Abstract
Human monoclonal antibodies (mAbs) against the central repeat and junction domain of Plasmodium falciparum circumsporozoite protein (PfCSP) have been studied extensively to guide malaria vaccine design compared to antibodies against the PfCSP C terminus. Here, we describe the molecular characteristics and protective potential of 73 germline and mutated human mAbs against the highly immunogenic PfCSP C‐terminal domain. Two mAbs recognized linear epitopes in the C‐terminal linker with sequence similarity to repeat and junction motifs, whereas all others targeted conformational epitopes in the α‐thrombospondin repeat (α‐TSR) domain. Specificity for the polymorphic Th2R/Th3R but not the conserved RII+/CS.T3 region in the α‐TSR was associated with IGHV3‐21/IGVL3‐21 or IGLV3‐1 gene usage. Although the C terminus specific mAbs showed signs of more efficient affinity maturation and class‐switching compared to anti‐repeat mAbs, live sporozoite binding and inhibitory activity was limited to a single C‐linker reactive mAb with cross‐reactivity to the central repeat and junction. The data provide novel insights in the human anti‐C‐linker and anti‐α‐TSR antibody response that support exclusion of the PfCSP C terminus from malaria vaccine designs.
Keywords: Affinity maturation, Antibodies, Malaria, PfCSP, Plasmodium
Subject Categories: Immunology; Microbiology, Virology & Host Pathogen Interaction; Structural Biology
Plasmodium falciparum circumsporozoite protein (PfCSP) is a promising malaria vaccine target. Antibodies against the PfCSP central repeat domain and N‐terminal junction mediate protection from the infection, but the role of antibodies against the C‐terminal domain (C‐CSP) remains controversial.

The paper explained.
Problem
RTS,S/AS01, the only available malaria vaccine shows limited efficacy. RTS,S/AS01 induces antibodies against the central repeat domain and C terminus of Plasmodium falciparum circumsporozoite protein (PfCSP). Anti‐repeat antibodies mediate protection from the infection but the role of antibodies against the C‐terminal domain (C‐CSP), which is immunodominant in recombinant PfCSP remains unclear.
Results
By generating a large panel of human monoclonal PfCSP antibodies, we demonstrate that C‐CSP specific antibodies lack parasite‐inhibitory activity independently of their C‐CSP fine epitope specificity. We show that high‐affinity binding C‐CSP specific antibodies are part of the naïve human B cell repertoire and undergo efficient affinity maturation. The most abundant C‐CSP specific antibodies targeted polymorphic epitopes in the α‐TSR domain and were frequently encoded by IGHV3‐21 genes.
Impact
The apparent lack of protective antibody epitopes in the PfCSP C terminus suggests that the design of a second generation PfCSP‐based malaria vaccine may benefit from focusing the humoral immune response on the repeat domain and junction while avoiding non‐protective antibody responses against C‐CSP.
Introduction
Plasmodium falciparum (Pf) malaria is a mosquito‐borne parasitic disease that is highly endemic in sub‐Saharan Africa, where it remains the primary cause of childhood illness and death (WHO, 2021). RTS,S/AS01 (Mosquirix™), the only marketed malaria vaccine, has been recommended for widespread use among children in areas with high to moderate Pf transmission.
RTS,S/AS01 is a subunit vaccine that targets Pf circumsporozoite protein (PfCSP), the major protein on the surface of sporozoites that are transmitted to humans during the blood meal of infected Anopheles mosquitoes (Swearingen et al, 2016). PfCSP consists of three domains: a poorly characterized N‐terminal domain (N‐CSP), an unstructured central repeat region with large numbers of repeating four amino‐acid motifs, and a C‐terminal domain (C‐CSP) with a linker (C‐linker) and α‐thrombospondin type‐1 repeat (α‐TSR) subdomain that is attached to the cell membrane by a GPI anchor (Dame et al, 1984; McCutchan et al, 1996; Wang et al, 2005; Doud et al, 2012). RTS,S/AS01 contains a genetic fusion protein of an N‐terminally truncated form of PfCSP from the Pf clone 3D7, derived from the laboratory strain NF54, and hepatitis B surface antigen (HbsAg), which is complexed with free HbsAg to form virus‐like particles and boost immunogenicity (Gordon et al, 1995; Stoute et al, 1997; Bojang et al, 2001; Casares et al, 2010; Agnandji et al, 2011). The PfCSP part of RTS,S comprises 18.5 of the 38 repeating asparagine (N)—alanine (A)—asparagine (N)—proline (P) motifs in the central repeat domain (Zavala et al, 1985; Stoute et al, 1997) and the complete C‐CSP. The characteristic NANP motifs are fully conserved in Pf parasites (with only slight variations in the number of repeating units) independent of their geographic origin and are long known to be the target epitopes of protective antibodies (Nardin et al, 1982; Zavala et al, 1983).
The successful clinical development of RTS,S/AS01 in contrast to the other numerous malaria vaccine candidates confirms the potency of PfCSP as a promising vaccine target. However, the overall efficacy is limited and protection is short‐lived (Agnandji et al, 2011; Asante et al, 2011; Olotu et al, 2011, 2016). To guide the design of a more efficacious PfCSP‐based malaria vaccine, recent studies have characterized large numbers of human monoclonal antibodies (mAbs) against PfCSP (Oyen et al, 2017; Triller et al, 2017; Kisalu et al, 2018; Murugan et al, 2018; Tan et al, 2018; Wang et al, 2020). Their molecular and functional characterization provided deep insights in the development and potency of anti‐repeat antibodies, and identified new epitopes of protective mAbs in the N‐terminal junction, with alternating NANP and NANP‐like (NPDP, NVDP) motifs that are not included in RTS,S.
In contrast to the large numbers of well‐characterized antibodies against the central repeat domain and N‐terminal junction, the role of antibodies against N‐CSP and C‐CSP remains controversial. N‐CSP is overall poorly immunogenic and undergoes proteolytic cleavage at region I (RI), a stretch of six amino acids that is highly conserved in all Plasmodium species (Coppi et al, 2005; Espinosa et al, 2015). Human monoclonal antibodies against N‐CSP distal to RI have not been described, and the few known mouse monoclonal antibodies show little sporozoite reactivity and parasite inhibitory activity (Herrera et al, 2015; Thai et al, 2020).
Although immunization with recombinant PfCSP induces dominant humoral responses against C‐CSP in animal models and humans (Genito et al, 2017; Cawlfield et al, 2019; Hutter et al, 2022), the limited number of human C‐CSP mAbs that have been described so far did not allow deep analyses of their gene features, epitope breadth or preferences, and parasite‐inhibitory activity. Two target sites of human monoclonal antibodies have been identified in the α‐TSR subdomain of C‐CSP: the highly polymorphic Th2R/Th3R region and the conserved RII+/CS.T3 region (Scally et al, 2018; Beutler et al, 2022). However, whether binding to these epitopes is associated with specific Ig genes, whether other epitopes are targeted by anti‐C‐CSP antibodies, and whether epitope specificity is linked to differences in parasite inhibition remains unclear. To address these questions, we characterized the molecular and functional properties, as well as binding‐specificity of a large panel of recombinant human monoclonal antibodies cloned from memory B cells of malaria‐naïve individuals immunized with aseptic, cryopreserved, radiation‐attenuated Pf sporozoites (PfSPZ Vaccine; Mordmuller et al, 2022).
Results
Immunization with radiation‐attenuated Plasmodium falciparum sporozoites (PfSPZ) induces antibody responses against C‐CSP
To identify individuals with antibody responses against C‐CSP, we measured the humoral response in sera from 12 malaria‐naive European volunteers 28 days after three direct venous inoculation (DVI) immunizations with 900,000 aseptic, purified, radiation‐attenuated Pf NF54 sporozoites (Sanaria® PfSPZ Vaccine; Mordmuller et al, 2022). Comparison of the IgG response against a peptide representing the central NANP repeat (NANP5) and against C‐CSP showed responses to both domains (Figs 1A and EV1A; Appendix Table S1).
Figure 1. B cell response against the PfCSP C terminus.
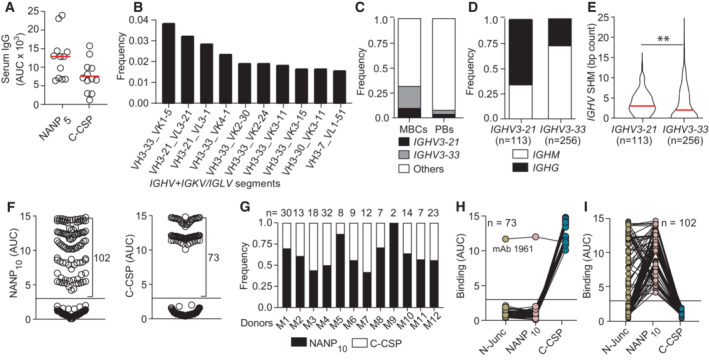
-
AAnti‐NANP5 and anti‐C‐CSP serum IgG. Circles represent ELISA area under the curve (AUC) values for individual donors (n = 12).
-
BTen most frequent Ig heavy and light chain V gene pairs in PfCSP‐reactive memory B cells (MBCs; n = 1,172).
-
CUsage frequency of IGHV3‐21, IGHV3‐33 and other IGHV genes in PfCSP‐reactive MBCs (n = 1,172) and plasmablasts (PBs; n = 2,380).
-
D, EIsotype distribution (D) and somatic hypermutation (SHM) count (E) in IGHV3‐21 and IGHV3‐33 genes from MBCs in (B, C).
-
FNANP10 (left) and C‐CSP (right) binding of FL‐CSP‐reactive mAbs (n = 177). Circles represent ELISA AUC values for each mAb.
-
GFrequency of NANP10 and C‐CSP reactive mAbs per donor. The number of tested mAbs is indicated.
-
H, ICross‐reactivity of C‐CSP (H) and NANP10 (I) reactive mAbs with the PfCSP N‐junc, NANP10 and C‐CSP; n indicates the number of mAbs tested.
Data information: Red lines in (A and E) indicate mean values. Black horizontal lines in (F), (H) and (I) indicate the threshold for binding. (E). The statistical significance in E was assessed by two‐tailed Mann–Whitney test: **P = 0.0017. Data in (A) are representative of two independent technical replicates. Data in (F, H and I) are obtained from three independent technical replicates and indicated as mean.
Source data are available online for this figure.
Figure EV1. Immunization with radiation‐attenuated Plasmodium falciparum sporozoites (PfSPZ Vac) induces strong antibody responses against the PfCSP C terminus.
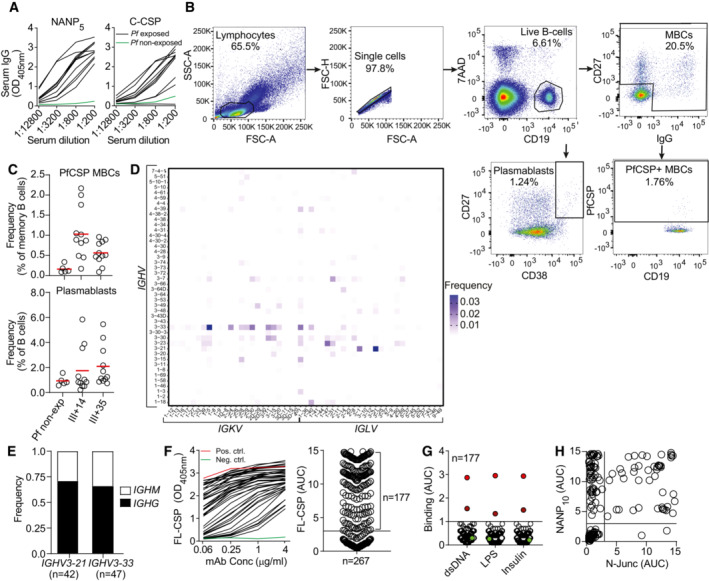
- Representative ELISA curves of serum IgG reactivity against PfCSP‐derived NANP5 peptide and C‐CSP domain.
- Flow‐cytometric single‐cell isolation strategy for PfCSP‐reactive memory B cells and plasmablasts from a representative immunized donor.
- Frequency of PfCSP‐reactive memory B cells (upper panel) and plasmablasts (lower panel) in PBMCs of immunized donors at the indicated time points (III + 14 and III + 35) and non‐immunized donors (Pf non‐exp).
