Abstract
Oocyte maturation arrest is one of the important causes of female infertility, but the genetic factors remain largely unknown. PABPC1L, a predominant poly(A)‐binding protein in Xenopus, mouse, and human oocytes and early embryos prior to zygotic genome activation, plays a key role in translational activation of maternal mRNAs. Here, we identified compound heterozygous and homozygous variants in PABPC1L that are responsible for female infertility mainly characterized by oocyte maturation arrest in five individuals. In vitro studies demonstrated that these variants resulted in truncated proteins, reduced protein abundance, altered cytoplasmic localization, and reduced mRNA translational activation by affecting the binding of PABPC1L to mRNA. In vivo, three strains of Pabpc1l knock‐in (KI) female mice were infertile. RNA‐sequencing analysis showed abnormal activation of the Mos‐MAPK pathway in the zygotes of KI mice. Finally, we activated this pathway in mouse zygotes by injecting human MOS mRNA, and this mimicked the phenotype of KI mice. Our findings reveal the important roles of PABPC1L in human oocyte maturation and add a genetic potential candidate gene to be screened for causes of infertility.
Keywords: female infertility, Mos‐MAPK, mRNA translational activation, oocyte maturation arrest, PABPC1L
Subject Categories: Genetics, Gene Therapy & Genetic Disease; Urogenital System
Seven rare variants in PABPC1L were identified in five infertile females mainly characterized by oocyte maturation arrest followed a recessive inheritance pattern. In vitro and in vivo functional studies confirmed the destructive effects and pathogenicity of PABPC1L variants.
The paper explained.
Problem
Oocyte maturation arrest is one of the common factors leading to repeated failures of assisted reproduction and female infertility. Studies have confirmed that genetic factors play a crucial role in oocyte maturation arrest, including TUBB8, PATL2, and TRIP13, but the underlying genetic factors for most affected individuals are still unknown.
Results
We identified compound heterozygous and homozygous pathogenic variants in PABPC1L that are responsible for female infertility mainly characterized by oocyte maturation arrest in five individuals from four independent families. PABPC1L encodes a poly(A)‐binding protein that plays a key role in translational activation of maternal mRNAs. In vitro studies demonstrated that pathogenic variants resulted in the functional impairment on PABPC1L, including truncated proteins, reduced protein abundance, altered cytoplasmic localization, and reduced mRNA translational activation by affecting the binding of PABPC1L to mRNA. In vivo, we constructed three strains of Pabpc1l KI mice corresponding to patient‐derived variants and found that female mice were infertile due to abnormal activation of the Mos‐MAPK pathway in zygotes. These results demonstrate the destructive effects and pathogenicity of the PABPC1L variants identified in affected individuals both in vitro and in vivo.
Impact
We identified and verified bi‐allelic pathogenic variants in PABPC1L that cause oocyte maturation arrest and female infertility. Our findings provide direct evidence for the important role of PABPC1L in human oocyte maturation and thus add a new marker for the genetic diagnosis of infertility patients in the clinic.
Introduction
Mammalian oocyte maturation involves two successive meiotic divisions, and these are accompanied by a series of molecular events, including germinal vesicle breakdown (GVBD), spindle assembly, extrusion of the first polar body (PB1), and finally arrest at meiotic metaphase II (MII) (Mihajlovic & FitzHarris, 2018). Only fully mature MII oocytes can be recognized and fertilized by sperm and undergo early embryonic development (Plachot & Mandelbaum, 1990). Oocyte maturation is therefore a prerequisite for successful human reproduction. In clinical practice, oocyte maturation arrest is a common cause of recurrent failure of assisted reproductive technology (ART), including in vitro fertilization (IVF), and intracytoplasmic sperm injection (ICSI), and such failure is characterized by the repeated production of immature oocytes with resulting infertility (Huang et al, 2018). Four types of maturation arrest have been described, namely, germinal vesicle (GV) arrest, metaphase I (MI) arrest, MII arrest, and mixed arrest (oocyte arrest at more than one stage) (Beall et al, 2010). In 2016, we found that pathogenic variants in the primate‐specific gene TUBB8 (MIM: 616768) cause human oocyte MI arrest by impairing oocyte spindle assembly (Feng et al, 2016). Subsequently, studies have shown that pathogenic variants in PATL2 (MIM: 614661) and TRIP13 (MIM: 604507) cause oocyte GV arrest and MI arrest, respectively (Maddirevula et al, 2017; Chen et al, 2017b; Zhang et al, 2020). Zona pellucida is an extracellular glycoprotein matrix composed of ZP1, ZP2, ZP3, and ZP4, which plays a vital role in oocyte maturation. Studies have reported that the variants in ZP1(MIM: 195000), ZP2 (MIM: 182888), and ZP3 (MIM: 182889) affect the formation of zona pellucida and result in female infertility (Huang et al, 2014; Liu et al, 2017; Dai et al, 2019), but there is no convincing evidence to prove that ZP4 (MIM: 613514) variant responsible for female infertility. Pathogenic variants in TUBB8, PATL2, and TRIP13 account for around 10% of infertility patients with oocyte maturation arrest (Feng et al, 2016; Huang et al, 2017; Maddirevula et al, 2017; Chen et al, 2017a, 2017b, 2019; Wang et al, 2018; Wu et al, 2019; Liu et al, 2020; Zhang et al, 2020; Cao et al, 2021; Yang et al, 2021; Yao et al, 2022). However, the genetic basis and the mechanisms involved in the majority of affected individuals remain unknown.
Oocyte maturation requires poly(A) tail‐modulated translational activation of dormant maternal mRNAs stored in the cytoplasm (Gebauer et al, 1994; Stebbins‐Boaz et al, 1996; Mendez et al, 2000). A specialized PABP (poly(A)‐binding protein) called PABPC1L (poly(A)‐binding protein cytoplasmic 1‐like) plays a key role in these processes. PABPC1L, also known as EPAB (embryonic poly(A)‐binding protein), was originally identified in Xenopus (Voeltz et al, 2001) and was found to be essential for Xenopus oocyte maturation (Friend et al, 2012). During Xenopus oocyte maturation, PABPC1L binds to the elongated poly(A) tail to stabilize polyadenylated mRNAs (Voeltz et al, 2001; Kim & Richter, 2007). Meanwhile, poly(A)‐associated PABPC1L promotes the dissociation of the translational suppressor Maskin from eIF4E and establishes eIF4G–eIF4E interactions for the initiation of translation (Cao & Richter, 2002). In mice, Pabpc1l is expressed in both immature GV and MI oocytes and mature MII oocytes, as well as in one‐cell and two‐cell embryos, and it is replaced by a somatic PABP (Pabpc1) after zygotic genome activation (ZGA) (Seli et al, 2005). Pabpc1l −/− female mice are infertile and unable to generate mature oocytes due to impaired translational activation of maternal mRNAs, including Ccnb1, c‐Mos, and Dazl, upon stimulation of oocyte maturation (Guzeloglu‐Kayisli et al, 2012). In addition, late antral follicles in the ovaries of Pabpc1l −/− mice exhibited defective cumulus expansion and ovulation (Guzeloglu‐Kayisli et al, 2012; Yang et al, 2016). Similar to the observations in Xenopus and mice, human PABPC1L is the predominant PABP in oocytes (Guzeloglu‐Kayisli et al, 2008). To date, PABPC1L has not been found to be associated with any diseases in humans.
In this study, we identified compound heterozygous and homozygous pathogenic variants in PABPC1L (GRCh37, GenBank: NM_001124756.2) that are responsible for female infertility mainly characterized by oocyte maturation arrest. All five affected individuals carried biallelic variants, including missense, stop gain, or frameshift insertions, and all the five followed a Mendelian recessive inheritance pattern. We investigated the functional mechanism of the corresponding pathogenic variants in HeLa cells and Pabpc1l knock‐in (KI) mice. Our findings suggest a causal relationship between PABPC1L and female infertility and reveal important roles for PABPC1L in human oocyte maturation.
Results
Clinical characteristics of the affected individuals
To investigate the genetic factors responsible for oocyte maturation arrest, we recruited 1,394 infertile female individuals diagnosed with oocyte maturation arrest. In this study, all five affected individuals had been diagnosed with primary infertility of unknown cause for several years despite their having normal menstrual cycles. Their male partners had normal sperm counts and normal sperm morphologic features and motility. The pedigrees of the individuals are shown in Fig 1A. Clinical characteristic of affected individuals are shown in Table 1. The affected individual in family 1 underwent one failed IVF attempt. All the retrieved oocytes were arrested at the GV or MI stages (Fig 1B). The affected individual in family 2 underwent one failed IVF attempt and one failed ICSI attempt. In her IVF cycle, all retrieved oocytes were immature. In her ICSI cycle, 16 oocytes were obtained, of which 13 oocytes were immature. In addition, two mature MII oocytes could be fertilized, but the embryos arrested at the 1‐cell and 3‐cell stages, respectively. The affected individual in family 3 underwent one failed IVF attempt in which 11 oocytes were retrieved. Nine were arrested at the GV or MI stage. One mature MII oocyte was successfully fertilized, but this embryo was arrested at the 2‐cell stage. The affected individuals in family 4 underwent one failed IVF attempt and one failed ICSI attempt. In her IVF cycle, two MII oocytes were successfully fertilized but failed to cleave, and the remaining mature oocytes failed to fertilize. In her ICSI cycle, eight oocytes were arrested at the MI stage and the other two MII oocytes failed to fertilize (Fig 1B). Her sister underwent one failed IVF attempt in which 10 MII oocytes were retrieved but could not be fertilized (Fig 1B).
Figure 1. Identification of pathogenic variants in PABPC1L and the morphology of normal and affected individuals' oocytes.
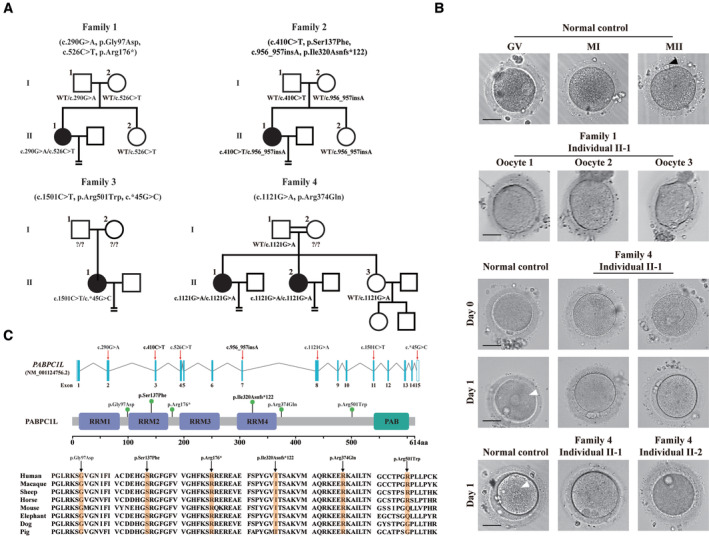
- Pedigrees of the four affected families. Squares indicate male members, circles indicate female members, black circles indicate the affected individuals, question marks indicate unavailable DNA samples, and equal signs (=) indicate infertility.
