32, 65]. This symmetry is broken when each cell migrates to a specific location in the midline during the L1 stage, after which the terminal P cells produce distinct lineages [65]. In wildtype hermaphrodites, P11-12 divide to produce anterior neuroblasts P11-12.a, which undergo three more rounds of division to each produce three motor neurons of the ventral cord: P11.a produces VA11, AS11, VD12 and P12.a produces VA12, VD13, and PDB [16, 66]. In egl-5 mutants, the P12.a descendant VA12 neuron ectopically expresses molecular markers of anterior VA neurons, transformation of a posterior to an anterior cell fate [67]. Such transformations are also widely observed in various cell types of flies and mice lacking Hox gene activity, and can be, at least partially, explained by posterior Hox genes repressing expression of anterior ones [5, 68]. That is indeed the case for egl-5; it represses lin-39 (Scr/Dfd/Hox3-5) in the P12.a descendant VA12 neuron [67].
Hox genes often function cell-autonomously to determine lineage patterns within their domain of expression [67, 69–71]. However, the underlying mechanism is more complex when one considers interactions between multiple Hox genes. For example, the overlapping of lin-39 and mab-5 expression domains in the hermaphrodite posterior midbody gives rise to specific P cell lineages, distinct from those in more anterior or posterior domains where either lin-39 or mab-5 is exclusively expressed [54–56, 72]. In wildtype males, LIN-39 specifies a serotonergic motor neuron fate in the descendants of P3-P8, while MAB-5 activity specifies a non-serotonergic interneuron fate in the descendants of P9-P11. In the absence of LIN-39, P7 and P8 generate non-serotonergic interneurons, reflecting the underlying activity of MAB-5 [54, 55]. Thus, in male-specific neurons of the ventral nerve cord, LIN-39 limits the activity but not the expression of mab-5 [53].
An additional facet to Hox function in lineage patterning is temporal segregation of Hox expression, i.e., some lineages require different Hox genes at different times. Both mab-5 and egl-5 expression are required in the P12 lineage (discussed above) but at different times [32, 53, 62]; mab-5 expression is required in P12 and its early descendants, but its expression terminates at the onset of egl-5 expression midway through P12 lineage progression [73]. This repression of mab-5 expression is egl-5-dependent [54], suggesting that Hox expression can be dynamic within lineages, consistent with studies in Drosophila, mouse, and zebrafish.
3.2.2. Patterning the sensory neurons of the male tail
In the C. elegans male, the V and T cell lineages generate the lateral epidermis and contribute neurons and other cells to sensory rays, the tail structures essential for copulation (Fig. 3 A). The anterior V cells (V1-V4) generate the seam cells that produce part of the cuticle, while the posterior V cells (V5-V6) generate the neuroblast lineages that produce cells for the sensory rays (Fig. 3 B–C). Each ray is a specialized sensillum that contains two sensory neurons, RnA and RnB, with distinct NT identity and connectivity (Fig. 3 D) [50, 74–77]. Rays also differentially express mab-5 and egl-5 (Fig. 3 E). In egl-5 mutants, posterior ray neurons (R3-5A, R3-5B) undergo a posterior-to-anterior transformation as they adopt the morphology and NT identity of anterior ray neurons (Fig. 3 F) [77]. Consistent with neuronal transformations in flies and mice, it was proposed that this transformation occurs due to egl-5 (posterior Hox) repressing mab-5 (mid-body Hox). In mab-5 mutants, ray neurons are absent altogether; V5-V6 descendants adopt the fate of their anterior homologs and produce the non-ray structures generated by V1-V4 cells (Fig. 3 F) [32, 62, 72]. In addition, overexpression of either mab-5 or egl-5 also leads to anterior-to-posterior transformations in ray identities (Fig. 3 F) [72, 78–80].
Figure 3. Posterior Hox genes are selectors of sensory rays in the male tail.
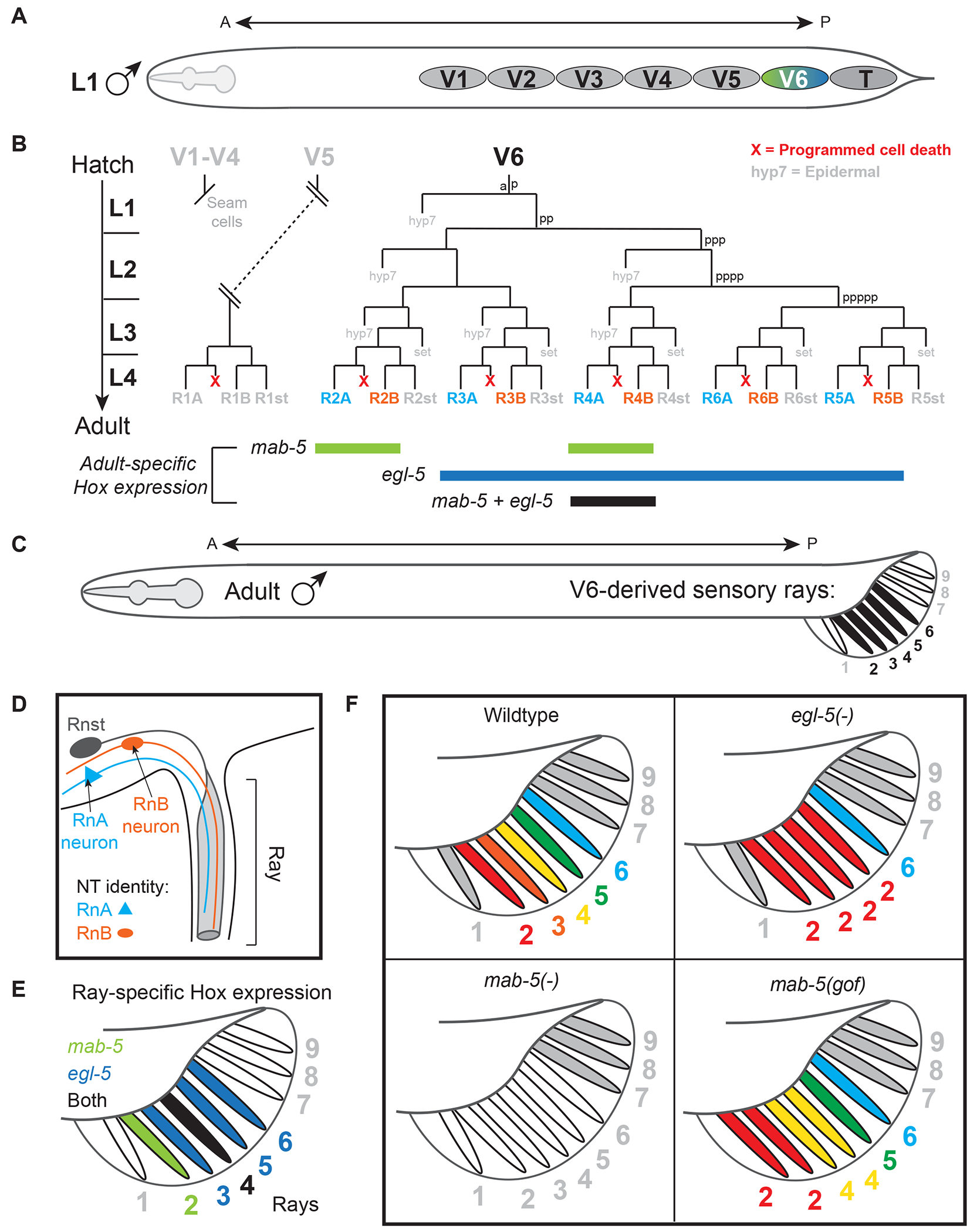
(A) Schematic of the V cells and T cell which generate the sensory rays in the copulatory male tail. (B) (top) Lineage diagrams of V5, which generates the R1A-R1B ray neurons, V6, which generates the R2A-R6A and R2B-R2A ray neurons; (bottom) Summary of hox expression across post-mitotic ray neurons in the adult. (C) Adult C. elegans male with V6-derived rays (ray 2-6) indicated in black. (D)The anatomy of a ray, including structure cell (Rnst) and both A- and B-type ray neurons color-coded blue and orange, respectively. (E) Expression of posterior Hox genes in V6-derived rays. (F) Summary of NT identity of rays in wildtype (top left), egl-5 (top right) and mab-5 (bottom left) null mutants, and in mab-5 gain-of-function (gof) mutants (bottom right).