- Paired Ig heavy and light chain V gene usage in PfCSP‐reactive memory B cells (n = 1,172).
- Isotype distribution of IGHV3‐21‐ and IGHV3‐33‐encoded mAbs.
- ELISA binding strength of FL‐CSP‐reactive mAbs to N‐Junc peptide vs NANP10.
Data information: Data in (C) represent one biological measurement obtained prior to cell sorting. Data in (F and H) indicate means from three independent technical replicates. Data in (G) are representative of two independent technical replicates.
To directly compare the anti‐C‐CSP and anti‐repeat antibody response at monoclonal level, we isolated single circulating PfCSP‐reactive memory B cells from all donors at 14 and 35 days after the third immunization (III + 14, III + 35; Fig EV1B and C) and amplified the paired immunoglobulin (Ig) heavy and light chain genes. Sequencing of the RT‐PCR Ig gene products showed that in all donors at both time points, the anti‐PfCSP response was dominated by memory B cells expressing VH3‐21 and VH3‐33 Igs, frequently in association with Vλ3‐21 or Vλ3‐1 and Vκ1‐5 light chains, respectively (Figs 1B and EV1D). Both IGHV genes were enriched in the PfCSP‐reactive memory B cell pool compared to circulating plasmablasts from the same individuals and time points that represent the B cell response to all sporozoite antigens (Fig 1C). The high abundance of VH3‐33 expressing PfCSP‐reactive memory B cells and frequent association with Vκ1‐5 suggested that these cells recognized the central repeat, similar to VH3‐33 antibodies induced by immunization with non‐irradiated sporozoites (Murugan et al, 2018). Compared to VH3‐33 antibodies, which were mostly IgM carrying a few somatic hypermutations (SHM), the VH3‐21 response was dominated by IgG cells with higher mean SHM counts (Fig 1D and E), indicating their participation in germinal center (GC) responses.
To determine how VH3‐21 usage was linked to PfCSP‐reactivity, we cloned and expressed 267 monoclonal antibodies (mAbs) covering the overall repertoire diversity of PfCSP‐reactive memory B cells, including 42 VH3‐21 and 47 VH3‐33 antibodies with similar isotype distribution (Fig EV1E; Appendix Table S2 and Dataset EV1). From this panel, 177 (66%) recombinant mAbs showed ELISA reactivity with full‐length PfCSP (FL‐CSP) and lacked binding to non‐related antigens, demonstrating their specificity for PfCSP (Fig EV1F and G). Of these FL‐CSP specific mAbs, 102 showed reactivity to a peptide representing the central NANP repeat (NANP10), and 73 mAbs recognized the C‐CSP domain (Fig 1F). Two mAbs lacked binding to NANP10 or C‐CSP, but bound a peptide covering the junction motifs (N‐junc) (Fig EV1H). NANP‐reactive mAbs were identified in all donors and C‐CSP‐reactive mAbs in all but one donor (Fig 1G).
To determine whether any of the C‐CSP‐reactive antibodies showed cross‐reactivity with the repeat domain including the N‐terminal junction, as previously reported (Murugan et al, 2020), we measured binding of the mAbs to NANP10 and the N‐junc peptide. With one exception (mAb 1961), all C‐CSP‐reactive mAbs were highly specific, whereas most of the NANP10‐binders showed cross‐reactivity to the junction as expected (Fig 1H and I). Thus, immunization with radiation‐attenuated sporozoites elicited PfCSP C‐CSP specific and repeat/junction cross‐reactive humoral and memory B cell responses associated with IGHV3‐21 and IGHV3‐33 gene signatures, respectively.
C‐CSP specific mAbs are frequently encoded by IGHV3‐21
To further characterize the panel of 73 C‐CSP specific mAbs, we determined their binding strength by surface plasmon resonance (SPR; Fig 2A). The mAbs recognized C‐CSP with a wide range of affinities (10−5 to 10−10 M). Ig gene sequence analysis showed that half (37/73) of all C‐CSP binders were encoded by IGHV3‐21 most of which paired with IGLV3‐21 or IGLV1‐3 genes, demonstrating the strong link between this gene combination and anti‐C‐CSP antibody reactivity (Figs 2B and EV2A). The other half of the anti‐C‐CSP antibodies used diverse gene combinations and showed no significant difference in affinity, isotype distribution or somatic mutation load compared to VH3‐21 mAbs (Figs 2B and EV2B–D). As expected, VH3‐33 heavy chains paired with Vκ1‐5 light chains were highly abundant among the NANP‐reactive mAbs, similar to VH3‐33/Vκ1‐5 anti‐repeat antibodies induced by immunization with non‐irradiated sporozoites (Fig EV2A and E).
Figure 2. C‐CSP specific mAbs are frequently encoded by IGHV3‐21 .
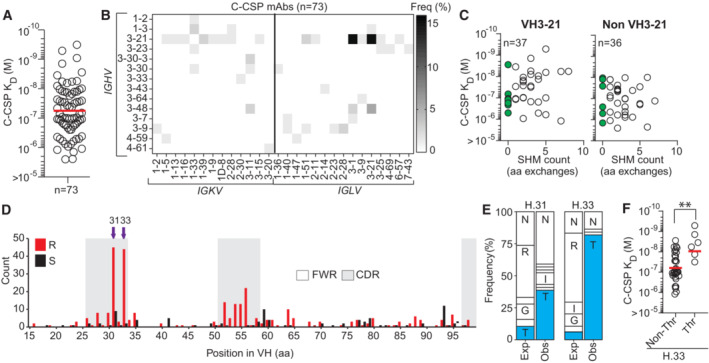
- SPR affinity of C‐CSP reactive mAbs.
- Frequency of mAbs encoded by the indicated IGHV and IGKV or IGLV pairs.
- VH SHM load of VH3‐21 and non‐VH3‐21 mAbs. mAbs with unmutated VH are highlighted in green; n indicates the number of mAbs tested.
- Amino acid (aa) VH replacement (red bars) and silent (black bars) SHM in VH3‐21 mAbs (n = 113). FWR, framework region; CDR, complementarity‐determining region.
- C‐CSP affinity for selected VH3‐21 mAbs with or without Thr mutation at position H.33.
Data information: The statistical significance in (F) was assessed by two‐tailed Mann–Whitney test: **P = 0.0019. Red horizontal lines in (A, F) indicate geometric mean values. Data in (A, C and F) are representative of two independent technical replicates.
Source data are available online for this figure.
Figure EV2. Molecular characteristics and affinity of C‐CSP specific mAbs.
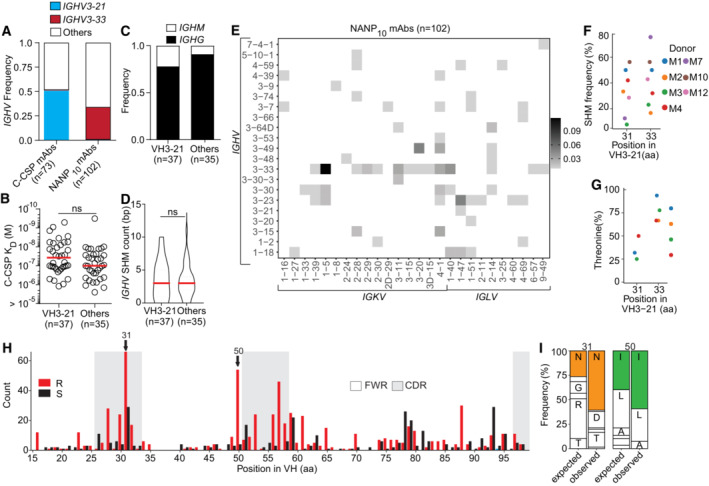
-
AIGHV3‐21 or IGHV3‐33 gene frequency in C‐CSP‐ and NANP10‐reactive mAbs.
-
B–DComparison of C‐CSP specific mAbs encoded by IGHV3‐21 and other IGHV genes. Anti‐C‐CSP SPR affinity (B), isotype distribution (C) and IGHV SHM count (D).
-
EFrequency of NANP10‐reactive mAbs encoded by the indicated IGHV + IGKV and IGHV + IGLV combinations (n = 102).
-
F, GPercentage of VH aa replacement SHM (F) and frequency of Thr at position H.31 and H.33 (G) in VH3‐21 mAbs at donor level.
-
HAmino acid (aa) VH replacement (red bars) and silent (black bars) SHM in VH3‐33 mAbs (n = 256). FWR, framework region; CDR, complementarity‐determining region.
-
IObserved (Obs) aa usage frequency at position H.31 and H.50 in VH3‐33 mAbs carrying a replacement mutation at these positions compared to the expected (Exp) neutral mutation model (Yaari et al, 2013; Gupta et al, 2015). Single‐letters indicate aa residues: A, Ala; D, Asp; G, Gly; I, Ile; L, Leu; N, Asn; R, Arg; T, Thr.
Data information: Data in (F and G) represent donors with at least 5 VH3‐21 mAbs and ≥ 4 mAbs with mutations at position H.33, respectively. Red lines in (B) and (D) indicate geometric and arithmetic mean values, respectively. ns, non‐significant, two‐tailed Mann–Whitney test (B, D). Data in (B) are representative of two independent technical replicates.
Although most C‐CSP specific antibodies were class‐switched and carried somatic mutations as signs of affinity maturation, we identified several VH3‐21 and non‐VH3‐21 germline antibodies with high C‐CSP affinity suggesting that the naïve human B cell repertoire contains numerous C‐CSP‐reactive precursor cells (Fig 2C). Sequence alignments of all VH3‐21 antibodies showed strong signs of selection indicated by enrichment of replacement mutations in CDRs compared to framework regions (FWR; Fig 2D). Almost half of the VH3‐21 antibodies carried replacement mutations at two positions in HCDR1 (H.31, H.33) and threonine was strongly enriched in these positions compared to the neutral mutation model (Figs 2D and E, and EV2F and G; Yaari et al, 2013; Gupta et al, 2015). VH3‐21 mAbs carrying the selected H.S33T mutation showed on average higher C‐CSP affinities than mAbs with the germline residue H.S33 or with non‐selected mutations demonstrating the role of the H.S33T exchange in affinity maturation (Fig 2F). The data shows a strong link of VH3‐21 gene segment usage with C‐CSP reactivity and provides direct evidence for effective affinity maturation of the anti‐C‐CSP response similar to the selection of H.S31N and H.V50I mutations in VH3‐33 antibodies (Fig EV2H and I), which mediate high affinity to repeating NANP motifs (Imkeller et al, 2018).
C‐CSP specific antibodies preferentially target two distinct conformational epitopes in the α‐TSR domain
To determine which of the two C‐terminal subregions (Fig 3A) the C‐CSP specific and C‐CSP cross‐reactive antibodies recognized, we measured binding to the C‐linker (aa 273–310) and the α‐TSR domain (aa 311–384) by ELISA (Figs 3B and EV3A). With two exceptions that recognized the C‐linker region (mAb 3764, and mAb 1961 with NANP10 and N‐junc cross‐reactivity; Fig 1H), all mAbs showed specificity for the α‐TSR domain.
Figure 3. Epitope specificity of C‐CSP specific mAbs.
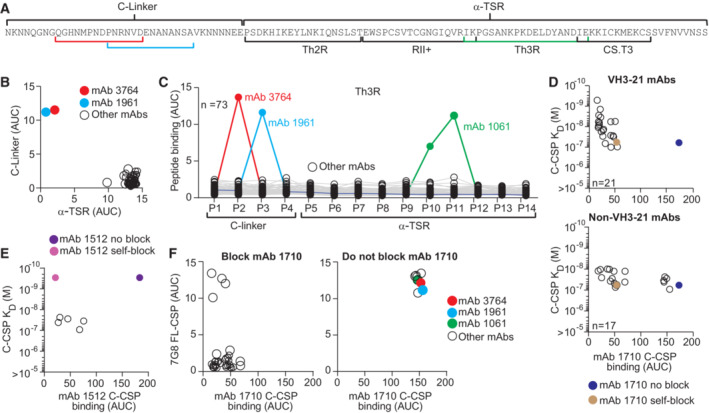
-
ANF54 PfCSP C‐CSP aa sequence. The C‐linker and α‐TSR domain with Th2R, RII+, Th3R and CS.T3 are indicated.
