Abstract
In recent years, lactate has been recognized as an important circulating energy substrate rather than only a dead-end metabolic waste product generated during glucose oxidation at low levels of oxygen. The term “aerobic glycolysis” has been coined to denote increased glucose uptake and lactate production despite normal oxygen levels and functional mitochondria. Hence, in “aerobic glycolysis,” lactate production is a metabolic choice, whereas in “anaerobic glycolysis,” it is a metabolic necessity based on inadequate levels of oxygen. Interestingly, lactate can be taken up by cells and oxidized to pyruvate and thus constitutes a source of pyruvate that is independent of insulin. Here, we show that the transcription factor Foxp1 regulates glucose uptake and lactate production in adipocytes and myocytes. Overexpression of Foxp1 leads to increased glucose uptake and lactate production. In addition, protein levels of several enzymes in the glycolytic pathway are upregulated, such as hexokinase 2, phosphofructokinase, aldolase, and lactate dehydrogenase. Using chromatin immunoprecipitation and real-time quantitative PCR assays, we demonstrate that Foxp1 directly interacts with promoter consensus cis-elements that regulate expression of several of these target genes. Conversely, knockdown of Foxp1 suppresses these enzyme levels and lowers glucose uptake and lactate production. Moreover, mice with a targeted deletion of Foxp1 in muscle display systemic glucose intolerance with decreased muscle glucose uptake. In primary human adipocytes with induced expression of Foxp1, we find increased glycolysis and glycolytic capacity. Our results indicate Foxp1 may play an important role as a regulator of aerobic glycolysis in adipose tissue and muscle.
Keywords: Foxp1, glucose uptake, lactate production, aerobic glycolysis, glucose tolerance test
In healthy nonmalignant cells, glucose is to a large extent converted to pyruvate and further oxidized in the mitochondria. Through a series of chemical reactions in the citric acid cycle and the electron transport chain, ATP will be generated. In malignant or rapidly dividing normal cells, a larger portion of glucose-derived pyruvate is instead reduced to form lactate (1). This takes place even when ample amounts of oxygen are present and is hence called aerobic glycolysis as opposed to anaerobic glycolysis—when lactate is produced out of necessity—when there is not enough oxygen to allow for mitochondrial oxidation of pyruvate, lactate is instead produced.
Interestingly, under physiological conditions, cells regulate how much of the glucose-derived pyruvate that will be further oxidized to acetyl-CoA and enter the TCA cycle and how much will be reduced to form lactate and transported out of the cell. Recent studies indicate that lactate is an important circulating energy substrate, especially during periods of fasting (2). Lactate produced in one cell type and secreted into circulation can be taken up and reduced to pyruvate by another cell type and further oxidized in the mitochondria to ATP. Hence, this process uncouples glycolysis from carbohydrate-driven mitochondrial energy production and allows for fine tuning of energy metabolism. This is an important feature of metabolic regulation in multiorgan organisms since it allows for coordination and synchronization of energy metabolism, using lactate as an intermediate energy donor, that optimizes the use and flow of energy. Only in recent years, this has been fully appreciated since systematic measurements of lactate flux are necessary to unmask this type of metabolic regulation (2, 3). In light of this, there is a renewed interest in investigating how lactate production is regulated.
Another interesting feature of lactate metabolism is for maintaining adequate glucose levels when glucose intake is low (4). During an overnight fasting period, up to 60 to 70% of hepatic gluconeogenesis derives from lactate mainly produced by muscle and adipose tissue (5, 6). Thus, lactate is an important energy substrate during periods of fasting when insulin levels are low, and insulin-dependent glucose uptake cannot supply adequate amounts of pyruvate—instead pyruvate derived from uptake of circulating lactate is used. Also, during insulin resistance in patients with type 2 diabetes, it is conceivable that lactate-derived pyruvate will be used to replace lower levels of glucose-derived pyruvate.
Previously, several genes have been implicated as regulators of glycolysis, in particular during tumorigenesis, for example, KLF14, MYC, and HIF-1 (7, 8). We identified the fasting-induced transcription factor Foxp1 as a transcriptional regulator of glycolysis. Foxp1 is member of the fork head box family of transcription factors that plays an important role in regulating the development of several organs, for example, heart, lung, and brain (9). Moreover, Foxp1 has been found to play important roles in an array of different physiological and pathophysiological processes such as B-cell development (10), repression of pro-apoptotic genes (11), macrophage development (12), and as a tumor suppressor (13). While the exact mode of action remains largely unknown, Foxp1 has also been implicated in a neurodevelopmental disorder known as FOXP1 syndrome (OMIM#605515) (14).
Interestingly, here we demonstrate that Foxp1 upregulates glucose uptake and lactate production in adipocytes and muscle cells, representing two important tissues in metabolic regulation and the pathogenesis of type 2 diabetes. This entails increased glycolysis and glycolytic capacity and increased levels of several enzymes of the glycolytic pathway. Suppression of Foxp1 induces the opposite phenotype. Based on these findings, we suggest that Foxp1 is a significant regulator of aerobic glycolysis in adipose tissue and muscle.
Results
Foxp1 regulates glucose uptake in 3T3-L1 adipocytes and L6 myotubes
To investigate how Foxp1 expression might regulate metabolism in two important peripheral tissues, one that store energy (adipose tissue) and one that expends energy (muscle), which also are relevant for the metabolic perturbations underlying type 2 diabetes, we subjected mice to a 16-h period without food but with free access to water. In such mice, we found that steady state levels of Foxp1 mRNA were upregulated in white adipose tissue (≈25%), interscapular brown adipose tissue (≈50%), and gastrocnemius muscle (≈100%; Fig. 1A). Foxp1 is also upregulated in skeletal muscles during the postexercise period during which myocytes replenish their energy stores but not during the exercise period (Fig. 1B). In line with these results, we investigated Foxp1 as a potential regulator of glucose uptake. 3T3-L1 adipocytes overexpressing FOXP1 display a significant increase in glucose uptake both during basal- and insulin-stimulated conditions (Fig. 1C). Most likely as a consequence of this, de novo triglyceride synthesis is significantly increased in 3T3-L1 adipocytes overexpressing FOXP1 (Fig. 1D). In 3T3-L1 adipocytes with shRNA-mediated knockdown of Foxp1, we demonstrate a significant reduction of glucose uptake (≈30%) during both basal- and insulin-stimulated conditions (Fig. 1E). In response to suppressed levels of Foxp1, there is a significant decrease in insulin-stimulated de novo triglyceride synthesis (Fig. 1F). In the myocyte cell line L6, in which FOXP1 overexpression was placed under the control of doxycycline (DOX) induction, cells increase their glucose uptake in response to DOX administration (Fig. 1G). To further investigate the role of Foxp1 on glucose uptake, we performed Western blots on extracts from 3T3-L1 adipocytes and L6 myotubes and found a robust upregulation of GLUT1 while GLUT4 appeared to be less regulated (Fig. S3, A–D).
Figure 1.
Regulation and metabolism in 3T3-L1 adipocytes and L6 myotubes overexpressing or knocked down for FOXP1.A, levels of Foxp1 mRNA in white adipose tissue (WAT), brown adipose tissue (BAT), and gastrocnemius skeletal muscle (GA) of mice with free access to food or after starvation for 16 h; ad libitum, n = 4; starved WAT, n = 4; starved BAT and GA, n = 4. Values calculated as fold differences compared to expression level in WAT at fed state. B, levels of Foxp1 mRNA in skeletal muscle (gastrocnemius), during and after exercise, n = 5. C and D, 3T3L1 adipocytes overexpressing FOXP1 and treated with or without insulin: C, glucose uptake, n = 4; D, triglyceride synthesis, n = 3. E and F, 3T3-L1 adipocytes with knockdown of Foxp1 using shRNAs targeting Foxp1 or empty vector and scrambled shRNA as controls, treated with or without insulin: E, glucose uptake, n = 4 per group; F, triglyceride synthesis, n = 4. G, glucose uptake in L6-differentiating myotubes at day 9 of differentiation under basal conditions. Expression of FOXP1 was induced by adding doxycycline at days −5, 0, 4, 7, and 8 after start of differentiation, n = 4. Day 0 denotes start of differentiation. All cells were harvested for analysis at day 9. Experiments replicated at least three times except for A and B (once). Representative experiments are shown. Data shown as mean ± s.d., n shows biological independent experiments. Unpaired two-sided Student’s t test was carried out; ∗∗∗p < 0.001, ∗∗p < 0.01, ∗p < 0.05.
