Abstract
Mycobacterium tuberculosis (Mtb) WhiB3 is an iron–sulfur cluster-containing transcription factor belonging to a subclass of the WhiB-Like (Wbl) family that is widely distributed in the phylum Actinobacteria. WhiB3 plays a crucial role in the survival and pathogenesis of Mtb. It binds to the conserved region 4 of the principal sigma factor (σA4) in the RNA polymerase holoenzyme to regulate gene expression like other known Wbl proteins in Mtb. However, the structural basis of how WhiB3 coordinates with σA4 to bind DNA and regulate transcription is unclear. Here we determined crystal structures of the WhiB3:σA4 complex without and with DNA at 1.5 Å and 2.45 Å, respectively, to elucidate how WhiB3 interacts with DNA to regulate gene expression. These structures reveal that the WhiB3:σA4 complex shares a molecular interface similar to other structurally characterized Wbl proteins and also possesses a subclass-specific Arg-rich DNA-binding motif. We demonstrate that this newly defined Arg-rich motif is required for WhiB3 binding to DNA in vitro and transcriptional regulation in Mycobacterium smegmatis. Together, our study provides empirical evidence of how WhiB3 regulates gene expression in Mtb by partnering with σA4 and engaging with DNA via the subclass-specific structural motif, distinct from the modes of DNA interaction by WhiB1 and WhiB7.
Keywords: Wbl family, WhiB3, transcription factor, iron–sulfur cluster, X-ray crystallography
The causative agent of tuberculosis, Mycobacterium tuberculosis (Mtb), continues to affect millions of people annually with high mortality, and the devastation caused by this pathogen is exacerbated by the ongoing COVID-19 and HIV epidemic (1, 2). The survival and persistence of Mtb in the host depends on a complex regulatory system to rapidly sense and respond to various assaults launched by the host immune system, such as acidic, oxidative, and nutritional stress. The seven members of WhiB-like (Wbl) family proteins found in Mtb, namely WhiB1-7, are key components in this regulatory system. Wbl proteins are a group of small iron–sulfur cluster ([4Fe-4S])-bound proteins first discovered in Streptomyces and exclusive to Actinobacteria (3, 4). Members of Wbl proteins play versatile and nonredundant roles in regulating biological processes and responding to various stresses Mtb encounters in the host, such as oxidative stress (WhiB1-7), cell division (WhiB2), acidic stress and nutritional starvation (WhiB3), virulence and reactivation (WhiB3, WhiB5, and WhiB6), and antibiotic resistance (WhiB2 and WhiB7) (5, 6, 7, 8). Among them, Mtb WhiB3 is one of the key global regulators involved in the early-stage response to acidic stress inside host macrophages (9, 10). It is exploited by Mtb to maintain redox and metabolic homeostasis in response to various host-generated redox stress, acidic stress, and carbon starvation and induced by hypoxia and nitric oxide in vitro (10, 11, 12, 13, 14, 15, 16, 17, 18). In Streptomyces, the WhiB3 ortholog, WhiD, is required for the late stage of sporulation (19, 20).
Correlated with their diverse roles in regulating gene expression, Mtb Wbl proteins share only 30 to 50% sequence identity and represent five different subclasses of Wbl proteins widely distributed in Actinobacteria. Two conserved motifs are found in all Wbl proteins—a [4Fe-4S]-cluster binding domain containing four conserved Cys and a “G[I/V]W[G/A]G” motif (the invariant residues are highlighted in bold fonts and the preferred residues are underlined; the same notations are used below), which is also referred to as the β turn (Fig. S1). The C terminus of Wbl proteins has been implicated in DNA binding since many Wbl proteins feature a cluster of basic residues in this region that is predicted to be in a helix-turn-helix fold (4). However, among the Wbl proteins in Mtb, only WhiB7 has a defined DNA-binding motif, the AT-hook (“RGRP”), in the C terminus. WhiB7 AT-hook preferably binds to the AT-rich sequence upstream of the −35 element in the target promoters. WhiB3 was also suggested to possess a C-terminal AT-hook–like motif (13). But these basic residues in the C-terminal WhiB3 are not conserved in the WhiB3 subclass (Fig. S1), and their role in WhiB3 binding to DNA has not been verified. Even less known is the N terminus of Wbl proteins prior to the Fe–S cluster–binding motif. This region varies substantially in length and sequence among the Wbl proteins (Fig. S1) and lacks information regarding its significance to the function of Wbl proteins.
Several Mtb Wbl proteins, including WhiB1, WhiB3, and WhiB7, have been shown to regulate gene expression in the [4Fe-4S]-bound (holo-) form by binding to the conserved region 4 in the σ70-family principal sigma factor σA (σA4) in the RNA polymerase (RNAP) holoenzyme (18, 21, 22). Other Mtb Wbl proteins except WhiB5 have also been reported to bind to σA4 in a [4Fe-4S]-dependent manner (23). Moreover, a recent study has shown that the WhiB3 ortholog of Streptomyces venezuelae also binds to region 4 of the principal sigma factor σHrdB, suggesting a shared mechanism of action by Wbl proteins in Actinobacteria (24).