- The morphology of normal and affected individuals' oocytes or zygotes by light microscopy. The normal oocytes extruded PB1 (black arrowhead) and entered MII. Oocytes from individual II‐1 of family 1 were arrested at the GV or MI stage. The normal MII oocytes formed pronuclei (white arrowhead) on day 1 after ART. However, MII oocytes from individuals II‐1 and II‐2 of family 4 failed to fertilize and form pronuclei on day 1 after ART. Scale bar represents 40 μm.
- Locations and conservation of variants in PABPC1L. Red arrows and green balls indicate the variants identified in this study. PABPC1L contains four different RNA recognition motifs (RRMs) and a C‐terminal protein domain (PABP).
Source data are available online for this figure.
Table 1.
Clinical characteristics of affected individuals and their retrieved oocytes.
| Individual | Age (Years) | Duration of infertility (Years) | IVF/ICSI cycles | Total number of oocytes retrieved | GV oocytes | MI oocytes | MII oocytes | Oocytes with abnormal morphology | Fertilized oocytes | Cleaved embryos |
|---|---|---|---|---|---|---|---|---|---|---|
| II‐1 in Family 1 | 25 | 4 | IVF | 7 | 5 | 2 | 0 | 0 | 0 | 0 |
| II‐1 in Family 2 | 33 | 7 | IVF | 21 | 5 | 8 | 0 | 8 | 0 | 0 |
| II‐1 in Family 2 | 33 | 7 | ICSI | 16 | 3 | 4 | 3 | 6 | 2 | 1 |
| II‐1 in Family 3 | 29 | 4 | IVF | 11 | 7 | 2 | 2 | 0 | 1 | 1 |
| II‐1 in Family 4 | 33 | 6 | IVF | 11 | 0 | 1 | 10 | 0 | 2 | 0 |
| II‐1 in Family 4 | 33 | 6 | ICSI | 11 | 0 | 8 | 2 | 1 | 0 | 0 |
| II‐2 in Family 4 | 29 | 3 | IVF | 15 | 0 | 4 | 10 | 1 | 0 | 0 |
Abbreviations are as follows: GV, germinal vesicle; MI, metaphase I; and MII, metaphase II.
Identification of bi‐allelic pathogenic variants in PABPC1L
We performed whole‐exome sequencing and bioinformatics analyses in accordance with our established protocol (see Material and Methods) to identify potential genetic causes of the oocyte maturation arrest observed in our patients. We first identified three infertile individuals from three independent families (families 1–3) harboring bi‐allelic variants in PABPC1L (GenBank: NM_001124756.2) (Fig 1A). No homozygous or compound heterozygous variants in PABPC1L were found in our control database. The affected individual in family 1 carried biallelic variants c.290G>A (p.Gly97Asp) and c.526C>T (p.Arg176*). The affected individual in family 2 carried the compound heterozygous variants c.410C>T (p.Ser137Phe) and c.956_957insA (p.Ile320Asnfs*122), whereas the affected individual in family 3 carried the variants c.1501C>T (p.Arg501Trp) and 3′UTR variant c.*45G>C. These results implied the genetic contribution of PABPC1L to oocyte maturation arrest and female infertility. We then performed mutational screening of PABPC1L in a cohort of 504 infertile individuals with abnormalities in fertilization and early embryonic development. We identified two affected sisters from family 4 who carried the same homozygous missense variant c.1121G>A (p.Arg374Gln). All identified variants were confirmed using Sanger sequencing (Fig EV1). The inheritance pattern in family 3 was unknown due to the unavailability of DNA samples from her parents, whereas the other three families followed a recessive inheritance pattern (Fig 1A). Specific information about the variants, including the genomic position, predicted damaging effect, and frequency, is provided in Table 2. PABPC1L contains 15 exons encoding a 614 amino acid protein, and the locations of the variants in the gene and protein structure are shown in Fig 1C. Most of the corresponding residues affected by the variants are conserved among different species, whereas the variant c.1501C>T (p.Arg501Trp) is only conserved in some species (Fig 1C). We measured the expression of PABPC1L mRNA in control samples and found that PABPC1L was highly expressed in human immature GV and MI oocytes, but was poorly expressed in mature MII oocyte, early embryos, and other somatic tissues (Fig EV2), suggesting an important role for PABPC1L in human oocyte maturation.
Figure EV1. Sanger sequencing confirmation of PABPC1L variants in four families.
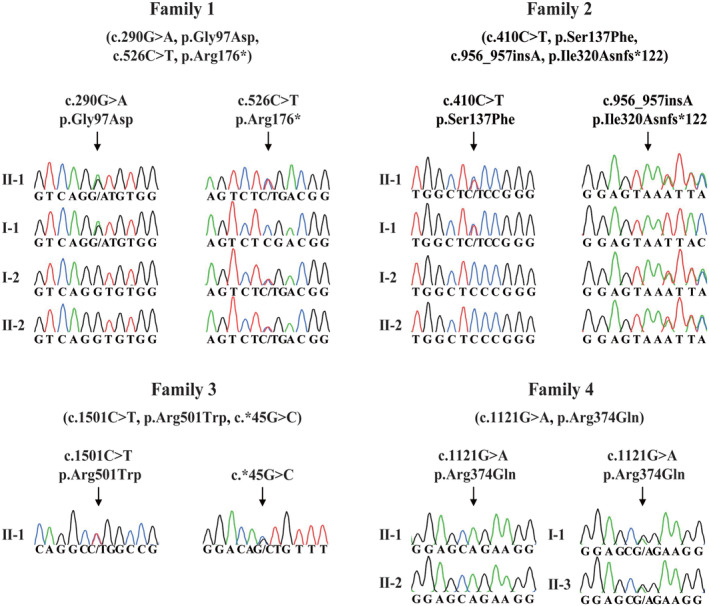
PABPC1L variants observed in the four families were confirmed using Sanger sequencing. Arrows indicate the mutation sites.Source data are available online for this figure.
Table 2.
PABPC1L pathogenic variants observed in the four families.
| Probands in families | Genomic position on chr20 (bp) | cDNA change | Protein change | Mutation type | SIFT a | PPH2 a | MutTas a | gnomAD b |
|---|---|---|---|---|---|---|---|---|
| Family 1 | 43541397 | c.290G>A | p.Gly97Asp | missense | D | P | D | NA |
| 43547569 | c.526C>T | p.Arg176* | stop gain | NA | NA | D | 4.2 × 10−6 | |
| Family 2 | 43545419 | c.410C>T | p.Ser137Phe | missense | D | P | D | 4.0 × 10−6 |
| 43552881 | c.956_957insA | p.Ile320Asnfs*122 | frameshift insertion | NA | NA | D | NA | |
| Family 3 | 43564088 | c.1501C>T | p.Arg501Trp | missense | D | P | P | 8.0 × 10−6 |
| 43567805 | c.*45G>C | – | missense | NA | NA | P | 7.2 × 10−4 | |
| Family 4 | 43559249 | c.1121G>A | p.Arg374Gln | missense | D | P | D | 1.2 × 10−5 |
PABPC1L—poly(A)‐binding protein cytoplasmic 1 like.
Mutation assessment by SIFT, PolyPhen‐2 (PPH2), and MutationTaster (MutTas). D, damaging; P, probably damaging; and NA, not available.
Allele frequency of corresponding mutations in the gnomAD database. NA, not available.
Figure EV2. Relative expression level of PABPC1L mRNA in different stages of human oocytes, embryos and several somatic tissues.
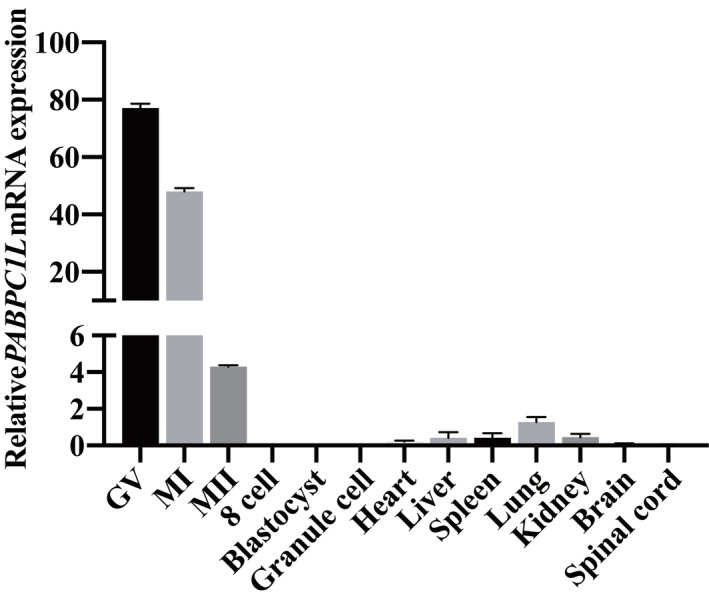
The relative expression of PABPC1L mRNA in different stages of human oocytes, early embryos, and several somatic tissues was measured by qRT–PCR and normalized to the expression of GAPDH mRNA (control). n = 3 biological replicates. Error bars denote the SD.Source data are available online for this figure.
Pathogenic variants in PABPC1L impaired protein expression and localization in HeLa cells
To evaluate the functional effects of the identified pathogenic variants in vitro, we first performed immunoblot analysis in HeLa cells transfected with HA‐labeled WT or mutant PABPC1L expression plasmids. Compared with WT, the c.526C>T (p.Arg176*) and c.956_957insA (p.Ile320Asnfs*122) variants resulted in truncated proteins and showed significantly reduced PABPC1L protein levels. The missense variants c.290G>A (p.Gly97Asp), c.410C>T (p.Ser137Phe), c.1501C>T (p.Arg501Trp), and c.1121G>A (p.Arg374Gln) did not lead to changes in protein levels (Fig 2A and B). Furthermore, to determine whether the 3′UTR variant c.*45G>C affects gene expression, we cloned WT and mutant 3′UTR segments into a dual luciferase reporter plasmid psiCHECK‐2 vector (Fig 2C). The luciferase assay in HeLa cells indicated that the WT 3′UTR significantly increased the reporter gene expression, whereas the mutant 3′UTR failed to up‐regulate the reporter gene expression compared to the empty vector (Fig 2D). This result suggests that the 3′UTR variant c.*45G>C may affect the expression of PABPC1L.
Figure 2. Effects of PABPC1L pathogenic variants on protein abundance and localization in HeLa cells.