FOXP1 induces aerobic glycolysis and lactate production
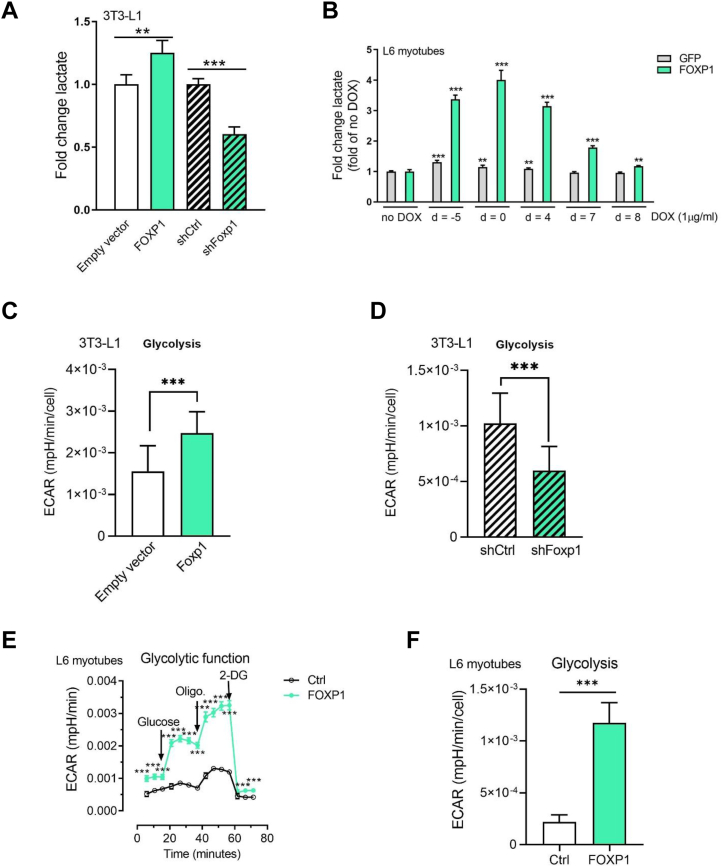
To investigate whether Foxp1-stimulated glucose uptake led to increased lactate production, we measured lactate secretion into the cell culture medium. As can be deduced from Figure 2A, there is a significant increase in lactate production in 3T3-L1 adipocytes overexpressing FOXP1, while suppression of Foxp1 leads to significantly lower lactate production. Using a DOX-dependent overexpression of FOXP1 in L6 myotubes, we demonstrated a significant increase in lactate production (Fig. 2B), which correlated with the length of FOXP1 overexpression and was most pronounced at induction time points around the day at which the cells reach confluence (d = 0). With the exception of d = −5, this agrees well with the glucose uptake data (Fig. 1G). We also confirmed the FOXP1 expression of the induced FOXP1 overexpression and knockdown in 3T3-L1 adipocytes using this DOX-inducible expression system (Fig. S1C).
Figure 2.
FOXP1 induces aerobic glycolysis and lactate production.A, secretion of lactate into medium by 3T3-L1 adipocytes overexpressing or knocked down for FOXP1, n = 4. B, secretion of lactate into medium by L6 myotubes with doxycycline-induced overexpression of FOXP1, n = 4. Expression of FOXP1 was induced by adding doxycycline at days −5, 0, 4, 7, and 8 after start of differentiation. Day 0 is denoted as start of differentiation. All cells were harvested for analysis at day 9. C and D, ECAR (glycolysis) in 3T3-L1 adipocytes overexpressing (C) or knocked down for (D) FOXP1. Empty vector, n = 31; FOXP1, n = 22; shCtrl, n = 24; shFoxp1 n = 24. E–H, glycolytic analysis of L6 myotubes with doxycycline-induced overexpression of FOXP1. E, kinetic ECAR response to glucose (10 mM), oligomycin (1 μM), and 2-DG (100 mM), (F) ECAR (glycolysis), (G) ECAR (glycolytic capacity), (H) ECAR (glycolytic reserve); E–H, n = 12. I, glycerol release in 3T3-L1 adipocytes with increased expression of FOXP1 with or without 250 μM cAMP, n = 4. J, glycerol release in 3T3-L1 adipocytes with knockdown of Foxp1 compared with shGFP, with or without 250 μM cAMP; n = 4. Experiments replicated at least three times. A representative experiment is shown. Data shown as mean ± s.d., n shows number of biological independent experiments. Unpaired two-sided Student’s t test was carried out, ∗∗∗p < 0.001; ∗∗p < 0.01. 2-DG, 2-deoxy-D-glucose; ECAR, extracellular acidification rate.
In a next step, we used a Seahorse system to study glycolysis in 3T3-L1 adipocytes. Glycolysis, measured as increase in extracellular acidification rate (ECAR) after addition of glucose, is increased in cells with both constitutive and DOX-inducible–overexpressing FOXP1 (Figs. 2C and S1A)) and decreased in cells with suppressed levels of Foxp1 (Figs. 2D and S1B). In L6 myotubes, we measured the following: glycolysis (Fig. 2, E and F); glycolytic capacity, the total drop in ECAR in response to addition of the glycolysis inhibitor 2-deoxy-D-glucose (2-DG; Fig. 2, E and G); and glycolytic reserve, the increase in ECAR seen after addiction of the synthetic mitochondrial uncoupler oligomycin (Oligo.; Fig. 2, E and H). Glycolysis, glycolytic capacity, and glycolytic reserve are all significantly increased in L6 cells overexpressing FOXP1 (Fig. 2, E and H). For both 3T3-L1 adipocytes and L6 myotubes, we demonstrate a robust and clear increase in glycolysis, approximately a 30% increase for adipocytes and a more than 3-fold increase for myotubes, which supports the previous findings of increased lactate production and strongly suggests that Foxp1 is a potent regulator of glycolysis in these cell types.
Interestingly, during both basal conditions and during cAMP stimulation, glycerol release was significantly increased in response to overexpression of FOXP1 and significantly reduced in response to the lowering of Foxp1 levels (Fig. 2, I and J).
Foxp1 regulates glycolytic gene products
To investigate protein levels of important enzymes in the glycolytic pathway, we used protein extracts form 3T3-L1 adipocytes with overexpression or knockdown of Foxp1. As can be deduced from Figure 3 (A–E), hexokinase II (HK2), phosphofructokinase (PFKM), pyruvate kinase m2 isoform (PKM-M2), aldolase (ALDOA), and lactate dehydrogenase A (LDHA) are all significantly regulated by Foxp1, apart from HK2 (p = 0.054) and pyruvate kinase m2 isoform (p = 0.056) levels during overexpression of FOXP1. While overexpression of FOXP1 induces expression of these glycolytic gene products, knockdown has the opposite effect. This establishes Foxp1 as a regulator of glycolytic gene products—a finding that is congruent with the results depicted in Figure 2. In a similar manner, Foxp1 also regulates glutamate dehydrogenase 1 that catalyzes the oxidative deamination of glutamate to alpha-ketoglutarate (Fig. 3F). This is interesting since the mitochondrial glutamine/glutamate pathway supports mitochondrial oxidative metabolism when pyruvate derived from glucose is used for lactate production (15). We also find regulation of pyruvate dehydrogenase kinase 1 and pyruvate dehydrogenase kinase 4 (PDK4; Fig. 3, G and H) which are both kinases that regulate pyruvate dehydrogenase and increased levels of these enzymes will promote lactate production (16). We also performed chromatin immunoprecipitation (ChIP) and real-time PCR assays to analyze if these regulated genes are direct Foxp1 targets. The results (Fig. S4) demonstrate that Foxp1 directly interacts with promoter consensus cis-elements that regulate the expression of several of the target genes, for example, Hk2, pyruvate kinase (Pkm), Aldoa, and Ldha. Hence, Foxp1 directly regulates several enzymes of importance for glycolysis and cellular adaptation to glycolytic lactate production (Fig. 3I).