Recent advances in the structural and biochemical characterization of the Wbl proteins have shed light on how Wbl proteins partner with σA to regulate gene expression. Mtb WhiB1 is the first Wbl protein that has been structurally characterized at the atomic level, first in the free holo-form by nuclear magnetic resonance and subsequently in the σA4-bound form by X-ray crystallography (22, 25). Together with the molecular and biochemical analyses, these studies reveal an unexpected molecular interface in the WhiB1:σA4 complex dominated by hydrophobic interactions and support a new molecular mechanism of transcription regulation by WhiB1 in Actinobacteria (25). Subsequently, the crystal structure of WhiB7 in complex with σA4 and its own promoter (PwhiB7) and the single-particle cryo-electron microscopy (cryo-EM) structure of WhiB7 bond to σA in RNAP and PwhiB7 were reported by our group and by the Campbell group, respectively (26, 27). The WhiB7 AT-hook binding site has the characteristics of A-track DNA (i.e., a short run of consecutive four or more adenine-thymine base pairs), which possesses distinct structural properties from canonical B-form DNA, including narrow minor grooves, high propeller twists, and DNA bending toward the AT-rich minor groove (see the review in (28)). Analysis of the 3D structures reveals the structural basis for how WhiB7 binding to the AT-rich region opens the minor groove, reversely bends the DNA in the direction opposite to the expected intrinsic bending of A-track DNAs, and orchestrates with σA for transcriptional regulation. Together, these studies provide an atomic view of how WhiB7 activates gene expression by coordinating DNA binding with σA4 via its AT-hook and unravel the WhiB7 subclass-specific structural features that enable WhiB7 to function differently from WhiB1 (26, 27).
WhiB3 is one of the most extensively investigated Mtb Wbl proteins owing to its importance for the pathogenesis of Mtb (10, 11, 12, 13, 14, 15, 16, 17, 18, 29). However, neither the DNA-binding motif nor the DNA-binding preference of WhiB3 has been determined to date. It remains enigmatic how WhiB3 binds to σA4 and DNA for transcriptional regulation. Here, we report crystal structures of the σA4-bound WhiB3 alone and in complex with DNA at 1.5 Å and 2.45 Å, respectively. Together, the results from our structural, biochemical, and functional analyses uncover an essential DNA-binding motif in WhiB3 and shed light on how WhiB3 coordinates with σA4 and interacts with DNA for transcriptional regulation. By structural comparison, we provide insights into how WhiB3 functions differently from WhiB1 and WhiB7 in Mtb by binding to the same site on σA4 and utilizing the subclass-specific structural motif for DNA binding.
Results
Crystal structure of the WhiB3:σA4-βtip complex
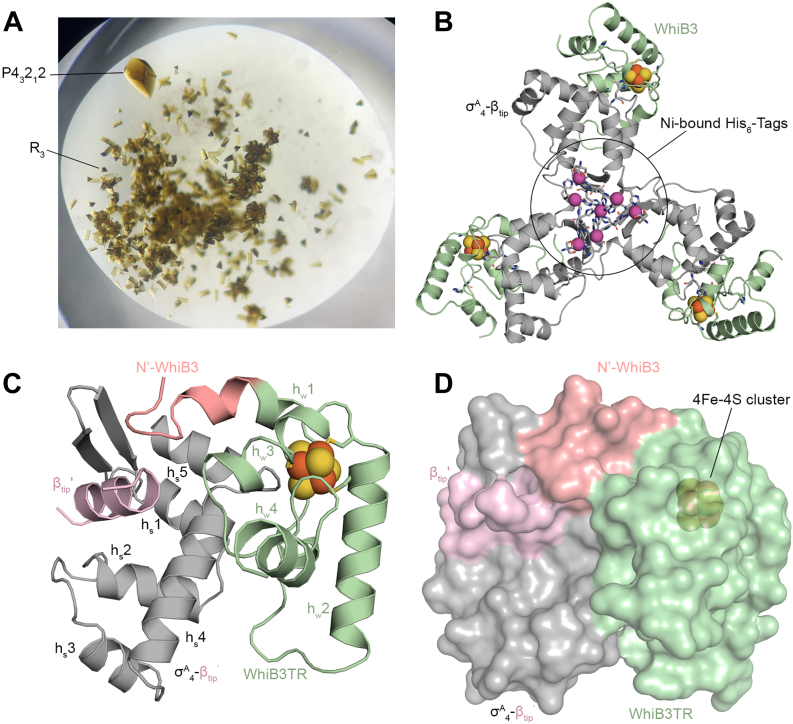
As described in the previous studies, a chimeric protein denoted σA4-βtip was used for the crystallographic characterization of the σA4-bound WhiB3 by fusing σA4 with the RNAP β-subunit flap tip helix (βtip) via an artificial linker to mimic the interaction between σA and the β subunit in the RNAP holoenzyme (see Experimental procedures) (26, 30, 31). We also generated a truncated WhiB3, denoted WhiB3TR, without the C-terminal residues (aa 91–102) containing the putative DNA binding motif to improve the protein stability and crystallizability. Phasing was performed using single-wavelength anomalous diffraction (SAD) of the iron–sulfur cluster containing four iron and four sulfur ions [4Fe-4S] cluster in the WhiB3:σA4-βtip complex (see Experimental procedures, Table 1 and Fig. S2). Two crystal forms were observed from the same crystallization drop, with the larger crystals in the P43212 form and the small ones in the R3 form (Fig. 1A). The final model of WhiB3:σA4-βtip was refined to 1.35 Å in the P43212 form and 1.5 Å in the R3 form.
Table 1.