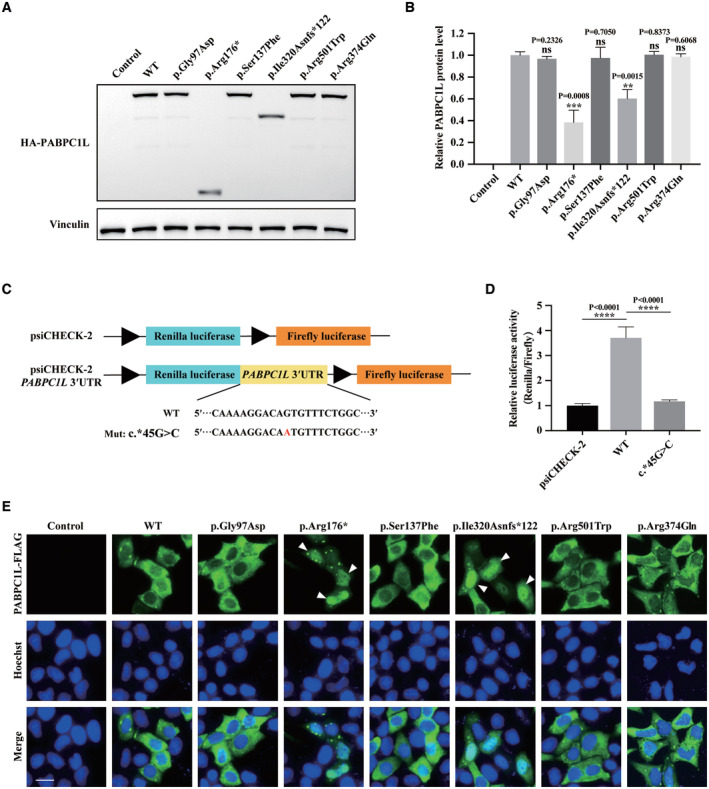
- The effects of the pathogenic variants on PABPC1L expression by immunoblot in HeLa cells. Empty vector was used as the control. Vinculin was used as the loading control.
- Quantitation of the Control, WT, and mutant PABPC1L protein levels shown in Fig 3A. n = 3 biological replicates. The statistics are analyzed by unpaired two‐tailed Student's t‐test. Data are shown as the mean and SD. **P < 0.005, ***P < 0.001.
- Schematic representation of dual luciferase reporter plasmid psiCHECK‐2 vectors containing the WT and mutant PABPC1L 3′UTR inserted downstream of the Renilla luciferase gene. The mutant nucleotide is marked in red.
- HeLa cells were transfected with luciferase reporter constructs. Empty vector was used as the control. Relative Renilla luciferase activity was determined and normalized to Firefly luciferase activity. n = 4 biological replicates. The statistics are analyzed by unpaired two‐tailed Student's t‐test. Data are shown as the mean and SD. ****P < 0.0001.
- HeLa cells were transfected with FLAG‐tagged full‐length WT or mutant PABPC1L expression plasmids and stained with an anti‐FLAG M2‐FITC antibody (green). Nuclei were labeled with Hoechst (blue). White arrowheads indicate that the p.Arg176* and p.Ile320Asnfs*122 mutant proteins were mostly mislocalized in the nucleus compared to the typical cytoplasmic localization for WT protein. Scale bar represents 20 μm.
Source data are available online for this figure.
It has been reported that PABPC1L is mainly localized in the cytoplasm in mouse oocytes (Uysal & Ozturk, 2019). The subcellular localization of proteins is important for their function, so to further investigate whether the identified variants affect the function of PABPC1L, immunofluorescence was performed using a commercial anti‐FLAG antibody in HeLa cells transfected with FLAG‐tagged WT or mutant PABPC1L expression plasmids. As shown in Fig 2E, the WT PABPC1L was mainly localized in the cytoplasm, while the variants c.526C>T (p.Arg176*) and c.956_957insA (p.Ile320Asnfs*122) resulted in mislocalization of most protein in the nucleus. The missense variants c.290G>A (p.Gly97Asp), c.410C>T (p.Ser137Phe), c.1501C>T (p.Arg501Trp), and c.1121G>A (p.Arg374Gln) had no effect on protein cytoplasmic localization. These results demonstrate that the identified pathogenic variants c.526C>T (p.Arg176*) and c.956_957insA (p.Ile320Asnfs*122) affect both the protein abundance and localization of PABPCL, while the 3′UTR variant c.*45G>C affects the protein expression of the reporter gene in HeLa cells.
Pathogenic variants in PABPC1L disrupted its mRNA translational activation and RNA‐binding ability in HeLa cells
PABPC1L is an RNA‐binding protein that promotes the translational activation of maternal RNA in Xenopus and mouse oocytes, which is required for oocyte maturation (Wilkie et al, 2005; Guzeloglu‐Kayisli et al, 2012). Because the missense variants c.290G>A (p.Gly97Asp), c.410C>T (p.Ser137Phe), c.1501C>T (p.Arg501Trp), and c.1121G>A (p.Arg374Gln) had no effect on protein level or localization, we speculated that these variants might influence the protein's ability to promote mRNA translational activation. Therefore, we compared the translational activation capacity of WT and mutant PABPC1L by transfecting a luciferase reporter together with WT or mutant PABPC1L expression plasmids in HeLa cells. All of the variants except c.1121G>A (p.Arg374Gln) resulted in significantly lower luciferase activity than the WT group (Fig 3A), while luciferase mRNA levels were not significantly different between the groups (Fig 3B). These results indicate that the missense variants c.290G>A (p.Gly97Asp), c.410C>T (p.Ser137Phe), and c.1501C>T (p.Arg501Trp) led to a significant decrease in mRNA translational activation in HeLa cells.
Figure 3. Effects of PABPC1L pathogenic variants on its mRNA translational activation and RNA‐binding ability in HeLa cells.
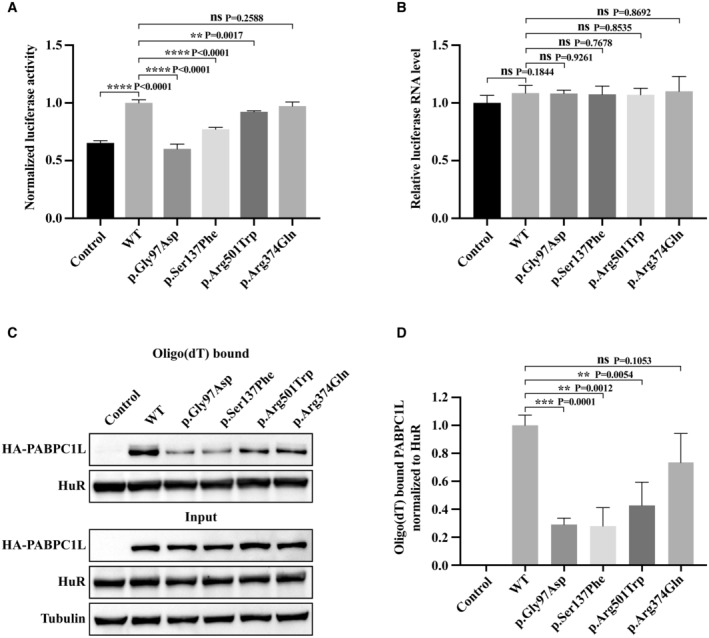
- HeLa cells were transfected with FLB reporter (Firefly luciferase fused to β‐globin) together with WT or mutant PABPC1L expression plasmids, and cell extracts were assayed for Firefly luciferase activity. Empty vector was used as the control. n = 4 biological replicates. The statistics are analyzed by unpaired two‐tailed Student's t‐test. Data are shown as the mean and SD. **P < 0.01, ****P < 0.0001.
- FLB mRNA levels were determined from the same cell extracts and normalized to the GAPDH mRNA level. n = 3 biological replicates. The statistics are analyzed by unpaired two‐tailed Student's t‐test. Data are shown as mean and SD. ns, not significant.
- Representative immunoblots for WT and mutant PABPC1L bound to RNA in HeLa cells. HuR was used as the internal control, and Tubulin was used as the loading control.
- Quantitation of the oligo‐dT–bound WT or mutant PABPC1L signal normalized to the HuR signal. n = 4 biological replicates. The statistics are analyzed by unpaired two‐tailed Student's t‐test. Data are shown as the mean and SD. **P < 0.01, ***P < 0.001, ns, not significant.
Source data are available online for this figure.
PABPC1L promotes the translational activation of mRNAs by interacting with the poly(A) tail of mRNA (Voeltz et al, 2001; Cao & Richter, 2002). To determine whether the missense variants affect PABPC1L's RNA‐binding ability, we performed RNA pull‐down experiments in HeLa cells. The results showed that the variants c.290G>A (p.Gly97Asp), c.410C>T (p.Ser137Phe), and c.1501C>T (p.Arg501Trp) had dramatically reduced RNA‐binding ability compared to WT (Fig 3C and D), indicating that these variants resulted in reduced mRNA translation activation by affecting the binding of PABPC1L to RNA. The variant c.1121G>A (p.Arg374Gln) also had a slight effect on RNA binding, but the difference was not significant (Fig 3C and D). The above in vitro data demonstrate that the identified pathogenic variants in PABPC1L have disruptive effects on protein function.
Pabpc1l KI female mice exhibit early embryonic arrest and infertility
To further explore the pathogenic role of PABPC1L variants in vivo, we utilized the CRISPR‐Cas9 system to generate Pabpc1l G97D, S137F, and R374Q KI mice corresponding to the pathogenic variants c.290G>A (p.Gly97Asp), c.410C>T (p.Ser137Phe), and c.1121G>A (p.Arg374Gln) identified in families 1, 2, and 4 respectively. We performed PCR and Sanger sequencing to confirm the mutated allele in the three KI mice (Fig EV3A). All three Pabpc1l KI mice were viable, but females were infertile (Fig 4A). To gain insights into the etiology of infertility in females, we first assessed the ovarian histology of unstimulated mature WT and Pabpc1l KI mice, and no differences were observed between them (Fig EV3B and C). Next, we performed phenotypic analysis of Pabpc1l KI females in terms of oocyte maturation, fertilization, and early embryonic development. Different from the phenotype observed in affected individuals with these three variants, Pabpc1l KI females ovulated mature MII oocytes after superovulation treatment (Fig EV3D). Furthermore, the fertilization rate of Pabpc1l KI oocytes was normal (Fig EV3E), and they formed pronuclei at 6 h after IVF (Fig 4C). However, in contrast to the WT zygotes, the cleavage of Pabpc1l KI zygotes was delayed, and most KI zygotes could cleavage in the subsequent in vitro culture, but all embryos eventually arrested at an early stage and failed to develop into blastocysts (Fig 4B and C). These results further indicated that the PABPC1L variants are indeed pathogenic for female fertility in the affected individuals and that there are species‐specific differences in the effects of mutant PABPC1L in humans and mice.
Figure EV3. Genotyping and phenotype analyses in homozygous Pabpc1l KI mice.
