Abstract
Photoacoustic tomography (PAT) is a newly developed medical imaging modality, which combines the advantages of pure optical imaging and ultrasound imaging, owning both high optical contrast and deep penetration depth. Very recently, PAT is studied in human brain imaging. Nevertheless, while ultrasound waves are passing through the human skull tissues, the strong acoustic attenuation and aberration will happen, which causes photoacoustic signals’ distortion. In this work, we use 180 T1 weighted magnetic resonance imaging (MRI) human brain volumes along with the corresponding magnetic resonance angiography (MRA) brain volumes, and segment them to generate the 2D human brain numerical phantoms for PAT. The numerical phantoms contain six kinds of tissues, which are scalp, skull, white matter, gray matter, blood vessel and cerebrospinal fluid. For every numerical phantom, Monte-Carlo based optical simulation is deployed to obtain the photoacoustic initial pressure based on optical properties of human brain. Then, two different k-wave models are used for the skull-involved acoustic simulation, which are fluid media model and viscoelastic media model. The former one only considers longitudinal wave propagation, and the latter model takes shear wave into consideration. Then, the PA sinograms with skull-induced aberration is taken as the input of U-net, and the skull-stripped ones are regarded as the supervision of U-net to train the network. Experimental result shows that the skull’s acoustic aberration can be effectively alleviated after U-net correction, achieving conspicuous improvement in quality of PAT human brain images reconstructed from the corrected PA signals, which can clearly show the cerebral artery distribution inside the human skull.
Keywords: Photoacoustic imaging, Deep learning, Human brain imaging, Skull acoustic aberration
1. Introduction
Cerebral vascular diseases have become one of the most dangerous killers in the world. Especially, stroke has ranked among the top ten pathogenies of death [1]. Effective and timely detection of the cerebral vascular stenosis/broken position or oxygen supply can greatly reduce the mortality of patients. Clinically, there are three major diagnosis methods to detect cerebral vascular disease. They are magnetic resonance angiography (MRA) [2], computed tomography angiography (CTA) [3] and digital subtraction angiography (DSA) [4]. Though the above three methods can reveal the clear vascular structure, they have their own drawbacks and limitations. The biggest drawback of MRA is its low imaging speed, which makes it less useful in diagnosis of acute patients. Although CTA saves much more scanning time, frequent exposing to radiation environment is still a big concern for many people. DSA is the golden standard of cerebral vascular diagnosis in real life. However, it needs high-dose radiography operation to show the perfusion process. The cost of DSA is also much more expensive than the other two, requiring hospitalization for 1–2 days. These limitations intrigue us to find new non-invasive and real-time imaging technologies.
As a hybrid imaging modality, photoacoustic (PA) imaging combines the advantages of both pure optical imaging (high optical contrast) and ultrasound imaging (deep penetration depth) [5]. Benefited from its vascular detection sensitivity and blood oxygen saturation quantification, PA imaging has become a promising non-invasive medical imaging modality in both preclinical and clinical application in recent decades. From now on, PA imaging has shown great potential in breast tumor classification, PA guided needle tip tracking, skin cancer detection and so on.
Considering that PA imaging plays an outstanding role in revealing vasculature, it seems that PA imaging can be competent in the cerebral vascular diagnosis task. Liming Nie et al. proved the feasibility that using a photon recycler can enable light to pass through the whole adult human skulls for the first time [6]. It starts the new era in human brain PA imaging. However, the aberration from wave reflection and refraction caused by skull tissue makes PA image reconstruction much difficult. In transcranial PA imaging, the targeted objects are cerebral vessels. The acoustic wavelength of the PA signals generated by micro vessels are at the same order as the size of skull pore, so the scattering mode is Mie scattering rather than Rayleigh scattering. In this scattering mode, some side-lobe signals are generated. The superimposition of main lobe and side lobes makes the reconstruction process more complicated [7]. In this regard, more works appear to correct aberration and make a big step towards the clinical PA human brain imaging. Chao Huang et al. proposed to combine PAT with the skull composition and morphology from adjunct X-ray CT modality to correct aberration [8]. This work performs well in both phantoms and monkey head skull-induced distortions. Leila Mohammadi et al. proposed a deterministic ray-tracing simulation framework [9]. Their model contains the attenuation and dispersion effects from wave reflection and refraction. They implement their work on the large 3D phantoms and get pretty good results with high computation speed. Shuai Na et al. proposed to take skull heterogeneity into account to build a three-layer human brain model [10]. With wave reflection and refraction coefficients modified, the proposed layered universal back-projection can correct the acoustic aberration. Then they prove the effectiveness on real ex vivo adult human skull with 64-element PAT system. Another way to solve the problem is model-based image reconstruction methods [11]. The wave propagation in heterogeneous and lossy medium is modeled and discretized to form a forward operator. Mitsuhashi et al. proposed a discrete forward operator and the corresponding adjoint operator for transcranial PACT image reconstruction based on elastic wave equations [12]. Poudel et al. proposed an optimization-based image reconstruction method [13]. The forward-adjoint operator pair is used to compute the regularizing penalty term, which is the least square estimates of the initial pressure distribution. However, this study didn’t consider the acoustic heterogeneities within the skull. Poudel et al. further developed the joint reconstruction method that recovers both the initial pressure distribution and the spatial distribution of skull acoustic parameters [14]. With a more accurate estimate of the acoustic properties, the initial pressure distribution with higher quality could be achieved.
All abovementioned works focus on building a mathematical model as accurate as possible to correct the influence caused by skull-distortion. However, to the best of our knowledge, applying data-driven approach to correct the acoustic aberration of human brain is still an untapped area to be explored. In this paper, we propose to build high-quality PA digital brain phantom, and use deep learning network (e.g. U-net [15]) to correct distorted PA signals. We list the contributions of this paper as follows:
-
•
We generate a realistic 2D human brain PA numerical phantom dataset for imaging and vascular disease diagnosis research based on MRA and T1 weighted images in IXI-dataset (https://brain-development.org/ixi-dataset/).
