Abstract
MicroRNAs (miRNA) are small non-coding RNA molecules that regulate posttranscriptional gene expression by repressing messengerRNA-targets. MiRNAs are abundant in many cell types and are secreted into extracellular fluids, protected from degradation by packaging in extracellular vesicles. These circulating miRNAs are easily accessible, disease-specific and sensitive to small changes, which makes them ideal biomarkers for diagnostic, prognostic, predictive or monitoring purposes. Specific miRNA signatures can be reflective of disease status and development or indicators of poor treatment response. This is especially important in malignant diseases, as the ease of accessibility of circulating miRNAs circumvents the need for invasive tissue biopsy. In osteogenesis, miRNAs can act either osteo-enhancing or osteo-repressing by targeting key transcription factors and signaling pathways. This review highlights the role of circulating and extracellular vesicle-derived miRNAs as biomarkers in bone-related diseases, with a specific focus on osteoporosis and osteosarcoma. To this end, a comprehensive literature search has been performed. The first part of the review discusses the history and biology of miRNAs, followed by a description of different types of biomarkers and an update of the current knowledge of miRNAs as biomarkers in bone related diseases. Finally, limitations of miRNAs biomarker research and future perspectives will be presented.
Keywords: microRNA, biomarker, bone, osteosarcoma, osteoporosis, extracellular vesicles, exosomes
1. Introduction
MicroRNAs (miRNAs) are small non-coding RNA molecules, about 19-24 nucleotides in length, that regulate posttranscriptional gene expression by targeting messengerRNAs (mRNAs) and thus repress or alter the translational process (1, 2). Each miRNA targets more than 100 genes and plays a role in multiple signaling pathways and biological processes (2). They can be secreted into extracellular fluids and act as messengers to mediate cell-cell communication (2).
The first miRNA-mRNA interaction has been described by Victor Ambros and Gary Ruvkun in 1993 (3, 4). They discovered lin-4 binding to the 3’ untranslated region of lin-14 mRNA and thereby regulating the level of LIN-14 protein, necessary for temporal development in Caenorhabditis elegans (3, 4). Since then, miRNAs have also been detected in flies, fish and mammals and are generally conserved amongst species (2, 5–10). With the discovery of miRNAs and target prediction facilitated by whole genome sequencing and bioinformatic approaches, researchers shifted their research interests towards functional characterizations of miRNA-mRNA interactions (11).
Given that miRNAs are tissue-specific and their expression patterns reflect cellular physiological processes, their importance in the context of human diseases became more and more apparent (1, 11). For instance, miRNAs can act as tumor suppressors or oncogenes, regulate cellular proliferation and apoptosis, modulate intracellular signaling and immune response (1, 11). Specifically, in oncologic diseases, miRNAs are involved in cell apoptosis, invasion and metastasis and disease progression and have diagnostic, prognostic and therapeutic use (11–20). Moreover, miRNAs have also been implied in viral disease (11, 21, 22), neurodegenerative disease (11, 23, 24) and immune disease (25, 26).
MiRNAs are also known to play an important role in epigenetic regulation of osteogenesis (27). By targeting key osteogenic regulators, e.g. runt-related transcription factor 2 (RUNX2), they can enhance or repress osteogenesis (27–37). Other miRNAs act by targeting genes and receptors of osteogenic signaling pathways, such as the Wnt- or Bone morphogenetic protein (BMP)-signaling pathway (38–48). Additionally, several miRNAs also regulate osteoclast differentiation and osteoblast-osteoclast communication (49, 50).
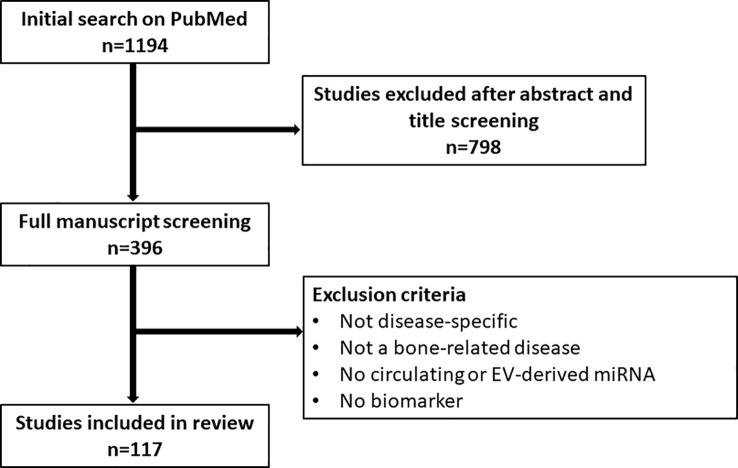
Considering that miRNAs are reflective of certain cellular and molecular processes, changes in their expression level are a useful indicator of disease, thus making them ideal biomarkers for diagnostic, predictive and prognostic purposes (1, 27, 51, 52). Additionally, miRNAs are packaged into extracellular vesicles (EVs) and released into the bloodstream, protected from degradation (27, 51). These circulating miRNAs and EV-miRNAs are easily harvested by liquid biopsy and thus represent an accessible tool to study miRNA signatures as biomarkers, circumventing the need for invasive tissue biopsy specifically in bone-related diseases (27, 51). Moreover, EV-miRNAs can also be harvested locally before being released into the bloodstream, e.g. in bile or cerebrospinal fluid, or can be linked to their tissue of origin based on detection of surface marker proteins, thus allowing for better diagnostic specificity and sensitivity (53). In this review, we highlight the role of circulating miRNAs as biomarkers in bone-related diseases, focusing on osteoporosis and osteosarcoma. To this end, a comprehensive literature search on PubMed has been performed. We applied the following search query and a filter for English language: (((((((((exosomes) OR (microvesicles)) OR (extracellular vesicles)) OR (circulating)) AND (miRNA)) OR (microRNA)) OR (miR)) AND (bone)) AND (biomarker)) NOT (Review). After an initial screening of titles and abstracts (n=1194), a more in-depth screening of the manuscript text was performed among the included 396 papers. Studies only discussing miRNA in a general osteogenesis context, but not disease-specific or studies not investigating a bone-related disease were excluded. Additionally, studies that did not discuss circulating or EV-derived miRNA or did not discuss their biomarker potential were excluded. The following review encompasses the remaining 117 studies ( Figure 1 ).
Figure 1.
Flow chart depicting the screening process for studies included in the review.
2. Disease biomarkers
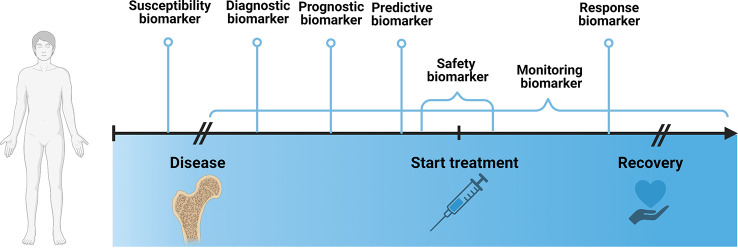
According to the Biomarkers, EndpointS, and other Tools (BEST)- glossary by the FDA-NIH Joint Leadership Council biomarkers are “a defined characteristic that is measured as an indicator of normal biological processes, pathogenic processes, or biological responses to an exposure or intervention, including therapeutic interventions”. They can be categorized into 7 different types, including susceptibility/risk, diagnostic, monitoring, prognostic, predictive, response and safety biomarker.
The susceptibility biomarker is a risk indicator for developing a disease before the onset of disease. In clinical practice, it is helpful to inform preventative care, such as making lifestyle or dietary changes or increasing the frequency of screening exams. Well known examples of susceptibility biomarkers are the Breast Cancer Gene 1/2 (BRCA1/2) mutations (54, 55), that indicate the likelihood of developing breast and ovarian cancer, and low-density lipoprotein cholesterol levels (56), that are accompanied by an increased risk for development of cardiovascular disease.
A diagnostic biomarker confirms a disease or a specific subtype of a disease, which may guide treatment decisions or potential enrollment in clinical trials. Common examples include the Glomerular filtration rate (GFR) to identify patients with chronic kidney disease (57), sweat chloride to identify individuals with cystic fibrosis (58) or HbA1c to diagnose diabetes mellitus (59).
A monitoring biomarker assesses the status of a disease or treatment response and is measured repeatedly. For instance, they are useful for assessment of disease progression, disease recurrence, changes in severity status of disease. Measurements are usually taken during defined periods of the disease, such as from diagnosis to treatment, e.g. to evaluate progression rate, during intervention, to assess treatment response or after treatment, to monitor recurrence. Common examples include Prostate-specific antigen (PSA), which is used to monitor disease status in patients with prostate cancer (60, 61) and International normalized ration (INR) or prothrombin time (PT) to evaluate anticoagulative treatment response (62).
A prognostic biomarker indicates the likelihood of a clinical event, disease recurrence or progression of disease, such as metastasis or overall survival, after onset of disease. They are measured at a defined baseline and are dependent on clinical setting, e.g. ongoing background treatment or stage of disease, and the endpoint of interest. They can inform decisions about treatment plans, for instance, plasma fibrinogen is used to identify patients with high risk for exacerbation of chronic obstructive pulmonary disease (63).
A predictive biomarker identifies patients who are more likely to have a defined outcome from a treatment and are useful for treatment selection. For instance, they can be characteristic to the patient’s constitution, e.g. a genetic marker, expression level of a specific protein in tissues or serum or mutations in tumor. On the other hand, predictive biomarkers can be dependent on the drug, which is often defined based on empiric evidence and pathophysiology of the drug. For example, in non-small cell lung cancer, squamous differentiation is predictive of a negative response to pemetrexed treatment (64). It is often difficult to distinguish between a prognostic and a predictive biomarker. In certain cases, a biomarker can also simultaneously be both, prognostic and predictive.
A response biomarker, e.g. a pharmacodynamic biomarker, indicates that a biological response has occurred after exposure to a treatment. A common example is circulating B lymphocytes to assess response to a B-lymphocyte stimulator in patients with systemic lupus erythematosus (65). In a clinical context, response biomarkers help guide decisions regarding dosing or continuation of a treatment, e.g. HbA1c to evaluate the response to antihyperglycemic agent in diabetes mellitus patients. Treatment plans can be modified according to the level of response.
Last, a safety biomarker is an indicator for toxicity of a treatment, which can be measured before or after treatment start and can predict, confirm or evaluate the extent of the adverse effect. Measurements are often taken repeatedly, to ensure adequate management of toxicity and adjustment of treatment plans, e.g. serum creatinine to monitor nephrotoxic effects of certain drugs (66). Additionally, if taken before start of the treatment, safety biomarkers can identify patients, who should not be given a specific treatment due to safety risk, such as deficiencies of metabolizing enzymes. An example includes the HLA-B* 1502 genotype to identify patients with an increased risk of developing Steven-Johnson syndrome upon treatment with carbamazepine (67).
Figure 2 establishes a timeline from before the onset of disease until after discovery and marks the timepoints at which a specific biomarker is measured.
Figure 2.
Schematic depicting different biomarker types in the context of disease development over time. Created with BioRender.com.
In the following, we aim to review current knowledge about miRNAs as biomarkers for bone-related diseases, specifically in osteoporosis and osteosarcoma.
3. MiRNA biomarkers in osteoporosis
Osteoporosis is the most common bone disease and affects over 10 million people in the United States and over 200 million women worldwide (68). An increase in osteoclast activity and a decrease in osteoblast activity leads to an imbalance in bone homeostasis and causes bone loss. The resulting decrease in bone density and bone mass results in bone fragility and increased risk of fracture (52, 68–70). In 2006, an estimated 8.9 million of osteoporotic fractures were recorded worldwide and 1 in 3 women and 1 in 5 men will experience a fracture after the age of 50 years (71, 72). Risk factors include postmenopausal estrogen deficiency, low physical activity, smoking, hormonal factors, genetic factors, nutrition and the use of specific drugs, e.g. glucocorticoids. Given our aging population and changing lifestyle habits, osteoporosis-related morbidity is expected to increase, which represents a major burden for the global health system (68, 72, 73). Thus, early identification of individuals at high risk for osteoporosis is important to allow for preventative care in the form of lifestyle interventions or pharmacologic treatments (68, 74). Current diagnostic methods include bone mineral density (BMD) measurements by dual-energy X-ray absorptiometry (DXA), which is recommended for all women 65 years and older (68). Bone turnover markers, such as alkaline phosphatase (ALP), osteocalcin (OCN) or parathyroid hormone (PTH), are valuable in risk assessment in combination with BMD measurements (68, 74). The US Preventive Services Task force (USPSTF) suggests questionnaire-based screening tools, e.g. Fracture Risk Assessment (FRAX), Osteoporosis Risk Assessment Instrument (ORAI) or Osteoporosis Index of Risk (OSIRIS), to assess increased osteoporotic risk in postmenopausal women younger than 65 years old.
However, DXA is expensive and comes with radiation exposure and bone turnover markers are unreliable, thus better screening methods are highly sought after. In the past decade, miRNA signatures have emerged as potential biomarkers for the early diagnosis of osteoporosis and risk prediction for fragility fractures.
Multiple authors have explored the potential of individual or a panel of miRNA as diagnostic biomarkers in postmenopausal osteoporosis (PMO) (52, 69, 75–84), in general patient cohorts including PMO and idiopathic osteoporosis (85–93) and in specific secondary osteoporosis cases, e.g. severe childhood-onset osteoporosis in patients with mutations in the Plastin 3 (PLS3) gene (94). Among the most investigated miRNAs are members of the miR-21-, miR-23-, miR-320-, miR-203- and miR-19-families. miR-203 is a known suppressor of bone formation by negatively regulating BMP-2 and distal-less homeobox 5 (DLX5) which are activators of key transcription factors for osteoblast differentiation like runt RUNX2 and Osterix (SP7) (79, 80, 94). miR-23 is also known to negatively regulate RUNX2 (92). miR-21 is one of the most studied microRNAs. Among its direct targets are members of the Transforming Growth Factor ß (TGFß)- and BMP-signaling pathways, RUNX2, osteocalcin, osteopontin, phosphatase and tensin homolog (PTEN) and SMAD7 (95). miR-21 is also involved in osteoclastogenesis by targeting osteoclast suppressor programmed cell death protein 4 (PDCD4) and osteoclast marker cathepsin K (CTSK) and receptor activator of nuclear factor ĸB ligand (RANKL) (92, 95), promoting osteoclast differentiation. miR-19b directly targets PTEN resulting in an increase of RUNX2 and phosphorylation of AKT (protein kinase B) and thus enhancing osteoblastogenesis (96, 97). Targets of miR-320 include BMP2, CTNNB1 (ß-catenin) and RUNX2, promoting osteoblast differentiation (98) ( Table 1 ).
Table 1.