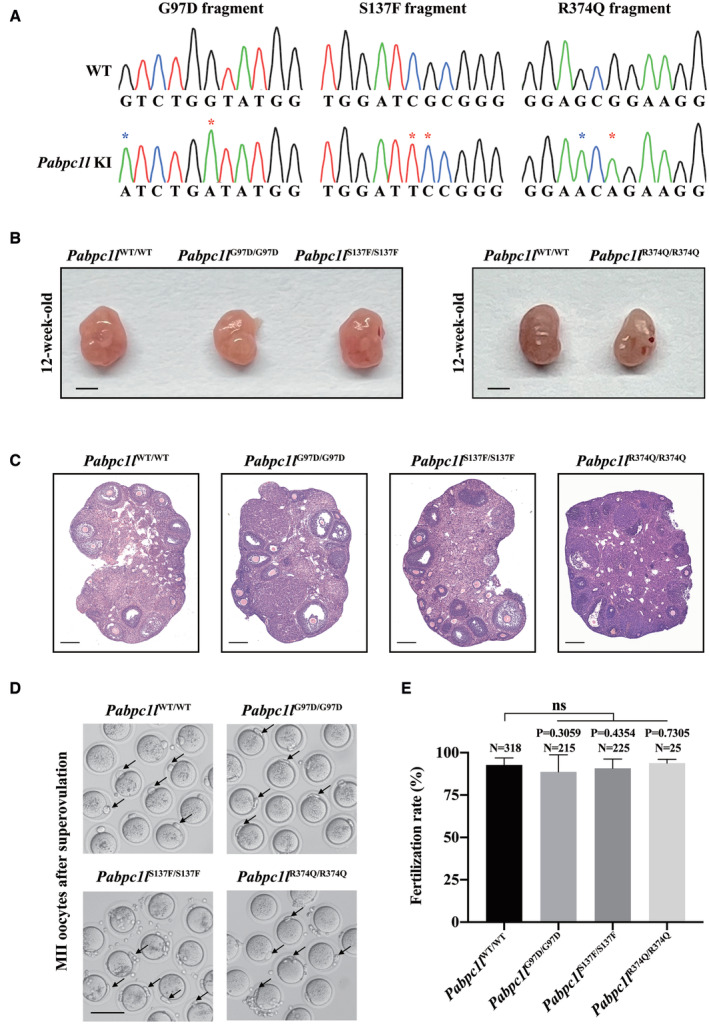
- Sanger sequencing showing the homozygous mutations in Pabpc1l KI mice. Red asterisks indicate the mutation sites, and the blue asterisk indicates the synonymous mutation site.
- Ovary morphology of 12‐week‐old WT and Pabpc1l KI mice. Scale bar represents 1 mm.
- Histological sections of ovaries from 12‐week‐old WT and Pabpc1l KI mice were stained with hematoxylin and eosin. Scale bars represent 200 μm.
- Representative images of superovulated oocytes of WT and Pabpc1l KI mice. The black arrows indicate PB1. Scale bar represents 100 μm.
- Quantitative analysis of the fertilization rate of WT and Pabpc1l KI mice. The number of analyzed embryos is indicated (N). n = 8 biological replicates for Pabpc1l WT/WT, Pabpc1l G97D/G97D, and Pabpc1l S137F/S137F mice. n = 2 biological replicates for Pabpc1l R374Q/R374Q mice. The statistics are analyzed by unpaired two‐tailed Student's t‐test. Data are shown as mean and SD. ns, not significant.
Figure 4. Pabpc1l KI female mice exhibit abnormal early embryonic development and infertility.
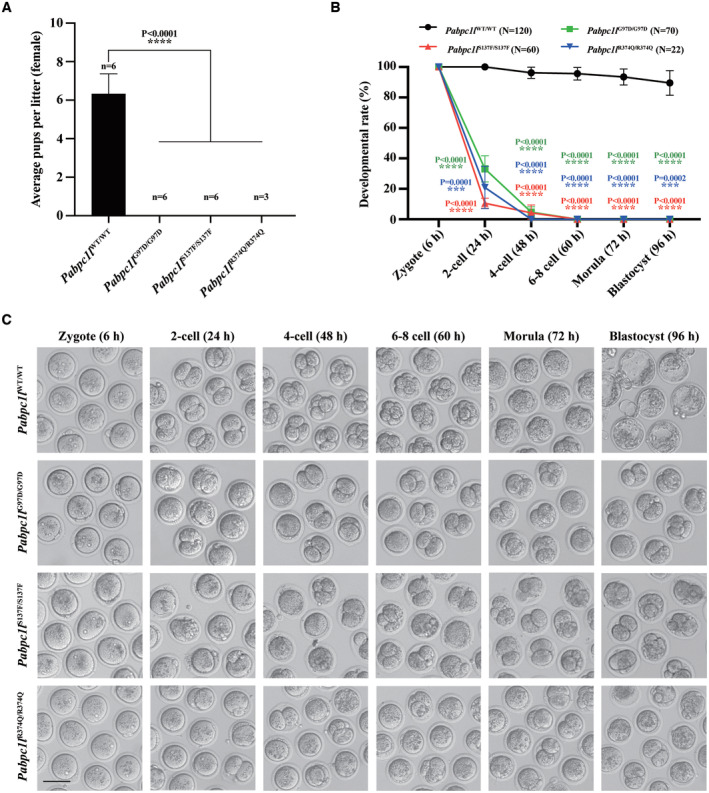
- The reproductive ability of female WT and Pabpc1l KI mice. n = 6 for Pabpc1l WT/WT, Pabpc1l G97D/G97D, and Pabpc1l S137F/S137F mice. n = 3 for Pabpc1l R374Q/R374Q mice. The statistics are analyzed by unpaired two‐tailed Student's t‐test. Data are shown as mean and SD. ****P < 0.0001.
- Quantification of early embryonic development at different times after IVF of superovulated oocytes from WT and Pabpc1l KI mice. The number of analyzed embryos is indicated (N). n = 4 biological replicates for Pabpc1l WT/WT, Pabpc1l G97D/G97D, and Pabpc1l S137F/S137F mice. n = 2 biological replicates for Pabpc1l R374Q/R374Q mice. The statistics are analyzed by unpaired two‐tailed Student's t‐test. Data are shown as the mean and SD. ***P < 0.001, ****P < 0.0001.
- Representative images of early embryonic development at different times after IVF of superovulated oocytes from WT and Pabpc1l KI mice. Scale bar represents 100 μm.
Source data are available online for this figure.
Abnormal activation of the Mos‐MAPK pathway in zygotes causes early embryonic arrest in Pabpc1l KI mice
To explore the mechanism of early embryonic arrest in Pabpc1l KI mice, we performed RNA‐seq using WT and Pabpc1l KI zygotes. Gene expression levels were assessed as FPKM, and all replicates showed high correlations (Fig 5A). Compared to WT zygotes, 1,933 transcripts were downregulated and 1,952 transcripts were upregulated in G97D zygotes, while 872 transcripts were downregulated and 618 transcripts were upregulated in S137F zygotes (Fig 5B). Furthermore, Venn diagrams showed that there were 344 co‐upregulated transcripts and 457 co‐downregulated transcripts in the two groups (G97D vs. WT and S137F vs. WT; Fig 5C). GO analysis revealed that the co‐upregulated genes were mainly involved in positive regulation of mitogen‐activated protein kinase (MAPK) cascade pathways, phosphorylation, embryonic development, etc, while the co‐downregulated genes were involved in translation, cell cycle, meiotic spindle organization, etc. (Fig 5D). To further investigate the molecular mechanism, we first analyzed the differentially expressed genes in G97D zygotes and mainly focused on oocyte‐specific genes. We found that Mos, an upstream activator of MAPK cascade (Dupre et al, 2011), was the only oocyte‐specific gene among the top 30 differentially expressed genes (Dataset EV1; Fig 5B). Then we checked the expression of Mos in S137F zygotes and observed that it was also up‐regulated (Dataset EV1; Fig 5B). The up‐regulation of Mos in KI zygotes is consistent with the activation of MAPK cascade revealed in GO analysis. The above RNA‐seq analysis results strongly suggest that Mos‐MAPK pathway is abnormally activated in KI zygote. In addition, studies have indicated that abnormal activation of Mos‐MAPK pathway in embryo led to cleavage arrest (Sagata et al, 1989; Haccard et al, 1993; Verlhac et al, 2000), which is similar to the phenotype of early embryonic arrest in KI mice. Therefore, we supposed that the abnormal up‐regulation of Mos expression and activation of the Mos‐MAPK pathway in KI zygotes might be the potential mechanism of early embryonic arrest in Pabpc1l KI mice.
Figure 5. RNA‐seq analysis of the transcriptome of WT and Pabpc1l KI zygotes.
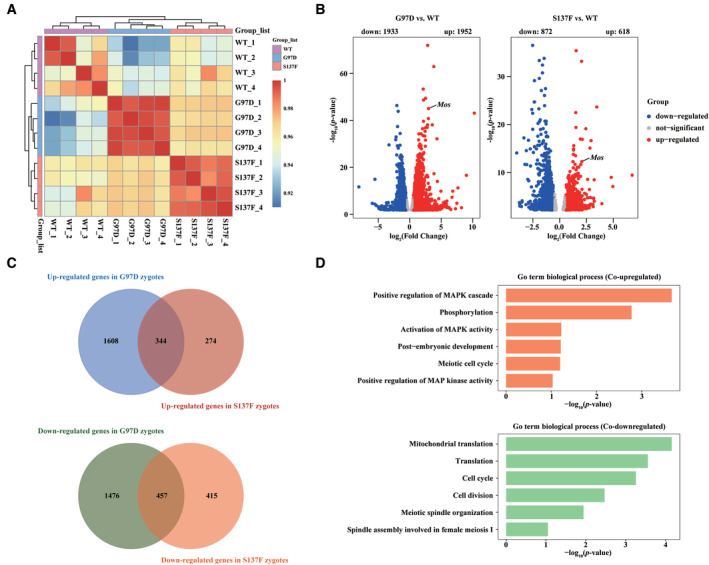
- Heatmap of the Spearman correlation coefficients in the RNA‐seq data obtained from WT, G97D, and S137F zygotes.
- Volcano plot of RNA‐seq data obtained from WT, G97D, and S137F zygotes. “Up” and “Down” are the upregulated and downregulated transcripts in KI zygotes compared with WT zygotes. The genes with upregulation and downregulation (abs[Fold Change] > 1.5, FDR <0.05) were labeled in red and blue, respectively.
- Venn diagram showing the number of co‐upregulated and co‐downregulated genes for G97D vs. WT and S137F vs. WT.
- GO analysis of co‐upregulated and co‐downregulated genes in G97D vs. WT and S137F vs. WT.
To determine whether Mos‐MAPK pathway is indeed abnormally activated in KI zygote, we first performed qRT‐PCR to confirm the RNA‐seq data. qRT‐PCR results showed that the expression levels of Mos and MAPK pathway‐related gene in G97D and S137F zygotes were indeed higher than those in WT zygotes, while the expression levels of Pabpc1l showed no differences between groups (Fig 6A). Phosphorylated ERK1/2 (pERK1/2) is a well‐known marker of MAPK pathway activation (Cao et al, 2021). To further confirm whether the MAPK pathway is activated in Pabpc1l KI zygotes, we collected the zygotes to examine the phosphorylation level of ERK1/2. As shown in Fig 6B and C, the pERK1/2 levels were significantly elevated in G97D, S137F, and R374Q zygotes compared with WT zygotes. These results demonstrate that the Mos‐MAPK pathway is indeed abnormally activated in Pabpc1l KI zygotes.
Figure 6. Pabpc1l pathogenic variants cause abnormal activation of the Mos‐MAPK pathway in zygotes and lead to early embryonic arrest in KI mice.