-
B, CELISA reactivity of C‐CSP‐reactive mAbs (n = 73) with the C‐linker and α‐TSR domain (B) and overlapping C‐CSP peptides (P1‐P14; Appendix Table S1 for sequences) (C).
-
DC‐CSP reactivity of the α‐TSR‐specific mAb 1710 (Scally et al, 2018) in a blocking ELISA with C‐CSP specific VH3‐21 (n = 21; upper panel) or non‐VH3‐21 (n = 17; lower panel) mAbs with the indicated SPR affinities. mAb 1710 binding without blocking (blue) and after self‐blocking (gold) is shown for comparison.
-
EC‐CSP reactivity of mAb 1512 (Beutler et al, 2022) in a blocking ELISA with mAbs (with the indicated SPR affinities; open circles) that do not block C‐CSP binding of mAb 1710 (from D lower panel). mAb 1512 binding without blocking (purple) and after self‐blocking (magenta) is shown for comparison.
-
FELISA cross‐reactivity with Pf‐7G8 FL‐CSP of CSP‐specific mAbs that block (left) or do not block (right) mAb 1710 binding to NF54 C‐CSP (D).
Data information: Highlighted mAbs (B, C and F) with their binding sites (highlighted in A) are mAb 3764 (C‐linker specific; red), mAb 1961 (C‐linker, repeat and junction cross‐reactive; light blue), mAb 1061 (α‐TSR peptide 10 and peptide 11 specific; green). Data in (B–F) are representative of two independent technical replicates.
Source data are available online for this figure.
Figure EV3. C‐CSP specific mAbs target preferentially two distinct conformational epitopes in the α‐TSR domain.
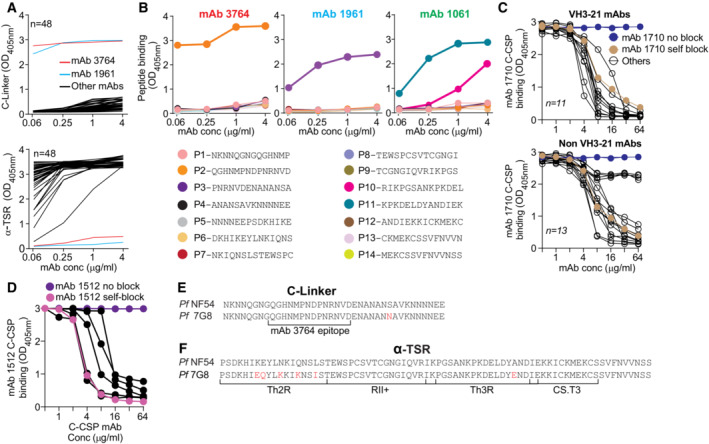
-
ARepresentative ELISA binding curves for all C‐CSP specific mAbs (black lines) to the C‐linker (upper panel) and α‐TSR (lower panel). The C‐linker specific mAb 3764 (red line) and C‐linker cross‐reactive mAb 1961 (light blue line) are highlighted.
-
BRepresentative ELISA binding curves to overlapping peptides (P1‐P14) covering the complete C‐CSP for mAb 3764 (left), mAb 1961 (middle) and mAb 1061 (right). Amino acid sequences of the peptides are indicated.
-
CRepresentative ELISA curves illustrating the ability of individual VH3‐21 (upper panel) and non‐VH3‐21 (lower panel) C‐CSP specific mAbs (black lines) to block binding of mAb 1710 (Scally et al, 2018) to C‐CSP. mAb 1710 C‐CSP binding without blocking (blue) and after self‐blocking (golden) is shown for comparison.
-
DELISA curves showing C‐CSP reactivity of mAb 1512 (Beutler et al, 2022) in a blocking ELISA with mAbs that do not block C‐CSP binding of mAb 1710 (black). mAb 1512 binding without blocking (purple) and after self‐blocking (magenta) is shown for comparison.
-
E, FAlignment of the PfCSP NF54 and 7G8 C‐linker (E) and α‐TSR (F) aa sequences. Amino acids that differ are highlighted in red.
To define whether any of the C‐CSP‐reactive antibodies recognized linear epitopes, we tested their binding to 14 overlapping peptides (P1‐14) covering the complete NF54 C‐CSP (Figs 3C and EV3B; Appendix Table S1). Only three mAbs showed reactivity in these assays: the C‐linker specific mAb 3764 with specificity for P2 spanning aa 281–294 (QGHNMPNDPNRNVD) and the α‐TSR‐reactive mAb 1061, which bound P10 and P11 covering aa 345–366 in Th3R region. These two peptides overlap by six aa (KPKDEL), suggesting that the mAb 1061 target epitope covers aa KPKDEL. The C‐CSP, junction and repeat cross‐reactive mAb 1961 showed reactivity with P3 (PNRNVDENANANSA) suggesting that C‐CSP binding of this mAb might be mediated via the NANA motif, similar to the previously reported C‐CSP, junction and repeat cross‐reactive mAb 3246 (Murugan et al, 2020). All other C‐CSP specific mAbs (70/73) lacked peptide reactivity indicating that they recognized conformational epitopes.
Similar to the previously reported anti‐α‐TSR mAb 1710 (Scally et al, 2018), many of these antibodies were encoded by IGHV3‐21, suggesting that these antibodies and mAb 1710 might recognize the same or overlapping epitopes in Th2R and Th3R. To address this question, we selected 21 IGHV3‐21‐encoded and 17 non‐IGHV3‐21 encoded C‐CSP specific mAbs with similar or higher C‐CSP affinity compared to mAb 1710 (≤ 10−7 M) and tested their ability to block mAb 1710 binding to C‐CSP by ELISA (Figs 3D and EV3C; Appendix Table S3). All 21 VH3‐21 and most of the non‐VH3‐21 (12/17) mAbs blocked binding of mAb 1710 demonstrating that the vast majority of anti‐C‐CSP antibodies shared specificity for the same or overlapping epitopes in the Th2R/Th3R. Only five of the 17 non‐VH3‐21 mAbs with diverse Ig genes did not interfere with mAb 1710 binding (Fig 3D). To determine their epitope specificity, we tested their ability to block mAb 1512, an RTS,S/AS01‐induced C‐CSP mAb with specificity for a conformational epitope in the conserved RII+/CS.T3 of the α‐TSR subdomain (Beutler et al, 2022). All five antibodies blocked mAb 1512 binding, demonstrating their specificity for the conserved RII+/CS.T3 epitope independent of their Ig gene usage (Figs 3E and EV3D).
The Th2R and to lesser extent the C‐linker and Th3R are highly polymorphic (Gandhi et al, 2012; Aragam et al, 2013). To evaluate whether the C‐CSP specific antibodies were able to accommodate these polymorphisms, we tested their ability to bind to PfCSP from 7G8, which differs from NF54 C‐CSP by seven aa: one aa in the C‐linker and six aa in the α‐TSR region (five in Th2R and one in Th3R; Fig EV3E and F). With four exceptions, none of the mAb 1710‐blocking mAbs recognized 7G8 PfCSP (Fig 3E). In contrast, mAbs 3764, 1961 and 1061 with linear peptide epitopes in the C‐linker and Th3R, respectively, and the five mAbs with reactivity to the conformational mAb 1512 epitope in RII+/CS.T3 showed cross‐reactivity with 7G8 (Fig 3F). Thus, only antibodies against the conserved linear or conformational C‐CSP epitopes not overlapping with the polymorphic mAb 1710 binding site showed 7G8 cross‐reactivity.
In summary, the vast majority of anti‐C‐CSP antibodies showed the same specificity as mAb 1710, demonstrating the strong immunogenicity of their polymorphic target epitopes in the α‐TSR domain. Binding to this site was strongly linked with IGHV3‐21 gene usage since all VH3‐21 mAbs bound this epitope. Antibodies against conserved epitopes in the α‐TSR domain or the C‐linker (conformational or linear) used diverse IgH and IgL combinations and showed cross‐reactivity to PfCSP from 7G8, but were overall rare.
C‐CSP specific antibodies fail to bind live sporozoites and lack parasite inhibitory activity
Differences in epitope specificity and Ig gene usage of antibodies might be linked to their parasite binding and inhibitory capacities. We selected 15 representative antibodies with high C‐CSP affinity against the linear and conformational epitopes in the C‐linker and α‐TSR domain and measured their ability to bind live fluorescent P. berghei sporozoites expressing PfCSP (PbPfCSP(mCherry), Fig EV4A; Appendix Table S4). Compared to the anti‐repeat mAb 2A10 (Triller et al, 2017), which showed strong binding at low concentration (1 μg/ml) in flow cytometry, none of the mAb 1710‐like or mAb 1512‐like anti‐α‐TSR mAbs recognized live sporozoites even at 100 μg/ml regardless of their epitope specificity and affinity (Figs 4A and EV4B), supporting the earlier observations that the α‐TSR domain on the surface of live sporozoites was not accessible for antibody binding (Scally et al, 2018; Wang et al, 2021). Weak binding was only observed for the C‐linker specific mAb 3764 at high (100 μg/ml) but not at low (1 μg/ml) concentration. However, the cross‐reactive mAb 1961 strongly bound to the sporozoites similar to the positive control anti‐repeat mAb 2A10 (Fig 4B–D). In agreement with their inability to bind live sporozoites, none of the anti‐C‐CSP specific antibodies showed Pf sporozoite traversal‐inhibitory capacity even at a high concentration (100 μg/ml), including the high‐affinity C‐linker specific mAb 3764, which weakly bound sporozoites (Fig 4E). Only the repeat and junction cross‐reactive mAb 1961 showed in vitro sporozoite‐inhibitory activity in the range of mAbs 2A10 and 317, a high affinity human anti‐NANP mAb (Oyen et al, 2017; Triller et al, 2017). Nevertheless, the capacity of mAb 1961 to protect from the development of parasitemia in vivo after passive transfer in mice was significantly lower than that of mAb 317 and similar to mAb 1710 (Figs 4F and EV4C; Appendix Table S5).
Figure EV4. Gating strategy to assess mAb binding to live PbPfCSP(mCherry) sporozoites.
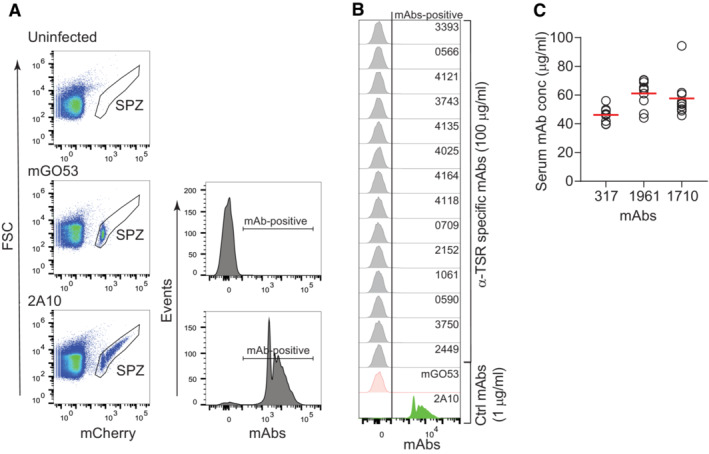
- Flow cytometric gating strategy to analyze the proportion of mAb‐positive PbPfCSP(mCherry) live sporozoites. Live mCherry‐positive sporozoites (SPZ) were gated. Salivary gland extracts from uninfected mosquitoes were used as sporozoite gating control. The sporozoite binding profile of the negative control mAb mGO53 (Wardemann et al, 2003) and the positive control anti‐NANP mAb 2A10 (Triller et al, 2017) are shown for comparison.
- Representative flow‐cytometric profiles of live PbPfCSP(mCherry) sporozoites recognized by α‐TSR‐specific mAbs (100 μg/ml) as determined by flow cytometry. The positive control anti‐NANP mAb 2A10 (1 μg/ml; Triller et al, 2017) and negative control non‐PfCSP reactive mAb mGO53 (1 μg/ml; Wardemann et al, 2003) are shown for comparison.