MiRNA biomarkers in osteoporosis.
| Bone-related disease | Author | MiRNA | Target genes of interest | Type of biomarker | Species | MiRNA source |
|---|---|---|---|---|---|---|
| Osteoporosis | Al-Rawaf et al. (69) | miR-148a miR-122-5p |
Diagnostic biomarker for PMO vs controls | Human | Serum | |
| Baloun et al. (52) | miR-1278 miR-24-1-5p miR-422a |
Diagnostic biomarker for PMO vs controls | Human | Serum | ||
| Bedene et al. (75) | miR-148a-3p | MAFB, PPAR, WNT1 | Diagnostic biomarker for PMO vs controls | Human | Plasma | |
| Chen et al. (76) | miR-21-5p miR-23a-3p miR-125-5p |
PDCD4, ASL, EIF4E3 (miR-21) RUNX2 (miR-23) PDGF (miR-125b) |
Diagnostic biomarker for PMO vs controls | Human | Serum | |
| Ciuffi et al. (85) | miR-23a-3p miR-21-5p miR-320a-3p |
Diagnostic biomarker for osteoporosis vs osteopenia vs healthy controls (miR-23a-3p, miR-21a-5p) Diagnostic biomarker for osteoporosis with fracture vs osteoporosis without fracture (miR-320a-3p) |
Human | Serum | ||
| Ding et al. (77) | Panel: miR-194-5p and five medical items (weight, age, left ventricular end systolic diameter, alanine aminotransferase, urine epithelial cell count) | Diagnostic biomarker for PMO vs osteopenia vs healthy controls | Human | Plasma | ||
| Ding et al. (86) | miR-100 | Diagnostic biomarker for osteoporosis vs healthy control | Human | Plasma | ||
| Garg et al. (99) | miR-23b-3p miR-140-3p miR-21-5p miR-122-5p miR-125-5p |
SMAD7, FGF18, SKP2, SPRY1/2, PDCD4, PTEN, RECK, GDF-5, SOX2, PLAP-1, ACVR2B (miR-21-5p) RUNX2, MRC2, CCND1, PTEN (miR-23b-3p) BMP2, IGF1R, RUNX2,SPARC, TSC22D3, VDR, PCP4 (miR-122-5p) BMPR1B, TRAF6 (miR-125b-5p) MCF2L, PTEN, BMP2 (miR-140-3p) |
Diagnostic biomarker for osteoporotic hip fractures vs non-osteoporotic hip fractures | Human | Plasma | |
| Heilmeier et al. (97) | Panel: miR-203a, miR-31-5p, miR-19b-1-5p, BMD measurements |
FOS, RUNX2, SMAD1 (miR-203a) PTEN, RUNX2 (miR-19b-1-5p) FZD3, RUNX2, SP7, SATB2 (miR-31-5p) |
Predictive biomarker for fracture risk in diabetic osteoporosis | Human | Serum | |
| Heilmeier et al. (100) | miR-382-3p, miR-188-3p, miR-942 | Diagnostic biomarker for PM osteoporotic fragility fracture vs postmenopausal patients without fractures | Human | Serum | ||
| Ismail et al. (78) | miR-208a-3p miR-155-5p miR-637 |
ETS1, ACVR1 (miR-208a-3p) SOCS1 (miR-155-5p) SP7 (miR-637) |
Diagnostic biomarker for PMO vs premenopausal osteoporosis vs healthy controls (miR-208a-3p) and for PMO vs healthy controls (miR-155-5p, miR-637) | Human | Serum | |
| Kerschan-Schindl et al. (79) | OsteomiR™ panel |
RUNX2, LRP5, ß-catenin (miR-375) Osteocalcin, CTX (miR-550a-3p) WNT10B (miR-152-3p) BMP2, DLX5, Osteocalcin (miR-203a) |
Diagnostic biomarker for PMO vs controls (miR-375) Diagnostic biomarker for postmenopausal osteoporotic fragility fractures vs osteoporosis without fracture (miR-203a) |
Human | Serum | |
| Kocijan et al. (101) | Panel: miR-152-3p, miR-30e-5p, miR-324-3p, miR-19b-3p, miR-335-5p, miR-19a-3p, miR-550a-3p, miR-186-5p, miR-532-5p, miR-93-5p, miR-378-5p, miR-320a, miR-16-5p, miR-215-5p, let-7b-5p, miR-29b-3p, miR-7-5p, miR-365a-3p |
DKK1 (miR-152-3p, miR-335-3p) LRP6 (miR-30e-5p) BMP2 (miR-140-5p) HDAC4, TGFß3, ACVR2A, CTNNBIP1, DUSP2 (miR-29b-2p) |
Diagnostic biomarker for idiopathic and postmenopausal osteoporotic fractures vs non-osteoporotic fracture controls | Human | Serum | |
| Kocijan et al. (80) | miR-203a | DLX5, RUNX2 | Diagnostic biomarker for PMO vs controls Monitoring biomarker for treatment response for zoledronate and teriparatide in PMO |
Human | Serum | |
| Ladang et al. (102) | OsteomiR™ panel | Predictive biomarker for fragility fracture risk | Human | Serum | ||
| Li et al. (81) | miR-21 miR-133a |
SPRY1 (miR-21) | Diagnostic biomarker for PM-osteopenia/osteoporosis vs controls | Human | Plasma | |
| Lincoln et al. (103) | miR-148a-3p | WNT1, WNT10B, KDM6B, DNMT1, IGF1, MAFB | Predictive biomarker for development of osteoporosis in patients with acute spinal cord injury | Human | Plasma | |
| Lu et al. (87) | miR-206 | HDAC4 | Diagnostic biomarker for osteoporosis vs control | Human | Serum | |
| Ma et al. (82) | miR-181c-5p miR-497-5p |
Diagnostic biomarker for PMO vs controls Monitoring biomarker for bisphosphonate and calcitriol treatment response in PMO |
Human | Serum | ||
| Mäkitie et al. (94) | Panel: miR-93-3p, miR-532-3p, miR-133a-3p, miR-301b-3p, miR-181c-5p, miR-203a-3p, miR-590-3p |
WNT1, LRP6, PTEN (miR-301b) DLX5, RUNX2 (miR-203a-3p) RANKL, MMP9, NF-ĸB, DKK1 (miR-218-5p) DKK1 (miR-203) |
Diagnostic biomarker for X-linked primary osteoporosis | Human | Serum | |
| Mandourah et al. (88) | miR-122-5p and/or miR-4516 |
BMP2K, FSHB, IGF1R, RUNX2, SPARC, TSC22D3, TSC22D3, VDR (both) CNR2, ALPL, ANKH, ESR1, LRP6 (miR-122-5p) CNR1, AR (miR-4516) |
Diagnostic biomarker for osteoporosis vs osteopenia vs control | Human | Serum and Plasma | |
| Meng et al. (83) | miR-194-5p |
COUP-TFII
TGFß-signaling pathway Wnt-signaling pathway |
Diagnostic biomarker for PM- osteopenia/osteoporosis vs controls | Human | Whole blood | |
| Messner et al. (104) | miR-454-3p, miR-584-5p, miR-101-3p, miR-191-5p, miR-26-5p, miR-32-5p, miR-4508 | Wnt- signaling pathway TGFß- signaling pathway |
Monitoring biomarker for denosumab treatment response in PMO | Human | Serum | |
| Nakashima et al. (89) | miR-195 miR-150 |
GIT1, BMP (miR-195) MMP14 (miR-150) |
Diagnostic biomarker for osteoporosis vs control | Human | Serum | |
| Panach et al. (105) | miR-122-5p miR-125b-5p miR-21-5p |
Diagnostic biomarker for osteoporotic bone fracture vs osteoarthritic control | Human | Serum | ||
| Pepe et al. (90) | miR-1246 miR-1224-5p |
Tetraspanin 5 (miR-1224-5p) | Diagnostic biomarker for osteoporosis vs osteopenia and control | Human | Plasma-exosomes | |
| Quillen et al. (98) | miR-197-3p miR-320a miR-320b miR-331-5p miR-423-5p |
FGB, IGFBP5, SOD1, SOD2 (miR-197-3p) SOD2 (miR-331-5p) GAPDH, MIF, MMP9, CTNNB1, RUNX2 (miR-320a) GAPDH (miR-423-5p) BMP2 (miR-320b) |
Susceptibility biomarker for bone loss | Baboon | Plasma | |
| Ramírez-Salazar et al. (91) | miR-140-3p miR-23b-3p |
AKT1, AKT2, AKT3, BMP2 FOXO3, GSK3B, IL6R, PRKACB, RUNX2, WNT5 | Diagnostic biomarker for osteopenia/osteoporosis vs control | Human | Serum | |
| Seeliger et al. (92) | Panel: miR-21, miR-23a, miR-24, miR-100, miR-125b |
PDCD4 (miR-21) RUNX2 (miR-23a, miR-24) BMPR2 (miR-100) |
Diagnostic biomarker for osteoporosis vs control | Human | Serum | |
| Shi et al. (106) | miR-324-3p miR-766-3p miR-1247-5p miR-330-5p miR-3124-5p |
WNT8B, FZD2, CSNK1E, DVL1, RAC3 (miR-324-3p) WNT1-A, FZD10, SENP2, VANGL1, WNT5B, PRKCA, NFATC2 (miR-776-3p) WNT9B, DVL3, CSNK2A2, APC2, MYCBP2, RNC1 (miR-1247-5p) WNT2B, FZD4, DVL3, PRKACA, APC2, BTRC, DVL1, PPP3CB (miR-330-5p) WNT10B, LRP6, CXXC4, CSNK2A1, GSK3B, CSN1A1L, FZD1, WNT5a, PLCB1, PRKCA (miR-3124-3p) |
Diagnostic biomarker for postmenopausal osteoporotic fragility fracture vs non-fracture control | Human | Serum-exosomes | |
| Shuai et al. (93) | 5 miRNA panel: miR-30c-2-3p, miR-199a-5p, miR-424-5p, miR-497-5p, miR-877-3p 4 miRNA panel: miR30c-2-3p, miR-877-3p, miR-199a-5p, miR-424-5p |
HIF1a pathway (miR-199a-5p) RUNX2 (miR-30c) BMP signaling pathway (miR-497) Smad7 signaling (miR-877-3p) |
Diagnostic biomarker for osteoporosis vs osteopenia and control | Human | Serum | |
| Weigl et al. (107) | miR-34a-5p, miR-31-5p, miR-30d-3p, miR-378a-5p (teriparatide and zolendronate) miR-375-3p, miR-183-5p, miR-203a-3p, miR-203b-3p (teriparatide) |
AGO3, MYC, SPRED1, MYCN, SON, NUFIP2, MDM4
Wnt signaling pathway Notch signaling pathway |
Monitoring biomarkers for teriparatide and zoledronate treatment response in ovariectomized Sprague-Dawley rats vs control | Rat | Serum | |
| Xu et al. (108) | miR-491-5p miR-485-3p |
Diagnostic biomarker for postmenopausal osteoporotic vertebral fracture vs postmenopausal controls without fracture | Human | Plasma | ||
| Xu et al. (109) | miR-27a-3p | SP7 | Diagnostic biomarker for osteoporosis vs controls | Human | Serum | |
| Yavropoulou et al. (110) | miR-124-3p miR-2861 miR-21-5p miR-23a-3p miR-29a-3p |
SPRY, PDCD4 (miR-21) RUNX2 (miR-23-3p) |
Diagnostic biomarker for postmenopausal osteopenic/osteoporotic vertebral fractures vs postmenopausal controls without fracture | Human | Serum | |
| Zarecki et al. (111) | miR-375 miR-532-3p miR-19b-3p miR-152-3p miR-23a-3p miR-335-5p miR-21-5p |
ESR1, ADCY1, ATF2, CALM1, PIK3R3, GNAQ, PIK3CA (miR-19b-3p) ITGA9, ITGA5, ITGA11, COL2A1, COL4A1, JARID2, INHBB, APC, WNT10B, IGF1, KLF4, MEIS1 (miR-152-3p) YAP1, SMAD7, LATS1, BMPR2 (miR-21-5p) |
Diagnostic biomarker for postmenopausal osteoporotic vertebral fracture vs postmenopausal controls without fracture | Human | Serum | |
| Zhao et al. (84) | miR-144-5p miR-506-3p miR-8068 miR-6841-3p |
YY1, VIM, YWHAE | Diagnostic biomarker for PMO vs control | Human | Serum |
OsteomiR™ panel: let-7b-5p, miR-127-3p, miR-133b, miR-141-3p, miR-143-3p, miR-144-5p, miR-152-3p, miR-17-5p, miR-188-5p, miR-19b-3p, miR-203a, miR-214-3p, miR-29b-3p, miR-31-5p, miR-320a, miR-335-5p, miR-375, miR-550a-3p, miR-582-5p; PMO: postmenopausal osteoporosis
Other authors sought to find biomarkers enabling diagnosis of osteoporotic fragility fractures. Previous studies aimed at identifying miRNA-biomarkers differentiating between patients with osteoporotic fragility fractures and patients without fractures (79, 85, 100, 106, 110, 111), patients with osteoporotic fractures versus (vs) patients with non-osteoporotic fractures (99, 101) and patients with osteoporotic fractures vs osteoarthritic controls without fractures (105) ( Table 1 ).
Few authors studied a population over time in a prospective study to investigate potential predictive biomarkers for osteoporotic fragility fracture. Heilmeier et al. (97) assessed 168 postmenopausal diabetic women over a mean follow-up of 5.8 ± 2.7 years. They demonstrated increased fracture risk in women with upregulated miR-203a and miR-31-5p serum levels, whereas low expression of miR-19b-1-5p was associated with lower fracture risk. The predictive potential of this 3 miRNA-signature was increased when combined with BMD measurements (97). Ladang et al. (102) evaluated the predictive potential of a signature set of 19 miRNAs in a longitudinal study with an osteopenic/osteoporotic patient cohort. They successfully demonstrated a positive predictive value of 68% and a sensitivity of 76% in predicting fragility fracture within three years before the event occurred. They concluded that the miRNAs panel might be more valuable than the ordinarily used FRAX (Fracture Risk Assessment Tool) in predicting fragility fractures (102) ( Table 1 ).
In a clinical setting, a monitoring biomarker may enable us to evaluate the status of treatment response. In 2020, Kocijan et al. (80) demonstrated upregulation of miR-203a in femoral head tissues and peripheral blood of ovariectomized Sprague-Dawley rats, which was reverted after 12 weeks of either resorptive treatment with zoledronate or osteo-anabolic treatment with teriparatide. Thus, besides being a diagnostic biomarker differentiating between ovariectomized rats and controls, miR-203a may also be a potential minimally invasive monitoring biomarker for treatment response (80). In 2021, the same group investigated the time dependent changes of miRNA serum expression levels during treatment in the same animal model (107). They presented two panels of miRNA biomarkers, miR-34a-5p, miR-31-5p, miR-30d-3p, miR-378a-5p and miR-375-3p, miR-183-5p, miR-203a-3p, miR-203b-3p, with upregulated serum expression levels over time, which was prevented by treatment with teriparatide or zoledronate and zoledronate only, respectively. In contrast to their previous study, the authors did not include miR-203a-3p in the zoledronate treatment response panel after demonstrating that the miR-203a-3p expression initially decreased during treatment, however, it increased again starting at 16 weeks of treatment (107). Ma et al. (82) investigated the biomarker potential of circulating miR-181c-5p and miR-497-5p, which were upregulated in the osteoporotic/osteopenic patient cohort vs healthy controls but decreased after anti-osteoporotic treatment with bisphosphonate and calcitriol, thereby representing a potential monitoring biomarker for treatment response. To assess denosumab treatment response in postmenopausal women, Messner et al. (104) demonstrated upregulation of a panel of 7 miRNAs, measured repeatedly over a period of 2 years, which was associated to increasing BMD and thereby representing a valuable monitoring biomarker set ( Table 1 ).
In most studies, miRNAs are evaluated after onset of a disease. Interestingly, Quillen et al. (98) investigated the expression levels of 5 miRNAs in a cohort of 147 healthy adult baboons. All miRNAs were negatively correlated with BMD, thus increased miRNA levels correlated with decreased BMD. Target prediction highlighted their role in extracellular matrix regulation, apoptosis and cell proliferation. The authors suggest the investigated miRNA panel might be indicative of a pre-metabolic shift in bone homeostasis, thus showing promise as a susceptibility biomarker for development of osteoporosis (98) ( Table 1 ).