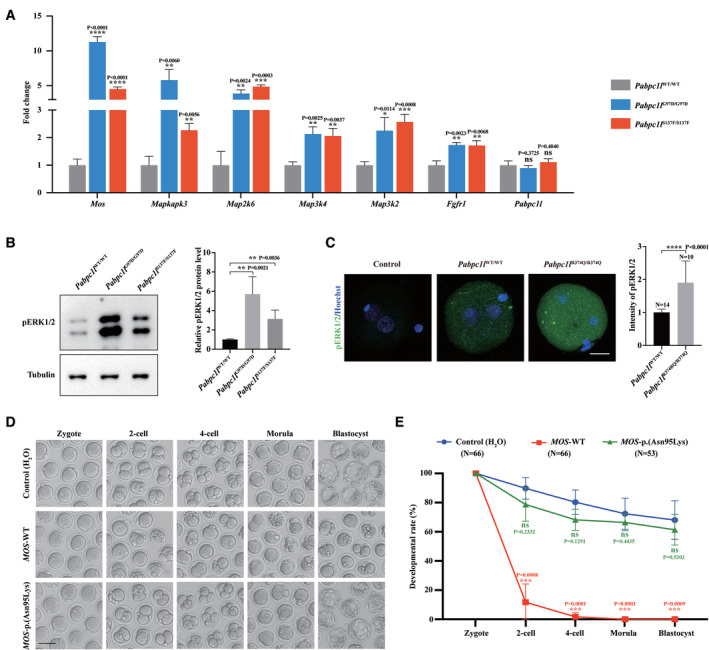
- qRT–PCR results verified the expression of upregulated Mos‐MAPK pathway‐related genes and Pabpc1l in the RNA‐seq data. n = 3 biological replicates. The statistics are analyzed by unpaired two‐tailed Student's t‐test. Data are shown as the mean and SD. *P < 0.01, **P < 0.005, ***P < 0.001, ****P < 0.0001, ns, not significant.
- Immunoblot analysis of pERK1/2 in WT, G97D, and S137F zygotes. Total protein from 50 zygotes was loaded in each lane. Tubulin was used as the loading control. n = 4 biological replicates. The statistics are analyzed by unpaired two‐tailed Student's t‐test. Data are shown as the mean and SD. **P < 0.005.
- Immunofluorescence analysis of pERK1/2 (green) in WT and R374Q zygotes. Hoechst (blue) was co‐stained to represent pronucleus. Scale bar represents 20 μm. The number of analyzed embryos is indicated (N). n = 2 biological replicates. The statistics are analyzed by unpaired two‐tailed Student's t‐test. Data are shown as the mean and SD. ****P < 0.0001.
- Representative images of early embryonic development at different times after WT zygotes were injected with H2O (Control), MOS‐WT, or MOS‐p.(Asn95Lys) cRNA. Scale bar represents 100 μm.
- Quantification of early embryonic development at different times after WT zygotes were injected with H2O (Control), MOS‐WT, or MOS‐p.(Asn95Lys) cRNA. The number of analyzed embryos is indicated (N). n = 3 biological replicates. The statistics are analyzed by unpaired two‐tailed Student's t‐test. Data are shown as the mean and SD. ***P < 0.001, ns, not significant.
Source data are available online for this figure.
The Mos‐MAPK pathway plays a crucial role in cytostatic factor‐induced oocyte MII arrest (Haccard et al, 1993; Colledge et al, 1994; Kosako et al, 1994). Previous studies in Xenopus and mice showed that injection of Mos mRNA into the blastomeres of two‐cell embryos could induce the abnormal activation of Mos‐MAPK pathway in blastomeres and eventually resulted in cleavage arrest. (Sagata et al, 1989; Verlhac et al, 2000). Thus, we microinjected zygotes with human MOS cRNA to activate the Mos‐MAPK pathway and observed the resulting embryonic development. Compared with the control group, injection of MOS‐WT cRNA caused early embryonic arrest (Fig 6D and E), mimicking the phenotype of Pabpc1l KI mice. However, injection of MOS‐p.(Asn95Lys) cRNA (a previously reported loss‐of‐function variant) (Zeng et al, 2022) did not result in the phenotype (Fig 6D and E). Together, these results indicate that PABPC1L pathogenic variants lead to increased Mos expression and abnormal activation of Mos‐MAPK pathway in zygotes, ultimately resulting in early embryonic arrest and female infertility in Pabpc1l KI mice.
Discussion
In this study, we identified different compound heterozygous and homozygous pathogenic variants in PABPC1L that are mainly responsible for human oocyte maturation arrest. In vitro studies showed that the truncating variants c.526C>T (p.Arg176*) and c.956_957insA (p.Ile320Asnfs*122) affected the expression pattern and localization of PABPC1L, while the missense variants c.290G>A (p.Gly97Asp), c.410C>T (p.Ser137Phe), and c.1501C>T (p.Arg501Trp) reduced its translation activation and RNA‐binding ability. Moreover, the 3′UTR variant c.*45G > C affected the protein expression of the luciferase reporter gene. Finally, we constructed three strains of Pabpc1l KI mice corresponding to patient‐derived variants and found that female mice were infertile. The Mos‐MAPK pathway was abnormally activated in the zygotes of KI mice, which may provide an explanation the infertile phenotype. These results demonstrate the destructive effects and pathogenicity of the PABPC1L variants identified in affected individuals both in vitro and in vivo.
Cytoplasmic PABPs (poly(A)‐binding proteins) are a family of multifunctional RNA‐binding proteins, including PABPC1, PABPC3, PABPC4, PABPC4L, PABPC5, and PABPC1L, that mediate mRNA metabolism at different stages of gametogenesis and early embryogenesis (Zhao & Fan, 2021). PABPC1L was initially discovered in 2001 as a novel ARE (AU‐rich element) binding protein in Xenopus oocyte extracts that regulates mRNA deadenylation, and it is the predominant PABP in Xenopus oocytes and early embryos (Voeltz et al, 2001). In the last two decades, the physiological function of the PABPC1L‐mediated translational activation of maternal mRNA in oocyte and early embryo has been well characterized in studies in Xenopus and mice. The amino acid sequence of PABPC1L is highly conserved in Xenopus and mammals, and PBAPC1L is also the predominant PABP in human oocytes and early embryos (Guzeloglu‐Kayisli et al, 2008). However, the physiological function of human PABPC1L is unknown. In this study, we showed that PABPC1L was highly expressed in human immature oocytes (Fig EV2) and that WT human PABPC1L could significantly stimulate the mRNA translation of reporter gene in vitro (Fig 3A). On the other hand, patient‐derived variants (c.290G>A (p.Gly97Asp), c.410C>T (p.Ser137Phe), and c.1501C>T (p.Arg501Trp)) disrupted the translational activation of PABPC1L (Fig 3A) and resulted in female infertility characterized by oocyte maturation arrest (Fig 1B and Table 1). Therefore, these results suggest that PABPC1L plays an important role in mRNA translational activation during human oocyte maturation and that it is essential for female fertility.
Previous studies have found that patients with different variants in the same oocyte maturation arrest‐related genes, including TUBB8, PATL2 and TRIP13, exhibit various phenotypes (Chen et al, 2019; Wu et al, 2019; Zhang et al, 2020). This may be due to different variants having different effects on protein function. In this study, all retrieved oocytes of the affected individual from family 1 were arrested at GV or MI stages (Fig 1B and Table 1). However, a few retrieved oocytes of the affected individuals in families 2 and 3 were able to mature and be fertilized, but they arrested at the 1‐cell to 3‐cell stages prior to ZGA (Table 1). PABPC1L plays a key role in the polyadenylation‐dependent translational activation of mRNAs, which is essential for oocyte maturation (Wakiyama et al, 2000; Guzeloglu‐Kayisli et al, 2012). Our functional experiment demonstrated that the pathogenic variant c.290G>A (p.Gly97Asp) from family 1 was significantly more disruptive to the translational activation ability of PABPC1L than variants c.410C>T (p.Ser137Phe) and c.1501C>T (p.Arg501Trp) from families 2 and 3 (Fig 3A). This suggests that the severity of the phenotype in affected individuals with PABPC1L variants depends on the degree of impairment of PABPC1L function resulting from the different variants. Furthermore, all embryos obtained from the affected individuals in families 2 and 3 were arrested at an early stage, suggesting that PABPC1L also play an important role in early embryonic development prior to ZGA.
For individuals II‐1 and II‐2 in consanguineous family 4, some oocytes obtained from ART were also immature and arrested at the MI stage. However, unlike the affected individuals in other families, most of their oocytes could extrude the PB1 to develop into mature MII oocytes but showed subsequent fertilization failure (Table 1). The two individuals carried the same homozygous variant c.1121G>A (p.Arg374Gln) in PABPC1L and showed the same infertile phenotype, while individual II‐3 carried the same heterozygous variant c.1121G>A (p.Arg374Gln) and had normal fertility. This result suggests that the c.1121G>A (p.Arg374Gln) variant may cause the phenotype of fertilization failure following a recessive inheritance pattern. The unique fertilization failure phenotype might be related to the observation that the c.1121G>A (p.Arg374Gln) variant had a slight effect on the RNA‐binding ability of PABPC1L (Fig 3B) but had no effect on mRNA translational activation (Fig 3A). To further confirm the pathogenicity of the variant c.1121G>A (p.Arg374Gln), we constructed Pabpc1l R374Q KI mice corresponding to this variant. Phenotypic evaluation found that Pabpc1l R374Q/R374Q female mice were infertile characterized by early embryonic arrest (Fig 4A–C), which was consistent with the other two KI mice. In addition, immunofluorescence analysis showed that the level of pERK1/2 in the R374Q KI zygotes was significantly increased (Fig 6C), indicating that the Mos‐MAPK pathway was also abnormally activated. These results fully demonstrate that the variant c.1121G>A (p.Arg374Gln) identified in family 4 has disruptive effects on protein function and ultimately leads to female infertility. We inferred that the biochemical mechanism of Arg374Gln in vitro may be different from other variants, but all variants have the similar pathological mechanism in vivo.
Pabpc1l knock‐out (KO) female mice were previously shown to be infertile due to impaired oocyte maturation (Guzeloglu‐Kayisli et al, 2012). In this study, we constructed three strains of Pabpc1l KI mouse models – Pabpc1l G97D/G97D Pabpc1l S137F/S137F, and Pabpc1l R374Q/R374Q – and unlike the phenotype of KO mice, the retrieved oocytes from KI mice matured and could be fertilized normally (Fig EV3D and E), but the embryos developed abnormally and most embryos were arrested at the 2‐cell stage prior to ZGA (Fig 4B and C). One possible explanation for this is that the mutant PABPC1L still has some function during oocyte maturation, but this activity is not sufficient for further early embryonic development after fertilization. Our Immunoblot and immunofluorescence analysis showed that the missense variants c.290G>A (p.Gly97Asp), c.410C>T (p.Ser137Phe), and c.1121G>A (p.Arg374Gln) had no effect on protein levels and cytoplasmic localization of PABPC1L (Fig 2A and E). In addition, RNA pull‐down assay indicated that the pathogenic variants c.290G>A (p.Gly97Asp) and c.410C>T (p.Ser137Phe) resulted in a significant decrease in the RNA‐binding ability of PABPC1L compared to WT; however, there was still a small amount of mutant PABPC1L bound to RNA (Fig 3C and D). This further suggests that the phenotypic severity of individuals with PABPC1L variants correlates with the relative loss of function of the variants.