Data collection and refinement statistics
| Proteins | WhiB3TR:σA4-βtip P43212 (phasing) | WhiB3TR:σA4-βtip P43212 | WhiB3TR:σA4-βtip R3 | WhiB3FL:σA4-βtip:PwhiB7 |
|---|---|---|---|---|
| Data collectiona | ||||
| Space group | P43212 | P43212 | R3 | P212121 |
| Cell dimensions | ||||
| a, b, c (Å) | 122.2, 122.2, 115.1 | 121.8, 121.8, 114.3 | 120.8, 120.8, 31.8 | 50.7, 72.9, 95.4 |
| α, β, γ (°) | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 | 90.0, 90.0, 120.0 | 90.0, 90.0, 90.0 |
| Wavelength (Å) | 1.7220 | 0.9795 | 0.9795 | 0.9787 |
| Resolution (Å) | 50–1.96 (1.99–1.96) | 50–1.35 (1.37–1.35) | 50–1.50 (1.53–1.50) | 50–2.45 (2.49–2.45) |
| Rmerge | 0.099 (1.094) | 0.082 (3.02) | 0.077 (2.16) | 0.128 (1.344) |
| I/σΙ | 48.1 (3.7) | 48.2 (1.2) | 45.4 (1.6) | 25.4 (1.2) |
| Completeness (%) | 100.0 (100.0) | 99.9 (100.0) | 99.8 (99.7) | 99.4 (94.6) |
| Multiplicityb | 49.8 (48.8) | 25.3 (24.7) | 18.1 (14.2) | 15.3 (6.9) |
| No. unique reflections | 62,861 (3092) | 187,184 (18,429) | 27,420 (2696) | 13,547 (617) |
| CC1/2(%) | 99.1 (95.3) | 99.9 (65.9) | 100.0 (74.1) | 98.7 (71.3) |
| Refinement | ||||
| Resolution (Å) | 50–1.35 (1.40–1.35) | 50–1.50 (1.56–1.50) | 50–2.45 (2.53–2.45) | |
| No. molecules per asymmetric unit | 3 | 1 | 1 | |
| Rwork/Rfree | 0.152/0.174 | 0.183/0.212 | 0.211/0.243 | |
| Included residue No. | ||||
| WhiB3 | 15–89; 15–89; 14–89c | 6–90 | 16–90 | |
| σA4 | 446–528 | 446–528 | 450–525 | |
| βtip | 815–826 | 815–827 | 815–826 | |
| His-tag | 1–6 | 1–6 | - | |
| No. atoms | 5398 | 1689 | 1974 | |
| Macromolecules | 4696 | 1555 | 1958 | |
| Ligand | 180 | 29 | 13 | |
| Water | 522 | 105 | 3 | |
| B-factors (Å2) | 30.1 | 41.3 | 80.7 | |
| Macromolecules | 28.4 | 41.2 | 76.6 | |
| Ligand | 38.3 | 38.8 | 79.3 | |
| Water | 42.5 | 43.9 | 66.2 | |
| r.m.s deviations | ||||
| Bond lengths (Å) | 0.012 | 0.01 | 0.018 | |
| Bond angles (°) | 1.68 | 1.29 | 2.29 | |
| Ramachandran statistics | ||||
| Favored regions (%) | 98.14 | 97.89 | 91.77 | |
| Allowed regions (%) | 1.30 | 1.58 | 8.23 | |
| Outliers (%) | 0.56 | 0.53 | 0 | |
| PDB code | 8CWT | 8CWR | 8CYF |
The highest resolution shell statistics are shown in parentheses.
For the data used for SAD phasing, the anomalous multiplicity was shown.
The three sets of the residues are for each of the three complexes in the asymmetric unit.
Figure 1.
Overview of the WhiB3TR:σA4-β-tipstructure.A, an optical image of the WhiB3TR:σA4-βtip crystals. The two forms of crystals, P43212 and R3, respectively, are shown in the same crystal drop. B, the trimeric structure of the WhiB3TR:σA4-βtip complexes in the R3 form. The trimer is centered on the His6-tags in the N terminus of σA4-βtip, which bind to Ni ions from the crystallization solution. The side chains of the His6-tag residues are shown in sticks, and the Ni atoms are shown in magenta spheres. C, cartoon representation of the WhiB3:σA4-βtipʹ complex in the R3 form, in which β-tip from the neighboring complex molecule (βtipʹ) is shown to reflect its interaction with σA4 and WhiB3. The four helices in WhiB3 and five helices in σA4 are labeled as hw1-4 and hs1-5, respectively. D, surface representation of the WhiB3TR: σA4-βtip complex, demonstrating the enclosed [4Fe-4S] cluster binding pocket in the complex. In all the structures, C atoms of WhiB3 are colored pale green with the N-terminus residues (aa 6–16) in salmon pink, σA4 in gray, and βtip in pink. The [4Fe-4S] clusters are shown in spheres, with Fe colored orange and S in yellow. [4Fe-4S] cluster, iron–sulfur cluster containing four iron and four sulfur ions; σA4-βtip, σA4 fused with βtip by an artificial linker; βtip, the C-terminal flap tip helix of the RNAP β-subunit; His6-tag, hexa histidine-tag; WhiB3TR, a truncated WhiB3 without C-terminal loop region (aa 91–102).
Like the other two σA4-bound Wbl proteins, the WhiB3:σA4-βtip complex exists as a single complex in solution determined by size-exclusion chromatography (25, 26). However, in the crystals, the His6-tags in the N-terminal σA4-βtip of three WhiB3:σA4-βtip complexes are “glued” together by multiple nickel ions from the crystallization solution, resulting in a trimer of the WhiB3:σA4-βtip complexes in both crystal forms (Figs. 1B and S2A). Three WhiB3:σA4-βtip complexes in the trimer form are found in the asymmetric unit of the P43212 structure. The R3 structure contains one WhiB3:σA4-βtip complex per asymmetric unit, as the trimer axis coincides with the crystallographic threefold axis. Correlated with the trimerization in the crystal form, we observed two structural re-arrangement in the WhiB3-bound σA4-βtip compared to that bound to WhiB7. First, the N-terminal residues (aa 446–456) of σA4 in the WhiB3:σA4-βtip trimer form a β-hairpin with the His6-tag residues, instead of being part of helix hs1 of σA4 as expected (Figs. 1B and S2B) (26, 31). Because the residues 446 to 456 of σA4 are far from the WhiB3 binding site, we do not anticipate this structural change affects the mode of WhiB3 binding to σA4. Second, βtip in the WhiB3:σA4-βtip complex, which was expected to form intramolecular contacts with σA4 in the σA4-βtip chimera, sticks into a neighboring protomer of the trimer and forms intermolecular interactions with σA4 in a second WhiB3:σA4-βtip complex (Figs. 1C and S2B). The resulting σA4-βtipʹ, however, resembles σA4-βtip observed in the WhiB7:σA4-βtip complex and the RNAP holoenzyme (Fig. S3C) (26, 31), and thus it is used for the following structural analysis.