-
•
We propose to use simulated PA sinograms obtained from realistic numerical human brain phantoms to train deep learning model to correct the skull-induced acoustic aberration. With recovered PA signals, the distortion is effectively alleviated in realistic reconstructed PA human brain images.
-
•
We use the generated numerical phantoms to prove the effectiveness of proposed method with good imaging quality.
The remainder of this paper is organized as follow: First, we will introduce some related work in Section 2. Next, details of our work are presented in Section 3. In Section 4, we show the experiment results. Finally, we draw conclusion in Section 5.
2. Related work
2.1. Photoacoustic numerical phantoms
The well-designed PA numerical phantoms are important for performing a simulation study, when lacking of enough experimental data, especially for PA imaging of human. Here we will summarize some PA numerical phantoms generation work and list parts of their database website links.
-
(1)
Human breast numerical phantoms: Lou et al. proposed to extract four parts (vessel, skin, fat and fibro glandular tissue) from contrast-enhanced magnetic resonance images (MRI) modality with threshold segmentation method [16]. But they only generate a few 2D numerical phantoms from 50 patients. Based on this work, Dantuma et al. proposed to add breast tumor tissue from mice additionally [17]. They also extend the 2D breast numerical phantoms to 3D, which makes simulation more realistic. Then Ma et al. proposed to extract four parts of breast (skin, fat, tumor and fibro glandular tissue) from mammography [18]. The first three tissues are manually segmented and the fibro glandular is segmented by deep neural network. This work can quickly generate large amounts of 2D breast numerical phantoms. But, lacking breast vessels greatly limits the usefulness of the dataset. Later, Han et al. proposed to extract four parts (skin, adipose, tumor and fibro glandular tissue) from a series of 2D US B-scan slices with manual segmentation [19]. The reconstructed 3D breast numerical phantoms are generated, but lacking vessel tissue either. After analyzing all the work above, Bao et al. proposed to generate a more realistic breast numerical phantoms including various kinds of breast types [20]. They use a software (VICTRE breast phantom) to generate a comprehensive breast model including skin, nipple, lactiferous duct, terminal duct lobular unit, interlobular gland tissue, fat, suspensory ligament, muscle, artery, and vein.
-
(2)
Human skin numerical phantoms: Lu et al. proposed to build two-layer skin model volumes (epidermis and underlying dermis) with random generation [21]. The structure is relatively simple but useful to optical properties analysis. Based on this, Lyu et al. proposed to extract vessel tissue from 3D lung CT scans with Frangi filter segmentation [22]. Then by embedding the vessel tissue to a three-layer cube (epidermis, dermis and hypodermis), they generate 3890 3D skin tissue numerical phantoms, which is available on the website: https://ieee-dataport.org/documents/3d-skin-tissue-vessel-models-medical-image-analysis.
-
(3)
Human brain numerical phantoms: Firouzi et al. proposed to generate a numerical brain phantom from MRI and MRA [23]. They segmented six kinds of biological tissue (skull, white matter, gray matter, cerebrospinal fluid, edema, tumor) with 3DSlicer software package from MRI and segmented vascular tissue with level-set algorithm from MRA. It is suitable to investigate the human brain tumor diseases instead of cerebral vascular diseases. Later, Na et al. proposed to generate 2D human brain numerical phantoms by embedding a small piece of vessel model to oval skull models, which are both manually designed [10]. These phantoms are relatively simple and discard lots of other brain tissue, like white matter.
2.2. Deep learning-based signal processing
Deep learning has achieved great success in signal processing (e.g. enhancement, denoising and recovery). Gutta et al. proposed to use a standard five layer fully connection neural network to enhance the bandwidth of PA signals [24]. The proposed deep neural network (DNN) can compensate the bandwidth loss caused by ultrasound transducers. With the bandwidth enhanced PA signals, they get the improved reconstructed PA images without significant computation burden. Based on this work, Awasthi et al. proposed to use convolutional neural network (CNN) to enhance the PA signals [25]. Combined with U-net (a well-known CNN structure) and proposed Elu activation function, the method can compensate both PA signal bandwidth and limited-view information loss. The results show that the method works well in PAT system with less root mean square error (RMSE) and higher signal-to-noise ratio (SNR) value. Ben Luijten et al. proposed to use DNN to learn an optimal set of apodization weights, which can adaptively enhance the ultrasound signals [26]. With back projection methods, the proposed deep learning-based adaptive signal processing framework achieves data-efficiency and robustness in ultrasound signal enhancement and image reconstruction. Allman et al. proposed to use CNN to distinguish PA source from reflection artifacts with the sampled PA signals [27]. This work greatly decreases the artifact influence in PA images with high accuracy.
3. Method
In this section, we will make detailed description about the proposed method. It includes five parts: human brain numerical phantoms generation, optical simulation, acoustic simulation, acoustic aberration correction using U-net, and PA image reconstruction. As illustrated in Fig. 1, detailed description of the proposed method will be introduced in following sections.
Fig. 1.
The whole process of proposed method.
3.1. PA human brain numerical phantom generation
We propose to generate the human brain numerical phantoms from other medical image modalities. Considering MRA can provide high contrast with rich vessel information, we choose 3D MRA images with T1 weighted MRI volume in IXI dataset to generate the 2D human brain numerical phantoms.