MOS is a serine/threonine kinase that activates the MAPK cascade through direct phosphorylation of the MAP kinase activator MEK (Posada et al, 1993). The Mos‐MAPK pathway is commonly activated during vertebrate oocyte maturation, but its function seems to be different among species. In Xenopus oocytes, the activation of the Mos‐MAPK pathway is necessary for triggering GVBD and for cytostatic factor‐induced MII arrest (Sagata et al, 1989; Haccard et al, 1993; Posada et al, 1993). However, Mos deletion in mouse oocytes has shown that the Mos‐MAPK pathway is not required for GVBD but is important for MII arrest and female fertility (Colledge et al, 1994; Choi et al, 1996; Hashimoto, 1996). In this study, we demonstrated that PABPC1L pathogenic variants lead to increased Mos expression and abnormal activation of the Mos‐MAPK pathway in mouse zygotes. Considering that the affected individuals with variants in PABPC1L showed either oocyte maturation arrest or fertilization failure, we believe that dysfunction of PABPC1L may lead to abnormal MOS‐MAPK pathway activity and ultimately affect human oocyte maturation and fertilization processes.
This study has some limitations. First, this study lacks experimental analysis of oocytes or embryos from affected individuals with PABPC1L variants, because we have not obtained valuable clinical samples from these patients. Second, although we have confirmed the pathogenicity of the PABPC1L variants and provided the exploration of the mechanism leading to phenotype, we have not found a possible treatment to overcome infertility. Previous studies have found that microinjection of Pabpc1l mRNA into Pabpc1l −/− preantral follicle‐enclosed oocytes rather than denuded GV oocytes could rescue oocyte maturation (Guzeloglu‐Kayisli et al, 2012; Lowther & Mehlmann, 2015). This strategy might provide a possible treatment option to overcome female infertility caused by PABPC1L dysfunction, but additional experiments should be pursed for evaluating effectiveness to embryonic development.
In this study, we identified five individuals with bi‐allelic variants in PABPC1L, which accounts for 0.26% of our cohort of 1,898 infertile woman (1,394 with oocyte maturation arrest and 504 with abnormalities in fertilization and early embryonic development). In addition, we also found other candidate pathogenic genes through bioinformatics analysis, but the pathogenicity of these mutant genes needs to be further verified by in vitro experiments and the generation of knockout or patient‐derived mutated animal models. Nevertheless, the genetic basis involved in the majority of affected individuals remain unknown, which requires extensive exploration using different research strategies. Previous studies have shown that gene‐based burden test is an effective strategy for finding novel pathogenic genes in many other disorders (Cirulli et al, 2015; Guo et al, 2018; Malik et al, 2021). By using the strategy, LHX8 and KPNA7 variants were identified to cause female infertility (Zhao et al, 2022; Wang et al, 2023). We believe that applying gene‐based burden test will reveal more genetic basis of female infertility. In addition, there is a good understanding of the functional impact of protein‐coding variants in female infertility (Jiao et al, 2021), but less understanding of variants in non‐coding regions, which accounts for 98% of the human genome. In the last decade, many studies have identified that variants in non‐coding regions cause different diseases (Turro et al, 2020; Wright et al, 2021; Wakeling et al, 2022), which suggests that the analysis of non‐coding regions in female infertility patients with unknown causes is also a powerful strategy to find novel pathogenic variants.
In conclusion, we identified bi‐allelic pathogenic variants in PABPC1L that cause oocyte maturation arrest and female infertility. Our findings provide direct evidence for the important role of PABPC1L in female reproduction and thus add a potential genetic candidate gene to be screened for causes of infertility patients in the clinic.
Materials and Methods
Clinical samples
In this study, five infertile individuals with pathogenic variants in PABPC1L and healthy controls were recruited from the Reproductive and Genetic Hospital of CITIC‐Xiangya, the Shanghai Ji Ai Genetics and IVF Institute affiliated with the Obstetrics and Gynecology Hospital of Fudan University, and the Reproductive Medicine Center of the Shaanxi Maternal and Child Care Service Center. A cohort of 504 infertile individuals with abnormalities in fertilization and early embryonic development was recruited from 62 collaborating hospitals. All blood samples were donated for the investigation after informed consent was obtained. The study was approved by the Ethics Committee of the Medical College of Fudan University and the Reproductive Study Ethics Committee of the hospitals. These experiments conformed to the principles set out in the WMA Declaration of Helsinki and the Department of Health and Human Services Belmont Report.
Whole‐exome sequencing and bioinformatics analysis
Genomic DNA was extracted from peripheral blood using the QIAamp DNA Blood Mini Kit (Qiagen), and whole‐exome sequencing was performed using the SeqCap EZ Exome Kit (Roche) on an Illumina HiSeq 3000 platform (Illumina). Sequencing analysis was compared with the human reference sequence (NCBI Genome build GRCh37). Variants were annotated with GRCh37 and the dbSNP (version 138) database. All candidate variants were filtered using the following criteria: (1) variants that conformed to the autosomal recessive inheritance pattern; (2) variants with a minor allele frequency less than 0.1% (for homozygous variants) or 1% (for compound heterozygous variants) in the public databases, including gnomAD, 1000 Genomes Project, or ExAC databases, and not existing in our control database; (3) variants predicted to be loss of function or to be damaging by SIFT, PolyPhen‐2, and MutationTaster; and (4) variants within genes that were highly expressed or specifically expressed in oocytes. The candidate pathogenic variants were subsequently verified by Sanger sequencing of the affected probands as well as all the available family members. The primers used are shown in Appendix Table S1.
PABPC1L expression analysis and real‐time quantitative PCR (qRT‐PCR)
Total RNA from control samples (including human GV, MI, and MII oocytes, Day 3 embryos (8‐cell), blastocysts, and other somatic tissues) were extracted with the RNeasy Mini Kit (Qiagen). Reverse transcription was performed with the PrimeScript RT Reagent Kit (Takara) according to the manufacturer's instructions. qRT‐PCR was performed with TB Green Premix Ex Taq (Takara) in triplicate on a LightCycler 480 II System (Roche). The expression of PABPC1L was normalized by comparison to the expression of an internal human GAPDH control. The qRT‐PCR primers for PABPC1L and GAPDH are shown in Appendix Table S1.
Construction of expression plasmids and transfection
The full‐length coding sequence and 3′‐untranslated region (3′UTR) of PABPC1L was amplified from control human oocyte cDNA. The coding sequence product was then cloned into the pcDNA3 vector containing the N‐HA tag or the pCMV6‐Entry vector containing the C‐FLAG tag, and the 3′UTR product was cloned into the psiCHECK‐2 vector. Site‐directed mutagenesis was performed to introduce the identified variants (p.Gly97Asp, p.Arg176*, p.Ser137Phe, p.Ile320Asnfs*122, p.Arg501Trp, p.Arg374Gln, and c.*45G>C) into the wild‐type (WT) vector according to the instructions of the KOD‐Plus‐Mutagenesis Kit (Toyobo Life Science). HeLa cells (Cell Bank of Shanghai Institute for Biological Sciences) were cultured in DMEM (Gibco) supplemented with 10% fetal bovine serum (Gibco) and 1% penicillin/streptomycin (Gibco) in a 5% CO2 atmosphere at 37°C. The WT and mutant expression plasmids were transfected into HeLa cells using the PolyJet In Vitro DNA Transfection Reagent (Signagen) according to a standard protocol.
Oligo(dT) pull‐down assay
HeLa cells were harvested 36 h post‐transfection and washed three times with cold PBS. Cells were lysed in NP‐40 lysis buffer with 1% protease inhibitor cocktail (Bimake) and RNase inhibitor (NEB). After quantification with the Bicinchoninic Acid Protein Assay Kit (Bo Cai Biological Technology), equal amounts of protein extracts were incubated with Oligo d(T)25 magnetic beads (NEB). After incubation at 4°C for 2 h, the beads were washed with lysis buffer three times. The bead‐bound proteins were eluted using sodium dodecyl sulfate (SDS) sample buffer.
Immunoblot analysis
All immunoblot samples were heated at 100°C for 10 min. Equal amounts of protein were separated by SDS‐polyacrylamide gel electrophoresis (EZBiolab) and transferred to nitrocellulose membranes (Pall Corporation). The membranes were blocked with 5% non‐fat milk diluted in PBS with 0.1% Tween 20 (PBST) for 1 h and then incubated at 4°C overnight with primary antibodies. The membranes were washed with PBST three times and incubated with secondary antibodies for 1 h at room temperature followed by washing again with PBST three times. Finally, enhanced chemiluminescence imaging was performed on a chemiluminescent imaging system (5200, Tanon). Quantitation of immunoblot results was performed with the ImageJ software (National Institutes of Health). The following primary antibodies were used for immunoblotting: antibodies against HA Tag (3724, CST), HuR (2582, CST), and pERK1/2 (4370, CST) were used at 1:1,000 dilution, and antibodies against Vinculin (13901, CST) and Tubulin (T6199, Sigma) were used at 1:3,000 dilution as the internal controls. The secondary antibodies were goat anti‐rabbit IgG (1:5,000 dilution, M21001L, Abmart) or goat anti‐mouse IgG (1:5,000 dilution, M21002L, Abmart) conjugated to horseradish peroxidase.
Immunofluorescent staining
HeLa cells were transfected with WT or mutant PABPCL expression plasmids, and cells were fixed for immunofluorescence after culturing for 36 h. Transfected cells were first treated with 4% paraformaldehyde (P6148, Sigma) for 30 min at room temperature, washed three times in PBS, and then blocked in a solution containing 1% bovine serum albumin (BSA) (B2064, Sigma) and 0.1% Triton X‐100 (T8787, Sigma) in PBS for 1 h at room temperature. The cells were washed again and then incubated with anti‐FLAG M2‐FITC antibody (1:200 dilution, F4049, Sigma) for 1 h at 37°C. After being washed three times, the cells were incubated with Hoechst (1:500 dilution, 33342, BD Biosciences) for 15 min at room temperature. Images were captured with the EVOS M7000 Microscope Imaging System (Thermo Fisher Scientific).
Mouse zygotes were fixed with 2% paraformaldehyde diluted in PBS for 30 min at room temperature and then permeabilized in PBS containing 0.5% Triton X‐100 for 20 min at room temperature. After incubation for 2 h in blocking buffer (3% BSA diluted in PBS with 0.1% Tween‐20 and 0.01% TritonX‐100) at room temperature, zygotes were stained with anti‐pERK1/2 (1:400, 4370, CST) primary antibodies diluted in blocking buffer for 1 h at 37°C. After three washes, samples were incubated with Alexa Fluor 488‐conjugated donkey anti‐rabbit (1:500, A21206, Invitrogen) secondary antibodies for 1 h at 37°C. Pronucleus was briefly stained with Hoechst (1:500, 33342, BD Biosciences) for 15 min at room temperature. The samples were in PBS and imaged using a laser scanning confocal microscopy (LSM880, Zeiss).
Luciferase assay
The luciferase assay was performed with the Dual‐Luciferase Reporter Assay System (Promega). WT and mutant PABPC1L 3′UTR were cloned into the C‐terminus of the Renilla luciferase gene in the psiCHECK™‐2 vector. Empty vector was used as the control, and Firefly luciferase expression served as the internal standard for transfection efficiency. HeLa cell extracts were prepared 36 h after transfection and assayed for the Firefly and Renilla luciferase activities. Renilla luciferase activity values were normalized to Firefly activity. Results are presented as the means obtained from three independent experiments.