- mAb serum concentrations in individual mice (n = 10 for mAb 317, n = 8 for mAb 1961, and n = 9 for mAb 1710) after passive transfer of the indicated monoclonal antibodies. The data in C only included mice that had detectable serum IgG concentrations of the indicated mAbs at the time of the challenge.
Figure 4. C‐CSP specific mAbs lack anti‐parasite activity.
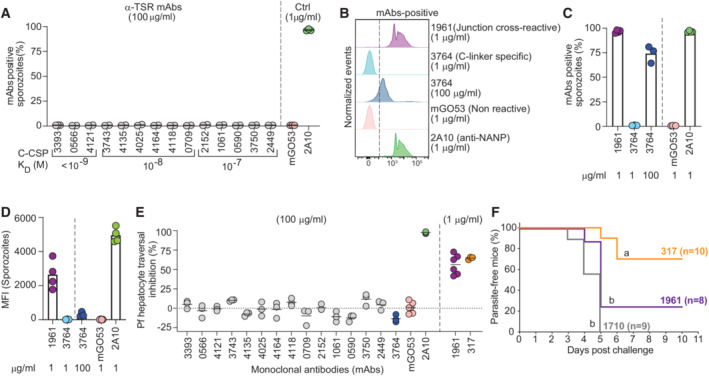
-
APercent of live PbPfCSP(mCherry) sporozoites recognized by α‐TSR‐specific mAbs (100 μg/ml) as determined by flow cytometry, with the respective affinities measured by SPR. The positive control anti‐NANP mAb 2A10 (1 μg/ml; Triller et al, 2017) and negative control non‐PfCSP reactive mAb mGO53 (1 μg/ml; Wardemann et al, 2003) are shown for comparison (n = 2–3).
-
B–DRepresentative flow cytometry profiles (B), percentage of mAb‐positive live PbPfCSP(mCherry) sporozoites (C), and mean fluorescence intensity (MFI) (D) for the junction cross‐reactive mAb 1961, C‐linker‐specific mAb 3764 and the control mAbs 2A10 and mGO53 at the indicated concentrations (n = 3–4).
-
EPf sporozoite hepatocyte traversal inhibitory capacity of the indicated C‐CSP reactive, C‐CSP cross‐reactive (1961), and control (2A10, mGO53 and anti‐NANP 317; Oyen et al, 2017) mAbs at the indicated concentrations (n = 3–6).
-
FCapacity of the indicated passively‐transferred antibodies (100 μg) to protect mice from parasitemia after the bite of three PbPfCSP(mCherry)‐infected mosquitoes (n = 10 for mAb 317; Oyen et al, 2017, n = 8 for mAb 1961, and n = 9 for mAb 1710; Scally et al, 2018). Data show the percentage of parasite‐free mice in two independent experiments (only mice with detectable serum IgG concentrations of the indicated mAbs at the time of the challenge are shown; Fig EV4C). Groups marked with the same letter were not statistically significantly different (Mantel–Cox log‐rank test, see also Appendix Table S5).
Data information: Circles in (A, C–E) indicate independent biological replicates.
Source data are available online for this figure.
Thus, regardless of their fine epitope conformational specificity and affinity, anti‐C‐CSP antibodies lacked measurable sporozoite‐binding capacity and in vitro parasite‐inhibitory activity even at 100‐fold higher concentrations than anti‐repeat and cross‐reactive mAbs. Only a single C‐CSP reactive mAb with cross‐reactivity to the repeat and N‐terminal junction mediated parasite inhibition in vitro but was less efficacious than the potent anti‐repeat mAb 317 in vivo.
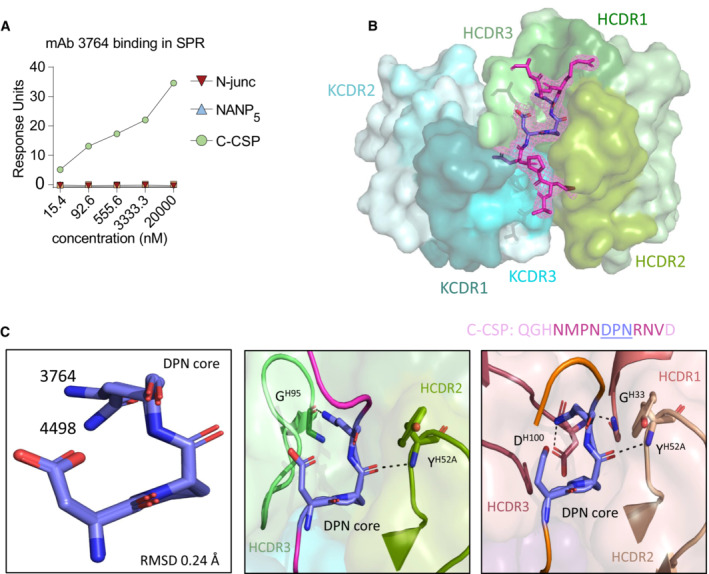
mAb 3764 binds a DPN core sequence in PfCSP281–294
The C‐linker specific mAb 3764 that weakly bound sporozoites was the only C‐CSP specific mAb (Fig EV5A) encoded by IGHV3‐33, a gene which is associated with germline NANP reactivity and also encodes for antibodies with cross‐reactivity to NPDP and NVDP motifs in the PfCSP N‐terminal junction (Murugan et al, 2020). The C‐CSP target peptide P2 of mAb 3764 (QGHNMPNDPNRNVD; Figs 3B and EV3B) showed sequence similarity with the N‐terminal junction (NPDPNAN). To define the molecular basis for the specific recognition of the C‐terminal linker region but not the N‐terminal junction, we solved the crystal structure of the 3764 Fab in complex with P2 to 2.36 Å resolution (Appendix Table S6). The 3764 Fab recognizes its epitope with all complementarity‐determining regions (CDRs) except Igκ CDR2 (KCDR2, Fig 5A). IgH CDR (HCDR) 2 and 3 form two hydrogen bonds with the core binding motif DPN (Fig 5A, inset I) contributing 274.7 Å2 of buried surface area (BSA). Comparison of this binding mode to that of mAb 4498, a VH3‐33 mAb that binds to the N‐terminal junction (peptide NANPNVDPNANP; PDB ID: 6ULF; Murugan et al, 2020) shows that both mAbs recognize the DPN core motif in a highly similar disposition (whole atom RMSD of 0.24 Å; Fig EV5B and C).
Figure EV5. Specificity of mAb 3764 towards the PfCSP C‐terminal region.
- mAb 3764 SPR affinity measurement against peptides covering the N‐term junction (N‐Junc), NANP5 or C‐CSP at various concentrations.
- Composite omit map electron density contoured at 1 σ (light magenta mesh) around the C‐CSP peptide (magenta).
- (Left) Superposition of the DPN core (deep blue) in crystal structures of mAb 3764 bound to the indicated C‐CSP peptide (magenta) and mAb 4498 (Murugan et al, 2020) bound to the NDN3 peptide (orange) (PDB ID: 6ULF). (Middle) mAb 3764 HCDR2 and 3 (pea and light green, respectively) are shown interacting with the C‐CSP peptide. (Right) mAb 4498 HCDR1, 2, and 3 (dark salmon, raspberry and wheat, respectively) are shown interacting with NDN3.
Data information: Data in (A) is a representative of two independent technical replicates.
Figure 5. Molecular characterization of mAb 3764 recognition of the PfCSP.
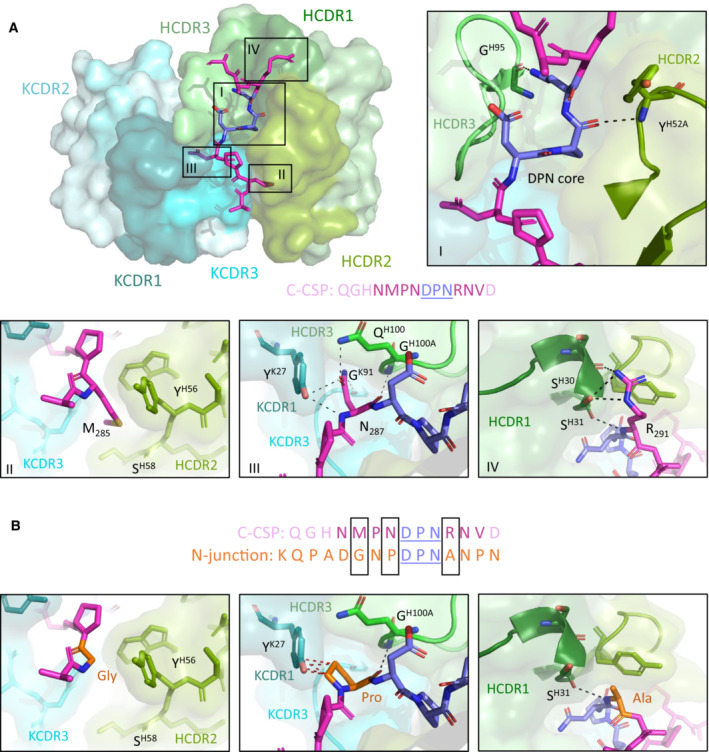
- Crystal structure showing the variable regions of the C‐linker specific mAb 3764 and P2, with the mAb 3764 heavy chain illustrated in green and the kappa chain in blue. HCDR1, 2 and 3 (dark, pea and light green, respectively) as well as KCDR1 and 3 (dark teal and aqua, respectively) contribute to epitope recognition, but KCDR2 (teal) does not. The central DPN core (deep blue) of the C‐CSP peptide (magenta) interacts with HCDR2 and 3 similarly to other VH3‐33 antibodies (inset I), forming hydrogen bonds (H‐bonds, shown with black dotted lines) with HCDR2 and 3. C‐CSP residues M285 (inset II), N287 (inset III) and R291 (inset IV) contribute substantial buried surface area (BSA) to the complex. (II) M285 does not contribute any H‐bonds, but does contribute to BSA. N287 (III) and R291 (IV) are responsible for hydrogen bonding with HCDR3, KCDR1 and KCDR3, and with HCDR1, respectively.
- Despite significant sequence similarity in the region of the DPN core between the C‐CSP peptide and the N‐junction peptide (sequence shown in orange), mAb 3764 does not bind to the N‐junction peptide. Molecular modeling replacing M285, N287 and R291 with Gly, Pro and Ala, respectively, reveals these alternate residues would result in a loss of hydrogen bonds, and reduced BSA and introduction of steric clashes (shown with red dotted lines).
In periphery of the DPN core motif, the surrounding C‐CSP P2 residues Met 285 (M285), Asp 287 (N287) and Arg 291 (R291) partake in important interactions: M285 contributes 160.3 Å2 of BSA at the interface with HCDR2 and Igκ CDR (KCDR) 3 (Fig 5A, inset II); N287 (Fig 5A, inset III) is responsible for five hydrogen bonds with HCDR3, KCDR1 and KCDR3 and contributes 156.8 Å2 of BSA; while R291 (Fig 5A, inset IV) contributes four hydrogen bonds with HCDR1 and 106.3 Å2 of BSA. Sequence analysis revealed that mAb 3764 had undergone limited somatic hypermutation, resulting in residues H.N56Y and H.Y58S in the HCDR2, which form part of the peptide‐binding interface and together contribute 70.8 Å2 of BSA around M285 (Fig 5A, inset II).
Given the sequence similarities between the core DPN in the C‐CSP and N‐junction peptides (Fig 5B), we investigated the molecular basis for mAb 3764‐binding specificity to C‐CSP. Modeling was performed to mutate the important peripheral C‐CSP peptide residues M285, N287 and R291 (see Fig 5A), which surround the DPN core, to their corresponding N‐junction peptide residues, Gly, Pro and Ala, respectively (Fig 5B). While it does not contribute any hydrogen bonds, the replacement of M285 with Gly would result in a much smaller BSA in the mAb 3764 HCDR2 and KCDR3 (compare Fig 5A inset II to Fig 5B left panel). Replacing N287 with Pro would result in the loss of four of the five hydrogen bonds with HCDR3, KCDR1 and KCDR3, as well as introducing steric hindrance with KCDR1 (compare Fig 5A inset III to Fig 5B center panel). Finally, Ala in place of Arg in position 291 would result in a loss of three of the four hydrogen bonds with HCDR1 (compare Fig 5A inset IV to Fig 5B right panel). Taken together, the crystal structure resolving PfCSP residues 284–293 in complex with mAb 3764, and molecular modeling, helped to explain the specificity of mAb 3764 for C‐CSP through interactions with the IgH (PfCSP284–292) and Igκ chains (PfCSP285–287 and PfCSP289), and the lack of cross‐reactivity with the DPN motif in the N‐terminal junction.