Figure 3.
FOXP1 regulates genes in glycolytic pathway.A–I, quantification of Western blot experiments in 3T3-L1 adipocytes overexpressing or knocked down for FOXP1 after normalization to actin levels. A, HK2, n = 4. B, PFKM, n = 4. C, PKM-M2, n = 3 for FOXP1 and empty vector, n = 4 for shFoxp1 and shCtrl. D, ALDOA, n = 3. E, LDHA, n = 3 for FOXP1 and empty vector, n = 4 for shFoxp1 and shCtrl. F, GLUD1, n = 3 for FOXP1 and empty vector, n = 4 for shFoxp1 and shCtrl. G, PDK1, n = 3 for FOXP1 and empty vector, n = 4 for shFoxp1 and shCtrl. H, PDK4, n = 4. I, schematic view of Foxp1 regulation sites in the glycolytic pathway. Three or four independent experiments were performed, with number of replicates are indicated above. Fold changes compared to the signal observed in the control cells (infected with empty vector or shCtrl) shown as mean ± s.d., n shows biological independent experiments. Significance of overexpression or knockdown versus control determined by one sample t test with hypothetical value as 1; ∗∗p < 0.01, ∗p< 0.05. ALDOA, aldolase; GLUD1, glutamate dehydrogenase 1; HK2, hexokinase; LDHA, lactate dehydrogenase A; PDK1, pyruvate dehydrogenase kinase 1; PDK4, pyruvate dehydrogenase kinase 4; PFKM, phosphofructokinase; PKM-M2, pyruvate kinase m2 isoform.
Regulation of Foxp1 at the transcriptome level
We next performed transcriptome analysis of 3T3-L1 adipocytes following overexpression and knockdown of Foxp1. There are 3536 and 3458 genes deregulated upon Foxp1 overexpression and knockdown, respectively (Fig. 4A). The differentially expressed genes (DEGs) are provided in Table S1. We also found that the glycolytic pathway genes (Pdk4, Hk2, Aldoa, and Slc2a4) are upregulated during FOXP1 overexpression, while their expression levels are reduced upon knockdown of Foxp1. Thus, the transcriptome data further confirm the role of Foxp1 in the regulation of glycolytic pathway genes (Fig. 4A). The majority of the deregulated genes in the Foxp1 overexpression experiment have an opposite pattern of regulation upon Foxp1 knockdown (Fig. 4, B and C). Out of 1350 genes commonly deregulated between Foxp1 overexpression and knockdown, 884 genes were showing an opposite pattern of regulation in knockdown and overexpression confirming that these genes are direct targets of Foxp1 (Fig. 4, B and C). From gene enrichment analysis, we found that pathways related to glycolysis, insulin signaling, triglyceride, and pyruvate metabolism are affected by commonly deregulated genes between Foxp1 overexpression and knockdown (Fig. 4D). The full list of functional analysis summary is shown in Table S2. This is consistent with our experimental data on Foxp1’s role in glycolytic pathway regulation and further confirms the role of Foxp1 in the regulation of glycolytic pathway genes.
Figure 4.
Regulation of Foxp1 at the transcriptome levels.A, volcano plots showing differentially expressed genes upon knockdown (KD) and overexpression (OE) of Foxp1. B, Venn diagram with overlapping differentially expressed genes (upregulated and downregulated) from Foxp1 knockdown (KD) and OE. C, expression status of overlapped differentially expressed gene from Foxp1 knockdown (control, n = 3; KD, n = 3) and overexpression (control, n = 3; OE, n = 3). D, pathways (KEGG) and Gene Ontology terms (GO_BP: Biological process; GO_CC: Cellular component; GO_MF: Molecular functions) enriched with commonly deregulated genes (n = 1350) from Foxp1 knockdown and overexpression. The numbers above individual bars represent percentage of genes from KEGG or gene ontology database matches with commonly deregulated genes (n = 1350).
Mice with reduced Foxp1 expression in muscle are glucose intolerant
We developed mice with a loxed Foxp1 allele and bred them with mice expressing cre under the myosin creatinine phosphokinase (MCK) promoter (Foxp1ΔMuscle). These mice have approximately a 50% reduction of Foxp1 mRNA and protein steady-state levels (Fig. 5, B and D). But when we isolated primary myoblasts from the mice and differentiated them to myocytes, then we can see there is a 90% reduction of Foxp1 protein expression in the cells from Foxp1ΔMuscle animals compared to control (Fig. 5C). Furthermore, expression of cre-recombinase in mouse embryonic fibroblast (MEF) adipocytes derived from mice lacking one (Foxp1fl/+-cre) or both Foxp1 alleles (Foxp1fl/fl-cre) leads to a dose-response reduction in Foxp1 expression, in magnitude, similar to the reduction seen in myocytes from Foxp1ΔMuscle animals (Fig. S2, A and B). When given an intra peritoneal glucose load, Foxp1ΔMuscle animals display significantly elevated blood glucose levels at 15, 30, 60, and 120 min after injection (Fig. 5A). Furthermore, when measuring glucose uptake in individual tissues of such mice, we found a significant reduction in glucose uptake for both the soleus and gastrocnemius muscle, while other tissues displayed no difference in glucose uptake (Fig. 5G). We also found significant reductions in the expression of Pdk4 (Fig. 5F) in the Foxp1ΔMuscle animals. These findings agree well with reduced glucose uptake in 3T3-L1 adipocytes when Foxp1 levels are reduced (Fig. 1E). There is no significant difference in body weight (Fig. 5E).
Figure 5.
Regulation of aerobic glycolysis by FOXP1 in vivo and in primary human cells.A, glucose tolerance test, n = 10 for WT, n = 15 for Foxp1ΔMUSCLE. B, expression of Foxp1 in soleus and extensor digitorium longus (EDL) muscle, n = 3 mice for each genotype. C and D, quantification of Western blot experiments in primary myocytes and skeletal muscle for FOXP1 level after normalization to actin, n = 4 to 5 for WT, n = 3 to 4 for Foxp1ΔMUSCLE. E, weight curves of the WT and Foxp1ΔMUSCLE mice. n = 7 for WT, n = 5 for Foxp1ΔMUSCLE. F, Pdk4 gene expression in the WT and Foxp1ΔMUSCLE mice. n = 3 mice for each genotype. G, glucose uptake; n = 4 mice for each group. H, histology of skeletal muscle. ATPase staining (left) or immunohistochemistry (IHC, right) for type 1 myosin; n = 3 mice for each genotype. I and J, bioenergetics analysis of human adipocytes from two donors. I, ECAR (glycolysis): Donor X GFP, n = 22 and FOXP1, n = 18; Donor Y GFP, n = 17 and FOXP1, n = 17. J, ECAR (glycolytic capacity): Donor X GFP, n = 22 and FOXP1, n = 18; Donor Y GFP, n = 17 and FOXP1, n = 17. Each graph shows mean ± s.d.; n shows biological independent experiments. Unpaired two-sided Student’s t test was carried out, ∗∗∗p < 0.001, ∗∗p < 0.01, ∗p< 0.05. ECAR, extracellular acidification rate; PDK4, pyruvate dehydrogenase kinase 4.
The ATPase staining and myosin immunohistochemistry patterns showed that there is no difference between the Foxp1ΔMuscle and the WT mice and that the muscle fibers appear normal in Foxp1ΔMuscle animals (Fig. 5H). To study Foxp1 as a regulator of glucose uptake in primary cells, we also used embryonic fibroblasts from mice with loxed Foxp1 alleles and control mice for ex vivo culturing and differentiation to adipocytes. When the DNA-binding domain of Foxp1, the so called fork head domain, was loxed out in response to overexpression of cre-recombinase, we observed a decreased glucose uptake which paralleled relative Foxp1 expression (Fig. S2, A and B).