The crystal structures of the WhiB3:σA4-βtip complex in the P43212 and R3 forms are essentially identical except for the N-terminal loop region, with an average Cα root-mean-square deviation of 0.27 Å for WhiB3 and 0.42 Å for σA4, respectively (Fig. S3, A and B). The R3 structure was used for the following structural analysis of the WhiB3:σA4-βtip complex because of the better-defined electron density of the N-terminal residues (aa 6–13) in WhiB3 (Fig. S3, D and E).
Molecular interface of the WhiB3:σA4-βtip complex compared to other σA4-bound Wbl proteins
The overall architecture of the WhiB3:σA4-βtip complex is comparable to the previously reported WhiB1:σA4 and WhiB7:σA4-βtip complexes. In all three cases, the molecular interface between the Wbl protein and σA4 is hinged at the [4Fe-4S] cluster (Fig. 1C) (25, 26). The 4Fe-4S cluster binding pocket in WhiB3:σA4-βtip is enclosed (Fig. 1D), similar to that of WhiB1:σA4 and in contrast to the solvent-accessible cluster in the case of WhiB7:σA4-βtip.
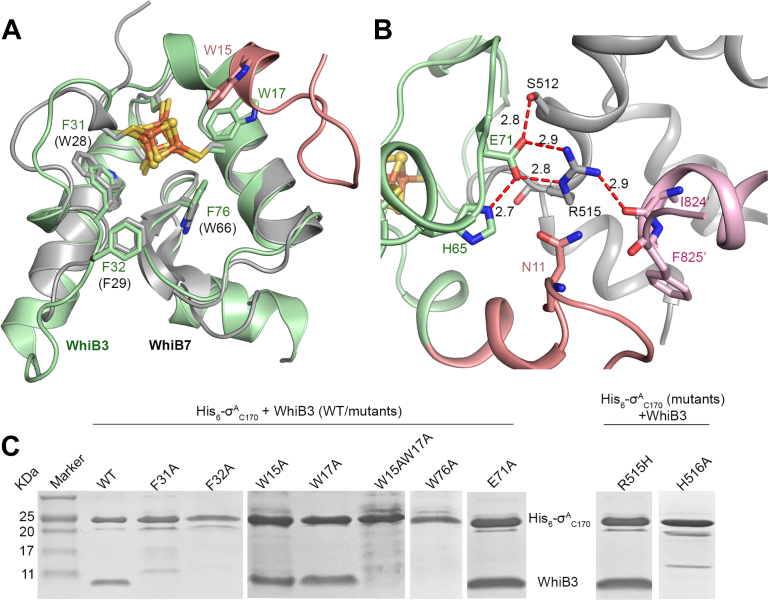
Complex formation between WhiB3 and σA4 is driven by the conserved aromatic residues near the [4Fe-4S] cluster binding pocket (Figs. 2A and S1B), as previously observed in the cases of WhiB1 and WhiB7 (25, 26). A single Ala substitution of F31, F32, or W76 in WhiB3 (corresponding to F17, F18, and W60 in WhiB1, respectively) or H516 in σA4 completely abolishes the complex formation in the pull-down assays (Fig. 2C). To our initial surprise, a W17A mutation in WhiB3 does not abolish σA4 binding in the pull-down assay (Figs. 2C and S4). W17 is invariant in the WhiB3 subclass, corresponding to the invariant W3 in WhiB1. It has been shown to play a crucial role in Fe–S cluster stability and complex formation in WhiB1:σA4, while the absence of a W3 counterpart in WhiB7 leads to a solvent-accessible Fe–S cluster with increased O2 sensitivity in the WhiB7:σA4 complex (25, 26). Subsequent sequence analysis reveals an additional conserved Trp in the WhiB3 subclass, W15, which is close to W17 and the Fe–S cluster binding pocket (Figs. 2A and S1B) and thus may compensate for the loss of W17 in the WhiB3 W17A mutant based on our pull-down assay. As shown in Figure. 2C and Fig. S4, although a single W15A mutation does not affect σA4 binding, the double mutation of W15A and W17 A in WhiB3 completely abolishes the interaction in the pull-down assays. The existence of redundant Trp residues in the N-terminal WhiB3 highlights their importance in the complex formation, consistent with the observations from our studies on WhiB1 and WhiB7 (25, 26).
Figure 2.
Molecular interface between WhiB3 σA4.A, a structural overlay of σA4-βtipʹ-bound WhiB3 (pale green, with the N-terminal region in salmon pink) and WhiB7 (PDB ID: 7KUG, gray). Only Wbl proteins are shown for clarity. B, close-up view of the hydrophilic network at the molecular interface of WhiB3 (pale green) and σA4 (gray). The βtipʹ residues interacting with σA4 in this region are shown in pink. C, SDS-PAGE analyses of the samples from the co-expression and affinity purification of tagless WhiB3 and His6-σAC170 (either wildtype [WT] or mutant as indicated). σA4-βtip, σA4 fused with βtip by an artificial linker.