Discussion
The characterization of our large panel of human mAbs provides insights in the gene usage characteristics, development, cellular origin, epitope specificity, binding mode and functional properties of C‐CSP reactive antibodies. Similar to the strong association between NANP‐repeat reactivity and IGHV3‐33/IGKV1‐5 gene usage, (Imkeller et al, 2018; Murugan et al, 2018; Tan et al, 2018), our data establish a link between C‐CSP‐reactivity and IGHV3‐21 and IGVL3‐21 or IGVL3‐1 gene usage for Th2R/Th3R specific antibodies, which might allow conclusions about the frequency of C‐CSP‐reactive cells based on Ig gene sequencing. The association between IGHV3‐21 gene usage and antibody specificity for the C‐CSP α‐TSR domain appears to be independent of whether the immune system responds to recombinant protein or whole sporozoites, since Ig genes encoding VH3‐21 anti‐C‐CSP mAbs have also been cloned from B cells isolated from RTS,S/AS01 and PfSPZ CVac vaccinees (Scally et al, 2018; Beutler et al, 2022). Nevertheless, the high IGHV3‐21 frequency that we observed after immunization with 3 doses of the PfSPZ Vaccine (900,000 radiation attenuated PfSPZ administered on days 1, 8, and 29) compared to immunization with the chemo‐attenuated PfSPZ‐CVac (Mordmuller et al, 2017) with strong dominance of IGHV3‐33 (Imkeller et al, 2018; Murugan et al, 2018; Tan et al, 2018) suggests that radiation, dose, or dosing interval might influence the anti‐sporozoite B cell response. The large number of VH3‐21 and non‐VH3‐21 B cells with Th2R/Th3R specificity including many that expressed unmutated antibodies indicates that the naïve B cell repertoire contains potent anti‐Th2R/Th3R precursors. The relative abundance of these cells might contribute to the apparent immunodominance of this epitope compared to the conserved RII+/CS.T3, which is targeted by antibodies with diverse gene combinations, and compared to non‐α‐TSR epitopes.
In contrast to antibodies against the α‐TSR domain, antibodies with reactivity to the C‐linker appear to be rare and target linear epitopes. The differences in abundance of these antibodies may be linked to differences in the frequency of naïve precursor B cells that recognize these epitopes and structural differences between the C‐linker region and α‐TSR domain or differences in epitope presentation and immunogenicity. Analysis of the crystal structure of mAb 3764 in complex with amino acid residues 284–293 provides first insights into how the C‐linker domain is recognized by a non‐cross‐reactive human anti‐PfCSP antibody. Despite sequence identity in the C‐linker with the N‐terminal junction, the structural data provides a molecular explanation for the specificity of mAb 3764 for C‐CSP. Steric constraints and suboptimal interactions with residues surrounding the core preclude cross‐reactivity with the DPN motif in the junction. The data support the notion that peripheral residues define the binding specificity and cross‐reactivity of antibodies beyond recognition of the core binding motif.
Despite the relative abundance of germline‐encoded α‐TSR‐reactive antibodies, B cells with this specificity seem to undergo efficient affinity maturation and IgG isotype switching in germinal center reactions. The relative abundance of IgG compared to IgM antibodies, higher numbers of selected somatic mutations in VH3‐21 mAbs compared to VH3‐33 mAbs, and enrichment of the affinity‐increasing H.T33 mutation, suggests that epitope specificity is linked to differences in cell differentiation pathways or cell differentiation kinetics. The large number of NANP motifs in the central repeat domain might mediate stronger BCR cross‐linking and B cell activation signals compared to the non‐repeating epitopes in the PfCSP C terminus, thereby affecting B cell activation, differentiation and germinal center selection (Imkeller et al, 2018). Alternatively, and not mutually exclusively, structure rather than valency differences between the disordered and highly flexible central repeat region compared to the structured C‐CSP domain may underlie these isotype and mutation‐load differences (MacRaild et al, 2018). Future studies will have to define the molecular and cellular mechanisms and determine whether epitope‐associated differences in affinity maturation between repeat and C‐CSP antibodies are also seen in response to immunization with recombinant PfCSP and RTS,S/AS01 or other antigens with repeating disordered domains that are characteristic of numerous parasite proteins.
Our study shows that the C‐CSP of irradiated sporozoites is highly immunogenic and identifies linear target epitopes in the C‐terminal linker and α‐TSR domain. However, in assays with live Pf sporozoites none of the C‐CSP epitopes seem to be the target of potent parasite‐inhibitory antibodies. Even the C‐linker specific mAb 3764, which showed weak parasite binding, lacked Pf‐inhibitory activity at a hundred‐fold higher concentration than antibodies against the central repeat domain, suggesting that modest C‐CSP binding is not sufficient to mediate protection. Although the C‐linker region may be slightly easier to reach for C‐CSP mAbs than the α‐TSR, which is directly anchored to the parasite surface, the central repeat and junction domains are clearly more accessible on the surface of live sporozoites. The direct comparison of C‐CSP and NANP antibodies demonstrates that the almost complete inaccessibility of the PfCSP C‐terminal domain strongly limits the direct parasite‐inhibitory activity and potency of antibodies with specificity for this domain.
We have shown previously that passive transfer of the Th2R/Th3R‐reactive mAb 1710 at high dose fails to protect mice from infection with PfCSP transgenic P. berghei parasites and the development of blood stage parasitemia (Scally et al, 2018; Murugan et al, 2020), findings that have recently been confirmed in an independent study (Wang et al, 2021). Since the epitopes of the Th2R/Th3R antibodies identified in this study overlap with the mAb 1710 target site (including many that share VH3‐21/Vλ3‐21 Ig gene combinations), we do not expect these mAbs to exhibit any direct parasite‐inhibitory activity in vivo. Given their lack of Pf sporozoite binding and in vitro parasite inhibition, we assume that the RII+/CS.T3 reactive antibodies would behave similarly. Some degree of in vivo protection from i.v. parasite challenge has recently been reported after high‐dose passive transfer of the RII+/CS.T3 and the Th2R/Th3R‐reactive mAbs 1512 and 236, respectively (Beutler et al, 2022). How the potency of these high‐affinity mAbs compares to anti‐repeat mAbs with similar binding strength has not been determined but given that mAbs 1512 and 236 target the same epitopes as the mAbs in our collection, we expect it to be overall low.
In response to RTS,S/AS01 immunization, anti‐NANP and anti‐C‐terminus antibody titers correlate with protection (Chaudhury et al, 2016, 2021; Dobano et al, 2019). To what degree the C‐CSP‐reactive serum antibodies recognize C‐CSP specifically or cross‐react with the repeat and junction has not been determined. Therefore, it is unclear whether protection is associated with C‐CSP specific or cross‐reactive antibodies or whether C‐CSP antibodies are simply a marker of the overall strength of the anti‐parasite response, while protection is mediated by anti‐repeat antibodies. Passive transfer of mAb 1961 shows that cross‐reactive mAbs can mediate low levels of parasite inhibition in vivo. Such antibodies likely confer some degree of protection by binding to repeat and junction epitopes that are readily accessible on the surface of live sporozoites and not to the hidden C‐CSP (Murugan et al, 2020). Cross‐reactive anti‐PfCSP mAbs have been shown to have overall higher affinity than antibodies that target the junction, repeat or C‐CSP specifically (Murugan et al, 2020). Thus, protection seems to be linked to high affinity C‐CSP cross‐reactive rather than C‐CSP specific antibodies. Nevertheless, we cannot exclude that C‐CSP specific antibodies gain access to the C‐terminus upon PfCSP binding by high affinity anti‐NANP or junction antibodies. PfCSP shedding induced by anti‐repeat/junction antibodies might uncover epitopes in the α‐TSR domain enabling C‐CSP‐antibody binding and parasite clearance. Although synergistic effects of anti‐repeat/junction and anti‐C‐CSP mAbs may not be easily detected in animal models with human monoclonal antibodies (Wang et al, 2021), antibody effector functions such as opsonization and phagocytosis or complement activation may contribute to parasite inhibition in humans. However, the high sequence diversity of the Th2R/Th3R domain among Pf parasite populations in natural settings will limit these indirect protective effects to antibodies against the conserved RII+/CS.T3 of the α‐TSR domain, and to protection from parasites with high sequence similarity to the vaccine strain.
Future studies will need to determine whether efforts to design a second generation PfCSP vaccine might benefit from suppressing or even abrogating the humoral response against C‐CSP, e.g. by boosting the anti‐repeat and junction response. However, exclusion of the complete domain, especially of the highly immunodominant α‐TSR, would eliminate the main T helper cell epitopes with likely strong negative effects on the quality and strength of the humoral response against the potent repeat and junction epitopes. Inclusion of linear peptide epitopes rather than the complete C‐CSP may be sufficient to provide efficient T cell help without inducing non‐protective humoral responses (Wahl et al, 2022). Alternatively, non‐PfCSP T cell epitopes could substitute for the loss of T cell help and promote affinity maturation of the PfCSP‐specific response (Ludwig et al, 2023).
In summary, with our mAb collection we demonstrate the lack of accessible C‐CSP epitopes on live sporozoites as targets for potent parasite‐inhibitory antibodies. Due to the high immunogenicity of the PfCSP C terminus, these observations are of direct relevance for the design of an improved PfCSP‐based malaria vaccine.
Materials and Methods
Clinical samples
Serum and peripheral blood mononuclear cells (PBMCs, obtained by Ficoll density gradient centrifugation) were collected from malaria‐naïve, healthy European volunteers who participated in the MAVACHE clinical trial and received three doses of 900,000 PfSPZ at 0, 1 and 4 weeks (Mordmuller et al, 2022). All study volunteers provided written informed consent. Ethical approval was obtained from the ethics committee of the medical faculty and the University Clinics of the University of Tübingen and regulatory approval by the Paul‐Ehrlich‐Institute. The study was conducted according to the principles of the Declaration of Helsinki. The study also conformed to the principles set out in the Department of Health and Human Services Belmont Report.
Cell lines
Human embryonic kidney HEK293T cells (Invitrogen, 0.3 × 106/ml) were cultured under rotating conditions (180 rpm) in 50 ml bioreactors with FreeStyle 293‐F medium at 37°C and 8% CO2. HC‐04 cells (BEI Resources MRA‐975; Sattabongkot et al, 2006) were cultured at 37°C and 5% CO2 in HC‐04 complete medium with 428.75 ml MEM (without l‐glutamine), 428.75 ml F‐12 Nutrient Mix (with l‐glutamine), 15 mM HEPES, 1.5 g/l NaHCO3, 2.5 mM l‐glutamine, 10% FCS and 1% penicillin/streptomycin solution.
Bacteria
MAX efficiency DH10B™ (ThermoFisher scientific; Cat. No: 18297010) competent cells were grown in LB medium for cultivation and in terrific broth for plasmid purification. Bacterial shaker at 37°C and 180 rpm was used for cultivation.
Proteins and peptides
Biotinylated NF54 FL‐CSP, non‐biotinylated 7G8 FL‐CSP and α‐TSR protein were expressed in HEK293F cells and purified by HisTrap HP (Cytiva) and size exclusion chromatography (Superdex 200 Increase 10/300 GL, Cytiva). FL‐CSP and C‐CSP were expressed in E.coli by the EMBL protein expression and purification core facility (Heidelberg, Germany). N‐junc, NANP5, NANP10, C‐linker and the 14 overlapping peptides (P1‐14) covering the complete NF54 PfCSP C‐terminus were synthesized by PSL Peptide Specialty Laboratories GmbH (Heidelberg, Germany). Sequences of peptides and proteins are listed in Appendix Table S1.