These studies support a role for Hox genes as ‘selectors’ - genes required to pattern the specific identity of a tissue or organ, a concept introduced by Antonio Garcia-Bellido in 1975 based on work in Drosophila wing disc development [81]. In the C. elegans male tail, egl-5 and mab-5 behave as selector genes of ray identity by controlling the region-specific features and the NT identity of sensory rays. The selector function of Hox in the nervous system appears highly conserved, as Hox1-4 also function as selectors of hindbrain patterning in vertebrates [82, 83].
3.3. Control of cell migration during nervous system development
During development, neuroblasts and post-mitotic neurons often migrate to reach their final destination. In C. elegans, the Q neuroblasts and their descendants (sensory and interneurons) represent a prime model for the study of Hox genes in cell migratory behavior. In late embryos and young larvae (larval stage 1, L1), two Q neuroblasts occupy the right (QR neuroblast) and left (QL) sides of the C. elegans body at a similar position along the A-P axis (Fig. 4 A). Descendants of each neuroblast migrate to a stereotypical position in the body, but in opposite directions. QL and its descendants migrate toward the posterior, whereas QR and its descendants migrate toward the anterior. The directionality of migration in Q neuroblast descendants depends on the activity of the midbody Hox genes lin-39 and mab-5, as demonstrated by seminal studies from the labs of Cynthia Kenyon and Bob Horvitz (Fig. 4 B).
Figure 4. Hox genes promote cell migration in the C. elegans nervous system.
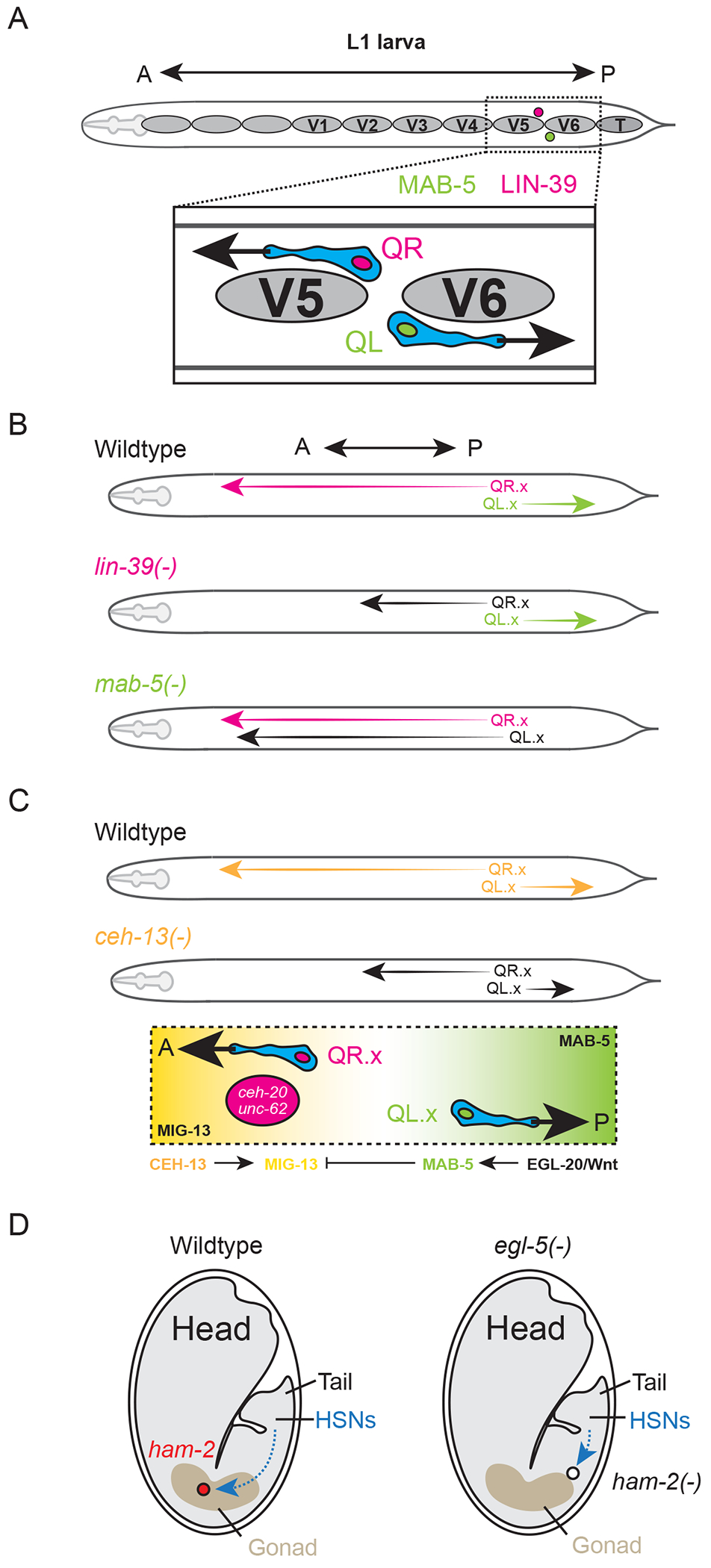
(A) Starting position of the QR (anteriorly polarized) and QL (posteriorly polarized) neuroblasts between V5 and V6 at L1 stage. (B) Summary of QR.x/QL.x neuroblast migration in wildtype (top) and in lin-39 and mab-5 null mutants (bottom). (C) Migration defects in ceh-13 null mutants and depiction of opposing gradients of MIG-13 (anterior) and MAB-5 (posterior) that guide the migration of QR.X and QL.X. (D) Summary of HSN migration path in wildtype (left) and egl-5 null mutants (right).
The descendants of QR (hereafter collectively referred to as QR.x) but not those of QL (QL.x) require lin-39 to successfully complete their anterior migration [55, 56]. Loss of lin-39 has no apparent effect on the posterior migration of QL.x. Rather, in QL.x, mab-5 is the key regulator and is both necessary and sufficient for posterior migration in these cells (Fig. 4 B) [84, 85]. How do the QR and QL neuroblasts come to express different Hox genes despite occupying similar positions along the A-P axis? The canonical Wnt/Beta-catenin signaling pathway becomes activated early and specifically in QL, ultimately leading to the activation of mab-5 [86–93]. QL requires an EGL-20/Wnt diffusion gradient along the A-P axis to activate mab-5 [87, 94]. This strategy appears specific to mab-5, as lin-39 regulates QR.x migration through a Wnt- independent mechanism [95].
The anterior Hox ceh-13 is also implicated in Q neuroblast migration. Unlike lin-39 and mab-5, ceh-13 mutants display defects in the migratory behaviors of both Q neuroblasts and their descendants [27]. In ceh-13 mutants, the migration defects are subtle in QL.x, but QR.x terminate their anterior migration early (Fig 4 C) [27]. What explains the differential severity of these migratory defects in ceh-13 mutants? Two Hox cofactors, ceh-20 (extradenticle/Pbx) and unc-62 (homothorax/Meis), are expressed only in QR.x, allowing QR.x descendants to respond to a migratory cue provided by the transmembrane protein MIG-13, an ortholog of Lrp12 in vertebrates [95, 96]. CEH-13 activates mig-13 in the anterior, while mab-5 inhibits its expression in the posterior (Fig. 4 C) [27, 96]. Interestingly, the mig-13 homolog in mice Lrp12 is expressed in migrating neurons during cortex development [97].