Generation of Pabpc1l KI mice
Pabpc1l KI mice were generated using the CRISPR‐Cas9 gene‐targeting approach. All animal operations were approved by the Shanghai Medical College of Fudan University. The guide RNA (gRNA) target site for mouse Pabpc1l close to the disease variant was selected using the CRISPR design tool (G97D‐gRNA: 5′‐GGC CTT CGG AAG TCT GGT ATG GG‐3′, S137F‐gRNA‐1: 5′‐TAC AAT GAA CAT GGA TCG CGG GG‐3′, S137F‐gRNA‐2: 5′‐ATA CAA TGA ACA TGG ATC GCG GG‐3′, R374Q‐gRNA‐1: 5′‐ACA GCG CAA GGA GGA GCG GAA GG‐3′, R374Q‐gRNA‐2: 5′‐TGG CAC AGC GCA AGG AGG AGC GG‐3′). Additionally, the donor template of G97D and R374Q contained the synonymous AAG to AAA p.K95 mutation and GAG to GAA p.E373 mutation to avoid gRNA recognition and excision. To produce the Cas9 mRNA, gRNA, and donor oligo, a T7 promoter was first fused to the Cas9 coding region of the px330 plasmid by PCR amplification. The T7‐Cas9 PCR product was purified and used as a template for in vitro transcription using the mMESSAGE mMACHINE T7 ULTRA Kit (Life Technologies). A T7 promoter was fused to the gRNA template by PCR amplification, and the T7‐gRNA PCR product was purified and used as a template for in vitro transcription using the MEGAshortscript T7 Kit (Life Technologies). Both the Cas9 mRNA and the gRNAs were purified using a MEGAclear kit (Life Technologies) and eluted in RNase‐free water. Oligo donors were obtained as ultramer DNA oligos from GenScript Corporation. Female C57BL/6 mice (Beijing Vital River Laboratory Animal Technology Co.) were superovulated by injecting 10 IU of pregnant mare serum gonadotropin (PMSG, Ningbo Second Hormone Factory), followed 48 h later by injection of 10 IU of human chorionic gonadotropin (hCG, Ningbo Second Hormone Factory). They were then mated to C57BL/6 males, and fertilized embryos were collected from the oviducts. Cas9 mRNA (100 ng/ml), gRNAs (50 ng/ml), and donor oligos (100 ng/ml) were mixed and injected into the cytoplasm of fertilized oocytes using an IM300 microinjector (Narishige). About 30 injected single‐cell embryos were transferred into the oviducts of pseudopregnant ICR females at 0.5 days after coitus. For the identification of Pabpc1l point mutation founders, DNA was extracted and the target fragment was amplified followed by sequence analysis. The primers are shown in Appendix Table S1.
Mouse oocyte collection, fertilization, and embryo culture in vitro
To obtain GV oocytes, ovaries were isolated from female mice (10–12 weeks old). GV oocytes were isolated from ovaries by puncturing the antral follicles with a fine needle on the stage of a dissecting microscope. For IVF, female mice (10–12 weeks old) were intraperitoneally injected with 10 IU of PMSG. After 46 h, mice were injected with 10 IU of hCG. After an additional 13 h, oocyte/cumulus masses were surgically isolated from the oviducts and were mixed with sperm in human tubal fluid medium (Nanjing Aibei Biotechnology Co.) for IVF. Zygotes were cultured in drops of K‐modified simplex optimized medium (Nanjing Aibei Biotechnology Co.) under oil at 37°C in an atmosphere of 5% CO2, and embryo development was monitored at regular intervals. All mouse experiments were reviewed and approved by the Shanghai Medical College of Fudan University.
RNA sequencing (RNA‐seq)
Each sample included five zygotes. RNA‐seq libraries were prepared using the cDNA Synthesis, Amplification and Library Generation Kit (E6420, NEB). RNA quality was examined by gel electrophoresis and with Qubit (Thermo). RNA samples were sequenced on the Illumina Novaseq 6000 instrument by Genergy Biotechnology Co. Ltd. The raw data were handled by trim galore software, and data quality was checked by FastQC v0.11.2. The read length was 2 × 150 bp. Clean reads were aligned to the mouse genome mm10 using STAR and StringTie. The expression levels of the genes were quantified using normalized fragments per kilobase million (FPKM). Differential gene expression analysis was performed by DESeq2 with read counts data. The thresholds for determining differentially expressed transcripts were abs[Fold Change] > 1.5 and FDR < 0.05. Venn software was used to identify the co‐upregulated and co‐downregulated transcripts in the two databases (G97D vs. WT and S137F vs. WT). Gene Ontology (GO) term analysis was performed using the DAVID online tool. The expression of Mos‐MAPK pathway‐related genes and Pabpc1l were determined using qRT‐PCR with mouse Actb expression as the internal control. The primers are shown in Appendix Table S1.
Statistical analyses
Experiments were performed with biological replicates as indicated in the Figure legends, and GraphPad Prism (GraphPad Software) was used to perform the statistical analyses. Differences were analyzed by Student's t‐tests when comparing experimental groups. Statistically significant values of P < 0.05, P < 0.01, P < 0.001, and P < 0.0001 are indicated by (*), (**), (***), and (****), respectively.
Author contributions
Lei Wang: Conceptualization; supervision; funding acquisition; writing – review and editing. Weijie Wang: Conceptualization; data curation; formal analysis; funding acquisition; investigation; visualization; methodology; writing – original draft; writing – review and editing. Jing Guo: Resources; investigation. Juanzi Shi: Resources; investigation. Qun Li: Data curation; formal analysis. Biaobang Chen: Data curation; formal analysis. Zhiqi Pan: Formal analysis; investigation. Ronggui Qu: Investigation. Jing Fu: Resources; investigation. Rong Shi: Resources; investigation. Xia Xue: Resources; investigation. Jian Mu: Methodology. Zhihua Zhang: Formal analysis. Tianyu Wu: Visualization. Wenjing Wang: Methodology. Lin Zhao: Validation. Qiaoli Li: Project administration. Lin He: Resources. Xiaoxi Sun: Resources. Qing Sang: Conceptualization; supervision; funding acquisition. Ge Lin: Resources; supervision.
Disclosure and competing interests statement
The authors declare no competing interests.
For more information
MutationTaster, http://mutationtaster.org/MutationTaster
OMIM, https://www.omim.org
PolyPhen‐2, http://genetics.bwh.harvard.edu/pph2/
DAVID online tool, https://david.ncifcrf.gov
Supporting information
Appendix
Expanded View Figures PDF
Dataset EV1
Source Data for Expanded View
PDF+
Source Data for Figure 1
Source Data for Figure 2
Source Data for Figure 3
Source Data for Figure 4
Source Data for Figure 6
Acknowledgements
We thank all the staff at the Reproductive and Genetic Hospital of CITIC‐Xiangya, the Shanghai Ji Ai Genetics and IVF Institute affiliated with the Obstetrics and Gynecology Hospital of Fudan University, and the Reproductive Medicine Center of the Shaanxi Maternal and Child Care Service Center. This work was supported by the National Natural Science Foundation of China (82288102), the National Key Research and Development Program of China (2021YFC2700100), the National Natural Science Foundation of China (32130029, 82171643, 81971450, 82201829), the Medical Engineering Fund of Fudan University (yg2021‐012), the Open Research Funds of the State Key Laboratory of Genetic Engineering, Fudan University (SKLGE‐2106), and the China Postdoctoral Science Foundation (2021M700829).
EMBO Mol Med (2023) 15: e17177
See also: X Ding & JC Schimenti (June 2023)
Contributor Information
Ge Lin, Email: linggf@hotmail.com.
Lei Wang, Email: wangleiwanglei@fudan.edu.cn.
Data availability
The processing RNA‐seq dataset supporting conclusion is included within the supplemental data. The raw sequence data reported in this paper have been deposited in the Genome Sequence Archive in National Genomics Data Center, China National Center for Bioinformation/Beijing Institute of Genomics, Chinese Academy of Sciences (GSA: CRA009502) that are publicly accessible at https://bigd.big.ac.cn/gsa/browse/CRA009502.