To better simulate realistic human brain, our PA numerical phantom consists of six parts, which are scalp, skull, vessel, gray matter, white matter and cerebrospinal fluid. However, segmenting human brain volumes without labels is a challenging task. In additional, segmentation via deep learning is barely impossible since the amount of data is limited. Consequently, we choose to combine traditional image processing methods with manual segmentation. In MRA, vessels show higher intensity compared with other tissues caused by contrast agent, which makes vessels can be easily segmented with thresholding or filtering methods. After comparing the segmentation results, we choose 3D Frangi filter to segment vessel part [28]. Because the segmentation process of the 3D volume (150 × 256 × 256) is time consuming, we only choose an upper slice (256 × 256) with the largest brain area to generate 2D numerical phantoms. Thus, we take a proper axial slice of each T1 weighted MRI volumes and use multiplicative intrinsic component optimization (MICO) algorithm [29] for rough segmentation. Simultaneously, some manual adjustments are implemented on the result obtained from MICO to acquire a more precise segmentation of T1 weighted human brain MRI image. Assuming that the PA signal propagation is also affected by scalp according to the tissues’ optical and acoustic properties, we manually add an additional layer of scalp outside the skull. Then we combine the 2D projection of 3D vessel information with other tissue parts to generate the final human brain numerical phantoms. The whole human brain numerical phantom generation process is illustrated in Fig. 2.
Fig. 2.
2D human brain numerical phantom generation process.
Considering light will pass through several layers of different types of tissues from top of the brain, we build pseudo-3D human brain numerical phantoms to make the optical simulation more realistic. Pseudo-3D means the numerical phantom is not generated from the real 3D MRA source. It is spliced with several 2D numerical phantom slices. As shown in Fig. 3, the light will go through the scalp layer (Layer 1: only with scalp), the skull layer (Layer 2: with scalp and skull), the medium layer (Layer 3: with scalp, skull and cerebrospinal fluid) to reach the final layer (Layer 4: complete 2D numerical human brain phantom that involves all 6 kinds of tissues). Layer 4 is the generated human brain numerical phantom illustrated in Fig. 2. Because different tissue has different size, we build the pseudo-3D human brain numerical phantom sizes 256 × 256 × 12 (Two Layer 1 on the top, five Layer 2 under Layer 1, four Layer 3 under Layer 2 and a single Layer 4 at the bottom). Vessel is within the human brain phantom in the layer 4 at the bottom which has 11 layers above it.
Fig. 3.
Four kinds of layers and their positions from the pseudo-3D human brain numerical phantoms.
3.2. Optical simulation
The optical fluence simulation is deployed using the 3D Monte Carlo method [30]. The optical properties of the six kinds of tissues are shown in Table 1 including optical absorption coefficients (μa), reduced scattering coefficients (μs), anisotropy (g) and refractive index (n). To more realistically simulate the variation of tissue’s optical properties among different human individuals, the value of optical absorption coefficients (μa) and reduced scattering coefficients (μs) assigned to tissues of different phantoms varies within a certain range. We use the MATLAB package MCXLAB to simulate the photons’ propagation inside the human brain numerical phantoms illuminated by the pulsed laser.
Table 1.
Optical properties of the tissues in human brain numerical phantom [31].
| μa (mm−1) | μs (mm−1) | g | n | |
|---|---|---|---|---|
| White matter | 0.014 ± 0.002 | 91.00 ± 13.65 | 0.90 | 1.37 |
| Gray matter | 0.036 ± 0.005 | 22.00 ± 3.30 | 0.90 | 1.37 |
| Cerebrospinal fluid | 0.004 ± 0.001 | 2.40 ± 0.36 | 0.90 | 1.33 |
| Scalp | 0.018 ± 0.003 | 19.00 ± 2.85 | 0.90 | 1.37 |
| Skull | 0.016 ± 0.002 | 16.00 ± 2.40 | 0.90 | 1.43 |
| Vessel | 0.238 ± 0.036 | 52.20 ± 7.80 | 0.99 | 1.40 |
Without loss of generality, our optical simulation setup based on 3D Monte Carlo method is as follows: 1) the shape of the light source is set to be planar; 2) the total number of photons to be simulated is 1 billion; 3) the position and incident vector of the source is set properly to make sure that light will cover all the regions of interest; 4) the starting time and the ending time of the simulation is set to 0 and 1 nanoseconds, respectively, and the time-gate width of the simulation is set to 0.01 nanoseconds. According to Table 1, the optical fluence through the six kinds of human brain tissues is computed and obtained given the pulsed laser excitation. In the Monte Carlo optical simulation, the vertical thickness of each layer in the generated pseudo-3D human brain numerical phantoms is 1 mm, which means the total vertical thickness of a single pseudo-3D numerical phantom is 12 mm.
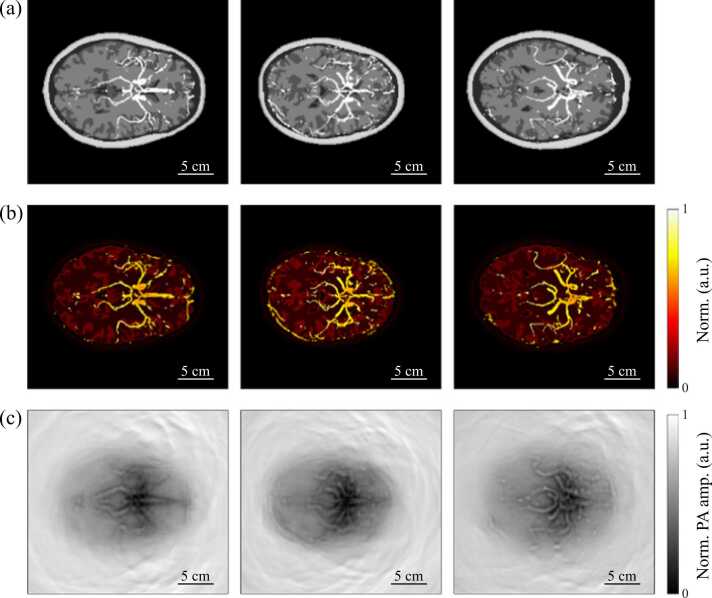
This optical simulation is run on a Linux server with 4 GTX 1080Ti GPUs. For each human brain numerical phantom, the photon propagation simulation process takes about 5 min to obtain an optical fluence volume. Only the optical fluence map of obtained optical fluence volumes’ 12th slice is needed for the acoustic simulation, since our human brain numerical phantoms are built to be pseudo 3D. Through multiplying the obtained optical fluence maps by the corresponding optical absorption coefficients (μa), we can get the initial acoustic pressure distribution which is used for the following acoustic simulation. Fig. 4(b) shows three images of normalized initial pressure distribution of the corresponding 2D numerical phantoms in Fig. 4(a).