In addition to neuroblast migration, Hox genes also control the migration of postmitotic neurons. The hermaphrodite-specific neurons (HSN class) are a bilaterally symmetric pair of motor neurons (HSNL/R) generated in the tail of the embryo [98]. Shortly after their birth, the HSNs begin anterior migration toward the hermaphrodite vulva, about halfway up the A-P axis. The posterior Hox protein EGL-5 is a key regulator of this process as well as many other aspects of HSN differentiation (Fig. 4 D) [98]. EGL-5 activates the expression of the zinc finger transcription factor ham-2 (HSN abnormal migration-2) [99]. HSNs lacking ham-2 terminate migration halfway to their final destination near the vulva [99]. Additionally, HSNs of egl-5 mutants fail to down regulate the expression of the pro-migratory zinc finger transcription factor, egl-43/EVI-1 [99]. Genes initially identified from Hox studies in C. elegans HSN migration (e.g., egl-43/evi-1, epi-1/laminin α5, unc-71/adam-13, and ham-1/stox1) have since been found to be necessary for migration of neural crest cells in vertebrates, suggesting the molecular mechanisms driving cell migration are deeply conserved [13, 14, 100].
4. Hox functions in late steps of C. elegans nervous system development
A striking one third of C. elegans neurons (93 of 302 neurons) maintain expression of at least one Hox gene in late developmental and adult stages [52]. By synthesizing information from multiple studies [23, 52, 67, 101, 102], we mapped the expression of each C. elegans Hox gene in the mature (L4) hermaphrodite nervous system with single-cell resolution (Fig. 5). All six C. elegans Hox genes are expressed in the nervous system. Their maintained expression suggests a continuous requirement for Hox in the nervous system at late developmental and adult stages of life. In fact, C. elegans has been instrumental in testing post-mitotic neuronal requirements for Hox function due to its short lifespan and powerful genetic tools, enabling Hox gene inactivation across different life stages. Such Hox requirements are less explored in flies and vertebrates. Below, we highlight recent studies on synapse maturation and neuronal terminal identity, which critically extend the Hox functional repertoire beyond early patterning.
Figure 5. Hox expression in the mature C. elegans nervous system.

(A) Anatomy of the mature C. elegans nervous system. (B) Expression matrix of all 6 Hox genes (rows) in every neuron (columns) in the mature nervous system, ordered from anterior to posterior. Left-right and dorsal-ventral pairs of neurons are merged into one column when Hox gene expression is identical. Matrices are broken into the anatomical groups in A. *ceh-13 expression pattern is inferred from data in [101]; single-cell expression pattern is pending for this gene.
4.1. Control of synapse formation/maturation in C. elegans
DA9, the ninth member of the DA class of cholinergic motor neurons, has been a powerful model to study synapse formation in C. elegans. The DA9 neuron is located close to the tail and its axon extends circumferentially to reach dorsal body wall muscles and form en passant neuromuscular synapses. In animals lacking activity of the posterior Hox gene egl-5, these DA9 synapses are generated onto more anteriorly located muscles when compared to wildtype animals, suggesting a synaptic specificity defect [67]. Split GFP reporter technology (GRASP) also revealed that the DA9 neurons of egl-5 mutant animals fail to maintain synaptic inputs from the AVG interneurons. Importantly, the AVG inputs are properly established at early larval stages but fail to be maintained only in adult egl-5 mutants, indicating a Hox requirement in synapse maintenance. Together, these findings suggest that the posterior Hox gene egl-5 controls both synaptic input and output of a posterior cholinergic motor neuron (DA9) in C. elegans. These observations are reminiscent of recent findings in Drosophila [103, 104] and mice [105], suggesting a conserved role for Hox proteins in the formation and maintenance of neuronal synapses.
4.2. Control of neuronal terminal identity by C. elegans Hox genes
The function of every neuronal circuit critically relies on the ability of its constituent neurons to communicate with each other via neurotransmitters (NTs) and/or neuropeptides, as well as to display neuron type-specific morphological and electrophysiological signatures. These abilities are defined by the continuous expression of NT biosynthesis proteins, ion channels, neuropeptides, NT receptors, gap junction proteins, and cell adhesion molecules. Genes coding for such proteins have been termed “terminal identity genes” [106, 107]. Because they are expressed continuously, from late developmental stages through adulthood, terminal identity genes determine the final (mature) identity and thus function of each neuron type. Emerging evidence suggests that all six C. elegans Hox genes are involved in the control of terminal identity of various neuron types [52, 61, 108, 109]. Here, we specifically focus on touch receptor neurons (section 5.2.1) and nerve cord motor neurons (section 5.2.2) because mechanistic studies have been performed on these cells, strongly supporting the idea that Hox genes are continuously required to establish (during development) and maintain (in the adult) neuronal terminal identity features.
4.2.1. Establishment of touch receptor terminal identity.
Six touch receptor neurons (TRNs) mediate sensory responses to light touch in C. elegans. TRNs are classified into four subtypes (classes). The bilaterally symmetrical pairs of ALM and PLM neurons are located in the midbody and tail region, respectively, whereas single AVM and PVM neurons are located in the midbody (Fig. 6 A). The TRNs synapse onto and provide input to various command interneuron classes (PVC, AVB, AVD, AVA), which stimulate downstream motor neurons, thus generating touch reflex responses.
Figure 6. Hox genes control neuronal terminal identity.

(A) (Top) Schematic of mature C. elegans with color-coded motor neurons and TRNs. Text label colors of motor neuron classes correspond to circles/cell bodies on the ventral surface of the animal. (Bottom) Hox expression domains are indicated with color code that is consistent with previous figures. (B) Schematic depicting anterior (CEH-13) and posterior (EGL-5) Hox genes collaborating with A/PLM terminal selector MEC-3 in sensory neurons to determine neuronal terminal identity. (C) Midbody (LIN-39, MAB-5) and posterior (EGL-5) Hox genes collaborate with UNC-3 to co-activate terminal identity genes in ventral cord motor neurons. (D) Midbody Hox genes and UNC-3 operate in a positive feedforward loop (FFL) to ensure robust expression of terminal identity genes in midbody motor neurons. (E) Hox (LIN-39) expression is maintained in motor neurons throughout life via positive autoregulation, which is balanced by negative UNC-3 feedback.
C. elegans animals lacking gene activity of the posterior Hox gene egl-5 are touch-insensitive at the tail, suggesting egl-5 controls the development of posteriorly located TRNs, the PLM neurons [110]. Indeed, egl-5 controls PLM morphological characteristics, such as neurite length, by repressing anterior Hox genes (lin-39, mab-5) and the Hox cofactors ceh-20 (extradenticle/Pbx) and unc-62 (homothorax/Meis) [111]. Further, egl-5 is necessary for the terminal identity and function of PLM neurons by activating the expression of various terminal identity genes, such as the gap junction-encoding gene inx-13 [111, 112] (Fig. 6 B). The case of egl-5 highlights a recurring theme of Hox gene function across model systems: posterior Hox genes repress the expression of anterior Hox to generate a distinct (novel) cell fate. This simple mechanism is often the underlying cause of cell fate transformations in Hox mutant animals, providing a conceptual framework for the evolution of novel cell types in the nervous system [5, 113]. Consistent with this idea, the PLM neurons in elg-5 mutants do acquire molecular and morphological features of anteriorly located TRNs, called ALM neurons [71].
In more anteriorly located TRNs, the ALM neurons, the anterior Hox gene ceh-13 regulates terminal identity; expression of a handful of ALM terminal identity genes (e.g., mec-4/SCNN1 sodium channel, mec-17/ATAT1 tubulin acetyltransferase) is reduced in ceh-13 mutant animals (Fig. 6 B) [111, 114]. Mechanistically, CEH-13 in ALM and EGL-5 in PLM act indirectly by controlling the levels of expression of mec-3, a LIM homeodomain transcription factor [111, 114]. MEC-3 is a terminal selector for both ALM and PLM neurons (Fig. 6 B) [115, 116]. Terminal selectors are transcription factors that determine the identity and function of specific neuron types by directly activating the expression of multiple terminal identity genes (e.g., NT biosynthesis proteins, ion channels, neuropeptides) [2]. The Hox proteins CEH-13 and EGL-5 increase the probability of transcriptional activation of the terminal selector gene mec-3 in ALM and PLM neurons, respectively, ensuring robustness of TRN terminal differentiation. This mechanism is also relevant for the problem of neuronal subtype diversification, which is evident in every nervous system. A common TRN fate is controlled by the terminal selector MEC-3, but specific ALM and PLM terminal identities are established through the activities of anterior (CEH-13) and posterior (EGL-5) Hox proteins (Fig. 6 B) [115, 116]. It is important to note that, in the context of TRNs, current evidence suggests that Hox proteins do not act as terminal selectors [111, 114].