Parasites, mosquitoes and mice
Anopheles coluzzii (Ngousso S1 strain; Harris et al, 2010) and transgenic Anopheles gambiae (7b strain; Pompon & Levashina, 2015) were maintained at 28°C 70–80% humidity 12/12 h day/night cycle. The P. falciparum NF54 clone (Pf) used in this study originated from Prof. Sauerwein's laboratory (RUMC, Nijmegen) and was regularly tested for Mycoplasma contamination. For P. falciparum infections, A. coluzzii mosquitoes were fed for 15 min via artificial midi feeders (Glass Instruments, The Netherlands) with NF54 gametocyte cultures and kept at 26°C 80% humidity 12/12 h day/night cycle in a secured BSL3 laboratory according to the national regulations (Landesamt für Gesundheit und Soziales, project number 297/13). Infected mosquitoes were offered an additional uninfected blood meal to boost sporozoite formation 8 days post infection (dpi). Sporozoites were dissected for traversal assay 5–7 days later as described below.
Anopheles gambiae 7b mosquitoes, an immunocompromised transgenic mosquito line derived from the G3 laboratory strain, were used for the production of transgenic P. berghei sporozoites expressing PfCSP and the reporter fluorescence protein mCherry (PbPfCSP(mCherry), Ludwig et al, 2023). Briefly, female CD1 mice (7–12‐weeks‐old) and female C57BL/6J mice (8‐weeks‐old) were bred in the MPIIB Experimental Animal Facility (Marienfelde, Berlin), housed in a pathogen‐free animal facility in accordance with the German Animal Protection Law (§8 Tierschutzgesetz) and approved by the Landesamt für Gesundheit und Soziales (LAGeSo), Berlin, Germany (project numbers 368/12 and H0335/17). A. gambiae 7b mosquitoes were fed on PbPfCSP(mCherry)‐infected female CD1 mice 3 days post passage (0.1–0.8% gametocytemia) for 30–45 min, and kept at 20°C 80% humidity 12/12 h day/night cycle. Infected mosquitoes were offered an additional uninfected blood meal to boost sporozoite formation 7 dpi, and after 11 days were used for C57BL/6J mice challenge experiments and live sporozoite FACS staining as described below.
Flow cytometry and isolation of PfCSP‐specific memory B cells
Frozen PBMCs were thawed, washed with RPMI 1640 (Gibco) and incubated with full‐length biotinylated NF54 PfCSP in the presence of the following mouse anti‐human antibodies at the indicated dilutions: CD27‐phycoerythrin (PE) (Clone ID: M‐T271; Cat. No: 555441) at 1:5, CD38‐ Brilliant Violet 605 (BV605) (Clone ID: HB7; Cat. No: 562665) at 1:20, IgG‐BV510 (Clone ID: G18‐145; Cat. No: 563247) at 1:20, IgM‐BV421 (Clone ID: G20‐127; Cat. No: 562618) at 1:20, IgD‐Allophycocyanin‐H7 (APC‐H7) (Clone ID: IA6‐2; Cat. No: 561305) at 1:20 (all from BD Biosciences), CD19‐BV786 (Clone ID: HIB19; Cat. No: 302240) at 1:10 and CD21‐ PE‐Cy7 (Clone ID: Bu32; Cat. No: 354912) at 1:20 (both from BioLegend). Biotin was detected using streptavidin FITC (Invitrogen; Cat. No: 11‐4317‐87) at 1:1,000 dilution. 7‐Aminoactinomycin D (7‐AAD) (Invitrogen; Cat No: A1310) at 1:400 was used as a marker for dead cells. Single‐cell sorting of 7AAD−PfCSP+CD19+ B cells gated positive for either IgG or CD27 as PfCSP memory B cells and of 7AAD−CD38+CD27+CD19+ cells as plasmablasts was performed using a FACS Aria II (BD Biosciences) with FACSDiva software (version 8.0.1) using the index sort option. Data were analyzed using FlowJo v.10.0.8 (Tree Star).
Ig gene amplification and sequence analysis
A high‐throughput robotic platform was used to amplify the IGH, IGK and IGL loci of Ig genes by RT‐PCR as previously reported (Wardemann & Busse, 2019). Briefly, after cDNA synthesis with random hexamers, a nested polymerase chain reaction (PCR) with barcoded primers in the second PCR was used to amplify the Ig heavy and light chain genes. Amplicons were pooled, purified, and sequenced using the MiSeq 2 × 300 base pair (bp) paired‐end sequencing platform (Illumina). Integrated indexed flow cytometry and sequence data integration as well as Ig gene annotation was performed using sciReptor version 1.0‐6‐g034c8ae (Busse et al, 2014; Imkeller et al, 2016). Downstream sequence analyses and data visualization were performed using GraphPad Prism (version 9.12) and R 4.2.2 and ggplot2.
Cloning of Ig genes and recombinant monoclonal antibodies production
Cloning of the Ig genes and recombinant antibody production were performed as previously described (Wardemann & Busse, 2019). Ig genes were amplified with V‐ and J‐specific primers containing restriction sites. The amplicons were cloned into human IGHG1 or IGKC or IGLC expression vectors (Addgene number 80795, 80796 and 99575, respectively). After co‐transfection of the Ig heavy and light encoding plasmid DNA into HEK293T cells (Invitrogen), the recombinant monoclonal antibodies (mAbs) were purified from the supernatant with protein G beads (GE Healthcare). Subsequent experiments with the mAbs, including functional assays, were performed in an unblinded manner.
Enzyme‐linked immunosorbent assay (ELISA)
ELISAs were performed as described (Triller et al, 2017; Murugan et al, 2020). In brief, 384‐well polystyrene plates (Corning) with high binding capacity were coated overnight (O/N) at 4°C with full‐length PfCSP (0.4 μg/ml), N‐junc, NANP10 (2 μg/ml), C‐CSP (1 μg/ml), Insulin (10 μg/ml; Sigma Aldrich), lipopolysaccharide (LPS; 10 μg/ml; Sigma Aldrich) and double‐stranded DNA (dsDNA; 20 μg/ml; Sigma Aldrich) in PBS. The plates were washed 3 times with PBS‐T (0.05% Tween20 in PBS) using a Tecan plate washer. ELISA plates were blocked at RT for 1 h with 4% BSA in PBS (serum ELISA), 1% BSA in PBS (mAb PfCSP ELISA) or blocking buffer (1× PBS, 0.05% (v/v) Tween20, 1 mM EDTA; mAb polyreactivity ELISA). Serum samples serially diluted at an initial dilution of 1:200 in 1% BSA with PBS, or mAbs at an initial concentration of 4 μg/ml (PfCSP ELISA) or 1 μg/ml (polyreactivity ELISA) in 1× PBS were loaded on the plate and incubated at RT for 1.5 h. Antibodies were detected with goat anti‐human IgG secondary antibody coupled to horseradish peroxidase (HRP) (Jackson Immuno Research; Cat No: 109‐035‐098) diluted at 1:1,000 in blocking buffer (1× PBS, 0.05% (v/v); Tween20, 1 mM EDTA) and then detected with 2,2′‐azino‐bis‐(3‐ethylbenzothiazoline‐6‐sulfonic acid) diammonium (ABTS) substrate (Roche Diagnostics; Cat No: 11112422001) diluted at 1:1,000 in H2O2. Optical density (OD) at 405 nm was determined using an M1000Pro plate reader (Tecan). Area under the curve (AUC) values were calculated using GraphPad Prism 9.1.2. Serum from placebo recipients was used as a negative control in serum ELISAs. mAbs 2A10 (Triller et al, 2017) and mGO53 (Wardemann et al, 2003) were used as positive and negative controls, respectively, in PfCSP ELISAs and mAbs ED38 (Meffre et al, 2004) and mGO53 as positive and negative controls, respectively, in polyreactivity ELISAs.
Blocking ELISA
Blocking ELISA was performed as described above with the following modifications. After 1.5 h incubation with the test mAbs (starting concentration of 64 μg/ml in 1× PBS and 1:2 dilution) at room temperature, biotinylated mAb 1710 (Scally et al, 2018) or mAb 1512 (Beutler et al, 2022) at 0.5 μg/ml was added to compete for an additional 15 min at room temperature. mAb 1710 or mAb 1512 binding was detected with streptavidin‐HRP diluted at 1:1,000 in blocking buffer (1× PBS, 0.05% (v/v) Tween20, 1 mM EDTA) and then detected with 2,2′‐azino‐bis‐(3‐ethylbenzothiazoline‐6‐sulfonic acid) diammonium (ABTS) substrate (Roche Diagnostics) diluted at 1:1,000 in H2O2. The optimal concentrations of the biotinylated 1710 and 1512 were determined by measuring their ability to self‐block their non‐biotinylated versions over a range of concentrations.
Surface Plasmon resonance (SPR)
SPR measurements were performed using a Biacore T200 (GE Health‐care) instrument docked with an S CM5 series sensor chip (GE Healthcare), as described (Murugan et al, 2018). In brief, anti‐human IgG antibodies were immobilized on the chip using an amine coupling‐based human antibody Fab capture kit. Hepes (10 mM) with 150 mM NaCl at pH 7.4 was used as running buffer. Equal concentrations of the sample antibody and isotype control (mGO53; Wardemann et al, 2003) were captured in the sample and reference flow cells, respectively. Running buffer was injected at a rate of 10 μl/min for 20 min to stabilize the flow cells. C‐CSP peptide at 0.015, 0.09, 0.55, 3.3 and 20 μM in running buffer was injected at a rate of 30 μl/min. The flow cells were regenerated with 3 M MgCl2. Data were fitted by steady‐state kinetic analysis using BIACORE T200 software V2.0.
mAb 3764 Fab production
mAb 3764 Ig heavy and light chain variable regions were cloned into custom pcDNA3.4 expression vectors upstream of human IgK constant and IgG1‐CH1 domains, and transiently co‐transfected into HEK293F cells. The resulting Fab was purified by Kappa‐select affinity chromatography (Cytiva), cation exchange chromatography (MonoS; Cytiva), and size exclusion chromatography (Superdex 200 Increase 10/300 GL; Cytiva).
Crystallization and structure determination
Purified 3764 Fab and PfCSP C‐terminal linker peptide (PfCSP281‐294) were mixed in a 1:3 molar ratio; and, in sparse matrix screening, crystals of the complex emerged from a 1:1 ratio mix with 19% (v/v) isopropanol, 19% (w/v) PEG 4000, 5% (v/v) glycerol, 0.095 M sodium citrate, pH 5.6. Crystals did not require cryopreservation before being flash frozen in liquid nitrogen. Data was collected at the 23‐ID‐B beamline at the Advanced Photon Source and manually processed using XDS (Kabsch, 2010). Molecular replacement using the heavy and light chains from the 4498 Fab (PDB ID: 6O24) was carried out in Phaser (McCoy et al, 2007). Iterative refinement was performed in Coot (Emsley et al, 2010) and phenix.refine (Adams et al, 2010). All software were accessed through SBGrid (Morin et al, 2013). The structure has been deposited in the PDB and is accessible under ID: 8FG0.
Pf sporozoite hepatocyte traversal assay in vitro
Traversal assays were performed as previously described (Triller et al, 2017; Murugan et al, 2018). Briefly, Pf sporozoites were isolated from the mosquito salivary glands 13–15 dpi in complete HC‐04 medium, pre‐incubated with mAb at the indicated concentrations for 30 min on ice and added to the human hepatocyte cell line (HC‐04, BEI Resources MRA‐975, Sattabongkot et al, 2006) for 2 h at 37°C and 5% CO2 in the presence of 0.5 mg/ml dextran‐rhodamine (Molecular Probes). Untreated Pf sporozoites were used to measure maximum traversal capacity. A chimeric version of mAb 2A10 (Zavala et al, 1983) with human IgG1 constant region (Triller et al, 2017), and mGO53 (Wardemann et al, 2003) were used as positive and negative controls, respectively. Cells were trypsinized, washed and fixed with 1% PFA/PBS before measuring dextran positivity using a FACS LSRFortessa instrument and FACSDiva software (BD Biosciences). Data analysis was performed by subtracting background (dextran positivity of cells treated with uninfected mosquito salivary gland material) and normalizing to maximum Pf traversal capacity (dextran positivity of cells without mAbs) using FlowJo V.10.8.2 (Tree Star).