Fig. 4.
Three examples of 2D human brain numerical phantoms along with their optical and acoustic simulation results. (a) 2D human brain numerical phantoms generated from different samples. (b) Normalized initial pressure distribution from 3D Monte Carlo optical simulation on phantoms in (a). (c) Normalized DAS Image reconstructions of skull-distorted PA signals obtained from fluid acoustic model using k-wave simulation.
3.3. Acoustic simulation
The acoustic simulation is conducted through the k-Wave toolbox developed by Treeby et al. [32]. Both fluid media acoustic model and viscoelastic media acoustic model are considered in this simulation. It deserves noting that shear waves is involved in viscoelastic acoustic simulation with Kelvin-Voigt model of viscoelasticity [33]. The parameters of k-Wave computational grid is set as follows: 1) the PML size is 17; 2) the number of grid points in the x direction (Nx) and the y direction (Ny) is 256 and 256 respectively; 3) the total grid size in the x direction (x) and y direction (y) is both 256 mm; 4) the grid point spacing in the x direction (dx) and y direction (dy) are both 1 mm.
The maximum and minimum speed of longitudinal acoustic waves (c), as well as the maximum and minimum medium density (d) of the different tissues in human brain numerical phantom, are presented in Table 2. Similar to the optical simulation, we let the speed of sound and density of tissues in different human brain numerical phantoms be the random values, which are between the maximum and minimum values for simulating the inter-individual variation of tissues in acoustic properties. In viscoelastic media acoustic model, speed of shear waves in skull is set to be between 1360 m/s and 1640 m/s [35], compressional and shear acoustic attenuation coefficients of skull are set to be 7.75 dB/(MHz2 cm) and 16.70 dB/(MHz2 cm) [36]. We use point-like ultrasound transducers with infinite bandwidth that minimize the measurement errors in the simulation, which help us to focus on the de-aberration of skull’s acoustic heterogeneity. The 256-elements ring array has a radius of 125 mm, and the brain phantom is placed in the center of the ring array.
Table 2.
Part of acoustic properties of the tissues involved in human brain numerical phantom [34].
| Cmin (m/s) | Cmax (m/s) | dmin (kg/m3) | dmax (kg/m3) | |
|---|---|---|---|---|
| White matter | 1532 | 1573 | 1040 | 1043 |
| Gray matter | 1500 | 1500 | 1039 | 1050 |
| Cerebrospinal fluid | 1502 | 1507 | 1007 | 1007 |
| Scalp | 1537 | 1720 | 1100 | 1125 |
| Skull | 2190 | 3360 | 1800 | 2100 |
| Vessel | 1559 | 1575 | 1056 | 1147 |
With 4 GTX 1080Ti GPUs, it takes about 0.3 s for fluid media acoustic simulation of each PA human brain numerical phantom, while viscoelastic media acoustic simulation will take about 1 min for each phantom. After obtaining the sensor data, we use the delay-and-sum (DAS) algorithm to reconstruct the original image. Fig. 4(c) shows the normalized images reconstructed from acoustic simulation results of fluid media model, in which we could clearly see that the reconstructed image is distorted due to the acoustic aberration of the skull tissue. To acquire the ideal distortion free PA sinograms as supervision term for deep learning-based algorithm, we manually remove the skull and scalp of the numerical human brain phantoms through setting the acoustic properties of these two tissues to be the same as the background, and then repeat the acoustic simulation process with fluid media model. Then, we can obtain the pseudo-ideal sensor data as the reference PA sinograms in which PA signals are free of acoustic aberration caused by skull and scalp. The skull-aberrated signals of two different acoustic simulation models are regarded as the raw data, which are used along with the reference PA sinograms for the following acoustic aberration correction study.
3.4. Acoustic aberration correction using U-Net
With the obtained PA sinogram from the acoustic simulation, we propose to use deep neural networks to correct the skull acoustic aberration, which is illustrated in Fig. 1 (step 5).
First, we have to pre-process the data and build training and testing datasets. With the acoustic simulation, we can get the paired data (PA sinogram with acoustic aberration and PA sinogram without acoustic aberration). To keep the consistency of dimension after each pooling operation, we manually extend the PA sinogram in training set to 256 × 3072 by adding zeros at the end of each channel signal.
In this work, we choose U-net for this task for three reasons. First, the input sinogram is similar in the first half of the data with outputs, and the end of the input sinogram is unwanted reflection aberration signals. So, residual structure is suitable to this work. Interestingly, U-net can be regarded as a big residual structure in some way. Second, the special skip-connection can extract multi-scale features to improve correction outputs. Third, compared with other CNN structures, such as AlexNet, U-net needs less training samples and can process images with less training time and computation cost. U-net structure used in this work is illustrated in Fig. 5.
Fig. 5.
The structure of U-net used in this paper.