4.2.2. Establishment and maintenance of motor neuron terminal identity.
Nine classes of motor neurons (MNs) are found in the C. elegans nerve cord of hermaphrodite animals. Based on neurotransmitter usage, they can be classified into cholinergic (SAB, DA, DB, VA, VB, AS, VC) and GABAergic (DD, VD) MNs (Fig. 6 A). The SAB, DA, DB, and DD neurons are generated embryonically, whereas the VA, VB, VC, VD, and AS neurons are generated post-embryonically [117]. The terminal identity of most cholinergic MN classes in the nerve cord (SAB, DA, DB, VA, VB, AS) critically depends on the terminal selector UNC-3, member of the conserved family of Collier/Olf/Ebf (COE) family of TFs [118–120]. Mechanistically, UNC-3 binds directly to the cis-regulatory region of multiple terminal identity genes (e.g., acetylcholine [ACh] biosynthesis proteins, ion channels, neuropeptides) and activates their transcription. The homeodomain TF UNC-30 (PITX) acts in analogous manner in GABAergic (DD, VD) MNs [121, 122].
Like anterior and posterior TRN subtypes, the study of nerve cord MNs offered critical insights into the role of Hox genes in neuronal subtype diversification. For example, the DA class consists of nine cholinergic MNs, which can be subdivided into three groups based on cell body position: (1) the anterior DA1 neuron is located anteriorly (at the retrovesicular ganglion), (2) the midbody DA2-7 neurons are located along the nerve cord, and (3) the posterior DA8-9 neurons are located at the posterior (preanal) ganglion (Fig. 6 C). Neurons of the remaining cholinergic (DB, VA, VB, AS, VC) and GABAergic (DD, VD) classes are organized in a similar manner along the nerve cord (Fig. 6 A). Moreover, anterior (e.g., DA1), midbody (e.g., DA2-7), and posterior (e.g., DA8-9) neurons do show distinct connectivity and expression profiles of terminal identity genes [123].
Hox genes control the terminal identity of mid-body and posterior cholinergic MNs via an intersectional strategy that involves the terminal selector UNC-3 [123]. For example, UNC-3 is expressed in all 9 DA neurons but collaborates with mid-body Hox genes lin-39 and mab-5 and the Hox cofactor ceh-20 (extradenticle/Pbx) in mid-body DA2-7 neurons to control expression of multiple terminal identity genes specific to these neurons (Fig. 6 C). Similarly, UNC-3 and the posterior Hox gene egl-5 determine the terminal identity of posterior DA9 neurons by co-activating a different set of terminal identity genes (Fig. 6 C). Biochemical evidence suggests that LIN-39 and MAB-5 – like UNC-3 – act directly by binding on the cis-regulatory region of terminal identity genes (unc-129, del-1, acr-2, dbl-1, unc-77, slo-2). The direct mode of action combined with their continuous expression in MNs during developmental and adult stages support the idea that, in mid-body cholinergic MNs (DA, DB, VA, VB, AS classes), Hox proteins, LIN-39 and MAB-5 act as bona fide terminal selectors. The anterior Hox gene ceh-13 is expressed in anterior MNs, but it remains unknown whether it controls of MN terminal identity.
A defining feature of terminal selectors is continuous requirement throughout life [106]. Are Hox genes required during adulthood to maintain the terminal identity and thereby the continuous functionality of nerve cord MNs? Protein depletion experiments using the auxin inducible degradation (AID) system demonstrated that the midbody Hox protein LIN-39 is indeed required in adult life to maintain expression of various terminal identity genes (e.g., ACh biosynthesis proteins, ion channels) of mid-body MNs [102, 124, 125]. Importantly, the terminal selectors LIN-39 and UNC-3 (Collier/Ebf) operate in a positive feedforward loop to ensure continuous and robust expression of terminal identity genes (Fig. 6 D). Of note, the continuous LIN-39 requirement in adult C. elegans MNs demonstrates a new function for Hox proteins, beyond their textbook roles in developmental patterning. Supporting this new role, a recent study in Drosophila demonstrated that the Hox gene Ultrabithorax (Ubx) is necessary for dopaminergic neuron identity and function in the adult [126].
How is Hox gene expression maintained in adult MNs? Feng et al. [125] identified a two-component mechanism for homeostatic control of lin-39 expression in adult MNs: (1) lin-39 positively and directly regulates its own expression, and (2) the transcriptional autoregulation of lin-39 is counterbalanced by negative UNC-3 feedback (Fig. 6 E). The same mechanism applies to mab-5 [125], the other mid-body Hox gene expressed in C. elegans nerve cord MNs. Future work in C. elegans and other model systems will determine whether transcriptional autoregulation and maintenance of neuronal terminal identity are conserved features of Hox gene function in the nervous system.
Recent work demonstrated that Hox genes can also act as terminal selectors in other MN classes that do not express unc-3. In GABAergic MNs, the midbody Hox genes lin-39 and mab-5 collaborate with the terminal selector unc-30 to control terminal identity gene expression during development [102]. In cholinergic VC neurons, LIN-39 is required not only to establish during development but also maintain in the adult the expression of multiple terminal identity genes (e.g., ACh biosynthesis components, NT receptors), corroborating the emerging notion that Hox proteins can act as terminal selectors in specific neuron types.
5. The ancestral function of Hox genes is likely neuronal
In 1998, Jean Deutsch and Herve Le Guyader proposed that the primordial function of Hox genes is to design and pattern the nervous system [127]. This hypothesis is largely based on (a) comparisons of Hox gene expression across species and (b) a lack of correlation between the number of Hox genes and the increase in morphological diversity (e.g., segmental differentiation) during evolution. In support of the first point, all extant bilaterians studied to date express Hox genes in the central nervous system (CNS). Further, some distantly related organisms, like leeches and amphioxus, which are separated in evolution by more than 500 million years, express Hox genes only in the CNS [127–130]. Consequently, this hypothesis postulates that the well-known functions of Hox genes in patterning various tissues outside the CNS along the A-P axis of the bilaterian body plan are derived functions [127, 131]. In their second point, Deutsch and Le Guyader acknowledge that the number and diversity of Hox genes can be similar between morphologically complex organisms (like vertebrates) and morphologically simpler species (like amphioxus). In this review, we explored the functions of Hox in nervous system development of an unsegmented (morphologically simple) animal, the nematode C. elegans. Like findings in complex model organisms, Hox genes are essential for neural patterning in C. elegans, lending support to the hypothesis that the ancestral function of Hox is neuronal. Importantly, Hox genes are involved at every level of nervous system development in C. elegans, from positioning neuroblasts in the embryo to maintaining neuronal terminal identity in the adult. Future studies are needed to determine whether the theme of a continuous requirement of Hox in the nervous system is widely applicable across bilaterians.