To validate FOXP1 as a regulator of glycolysis in human primary adipocytes, we show that overexpression of FOXP1 in such cells increases both glycolysis and glycolytic capacity (Fig. 5, I and J).
Discussion
Lactate has been regarded as a metabolic end product typically generated when there is not enough oxygenation to allow for full mitochondrial oxidation, for example, during physical exercise, instead cytosolic glycolysis will reduce pyruvate to form lactate. Interestingly, it has been known for many years that lactate can also be taken up and oxidized by tissues such as the myocardium skeletal muscle (17, 18). Recently, there has been renewed interest in circulating lactate as an important source of energy. When systematically investigating lactate flux, using 13C-labeled nutrients, Hui et al.(2) discovered that 13C-lactate extensively labeled TCA cycle intermediates in all tissues examined. This is particularly important during fasting when in all tissues, except for the brain, the contribution of glucose to tissue TCA metabolism is primarily indirect via circulating lactate (2). This important finding, revealed by applying flux measurements, highlights the importance of lactate as a previously not fully recognized energy substrate.
Peripheral tissues such as adipose tissue and skeletal muscle have a high rate of lactate production, especially during shorter periods of fasting when it is a dominating substrate for liver and kidney gluconeogenesis (6, 19). Interestingly, lactate is taken up and oxidized to pyruvate and further metabolized to glucose essentially by reversing many of the reactions found in glycolysis. Thus, lactate constitutes an insulin-independent source of pyruvate that cells can utilize when glucose levels are low.
Here, we show that Foxp1 is induced during a shorter period of fasting/starvation (mice were without food for 16 h) and postexercise (Fig. 1, A and B). In cultured adipocytes and myotubes, we can show that increased expression of FOXP1 will increase glucose uptake (Fig. 1, C and G) and lactate production (Fig. 2, A and B). Moreover, we find that this is most likely derived from increased protein levels of several glycolytic enzymes (Fig. 3) together with an induced rate of glycolysis (Fig. 2, C–H). Notably, in 3T3-L1 adipocytes, increased FOXP1 expression upregulates glycerol release during both basal- and cAMP-stimulated conditions, and glycerol release is decreased when Foxp1 levels are lowered (Fig. 2, I and J). We would like to speculate that this reflects a need for fatty acids to provide the TCA cycle with substrate under conditions when pyruvate is reduced to form lactate instead of entering the mitochondria.
Another interesting finding is that the pathway analysis (Fig. 4D) identifies collagen-containing extracellular matrix pathways as the second and fourth most regulated set of pathways. This is an interesting observation in the light of recent findings that glycosaminoglycan synthesis, e.g., hyaluronan synthesis, is to a great extent depend on nutrients availability (20). Moreover, extracellular matrix remodeling has been shown to correlate with the regulation of glycolysis in cancer and embryogenesis (21).
We have previously shown that the transcription factors Foxk1 and Foxk2, members of the same family as Foxp1—the fork head family of transcription factors—also regulate glycolysis and lactate production (22). Based on these findings, we would like to suggest that transcriptional regulation of genes encoding glycolytic enzymes may play a previously not fully recognized role in regulating energy metabolism. Furthermore, when glucose levels are low, or cells/tissues are highly insulin resistant, circulating lactate can be utilized as an insulin-independent source of pyruvate for TCA cycle–dependent oxidation and subsequent ATP production. It is tempting to speculate that particularly during insulin resistance an alternative source, to that of glucose, for pyruvate production, might be especially important in cells with diminishing glucose uptake so that they can maintain proper energy production.
Experimental procedures
No statistical methods were used to predetermine sample size. The experiments were not randomized, and the investigators were not blinded to allocation during experiments and outcome assessment.
Reagents
Isobutylmethylxanthine, troglitazone, puromycin, dexamethasone, cytochalasin B, bovine insulin, Matrigel Basement Membrane Matrix, bromoadenosine 3′,5′-cyclic monophosphate sodium salt, Free Glycerol Determination Kit, TRI reagent, polybrene (hexadimethrine bromide), 2-DG, FCCP, oligomycin, bovine serum albumin (BSA) free fatty acid, palmitate, and cAMP were obtained from Sigma-Aldrich (Merck). DOX was obtained from Clontech Laboratories (Takara). Dulbecco’s modified Eagle’s medium (DMEM), F-10 Nutrient Mixture, minimum essential medium (MEM), heat-inactivated fetal bovine serum (FBS), geneticin (G418), penicillin/streptomycin solution, Lipofectamin 2000, NuPAGE Novex Bis-Tris gels, LDS sample buffer, NuPage MOPS SDS running buffer, and NuPage transfer buffers were obtained from Invitrogen (Thermo Fisher Scientific Inc). Polyvinylidene difluoride membranes were obtained from Amersham Biosciences. RNeasy mini kit was obtained from Qiagen. PhosSTOP phosphatase inhibitor cocktail, complete protease inhibitor cocktail, first-strand cDNA synthesis kit, and collagenase A and collagenase II were obtained from Roche Life Science. SuperSignal West Pico, SuperSignal West Dura chemiluminescent substrate, Magnetic ChIP Kit, and BCA protein assay kit were obtained from Pierce (Thermo Fisher Scientific). Radiochemicals, 2-[1,2-3H(N)]-deoxy-D-glucose, and d-[14C(U)]-glucose were obtained from PerkinElmer. EnzyChrom L-lactate assay kit (ECLC-1000) was from BioAssay Systems.
Antibodies
Antibodies against GLUT-1 (07-1401) and GLUT-4 (07-1404) were obtained from Millipore. Antibodies against PKM2 (4053), HK2 (2867), LDHA (2012), FOXP1 (4402), anti-mouse IgG-HRP (7076), and anti-rabbit IgG-HRP (7074) were obtained from Cell Signaling Technology. Antibodies against pyruvate dehydrogenase kinase 4 (PDK4) (ab214938), pyruvate dehydrogenase kinase 1 (PDK1) (ab202468), aldolase (ALDOA) (ab169544), phosphofructokinase (PFKM) (ab154804), glutamate dehydrogenase 1 (GLUD1) (ab166618), and β-ACTIN-HRP (ab8226) were from Abcam.
Antibodies against myosin (skeletal, slow type I) (M8421), FLAG–M2-HRP (A8592), and FLAG (F7425) were obtained from Sigma-Aldrich. Vector M.O.M. Immunodetection Kit (PK-2200) was obtained from Vector Laboratories.
Plasmids
Full-length human FOXP1 isoform α protein (NM_001244814.2) with C-terminal triple Flag-tag (3×FLAG) was created in pcDNA3.1/Hygro(+) vector and subcloned into retroviral pBabe-puro, pRetroX-TRE3G, and lentiviral pLVX-puro vectors for constitutive or DOX-inducible expression of proteins. For stable knockdown of gene shRNA to rodent, Foxp1 was cloned into a retroviral pRS (OriGene) and lentiviral Tet-pLKO-puro (Addgene (#21915)) vectors. Targeting sequence was as follows: shFoxp1-1, GGTAACCCTTCCCTTATTAAA. Empty vector and noneffective scrambled shRNA (shCtrl; Origine, TR30003) were used as controls. Designed shRNA resulted in the reduction of endogenous level of Foxp1 mRNA to approximately 30% of WT levels.