Both the WhiB3 and WhiB7 subclasses contain a similar triplet motif (“EPY” in WhiB3 and “EPW” in WhiB7, respectively) immediately upstream of the β-turn, in which Glu forms hydrophilic interactions with σA4 (Fig. S1B) (26). However, these motifs contribute differently to the complex formation. E61 in the “EPW” motif of WhiB7 orchestrates a hydrogen bond network at the molecular interface of the WhiB7:σA4 complex and is required for σA4 binding in vitro and the WhiB7-dependent antibiotic resistance in mycobacteria (21, 26). In contrast, E71 in the “EPY” motif of WhiB3 is part of the multicentered polar interaction networks involving N11, H65, and E71 in WhiB3, S512, and R515 in σA4, as well as the backbone O of I824 and F825 in βtip (Fig. 2B). Accordingly, either a WhiB3-E71A or σA-R515H mutant does not affect the complex formation of WhiB3:σA4 in the pull-down assays (Fig. 2C). Notably, the σA4-R515H mutant was previously shown to affect the interaction between σA4 and WhiB3 in yeast two-hybrid assay (18). The discrepancy observed here might be due to the different sensitivity between the two techniques. Nonetheless, the results from our pull-down assays indicate that the contribution of the polar interactions between WhiB3 E71 and σA R515 to the formation of the WhiB3:σA4 complex is not as significant as the hydrophobic interactions between the conserved aromatic residues as described above.
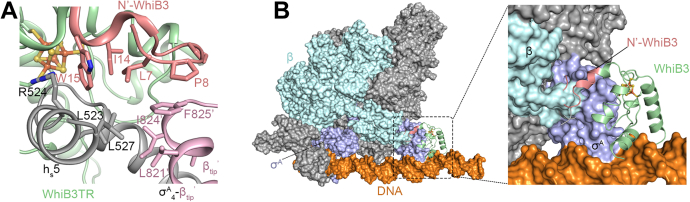
The N-terminal residues of WhiB3 before the first Cys in the Fe–S cluster binding motif are more conserved relative to WhiB7, while WhiB1 has an unusually short N terminus (Fig. S1) (25, 26). In the R3 crystal structure, the N-terminal residues 6 to 16 of WhiB3 interact with both σA4 and βtip outside the Fe–S cluster binding pocket (Fig. 3). The buried surface area between WhiB3 and σA4-βtip (1075.3 Å2) is significantly larger than that of the WhiB1:σA4 complex (645 ± 27 Å2) and WhiB7:σA4-βtip complex (576 ± 3 Å2) (see Experimental procedures) (26). The net increase in the surface contact between WhiB3 and σA4-βtip is mainly attributed to the N-terminal residues 6 to 16 of WhiB3, which has a buried surface area of 372.1 Å2 with σA4-βtip. In particular, L7, P8, I14, and W15 of WhiB3 form a hydrophobic core with both σA4 and βtip (Figs. 3A and S1B). When superposing the WhiB3:σA4-βtip structure onto the cryo-EM structure of the WhiB7–RNAP–DNA complex in the closed state (W-RPc, PDB ID: 7KIM) (27), the N-terminal WhiB3 is expected to interact with both σA and the β-subunit of RNAP (Fig. 3B). Interestingly, the N terminus of WhiB3 points to an opposite direction relative to that of the N-terminal WhiB7, which also extends into the RNA polymerase but interacts with the βʹ-subunit of RNAP in the WhiB7-RNAP-DNA complex (Fig. S5) (27). The observation that the conserved N-terminal WhiB3 forms both hydrophobic and hydrophilic interactions with σA4 and βtip in the structural analysis (Figs. 2B and 3A) warrants further investigation of its effects on the WhiB3 binding to RNAP and transcriptional regulation.
Figure 3.
Interactions between N-terminal WhiB3 and σA4-βtip.A, a hydrophobic interface among N-terminal residues (aa 6–11) of WhiB3 (pale green with N-terminal residues in salmon pink), σA4 (gray), and βtip (pink). βtip from the neighboring complex molecule (βtipʹ) is shown to reflect its interaction with σA4 and WhiB3 in the WhiB3TR:σA4-βtip complex. The hydrophobic residues at the molecular interface are shown in sticks. B, an overlay of σA4 in the WhiB3:σA4-βtip complex with the WhiB7-RNAP-DNA complex in the closed state (PDB ID: 7KIM, σA colored purple blue, the α-subunits in gray, the β-subunit in cyan, and DNA in orange). The WhiB3:σA4-βtip complex is shown in the cartoon representations, and the WhiB7–RNAP–DNA complex is shown in the surface representation. Only WhiB3 (pale green with the N-terminal WhiB3 in salmon red) in the WhiB3:σA4-βtip complex is shown for clarity. By comparison, the N-terminal WhiB7 points toward an opposite direction relative to the N terminus of WhiB3 and interacts with the βʹ-subunit of RNAP (Fig. S5). σA4-βtip, σA4 fused with βtip by an artificial linker; RNAP, RNA polymerase; WhiB3TR, a truncated WhiB3 without C-terminal loop region (aa 91–102).