Acknowledgements
We would like to thank several members of the Kratsios lab (Manasa Prahlad, Honorine Destain, Weidong Feng, Filipe Marques) for helpful discussions and edits pertaining to this manuscript. This work was supported by two NIH grants to P.K (R01 NS116365-01, R01 NS118078-01).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- [1].Zappaterra MW, Lehtinen MK, The cerebrospinal fluid: regulator of neurogenesis, behavior, and beyond, Cellular and Molecular Life Sciences 69(17) (2012) 2863–2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Hobert O, Kratsios P, Neuronal identity control by terminal selectors in worms, flies, and chordates, Current Opinion in Neurobiology 56 (2019) 97–105. [DOI] [PubMed] [Google Scholar]
- [3].Hobert O, Chapter Twenty-Five - Terminal Selectors of Neuronal Identity, in: Wassarman PM (Ed.), Current Topics in Developmental Biology, Academic Press; 2016, pp. 455–475. [DOI] [PubMed] [Google Scholar]
- [4].Di Bonito M, Glover JC, Studer M, Hox genes and region-specific sensorimotor circuit formation in the hindbrain and spinal cord, Developmental Dynamics 242(12) (2013) 1348–1368. [DOI] [PubMed] [Google Scholar]
- [5].Philippidou P, Dasen JS, Hox Genes: Choreographers in Neural Development, Architects of Circuit Organization, Neuron 80(1) (2013) 12–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Parker HJ, Krumlauf R, A Hox gene regulatory network for hindbrain segmentation, Current Topics in Developmental Biology 139 (2020) 169–203. [DOI] [PubMed] [Google Scholar]
- [7].Lemons D, McGinnis W, Genomic Evolution of Hox Gene Clusters, Science 313(5795) (2006) 1918–1922. [DOI] [PubMed] [Google Scholar]
- [8].Lewis EB, A Gene Complex Controlling Segmentation in Drosophila, in: Lipshitz HD (Ed.), Genes, Development and Cancer: The Life and Work of Edward B. Lewis, Springer US, Boston, MA, 2004, pp. 205–217. [Google Scholar]
- [9].Lawrence PA, Morata G, Homeobox genes: Their function in Drosophila segmentation and pattern formation, Cell 78(2) (1994) 181–189. [DOI] [PubMed] [Google Scholar]
- [10].Estacio-Gómez A, Díaz-Benjumea FJ, Roles of Hox genes in the patterning of the central nervous system of Drosophila, Fly 8(1) (2013) 26–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Gummalla M, Galetti S, Maeda RK, Karch F, Hox gene regulation in the central nervous system of Drosophila, Frontiers in Cellular Neuroscience 8 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Nolte C, Krumlauf R, Expression of Hox genes in the Nervous System of Vertebrates, Springer, New York, NY: 2007, pp. 14–41. [Google Scholar]
- [13].Prince V, Lumsden A, Hoxa-2 expression in normal and transposed rhombomeres: independent regulation in the neural tube and neural crest, Development 120(4) (1994) 911–923. [DOI] [PubMed] [Google Scholar]
- [14].Saldivar JR, Krull CE, Krumlauf R, Ariza-McNaughton L, Bronner-Fraser M, Rhombomere of origin determines autonomous versus environmentally regulated expression of Hoxa-3 in the avian embryo, Development 122(3) (1996) 895–904. [DOI] [PubMed] [Google Scholar]
- [15].Trainor PA, Krumlauf R, Patterning the cranial neural crest: Hinbrain segmentation and hox gene plasticity, Nature Reviews Neuroscience 1(2) (2000) 116–124. [DOI] [PubMed] [Google Scholar]
- [16].Hall DH, Russell RL, The posterior nervous system of the nematode Caenorhabditis elegans: serial reconstruction of identified neurons and complete pattern of synaptic interactions, The Journal of Neuroscience 11(1) (1991) 1 LP–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Ward S, Thomson N, White JG, Brenner S, Electron microscopical reconstruction of the anterior sensory anatomy of the nematode caenorhabditis elegans, Journal of Comparative Neurology 160(3) (1975) 313–337. [DOI] [PubMed] [Google Scholar]
- [18].Ware RW, Clark D, Crossland K, Russell RL, The nerve ring of the nematode Caenorhabditis elegans: Sensory input and motor output, Journal of Comparative Neurology 162(1) (1975) 71–110. [Google Scholar]
- [19].White JG, Southgate E, Thomson JN, Brenner S, The Structure of the Nervous System of the Nematode Caenorhabditis elegans, Philos Trans R Soc Lond Biol Sci 314(1165) (1986) 1–340. [DOI] [PubMed] [Google Scholar]
- [20].Durbin RM, Studies in the Development and Organisation of the Nervous System of Caenorhabditis Elegans, 1987, pp. 121–121. [Google Scholar]
- [21].Chen BL, Hall DH, Chklovskii DB, Wiring optimization can relate neuronal structure and function, Proceedings of the National Academy of Sciences of the United States of America 103(12) (2006) 4723–4728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Cook SJ, Jarrell TA, Brittin CA, Wang Y, Bloniarz AE, Yakovlev MA, Nguyen KCQ, Tang LTH, Bayer EA, Duerr JS, Bülow HE, Hobert O, Hall DH, Emmons SW, Whole-animal connectomes of both Caenorhabditis elegans sexes, Nature 571(7763) (2019) 63–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Taylor SR, Santpere G, Weinreb A, Barrett A, Reilly MB, Xu C, Varol E, Oikonomou P, Glenwinkel L, McWhirter R, Poff A, Basavaraju M, Rafi I, Yemini E, Cook SJ, Abrams A, Vidal B, Cros C, Tavazoie S, Sestan N, Hammarlund M, Hobert O, Miller DM 3rd, Molecular topography of an entire nervous system, Cell 184(16) (2021) 4329–4347.e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Bürglin TR, Affolter M, Homeodomain proteins: an update, Chromosoma 125(3) (2016) 497–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Hobert O, Homeobox genes and the specification of neuronal identity, Nature Reviews Neuroscience 22(10) (2021) 627–636. [DOI] [PubMed] [Google Scholar]
- [26].Aboobaker A, Blaxter M, Hox gene evolution in nematodes: novelty conserved, Current Opinion in Genetics & Development 13(6) (2003) 593–598. [DOI] [PubMed] [Google Scholar]
- [27].Tihanyi B, Vellai T, Regős Á, Ari E, Müller F, Takács-Vellai K, The C. elegans Hox gene ceh-13 regulates cell migration and fusion in a non-colinear way. Implications for the early evolution of Hoxclusters, BMC Developmental Biology 10(1) (2010) 78–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Sulston J, Du Z, Thomas K, Wilson R, Hillier L, Staden R, Halloran N, Green P, Thierry-Mieg J, Qiu L, Dear S, Coulson A, Craxton M, Durbin R, Berks M, Metzstein M, Hawkins T, Ainscough R, Waterston R, The C. elegans genome sequencing project: a beginning, Nature 356 (1992) 37–41. [DOI] [PubMed] [Google Scholar]
- [29].Ellis HM, Horvitz HR, Genetic control of programmed cell death in the nematode C. elegans, Cell 44(6) (1986) 817–829. [DOI] [PubMed] [Google Scholar]
- [30].Fixsen W, Sternberg P, Ellis H, Horvitz R, Genes That Affect Cell Fates during the Development of Caenorhabditis elegans, Cold Spring Harbor Symposia on Quantitative Biology 50(1985) 99–104. [DOI] [PubMed] [Google Scholar]
- [31].Hodgkin J, MALE PHENOTYPES AND MATING EFFICIENCY IN CAENORHABDITIS ELEGANS, Genetics 103(1) (1983) 43–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Chisholm A, Control of cell fate in the tail region of C. elegans by the gene egl-5, Development 111(4) (1991) 921–932. [DOI] [PubMed] [Google Scholar]