3.5. PA image reconstruction
The PA sonograms (with acoustic aberration correction) are used to reconstruct PA images. Because the U-net has eliminated the skull impact, we can regard the sensor data is generated from homogeneous medium. Then we can apply a lot of classical reconstruction algorithms, such as delay-and-sum, time reversal, model-based reconstruction and so on. In order to guarantee the reconstruction speed, we choose DAS as the PA image reconstruction algorithm in this work.
4. Experiment and results
We choose a subset of IXI dataset for experiment that is from Hammersmith Hospital (HH), which are used to generate 180 sets of human brain PA sonograms with size of 256 × 3000. The obtained sonograms are randomly divided into 120 training sets and 60 testing sets. Since the sensors in acoustic simulation is set to be circular and data in each channel of the obtained PA sonograms is from the corresponding sensor, we uniformly separate each extended PA sinogram in the training set into 8 segments with 32 channels each based on sensor positions. Then we sequentially choose 16 channels from each segment and concatenate the chosen data together to get a 128 × 3000 sub-sinogram. By overlapping channels in each segment, we can have 16 combinations of sub-sinogram from an original extended PA sinogram. The data augmentation eventually gives 1920 PA sinograms (128 × 3072) as training set. PA signals obtained from fluid media acoustic model and viscoelastic media acoustic model are trained separately but with the same U-net structure and settings. The iteration is set to 400 epochs and batch size is set to 16. We choose ADAM optimizer and set the learning rate to 0.001. Mean square error is used as the loss in U-net training process. The training process takes 5 min per epoch and is conducted on 4 NVIDIA TITAN RTX GPU with 24 GB of RAM each. Testing process is conducted on a single NVIDIA TITAN RTX GPU and takes about 0.3 s for correcting one PA sinogram.
The normalized fluid media acoustic model’s testing results of three samples shown in Fig. 4(a) are presented in Fig. 6. We could see that the corrected sensor data is much more similar to the reference PA signals. Since we want to make the corrections that are based on the physical features of the PA signals rather than the texture feature of images, the U-net used in this paper is trained with PA sinograms instead of the reconstructed PA images. In order to figure out whether the imaging quality is also improved with U-net correction, we use DAS to reconstruct the image from the PA sonograms presented in Fig. 6. Fig. 7 shows the reconstructed gray-scale human brain PA images with normalization. Compared with the reconstructed images without correction, we can clearly see the main cerebral vessels’ shape and the cerebrovascular distribution from the reconstructed images of the U-net corrected results.
Fig. 6.
Three examples of normalized PA sinogram in acoustic aberration correction process. The top row shows the PA sinograms which are used as supervision during U-net training. The second row are the inputs of U-net that are PA sinograms with acoustic aberration from skull. The U-net corrected PA sonograms are at the bottom row.
Fig. 7.
Corresponding normalized human brain images reconstructed from PA sinograms with delay-and-sum (DAS) algorithms.
To better demonstrate the improvement on signal and image quality, we use structural similarity (SSIM), peak signal-to-noise ratio (PSNR) and mean absolute error (MAE) of the normalized PA sinograms and normalized grayscale DAS reconstructed PA images for quantitative evaluation. As shown in Table 3, three indices are greatly improved in comparison of both PA sinogram and reconstructed image. Furthermore, the largest improvement is on the structural similarity, which means U-net can effectively help identify the anatomical structure of main cerebral vessel without removing the skull.
Table 3.
Quantitative comparison of results from fluid media acoustic model with three different evaluation indices.
| SSIM | PSNR | MAE | |
|---|---|---|---|
| Original (Normalized PA sinogram) |
0.526 | 19.871 | 0.083 |
| U-net corrected (Normalized PA sinogram) |
0.832 | 25.638 | 0.044 |
| Original (Normalized PA image) |
0.718 | 24.099 | 0.052 |
| U-net corrected (Normalized PA image) |
0.939 | 33.424 | 0.012 |
For results of viscoelastic media model, the quantitative evaluation is presented in Table 4. Similar to the results of fluid media model, indices of both PA sinograms and PAT reconstruction images are conspicuously improved after the correction with U-net. The corresponding normalized PA sinograms and normalized PAT images reconstructed with DAS are illustrated in Fig. 8. Five normalized PA signals taken from the first channel of PA sinograms acquired from the rightest object in Fig. 4(a) in different status are shown in Fig. 9, in which we can see that U-net filters out the redundant part of the PA signals and corrects the signal waveform, while the remained PA signals are still slightly distorted compared with the reference PA signal.
Table 4.
Quantitative comparison of results from viscoelastic media acoustic model with three different evaluation indices.
| SSIM | PSNR | MAE | |
|---|---|---|---|
| Original (Normalized PA sinogram) |
0.499 | 21.058 | 0.066 |
| U-net corrected (Normalized PA sinogram) |
0.805 | 25.015 | 0.045 |
| Original (Normalized PA image) |
0.732 | 25.315 | 0.042 |
| U-net corrected (Normalized PA image) |
0.921 | 31.950 | 0.014 |
Fig. 8.
Normalized PA sinograms and normalized DAS reconstructed human brain PAT images from viscoelastic media acoustic model.
Fig. 9.
Normalized PA signals taken from the first channel of the right-column sample shown in Fig. 4(a). (a) Reference PA signal. (b) PA signal with skull aberration obtained from fluid media acoustic model. (c) PA signal with skull aberration obtained from viscoelastic media acoustic model. (d) PA signal in (b) after U-net correction. (e) PA signal in (c) after U-net correction.
5. Discussion and conclusion
Previously reported model-based inversion can iteratively recover the optical absorption distribution by overcoming the acoustic aberration, which however is time-consuming and sometimes suffers convergence issue. Deep learning based method can be quite fast when it is well trained, which is more suitable for real-time imaging applications.