References
- Beall S, Brenner C, Segars J (2010) Oocyte maturation failure: a syndrome of bad eggs. Fertil Steril 94: 2507–2513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Q, Richter JD (2002) Dissolution of the maskin‐eIF4E complex by cytoplasmic polyadenylation and poly(a)‐binding protein controls cyclin B1 mRNA translation and oocyte maturation. EMBO J 21: 3852–3862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Q, Zhao C, Wang C, Cai L, Xia M, Zhang X, Han J, Xu Y, Zhang J, Ling X et al (2021) The recurrent mutation in PATL2 inhibits its degradation thus causing female infertility characterized by oocyte maturation defect through regulation of the Mos‐MAPK pathway. Front Cell Dev Biol 9: 628649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Li B, Li D, Yan Z, Mao X, Xu Y, Mu J, Li Q, Jin L, He L et al (2017a) Novel mutations and structural deletions in TUBB8: expanding mutational and phenotypic spectrum of patients with arrest in oocyte maturation, fertilization or early embryonic development. Hum Reprod 32: 457–464 [DOI] [PubMed] [Google Scholar]
- Chen B, Zhang Z, Sun X, Kuang Y, Mao X, Wang X, Yan Z, Li B, Xu Y, Yu M et al (2017b) Biallelic mutations in PATL2 cause female infertility characterized by oocyte maturation arrest. Am J Hum Genet 101: 609–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Wang W, Peng X, Jiang H, Zhang S, Li D, Li B, Fu J, Kuang Y, Sun X et al (2019) The comprehensive mutational and phenotypic spectrum of TUBB8 in female infertility. Eur J Hum Genet 27: 300–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi T, Fukasawa K, Zhou R, Tessarollo L, Borror K, Resau J, Vande Woude GF (1996) The Mos/mitogen‐activated protein kinase (MAPK) pathway regulates the size and degradation of the first polar body in maturing mouse oocytes. Proc Natl Acad Sci U S A 93: 7032–7035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirulli ET, Lasseigne BN, Petrovski S, Sapp PC, Dion PA, Leblond CS, Couthouis J, Lu YF, Wang Q, Krueger BJ et al (2015) Exome sequencing in amyotrophic lateral sclerosis identifies risk genes and pathways. Science 347: 1436–1441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colledge WH, Carlton MB, Udy GB, Evans MJ (1994) Disruption of c‐Mos causes parthenogenetic development of unfertilized mouse eggs. Nature 370: 65–68 [DOI] [PubMed] [Google Scholar]
- Dai C, Hu L, Gong F, Tan Y, Cai S, Zhang S, Dai J, Lu C, Chen J, Chen Y et al (2019) ZP2 pathogenic variants cause in vitro fertilization failure and female infertility. Genet Med 21: 431–440 [DOI] [PubMed] [Google Scholar]
- Dupre A, Haccard O, Jessus C (2011) Mos in the oocyte: how to use MAPK independently of growth factors and transcription to control meiotic divisions. J Signal Transduct 2011: 350412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng R, Sang Q, Kuang Y, Sun X, Yan Z, Zhang S, Shi J, Tian G, Luchniak A, Fukuda Y et al (2016) Mutations in TUBB8 and human oocyte meiotic arrest. N Engl J Med 374: 223–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friend K, Brook M, Bezirci FB, Sheets MD, Gray NK, Seli E (2012) Embryonic poly(a)‐binding protein (ePAB) phosphorylation is required for Xenopus oocyte maturation. Biochem J 445: 93–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebauer F, Xu W, Cooper GM, Richter JD (1994) Translational control by cytoplasmic polyadenylation of c‐Mos mRNA is necessary for oocyte maturation in the mouse. EMBO J 13: 5712–5720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo MH, Plummer L, Chan YM, Hirschhorn JN, Lippincott MF (2018) Burden testing of rare variants identified through exome sequencing via publicly available control data. Am J Hum Genet 103: 522–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzeloglu‐Kayisli O, Pauli S, Demir H, Lalioti MD, Sakkas D, Seli E (2008) Identification and characterization of human embryonic poly(a) binding protein (EPAB). Mol Hum Reprod 14: 581–588 [DOI] [PubMed] [Google Scholar]
- Guzeloglu‐Kayisli O, Lalioti MD, Aydiner F, Sasson I, Ilbay O, Sakkas D, Lowther KM, Mehlmann LM, Seli E (2012) Embryonic poly(a)‐binding protein (EPAB) is required for oocyte maturation and female fertility in mice. Biochem J 446: 47–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haccard O, Sarcevic B, Lewellyn A, Hartley R, Roy L, Izumi T, Erikson E, Maller JL (1993) Induction of metaphase arrest in cleaving Xenopus embryos by MAP kinase. Science 262: 1262–1265 [DOI] [PubMed] [Google Scholar]
- Hashimoto N (1996) Role of c‐Mos proto‐oncogene product in the regulation of mouse oocyte maturation. Horm Res 46: 11–14 [DOI] [PubMed] [Google Scholar]
- Huang HL, Lv C, Zhao YC, Li W, He XM, Li P, Sha AG, Tian X, Papasian CJ, Deng HW et al (2014) Mutant ZP1 in familial infertility. N Engl J Med 370: 1220–1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Tong X, Luo L, Zheng S, Jin R, Fu Y, Zhou G, Li D, Liu Y (2017) Mutation analysis of the TUBB8 gene in nine infertile women with oocyte maturation arrest. Reprod Biomed Online 35: 305–310 [DOI] [PubMed] [Google Scholar]
- Huang L, Tong X, Wang F, Luo L, Jin R, Fu Y, Zhou G, Li D, Song G, Liu Y et al (2018) Novel mutations in PATL2 cause female infertility with oocyte germinal vesicle arrest. Hum Reprod 33: 1183–1190 [DOI] [PubMed] [Google Scholar]
- Jiao SY, Yang YH, Chen SR (2021) Molecular genetics of infertility: loss‐of‐function mutations in humans and corresponding knockout/mutated mice. Hum Reprod Update 27: 154–189 [DOI] [PubMed] [Google Scholar]
- Kim JH, Richter JD (2007) RINGO/cdk1 and CPEB mediate poly(a) tail stabilization and translational regulation by ePAB. Genes Dev 21: 2571–2579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosako H, Gotoh Y, Nishida E (1994) Requirement for the MAP kinase kinase/MAP kinase cascade in Xenopus oocyte maturation. EMBO J 13: 2131–2138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Li K, Bai D, Yin J, Tang Y, Chi F, Zhang L, Wang Y, Pan J, Liang S et al (2017) Dosage effects of ZP2 and ZP3 heterozygous mutations cause human infertility. Hum Genet 136: 975–985 [DOI] [PubMed] [Google Scholar]
- Liu Z, Zhu L, Wang J, Luo G, Xi Q, Zhou X, Li Z, Yang X, Duan J, Jin L et al (2020) Novel homozygous mutations in PATL2 lead to female infertility with oocyte maturation arrest. J Assist Reprod Genet 37: 841–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowther KM, Mehlmann LM (2015) Embryonic poly(a)‐binding protein is required during early stages of mouse oocyte development for chromatin organization, transcriptional silencing, and meiotic competence. Biol Reprod 93: 43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddirevula S, Coskun S, Alhassan S, Elnour A, Alsaif HS, Ibrahim N, Abdulwahab F, Arold ST, Alkuraya FS (2017) Female infertility caused by mutations in the oocyte‐specific translational repressor PATL2. Am J Hum Genet 101: 603–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik R, Beaufort N, Frerich S, Gesierich B, Georgakis MK, Rannikmae K, Ferguson AC, Haffner C, Traylor M, Ehrmann M et al (2021) Whole‐exome sequencing reveals a role of HTRA1 and EGFL8 in brain white matter hyperintensities. Brain 144: 2670–2682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez R, Hake LE, Andresson T, Littlepage LE, Ruderman JV, Richter JD (2000) Phosphorylation of CPE binding factor by Eg2 regulates translation of c‐Mos mRNA. Nature 404: 302–307 [DOI] [PubMed] [Google Scholar]
- Mihajlovic AI, FitzHarris G (2018) Segregating chromosomes in the mammalian oocyte. Curr Biol 28: R895–R907 [DOI] [PubMed] [Google Scholar]
- Plachot M, Mandelbaum J (1990) Oocyte maturation, fertilization and embryonic growth in vitro. Br Med Bull 46: 675–694 [DOI] [PubMed] [Google Scholar]
- Posada J, Yew N, Ahn NG, Vande Woude GF, Cooper JA (1993) Mos stimulates MAP kinase in Xenopus oocytes and activates a MAP kinase kinase in vitro. Mol Cell Biol 13: 2546–2553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagata N, Watanabe N, Vande Woude GF, Ikawa Y (1989) The c‐Mos proto‐oncogene product is a cytostatic factor responsible for meiotic arrest in vertebrate eggs. Nature 342: 512–518 [DOI] [PubMed] [Google Scholar]
- Seli E, Lalioti MD, Flaherty SM, Sakkas D, Terzi N, Steitz JA (2005) An embryonic poly(a)‐binding protein (ePAB) is expressed in mouse oocytes and early preimplantation embryos. Proc Natl Acad Sci U S A 102: 367–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbins‐Boaz B, Hake LE, Richter JD (1996) CPEB controls the cytoplasmic polyadenylation of cyclin, Cdk2 and c‐Mos mRNAs and is necessary for oocyte maturation in Xenopus. EMBO J 15: 2582–2592 [PMC free article] [PubMed] [Google Scholar]
- Turro E, Astle WJ, Megy K, Graf S, Greene D, Shamardina O, Allen HL, Sanchis‐Juan A, Frontini M, Thys C et al (2020) Whole‐genome sequencing of patients with rare diseases in a national health system. Nature 583: 96–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uysal F, Ozturk S (2019) Embryonic poly(a)‐binding protein is differently expressed and interacts with the messenger RNAs in the mouse oocytes and early embryos. J Cell Biochem 120: 4694–4709 [DOI] [PubMed] [Google Scholar]
- Verlhac MH, Lefebvre C, Kubiak JZ, Umbhauer M, Rassinier P, Colledge W, Maro B (2000) Mos activates MAP kinase in mouse oocytes through two opposite pathways. EMBO J 19: 6065–6074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voeltz GK, Ongkasuwan J, Standart N, Steitz JA (2001) A novel embryonic poly(a) binding protein, ePAB, regulates mRNA deadenylation in Xenopus egg extracts. Genes Dev 15: 774–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakeling MN, Owens NDL, Hopkinson JR, Johnson MB, Houghton JAL, Dastamani A, Flaxman CS, Wyatt RC, Hewat TI, Hopkins JJ et al (2022) Non‐coding variants disrupting a tissue‐specific regulatory element in HK1 cause congenital hyperinsulinism. Nat Genet 54: 1615–1620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakiyama M, Imataka H, Sonenberg N (2000) Interaction of eIF4G with poly(a)‐binding protein stimulates translation and is critical for Xenopus oocyte maturation. Curr Biol 10: 1147–1150 [DOI] [PubMed] [Google Scholar]
- Wang AC, Zhang YS, Wang BS, Zhao XY, Wu FX, Zhai XH, Sun JX, Mei SY (2018) Mutation analysis of the TUBB8 gene in primary infertile women with arrest in oocyte maturation. Gynecol Endocrinol 34: 900–904 [DOI] [PubMed] [Google Scholar]
- Wang W, Miyamoto Y, Chen B, Shi J, Diao F, Zheng W, Li Q, Yu L, Li L, Xu Y et al (2023) Karyopherin alpha deficiency contributes to human preimplantation embryo arrest. J Clin Invest 133: e159951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkie GS, Gautier P, Lawson D, Gray NK (2005) Embryonic poly(a)‐binding protein stimulates translation in germ cells. Mol Cell Biol 25: 2060–2071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright CF, Quaife NM, Ramos‐Hernandez L, Danecek P, Ferla MP, Samocha KE, Kaplanis J, Gardner EJ, Eberhardt RY, Chao KR et al (2021) Non‐coding region variants upstream of MEF2C cause severe developmental disorder through three distinct loss‐of‐function mechanisms. Am J Hum Genet 108: 1083–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Chen H, Li D, Song D, Chen B, Yan Z, Lyu Q, Wang L, Kuang Y, Li B et al (2019) Novel mutations in PATL2: expanding the mutational spectrum and corresponding phenotypic variability associated with female infertility. J Hum Genet 64: 379–385 [DOI] [PubMed] [Google Scholar]
- Yang CR, Lowther KM, Lalioti MD, Seli E (2016) Embryonic poly(a)‐binding protein (EPAB) is required for granulosa cell EGF signaling and cumulus expansion in female mice. Endocrinology 157: 405–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P, Yin C, Li M, Ma S, Cao Y, Zhang C, Chen T, Zhao H (2021) Mutation analysis of tubulin beta 8 class VIII in infertile females with oocyte or embryonic defects. Clin Genet 99: 208–214 [DOI] [PubMed] [Google Scholar]
- Yao Z, Zeng J, Zhu H, Zhao J, Wang X, Xia Q, Li Y, Wu L (2022) Mutation analysis of the TUBB8 gene in primary infertile women with oocyte maturation arrest. J Ovarian Res 15: 38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y, Shi J, Xu S, Shi R, Wu T, Li H, Xue X, Zhu Y, Chen B, Sang Q et al (2022) Bi‐allelic mutations in MOS cause female infertility characterized by preimplantation embryonic arrest. Hum Reprod 37: 612–620 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Li B, Fu J, Li R, Diao F, Li C, Chen B, Du J, Zhou Z, Mu J et al (2020) Bi‐allelic missense pathogenic variants in TRIP13 cause female infertility characterized by oocyte maturation arrest. Am J Hum Genet 107: 15–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao LW, Fan HY (2021) Revisiting poly(a)‐binding proteins: multifaceted regulators during gametogenesis and early embryogenesis. Bioessays 43: e2000335 [DOI] [PubMed] [Google Scholar]
- Zhao L, Li Q, Kuang Y, Xu P, Sun X, Meng Q, Wang W, Zeng Y, Chen B, Fu J et al (2022) Heterozygous loss‐of‐function variants in LHX8 cause female infertility characterized by oocyte maturation arrest. Genet Med 24: 2274–2284 [DOI] [PubMed] [Google Scholar]