- [33].Nelson MD, Zhou E, Kiontke K, Fradin H, Maldonado G, Martin D, Shah K, Fitch DHA, A Bow-Tie Genetic Architecture for Morphogenesis Suggested by a Genome-Wide RNAi Screen in Caenorhabditis elegans, PLOS Genetics 7(3) (2011) e1002010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Hori S, Oda S, Suehiro Y, lino Y, Mitani S, OFF-responses of interneurons optimize avoidance behaviors depending on stimulus strength via electrical synapses, PLOS Genetics 14(6) (2018)e1007477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Brunschwig K, Wittmann C, Schnabel R, Burglin TR, Tobler H, Muller F, Anterior organization of the Caenorhabditis elegans embryo by the labial-like Hox gene ceh-13, Development 126(7) (1999) 1537–1546. [DOI] [PubMed] [Google Scholar]
- [36].Van Auken K, Weaver DC, Edgar LG, Wood WB, Caenorhabditis elegans embryonic axial patterning requires two recently discovered posterior-group Hox genes, Proceedings of the National Academy of Sciences of the United States of America 97(9) (2000) 4499–4503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Maurange C, Gould AP, Brainy but not too brainy: starting and stopping neuroblast divisions in Drosophila, Trends in Neurosciences 28(1) (2005) 30–36. [DOI] [PubMed] [Google Scholar]
- [38].Gouti M, Gavalas A, Hoxb1 Controls Cell Fate Specification and Proliferative Capacity of Neural Stem and Progenitor Cells, Stem Cells 26(8) (2008) 1985–1997. [DOI] [PubMed] [Google Scholar]
- [39].Bello BC, Hirth F, Gould AP, A Pulse of the Drosophila Hox Protein Abdominal-A Schedules the End of Neural Proliferation via Neuroblast Apoptosis, Neuron 37(2) (2003) 209–219. [DOI] [PubMed] [Google Scholar]
- [40].Economides KD, Zeltser L, Capecchi MR, Hoxb13 mutations cause overgrowth of caudal spinal cordand tail vertebrae, Developmental Biology 256(2) (2003) 317–330. [DOI] [PubMed] [Google Scholar]
- [41].Wittmann C, Bossinger O, Goldstein B, Fleischmann M, Kohler R, Brunschwig K, Tobler H, Muller F, The expression of the C. elegans labial-like Hox gene ceh-13 during early embryogenesis relies on cell fate and on anteroposterior cell polarity, Development 124(21) (1997) 4193–4200. [DOI] [PubMed] [Google Scholar]
- [42].Murray JI, Preston E, Crawford JP, Rumley JD, Amom P, Anderson BD, Sivaramakrishnan P, Patel SD, Bennett BA, Lavon TD, Hsiao E, Peng F, Zacharias AL, The anterior Hox gene ceh-13 and elt-1/GATA activate the posterior Hox genes nob-1 and php-3 to specify posterior lineages in the C. elegans embryo, PLOS Genetics 18(5) (2022) e1010187–e1010187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Sulston JE, Schierenberg E, White JG, Thomson JN, The embryonic cell lineage of the nematode Caenorhabditis elegans, Developmental Biology 100(1) (1983) 64–119. [DOI] [PubMed] [Google Scholar]
- [44].Cowing D, Kenyon C, Correct Hox gene expression established independently of position in Caenorhabditis elegans, Nature 382(6589) (1996) 353–356. [DOI] [PubMed] [Google Scholar]
- [45].Arata Y, Kouike H, Zhang Y, Herman MA, Okano H, Sawa H, Wnt Signaling and a Hox Protein Cooperatively Regulate PSA-3/Meis to Determine Daughter Cell Fate after Asymmetric Cell Division in C. elegans, Developmental Cell 11(1) (2006) 105–115. [DOI] [PubMed] [Google Scholar]
- [46].Wagmaister JA, Gleason JE, Eisenmann DM, Transcriptional upregulation of the C. elegans Hox gene lin-39 during vulval cell fate specification, Mechanisms of Development 123(2) (2006) 135–150. [DOI] [PubMed] [Google Scholar]
- [47].Ji N, Middelkoop Teije C., Mentink Remco A., Betist Marco C., Tonegawa S, Mooijman D, Korswagen Hendrik C., van Oudenaarden A, Feedback Control of Gene Expression Variability in the Caenorhabditis elegans Wnt Pathway, Cell 155(4) (2013) 869–880. [DOI] [PubMed] [Google Scholar]
- [48].Nordström U, Maier E, Jessell TM, Edlund T, An Early Role for Wnt Signaling in Specifying Neural Patterns of Cdx and Hox Gene Expression and Motor Neuron Subtype Identity, PLOS Biology 4(8) (2006) e252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Streit A, Kohler R, Marty T, Belfiore M, Takacs-Vellai K, Vigano MA, Schnabel R, Affolter M, Müller F, Conserved Regulation of the Caenorhabditis elegans labial/Hox1 Gene ceh-13, Developmental Biology 242(2) (2002) 96–108. [DOI] [PubMed] [Google Scholar]
- [50].Sulston JE, Horvitz HR, Post-embryonic cell lineages of the nematode, Caenorhabditis elegans, Developmental Biology 56(1) (1977) 110–156. [DOI] [PubMed] [Google Scholar]
- [51].Kenyon CJ, Austin J, Costa M, Cowing DW, Harris JM, Honigberg L, Hunter CP, Maloof JN, Muller-Immerglük MM, Salser SJ, Waring DA, Wang BB, Wrischnik LA, The Dance of the Hox Genes: Patterning the Anteroposterior Body Axis of Caenorhabditis elegans, Cold Spring Harbor Symposia on Quantitative Biology 62 (1997) 293–305. [PubMed] [Google Scholar]
- [52].Zheng C, Lee HMT, Pham K, Nervous system-wide analysis of Hox regulation of terminal neuronal fate specification in Caenorhabditis elegans, PLOS Genetics 18(2) (2022) e1010092–e1010092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Salser SJ, Kenyon C, Patterning C. elegans: homeotic cluster genes, cell fates and cell migrations, Trends in Genetics 10(5) (1994) 159–164. [DOI] [PubMed] [Google Scholar]
- [54].Salser SJ, Loer CM, Kenyon C, Multiple HOM-C gene interactions specify cell fates in the nematode central nervous system, Genes & Development 7(9) (1993) 1714–1724. [DOI] [PubMed] [Google Scholar]
- [55].Clark SG, Chisholm AD, Horvitz HR, Control of cell fates in the central body region of C. elegans by the homeobox gene lin-39, Cell 74(1) (1993) 43–55. [DOI] [PubMed] [Google Scholar]
- [56].Wang BB, Müller-Immergluck MM, Austin J, Robinson NT, Chisholm A, Kenyon C, A homeotic gene cluster patterns the anteroposterior body axis of C. elegans, Cell 74(1) (1993) 29–42. [DOI] [PubMed] [Google Scholar]
- [57].Liu H, Strauss TJ, Potts MB, Cameron S, Direct regulation of egl-1 and of programmed cell death by the Hox protein MAB-5 and by CEH-20, a C. elegans homolog of Pbx1, Development 133(4) (2006) 641–650. [DOI] [PubMed] [Google Scholar]
- [58].Bürglin TR, Analysis of TALE superclass homeobox genes (MEIS, PBC, KNOX, Iroquois, TGIF) reveals a novel domain conserved between plants and animals, Nucleic Acids Research 25(21) (1997) 4173–4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Potts MB, Wang DP, Cameron S, Trithorax, Hox, and TALE-class homeodomain proteins ensure cell survival through repression of the BH3-only gene egl-1, Developmental Biology 329(2) (2009) 374–385. [DOI] [PubMed] [Google Scholar]
- [60].Mann RS, Lelli KM, Joshi R, Chapter 3 Hox Specificity: Unique Roles for Cofactors and Collaborators, Current Topics in Developmental Biology, Academic Press; 2009, pp. 63–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Kalis AK, Kissiov DU, Kolenbrander ES, Palchick Z, Raghavan S, Tetreault BJ, Williams E, Loer CM, Wolff JR, Patterning of sexually dimorphic neurogenesis in the Caenorhabditis elegans ventral cord by Hox and TALE homeodomain transcription factors, Developmental Dynamics 243(1) (2014) 159–171. [DOI] [PubMed] [Google Scholar]
- [62].Kenyon C, A gene involved in the development of the posterior body region of C. elegans, Cell 46(3) (1986) 477–487. [DOI] [PubMed] [Google Scholar]
- [63].Rivas ML, Espinosa-Vázquez JM, Sambrani N, Greig S, Merabet S, Graba Y, Hombría JC-G, Antagonism versus cooperativity with TALE cofactors at the base of the functional diversification of Hox protein function, PLoS genetics 9(2) (2013) e1003252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Shen W-F, Rozenfeld S, Lawrence HJ, Largman C, The Abd-B-like Hox Homeodomain Proteins Can Be Subdivided by the Ability to Form Complexes with Pbx1a on a Novel DNA Target *, Journal of Biological Chemistry 272(13) (1997) 8198–8206. [DOI] [PubMed] [Google Scholar]