In this paper, we built PA digital brain dataset, and proposed to correct the human skull acoustic aberration through deep learning method for human brain PAT imaging. We designed and made 180 human brain numerical phantoms generated from MRA and T1 weighted MRI modality. Optical and acoustic simulations were performed on our human brain numerical phantoms to simulate the PA signal generation process. Based on pseudo-3D human brain numerical phantoms manually generated from 2D numerical phantoms, 3D Monte Carlo optical simulation was used to acquire the optical fluence maps for calculating the initial pressure distribution used in the acoustic simulation. In the acoustic simulation part, we designed two experiments to simulate the PA signals propagation process in human brain with and without distortion of skull. During the simulation with skull, we use heterogeneous acoustic properties of tissues for obtaining the sensor data with skull aberration. Moreover, both fluid media model and viscoelastic media model are taken into consideration. Then, the augmented PA sinograms with skull aberration were used as input for training the U-net model that is supervised with reference PA data acquired in the skull-free acoustic simulation. The result shows that the deep learning assisted method can effectively improve the quality of reconstructed PAT images and correct the acoustic aberration caused by human skull with and without taking the propagation of shear waves into consideration.
Compared with methods based on physical model, deep learning assisted methods are with high-efficiency. However, there still exist some limitations when facing the clinical situation such as lack of high quality data, poor interpretability and so on. In the future work, we will continue exploring deep learning assisted methods that are used to improve the PAT image quality and optimize the experiment designs to make it better mimic realistic circumstances. In addition, our further works may be also validated by ex vivo experiments using a monkey (Macaca fascicularis)’s skull, and in vivo experiment in monkey and human brain in the long term.
Declaration of Competing Interest
The authors declare no conflicts of interest.
Acknowledgments
This research was funded National Natural Science Foundation of China (61805139), United Imaging Intelligence (2019X0203-501-02), Shanghai Clinical Research and Trial Center (2022A0305-418-02), and Double First-Class Initiative Fund of ShanghaiTech University (2022X0203-904-04). We also want to thank IXI-dataset for providing high quality human brain images (https://brain-development.org/ixi-dataset/). The authors declare there is no conflict of interest.
Biographies

Fan Zhang received his bachelor degree in Computer Science from ShanghaiTech University in 2022. And he is now a graduate student in ShanghaiTech University. His current research is focused on photoacoustic imaging and deep learning.

Jiadong Zhang received the B.S. degree from the Faculty of Electronic Information and Electrical Engineering, Dalian University of Technology, Dalian, China, in 2020. He is currently pursuing the M.S. degree with the School of Biomedical Engineering, ShanghaiTech University, Shanghai, China. His research interests include artificial intelligence and medical image analysis.

Yuting Shen received the B.S. degree in computer science from ShanghaiTech University, Shanghai, China, in 2021, where she is currently pursuing the master’s degree with the School of Information Science and Technology. Her research interests are photoacoustic imaging reconstruction algorithms and image processing and optimization.

Zijian Gao received the B.S. degree in automation from Civil Aviation University of China, Tianjin, China, in 2020. He is currently pursuing the M.S. degree with the School of Information Science and Technology, ShanghaiTech University, Shanghai, China. His research interests are novel photoacoustic imaging system. He proposed palm-photoacoustic system for application of low-cost, low power and small-size photoacoustic and MRI system.

Changchun Yang received his bachelor’s degree in computer science from Huazhong University of Science and Technology in 2018 and his master’s degree in computer science in ShanghaiTech University in 2021. And he is pursuing his Ph.D. in Delft University of Technology. His research interest is interpretable representation learning for medical image analysis, especially for quantitative cardiac MRI.

Mingtao Liang received the B.E. degree from the School of Information Science and Technology, Shanghaitech University, Shanghai, China, in 2022. He is going to pursue the Master of Science in Analytics at Georgia Tech Shenzhen Institute, Shenzhen, China. His research interests include deep learning for photoacoustic imaging.

Feng Gao received his bachelor degree at Xi'an University of Posts and Telecommunications in 2009 and his master degree at XIDIAN University in 2012. From 2012 to 2017, he worked as a Digital Hardware Development Engineer in ZTE Microelectronics Research Institute. From 2017 to 2019, he worked as IC Development Engineer in HISI Semiconductor. During this period, he completed project delivery of multiple media subsystems as IP development director. Various kinds of SOC chips which he participated in R & D have entered into volume production and the corresponding products have been sold well in market. In October 2019, he participated in the Hybrid Imaging System Laboratory of ShanghaiTech University (hislab, website: www.hislab.cn). His research interests are image processing and digital circuit design. In addition, he devotes his efforts to the development of underlying technology of photoacoustic imaging and promote the clinical application of photoacoustic technology.

Li Liu received his Ph.D. degree in Biomedical Engineering from the University of Bern, Switzerland, and then worked as a postdoctoral fellow at the University of Bern and the Chinese University of Hong Kong. He held the position of assistant professor in the School of Biomedical Engineering at Shenzhen University since 2016. In 2019 he joined CUHK as a faculty member. He has served as Program and Publication Chair of many international conferences including Publication Chair of IEEE ICIA 2017, 2018 and Program Chair of ROBIO 2019, Video Chair of IEEE ICRA 2021. He served as Associate Editor of Biomimetic Intelligence and Robotics (BIROB) since 2021. He is a recipient of the Distinguished Doctorate Dissertation Award, Swiss Institute of Computer Assisted Surgery (2016), the MICCAI Student Travel Award (2014) and the Best Paper Award of IEEE ICIA (2009).

Hulin Zhao received Ph.D. degree from the Secondary Military Medical University in 1998, and became an attending physician in 2003, then deputy chief physician in 2014. Now he is working in the Department of Neurosurgery of PLA General Hospital as deputy director of functional neurosurgery department. He is actively engaged in the surgical treatment of epilepsy, Parkinson's disease and pain. He is now the member of the Beijing Anti-epileptic Association, the member of the Neuromicroinvasive Treatment Professional Committee of the Chinese Association of Research Hospitals, and the Neurological Injury Branch of the Medical Health Promotion Association.