Pb‐PfCSP(mCherry) live sporozoite flow cytometry
Plasmodium berghei PbPfCSP(mCherry) (Ludwig et al, 2023) sporozoites were isolated from mosquito salivary glands 18 dpi in HC‐04 complete medium, incubated for 30 min on ice in siliconized tubes with the indicated concentrations of mAbs (150,000 sporozoites in total volume of 100 μl 1% FCS/PBS). Sporozoites were washed with 1.5 ml of 1% FCS/PBS by centrifugation at 9,300 g for 4 min at 4°C. The sporozoite pellet was resuspended in 100 μl 1% FCS/PBS containing anti‐human IgG1‐Cy5 (clone HP6001, 2 μg/ml, Central Laboratory Facility Deutsches Rheuma‐Forschungszentrum, Berlin) and incubated on ice for 30 min. The sporozoite suspension was washed again, and the pellet was re‐suspended in 200 μl 1% FCS/PBS. Sporozoites stained with antibodies were detected by endogenous mCherry expression and analyzed for Cy5 signal in a FACS LSRFortessa instrument with FACSDiva software (BD Biosciences). Salivary gland material from uninfected mosquitoes was used as sporozoite gating control (Fig EV4A). Data were analyzed using FlowJo V.10.8.2 (Tree Star).
Plasmodium berghei PbPfCSP(mCherry) mouse challenge model by mosquito bites
Female C57BL/6J mice were passively immunized by intraperitoneal injection of 100 μg mAb in 100–200 μl PBS. Mice were randomly assigned to different groups. After 20 h, mice were exposed to the bites of three PbPfCSP(mCherry) salivary gland‐infected mosquitoes as described elsewhere (Ludwig et al, 2023). Briefly, PbPfCSP(mCherry)‐infected mosquitoes were put to sleep on ice at 17 dpi and selected for mCherry signal in the salivary glands under a fluorescence stereo microscope (Leica, M205 FA). Single salivary gland‐positive mosquitoes were transferred into individual containers and starved overnight. The next day (18 dpi), anesthetized naïve C57BL/6J mice were exposed for 10 min to three bites of the selected PbPfCSP(mCherry)‐infected mosquitoes. Peripheral whole blood was collected from the submandibular vein 2–3 h after challenge to obtain sera. Serum mAb concentrations were determined by ELISA. Mice with undetectable levels of mAbs in the serum were excluded from the analysis. From days 3 to 7 and on day 10 post mosquito bite, parasitemia (mCherry‐positive red blood cells/total red blood cells) was measured by flow cytometry (LSRFortessa, BD Biosciences), and confirmed by Giemsa‐stained thin blood smears. All infected mice were euthanized on day 7 post mosquito bite, before the occurrence of malaria symptoms. FACS data were analyzed by FlowJo V.10.8.2 and the pre‐patency period was declared on the first day when parasitemia values were above the background signal of negative mice.
Statistics
Statistical analysis was done with GraphPad Prism (version 9.12) using a two‐tailed non‐parametric Mann–Whitney test assuming non‐normal distribution of the data and two‐tailed Mantel‐Cox log‐rank test for the in vivo experiment, as described in the figure legends. In vitro and in vivo experiments were performed in at least two independent technical and biological replicates, respectively. Mice were randomly assigned to their respective experimental groups from the same batches. No experiment was done blindly. P values lower than 0.05 were regarded as statistically significant, with asterisks used to indicate the level of significance (*P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001), as described in the figure legends.
Author contributions
Opeyemi Ernest Oludada: Formal analysis; investigation; visualization; writing – original draft; writing – review and editing. Giulia Costa: Formal analysis; investigation; visualization; writing – original draft; writing – review and editing. Clare Burn Aschner: Formal analysis; investigation; visualization; writing – original draft; writing – review and editing. Anna S Obraztsova: Data curation; formal analysis; visualization. Katherine Prieto: Investigation. Caterina Canetta: Investigation. Stephen L Hoffman: Resources. Peter G Kremsner: Resources. Benjamin Mordmüeller: Resources. Rajagopal Murugan: Conceptualization; writing – review and editing. Jean‐Philippe Julien: Conceptualization; funding acquisition; writing – original draft; writing – review and editing. Elena A Levashina: Conceptualization; funding acquisition; writing – original draft; writing – review and editing. Hedda Wardemann: Conceptualization; funding acquisition; writing – original draft; writing – review and editing.
Disclosure and competing interests statement
The authors declare the following competing interests: S.L.H is a salaried employee of Sanaria Inc. and has a financial interest in Sanaria Inc. All other authors declare no financial or commercial conflict of interest.
For more information
Supporting information
Appendix
Expanded View Figures PDF
Dataset EV1
PDF+
Source Data for Figure 1
Source Data for Figure 2
Source Data for Figure 3
Source Data for Figure 4
Acknowledgements
The authors thank Dorien Foster, Julia Gärtner, Christine Niesik, Claudia Winter (DKFZ) and Hadenal Gordon, Cornelia Kreschel, Manuela Andres, Daniel Eyermann and Liane Spohr (Max Planck Institute for Infection Biology, Berlin) for experimental assistance, Christian E. Busse (DKFZ) for bioinformatic support, the EMBL Protein Expression and Purification Core Facility and the DKFZ/ EMBL/Heidelberg University Chemical Biology Core Facility, especially P. Sehr, and the Deutsches Rheuma‐Forschungszentrum Flow Cytometry Core Facility for technical assistance and services. The work was supported by the Bill and Melinda Gates Foundation (OPP1179906; J.P.J, H.W. and E.A.L.), by the CIFAR Azrieli Global Scholar program (J.P.J.), the Ontario Early Researcher Award program (J.P.J.), and the Canada Research Chair program (J.P.J.). C.B.A was supported by a by Hospital for Sick Children Restracomp Postdoctoral Fellowship and a Banting Postdoctoral Fellowship. X‐ray diffraction experiments were performed at GM/CA@APS, which has been funded in whole or in part with federal funds from the National Cancer Institute (ACB‐12002) and the National Institute of General Medical Sciences (AGM‐12006). The Eiger 16 M detector at GM/CA‐XSD was funded by NIH grant S10 OD012289. This research used resources of the Advanced Photon Source, a US Department of Energy (DOE) Office of Science user facility operated for the DOE Office of Science by Argonne National Laboratory under contract DE‐AC02‐06CH11357. Manufacture of PfSPZ Vaccine was funded in part by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under SBIR award numbers 5R44AI058375 and 5R44AI055229.
EMBO Mol Med (2023) 15: e17454
See also: J Murdoch & J Baum (June 2023)
Contributor Information
Jean‐Philippe Julien, Email: jean-philippe.julien@sickkids.ca.
Elena A Levashina, Email: levashina@mpiib-berlin.mpg.de.
Hedda Wardemann, Email: h.wardemann@dkfz.de.
Data availability
The datasets and material that were generated in this study can be obtained from the corresponding authors under standard transfer agreements. Datasets are deposited on ENA with accession ID: PRJEB60486, Zenodo DOI: https://doi.org/10.5281/zenodo.7708746. The crystal structure has been deposited in the Protein Data Bank at www.rcsb.org under PDB ID code 8FG0 (http://identifiers.org/pdb/8FG0).
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse‐Kunstleve RW et al (2010) PHENIX: a comprehensive python‐based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66: 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agnandji ST, Lell B, Soulanoudjingar SS, Fernandes JF, Abossolo BP, Conzelmann C, Methogo BG, Doucka Y, Flamen A, Mordmuller B et al (2011) First results of phase 3 trial of RTS,S/AS01 malaria vaccine in African children. N Engl J Med 365: 1863–1875 [DOI] [PubMed] [Google Scholar]
- Aragam NR, Thayer KM, Nge N, Hoffman I, Martinson F, Kamwendo D, Lin FC, Sutherland C, Bailey JA, Juliano JJ (2013) Diversity of T cell epitopes in Plasmodium falciparum circumsporozoite protein likely due to protein‐protein interactions. PLoS One 8: e62427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asante KP, Abdulla S, Agnandji S, Lyimo J, Vekemans J, Soulanoudjingar S, Owusu R, Shomari M, Leach A, Jongert E et al (2011) Safety and efficacy of the RTS,S/AS01E candidate malaria vaccine given with expanded‐programme‐on‐immunisation vaccines: 19 month follow‐up of a randomised, open‐label, phase 2 trial. Lancet Infect Dis 11: 741–749 [DOI] [PubMed] [Google Scholar]
- Beutler N, Pholcharee T, Oyen D, Flores‐Garcia Y, MacGill RS, Garcia E, Calla J, Parren M, Yang L, Volkmuth W et al (2022) A novel CSP C‐terminal epitope targeted by an antibody with protective activity against Plasmodium falciparum . PLoS Pathog 18: e1010409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bojang KA, Milligan PJ, Pinder M, Vigneron L, Alloueche A, Kester KE, Ballou WR, Conway DJ, Reece WH, Gothard P et al (2001) Efficacy of RTS,S/AS02 malaria vaccine against Plasmodium falciparum infection in semi‐immune adult men in The Gambia: a randomised trial. Lancet 358: 1927–1934 [DOI] [PubMed] [Google Scholar]
- Busse CE, Czogiel I, Braun P, Arndt PF, Wardemann H (2014) Single-cell based high-throughput sequencing of full-length immunoglobulin heavy and light chain genes. Eur J Immunol 44: 597–603 [DOI] [PubMed] [Google Scholar]
- Casares S, Brumeanu TD, Richie TL (2010) The RTS,S malaria vaccine. Vaccine 28: 4880–4894 [DOI] [PubMed] [Google Scholar]
- Cawlfield A, Genito CJ, Beck Z, Bergmann‐Leitner ES, Bitzer AA, Soto K, Zou X, Hadiwidjojo SH, Gerbasi RV, Mullins AB et al (2019) Safety, toxicity and immunogenicity of a malaria vaccine based on the circumsporozoite protein (FMP013) with the adjuvant army liposome formulation containing QS21 (ALFQ). Vaccine 37: 3793–3803 [DOI] [PubMed] [Google Scholar]
- Chaudhury S, Ockenhouse CF, Regules JA, Dutta S, Wallqvist A, Jongert E, Waters NC, Lemiale F, Bergmann‐Leitner E (2016) The biological function of antibodies induced by the RTS,S/AS01 malaria vaccine candidate is determined by their fine specificity. Malar J 15: 301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhury S, MacGill RS, Early AM, Bolton JS, King CR, Locke E, Pierson T, Wirth DF, Neafsey DE, Bergmann‐Leitner ES (2021) Breadth of humoral immune responses to the C‐terminus of the circumsporozoite protein is associated with protective efficacy induced by the RTS,S malaria vaccine. Vaccine 39: 968–975 [DOI] [PubMed] [Google Scholar]
- Coppi A, Pinzon‐Ortiz C, Hutter C, Sinnis P (2005) The Plasmodium circumsporozoite protein is proteolytically processed during cell invasion. J Exp Med 201: 27–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dame JB, Williams JL, McCutchan TF, Weber JL, Wirtz RA, Hockmeyer WT, Maloy WL, Haynes JD, Schneider I, Roberts D et al (1984) Structure of the gene encoding the immunodominant surface antigen on the sporozoite of the human malaria parasite Plasmodium falciparum . Science 225: 593–599 [DOI] [PubMed] [Google Scholar]
- Dobano C, Sanz H, Sorgho H, Dosoo D, Mpina M, Ubillos I, Aguilar R, Ford T, Diez‐Padrisa N, Williams NA et al (2019) Concentration and avidity of antibodies to different circumsporozoite epitopes correlate with RTS,S/AS01E malaria vaccine efficacy. Nat Commun 10: 2174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doud MB, Koksal AC, Mi LZ, Song G, Lu C, Springer TA (2012) Unexpected fold in the circumsporozoite protein target of malaria vaccines. Proc Natl Acad Sci U S A 109: 7817–7822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K (2010) Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66: 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinosa DA, Gutierrez GM, Rojas‐Lopez M, Noe AR, Shi L, Tse SW, Sinnis P, Zavala F (2015) Proteolytic cleavage of the Plasmodium falciparum circumsporozoite protein is a target of protective antibodies. J Infect Dis 212: 1111–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandhi K, Thera MA, Coulibaly D, Traore K, Guindo AB, Doumbo OK, Takala‐Harrison S, Plowe CV (2012) Next generation sequencing to detect variation in the Plasmodium falciparum circumsporozoite protein. Am J Trop Med Hyg 86: 775–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genito CJ, Beck Z, Phares TW, Kalle F, Limbach KJ, Stefaniak ME, Patterson NB, Bergmann‐Leitner ES, Waters NC, Matyas GR et al (2017) Liposomes containing monophosphoryl lipid A and QS‐21 serve as an effective adjuvant for soluble circumsporozoite protein malaria vaccine FMP013. Vaccine 35: 3865–3874 [DOI] [PubMed] [Google Scholar]