Cell culture
293T, 3T3-L1, and L6 cells were obtained from American Type Culture Collection (ATCC). All mammalian cell cultures were incubated at 37 °C with 5% CO2 in medium containing 10% FBS in the presence of penicillin and streptomycin. 293T, 3T3-L1 cells, and MEFs were grown in high-glucose DMEM. L6 cells were maintained in MEM. MEFs from WT, Foxp1fl/+, and Foxp1fl/fl mice were isolated, cultured, and differentiated into adipocytes as described previously (23). Lipofectamin 2000 (LifeTechnologies) was used for mammalian cell transfections according to the manufacturer’s instructions. MEFs were infected with pHR-MMPCreGFP (PlasmID Repository at Harvard Medical School) as previously described (24). 3T3-L1 and L6 cells (ATCC) were maintained and differentiated as described (25, 26). In brief, for differentiation into adipocytes, 2 days post confluent, 3T3-L1 cells (day 0) were stimulated with differentiation medium (DMEM, 10% FBS, 500 μM 3-isobutyl-1-methylxanthine, 1 μM dexamethasone, 5 μg/ml insulin, 1 μM troglitazone). New differentiation medium was added on day 2. Differentiation medium was exchanged to only insulin-containing medium at day 4 to accomplish full differentiation (day 6). Cells were kept in normal growth medium from day 6 and medium was changed every other day. Myogenic differentiation of L6 cells was initiated by culturing cells in medium containing 2% FBS. 3T3-L1 or MEF-derived adipocytes at days 10 to 12 of differentiation and L6 myotubes at day 9 of differentiation were used in the experiments. Before all metabolic experiments, L6 myotubes were washed from serum and incubated overnight in MEM medium. Tet-On 3G Systems (Clontech Laboratories) or TET-pLKO (Addgene) were used to investigate the effect of modulation of FOXP1 expression in fully differentiated adipocytes or myoblasts. All cells in experiments with DOX were plated simultaneously. Day = denotes start of differentiation. Control conditions (“noDOX”) denote cells that were treated just like the other cells, but they were never exposed to DOX. Cells were differentiated as described above, and expression of FOXP1 or shRNA to Foxp1 was induced at different days of differentiation by addition of DOX (1 μg/ml) to the growth medium at indicated time points. DOX was then kept in the medium until the end of the experiment. The minimum time of DOX induction before all metabolic assays was 4 days for 3T3-L1 adipocytes and one day for L6 myotubes. Metabolic measurements and samples collection were performed concurrently in all cells at day 9 of differentiation for myoblasts and day 11 for 3T3-L1 adipocytes.
Human primary adipocytes
Primary human adipocytes used in bioenergetics experiments were collected from patients undergoing elective surgery at Sahlgrenska University Hospital in Gothenburg. All study subjects received oral and written information and gave written informed consent for the use of the tissue. The studies were approved by The Regional Ethical Review Board in Gothenburg. All procedures performed in studies involving human participants were in accordance with the ethical standards of the national and institutional research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. All subjects complied with ethical regulations. For adipocyte differentiation, 90% confluent cells were treated with DMEM/F12 with 3% FBS (PAA, Gold) supplemented with 500 μM 3-isobutyl-1-methylxanthine, 500 nM dexamethasone, 0.10 μM insulin, and 1 μM pioglitazone. Media were changed every other day until cells were fully differentiated (day 7). At this point, lentiviral transduction with vectors carrying FOXP1–3×Flag or control EGFP was performed. Cells were kept in maintenance medium (DMEM/F12, 3% FBS, 100 mM dexamethasone, 200 nM insulin) for an extra 7 days. After differentiation was completed, cells were cultured in growth medium (PromoCell, C-27437 or Lonza, CC-3245) for 1 day (day 1 postdifferentiation) before Seahorse analysis (Seahorse Bioscience).
Bioenergetic analysis
ECAR were measured using XF96 Analyzer (Seahorse Bioscience). All media and compounds were prepared according to the manufacturers’ instructions unless indicated otherwise. A base medium was used for the assays described in this study directly or supplemented with substrates and cofactors as previously described (27).
In vitro metabolic assays
Glucose uptake in 3T3-L1– and MEF-derived adipocytes were measured by the analysis of 2-[1,2-3H(N)]-deoxy-D-glucose (0.5 μCi/well) uptake on day 11 of postdifferentiation as previously described (28). In brief, fully differentiated cells were washed and serum-starved for 2 h in Krebs-Ringer Phosphate buffer supplemented with 0.2% BSA followed by 15 min incubation with or without 100 nM insulin. For glucose uptake, 2-deoxy-d-glucose [1,2-3H(N)] (0.5 μCi/well) and 50 nM cold 2-DG were added to the cells for 10 min. Cells were washed three times with cold PBS and harvested in PBS supplemented with 1% Triton-X-100. Radioactivity was determined in a liquid scintillation counter (Beckman LS 7000 Coulter). Passive diffusion of glucose was analyzed in cells treated with 25 μM cytochalasin B and subtracted from values for glucose uptake. The values were normalized to the total cellular protein level. Each experiment was performed in triplicate or quadruplicate.
De novo lipogenesis was assessed in the cells by analysis of D-[14C(U)]-glucose incorporation into lipids as described previously (29). Assays were performed in Krebs-Ringer Phosphate buffer containing 5 mM glucose and 0.2% BSA in the presence of 1 μCi/well of D-[14C(U)]-glucose. The values were normalized to the total cellular protein level.
Lipolysis was measured in 3T3-L1 adipocytes on day 10 of differentiation. Cells were washed with PBS and incubated in DMEM medium containing 2% BSA with or without 250 μM cAMP. The supernatant was collected after 2 h of incubation at 37 °C, and glycerol content was analyzed using a colorimetric assay (Free Glycerol Determination Kit) by measuring absorbance at 570 nm on a TEXAS plate reader. Glycerol values were normalized to the total cellular protein level.
Lactate release into medium was measured using an EnzyChrom L-lactate assay kit (ECLC-1000) according to the manufacturer’s protocol. 3T3-L1 adipocytes and L6 myotubes on day 10 or day 8 of differentiation were washed once and incubated for an additional 24 h in corresponding fresh growth medium. Medium was collected on ice, centrifuged for 10 min at 2500 r.p.m. at 4 °C, and stored at −80 °C before analysis. The values were normalized to the total cellular protein level.
RNA and protein analysis
For western blots, cells and animal tissues were lysed in RIPA buffer (50 mM Tris–HCl, pH 8.0, 1% Triton X-100, 0.1% SDS, 0.5% sodium deoxycholate, 150 mM NaCl, 1 mM EDTA), supplemented with protease and phosphatase inhibitor cocktails. Protein concentrations were analyzed using the BCA protein assay kit. Proteins were separated by SDS–PAGE on NuPAGE Novex 4–12% Bis-Tris protein gels and transferred to polyvinylidene difluoride membranes. Specific proteins were detected with the indicated primary and horseradish peroxidase–coupled secondary antibody. Signal was visualized with SuperSignal West Dura or SuperSignal West Pico chemiluminescent substrates on a LAS-4000 Luminescent Image Analyzer Ver 2.0 (FujiFilm). The density of the bands was quantified using Multi Gauge V 3.1 (FujiFilm) software, and the results were expressed as fold changes compared to the signal observed in the control cells (infected with empty vector or shCtrl) after normalization to β-actin.
Total RNA from cells and animal tissues was extracted using TRI or RLT reagents and isolated using RNeasy mini kit according to the manufacturer’s instructions. DNAse treatment on columns was performed according to the manufacturer’s protocol. Reverse transcription of 1 μg of total RNA was carried out using the First-Strand cDNA Synthesis Kit. The expression levels of specific mRNAs were quantified by real-time PCR using Power SYBR green PCR Master Mix on an ABI Prism 7900 Sequence Detection System (Applied Biosystems, Thermo Fisher Scientific) and normalized to the level of 36b4 (also known as Rplp0). All samples were analyzed in triplicate or quadruplicate, and mean values were calculated. The sequence for primers used was as follows: Foxp1, forward: TGGTTCACACGAATGTTTGC, reverse: GTGGCTGCTCTGCATGTTT; 36b4, forward, GCTCAGAACACTGGTCTAGG, reverse, GCACATCACTCAGAATTTCAA.
Animals
All animal procedures were approved by the Ethical Committee for Animal Experiments at the University of Gothenburg, which adheres to the principles and guidelines established by the European Convention for the Protection of Laboratory Animals. C57BL/6 mice used for breeding, in starvation, and exercise experiments were obtained from Charles River Laboratories.