Using a panel of biomarker vs single biomarkers increases specificity and sensitivity and allows for some variation, thus current trends in biomarker potential of miRNA go towards identification of disease-signature sets. Seeliger et al. (92) were among the first to identify a signature panel of circulating miRNA able to differentiate between osteoporotic and non-osteoporotic patients. They first performed miRNA PCR arrays in serum samples from patients with hip fractures, which were further classified in osteoporotic and non-osteoporotic sample. They identified 83 miRNAs, which were validated in a separate analysis using serum samples from osteoporotic and non-osteoporotic patients. Amongst those, 9 miRNAs were significantly upregulated in osteoporotic conditions. The authors proceeded in further validating these candidates in osteoporotic bone tissue. They conclude their study presenting a panel of 5 miRNAs (miR-21, miR-23a, miR-24, miR-25, miR-100, miR-125b) accurately diagnosing osteoporosis (92). The company TAmiRNA developed an osteomiR™ kit, that allows measurements of serum/plasma expression levels of 19 miRNAs, that have been extensively tested and validated in bone pathophysiology and bone-related diseases (80, 89, 100, 105, 111, 112). Kerschan-Schindl et al. (79) identified 4 clusters of miRNAs within the osteomiR™ set. Combining individual miRNA from different sets was shown to be relevant to the classification of osteoporosis according to the WHO definition and fracture-based classification (Major osteoporotic Fractures MOFx), with miR-375 as a main contributor for the former and miR-203 for the latter, respectively (79). Kocijan et al. (101) isolated a set of 19 miRNAs in osteoporotic patients vs healthy controls. Within the set, a panel of 8 miRNAs (miR-152-3p, miR-30e-5p, miR-140-5p, miR-324-3p, miR19b-3p, miR-335-5p, miR-19a-3p, miR-550a-3p) represent potential diagnostic biomarkers discriminating between patients with osteoporotic fractures vs non-osteoporotic fracture controls, regardless of age or sex. 7 of the 19 tested miRNAs are included in the osteomiR™ signature ( Table 1 ).
4. MiRNA biomarkers in osteosarcoma
Osteosarcoma is the most common primary bone tumor occurring predominantly in children and young adults. Primary sites include the metaphyses of long bone, e.g. distal femur, proximal tibia or proximal humerus. By the time of diagnosis, about 15-25% of patients present with distant metastasis mostly to the lung, but also in bone and rarely in lymph nodes. Symptoms include local pain, swelling and limitation of movement. Diagnostic methods rely on traditional x-ray imaging techniques, often accompanied by magnetic resonance imaging (MRI) to detect soft tissue invasion and skip lesions, followed by bone tissue biopsy (113–116). Treatment requires a multidisciplinary approach including combination chemotherapy and complete surgical resection. Osteosarcoma is an aggressive tumor with a 5-year survival rate of 60-70%, which is drastically reduced to 10-40% in patients with relapsing disease or metastatic status at time of diagnosis (114, 117). Due to the difficulty in early assessment of relapse or diagnosis of minimal residual disease by conventional imaging techniques, biomarkers for monitoring of tumor response and surveillance of recurrence are highly sought after (115).
Multiple authors identify singular miRNAs as diagnostic biomarkers differentiating between osteosarcoma patients and a control group (118–141). Selection of miRNAs is based on previously established associations to different cancer types (118, 120–128, 131, 133, 135, 136, 139), which is an approach that drastically limits the potential candidate miRNAs. Other authors have focused on more comprehensive screening methods for potential biomarker selection, followed by validation by quantitative RT-PCR (138, 142, 143). Using this approach, Fujiwara et al. (142) demonstrated superiority of miR-25-3p as a diagnostic biomarker compared to the conventional biomarker ALP. Other authors created miRNA-mRNA regulatory networks using pre-screened miRNAs and their predicted targets. Candidate miRNAs were selected based on targeted hub genes [miR-199a-5p (137)] ( Table 2 ).
Table 2.
MiRNA biomarkers in osteosarcoma.
| Bone-related disease | Author | MiRNA | Target genes of interest | Type of biomarker | Species | MiRNA source |
|---|---|---|---|---|---|---|
| Osteosarcoma | Allen-Rhoades et al. (115) | Panel: miR-205-5p, miR-335-5p, miR-574-3p, miR-214 |
LZTS1 (miR-214) Wnt-signaling pathway, SOX9 (miR-335-5p, miR-574-3p) |
Diagnostic biomarker (all) Prognostic biomarker for overall survival (miR-214) Monitoring biomarker for disease development (all) |
Mouse and human | Plasma |
| Asano et al. (1) | Panel: Index VI (miR-4736, miR-6836-3p, miR-4281, miR-762, miR-658, miR-4649-5p, miR-4665-3p) | Diagnostic biomarker for sarcomas (including bone sarcomas) | Human | Serum | ||
| Cao et al. (118) | miR-326 | BCL2 | Diagnostic biomarker Prognostic biomarker for distant metastasis |
Human | Serum | |
| Cong et al. (119) | miR-124 | Diagnostic biomarker Prognostic biomarker for overall survival |
Human | Serum | ||
| Cuscino et al. (116) | 8 novel candidate miRNA | Diagnostic biomarker | Human | Osteosarcoma cell line-derived exosomes | ||
| Dailey et al. (144) | Panel: miR-23a-3p, miR-30c-5p | Prognostic biomarker for disease-free interval | Canine | Serum | ||
| Diao et al. (120) | miR-22 | Diagnostic biomarker Prognostic biomarker for outcome (large tumor size, advanced clinical stages, distant metastasis) Predictive biomarker for chemosensitivity |
Human | Plasma | ||
| Dong et al. (121) | miR-223 | Diagnostic biomarker Prognostic biomarker for distant metastasis, advanced clinical stage and overall survival |
Human | Serum | ||
| Fujiwara et al. (142) | miR-25-3p | Diagnostic biomarker Prognostic biomarker for distant metastasis |
Human Mouse |
Serum | ||
| Gong et al. (145) | miR-675 | CALN1 | Prognostic biomarker for distant metastasis | Human | Plasma | |
| Heishima et al. (146) | Panel: miR-214, miR-126 and ALP | Prognostic biomarker for metastasis, disease free survival and overall survival | Canine | Plasma | ||
| Hong et al. (147) | miR-29a miR-29b |
Prognostic biomarker for disease free survival and overall survival | Human | Serum | ||
| Hua et al. (122) | miR-Let7A | Diagnostic biomarker Prognostic biomarker for overall survival |
Human | Blood | ||
| Hua et al. (123) | miR-21 | PDCD4 | Diagnostic biomarker Predictive biomarker for chemosensitivity |
Human | Serum | |
| Huang et al. (143) | miR-663 | Diagnostic biomarker | Human | Plasma | ||
| Huang et al. (148) | Panel: miR-487a, miR-493-5p, miR-501-3p, miR-502-5p) | Diagnostic biomarker | Human | Serum | ||
| Jerez et al. (149) | miR-21-5p miR-143-3p miR-148a-3p miR-181-5p |
MAPK1, NRAS, FRS2, PRCKE, BCL2, QKI | Prognostic biomarker for metastasis | Human | Osteosarcoma cell line-derived extracellular vesicles | |
| Kosela-Paterczyk et al. (150) | Panel: miR-133a, miR-223-3p, miR-450b-5p, miR-548q | Diagnostic biomarker | Human | Serum | ||
| Li et al. (151) | miR-542-3p | Prognostic biomarker for tumor progression and overall survival | Human | Serum | ||
| Lian et al. (152) | Panel: miR-195-5p, miR-199a-3p, miR-320a, miR-374-5p |
FASN (miR-195-5p) p53 signaling pathway (miR-199-3p) |
Diagnostic biomarker (4 miRNA set) Prognostic biomarker for metastasis (miR-195-5p, miR-199a-3p) Monitoring biomarker for tumor response after surgery |
Human | Plasma | |
| Lian et al. (124) | miR-34a | Diagnostic biomarker Prognostic biomarker metastasis, recurrence and overall survival Predictive biomarker for chemosensitivity |
Human | Serum | ||
| Liu et al. (125) | miR-375 | Diagnostic biomarker Prognostic biomarker for tumor stage and metastasis Predictive biomarker for chemosensitivity |
Human | Serum | ||
| Luo et al. (153) | miR-337-3p miR-484 miR-582 miR-3677 |
Diagnostic biomarker Prognostic biomarker for tumor stage, metastasis and overall survival Monitoring biomarker for surgical treatment response |
Human | Serum | ||
| Ma et al. (126) | miR-148a | Diagnostic biomarker Prognostic biomarker for tumor size, metastasis, overall survival and disease-specific survival |
Human | Plasma | ||
| Monterde-Cruz et al. (140) | miR-215-5p miR-642-5p |
RAB2A, RB1, BLCAP, CCNT2 (miR-215-5p) MAPK-signaling pathway, TGFß-signaling pathways (miR-642a-5p) |
Diagnostic biomarker | Human | Serum | |
| Nakka et al. (141) | miR-21 miR-221 miR-106a |
RECK, PTEN (miR-21) HDAC6, DNMT3b, NOSTRIN, E-cadherin, uPAR7b, PTEN, KIP1, CDKN1B (miR-221) |
Diagnostic biomarker | Human | Plasma | |
| Niu et al. (127) | miR-95-3p | Diagnostic biomarker Prognostic biomarker for overall survival |
Human | Serum | ||
| Ouyang et al. (154) | Panel: miR-21, miR-199a-3p, miR-143 | Diagnostic biomarker | Human | Serum | ||
| Pang et al. (128) | miR-497 | Diagnostic biomarker Prognostic biomarker for advanced stage, distant metastasis Predictive biomarker for chemosensitivity |
Human | Serum | ||
| Shi et al. (129) | miR-194 | Multiple oncogenic target genes, e.g. HIF-1a, YAP1, AKT2 | Diagnostic biomarker Prognostic biomarker for advanced clinical stage, metastasis and overall survival Monitoring biomarker for treatment response (surgery) |
Human | Serum | |
| Tian et al. (130) | miR-34b | Diagnostic biomarker Prognostic biomarker for metastasis |
Human | Plasma | ||
| Wang et al. (131) | miR-491 | CRYAB | Diagnostic biomarker Prognostic biomarker for lung metastasis and overall survival Predictive biomarker for chemosensitivity |
Human | Serum | |
| Wang et al. (132) | miR-191 | Diagnostic biomarker Prognostic biomarker for advanced clinical stage, distant metastasis, disease-free survival and overall survival |
Human | Serum | ||
| Xie et al. (133) | miR-26a-5p | ITGB8, HOXA5 | Diagnostic biomarker Prognostic biomarker for advanced stage, metastasis and overall survival |
Human | Serum | |
| Yang et al. (155) | miR-429 miR-143-3p |
Diagnostic biomarker Prognostic biomarker for advanced stage, distant metastasis and overall survival |
Human | Serum | ||
| Yang et al. (134) | miR-221 | RECK, ARHI, DVL2 | Diagnostic biomarker Prognostic biomarker for distant metastasis, advanced clinical stage, disease-free survival and overall survival |
Human | Serum | |
| Yao et al. (135) | miR-101 | RAC1, USP22, VEGF-C, Girdin, COX-2, EZH2, SOCS-2, ZEB1, ZEB2 | Diagnostic biomarker Prognostic biomarker for advanced clinical stage, distant metastasis, disease-free survival and overall survival Monitoring biomarker for treatment response in non-metastatic patients |
Human | Serum | |
| Yuan et al. (136) | miR-21 | Prognostic biomarker for advanced stage and overall survival Predictive biomarker for chemosensitivity |
Human | Serum | ||
| Zhang et al. (156) | miR-133b miR-206 |
EGFR, MCL1, FSCN1, c-Met, BCL2L2 (miR-133b) | Diagnostic biomarker Prognostic biomarker for high tumor grade, metastasis, recurrence, disease-free survival and overall survival |
Human | Serum | |
| Zhang et al. (137) | miR-199a-5p | VEGFA | Diagnostic biomarker | Human | Osteosarcoma cell line-derived exosomes Plasma |
|
| Zhou et al. (138) | miR-199a-5p | Diagnostic biomarker Monitoring biomarker for surgical treatment response and tumor status |
Human | Serum | ||
| Zhou et al. (139) | miR-421 | Diagnostic biomarker Prognostic biomarker for overall survival |
Human | Serum |
High biovariability and small sample sizes due to the rarity of the disease make biomarker identification in osteosarcoma difficult. Multiple authors have focused on multi-panel miRNA signatures to increase biomarker accuracy and allow for individual variations (1, 115, 116, 148, 150, 152–154). Asano et al. (1) screened serum miRNAs in over a 1000 sarcoma samples, including osteosarcoma. They identified a classifier Index VI differentiating between sarcomas and benign tumors and healthy controls. The group validated their findings in a smaller subset of patients and also comparing to patients with other malignancies, e.g. lung cancer. Although the discriminatory ability of Index VI was high in bone sarcomas, especially in the early stages, the authors point out that the Index does not discriminate between histological subtypes and that without confirmatory studies, it should only be used for diagnosis of malignant vs benign tumors (1). In 2015, Allen-Rhoades et al. (115) developed a mouse model for osteosarcoma and chose a signature set of four candidate miRNAs differentiating between affected mice and their healthy controls. The authors validated their findings in human plasma samples and thereby demonstrated cross-species application of their model (115) ( Table 2 ).
Additionally, multiple miRNAs have been suggested as prognostic biomarkers in osteosarcoma. High/low expression levels of specific miRNAs have been associated with advanced tumor stage, positive metastasis, higher recurrence and/or shorter overall survival (115, 124–136, 139, 141, 142, 144, 145, 147, 151, 152, 154–156). Similarly, multiple authors demonstrated superiority of using a miRNA panel with or without combination with clinical markers for prognostic accuracy compared to individual miRNAs (146, 149, 153) ( Table 2 ).
Monitoring biomarkers in osteosarcoma are useful to assess adequate response to surgical resection or chemotherapy and to screen for recurrence. Some authors identified upregulated miRNA expression levels preoperatively, that decreased upon surgical tumor resection (138, 142, 152) and vice versa, respectively (129, 135, 153). Considering that these miRNAs react to changes in tumor status, they represent valuable candidates for monitoring surgical treatment response and detecting residue and recurrence. In addition to surgery, combination chemotherapy is required for a curative approach in treating osteosarcoma (114). Fujiwara et al. (142) evaluated the potential of miR-25-3p to monitor tumor response in a case series and were able to show decreasing expression levels after surgical tumor resection and during/after neoadjuvant combination chemotherapy, unlike ALP levels, which plateaued during treatment. In a mouse model of osteosarcoma, Allen-Rhoades et al. (115) demonstrated detectable changes in expression levels of a previously identified 4-miRNA signature set coinciding with tumor formation 14 weeks after transplantation of osteosarcoma cells. Additionally, they measured expression level of a 4-miRNA set in placebo treated and Doxorubicin-treated mice with osteosarcoma and were able to demonstrate reduced magnitude of alteration in miRNA plasma expression levels in the treated mice, suggesting that these miRNAs may be a valuable monitoring marker for tumor development and chemo-response ( Table 2 ).
Predictive biomarkers may guide treatment decisions in osteosarcoma as certain preconditions, e.g. specific miRNA up- or downregulation, may indicate the likelihood of a patient to respond well or poorly to a chemotherapy. For instance, patients with lower levels of certain miRNAs before chemotherapy, such as miR-34 (124) demonstrated good treatment response compared to patients with initially high miRNA expression levels. In contrast, patients with low levels of miR-375 (125), miR-497 (128), miR-491 (131) or miR-21 (136) before neoadjuvant chemotherapy showed a poor treatment response ( Table 2 ).