Fei Gao received his bachelor degree in Microelectronics from Xi’an JiaoTong University in 2009, and Ph.D. degree in Electrical and Electronic Engineering from Nanyang Technological University, Singapore in 2015. He joined School of Information Science and Technology, ShanghaiTech University as an assistant professor in Jan. 2017, and established Hybrid Imaging System Laboratory (HISLab: www.hislab.cn). During his Ph.D. study, he has received Chinese Government Award for Outstanding Self-Financed Students Abroad (2014). His Ph.D. thesis was selected as Springer Thesis Award 2016. He is currently serving as editorial board member of Photoacoustics. He has published about 150 journal and conference papers with 2400 + citations. His interdisciplinary research topics include photoacoustic (PA) imaging physics (proposed passive PA effect, PA resonance imaging, phase-domain PA sensing, pulsed-CW hybrid nonlinear PA imaging, TRPA-TRUE focusing inside scattering medium, etc.), biomedical circuits and systems (proposed miniaturization methods of laser source and ultrasound sensors, delay-line based DAQ system, hardware acceleration for PA imaging, etc.), algorithm and AI (proposed frameworks such as Ki-GAN, AS-Net, Y-Net, EDA-Net, DR2U-Net, etc,), as well as close collaboration with doctors to address unmet clinical needs (Some prototypes are under clinical trials).
Contributor Information
Li Liu, Email: liliu@cuhk.edu.hk.
Hulin Zhao, Email: zhaohulin_90@sohu.com.
Fei Gao, Email: gaofei@shanghaitech.edu.cn.
Data Availability
Data will be made available on request.
References
- 1.Donkor E.S. Stroke in the century: a snapshot of the burden, epidemiology, and quality of life. Stroke Res. Treat. 2018;2018 doi: 10.1155/2018/3238165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dumoulin C.L., Hart Jr H. Magnetic resonance angiography. Radiology. 1986;161(3):717–720. doi: 10.1148/radiology.161.3.3786721. [DOI] [PubMed] [Google Scholar]
- 3.Achenbach S. Computed tomography coronary angiography. J. Am. Coll. Cardiol. 2006;48(10):1919–1928. doi: 10.1016/j.jacc.2006.08.012. [DOI] [PubMed] [Google Scholar]
- 4.Brody W.R. Digital subtraction angiography. IEEE Trans. Nucl. Sci. 1982;29(3):1176–1180. [Google Scholar]
- 5.Xia J., Yao J., Wang L.V. Photoacoustic tomography: principles and advances. Prog. Electromagn. Res. 2014;147:1–22. doi: 10.2528/pier14032303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nie L., Cai X., Maslov K., Garcia-Uribe A., Anastasio M.A., Wang L.V. Photoacoustic tomography through a whole adult human skull with a photon recycler. J. Biomed. Opt. 2012;17(11) doi: 10.1117/1.JBO.17.11.110506. (110506-110506) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gao Y., Xu W., Chen Y., Xie W., Cheng Q. Deep learning-based photoacoustic imaging of vascular network through thick porous media. IEEE Trans. Med. Imaging. 2022;41(8):2191–2204. doi: 10.1109/TMI.2022.3158474. [DOI] [PubMed] [Google Scholar]
- 8.Huang C., Nie L., Schoonover R.W., Guo Z., Schirra C.O., Anastasio M.A., Wang L.V. Aberration correction for transcranial photoacoustic tomography of primates employing adjunct image data. J. Biomed. Opt. 2012;17(6) doi: 10.1117/1.JBO.17.6.066016. (066016-066016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mohammadi L., Behnam H., Tavakkoli J., Avanaki M.R. Skull’s photoacoustic attenuation and dispersion modeling with deterministic ray-tracing: towards real-time aberration correction. Sensors. 2019;19(2):345. doi: 10.3390/s19020345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Na S., Yuan X., Lin L., Isla J., Garrett D., Wang L.V. Transcranial photoacoustic computed tomography based on a layered back-projection method. Photoacoustics. 2020;20 doi: 10.1016/j.pacs.2020.100213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Na S., Wang L.V. Photoacoustic computed tomography for functional human brain imaging. Biomed. Opt. Express. 2021;12(7):4056–4083. doi: 10.1364/BOE.423707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mitsuhashi K., Poudel J., Matthews T.P., Garcia-Uribe A., Wang L.V., Anastasio M.A. A forward-adjoint operator pair based on the elastic wave equation for use in transcranial photoacoustic computed tomography. SIAM J. Imaging Sci. 2017;10(4):2022–2048. doi: 10.1137/16M1107619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Poudel J., Na S., Wang L.V., Anastasio M.A. Iterative image reconstruction in transcranial photoacoustic tomography based on the elastic wave equation. Phys. Med. Biol. 2020;65(5) doi: 10.1088/1361-6560/ab6b46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Poudel J., Anastasio M.A. Joint reconstruction of initial pressure distribution and spatial distribution of acoustic properties of elastic media with application to transcranial photoacoustic tomography. Inverse Probl. 2020;36(12) [Google Scholar]
- 15.O. Ronneberger, P. Fischer, T. Brox, U-net: convolutional networks for biomedical image segmentation, in: Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015: 18th International Conference, Munich, Germany, October 5–9, 2015, Proceedings, Part III 18, Springer, 2015, pp. 234–41.