DNA binding by the WhiB3:σA4-βtip complex
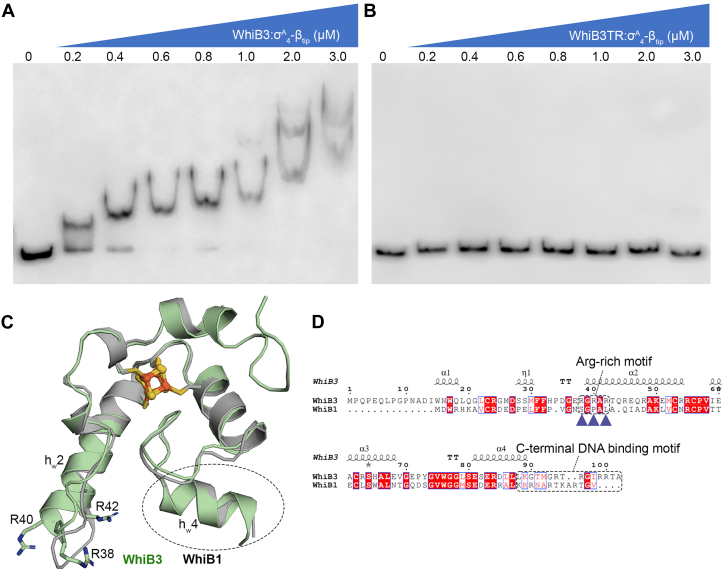
A prior study by Singh et al. (12) has shown that WhiB3 binds to the promoter of the pks2 and pks3 genes to regulate the biosynthesis of major complex polyketides in Mtb. In this study, the oxidized but not the reduced WhiB3 in the cluster-free (apo-) form was found to bind DNA with high affinity but low specificity, and the interaction is sensitive to high concentrations of NaCl in the electrophoretic mobility shift assays (EMSAs). Holo-WhiB3 was also bound to DNA in the EMSAs but with lower affinity (with an observed shift at ∼0.8 μM), resulting in only a marginal DNA mobility shift. Since the previous studies on WhiB7 have shown that the DNA binding motifs in both WhiB7 and σA4 coordinate DNA binding (26, 27), we thus tested whether that is also the case for WhiB3 by the EMSA using the pks3 promoter as previously reported (12). As shown in Figure 4A, the WhiB3:σA4-βtip complex binds to the pks3 promoter at concentrations as low as 0.2 μM in the EMSA, of which the binding affinity is higher than previously reported for holo-WhiB3 alone and thus implies a cooperative DNA binding by WhiB3 and σA4 similarly to WhiB7 (12). The multiple DNA shifts observed in the EMSAs with a 316-bp Ppks3 DNA, but not with a shorter DNA (22 bp), indicate that WhiB3:σA4-βtip binds to different sites on the long Ppks3 DNA (Figs. 4A and S6A), reconciling with the low DNA specificity of WhiB3. We also find by the EMSAs that the C-terminal WhiB3 (aa 91–102) is important for DNA binding, as deletion of these residues abolishes DNA binding under the experimental conditions (Fig. 4B).
Figure 4.
Effect of the C-terminal WhiB3 on DNA binding.A and B, the EMSAs of the WhiB3:σA4-β-tip complex with the pks3 promoter, either with wildtype WhiB3 protein (A) or the truncated WhiB3 without the C-terminal restudies 91 to 102 (WhiB3TR, B). C, an overlay of WhiB3 (pale green) in the WhiB3:σA4-βtip with the σA4-bound WhiB1 (PDB ID: 6ONO, gray). Only Wbl proteins are shown for clarity. D, sequence alignment between Mtb WhiB3 and WhiB1. The putative C-terminal DNA binding motifs in the Wbl proteins and the conserved Arg-rich motif specific to the WhiB3 subclass are highlighted by dashed rectangles. The two invariant Arg residues (R38 and R42) in the conserved Arg-rich motif of WhiB3 and absent in WhiB1, are indicated by blue triangles (Fig. S1). R40 is a variant in the WhiB3 subclass. σA4-βtip, σA4 fused with βtip by an artificial linker; EMSA, electrophoretic mobility shift assay.
The observed DNA binding activity of σA4-bound WhiB3 is in stark contrast with WhiB1. Thus far, only apo-WhiB1 has been shown to bind to its own promoter and several other target genes, and the basic residues (such as R74) in the C-terminal loop are required for DNA binding (22, 25, 32, 33). Neither holo-WhiB1 nor the WhiB1:σA4 complex has shown DNA binding activity despite the extensive efforts. Our initial structural analysis did not uncover the structural basis that may account for the differences in DNA binding between σA4-bound WhiB3 and WhiB1. The σA4-bound WhiB3 and WhiB1 are strikingly similar, with an average Cα root-mean-square deviation of 0.54 in the 3D structural overlay (Fig. 4C). In particular, the structural arrangement of helix hw4 immediately adjacent to the putative C-terminal DNA-binding motif is essentially identical in the two structures, and an equal number of basic residues (four Arg and one Lys) is found in the C-terminal loop of WhiB1 and WhiB3 (Fig. 4, C and D). However, a closer examination of the WhiB3 subclass-specific sequences reveals an Arg-rich motif (corresponding to 38-RGRAR-42 in Mtb WhiB3) near the N-terminal helix hw2 that points toward the interface with DNA (Figs. 4, C and D and S1B). The role of this motif for WhiB3 binding to DNA has not yet been examined.
Characterization of the conserved Arg-rich DNA-binding motif in WhiB3
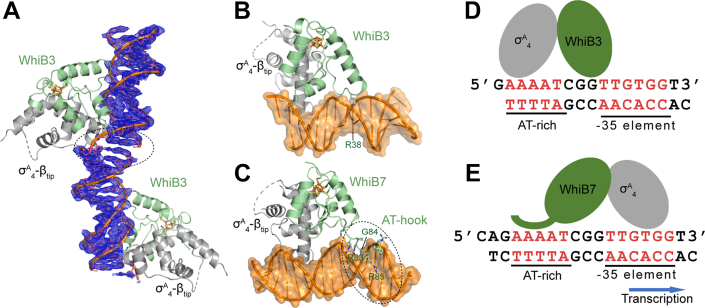
To reveal the structural basis of the WhiB3:σA4-βtip complex binding to DNA, we attempted cocrystallization of WhiB3:σA4-βtip with the promoter DNA of either pks3, apt, or whiB6, all of which have been previously suggested to be under the regulation of WhiB3 (10, 12, 16, 34). Unfortunately, none of the hits from the crystallization screen led to high-resolution diffraction data for confident structure determination. Since WhiB3 possesses a putative AT-hook-like motif, out of curiosity, we tested and confirmed that the WhiB3:σA4-βtip complex binds to the whiB7 promoter DNA (PwhiB7) (Fig. S6B). We then cocrystallized the WhiB3:σA4-βtip complex with PwhiB7 that was used in the crystallographic study of the WhiB7:σA4-βtip:PwhiB7 complex (26), which enabled us to collect high-quality diffraction data from a WhiB3:σA4-βtip:PwhiB7 crystal and refined the crystal structure at 2.45-Å resolution with the final Rfree/Rwork value of 0.211/0.243 (see Experimental procedures). The well-defined electron density map allows us to unambiguously assign the nucleotides in the DNA helix of the complex crystal structure (Fig. 5A).