5. MiRNA biomarkers in other bone-related diseases
Besides osteosarcoma, Ewing Sarcoma is the second most common bone and soft tissue tumor in children and adolescents. The Ewing’s sarcoma family of tumors (ESFT) includes Ewing’s sarcoma, Askin tumor and peripheral primitive neuroectodermal tumor. At the time of diagnosis, about 25% of patients have detectable metastasis, primarily in the lung, bone and bone marrow. Treatment includes combination chemotherapy and surgical resection (157). Using miRNA sequencing, Crow et al. (158) identified a disease-specific signature set of 62 exosomal miRNAs able to differentiate between ESFT and non-ESFT samples. Similarly, Kosela-Patercyzk et al. (150) and Nie et al. (159) identified a 4 miRNA panel and a singular miRNA marker, respectively, as diagnostic biomarkers for ESFT. In chondrosarcoma, a type of bone sarcoma, miR-145 (160) was identified as a diagnostic biomarker for early detection, which is important due to limited treatment options. However, studies investigating miRNA biomarkers for these types of bone sarcomas are rare ( Table 3 ).
Table 3.
MiRNA biomarkers in other bone-related diseases.
| Bone-related disease | Author | MiRNA | Target genes of interest | Type of biomarker | Species | MiRNA source |
|---|---|---|---|---|---|---|
| Osteoarthritis | Ali et al. (161) | miR-320b miR-320c miR-320d miR-320e |
14-3-3 gene family | Diagnostic biomarker for fast-progressing OA | Human | Plasma |
| Pertusa et al. (162) | miR-497 | SMURF2 | Diagnostic biomarker | Human | Serum | |
| Wan et al. (163) | miR-136 | IL17 | Diagnostic biomarker | Human | Plasma | |
| Xia et al. (164) | miR-181-5p | TNFA | Diagnostic biomarker | Human | Peripheral blood | |
| Ewing Sarcoma Family of Tumors (ESFT) | Crow et al. (158) | Panel of 62 miRNAs (see full list in original paper) | Diagnostic biomarker | Human | ESFT cell line-derived extracellular vesicles Serum |
|
| Kosela-Paterczyk et al. (150) | Panel: miR-424-5p, miR-3173-3p, miR-142-3p, miR-4746-5p | Diagnostic biomarker | Human | Serum | ||
| Nie et al. (159) | miR-125b | Diagnostic biomarker | Human | Serum | ||
| Chrondrosarcoma | Urdinez et al. (160) | miR-145 | FSCN1 | Diagnostic biomarker | Human | Plasma |
| Multiple Myeloma Bone Disease | Papanota et al. (165) | Panel: let-7b-5p, miR-143-3p, miR-17-5p miR-214-3p, miR-335-5p |
ATF4, SP7, FGFR1, PTEN (miR-214-3p) SP7 (miR-143-3p) TGFBR1, IGF1R, MYC (let-7a-5p) DKK1, IGF1R (miR-335-5p) SMAD5, BMP2 (miR-17-5p) |
Diagnostic biomarker Prognostic biomarker for progression-free survival (let-7b-5p, miR-335-5p) |
Human | Plasma |
| Moura et al. (166) | miR-29c-3p | Diagnostic biomarker | Human | Plasma | ||
| Hao et al. (167) | miR-214 miR-135b |
PTEN (miR-214) | Prognostic biomarker for bone disease in Multiple Myeloma patients Predictive biomarker for bisphosphonate treatment response (miR-214) |
Human | Serum | |
| Bone metastasis | Fang et al. (168) | miR-214 | PTEN | Prognostic biomarker for bone metastasis in PCa | Human | Serum |
| Guo et al. (169) | miR-205 | Prognostic biomarker for bone metastasis in PCa | Human | Serum | ||
| Lu et al. (170) | miR-125a-3p miR-330-3p miR-339-5p miR-613 |
KRAS, RHOA, BDNF, HNRNPA1, BRCA1, FYN, BCL2L11, NRG1, SETX, FXR1, BACE1, CDC37, TIMP3, PDCD4, SGK1 | Prognostic biomarker for bone metastasis in PCa | Human | Plasma-derived exosomes | |
| Peng et al. (171) | miR-218-5p | TRAF1, TRAF2, TRAF5 | Prognostic biomarker for bone metastasis in PCa | Human | Serum | |
| Rode et al. (172) | miR-425-5p | HSPB8 | Prognostic biomarker for bone metastasis in PCa | Human | Prostate cancer cell-line derived exosomes | |
| Wa et al. (173) | miR-204-5p | TRAF1, TAB3, MAP3K3 | Prognostic biomarker for bone metastasis in PCa | Human | Serum | |
| Wang et al. (174) | miR-181-5p | Prognostic biomarker for bone metastasis in PCa | Human | Serum-exosomes | ||
| Zhang et al. (175) | miR-141 | Prognostic biomarker for bone metastasis in PCa | Human | Serum | ||
| Wu et al. (176) | miR-19a | PTEN | Prognostic biomarker for bone metastasis in breast cancer | Human | Bone-metastatic estrogen-receptor positive breast cancer cell-derived exosomes | |
| Zhao et al. (177) | miR-10b | Prognostic biomarker for bone metastasis in breast cancer | Human | Serum | ||
| Xiang et al. (178) | miR-34a | TGIF2 | Prognostic biomarker for bone metastasis in hepatocellular carcinoma | Human | Serum | |
| Valencia et al. (179) | miR-326 | Monitoring biomarker for metastatic progression in bone metastasis in lung cancer | Mouse | Serum | ||
| Xu et al. (180) | miR-139-5p | NOTCH1 | Prognostic biomarker for bone metastasis in lung cancer | Human | Serum | |
| Yang et al. (181) | Panel of 144 miRNA (Cluster B, see (181)), e.g. miR-574-5p, miR-328-3p, miR-423-3p | Wnt/ß-catenin signaling pathway | Prognostic biomarker for bone metastasis in lung cancer | Human | Plasma-derived exosomes | |
| Zeng et al. (182) | miR-31-3p | FOXO1 | Prognostic biomarker for bone metastasis in lung cancer | Human | Serum | |
| Fracture healing | Bourgery et al. (183) | miR-451 miR-328-3p miR-133a-3p miR-375-3p miR-423-5p miR-150-5p |
P38 MAPK (miR-451) Axin1, PTEN (miR-328-3p) Egfr, Fgfr1, Igfr1, Met (miR-133a-3p) Lrp5, Ctnnp, Brd4 (miR-375-3p) Tnip2, Cdkn1a, Igf2bp1 (miR-423-5p) Vegf, Socs1, Rab9, Mmp14, Slc2a1, Elk1 (MiR-150-5p) |
Monitoring biomarker for adequate fracture healing | Mouse | Serum |
| Dietz et al. (184) | miR-223 | Prognostic biomarker for development of CRPS after fracture | Human | Serum-exosomes | ||
| Xiong et al. (185) | miR-193a-3p | MAPK10 | Prognostic biomarker for fracture non-union | Human | Serum | |
| Adolescent idiopathic scoliosis | García-Giménez et al. (186) | Panel: miR-122-5p, miR-27a-5p, miR-223-5p, miR-1306-3p | Wnt/ß-catenin pathway, BMP4 (miR-122-5p) APC, PPARG, CEBPA (miR-27a) FGF2, PCGF3, BMI1, PU.1, RANKL, NFATc1, TRAP, c-Jun, cathepsin K, NFIA (miR-223-5p) |
Diagnostic biomarker | Human | Plasma |
| Wang et al. (187) | miR-151a-3p | GREM1 | Diagnostic biomarker | Human | Plasma | |
| Osteonecrosis of the femoral head (ONFH) | Hong et al. (188) | miR-127-3p miR-628-3p miR-1 miR-885-5p miR-483-3p miR-483-5p |
IGF2, PDGFA, RUNX2, PTEN, VEGF | Diagnostic biomarker for alcohol-induced ONFH | Human | Serum |
| Kao et al. (189) | miR-18a miR-19a miR-138-1 miR-1290 miR-3609 |
TP53
SERBP1 |
Diagnostic biomarker | Human | Blood | |
| Liu et al. (190) | miR-93-5p miR-320a |
Diagnostic marker | Human | Serum | ||
| Fibrous dysplasia of the bone | Legrand et al. (191) | miR-25-3p miR-93-5p miR-182-5p miR-324-5p miR-363-3p miR-451a |
IL6ST (miR-25-3p, miR-363-3p) FOXO1 (miR-324-5p) IL6R (miR-451a) GNAS, FOXO3, PDGFB, ESR2, BMP2 (miR-93-5p) |
Diagnostic biomarker | Human | Serum |
| Developmental dysplasia of the hip | Luo et al. (192) | miR-140 and 25-hydroxyvitamin D status | Diagnostic biomarker | Human | Serum | |
| Renal osteodystrophy | Nickolas et al. (193) | Panel: miR-30b, miR-30c, miR-125b, miR-155 | Diagnostic biomarker low vs non-low bone turnover | Human | Serum |
Multiple Myeloma is a hematological malignancy of the bone marrow. One of the more severe complications of Multiple Myeloma is bone disease, which is defined by osteolytic lesions or osteoporotic fractures due to clonal plasma cell disorder. Even though imaging techniques are improving quickly, early diagnosis of bone disease using non-invasive techniques would be very beneficial to guide treatment decisions (194). To our knowledge, three studies have investigated circulating miRNA as biomarkers for multiple myeloma bone disease, using a 5 miRNA-panel (165), miR-29c-3p (166), and miR-214 and miR-135b (167) as individual biomarkers, respectively. Out of the 5 miRNA-panel, two miRNAs (let-7b-5p and miR-335-5p) were also identified as a prognostic biomarker for progression-free survival (165). Hao et al. (167) identified miR-214 as a predictive biomarker for good response to bisphosphonate treatment ( Table 3 ).
Bone homeostasis is maintained by a constant remodeling process and relies on a balance of osteoblast and osteoclast activity. Solid tumors often metastasize to bone and over 70% of metastatic prostate and breast cancer patients experience bone metastasis. Tumor cells mainly infiltrate the endosteal and perivascular niches and are thought to remain in a state of dormancy often up to many years. Upon activation of tumor cells, cross-talk between the bone modulating cells leads to a release of growth factors and tumor cell proliferation is promoted by a feed-forward loop. Symptoms of bone metastasis include pain and increased fracture risk. Tumors are often considered incurable once bone metastasis has occurred and current diagnostic methods expose patients to radiation, thus biomarkers for early detection and prediction of patients at risk are highly sought after (176, 195). Multiple miRNA biomarkers have been identified for detection of bone metastasis in prostate cancer (168–175), breast cancer (176, 177), hepatocellular carcinoma (178) and lung cancer (180–182). Additionally, in lung cancer bone metastasis, Valencia et al. (179) identified miR-326 as a valuable biomarker to monitor metastatic progression and tumor burden. It correlated with the conventional bone turnover marker PINP (procollagen I amino-terminal pro-peptide (179) ( Table 3 ).
Osteoarthritis is one of the leading causes of disability in adults worldwide and is a socioeconomic and financial burden. It is a metabolic inflammatory disease that leads to progressive cartilage degeneration. Major symptoms include chronic pain, stiffness and loss of mobility (196). MiRNAs have been investigated as diagnostic biomarkers in osteoarthritis (162–164). Additionally, Ali et al. (161) has identified the miR-320 family as potential diagnostic biomarker for fast-progressing osteoarthritis.
Long-bone fracture healing occurs in stage starting with hematoma formation, inflammation, migration and differentiation of mesenchymal stromal cells (MSCs), bone formation and angiogenesis and bone remodeling. During endochondral ossification, hypertrophic chondrocytes turn into osteoblasts. This process is tightly controlled by miRNAs (183). Any disruption of this complex process can lead to fracture non-union, arthrosis and chronic pain syndromes. Bourgery et al. (183) analyzed the serum expression levels of circulating miRNAs during fracture healing in a mouse model. They identified differentially expressed miRNAs over a follow-up period of 14 days compared to the baseline and were able to establish a potential miRNA signature for monitoring adequate fracture healing (183). Similarly, Xiong et al. (185) demonstrated miR-193a-3p as a potential prognostic biomarker indication fracture non-union. Development of chronic regional pain syndrome (CRPS) after fracture is often difficult to diagnose with potentially devastating consequences for the patient. Dietz et al. (184) identified miR-223-5p as a potential prognostic biomarker for development of CRPS after bone fracture ( Table 3 ).
Other bone-related diseases that have been studied in the context of circulating miRNA expression levels include adolescent idiopathic scoliosis (186, 187), osteonecrosis of the femoral head (188–190), fibrous dysplasia of the bone (191), developmental dysplasia of the hip (192) and renal osteodystrophy (193) ( Table 3 ).
6. Discussion
This review discusses the potential use of circulating and EV-derived miRNA as biomarkers of bone-related disease. Although this is a promising approach for diagnostic, monitoring, prognostic and predictive purposes, this area of research is still in early stages and has many limitations.
One of the main criticisms of circulating miRNAs as biomarker of disease is its lack of reproducibility, low specificity and high intra- and inter-assay variability. In the studies discussed in this review, there is only minimal overlap of biomarker candidates in osteoporosis, even less so in osteosarcoma studies. Moreover, some studies are even directly contradictory, e.g. Yang et al. (134) demonstrated high levels of miR-221 associated with poor prognosis and Nakka et al. (141) reported the reversed situation. These inconsistencies are due to lack of standardization of pre-analytical and analytical conditions and unaccounted differences in patient population, e.g. sex, age and existing preconditions (53, 197). For instance, centrifugation conditions can impact measurements of miRNA expression in plasma or serum samples and differences in miRNA purification methods accounts for 77-92% of intra-assay variation in miRNA quantification (197). Additionally, detection methods for circulating miRNA include Next-Generation Sequencing (NGS), real-time PCR, miRNA microarray, and less frequently, Northern blot analysis and in situ hybridization (198). To assure consistency between studies, miRNA detection should be accompanied by an adequate normalization strategy. Usually, this can be done by using a reference marker, most commonly spike-in cel-miR-39 or endogenous miR-16 during RNA extraction. A combination of spike-in and endogenous reference markers is preferred (197–199). The osteomiR™ panel contains three spike-in controls as well as two endogenous reference markers for hemolysis (79). In osteosarcoma, Allen-Rhoades et al. (115) investigated cross-species application of miRNA biomarkers and identified disease-specific endogenous reference plasma miRNAs in mouse and human.
Moreover, selection of biomarker candidates is inconsistent and unreliable, leading to low specificity. Many authors do not perform comprehensive screening methods to select potential miRNA biomarker candidates, but rather rely on previously published literature selecting miRNAs that have been shown to play a role in related diseases. To identify novel candidates that are specific to the investigated disease, comprehensive screening methods need to be performed and potential biomarkers should be selected based on differential expression levels at multiple different time points and their known or potential biological and molecular function. For instance, miR-21 is a widely known oncogenic miRNA implicated in tumorigenesis of multiple entities. Overexpression of miR-21 has been demonstrated in multiple tumors, including glioblastoma, ovarian cancer, B-cell lymphoma, hepatocellular carcinoma, lung cancer and breast cancer and is associated with increased proliferation and invasion and decreased apoptosis (200, 201).Target genes include tumor suppressor genes, e.g. PTEN, programmed cell death 4 (PDCD4), SMAD7 and tropomyosin (TPM1). Thus, miR-21 can be valuable as a monitoring biomarker for tumor progression or treatment response or as part of a diagnostic panel of miRNA, as suggested by many authors (136, 141, 149, 154), but its value as an individual diagnostic biomarker is limited (53).The Receiver Operating Characteristic (ROC) approach allows calculation of sensitivity and specificity and generation of an area under the curve (AUC) which is an indicator for how well the biomarker differentiates between a disease and its control (202). Many, but not all studies, discussed in this review specify AUC values and accepted cut-off values are not standardized and/or inconsistent, leading to low specificity of biomarker candidates (115). Additionally, according to the BEST Glossary by the FDA-NIH Biomarker Working Group, the test conditions play an important role in biomarker validation. For instance, a single measurement of blood measure is not sufficient for diagnosis of hypertension. The studies discussed in this review often measure expression levels of the candidate miRNA at only one defined time point. However, many miRNA expressions change with disease progression, which might explain some contradictions. Generally, given the heterogeneity of disease, using a panel compared to individual biomarkers is advisable (198). Further, positive and negative predictive values are dependent on the prevalence of disease in the population and these values were only rarely discussed in the presented studies (102, 151).