Generation of Foxp1fl/fl mice
The mouse genomic DNA of the Foxp1 locus was obtained from a 129S7/SvEv mouse BAC clone (Source BioScience LifeSciences) by a recombineering method (30), in which loxP sites were inserted to flank exons 13 and 14 of Foxp1 (ENSMUST00000113326.8), which encode the highly conserved DNA-binding fork head domain of the gene. ES clones without neor cassette were identified by Southern blot hybridization of XbaI-digested genomic DNA. Correctly targeted embryonic stem (ES) cells for Foxp1fl/+ were karyotyped before injection into blastocysts isolated from superovulated C57BL/6 mice. Offspring were backcrossed with C57BL/6 mice for at least eight generations. Tail DNA of the offspring was genotyped by PCR. The following primers were used for PCR genotyping of the locus: Foxp1 forward: CCTCTCCAAGTCTGCCTCAG, reverse: TGATAGCATTTCACGGTCCA.
Generation of Foxp1ΔMUSCLE mice
Foxp1fl/fl mice were bred with mice hemizygous for the Cre-recombinase gene under the control of the Mck-promoter (Stock #006475, The Jackson Laboratory). Offspring were further bred to achieve Mck-Cre;Foxp1fl/fl (shown as FoxpΔMUSCLE), which in experiments were compared to Foxp1fl/fl mice (shown as WT). Primers used for genotyping were:
Mck-Cre forward: ACAAAAGGTTTTGCCCTCCT, reverse: GTGAAACAGCATTGCTGTCACTT; Foxp1fl/fl forward: CCTCTCCAAGTCTGCCTCAG, reverse: TGATAGCATTTCACGGTCCA.
In vivo assays
In the fasting/starvation experiment, C57BL/6J mice were starved for 16 h (17.00–09.00) with free access to water. Dissected subcutaneous white adipose tissue, interscapular brown adipose tissue, and skeletal muscle tissue were used for RNA preparation and cDNA synthesis, as outlined under “RNA and protein analysis”. A glucose tolerance test was conducted on 3-month-old FoxpΔMUSCLE and WT mice fed with normal chow diet. Mice were fasted for 4 h before a single dose of D-glucose (20% solution in PBS, 10 ml/kg) was injected intraperitoneally. At 0, 15, 30, 60, 90, and 120 min after injection, blood was drawn from tails of mice, and glucose concentrations were determined using an ACCU-CHEK Compact Plus glucose meter (Roche). All analyses were performed according to the manufacturers’ protocols. Tissue glucose uptake was assessed in vivo by measuring uptake of 2-[3H]-deoxy-D-glucose as described (22). All animal procedures were approved by the Ethical Committee for Animal Experiments at the University of Gothenburg, which adheres to the principles and guidelines established by the European Convention for the Protection of Laboratory Animals.
Isolation of SVF progenitor cells and adipocytes
Female C57BL/6 mice were used to obtain primary adipose cells and stromal–vascular fraction (SVF) progenitor cells, essentially as described previously (31). In brief, the epididymal fat pads were isolated, washed in DMEM, and digested using collagenase A (1 mg/g tissue) at 37 °C for 1 h in DMEM containing 4% BSA. Samples were filtered through a 250 μm nylon mesh, and adipose cells and SVF were separated by centrifugation at 700g for 7 min at room temperature. Both cell fractions were washed three times with DMEM containing 4% BSA and directly used for RNA isolation.
Isolation, culturing, and differentiation of primary myoblasts
FoxpΔMUSCLE and WT mice were used to obtain primary myoblasts that were isolated and cultured as previously described (32). In brief, the skeletal muscles were isolated and digested using collagenase II (400 U/ml) at 37 °C for 1 h. Samples were filtered and centrifuged at 1400g for 5 min at room temperature. Cells were washed, filtered, and cultured on Matrigel precoated dishes in myoblast growth medium. Differentiation of primary myoblasts to myocytes was carried on as previously described (32).
Muscle fiber type quantification
Skeletal muscle was collected from the hind legs of adult (3–6-month-old)Foxp1ΔMUSCLE mice, rapidly frozen in isopentane cooled in liquid nitrogen in a drop of OCT Cryomount. The frozen tissue was sectioned at 10 μm on a cryostat, and sections were collected onto Superfrost slides. ATPase staining was performed essentially as previously described (33); anti-myosin (skeletal, slow type I) antibody was used together with a Vector M.O.M. Immunodetection Kit according to the manufacturer’s protocol. In the soleus muscle, the ATPase-stained muscle fibers could be classified into three different types based on the degree of staining. The number of each fiber type was counted in equally sized areas of cross-sections from three different longitudinal levels of the skeletal muscle.
ChIP-qPCR
ChIP experiment was performed using Pierce Magnetic ChIP Kit with modified lysis and immunoprecipitation procedures. In brief, 3T3-L1 cells infected with empty control vector or 3×FLAG-tagged FOXP1 expression vector were used in the experiment. At day 5 of differentiation, cells were fixed with 1% formaldehyde for 10 min at room temperature, followed by 5 min incubation with 1x (final concentration, Pierce kit) glycine solution. After washing and harvesting, fixed cells were lysed in the lysis buffer (50 mM Tris–HCl pH 8.0, 5 mM EDTA, 1% SDS, protease inhibitors) on ice for 10 min. Cell lysis was sonicated with PIXUL Multi-Sample Sonicator (Active Motif) at a processing time of 40 min to obtain 200 to 1000 bp chromatin fragments. ChIP was performed using normal rabbit IgG antibody (Pierce kit) and anti-Flag antibody (F7425, Sigma-Aldrich) and ChIP Grade Protein A/G Magnetic Beads (Pierce kit) according to the manufacturer’s protocol. After incubation, the magnetic beads were washed sequentially by wash buffer I (20 mM Tris–HCl pH 8.0, 2 mM EDTA, 1% Triton X-100, 150 mM NaCl, 0.1% SDS), wash buffer II (20 mM Tris–HCl pH 8.0, 2 mM EDTA, 1% Triton X-100, 500 mM NaCl, 0.1% SDS), wash buffer III (10 mM Tris–HCl pH 8.0, 1 mM EDTA, 0.25 M LiCl, 1% NP-40, 1% sodium deoxycholate), and Tris-EDTA (10 mM Tris-HCl pH 8.0, 1mM EDTA) buffer. After washing, the magnetic beads were eluted twice by 200 μl elution buffer (25 mM Tris–HCl, 10 mM EDTA, 0.5% SDS). The ChIP DNA was reverse cross-linked and purified according to the manual from the Pierce Magnetic ChIP Kit protocol. The potential fork head gene-binding sites to the target genes from Figure 3 were predicted as described (22) and were verified by real-time PCR. Primers for verification are listed in Table S3.
RNA-seq analysis of Foxp1 knockdown and overexpression
The paired-end reads with strand-specific library (fr-first strand) preparation of Foxp1 knockdown (n = 6), and Foxp1 overexpression (n = 6) were aligned using HISAT2 (v2.1.0) (34) against reference ensemble mouse genome GRCm38 (35) with 84 to 85% alignment rate. Obtained alignment files are indexed and sorted using SAMtools (v1.5) (36). These alignment files were further used for gene quantification using ensemble gene annotation corresponding to GRCm38 genome. featureCounts from subread (v1.5.3) (37) package was used for assigning high quality uniquely mapped reads to the gene features with paired-end and strand-specific parameters (-p -s 2 -B --minOverlap 10 -Q 30 --ignoreDup). Bioconductor package edgeR (R v4.0) (38) was used to obtain DEGs between different biological conditions. The significant differential genes were filtered based on adjusted p-value less than 0.001. Obtained DEGs were subjected to functional enrichment analysis against KEGG and gene ontology databases using GeneSCF (v1.1-p2). The significant differential genes were filtered based on adjusted p-value less than 0.001. Obtained DEGs were subjected to functional enrichment analysis against KEGG and gene ontology databases using GeneSCF (v1.1-p2) (39). Significant pathways were filtered using p-value less than 0.05.
Data availability
The raw data from all sequencing samples related to this study is deposited in GEO repository under accession GSE179031.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We thank G. Petersson for technical support. All computations were performed on resources provided by Uppsala Multidisciplinary Center for Advanced Computational Science (UPPMAX) high-performance computing (HPC), which is part of the Swedish National Infrastructure for Computing (SNIC).