- [65].Sulston JE, White JG, Regulation and cell autonomy during postembryonic development of Caenorhabditis elegans, Developmental Biology 78(2) (1980) 577–597. [DOI] [PubMed] [Google Scholar]
- [66].Jiang LI, Sternberg PW, Interactions of EGF, Wnt and HOM-C genes specify the P12 neuroectoblast fate in C. elegans, Development 125(12) (1998) 2337–2347. [DOI] [PubMed] [Google Scholar]
- [67].Kratsios P, Kerk SY, Catela C, Liang J, Vidal B, Bayer EA, Feng W, De La Cruz ED, Croci L, Consalez GG, Mizumoto K, Hobert O, An intersectional gene regulatory strategy defines subclass diversity of C. elegans motor neurons, eLife 6 (2017) e25751–e25751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Hombría JC-G, Lovegrove B, Beyond homeosis—HOX function in morphogenesis and organogenesis, Differentiation 71(8) (2003) 461–476. [DOI] [PubMed] [Google Scholar]
- [69].Miguel-Aliaga I, Thor S, Segment-specific prevention of pioneer neuron apoptosis by cell-autonomous, postmitotic Hox gene activity, Development 131(24) (2004) 6093–6105. [DOI] [PubMed] [Google Scholar]
- [70].Becker H, Renner S, Technau GM, Berger C, Cell-Autonomous and Non-cell-autonomous Function of Hox Genes Specify Segmental Neuroblast Identity in the Gnathal Region of the Embryonic CNS in Drosophila, PLOS Genetics 12(3) (2016) e1005961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Zheng C, Diaz-Cuadros M, Chalfie M, Hox Genes Promote Neuronal Subtype Diversification through Posterior Induction in Caenorhabditis elegans, Neuron 88(3) (2015) 514–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Chow KL, Emmons SW, HOM-C/Hox genes and four interacting loci determine the morphogenetic properties of single cells in the nematode male tail, Development 120(9) (1994) 2579–2592. [DOI] [PubMed] [Google Scholar]
- [73].Ferreira HB, Zhang Y, Zhao C, Emmons SW, Patterning ofCaenorhabditis elegansPosterior Structures by theAbdominal-BHomolog,egl-5, Developmental Biology 207(1) (1999) 215–228. [DOI] [PubMed] [Google Scholar]
- [74].Lints R, Emmons SW, Patterning of dopaminergic neurotransmitter identity among Caenorhabditis elegans ray sensory neurons by a TGFbeta family signaling pathway and a Hox gene, Development 126(24) (1999) 5819–5831. [DOI] [PubMed] [Google Scholar]
- [75].Schinkmann K, Li C, Localization of FMRF amide-like peptides in Caenorhabditis elegans, Journal of Comparative Neurology 316(2) (1992) 251–260. [DOI] [PubMed] [Google Scholar]
- [76].Sulston JE, Albertson DG, Thomson JN, The Caenorhabditis elegans male: Postembryonic development of nongonadal structures, Developmental Biology 78(2) (1980) 542–576. [DOI] [PubMed] [Google Scholar]
- [77].Lints R, Jia L, Kim K, Li C, Emmons SW, Axial patterning of C elegans male sensilla identities by selector genes, Developmental Biology 269(1) (2004) 137–151. [DOI] [PubMed] [Google Scholar]
- [78].Salser SJ, Kenyon C, A C. elegans Hox gene switches on, off, on and off again to regulate proliferation, differentiation and morphogenesis, Development 122(5) (1996) 1651–1661. [DOI] [PubMed] [Google Scholar]
- [79].Zhang H, Emmons SW, The novel C elegans gene sop-3 modulates Wnt signaling to regulate Hox gene expression, Development 128(5) (2001) 767–777. [DOI] [PubMed] [Google Scholar]
- [80].Zhang H, Azevedo RBR, Lints R, Doyle C, Teng Y, Haber D, Emmons SW, Global regulation of Hox gene expression in C. elegans by a SAM domain protein, Developmental Cell 4(6) (2003) 903–915. [DOI] [PubMed] [Google Scholar]
- [81].Garcia-Bellido A, Genetic Control of Wing Disc Development in Drosophila, Ciba Foundation Symposium 29 - Cell Patterning 1975, pp. 161–182. [DOI] [PubMed] [Google Scholar]
- [82].Narita Y, F.M.B.T.C.T.i.D.B. Rijli, Chapter 5 Hox Genes in Neural Patterning and Circuit Formation in the Mouse Hindbrain, Academic Press; 2009, pp. 139–167. [DOI] [PubMed] [Google Scholar]
- [83].Rossel M, Capecchi MR, Mice mutant for both Hoxa1 and Hoxb1 show extensive remodeling of the hindbrain and defects in craniofacial development, Development 126(22) (1999) 5027–5040. [DOI] [PubMed] [Google Scholar]
- [84].Salser SJ, Kenyon C, Activation of a C. elegans Antennapedia homologue in migrating cells controls their direction of migration, Nature 355(6357) (1992) 255–258. [DOI] [PubMed] [Google Scholar]
- [85].Harris J, Honigberg L, Robinson N, Kenyon C, Neuronal cell migration in C. elegans: regulation of Hox gene expression and cell position, Development 122(10) (1996) 3117–3131. [DOI] [PubMed] [Google Scholar]
- [86].Maloof JN, Whangbo J, Harris JM, Jongeward GD, Kenyon C, A Wnt signaling pathway controls hox gene expression and neuroblast migration in C. elegans, Development 126(1) (1999) 37–49. [DOI] [PubMed] [Google Scholar]
- [87].Whangbo J, Kenyon C, A Wnt Signaling System that Specifies Two Patterns of Cell Migration in C. elegans, Molecular Cell 4(5) (1999) 851–858. [DOI] [PubMed] [Google Scholar]
- [88].Korswagen HC, Herman MA, Clevers HC, Distinct β-catenins mediate adhesion and signalling functions in C. elegans, Nature 406(6795) (2000) 527–532. [DOI] [PubMed] [Google Scholar]
- [89].Herman M, C. elegans POP-1/TCF functions in a canonical Wnt pathway that controls cell migration and in a noncanonical Wnt pathway that controls cell polarity, Development (Cambridge, England) 128(4) (2001) 581–590. [DOI] [PubMed] [Google Scholar]
- [90].Korswagen HC, Coudreuse DYM, Betist MC, van de Water S, Zivkovic D, Clevers HC, The Axin-like protein PRY-1 is a negative regulator of a canonical Wnt pathway in C. elegans, Genes & development 16(10) (2002) 1291–1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Walston T, Guo C, Proenca R, Wu M, Herman M, Hardin J, Hedgecock E, mig-5/Dsh controls cell fate determination and cell migration in C. elegans, Developmental biology 298(2) (2006) 485–497. [DOI] [PubMed] [Google Scholar]
- [92].Oosterveen T, Coudreuse DYM, Yang P-T, Fraser E, Bergsma J, Dale TC, Korswagen HC, Two functionally distinct Axin-like proteins regulate canonical Wnt signaling in C. elegans, Developmental biology 308(2) (2007) 438–448. [DOI] [PubMed] [Google Scholar]
- [93].Silhankova M, Korswagen HC, Migration of neuronal cells along the anterior–posterior body axis of C. elegans: Wnts are in control, Current Opinion in Genetics & Development 17(4) (2007) 320–325. [DOI] [PubMed] [Google Scholar]
- [94].Coudreuse DYM, Roël G, Betist MC, Destrée O, Korswagen HC, Wnt Gradient Formation Requires Retromer Function in Wnt-Producing Cells, Science 312(5775) (2006) 921–924. [DOI] [PubMed] [Google Scholar]
- [95].Yang L, Sym M, Kenyon C, The roles of two C. elegans HOX co-factor orthologs in cell migration and vulva development, Development (Cambridge, England) 132(6) (2005) 1413–1428. [DOI] [PubMed] [Google Scholar]
- [96].Sym M, Robinson N, Kenyon C, MIG-13 Positions Migrating Cells along the Anteroposterior Body Axis of C. elegans, Cell 98(1) (1999) 25–36. [DOI] [PubMed] [Google Scholar]
- [97].Schneider S, Gulacsi A, Hatten ME, Lrp12/Mig13a Reveals Changing Patterns of Preplate Neuronal Polarity during Corticogenesis that Are Absent in Reeler Mutant Mice, Cerebral Cortex 21(1) (2011) 134–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Desai C, Garriga G, McLntire SL, Horvitz HR, A genetic pathway for the development of the Caenorhabditis elegans HSN motor neurons, Nature 336(6200) (1988) 638–646. [DOI] [PubMed] [Google Scholar]