The majority of circulating biomarkers in blood is released by blood or endothelial cell and is thus not directly derivative of a specific diseased tissue, e.g. a tumor, and does not have biomarker potential. Even though it is well known, that tumors secrete miRNA packaged in EVs into the extracellular fluid, the fraction of these tumor-derived miRNAs compared to the larger fraction of the physiologically present endothelial/blood cell derived miRNAs is small and depends on the size of the tissue, access to variation and a large enough magnitude of differential expression compared to healthy tissue. Circulating miRNA biomarkers are thus not directly tumor- or disease-derivative, but rather a physiological response to the presence of a neoplastic formation or other disease. Moreover, miRNA expression levels vary greatly depending on sex, ethnicity, lifestyle and sample cell type (199). In contrast, EV-miRNAs are expressed stably and protected from degradation. Additionally, they express surface markers, that are highly specific to their tissue of origin. New isolation and purification methods allow for harvesting of EVs based on expression of surface marker proteins (53, 197–199). Hereby, EVs and their cargo could be directly linked to the releasing tissue and allow for differentiation of small disease-dependent miRNA from the larger fraction of the physiological miRNA secretome (53). Thus, EV-derived miRNA content is more tissue-specific and selective analysis harbors potential to improve the specificity of circulating EV-miRNA biomarker candidates.
7. Conclusions and future perspectives
In the era of “personalized/precision medicine”, discovery of new biomarkers takes a growing place. An ideal biomarker needs to be easily accessible, highly specific, and preferably sensitive in early detection of disease and changes in disease status due to progression or treatment (198). Given that circulating miRNA are secreted in extracellular fluids and are thereby easily harvested by minimally invasive liquid biopsy, are disease- and tissue-specific and reflective of even small changes in disease status, they technically meet the criteria of an ideal biomarker. However, technical limitations like lack of standardization of screening methods, selection and analysis leads to low reproducibility and severely undermines the value of the currently existing data. However, considering that this area of research is still in its early stages, adequate standardization of techniques, analysis and interpretation of results and comprehensive description of applied methods can help support large-scale validation studies of the promising, but preliminary, data known today. Additionally, selective analysis of EV-miRNA based on surface marker proteins represents a promising approach to further optimize specificity of the miRNA biomarker candidates.
Author contributions
JH was responsible for conception of the study, data collection and analysis and drafting the manuscript. ML was responsible for conception of the study and manuscript drafting and revision. NQ was responsible for conception of the study, drafting the article and final revision. All authors contributed to the article and approved the submitted version.
Funding Statement
JH is supported by the German Research Foundation (HU 2817/1-1). ML is supported by NIH Grants 1R01GM136659-01, 1R01DE027323-01A1 and 1R01DE027346-01A1.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- 1. Asano N, Matsuzaki J, Ichikawa M, Kawauchi J, Takizawa S, Aoki Y, et al. A serum microRNA classifier for the diagnosis of sarcomas of various histological subtypes. Nat Commun (2019) 10(1):1299. doi: 10.1038/s41467-019-09143-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. O'Brien J, Hayder H, Zayed Y, Peng C. Overview of MicroRNA biogenesis, mechanisms of actions, and circulation. Front Endocrinol (Lausanne) (2018) 9:402. doi: 10.3389/fendo.2018.00402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lee RC, Feinbaum RL, Ambros V. The c. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell (1993) 75(5):843–54. doi: 10.1016/0092-8674(93)90529-y [DOI] [PubMed] [Google Scholar]
- 4. Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in c. elegans Cell (1993) 75(5):855–62. doi: 10.1016/0092-8674(93)90530-4 [DOI] [PubMed] [Google Scholar]
- 5. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell (2004) 116(2):281–97. doi: 10.1016/S0092-8674(04)00045-5 [DOI] [PubMed] [Google Scholar]
- 6. Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature (2000) 408(6808):86–9. doi: 10.1038/35040556 [DOI] [PubMed] [Google Scholar]
- 7. Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr Biol (2002) 12(9):735–9. doi: 10.1016/S0960-9822(02)00809-6 [DOI] [PubMed] [Google Scholar]
- 8. Lagos-Quintana M, Rauhut R, Meyer J, Borkhardt A, Tuschl T. New microRNAs from mouse and human. Rna (2003) 9(2):175–9. doi: 10.1261/rna.2146903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Aravin AA, Lagos-Quintana M, Yalcin A, Zavolan M, Marks D, Snyder B, et al. The small RNA profile during drosophila melanogaster development. Dev Cell (2003) 5(2):337–50. doi: 10.1016/S1534-5807(03)00228-4 [DOI] [PubMed] [Google Scholar]
- 10. Lim LP, Glasner ME, Yekta S, Burge CB, Bartel DP. Vertebrate microRNA genes. Science (2003) 299(5612):1540. doi: 10.1126/science.1080372 [DOI] [PubMed] [Google Scholar]
- 11. Li Y, Kowdley KV. MicroRNAs in common human diseases. Genomics Proteomics Bioinf (2012) 10(5):246–53. doi: 10.1016/j.gpb.2012.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Iorio MV, Croce CM. MicroRNA dysregulation in cancer: diagnostics, monitoring and therapeutics. a comprehensive review. EMBO Mol Med (2012) 4(3):143–59. doi: 10.1002/emmm.201100209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. (2002) 99(24):15524–9. doi: 10.1073/pnas.242606799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res (2005) 65(16):7065–70. doi: 10.1158/0008-5472.CAN-05-1783 [DOI] [PubMed] [Google Scholar]
- 15. Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell (2006) 9(3):189–98. doi: 10.1016/j.ccr.2006.01.025 [DOI] [PubMed] [Google Scholar]
- 16. Cho WC, Chow AS, Au JS. MiR-145 inhibits cell proliferation of human lung adenocarcinoma by targeting EGFR and NUDT1. RNA Biol (2011) 8(1):125–31. doi: 10.4161/rna.8.1.14259 [DOI] [PubMed] [Google Scholar]
- 17. Shimizu S, Takehara T, Hikita H, Kodama T, Miyagi T, Hosui A, et al. The let-7 family of microRNAs inhibits bcl-xL expression and potentiates sorafenib-induced apoptosis in human hepatocellular carcinoma. J Hepatol (2010) 52(5):698–704. doi: 10.1016/j.jhep.2009.12.024 [DOI] [PubMed] [Google Scholar]
- 18. Fang Y, Xue JL, Shen Q, Chen J, Tian L. MicroRNA-7 inhibits tumor growth and metastasis by targeting the phosphoinositide 3-kinase/Akt pathway in hepatocellular carcinoma. Hepatology (2012) 55(6):1852–62. doi: 10.1002/hep.25576 [DOI] [PubMed] [Google Scholar]
- 19. Gao P, Xing AY, Zhou GY, Zhang TG, Zhang JP, Gao C, et al. The molecular mechanism of microRNA-145 to suppress invasion-metastasis cascade in gastric cancer. Oncogene (2013) 32(4):491–501. doi: 10.1038/onc.2012.61 [DOI] [PubMed] [Google Scholar]
- 20. Peng X, Guo W, Liu T, Wang X, Tu X, Xiong D, et al. Identification of miRs-143 and -145 that is associated with bone metastasis of prostate cancer and involved in the regulation of EMT. PloS One (2011) 6(5):e20341. doi: 10.1371/journal.pone.0020341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ostrycharz E, Hukowska-Szematowicz B. Micro-players of great significance-host microRNA signature in viral infections in humans and animals. Int J Mol Sci (2022) 23(18):10536. doi: 10.3390/ijms231810536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nahand JS, Mahjoubin-Tehran M, Moghoofei M, Pourhanifeh MH, Mirzaei HR, Asemi Z, et al. Exosomal miRNAs: novel players in viral infection. Epigenomics (2020) 12(4):353–70. doi: 10.2217/epi-2019-0192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Blount GS, Coursey L, Kocerha J. MicroRNA networks in cognition and dementia. Cells (2022) 11(12):1882. doi: 10.3390/cells11121882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Godlewski J, Lenart J, Salinska E. MicroRNA in brain pathology: neurodegeneration the other side of the brain cancer. Noncoding RNA (2019) 5(1):20. doi: 10.3390/ncrna5010020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Weidner J, Bartel S, Kılıç A, Zissler UM, Renz H, Schwarze J, et al. Spotlight on microRNAs in allergy and asthma. Allergy (2021) 76(6):1661–78. doi: 10.1111/all.14646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mirzaei R, Zamani F, Hajibaba M, Rasouli-Saravani A, Noroozbeygi M, Gorgani M, et al. The pathogenic, therapeutic and diagnostic role of exosomal microRNA in the autoimmune diseases. J Neuroimmunol (2021) 358:577640. doi: 10.1016/j.jneuroim.2021.577640 [DOI] [PubMed] [Google Scholar]
- 27. Hensley AP, McAlinden A. The role of microRNAs in bone development. Bone (2021) 143:115760. doi: 10.1016/j.bone.2020.115760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chan WCW, Tan Z, To MKT, Chan D. Regulation and role of transcription factors in osteogenesis. Int J Mol Sci (2021) 22(11):5445. doi: 10.3390/ijms22115445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chen R, Qiu H, Tong Y, Liao F, Hu X, Qiu Y, et al. MiRNA-19a-3p alleviates the progression of osteoporosis by targeting HDAC4 to promote the osteogenic differentiation of hMSCs. Biochem Biophys Res Commun (2019) 516(3):666–72. doi: 10.1016/j.bbrc.2019.06.083 [DOI] [PubMed] [Google Scholar]
- 30. Tan K, Peng YT, Guo P. MiR-29a promotes osteogenic differentiation of mesenchymal stem cells via targeting HDAC4. Eur Rev Med Pharmacol Sci (2018) 22(11):3318–26. doi: 10.26355/eurrev_201806_15151 [DOI] [PubMed] [Google Scholar]
- 31. Gao XL, Cao MG, Ai GG, Hu YB. Mir-98 reduces the expression of HMGA2 and promotes osteogenic differentiation of mesenchymal stem cells. Eur Rev Med Pharmacol Sci (2018) 22(11):3311–7. doi: 10.26355/eurrev_201806_15150 [DOI] [PubMed] [Google Scholar]
- 32. Xu D, Gao Y, Hu N, Wu L, Chen Q. miR-365 ameliorates dexamethasone-induced suppression of osteogenesis in MC3T3-E1 cells by targeting HDAC4. Int J Mol Sci (2017) 18(5):977. doi: 10.3390/ijms18050977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yan X, Wang H, Li Y, Jiang Y, Shao Q, Xu W. MicroRNA−92a overexpression promotes the osteogenic differentiation of bone mesenchymal stem cells by impeding Smad6−mediated runt−related transcription factor 2 degradation. Mol Med Rep (2018) 17(6):7821–6. doi: 10.3892/mmr.2018.8829 [DOI] [PubMed] [Google Scholar]
- 34. Lin Z, He H, Wang M, Liang J. MicroRNA-130a controls bone marrow mesenchymal stem cell differentiation towards the osteoblastic and adipogenic fate. Cell Prolif (2019) 52(6):e12688. doi: 10.1111/cpr.12688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kong L, Zuo R, Wang M, Wang W, Xu J, Chai Y, et al. Silencing MicroRNA-137-3p, which targets RUNX2 and CXCL12 prevents steroid-induced osteonecrosis of the femoral head by facilitating osteogenesis and angiogenesis. Int J Biol Sci (2020) 16(4):655–70. doi: 10.7150/ijbs.38713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Peng W, Zhu S, Li X, Weng J, Chen S. miR-27b-3p suppressed osteogenic differentiation of maxillary sinus membrane stem cells by targeting Sp7. Implant Dent (2017) 26(4):492–9. doi: 10.1097/ID.0000000000000637 [DOI] [PubMed] [Google Scholar]
- 37. Zhang SY, Gao F, Peng CG, Zheng CJ, Wu MF. miR-485-5p promotes osteoporosis via targeting osterix. Eur Rev Med Pharmacol Sci (2018) 22(15):4792–9. doi: 10.26355/eurrev_201808_15613 [DOI] [PubMed] [Google Scholar]
- 38. Hu H, Zhao C, Zhang P, Liu Y, Jiang Y, Wu E, et al. miR-26b modulates OA induced BMSC osteogenesis through regulating GSK3β/β-catenin pathway. Exp Mol pathol (2019) 107:158–64. doi: 10.1016/j.yexmp.2019.02.003 [DOI] [PubMed] [Google Scholar]
- 39. Lee EJ, Kim SM, Choi B, Kim EY, Chung YH, Lee EJ, et al. Interleukin-32 gamma stimulates bone formation by increasing miR-29a in osteoblastic cells and prevents the development of osteoporosis. Sci Rep (2017) 7:40240. doi: 10.1038/srep40240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tang X, Lin J, Wang G, Lu J. MicroRNA-433-3p promotes osteoblast differentiation through targeting DKK1 expression. PloS One (2017) 12(6):e0179860. doi: 10.1371/journal.pone.0179860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hwang S, Park SK, Lee HY, Kim SW, Lee JS, Choi EK, et al. miR-140-5p suppresses BMP2-mediated osteogenesis in undifferentiated human mesenchymal stem cells. FEBS Lett (2014) 588(17):2957–63. doi: 10.1016/j.febslet.2014.05.048 [DOI] [PubMed] [Google Scholar]
- 42. Zhang Y, Wei QS, Ding WB, Zhang LL, Wang HC, Zhu YJ, et al. Increased microRNA-93-5p inhibits osteogenic differentiation by targeting bone morphogenetic protein-2. PloS One (2017) 12(8):e0182678. doi: 10.1371/journal.pone.0182678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Liu K, Jing Y, Zhang W, Fu X, Zhao H, Zhou X, et al. Silencing miR-106b accelerates osteogenesis of mesenchymal stem cells and rescues against glucocorticoid-induced osteoporosis by targeting BMP2. Bone (2017) 97:130–8. doi: 10.1016/j.bone.2017.01.014 [DOI] [PubMed] [Google Scholar]
- 44. Lu XD, Han WX, Liu YX. Suppression of miR-451a accelerates osteogenic differentiation and inhibits bone loss via Bmp6 signaling during osteoporosis. BioMed Pharmacother (2019) 120:109378. doi: 10.1016/j.biopha.2019.109378 [DOI] [PubMed] [Google Scholar]
- 45. Zhang Y, Li S, Yuan S, Zhang H, Liu J. MicroRNA-23a inhibits osteogenesis of periodontal mesenchymal stem cells by targeting bone morphogenetic protein signaling. Arch Oral Biol (2019) 102:93–100. doi: 10.1016/j.archoralbio.2019.04.001 [DOI] [PubMed] [Google Scholar]