Author contributions
H. M., V. S., W. Z., F. M., H. P., I. A., H. H., S. Z., S. B., C. K., and S. E. methodology; H. M., V. S., W. Z., F. M., H. P., I. A., H. H., S. Z., S. B., and C. K. investigation; S. S. formal analysis; S. E. writing–original draft; S. E. project administration.
Funding and additional information
S. E. is supported by the Swedish Research Council (2019-00773), The Knut and Alice Wallenberg Foundation, Sahlgrenska’s University Hospital (LUA-ALF), and Novo Nordisk Foundation.
Reviewed by members of the JBC Editorial Board. Edited by Qi-Qun Tang
Supporting information
Differentially expressed gene lists.
Full list of functional analysis summary.
Real-time PCR primers for ChIP-qPCR.
Metabolic profile induced by FOXP1.A, ECAR (glycolysis) in 3T3-L1 adipocytes with doxycycline-induced overexpressing for FOXP1. Empty vector n = 11, FOXP1 n = 9. B, ECAR (glycolysis) in 3T3-L1 adipocytes with doxycycline-induced knockdown of FOXP1. shCtrl n = 12, shFoxp1 n = 10. C, Western blot experiments showing overexpression and knockdown of FOXP1. Representative experiments shown. Data shown as mean ± s.d., n shows biological independent experiments. Unpaired two-sided Student’s t-test; ∗∗∗P < 0.001, ∗∗P < 0.01, ∗P < 0.05.
Glucose uptake in primary mouse adipocytes.A, Foxp1 mRNA levels in MEF adipocytes derived from wild-type mice or mice lacking one Foxp1 allele (Foxp1fl/+-cre) or both Foxp1 alleles (Foxp1fl/fl-cre) after infection with retrovirally derived cre-recombinase; n = 4. B, Glucose uptake in MEF adipocytes from wild-type, Foxp1fl/+-cre or Foxp1fl/fl-cre mice; n = 4. Experiments replicated at least three times except for B (twice). Unpaired two-sided Student’s t-test; ∗∗∗P < 0.001, ∗∗P < 0.01, ∗P < 0.05.
FOXP1 regulates GLUT1 protein levels in 3T3-L1 adipocytes and L6 myotubes.A, Western blot of GLUT1 and GLUT4 in 3T3-L1 adipocytes (on day 11) in response to doxycycline-induced overexpression of FOXP1 (d = 6). B, Western blot of GLUT1 and GLUT4 and proteins in L6 myotubes with doxycycline-induced overexpression of FOXP1. C, Western blot of FOXP1 and FLAG in 3T3-L1 adipocytes in response to doxycycline-induced overexpression by different exposure time to detect the endogenous FOXP1 protein. D, Western blot of FOXP1 and FLAG in L6 myotubes in response to doxycycline-induced overexpression by different exposure time to detect the endogenous FOXP1 protein. Representative experiments from three independent replicate samples are shown.
ChIP-qPCR validation of Foxp1 target genes.A–G, Schematic of predicted binding sites and real-time PCR results for Foxp1 target genes: Hk2 (A), Pfkm (B), Pkm (C), Aldoa (D), Ldha (E), Pdk1 (F) and Pkd4 (G). Cycle threshold (Ct) values of each group were subtracted by the value of empty vector control with anti-IgG antibody (which means fold change to control as 1). The average value to control with anti-IgG of three technical replicates was shown in the figures. Unpaired two-sided Student’s t-test; ∗∗∗P < 0.001, ∗∗P < 0.01, ∗P < 0.05.
References
- 1.Vander Heiden M.G., Cantley L.C., Thompson C.B. Understanding the warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hui S., Ghergurovich J.M., Morscher R.J., Jang C., Teng X., Lu W., et al. Glucose feeds the TCA cycle via circulating lactate. Nature. 2017;551:115–118. doi: 10.1038/nature24057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jang C., Hui S., Zeng X., Cowan A.J., Wang L., Chen L., et al. Metabolite exchange between mammalian organs quantified in Pigs. Cell Metab. 2019;30:594–606.e593. doi: 10.1016/j.cmet.2019.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cori C.F. The glucose-lactic acid cycle and gluconeogenesis. Curr. Top. Cell. Regul. 1981;18:377–387. [PubMed] [Google Scholar]
- 5.Thacker S.V., Nickel M., DiGirolamo M. Effects of food restriction on lactate production from glucose by rat adipocytes. Am. J. Physiol. 1987;253:E336–E342. doi: 10.1152/ajpendo.1987.253.4.E336. [DOI] [PubMed] [Google Scholar]
- 6.DiGirolamo M., Newby F.D., Lovejoy J. Lactate production in adipose tissue: a regulated function with extra-adipose implications. FASEB J. 1992;6:2405–2412. doi: 10.1096/fasebj.6.7.1563593. [DOI] [PubMed] [Google Scholar]
- 7.Wu G., Yuan S., Chen Z., Chen G., Fan Q., Dong H., et al. The KLF14 transcription factor regulates glycolysis by downregulating LDHB in colorectal cancer. Int. J. Biol. Sci. 2019;15:628–635. doi: 10.7150/ijbs.30652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yeung S.J., Pan J., Lee M.H. Roles of p53, MYC and HIF-1 in regulating glycolysis - the seventh hallmark of cancer. Cell. Mol. Life Sci. 2008;65:3981–3999. doi: 10.1007/s00018-008-8224-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shu W., Yang H., Zhang L., Lu M.M., Morrisey E.E. Characterization of a new subfamily of winged-helix/forkhead (Fox) genes that are expressed in the lung and act as transcriptional repressors. J. Biol. Chem. 2001;276:27488–27497. doi: 10.1074/jbc.M100636200. [DOI] [PubMed] [Google Scholar]
- 10.Hu H., Wang B., Borde M., Nardone J., Maika S., Allred L., et al. Foxp1 is an essential transcriptional regulator of B cell development. Nat. Immunol. 2006;7:819–826. doi: 10.1038/ni1358. [DOI] [PubMed] [Google Scholar]
- 11.van Boxtel R., Gomez-Puerto C., Mokry M., Eijkelenboom A., van der Vos K.E., Nieuwenhuis E.E., et al. FOXP1 acts through a negative feedback loop to suppress FOXO-induced apoptosis. Cell Death Differ. 2013;20:1219–1229. doi: 10.1038/cdd.2013.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shi C., Sakuma M., Mooroka T., Liscoe A., Gao H., Croce K.J., et al. Down-regulation of the forkhead transcription factor Foxp1 is required for monocyte differentiation and macrophage function. Blood. 2008;112:4699–4711. doi: 10.1182/blood-2008-01-137018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koon H.B., Ippolito G.C., Banham A.H., Tucker P.W. FOXP1: a potential therapeutic target in cancer. Expert Opin. Ther. Targets. 2007;11:955–965. doi: 10.1517/14728222.11.7.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pariani M.J., Spencer A., Graham J.M., Jr., Rimoin D.L. A 785kb deletion of 3p14.1p13, including the FOXP1 gene, associated with speech delay, contractures, hypertonia and blepharophimosis. Eur. J. Med. Genet. 2009;52:123–127. doi: 10.1016/j.ejmg.2009.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yoo H.C., Yu Y.C., Sung Y., Han J.M. Glutamine reliance in cell metabolism. Exp. Mol. Med. 2020;52:1496–1516. doi: 10.1038/s12276-020-00504-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Atas E., Oberhuber M., Kenner L. The implications of PDK1-4 on tumor energy metabolism, aggressiveness and therapy resistance. Front. Oncol. 2020;10 doi: 10.3389/fonc.2020.583217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goodale W.T., Olson R.E., Hackel D.B. The effects of fasting and diabetes mellitus on myocardial metabolism in man. Am. J. Med. 1959;27:212–220. doi: 10.1016/0002-9343(59)90341-9. [DOI] [PubMed] [Google Scholar]
- 18.Jorfeldt L. Metabolism of L(plus)-lactate in human skeletal muscle during exercise. Acta Physiol. Scand. Suppl. 1970;338:1–67. [PubMed] [Google Scholar]
- 19.Brooks G.A. Lactate production under fully aerobic conditions: the lactate shuttle during rest and exercise. Fed. Proc. 1986;45:2924–2929. [PubMed] [Google Scholar]