- Gordon DM, McGovern TW, Krzych U, Cohen JC, Schneider I, LaChance R, Heppner DG, Yuan G, Hollingdale M, Slaoui M et al (1995) Safety, immunogenicity, and efficacy of a recombinantly produced Plasmodium falciparum circumsporozoite protein‐hepatitis B surface antigen subunit vaccine. J Infect Dis 171: 1576–1585 [DOI] [PubMed] [Google Scholar]
- Gupta NT, Vander Heiden JA, Uduman M, Gadala‐Maria D, Yaari G, Kleinstein SH (2015) Change‐O: a toolkit for analyzing large‐scale B cell immunoglobulin repertoire sequencing data. Bioinformatics 31: 3356–3358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris C, Lambrechts L, Rousset F, Abate L, Nsango SE, Fontenille D, Morlais I, Cohuet A (2010) Polymorphisms in Anopheles gambiae immune genes associated with natural resistance to Plasmodium falciparum . PLoS Pathog 6: e1001112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera R, Anderson C, Kumar K, Molina‐Cruz A, Nguyen V, Burkhardt M, Reiter K, Shimp R Jr, Howard RF, Srinivasan P et al (2015) Reversible conformational change in the Plasmodium falciparum Circumsporozoite protein masks its adhesion domains. Infect Immun 83: 3771–3780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutter JN, Robben PM, Lee C, Hamer M, Moon JE, Merino K, Zhu L, Galli H, Quinn X, Brown DR et al (2022) First‐in‐human assessment of safety and immunogenicity of low and high doses of Plasmodium falciparum malaria protein 013 (FMP013) administered intramuscularly with ALFQ adjuvant in healthy malaria‐naive adults. Vaccine 40: 5781–5790 [DOI] [PubMed] [Google Scholar]
- Imkeller K, Arndt PF, Wardemann H, Busse CE (2016) sciReptor: analysis of single-cell level immunogobulin repertoires. BMC Bioinformatics 17: 67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imkeller K, Scally SW, Bosch A, Marti GP, Costa G, Triller G, Murugan R, Renna V, Jumaa H, Kremsner PG et al (2018) Antihomotypic affinity maturation improves human B cell responses against a repetitive epitope. Science 360: 1358–1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W (2010) XDS. Acta Crystallogr D Biol Crystallogr 66: 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisalu NK, Idris AH, Weidle C, Flores‐Garcia Y, Flynn BJ, Sack BK, Murphy S, Schon A, Freire E, Francica JR et al (2018) A human monoclonal antibody prevents malaria infection by targeting a new site of vulnerability on the parasite. Nat Med 24: 408–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig J, Scally SW, Costa G, Hoffmann S, Murugan R, Lossin J, Prieto K, Obraztcova A, Lobeto N, Franke-Fayard B et al (2023) Glycosylated nanoparticle-based PfCSP vaccine confers long-lasting antibody responses and sterile protection in mouse malaria model. NPJ Vaccines 8: 52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacRaild CA, Seow J, Das SC, Norton RS (2018) Disordered epitopes as peptide vaccines. Pept Sci 110: e24067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse‐Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ (2007) Phaser crystallographic software. J Appl Crystallogr 40: 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCutchan TF, Kissinger JC, Touray MG, Rogers MJ, Li J, Sullivan M, Braga EM, Krettli AU, Miller LH (1996) Comparison of circumsporozoite proteins from avian and mammalian malarias: biological and phylogenetic implications. Proc Natl Acad Sci U S A 93: 11889–11894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meffre E, Schaefer A, Wardemann H, Wilson P, Davis E, Nussenzweig MC (2004) Surrogate light chain expressing human peripheral B cells produce self‐reactive antibodies. J Exp Med 199: 145–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mordmuller B, Surat G, Lagler H, Chakravarty S, Ishizuka AS, Lalremruata A, Gmeiner M, Campo JJ, Esen M, Ruben AJ et al (2017) Sterile protection against human malaria by chemoattenuated PfSPZ vaccine. Nature 542: 445–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mordmuller B, Sulyok Z, Sulyok M, Molnar Z, Lalremruata A, Calle CL, Bayon PG, Esen M, Gmeiner M, Held J et al (2022) A PfSPZ vaccine immunization regimen equally protective against homologous and heterologous controlled human malaria infection. NPJ Vaccines 7: 100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin A, Eisenbraun B, Key J, Sanschagrin PC, Timony MA, Ottaviano M, Sliz P (2013) Collaboration gets the most out of software. Elife 2: e01456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murugan R, Buchauer L, Triller G, Kreschel C, Costa G, Pidelaserra Marti G, Imkeller K, Busse CE, Chakravarty S, Sim BKL et al (2018) Clonal selection drives protective memory B cell responses in controlled human malaria infection. Sci Immunol 3: eaap8029 [DOI] [PubMed] [Google Scholar]
- Murugan R, Scally SW, Costa G, Mustafa G, Thai E, Decker T, Bosch A, Prieto K, Levashina EA, Julien JP et al (2020) Evolution of protective human antibodies against Plasmodium falciparum circumsporozoite protein repeat motifs. Nat Med 26: 1135–1145 [DOI] [PubMed] [Google Scholar]
- Nardin EH, Nussenzweig V, Nussenzweig RS, Collins WE, Harinasuta KT, Tapchaisri P, Chomcharn Y (1982) Circumsporozoite proteins of human malaria parasites Plasmodium falciparum and Plasmodium vivax . J Exp Med 156: 20–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olotu A, Lusingu J, Leach A, Lievens M, Vekemans J, Msham S, Lang T, Gould J, Dubois MC, Jongert E et al (2011) Efficacy of RTS,S/AS01E malaria vaccine and exploratory analysis on anti‐circumsporozoite antibody titres and protection in children aged 5–17 months in Kenya and Tanzania: a randomised controlled trial. Lancet Infect Dis 11: 102–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olotu A, Fegan G, Wambua J, Nyangweso G, Leach A, Lievens M, Kaslow DC, Njuguna P, Marsh K, Bejon P (2016) Seven‐year efficacy of RTS,S/AS01 malaria vaccine among young African children. N Engl J Med 374: 2519–2529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyen D, Torres JL, Wille‐Reece U, Ockenhouse CF, Emerling D, Glanville J, Volkmuth W, Flores‐Garcia Y, Zavala F, Ward AB et al (2017) Structural basis for antibody recognition of the NANP repeats in Plasmodium falciparum circumsporozoite protein. Proc Natl Acad Sci U S A 114: E10438–E10445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pompon J, Levashina EA (2015) A new role of the mosquito complement‐like Cascade in male fertility in Anopheles gambiae . PLoS Biol 13: e1002255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattabongkot J, Yimamnuaychoke N, Leelaudomlipi S, Rasameesoraj M, Jenwithisuk R, Coleman RE, Udomsangpetch R, Cui L, Brewer TG (2006) Establishment of a human hepatocyte line that supports in vitro development of the exo‐erythrocytic stages of the malaria parasites Plasmodium falciparum and P. vivax . Am J Trop Med Hyg 74: 708–715 [PubMed] [Google Scholar]
- Scally SW, Murugan R, Bosch A, Triller G, Costa G, Mordmuller B, Kremsner PG, Sim BKL, Hoffman SL, Levashina EA et al (2018) Rare PfCSP C‐terminal antibodies induced by live sporozoite vaccination are ineffective against malaria infection. J Exp Med 215: 63–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoute JA, Slaoui M, Heppner DG, Momin P, Kester KE, Desmons P, Wellde BT, Garcon N, Krzych U, Marchand M (1997) A preliminary evaluation of a recombinant circumsporozoite protein vaccine against Plasmodium falciparum malaria. RTS,S Malaria Vaccine Evaluation Group. N Engl J Med 336: 86–91 [DOI] [PubMed] [Google Scholar]
- Swearingen KE, Lindner SE, Shi L, Shears MJ, Harupa A, Hopp CS, Vaughan AM, Springer TA, Moritz RL, Kappe SH et al (2016) Interrogating the Plasmodium Sporozoite surface: identification of surface‐exposed proteins and demonstration of glycosylation on CSP and TRAP by mass spectrometry‐based proteomics. PLoS Pathog 12: e1005606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan J, Sack BK, Oyen D, Zenklusen I, Piccoli L, Barbieri S, Foglierini M, Fregni CS, Marcandalli J, Jongo S et al (2018) A public antibody lineage that potently inhibits malaria infection through dual binding to the circumsporozoite protein. Nat Med 24: 401–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thai E, Costa G, Weyrich A, Murugan R, Oyen D, Flores‐Garcia Y, Prieto K, Bosch A, Valleriani A, Wu NC et al (2020) A high‐affinity antibody against the CSP N‐terminal domain lacks Plasmodium falciparum inhibitory activity. J Exp Med 217: e20200061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triller G, Scally SW, Costa G, Pissarev M, Kreschel C, Bosch A, Marois E, Sack BK, Murugan R, Salman AM et al (2017) Natural parasite exposure induces protective human anti‐malarial antibodies. Immunity 47: 1197–1209.e10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl I, Obraztsova AS, Puchan J, Hundsdorfer R, Chakravarty S, Sim BKL, Hoffman SL, Kremsner PG, Mordmuller B, Wardemann H (2022) Clonal evolution and TCR specificity of the human TFH cell response to Plasmodium falciparum CSP. Sci Immunol 7: eabm9644 [DOI] [PubMed] [Google Scholar]
- Wang Q, Fujioka H, Nussenzweig V (2005) Mutational analysis of the GPI‐anchor addition sequence from the circumsporozoite protein of Plasmodium . Cell Microbiol 7: 1616–1626 [DOI] [PubMed] [Google Scholar]
- Wang LT, Pereira LS, Flores‐Garcia Y, O'Connor J, Flynn BJ, Schon A, Hurlburt NK, Dillon M, Yang ASP, Fabra‐Garcia A et al (2020) A potent anti‐malarial human monoclonal antibody targets circumsporozoite protein minor repeats and neutralizes sporozoites in the liver. Immunity 53: 733–744.e8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LT, Pereira LS, Kiyuka PK, Schon A, Kisalu NK, Vistein R, Dillon M, Bonilla BG, Molina‐Cruz A, Barillas‐Mury C et al (2021) Protective effects of combining monoclonal antibodies and vaccines against the Plasmodium falciparum circumsporozoite protein. PLoS Pathog 17: e1010133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wardemann H, Busse CE (2019) Expression cloning of antibodies from single human B cells. Methods Mol Biol 1956: 105–125 [DOI] [PubMed] [Google Scholar]
- Wardemann H, Yurasov S, Schaefer A, Young JW, Meffre E, Nussenzweig MC (2003) Predominant autoantibody production by early human B cell precursors. Science 301: 1374–1377 [DOI] [PubMed] [Google Scholar]
- WHO (2021) World malaria report 2021, p 2021. Geneva: World Health Organization; [Google Scholar]
- Yaari G, Vander Heiden JA, Uduman M, Gadala‐Maria D, Gupta N, Stern JN, O'Connor KC, Hafler DA, Laserson U, Vigneault F et al (2013) Models of somatic hypermutation targeting and substitution based on synonymous mutations from high‐throughput immunoglobulin sequencing data. Front Immunol 4: 358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavala F, Cochrane AH, Nardin EH, Nussenzweig RS, Nussenzweig V (1983) Circumsporozoite proteins of malaria parasites contain a single immunodominant region with two or more identical epitopes. J Exp Med 157: 1947–1957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavala F, Tam JP, Hollingdale MR, Cochrane AH, Quakyi I, Nussenzweig RS, Nussenzweig V (1985) Rationale for development of a synthetic vaccine against Plasmodium falciparum malaria. Science 228: 1436–1440 [DOI] [PubMed] [Google Scholar]