- 46. Liu H, Zhong L, Yuan T, Chen S, Zhou Y, An L, et al. MicroRNA-155 inhibits the osteogenic differentiation of mesenchymal stem cells induced by BMP9 via downregulation of BMP signaling pathway. Int J Mol Med (2018) 41(6):3379–93. doi: 10.3892/ijmm.2018.3526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhang HG, Wang XB, Zhao H, Zhou CN. MicroRNA-9-5p promotes osteoporosis development through inhibiting osteogenesis and promoting adipogenesis via targeting Wnt3a. Eur Rev Med Pharmacol Sci (2019) 23(2):456–63. doi: 10.26355/eurrev_201901_16855 [DOI] [PubMed] [Google Scholar]
- 48. Duan L, Zhao H, Xiong Y, Tang X, Yang Y, Hu Z, et al. miR-16-2* interferes with WNT5A to regulate osteogenesis of mesenchymal stem cells. Cell Physiol Biochem (2018) 51(3):1087–102. doi: 10.1159/000495489 [DOI] [PubMed] [Google Scholar]
- 49. Inoue K, Ng C, Xia Y, Zhao B. Regulation of osteoclastogenesis and bone resorption by miRNAs. Front Cell Dev Biol (2021) 9:651161. doi: 10.3389/fcell.2021.651161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hrdlicka HC, Lee SK, Delany AM. MicroRNAs are critical regulators of osteoclast differentiation. Curr Mol Biol Rep (2019) 5(1):65–74. doi: 10.1007/s40610-019-0116-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hackl M, Heilmeier U, Weilner S, Grillari J. Circulating microRNAs as novel biomarkers for bone diseases - complex signatures for multifactorial diseases? Mol Cell Endocrinol (2016) 432:83–95. doi: 10.1016/j.mce.2015.10.015 [DOI] [PubMed] [Google Scholar]
- 52. Baloun J, Pekacova A, Wenchich L, Hruskova H, Senolt L, Svec X, et al. Menopausal transition: prospective study of estrogen status, circulating MicroRNAs, and biomarkers of bone metabolism. Front Endocrinol (Lausanne) (2022) 13:864299. doi: 10.3389/fendo.2022.864299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Witwer KW. Circulating microRNA biomarker studies: pitfalls and potential solutions. Clin Chem (2015) 61(1):56–63. doi: 10.1373/clinchem.2014.221341 [DOI] [PubMed] [Google Scholar]
- 54. Thorlacius S, Struewing JP, Hartge P, Olafsdottir GH, Sigvaldason H, Tryggvadottir L, et al. Population-based study of risk of breast cancer in carriers of BRCA2 mutation. Lancet (London England) (1998) 352(9137):1337–9. doi: 10.1016/S0140-6736(98)03300-5 [DOI] [PubMed] [Google Scholar]
- 55. Struewing JP, Hartge P, Wacholder S, Baker SM, Berlin M, McAdams M, et al. The risk of cancer associated with specific mutations of BRCA1 and BRCA2 among ashkenazi jews. N Engl J Med (1997) 336(20):1401–8. doi: 10.1056/NEJM199705153362001 [DOI] [PubMed] [Google Scholar]
- 56. Greenland P, Alpert JS, Beller GA, Benjamin EJ, Budoff MJ, Fayad ZA, et al. 2010 ACCF/AHA guideline for assessment of cardiovascular risk in asymptomatic adults: executive summary: a report of the American college of cardiology Foundation/American heart association task force on practice guidelines. Circulation (2010) 122(25):2748–64. doi: 10.1016/j.jacc.2010.09.001 [DOI] [PubMed] [Google Scholar]
- 57. Levey AS, Coresh J, Balk E, Kausz AT, Levin A, Steffes MW, et al. K/DOQI clinical practice guidelines for chronic kidney disease: evaluation, classification, and stratification. Am J Kidney Dis (2002) 39(2 Suppl 1):S1–266. doi: 10.7326/0003-4819-139-2-200307150-00013 [DOI] [PubMed] [Google Scholar]
- 58. Farrell PM, Rosenstein BJ, White TB, Accurso FJ, Castellani C, Cutting GR, et al. Guidelines for diagnosis of cystic fibrosis in newborns through older adults: cystic fibrosis foundation consensus report. J Pediatr (2008) 153(2):S4–s14. doi: 10.1016/j.jpeds.2008.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. American Diabetes Association Professional Practice Committee . 2. classification and diagnosis of diabetes: standards of medical care in diabetes-2019. Diabetes Care (2019) 42(Suppl 1):S13–s28. doi: 10.2337/dc22-S002 [DOI] [PubMed] [Google Scholar]
- 60. Sandler HM, Eisenberger MA. Assessing and treating patients with increasing prostate specific antigen following radical prostatectomy. J Urol (2007) 178(3 Pt 2):S20–4. doi: 10.1016/j.juro.2007.04.034 [DOI] [PubMed] [Google Scholar]
- 61. Freedland SJ, Moul JW. Prostate specific antigen recurrence after definitive therapy. J Urol (2007) 177(6):1985–91. doi: 10.1016/j.juro.2007.01.137 [DOI] [PubMed] [Google Scholar]
- 62. Holbrook A, Schulman S, Witt DM, Vandvik PO, Fish J, Kovacs MJ, et al. Evidence-based management of anticoagulant therapy: antithrombotic therapy and prevention of thrombosis, 9th ed: American college of chest physicians evidence-based clinical practice guidelines. Chest (2012) 141(2 Suppl):e152S–e84S. doi: 10.1378/chest.11-2295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Miller BE, Tal-Singer R, Rennard SI, Furtwaengler A, Leidy N, Lowings M, et al. Plasma fibrinogen qualification as a drug development tool in chronic obstructive pulmonary disease. perspective of the chronic obstructive pulmonary disease biomarker qualification consortium. Am J Respir Crit Care Med (2016) 193(6):607–13. doi: 10.1164/rccm.201509-1722PP [DOI] [PubMed] [Google Scholar]
- 64. Scagliotti G, Hanna N, Fossella F, Sugarman K, Blatter J, Peterson P, et al. The differential efficacy of pemetrexed according to NSCLC histology: a review of two phase III studies. Oncologist (2009) 14(3):253–63. doi: 10.1634/theoncologist.2008-0232 [DOI] [PubMed] [Google Scholar]
- 65. Stohl W, Hilbert DM. The discovery and development of belimumab: the anti-BLyS-lupus connection. Nat Biotechnol (2012) 30(1):69–77. doi: 10.1038/nbt.2076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Wasung ME, Chawla LS, Madero M. Biomarkers of renal function, which and when? Clin Chim Acta (2015) 438:350–7. doi: 10.1016/j.cca.2014.08.039 [DOI] [PubMed] [Google Scholar]
- 67. Chung WH, Hung SI, Hong HS, Hsih MS, Yang LC, Ho HC, et al. Medical genetics: a marker for stevens-Johnson syndrome. Nature (2004) 428(6982):486. doi: 10.1038/428486a [DOI] [PubMed] [Google Scholar]
- 68. Lane NE. Epidemiology, etiology, and diagnosis of osteoporosis. Am J Obstet Gynecol (2006) 194(2 Suppl):S3–11. doi: 10.1016/j.ajog.2005.08.047 [DOI] [PubMed] [Google Scholar]
- 69. Al-Rawaf HA, Alghadir AH, Gabr SA. Circulating MicroRNA expression, vitamin d, and hypercortisolism as predictors of osteoporosis in elderly postmenopausal women. Dis Markers (2021) 2021:3719919. doi: 10.1155/2021/3719919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Johnston CB, Dagar M. Osteoporosis in older adults. Med Clin North Am (2020) 104(5):873–84. doi: 10.1016/j.mcna.2020.06.004 [DOI] [PubMed] [Google Scholar]
- 71. Johnell O, Kanis JA. An estimate of the worldwide prevalence and disability associated with osteoporotic fractures. Osteoporos Int (2006) 17(12):1726–33. doi: 10.1007/s00198-006-0172-4 [DOI] [PubMed] [Google Scholar]
- 72. Reid IR. A broader strategy for osteoporosis interventions. Nat Rev Endocrinol (2020) 16(6):333–9. doi: 10.1038/s41574-020-0339-7 [DOI] [PubMed] [Google Scholar]
- 73. Reginster JY, Burlet N. Osteoporosis: a still increasing prevalence. Bone (2006) 38(2 Suppl 1):S4–9. doi: 10.1016/j.bone.2005.11.024 [DOI] [PubMed] [Google Scholar]
- 74. Huai Y, Zhang W, Chen Z, Zhao F, Wang W, Dang K, et al. A comprehensive analysis of MicroRNAs in human osteoporosis. Front Endocrinol (Lausanne) (2020) 11:516213. doi: 10.3389/fendo.2020.516213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Bedene A, Mencej Bedrač S, Ješe L, Marc J, Vrtačnik P, Preželj J, et al. MiR-148a the epigenetic regulator of bone homeostasis is increased in plasma of osteoporotic postmenopausal women. Wien Klin Wochenschr (2016) 128(Suppl 7):519–26. doi: 10.1007/s00508-016-1141-3 [DOI] [PubMed] [Google Scholar]
- 76. Chen Z, Bemben MG, Bemben DA. Bone and muscle specific circulating microRNAs in postmenopausal women based on osteoporosis and sarcopenia status. Bone (2019) 120:271–8. doi: 10.1016/j.bone.2018.11.001 [DOI] [PubMed] [Google Scholar]
- 77. Ding H, Meng J, Zhang W, Li Z, Li W, Zhang M, et al. Medical examination powers miR-194-5p as a biomarker for postmenopausal osteoporosis. Sci Rep (2017) 7(1):16726. doi: 10.1038/s41598-017-17075-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Ismail SM, El Boghdady NA, Hamoud HS, Shabayek MI. Evaluation of circulating miRNA-208a-3p, miRNA-155-5p and miRNA-637 as potential non-invasive biomarkers and the possible mechanistic insights into pre- and postmenopausal osteoporotic females. Arch Biochem Biophys (2020) 684:108331. doi: 10.1016/j.abb.2020.108331 [DOI] [PubMed] [Google Scholar]
- 79. Kerschan-Schindl K, Hackl M, Boschitsch E, Föger-Samwald U, Nägele O, Skalicky S, et al. Diagnostic performance of a panel of miRNAs (OsteomiR) for osteoporosis in a cohort of postmenopausal women. Calcified Tissue Int (2021) 108(6):725–37. doi: 10.1007/s00223-020-00802-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Kocijan R, Weigl M, Skalicky S, Geiger E, Ferguson J, Leinfellner G, et al. MicroRNA levels in bone and blood change during bisphosphonate and teriparatide therapy in an animal model of postmenopausal osteoporosis. Bone (2020) 131:115104. doi: 10.1016/j.bone.2019.115104 [DOI] [PubMed] [Google Scholar]
- 81. Li H, Wang Z, Fu Q, Zhang J. Plasma miRNA levels correlate with sensitivity to bone mineral density in postmenopausal osteoporosis patients. Biomarkers (2014) 19(7):553–6. doi: 10.3109/1354750X.2014.935957 [DOI] [PubMed] [Google Scholar]
- 82. Ma J, Lin X, Chen C, Li S, Zhang S, Chen Z, et al. Circulating miR-181c-5p and miR-497-5p are potential biomarkers for prognosis and diagnosis of osteoporosis. J Clin Endocrinol Metab (2020) 105(5):1445–60. doi: 10.1210/clinem/dgz300 [DOI] [PubMed] [Google Scholar]
- 83. Meng J, Zhang D, Pan N, Sun N, Wang Q, Fan J, et al. Identification of miR-194-5p as a potential biomarker for postmenopausal osteoporosis. PeerJ (2015) 3:e971. doi: 10.7717/peerj.971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Zhao SL, Wen ZX, Mo XY, Zhang XY, Li HN, Cheung WH, et al. Bone-Metabolism-Related serum microRNAs to diagnose osteoporosis in middle-aged and elderly women. Diagn (Basel) (2022) 12(11):2872. doi: 10.3390/diagnostics12112872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Ciuffi S, Marini F, Fossi C, Donati S, Giusti F, Botta A, et al. Circulating MicroRNAs as biomarkers of osteoporosis and fragility fractures. J Clin Endocrinol Metab (2022) 107(8):2267–85. doi: 10.1210/clinem/dgac293 [DOI] [PubMed] [Google Scholar]
- 86. Ding W, Ding S, Li J, Peng Z, Hu P, Zhang T, et al. Aberrant expression of miR-100 in plasma of patients with osteoporosis and its potential diagnostic value. Clin Lab (2019) 65(9). doi: 10.7754/Clin.Lab.2019.190327 [DOI] [PubMed] [Google Scholar]
- 87. Lu Z, Wang D, Wang X, Zou J, Sun J, Bi Z. MiR-206 regulates the progression of osteoporosis via targeting HDAC4. Eur J Med Res (2021) 26(1):8. doi: 10.1186/s40001-021-00480-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Mandourah AY, Ranganath L, Barraclough R, Vinjamuri S, Hof RV, Hamill S, et al. Circulating microRNAs as potential diagnostic biomarkers for osteoporosis. Sci Rep (2018) 8(1):8421. doi: 10.1038/s41598-018-26525-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Nakashima H, Ando K, Kobayashi K, Seki T, Ishizuka S, Fujii R, et al. Associations of serum MicroRNA with bone mineral density in community-dwelling subjects: the yakumo study. BioMed Res Int (2020) 2020:5047243. doi: 10.1155/2020/5047243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Pepe J, Rossi M, Battafarano G, Vernocchi P, Conte F, Marzano V, et al. Characterization of extracellular vesicles in osteoporotic patients compared to osteopenic and healthy controls. J Bone Miner Res (2022) 37(11):2186–200. doi: 10.1002/jbmr.4688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Ramírez-Salazar EG, Carrillo-Patiño S, Hidalgo-Bravo A, Rivera-Paredez B, Quiterio M, Ramírez-Palacios P, et al. Serum miRNAs miR-140-3p and miR-23b-3p as potential biomarkers for osteoporosis and osteoporotic fracture in postmenopausal Mexican-mestizo women. Gene (2018) 679:19–27. doi: 10.1016/j.gene.2018.08.074 [DOI] [PubMed] [Google Scholar]
- 92. Seeliger C, Karpinski K, Haug AT, Vester H, Schmitt A, Bauer JS, et al. Five freely circulating miRNAs and bone tissue miRNAs are associated with osteoporotic fractures. J Bone Miner Res (2014) 29(8):1718–28. doi: 10.1002/jbmr.2175 [DOI] [PubMed] [Google Scholar]
- 93. Shuai Y, Liao L, Su X, Sha N, Li X, Wu Y, et al. Circulating microRNAs in serum as novel biomarkers for osteoporosis: a case-control study. Ther Adv Musculoskelet Dis (2020) 12:1759720x20953331. doi: 10.1177/1759720X20953331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Mäkitie RE, Hackl M, Weigl M, Frischer A, Kämpe A, Costantini A, et al. Unique, gender-dependent serum microRNA profile in PLS3 gene-related osteoporosis. J Bone Miner Res (2020) 35(10):1962–73. doi: 10.1002/jbmr.4097 [DOI] [PubMed] [Google Scholar]
- 95. Smieszek A, Marcinkowska K, Pielok A, Sikora M, Valihrach L, Marycz K. The role of miR-21 in osteoblasts-osteoclasts coupling In Vitro . Cells (2020) 9(2):479. doi: 10.3390/cells9020479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Sun M, Hu L, Wang S, Huang T, Zhang M, Yang M, et al. Circulating MicroRNA-19b identified from osteoporotic vertebral compression fracture patients increases bone formation. J Bone Miner Res (2020) 35(2):306–16. doi: 10.1002/jbmr.3892 [DOI] [PubMed] [Google Scholar]
- 97. Heilmeier U, Hackl M, Schroeder F, Torabi S, Kapoor P, Vierlinger K, et al. Circulating serum microRNAs including senescent miR-31-5p are associated with incident fragility fractures in older postmenopausal women with type 2 diabetes mellitus. Bone (2022) 158:116308. doi: 10.1016/j.bone.2021.116308 [DOI] [PubMed] [Google Scholar]