- 20.Caon I., Parnigoni A., Viola M., Karousou E., Passi A., Vigetti D. Cell energy metabolism and hyaluronan synthesis. J. Histochem. Cytochem. 2021;69:35–47. doi: 10.1369/0022155420929772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sullivan W.J., Mullen P.J., Schmid E.W., Flores A., Momcilovic M., Sharpley M.S., et al. Extracellular matrix remodeling regulates glucose metabolism through TXNIP Destabilization. Cell. 2018;175:117–132.e121. doi: 10.1016/j.cell.2018.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sukonina V., Ma H., Zhang W., Bartesaghi S., Subhash S., Heglind M., et al. FOXK1 and FOXK2 regulate aerobic glycolysis. Nature. 2019;566:279–283. doi: 10.1038/s41586-019-0900-5. [DOI] [PubMed] [Google Scholar]
- 23.Cederberg A., Grande M., Rhedin M., Peng X.R., Enerback S. In vitro differentiated adipocytes from a Foxc2 reporter knock-in mouse as screening tool. Transgenic Res. 2009;18:889–897. doi: 10.1007/s11248-009-9280-1. [DOI] [PubMed] [Google Scholar]
- 24.Silver D.P., Livingston D.M. Self-excising retroviral vectors encoding the Cre recombinase overcome Cre-mediated cellular toxicity. Mol. Cell. 2001;8:233–243. doi: 10.1016/s1097-2765(01)00295-7. [DOI] [PubMed] [Google Scholar]
- 25.Ohsumi J., Sakakibara S., Yamaguchi J., Miyadai K., Yoshioka S., Fujiwara T., et al. Troglitazone prevents the inhibitory effects of inflammatory cytokines on insulin-induced adipocyte differentiation in 3T3-L1 cells. Endocrinology. 1994;135:2279–2282. doi: 10.1210/endo.135.5.7956951. [DOI] [PubMed] [Google Scholar]
- 26.Lee S., Lim H.J., Park H.Y., Lee K.S., Park J.H., Jang Y. Berberine inhibits rat vascular smooth muscle cell proliferation and migration in vitro and improves neointima formation after balloon injury in vivo. Berberine improves neointima formation in a rat model. Atherosclerosis. 2006;186:29–37. doi: 10.1016/j.atherosclerosis.2005.06.048. [DOI] [PubMed] [Google Scholar]
- 27.Pike Winer L.S., Wu M. Rapid analysis of glycolytic and oxidative substrate flux of cancer cells in a microplate. PLoS One. 2014;9 doi: 10.1371/journal.pone.0109916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shi J., Kandror K.V. Study of glucose uptake in adipose cells. Methods Mol. Biol. 2008;456:307–315. doi: 10.1007/978-1-59745-245-8_23. [DOI] [PubMed] [Google Scholar]
- 29.Jensen T.C., Crosson S.M., Kartha P.M., Brady M.J. Specific desensitization of glycogen synthase activation by insulin in 3T3-L1 adipocytes. Connection between enzymatic activation and subcellular localization. J. Biol. Chem. 2000;275:40148–40154. doi: 10.1074/jbc.M004902200. [DOI] [PubMed] [Google Scholar]
- 30.Liu P., Jenkins N.A., Copeland N.G. A highly efficient recombineering-based method for generating conditional knockout mutations. Genome Res. 2003;13:476–484. doi: 10.1101/gr.749203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hauner H., Skurk T., Wabitsch M. Cultures of human adipose precursor cells. Methods Mol. Biol. 2001;155:239–247. doi: 10.1385/1-59259-231-7:239. [DOI] [PubMed] [Google Scholar]
- 32.Hindi L., McMillan J.D., Afroze D., Hindi S.M., Kumar A. Isolation, culturing, and differentiation of primary myoblasts from skeletal muscle of adult mice. Bio Protoc. 2017;7 doi: 10.21769/BioProtoc.2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ogilvie R.W., Feeback D.L. A metachromatic dye-ATPase method for the simultaneous identification of skeletal muscle fiber types I, IIA, IIB and IIC. Stain Technol. 1990;65:231–241. doi: 10.3109/10520299009105613. [DOI] [PubMed] [Google Scholar]
- 34.Kim D., Paggi J.M., Park C., Bennett C., Salzberg S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019;37:907–915. doi: 10.1038/s41587-019-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Howe K.L., Achuthan P., Allen J., Allen J., Alvarez-Jarreta J., Amode M.R., et al. Ensembl 2021. Nucleic Acids Res. 2021;49:D884–D891. doi: 10.1093/nar/gkaa942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Danecek P., Bonfield J.K., Liddle J., Marshall J., Ohan V., Pollard M.O., et al. Twelve years of SAMtools and BCFtools. Gigascience. 2021;10 doi: 10.1093/gigascience/giab008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liao Y., Smyth G.K., Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 38.McCarthy D.J., Chen Y., Smyth G.K. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012;40:4288–4297. doi: 10.1093/nar/gks042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Subhash S., Kanduri C. GeneSCF: a real-time based functional enrichment tool with support for multiple organisms. BMC Bioinformatics. 2016;17:365. doi: 10.1186/s12859-016-1250-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Differentially expressed gene lists.
Full list of functional analysis summary.
Real-time PCR primers for ChIP-qPCR.
Metabolic profile induced by FOXP1.A, ECAR (glycolysis) in 3T3-L1 adipocytes with doxycycline-induced overexpressing for FOXP1. Empty vector n = 11, FOXP1 n = 9. B, ECAR (glycolysis) in 3T3-L1 adipocytes with doxycycline-induced knockdown of FOXP1. shCtrl n = 12, shFoxp1 n = 10. C, Western blot experiments showing overexpression and knockdown of FOXP1. Representative experiments shown. Data shown as mean ± s.d., n shows biological independent experiments. Unpaired two-sided Student’s t-test; ∗∗∗P < 0.001, ∗∗P < 0.01, ∗P < 0.05.
Glucose uptake in primary mouse adipocytes.A, Foxp1 mRNA levels in MEF adipocytes derived from wild-type mice or mice lacking one Foxp1 allele (Foxp1fl/+-cre) or both Foxp1 alleles (Foxp1fl/fl-cre) after infection with retrovirally derived cre-recombinase; n = 4. B, Glucose uptake in MEF adipocytes from wild-type, Foxp1fl/+-cre or Foxp1fl/fl-cre mice; n = 4. Experiments replicated at least three times except for B (twice). Unpaired two-sided Student’s t-test; ∗∗∗P < 0.001, ∗∗P < 0.01, ∗P < 0.05.
FOXP1 regulates GLUT1 protein levels in 3T3-L1 adipocytes and L6 myotubes.A, Western blot of GLUT1 and GLUT4 in 3T3-L1 adipocytes (on day 11) in response to doxycycline-induced overexpression of FOXP1 (d = 6). B, Western blot of GLUT1 and GLUT4 and proteins in L6 myotubes with doxycycline-induced overexpression of FOXP1. C, Western blot of FOXP1 and FLAG in 3T3-L1 adipocytes in response to doxycycline-induced overexpression by different exposure time to detect the endogenous FOXP1 protein. D, Western blot of FOXP1 and FLAG in L6 myotubes in response to doxycycline-induced overexpression by different exposure time to detect the endogenous FOXP1 protein. Representative experiments from three independent replicate samples are shown.
ChIP-qPCR validation of Foxp1 target genes.A–G, Schematic of predicted binding sites and real-time PCR results for Foxp1 target genes: Hk2 (A), Pfkm (B), Pkm (C), Aldoa (D), Ldha (E), Pdk1 (F) and Pkd4 (G). Cycle threshold (Ct) values of each group were subtracted by the value of empty vector control with anti-IgG antibody (which means fold change to control as 1). The average value to control with anti-IgG of three technical replicates was shown in the figures. Unpaired two-sided Student’s t-test; ∗∗∗P < 0.001, ∗∗P < 0.01, ∗P < 0.05.
Data Availability Statement
The raw data from all sequencing samples related to this study is deposited in GEO repository under accession GSE179031.