- 16.Lou Y., Zhou W., Matthews T.P., Appleton C.M., Anastasio M.A. Generation of anatomically realistic numerical phantoms for photoacoustic and ultrasonic breast imaging. J. Biomed. Opt. 2017;22(4) doi: 10.1117/1.JBO.22.4.041015. (041015-041015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dantuma M., van Dommelen R., Manohar S. Semi-anthropomorphic photoacoustic breast phantom. Biomed. Opt. Express. 2019;10(11):5921–5939. doi: 10.1364/BOE.10.005921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Y. Ma, C. Yang, J. Zhang, Y. Wang, F. Gao, F. Gao, Human breast numerical model generation based on deep learning for photoacoustic imaging, in: Proceedings of the 2020 42nd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), IEEE, 2020, pp. 1919–22. [DOI] [PubMed]
- 19.Han T., Yang M., Yang F., Zhao L., Jiang Y., Li C. A three-dimensional modeling method for quantitative photoacoustic breast imaging with handheld probe. Photoacoustics. 2021;21 doi: 10.1016/j.pacs.2020.100222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bao Y., Deng H., Wang X., Zuo H., Ma C. Development of a digital breast phantom for photoacoustic computed tomography. Biomed. Opt. Express. 2021;12(3):1391–1406. doi: 10.1364/BOE.416406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu J.Q., Hu X.-H., Dong K. Modeling of the rough-interface effect on a converging light beam propagating in a skin tissue phantom. Appl. Opt. 2000;39(31):5890–5897. doi: 10.1364/ao.39.005890. [DOI] [PubMed] [Google Scholar]
- 22.T. Lyu, C. Yang, F. Gao, F. Gao, 3D Photoacoustic simulation of human skin vascular for quantitative image analysis, in: Proceedings of the 2021 IEEE International Ultrasonics Symposium (IUS), IEEE, 2021, pp. 1–3.
- 23.Firouzi K., Saffari N. Numerical modeling of photoacoustic imaging of brain tumors. J. Acoust. Soc. Am. 2011;129(4) (2611-2611) [Google Scholar]
- 24.Gutta S., Kadimesetty V.S., Kalva S.K., Pramanik M., Ganapathy S., Yalavarthy P.K. Deep neural network-based bandwidth enhancement of photoacoustic data. J. Biomed. Opt. 2017;22(11) doi: 10.1117/1.JBO.22.11.116001. (116001-116001) [DOI] [PubMed] [Google Scholar]
- 25.Awasthi N., Jain G., Kalva S.K., Pramanik M., Yalavarthy P.K. Deep neural network-based sinogram super-resolution and bandwidth enhancement for limited-data photoacoustic tomography. IEEE Trans. Ultrason. Ferroelectr. Freq. Control. 2020;67(12):2660–2673. doi: 10.1109/TUFFC.2020.2977210. [DOI] [PubMed] [Google Scholar]
- 26.Luijten B., Cohen R., de Bruijn F.J., Schmeitz H.A., Mischi M., Eldar Y.C., van Sloun R.J. Adaptive ultrasound beamforming using deep learning. IEEE Trans. Med. Imaging. 2020;39(12):3967–3978. doi: 10.1109/TMI.2020.3008537. [DOI] [PubMed] [Google Scholar]
- 27.Allman D., Reiter A., Bell M.A.L. Photoacoustic source detection and reflection artifact removal enabled by deep learning. IEEE Trans. Med. Imaging. 2018;37(6):1464–1477. doi: 10.1109/TMI.2018.2829662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.A.F. Frangi, W.J. Niessen, K.L. Vincken, M.A. Viergever, Multiscale vessel enhancement filtering, In: Proceedings of the Medical Image Computing and Computer-Assisted Intervention—MICCAI’98: First International Conference Cambridge, MA, USA, October 11–13, 1998 Proceedings 1, Springer, 1998, pp. 130–7.
- 29.Li C., Gore J.C., Davatzikos C. Multiplicative intrinsic component optimization (MICO) for MRI bias field estimation and tissue segmentation. Magn. Reson. Imaging. 2014;32(7):913–923. doi: 10.1016/j.mri.2014.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fang Q., Boas D.A. Monte Carlo simulation of photon migration in 3D turbid media accelerated by graphics processing units. Opt. Express. 2009;17(22):20178–20190. doi: 10.1364/OE.17.020178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li T., Gong H., Luo Q. Visualization of light propagation in visible Chinese human head for functional near-infrared spectroscopy. J. Biomed. Opt. 2011;16(4):045001–045001-6. doi: 10.1117/1.3567085. [DOI] [PubMed] [Google Scholar]
- 32.Treeby B.E., Cox B.T. k-Wave: MATLAB toolbox for the simulation and reconstruction of photoacoustic wave fields. J. Biomed. Opt. 2010;15(2):021314–021314-12. doi: 10.1117/1.3360308. [DOI] [PubMed] [Google Scholar]
- 33.B.E. Treeby, J. Jaros, D. Rohrbach, B. Cox, Modelling elastic wave propagation using the k-wave matlab toolbox, in: Proceedings of the 2014 IEEE International Ultrasonics Symposium, IEEE, 2014, pp. 146–9.
- 34.P. Hasgall, F. Di Gennaro, C. Baumgartner, E. Neufeld, B. Lloyd, M. Gosselin, D. Payne, A. Klingenböck, N. Kuster, IT’IS Database for thermal and electromagnetic parameters of biological tissues, Version 4.0, 2018.
- 35.White P.J., Clement G.T., Hynynen K. Longitudinal and shear mode ultrasound propagation in human skull bone. Ultrasound Med. Biol. 2006;32(7):1085–1096. doi: 10.1016/j.ultrasmedbio.2006.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Treeby B.E., Cox B.T. Modeling power law absorption and dispersion in viscoelastic solids using a split-field and the fractional Laplacian. J. Acoust. Soc. Am. 2014;136(4):1499–1510. doi: 10.1121/1.4894790. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.