- 98. Quillen EE, Foster J, Sheldrake A, Stainback M, Glenn J, Cox LA, et al. Circulating miRNAs associated with bone mineral density in healthy adult baboons. J Orthop Res (2022) 40(8):1827–33. doi: 10.1002/jor.25215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Garg B, Malhotra R, Mittal S, Kumar A, Mehta N, Malik G, et al. Differential miRNA expression in osteoporotic elderly patients with hip fractures compared to young patients. Indian J Orthop (2022) 56(3):399–411. doi: 10.1007/s43465-021-00561-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Heilmeier U, Hackl M, Skalicky S, Weilner S, Schroeder F, Vierlinger K, et al. Serum miRNA signatures are indicative of skeletal fractures in postmenopausal women with and without type 2 diabetes and influence osteogenic and adipogenic differentiation of adipose tissue-derived mesenchymal stem cells in vitro. J Bone Miner Res (2016) 31(12):2173–92. doi: 10.1002/jbmr.2897 [DOI] [PubMed] [Google Scholar]
- 101. Kocijan R, Muschitz C, Geiger E, Skalicky S, Baierl A, Dormann R, et al. Circulating microRNA signatures in patients with idiopathic and postmenopausal osteoporosis and fragility fractures. J Clin Endocrinol Metab (2016) 101(11):4125–34. doi: 10.1210/jc.2016-2365 [DOI] [PubMed] [Google Scholar]
- 102. Ladang A, Beaudart C, Locquet M, Reginster JY, Bruyère O, Cavalier E. Evaluation of a panel of MicroRNAs that predicts fragility fracture risk: a pilot study. Calcified Tissue Int (2020) 106(3):239–47. doi: 10.1007/s00223-019-00628-8 [DOI] [PubMed] [Google Scholar]
- 103. Lincoln S, Morse LR, Troy K, Mattson N, Nguyen N, Battaglino RA. MicroRNA-148a-3p is a candidate mediator of increased bone marrow adiposity and bone loss following spinal cord injury. Front Endocrinol (Lausanne) (2022) 13:910934. doi: 10.3389/fendo.2022.910934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Messner Z, Carro-Vazquez D, Haschka J, Grillari J, Resch H, Muschitz C, et al. Circulating miRNAs respond to denosumab treatment after two years in postmenopausal women with osteoporosis. J Clin Endocrinol Metab (2022) 108(5):1154–65. doi: 10.1210/clinem/dgac667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Panach L, Mifsut D, Tarín JJ, Cano A, García-Pérez M. Serum circulating MicroRNAs as biomarkers of osteoporotic fracture. Calcified Tissue Int (2015) 97(5):495–505. doi: 10.1007/s00223-015-0036-z [DOI] [PubMed] [Google Scholar]
- 106. Shi H, Jiang X, Xu C, Cheng Q. MicroRNAs in serum exosomes as circulating biomarkers for postmenopausal osteoporosis. Front Endocrinol (Lausanne) (2022) 13:819056. doi: 10.3389/fendo.2022.819056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Weigl M, Kocijan R, Ferguson J, Leinfellner G, Heimel P, Feichtinger X, et al. Longitudinal changes of circulating miRNAs during bisphosphonate and teriparatide treatment in an animal model of postmenopausal osteoporosis. J Bone Miner Res (2021) 36(6):1131–44. doi: 10.1002/jbmr.4276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Xu J, Li M, Pei W, Ding J, Pan Y, Peng H, et al. Reduced circulating levels of miR-491-5p and miR-485-3p are associated with the occurrence of vertebral fractures in postmenopausal women with osteoporosis. Genet Res (Camb). (2022) 2022:3838126. doi: 10.1155/2022/3838126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Xu Y, Li D, Zhu Z, Li L, Jin Y, Ma C, et al. miR−27a−3p negatively regulates osteogenic differentiation of MC3T3−E1 preosteoblasts by targeting osterix. Mol Med Rep (2020) 22(3):1717–26. doi: 10.3892/mmr.2020.11246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Yavropoulou MP, Anastasilakis AD, Makras P, Tsalikakis DG, Grammatiki M, Yovos JG. Expression of microRNAs that regulate bone turnover in the serum of postmenopausal women with low bone mass and vertebral fractures. Eur J endocrinol (2017) 176(2):169–76. doi: 10.1530/EJE-16-0583 [DOI] [PubMed] [Google Scholar]
- 111. Zarecki P, Hackl M, Grillari J, Debono M, Eastell R. Serum microRNAs as novel biomarkers for osteoporotic vertebral fractures. Bone (2020) 130:115105. doi: 10.1016/j.bone.2019.115105 [DOI] [PubMed] [Google Scholar]
- 112. Weilner S, Skalicky S, Salzer B, Keider V, Wagner M, Hildner F, et al. Differentially circulating miRNAs after recent osteoporotic fractures can influence osteogenic differentiation. Bone (2015) 79:43–51. doi: 10.1016/j.bone.2015.05.027 [DOI] [PubMed] [Google Scholar]
- 113. Ritter J, Bielack SS. Osteosarcoma. Ann Oncol (2010) 21(Suppl 7):vii320–5. doi: 10.1093/annonc/mdq276 [DOI] [PubMed] [Google Scholar]
- 114. Beird HC, Bielack SS, Flanagan AM, Gill J, Heymann D, Janeway KA, et al. Osteosarcoma. Nat Rev Dis Primers (2022) 8(1):77. doi: 10.1038/s41572-022-00409-y [DOI] [PubMed] [Google Scholar]
- 115. Allen-Rhoades W, Kurenbekova L, Satterfield L, Parikh N, Fuja D, Shuck RL, et al. Cross-species identification of a plasma microRNA signature for detection, therapeutic monitoring, and prognosis in osteosarcoma. Cancer Med (2015) 4(7):977–88. doi: 10.1002/cam4.438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Cuscino N, Raimondi L, De Luca A, Carcione C, Russelli G, Conti L, et al. Gathering novel circulating exosomal microRNA in osteosarcoma cell lines and possible implications for the disease. Cancers (2019) 11(12):1924. doi: 10.3390/cancers11121924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Andersen GB, Knudsen A, Hager H, Hansen LL, Tost J. miRNA profiling identifies deregulated miRNAs associated with osteosarcoma development and time to metastasis in two large cohorts. Mol Oncol (2018) 12(1):114–31. doi: 10.1002/1878-0261.12154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Cao L, Wang J, Wang PQ. MiR-326 is a diagnostic biomarker and regulates cell survival and apoptosis by targeting bcl-2 in osteosarcoma. BioMed Pharmacother (2016) 84:828–35. doi: 10.1016/j.biopha.2016.10.008 [DOI] [PubMed] [Google Scholar]
- 119. Cong C, Wang W, Tian J, Gao T, Zheng W, Zhou C. Identification of serum miR-124 as a biomarker for diagnosis and prognosis in osteosarcoma. Cancer biomark (2018) 21(2):449–54. doi: 10.3233/CBM-170672 [DOI] [PubMed] [Google Scholar]
- 120. Diao ZB, Sun TX, Zong Y, Lin BC, Xia YS. Identification of plasma microRNA-22 as a marker for the diagnosis, prognosis, and chemosensitivity prediction of osteosarcoma. J Int Med Res (2020) 48(12):300060520967818. doi: 10.1177/0300060520967818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Dong J, Liu Y, Liao W, Liu R, Shi P, Wang L. miRNA-223 is a potential diagnostic and prognostic marker for osteosarcoma. J Bone Oncol (2016) 5(2):74–9. doi: 10.1016/j.jbo.2016.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Hua J, Liu D, Cao L, Wang D, Wu T, Lin F, et al. Diagnostic and prognostic values of blood microRNA-Let7A for osteosarcoma. J Bone Oncol (2018) 12:65–8. doi: 10.1016/j.jbo.2018.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Hua Y, Jin Z, Zhou F, Zhang YQ, Zhuang Y. The expression significance of serum MiR-21 in patients with osteosarcoma and its relationship with chemosensitivity. Eur Rev Med Pharmacol Sci (2017) 21(13):2989–94. [PubMed] [Google Scholar]
- 124. Lian H, Zhou Y, Sun Z, Liu K. MicroRNA34a is associated with chemotherapy resistance, metastasis, recurrence, survival, and prognosis in patient with osteosarcoma. Med (Baltimore) (2022) 101(38):e30722. doi: 10.1097/MD.0000000000030722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Liu W, Zhao X, Zhang YJ, Fang GW, Xue Y. MicroRNA-375 as a potential serum biomarker for the diagnosis, prognosis, and chemosensitivity prediction of osteosarcoma. J Int Med Res (2018) 46(3):975–83. doi: 10.1177/0300060517734114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Ma W, Zhang X, Chai J, Chen P, Ren P, Gong M. Circulating miR-148a is a significant diagnostic and prognostic biomarker for patients with osteosarcoma. Tumour Biol (2014) 35(12):12467–72. doi: 10.1007/s13277-014-2565-x [DOI] [PubMed] [Google Scholar]
- 127. Niu J, Sun Y, Guo Q, Niu D, Liu B. Serum miR-95-3p is a diagnostic and prognostic marker for osteosarcoma. Springerplus (2016) 5(1):1947. doi: 10.1186/s40064-016-3640-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Pang PC, Shi XY, Huang WL, Sun K. miR-497 as a potential serum biomarker for the diagnosis and prognosis of osteosarcoma. Eur Rev Med Pharmacol Sci (2016) 20(18):3765–9. [PubMed] [Google Scholar]
- 129. Shi L, Xie C, Zhu J, Chen X. Downregulation of serum miR-194 predicts poor prognosis in osteosarcoma patients. Ann Diagn Pathol (2020) 46:151488. doi: 10.1016/j.anndiagpath.2020.151488 [DOI] [PubMed] [Google Scholar]
- 130. Tian Q, Jia J, Ling S, Liu Y, Yang S, Shao Z. A causal role for circulating miR-34b in osteosarcoma. Eur J Surg Oncol (2014) 40(1):67–72. doi: 10.1016/j.ejso.2013.08.024 [DOI] [PubMed] [Google Scholar]
- 131. Wang SN, Luo S, Liu C, Piao Z, Gou W, Wang Y, et al. miR-491 inhibits osteosarcoma lung metastasis and chemoresistance by targeting αB-crystallin. Mol Ther (2017) 25(9):2140–9. doi: 10.1016/j.ymthe.2017.05.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Wang T, Ji F, Dai Z, Xie Y, Yuan D. Increased expression of microRNA-191 as a potential serum biomarker for diagnosis and prognosis in human osteosarcoma. Cancer biomark (2015) 15(5):543–50. doi: 10.3233/CBM-150493 [DOI] [PubMed] [Google Scholar]
- 133. Xie XY, Chen XM, Shi L, Liu JW. Increased expression of microRNA-26a-5p predicted a poor survival outcome in osteosarcoma patients: an observational study. Med (Baltimore) (2021) 100(12):e24765. doi: 10.1097/MD.0000000000024765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Yang Z, Zhang Y, Zhang X, Zhang M, Liu H, Zhang S, et al. Serum microRNA-221 functions as a potential diagnostic and prognostic marker for patients with osteosarcoma. BioMed Pharmacother (2015) 75:153–8. doi: 10.1016/j.biopha.2015.07.018 [DOI] [PubMed] [Google Scholar]
- 135. Yao ZS, Li C, Liang D, Jiang XB, Tang JJ, Ye LQ, et al. Diagnostic and prognostic implications of serum miR-101 in osteosarcoma. Cancer biomark (2018) 22(1):127–33. doi: 10.3233/CBM-171103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Yuan J, Chen L, Chen X, Sun W, Zhou X. Identification of serum microRNA-21 as a biomarker for chemosensitivity and prognosis in human osteosarcoma. J Int Med Res (2012) 40(6):2090–7. doi: 10.1177/030006051204000606 [DOI] [PubMed] [Google Scholar]
- 137. Zhang L, Cao H, Gu G, Hou D, You Y, Li X, et al. Exosomal MiR-199a-5p inhibits tumorigenesis and angiogenesis by targeting VEGFA in osteosarcoma. Front Oncol (2022) 12:884559. doi: 10.3389/fonc.2022.884559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Zhou G, Lu M, Chen J, Li C, Zhang J, Chen J, et al. Identification of miR-199a-5p in serum as noninvasive biomarkers for detecting and monitoring osteosarcoma. Tumour Biol (2015) 36(11):8845–52. doi: 10.1007/s13277-015-3421-3 [DOI] [PubMed] [Google Scholar]
- 139. Zhou S, Wang B, Hu J, Zhou Y, Jiang M, Wu M, et al. miR-421 is a diagnostic and prognostic marker in patients with osteosarcoma. Tumour Biol (2016) 37(7):9001–7. doi: 10.1007/s13277-015-4578-5 [DOI] [PubMed] [Google Scholar]
- 140. Monterde-Cruz L, Ramírez-Salazar EG, Rico-Martínez G, Linares-González LM, Guzmán-González R, Delgado-Cedillo E, et al. Circulating miR-215-5p and miR-642a-5p as potential biomarker for diagnosis of osteosarcoma in Mexican population. Hum Cell (2018) 31(4):292–9. doi: 10.1007/s13577-018-0214-1 [DOI] [PubMed] [Google Scholar]
- 141. Nakka M, Allen-Rhoades W, Li Y, Kelly AJ, Shen J, Taylor AM, et al. Biomarker significance of plasma and tumor miR-21, miR-221, and miR-106a in osteosarcoma. Oncotarget (2017) 8(57):96738–52. doi: 10.18632/oncotarget.18236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Fujiwara T, Uotani K, Yoshida A, Morita T, Nezu Y, Kobayashi E, et al. Clinical significance of circulating miR-25-3p as a novel diagnostic and prognostic biomarker in osteosarcoma. Oncotarget (2017) 8(20):33375–92. doi: 10.18632/oncotarget.16498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Huang C, Sun Y, Ma S, Vadamootoo AS, Wang L, Jin C. Identification of circulating miR-663a as a potential biomarker for diagnosing osteosarcoma. Pathol Res Pract (2019) 215(6):152411. doi: 10.1016/j.prp.2019.04.003 [DOI] [PubMed] [Google Scholar]
- 144. Dailey DD, Hess AM, Bouma GJ, Duval DL. MicroRNA expression changes and integrated pathways associated with poor outcome in canine osteosarcoma. Front Vet Sci (2021) 8:637622. doi: 10.3389/fvets.2021.637622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Gong L, Bao Q, Hu C, Wang J, Zhou Q, Wei L, et al. Exosomal miR-675 from metastatic osteosarcoma promotes cell migration and invasion by targeting CALN1. Biochem Biophys Res Commun (2018) 500(2):170–6. doi: 10.1016/j.bbrc.2018.04.016 [DOI] [PubMed] [Google Scholar]
- 146. Heishima K, Meuten T, Yoshida K, Mori T, Thamm DH. Prognostic significance of circulating microRNA-214 and -126 in dogs with appendicular osteosarcoma receiving amputation and chemotherapy. BMC Vet Res (2019) 15(1):39. doi: 10.1186/s12917-019-1776-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Hong Q, Fang J, Pang Y, Zheng J. Prognostic value of the microRNA-29 family in patients with primary osteosarcomas. Med Oncol (2014) 31(8):37. doi: 10.1007/s12032-014-0037-1 [DOI] [PubMed] [Google Scholar]
- 148. Huang C, Wang Q, Ma S, Sun Y, Vadamootoo AS, Jin C. A four serum-miRNA panel serves as a potential diagnostic biomarker of osteosarcoma. Int J Clin Oncol (2019) 24(8):976–82. doi: 10.1007/s10147-019-01433-x [DOI] [PubMed] [Google Scholar]
- 149. Jerez S, Araya H, Hevia D, Irarrázaval CE, Thaler R, van Wijnen AJ, et al. Extracellular vesicles from osteosarcoma cell lines contain miRNAs associated with cell adhesion and apoptosis. Gene (2019) 710:246–57. doi: 10.1016/j.gene.2019.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Kosela-Paterczyk H, Paziewska A, Kulecka M, Balabas A, Kluska A, Dabrowska M, et al. Signatures of circulating microRNA in four sarcoma subtypes. J Cancer (2020) 11(4):874–82. doi: 10.7150/jca.34723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Li Q, Song S, Ni G, Li Y, Wang X. Serum miR-542-3p as a prognostic biomarker in osteosarcoma. Cancer biomark (2018) 21(3):521–6. doi: 10.3233/CBM-170255 [DOI] [PubMed] [Google Scholar]