- [99].Baum PD, Guenther C, Frank CA, Pham BV, Garriga G, The Caenorhabditis elegans gene ham-2 links Hox patterning to migration of the HSN motor neuron, Genes & Development 13(4) (1999) 472–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Kee Y, Hwang BJ, Sternberg PW, Bronner-Fraser M, Evolutionary conservation of cell migration genes: from nematode neurons to vertebrate neural crest, Genes & Development 21(4) (2007) 391–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Reilly MB, Cros C, Varol E, Yemini E, Hobert O, Unique homeobox codes delineate all the neuron classes of C. elegans, Nature 584(7822) (2020) 595–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Feng W, Li Y, Dao P, Aburas J, Islam P, Elbaz B, Kolarzyk A, Brown AE, Kratsios P, A terminal selector prevents a Hox transcriptional switch to safeguard motor neuron identity throughout life, Elife 9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Friedrich J, Sorge S, Bujupi F, Eichenlaub MP, Schulz NG, Wittbrodt J, Lohmann I, Hox Function Is Required for the Development and Maintenance of the Drosophila Feeding Motor Unit, Cell Reports 14(4) (2016) 850–860. [DOI] [PubMed] [Google Scholar]
- [104].Joshi R, Sipani R, Bakshi A, Roles of Drosophila Hox Genes in the Assembly of Neuromuscular Networks and Behavior, Front Cell Dev Biol 9 (2021) 786993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Feng W, Li Y, Kratsios P, Emerging Roles for Hox Proteins in the Last Steps of Neuronal Development in Worms, Flies, and Mice, Front. Neurosci 15(801791) (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Hobert O, Regulatory logic of neuronal diversity: terminal selector genes and selector motifs, Proc Natl Acad Sci U S A 105(51) (2008) 20067–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Hobert O, Kratsios P, Neuronal identity control by terminal selectors in worms, flies, and chordates, Curr Opin Neurobiol 56 (2019) 97–105. [DOI] [PubMed] [Google Scholar]
- [108].Kalis AK, Sterrett MC, Armstrong C, Ballmer A, Burkstrand K, Chilson E, Emlen E, Ferrer E, Loeb S, Olin T, Tran K, Wheeler A, Ross Wolff J, Hox proteins interact to pattern neuronal subtypes in Caenorhabditis elegans males, Genetics 220(4) (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Reilly MB, Tekieli T, Cros C, Aguilar GR, Lao J, Toker IA, Vidal B, Leyva-Díaz E, Bhattacharya A, Cook SJ, Smith JJ, Kovacevic I, Gulez B, Fernandez RW, Bradford EF, Ramadan YH, Kratsios P, Bao Z, Hobert O, Widespread employment of conserved C. elegans homeobox genes in neuronal identity specification, PLOS Genetics 18(9) (2022) e1010372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Chisholm A, Control of cell fate in the tail region of C. elegans by the gene egl-5, Development 111(4) (1991) 921–32. [DOI] [PubMed] [Google Scholar]
- [111].Zheng C, Diaz-Cuadros M, Chalfie M, Hox Genes Promote Neuronal Subtype Diversification through Posterior Induction in Caenorhabditis elegans, Neuron 88(3) (2015) 514–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Toker AS, Teng Y, Ferreira HB, Emmons SW, Chalfie M, The Caenorhabditis elegans spalt-like gene sem-4 restricts touch cell fate by repressing the selector Hox gene egl-5 and the effector gene mec-3, Development 130(16) (2003) 3831–40. [DOI] [PubMed] [Google Scholar]
- [113].Arlotta P, Hobert O, Homeotic Transformations of Neuronal Cell Identities, Trends in Neurosciences 38(12) (2015) 751–762. [DOI] [PubMed] [Google Scholar]
- [114].Zheng C, Jin FQ, Chalfie M, Hox Proteins Act as Transcriptional Guarantors to Ensure Terminal Differentiation, Cell Rep 13(7) (2015) 1343–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Way JC, Chalfie M, mec-3, a homeobox-containing gene that specifies differentiation of the touch receptor neurons in C. elegans, Cell 54(1) (1988) 5–16. [DOI] [PubMed] [Google Scholar]
- [116].Duggan A, Ma C, Chalfie M, Regulation of touch receptor differentiation by the Caenorhabditis elegans mec-3 and unc-86 genes, Development 125(20) (1998) 4107–4119. [DOI] [PubMed] [Google Scholar]
- [117].Von Stetina SE, Treinin M, Miller DM 3rd, The motor circuit, Int Rev Neurobiol 69 (2006) 125–67. [DOI] [PubMed] [Google Scholar]
- [118].Kratsios P, Pinan-Lucarre B, Kerk SY, Weinreb A, Bessereau JL, Hobert O, Transcriptional coordination of synaptogenesis and neurotransmitter signaling, Curr Biol 25(10) (2015) 1282–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Prasad B, Karakuzu O, Reed RR, Cameron S, unc-3-dependent repression of specific motor neuron fates in Caenorhabditis elegans, Dev Biol 323(2) (2008) 207–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Prasad BC, Ye B, Zackhary R, Schrader K, Seydoux G, Reed RR, unc-3, a gene required for axonal guidance in Caenorhabditis elegans, encodes a member of the O/E family of transcription factors, Development 125(8) (1998) 1561–8. [DOI] [PubMed] [Google Scholar]
- [121].Eastman C, Horvitz HR, Jin Y, Coordinated transcriptional regulation of the unc-25 glutamic acid decarboxylase and the unc-47 GABA vesicular transporter by the Caenorhabditis elegans UNC-30 homeodomain protein, J Neurosci 19(15) (1999) 6225–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Jin Y, Hoskins R, Horvitz HR, Control of type-D GABAergic neuron differentiation by C. elegans UNC-30 homeodomain protein, Nature 372(6508) (1994) 780–3. [DOI] [PubMed] [Google Scholar]
- [123].Kratsios P, Kerk SY, Catela C, Liang J, Vidal B, Bayer EA, Feng W, De La Cruz ED, Croci L, Consalez GG, Mizumoto K, Hobert O, An intersectional gene regulatory strategy defines subclass diversity of C. elegans motor neurons, Elife 6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Li Y, Osuma A, Correa E, Okebalama MA, Dao P, Gaylord O, Aburas J, Islam P, Brown AE, Kratsios P, Establishment and maintenance of motor neuron identity via temporal modularity in terminal selector function, Elife 9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Feng W, Destain H, Smith JJ, Kratsios P, Maintenance of neurotransmitter identity by Hox proteins through a homeostatic mechanism, Nature Communications 13(1) (2022) 6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Raouf Issa CRAA, A novel post-developmental role of the Hox genes underlies normal adult behaviour, bioRxiv (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Deutsch J, Le Guyader H, The neuronal zootype. An hypothesis, Comptes Rendus de l’Académie des Sciences - Series III - Sciences de la Vie 321(9) (1998) 713–719. [DOI] [PubMed] [Google Scholar]
- [128].Wysocka-Diller JW, Aisemberg GO, Baumgarten M, Levine M, Macagno ER, Characterization of a homologue of bithorax-complex genes in the leech Hirudo medicinalis, Nature 341(6244) (1989) 760–763. [DOI] [PubMed] [Google Scholar]
- [129].Wong V, Aisemberg G, Gan W, Macagno E, The leech homeobox gene Lox4 may determine segmental differentiation of identified neurons, The Journal of Neuroscience 15(8) (1995) 5551–5559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Holland PW, Holland LZ, Williams NA, Holland ND, An amphioxus homeobox gene: sequence conservation, spatial expression during development and insights into vertebrate evolution, Development 116(3) (1992) 653–661. [DOI] [PubMed] [Google Scholar]
- [131].Deutsch JS, Hox genes: studies from the 20th to the 21st century. Preface, Adv Exp Med Biol 689 (2010) ix–xi. [PubMed] [Google Scholar]