- 152. Lian F, Cui Y, Zhou C, Gao K, Wu L. Identification of a plasma four-microRNA panel as potential noninvasive biomarker for osteosarcoma. PloS One (2015) 10(3):e0121499. doi: 10.1371/journal.pone.0121499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Luo H, Ye Z. Identification of serum miR-337-3p, miR-484, miR-582, and miR-3677 as promising biomarkers for osteosarcoma. Clin Lab (2021) 67(4). doi: 10.7754/Clin.Lab.2020.200455 [DOI] [PubMed] [Google Scholar]
- 154. Ouyang L, Liu P, Yang S, Ye S, Xu W, Liu X. A three-plasma miRNA signature serves as novel biomarkers for osteosarcoma. Med Oncol (2013) 30(1):340. doi: 10.1007/s12032-012-0340-7 [DOI] [PubMed] [Google Scholar]
- 155. Yang L, Li H, Huang A. MiR-429 and MiR-143-3p function as diagnostic and prognostic markers for osteosarcoma. Clin Lab (2020) 66(10). doi: 10.7754/Clin.Lab.2020.191237 [DOI] [PubMed] [Google Scholar]
- 156. Zhang C, Yao C, Li H, Wang G, He X. Serum levels of microRNA-133b and microRNA-206 expression predict prognosis in patients with osteosarcoma. Int J Clin Exp Pathol (2014) 7(7):4194–203. [PMC free article] [PubMed] [Google Scholar]
- 157. Balamuth NJ, Womer RB. Ewing's sarcoma. Lancet Oncol (2010) 11(2):184–92. doi: 10.1016/S1470-2045(09)70286-4 [DOI] [PubMed] [Google Scholar]
- 158. Crow J, Samuel G, Farrow E, Gibson M, Johnston J, Guest E, et al. MicroRNA content of Ewing sarcoma derived extracellular vesicles leads to biomarker potential and identification of a previously undocumented EWS-FLI1 translocation. biomark Insights (2022) 17:11772719221132693. doi: 10.1177/11772719221132693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Nie CL, Ren WH, Ma Y, Xi JS, Han B. Circulating miR-125b as a biomarker of ewing's sarcoma in Chinese children. Genet Mol Res (2015) 14(4):19049–56. doi: 10.4238/2015.December.29.12 [DOI] [PubMed] [Google Scholar]
- 160. Urdinez J, Boro A, Mazumdar A, Arlt MJ, Muff R, Botter SM, et al. The miR-143/145 cluster, a novel diagnostic biomarker in chondrosarcoma, acts as a tumor suppressor and directly inhibits fascin-1. J Bone Miner Res (2020) 35(6):1077–91. doi: 10.1002/jbmr.3976 [DOI] [PubMed] [Google Scholar]
- 161. Ali SA, Espin-Garcia O, Wong AK, Potla P, Pastrello C, McIntyre M, et al. Circulating microRNAs differentiate fast-progressing from slow-progressing and non-progressing knee osteoarthritis in the osteoarthritis initiative cohort. Ther Adv Musculoskelet Dis (2022) 14:1759720x221082917. doi: 10.1177/1759720X221082917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Pertusa C, Tarín JJ, Cano A, García-Pérez M, Mifsut D. Serum microRNAs in osteoporotic fracture and osteoarthritis: a genetic and functional study. Sci Rep (2021) 11(1):19372. doi: 10.1038/s41598-021-98789-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Wan L, Zhao Q, Niu G, Xiang T, Ding C, Wang S. Plasma miR-136 can be used to screen patients with knee osteoarthritis from healthy controls by targeting IL-17. Exp Ther Med (2018) 16(4):3419–24. doi: 10.3892/etm.2018.6625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Xia S, Tian H, Fan L, Zheng J. Peripheral blood miR-181-5p serves as a marker for screening patients with osteoarthritis by targeting TNFα. Clin Lab (2017) 63(11):1819–25. doi: 10.7754/Clin.Lab.2017.170521 [DOI] [PubMed] [Google Scholar]
- 165. Papanota AM, Tsiakanikas P, Kontos CK, Malandrakis P, Liacos CI, Ntanasis-Stathopoulos I, et al. A molecular signature of circulating MicroRNA can predict osteolytic bone disease in multiple myeloma. Cancers (2021) 13(15):3877. doi: 10.3390/cancers13153877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Moura SR, Abreu H, Cunha C, Ribeiro-Machado C, Oliveira C, Barbosa MA, et al. Circulating microRNAs correlate with multiple myeloma and skeletal osteolytic lesions. Cancers (2021) 13(21):5258. doi: 10.3390/cancers13215258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Hao M, Zang M, Zhao L, Deng S, Xu Y, Qi F, et al. Serum high expression of miR-214 and miR-135b as novel predictor for myeloma bone disease development and prognosis. Oncotarget (2016) 7(15):19589–600. doi: 10.18632/oncotarget.7319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Fang Y, Qiu J, Jiang ZB, Xu SR, Zhou ZH, He RL. Increased serum levels of miR-214 in patients with PCa with bone metastasis may serve as a potential biomarker by targeting PTEN. Oncol letters (2019) 17(1):398–405. doi: 10.3892/ol.2018.9522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Guo X, Han T, Hu P, Guo X, Zhu C, Wang Y, et al. Five microRNAs in serum as potential biomarkers for prostate cancer risk assessment and therapeutic intervention. Int Urol Nephrol (2018) 50(12):2193–200. doi: 10.1007/s11255-018-2009-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Lu Z, Hou J, Li X, Zhou J, Luo B, Liang S, et al. Exosome-derived miRNAs as potential biomarkers for prostate bone metastasis. Int J Gen Med (2022) 15:5369–83. doi: 10.2147/IJGM.S361981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Peng P, Chen T, Wang Q, Zhang Y, Zheng F, Huang S, et al. Decreased miR-218-5p levels as a serum biomarker in bone metastasis of prostate cancer. Oncol Res Treat (2019) 42(4):165–85. doi: 10.1159/000495473 [DOI] [PubMed] [Google Scholar]
- 172. Rode MP, Silva AH, Cisilotto J, Rosolen D, Creczynski-Pasa TB. miR-425-5p as an exosomal biomarker for metastatic prostate cancer. Cell Signal (2021) 87:110113. doi: 10.1016/j.cellsig.2021.110113 [DOI] [PubMed] [Google Scholar]
- 173. Wa Q, Huang S, Pan J, Tang Y, He S, Fu X, et al. miR-204-5p represses bone metastasis via inactivating NF-κB signaling in prostate cancer. Mol Ther Nucleic Acids (2019) 18:567–79. doi: 10.1016/j.omtn.2019.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Wang Y, Fang YX, Dong B, Du X, Wang J, Wang X, et al. Discovery of extracellular vesicles derived miR-181a-5p in patient's serum as an indicator for bone-metastatic prostate cancer. Theranostics (2021) 11(2):878–92. doi: 10.7150/thno.49186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Zhang HL, Qin XJ, Cao DL, Zhu Y, Yao XD, Zhang SL, et al. An elevated serum miR-141 level in patients with bone-metastatic prostate cancer is correlated with more bone lesions. Asian J Androl (2013) 15(2):231–5. doi: 10.1038/aja.2012.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Wu K, Feng J, Lyu F, Xing F, Sharma S, Liu Y, et al. Exosomal miR-19a and IBSP cooperate to induce osteolytic bone metastasis of estrogen receptor-positive breast cancer. Nat Commun (2021) 12(1):5196. doi: 10.1038/s41467-021-25473-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Zhao FL, Hu GD, Wang XF, Zhang XH, Zhang YK, Yu ZS. Serum overexpression of microRNA-10b in patients with bone metastatic primary breast cancer. J Int Med Res (2012) 40(3):859–66. doi: 10.1177/147323001204000304 [DOI] [PubMed] [Google Scholar]
- 178. Xiang ZL, Zhao XM, Zhang L, Yang P, Fan J, Tang ZY, et al. MicroRNA-34a expression levels in serum and intratumoral tissue can predict bone metastasis in patients with hepatocellular carcinoma. Oncotarget (2016) 7(52):87246–56. doi: 10.18632/oncotarget.13531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Valencia K, Martín-Fernández M, Zandueta C, Ormazábal C, Martínez-Canarias S, Bandrés E, et al. miR-326 associates with biochemical markers of bone turnover in lung cancer bone metastasis. Bone (2013) 52(1):532–9. doi: 10.1016/j.bone.2012.10.033 [DOI] [PubMed] [Google Scholar]
- 180. Xu S, Yang F, Liu R, Li X, Fan H, Liu J, et al. Serum microRNA-139-5p is downregulated in lung cancer patients with lytic bone metastasis. Oncol Rep (2018) 39(5):2376–84. doi: 10.3892/or.2018.6316 [DOI] [PubMed] [Google Scholar]
- 181. Yang XR, Pi C, Yu R, Fan XJ, Peng XX, Zhang XC, et al. Correlation of exosomal microRNA clusters with bone metastasis in non-small cell lung cancer. Clin Exp Metastasis (2021) 38(1):109–17. doi: 10.1007/s10585-020-10062-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Zeng X, Liu D, Peng G, Liu J, Yang H. MiroRNA-31-3p promotes the invasion and metastasis of non-Small-Cell lung cancer cells by targeting forkhead box 1 (FOXO1). Comput Math Methods Med (2022) 2022:4597087. doi: 10.1155/2022/4597087 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 183. Bourgery M, Ekholm E, Hiltunen A, Heino TJ, Pursiheimo JP, Bendre A, et al. Signature of circulating small non-coding RNAs during early fracture healing in mice. Bone Rep (2022) 17:101627. doi: 10.1016/j.bonr.2022.101627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Dietz C, Müller M, Reinhold AK, Karch L, Schwab B, Forer L, et al. What is normal trauma healing and what is complex regional pain syndrome I? an analysis of clinical and experimental biomarkers. Pain (2019) 160(10):2278–89. doi: 10.1097/j.pain.0000000000001617 [DOI] [PubMed] [Google Scholar]
- 185. Xiong Y, Cao F, Chen L, Yan C, Zhou W, Chen Y, et al. Identification of key microRNAs and target genes for the diagnosis of bone nonunion. Mol Med Rep (2020) 21(4):1921–33. doi: 10.3892/mmr.2020.10996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. García-Giménez JL, Rubio-Belmar PA, Peiró-Chova L, Hervás D, González-Rodríguez D, Ibañez-Cabellos JS, et al. Circulating miRNAs as diagnostic biomarkers for adolescent idiopathic scoliosis. Sci Rep (2018) 8(1):2646. doi: 10.1038/s41598-018-21146-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Wang Y, Zhang H, Yang G, Xiao L, Li J, Guo C. Dysregulated bone metabolism is related to high expression of miR-151a-3p in severe adolescent idiopathic scoliosis. BioMed Res Int (2020) 2020:4243015. doi: 10.1155/2020/4243015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Hong G, Han X, He W, Xu J, Sun P, Shen Y, et al. Analysis of circulating microRNAs aberrantly expressed in alcohol-induced osteonecrosis of femoral head. Sci Rep (2019) 9(1):18926. doi: 10.1038/s41598-019-55188-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Kao GS, Tu YK, Sung PH, Wang FS, Lu YD, Wu CT, et al. MicroRNA-mediated interacting circuits predict hypoxia and inhibited osteogenesis of stem cells, and dysregulated angiogenesis are involved in osteonecrosis of the femoral head. Int Orthop (2018) 42(7):1605–14. doi: 10.1007/s00264-018-3895-x [DOI] [PubMed] [Google Scholar]
- 190. Liu GZ, Chen C, Kong N, Tian R, Li YY, Li Z, et al. Identification of potential miRNA biomarkers for traumatic osteonecrosis of femoral head. J Cell Physiol (2020) 235(11):8129–40. doi: 10.1002/jcp.29467 [DOI] [PubMed] [Google Scholar]
- 191. Legrand MA, Millet M, Merle B, Rousseau JC, Hemmendinger A, Gineyts E, et al. A signature of circulating miRNAs associated with fibrous dysplasia of bone: the mirDys study. J Bone Miner Res (2020) 35(10):1881–92. doi: 10.1002/jbmr.4111 [DOI] [PubMed] [Google Scholar]
- 192. Luo W, Dong Y, Hu T, Liu D, Wei X, Ma W, et al. 25(OH)D status and expression of miR-140 in the serum of patients with developmental dysplasia of the hip. Nutrition (2021) 81:110896. doi: 10.1016/j.nut.2020.110896 [DOI] [PubMed] [Google Scholar]
- 193. Nickolas TL, Chen N, McMahon DJ, Dempster D, Zhou H, Dominguez J, 2nd, et al. A microRNA approach to discriminate cortical low bone turnover in renal osteodystrophy. JBMR Plus (2020) 4(5):e10353. doi: 10.1002/jbm4.10353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Rajkumar SV, Dimopoulos MA, Palumbo A, Blade J, Merlini G, Mateos MV, et al. International myeloma working group updated criteria for the diagnosis of multiple myeloma. Lancet Oncol (2014) 15(12):e538–48. doi: 10.1016/S1470-2045(14)70442-5 [DOI] [PubMed] [Google Scholar]
- 195. Fornetti J, Welm AL, Stewart SA. Understanding the bone in cancer metastasis. J Bone Miner Res (2018) 33(12):2099–113. doi: 10.1002/jbmr.3618 [DOI] [PubMed] [Google Scholar]
- 196. Abramoff B, Caldera FE. Osteoarthritis: pathology, diagnosis, and treatment options. Med Clin North Am (2020) 104(2):293–311. doi: 10.1016/j.mcna.2019.10.007 [DOI] [PubMed] [Google Scholar]
- 197. Binderup HG, Madsen JS, Heegaard NHH, Houlind K, Andersen RF, Brasen CL. Quantification of microRNA levels in plasma - impact of preanalytical and analytical conditions. PloS One (2018) 13(7):e0201069. doi: 10.1371/journal.pone.0201069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. Condrat CE, Thompson DC, Barbu MG, Bugnar OL, Boboc A, Cretoiu D, et al. miRNAs as biomarkers in disease: latest findings regarding their role in diagnosis and prognosis. Cells (2020) 9(2):276. doi: 10.3390/cells9020276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199. Mori MA, Ludwig RG, Garcia-Martin R, Brandão BB, Kahn CR. Extracellular miRNAs: from biomarkers to mediators of physiology and disease. Cell Metab (2019) 30(4):656–73. doi: 10.1016/j.cmet.2019.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200. Pfeffer SR, Yang CH, Pfeffer LM. The role of miR-21 in cancer. Drug Dev Res (2015) 76(6):270–7. doi: 10.1002/ddr.21257 [DOI] [PubMed] [Google Scholar]
- 201. Chi LH, Cross RSN, Redvers RP, Davis M, Hediyeh-zadeh S, Mathivanan S, et al. MicroRNA-21 is immunosuppressive and pro-metastatic via separate mechanisms. Oncogenesis (2022) 11(1):38. doi: 10.1038/s41389-022-00413-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202. English PA, Williams JA, Martini JF, Motzer RJ, Valota O, Buller RE. A case for the use of receiver operating characteristic analysis of potential clinical efficacy biomarkers in advanced renal cell carcinoma. Future Oncol (2016) 12(2):175–82. doi: 10.2217/fon.15.290 [DOI] [PMC free article] [PubMed] [Google Scholar]