Figure 5.
Comparison of the PwhiB7binding site between WhiB3:σA4-β-tipand WhiB7:σA4-β-tip.A, simulated-annealing composite omit map around the whiB7 promoter (PwhiB7) in the crystal structure of two adjacent WhiB3:σA4-βtip:PwhiB7 complex molecules, contoured at 1.0 σ. The gap in the electron density map between the two adjacent PwhiB7 DNA molecules is highlighted by a black dash circle, indicative of the correct assignment of the PwhiB7 DNA. B and C, a side-by-side comparison of the crystal structures of WhiB3:σA4-βtip:PwhiB7 and WhiB7:σA4-βtip:PwhiB7 (PDB ID: 7KUF), respectively. The Wbl residues (R38 of WhiB3; R83-G84-R85 of WhiB7) inserted into the minor groove of the DNA helix are labeled. D and E, cartoon illustrations of the different modes of PwhiB7 binding by WhiB3:σA4-βtip and WhiB7:σA4-βtip, respectively. The blue arrow indicate the direction of the transcription in WhiB7:σA4-βtip:PwhiB7. In all the structures, WhiB3 and WhiB7 are colored pale green, σA4-βtip in gray, and the PwhiB7 DNA in orange. σA4-βtip, σA4 fused with βtip by an artificial linker.
A structural comparison shows that although both WhiB3 and σA4 in the WhiB3:σA4-βtip:PwhiB7 structure are involved in DNA binding like the case of WhiB7, they are dramatically different in how the Wbl-bound σA4 interacts with PwhiB7 (Fig. S5, B–E) (26, 27). σA4 of the WhiB3:σA4 complex orients about 180° relative to the WhiB7-bound σA4 along with the PwhiB7 DNA and inserts into the major groove upstream of the AT-rich WhiB7 binding site instead of the expected −35 element. The σA4 binding site to the −35 element of PwhiB7 has been previously confirmed in both the crystallographic and cryo-EM studies (26, 27). Therefore, the observation that σA4 in the WhiB3:σA4-βtip:PwhiB7 complex binds to a physiologically irrelevant site of PwhiB7 opposite to the direction of transcription initiation indicates that PwhiB7 is unlikely a target of WhiB3. However, the crystal structure of the WhiB3:σA4-βtip:PwhiB7 complex provides valuable insights into the general features of how WhiB3 interacts with DNA, which is the primary focus of our structural analysis below.
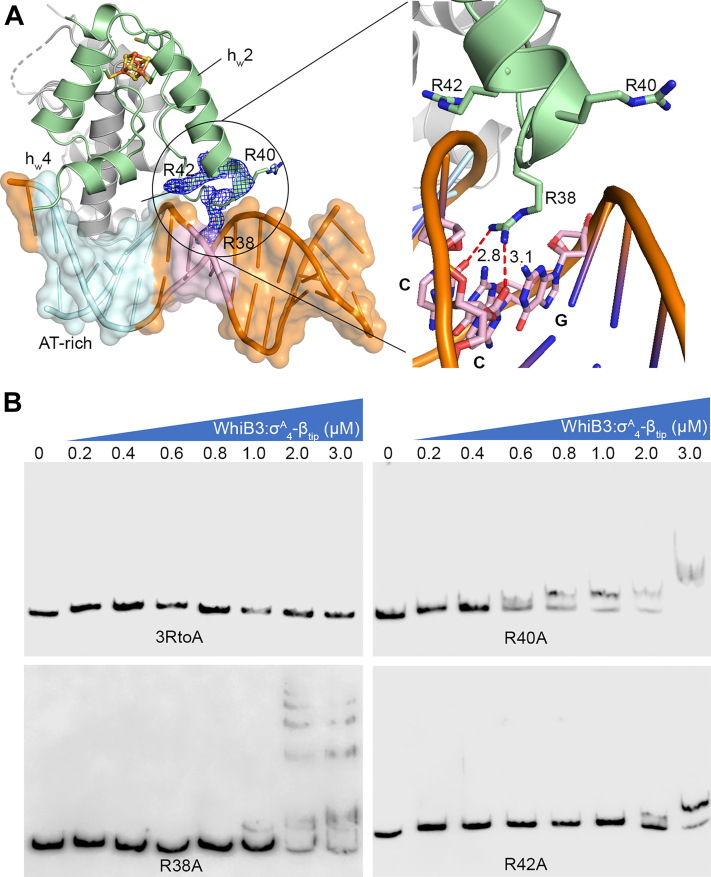
Our structural and biochemical analyses show that WhiB3 engages with DNA differently from WhiB7, and the newly identified Arg-rich motif in WhiB3 as described above plays a central role in DNA binding (Table S2; Figs. 5 and S7) (26, 27). For the case of WhiB7, the central RGR motif of the WhiB7 AT-hook lies in the minor groove of the A-track sequence upstream of the −35 element and forms both hydrophilic and hydrophobic interactions with the A/T nucleotides as well as the sugar and phosphate groups of the DNA. Although a “RGR” sequence is also present in the Arg-rich motif (“RGRAR”) of Mtb WhiB3, the RGR sequence is not conserved in the WhiB3 subclass and only R38 in the motif inserts vertically into the minor groove of the junction between the WhiB7 binding site and the −35 element. R38 forms polar contacts with the nucleotides of 2 GC pairs (Table S2; Figs. 6, A and B; Fig. S7), in contrast to the preference of AT-rich DNA by the WhiB7 AT-hook. The other conserved residue in the Arg-rich motif, R42, is near the edge of the DNA duplex (∼4 Å away) and thus may form hydrophilic interactions with the phosphate backbone in solution. The variant residue R40 is also near the DNA helix. However, it is unclear how it interacts with DNA because of the poor electron density of the side chain, implying a nonspecific interaction with the PwhiB7 DNA and echoing the low conservation of this residue in the WhiB3 subclass. We notice reduced DNA contacts for the residues in the WhiB3-bound σA4 (e.g., R478 and T488) compared to that bound to WhiB7, coincident with the observation that WhiB3-bound σA4 being displaced off the −35 binding site of PwhiB7. The differences observed in how σA4 -bound WhiB3 and WhiB7 bind to PwhiB7 are in agreement with our analysis of the PwhiB7 DNA structure between the two complexes. As shown in Fig. S8, the central minor-groove width (∼3.5 Å) around the AT-rich region of PwhiB7 in WhiB3:σA4-βtip:PwhiB7 is characteristic of A-track DNAs (35, 36) and is significantly narrower than that of WhiB7:σA4-βtip:PwhiB7 (∼7 Å) where WhiB7-bound to the minor groove of the AT-rich region. Additionally, the break of base–base stacking between the −36 and −37 nucleotide, indicating DNA bending as a result of the cooperative action of WhiB7 binding to the AT-rich region and σA4 binding to the adjacent −35 hexamer (26), is absent from PwhiB7 in the WhiB3:σA4-βtip:PwhiB7 complex (Fig. S7). However, it should be noted that since σA4 binds to the major groove immediately upstream of the AT-rich region, and PwhiB7 in the WhiB3:σA4-βtip:PwhiB7 is 2-bp shorter than in WhiB7:σA4-βtip:PwhiB7 near the AT-rich region, we cannot unequivocally attribute the cause of the observed differences.
Figure 6.
Identification of a conserved Arg-rich DNA binding motif in WhiB3.A, highlights of the hydrophilic interactions between the conserved Arg-rich motif of WhiB3 and DNA in the WhiB3:σA4-βtip:PwhiB7 complex. The AT-rich DNA sequence in PwhiB7 is colored cyan, with the rest of the DNA in orange, WhiB3 in pale green, and σA4-βtip in gray. The three Arg residues (i.e., R38, R40 and R42) in the conserved Arg-rich motif of Mtb WhiB3 are shown in stick representations, with the 2Fo-Fc density map contoured at 1.0 σ. B, EMSAs of the σA4-βtip-bound WhiB3 (wildtype or mutant as indicated) with the pks3 promoter DNA. All the three Arg residues are substituted by Ala in the WhiB3-3RtoA mutant. σA4-βtip, σA4 fused with βtip by an artificial linker; EMSA, electrophoretic mobility shift assay.
Results from the EMSAs confirm our structural analysis that all three Arg residues in the conserved Arg-rich motif play a role in WhiB3 binding to the pks3 promoter, while the difference in their contributions is noted (Fig. 6B). A triple Arg-to-Ala mutation of the Arg-rich motif (3RtoA) completely abolishes DNA binding in the EMSAs. Substitution of each Arg by an Ala significantly affects WhiB3 binding to the pks3 promoter, with the effect of the R40A mutation relatively weaker than the other two (i.e., R38A and R42A) consistent with the structural and sequence analysis as mentioned above. The C-terminal WhiB3 residues (aa 91–102), which are required for DNA binding in the EMSA (Fig. 4B), are not visible in the electron density map. It is possible that these C-terminal residues interact with DNA through nonspecific polar contacts and thus result in the ill-defined electron density in the crystal structure.
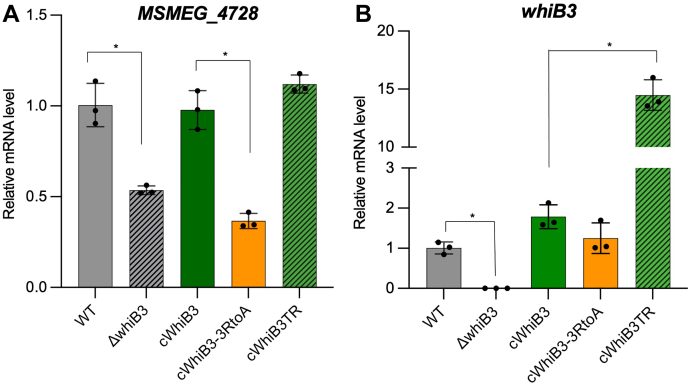
Consistent with the structural and biochemical analyses, our reverse transcription-quantitative polymerase chain reaction (RT-qPCR) study indicates that the conserved Arg-rich motif (“RGRAR”) is required for the WhiB3-dependent transcriptional regulation in Mycobacterium smegmatis (Msm) (see Experimental Procedures) (Fig. 7). As the previous study suggested (29), our results show that WhiB3 regulates the expression of MSMEG_4728 in Msm, which encodes a putative polyketide synthase-associated protein. The reduction of the mRNA levels of MSMEG_4728 in the whiB3 deletion mutant (ΔwhiB3) can be complemented by the wildtype WhiB3 but not by the WhiB3-3RtoA mutant, indicating the essential role of the conserved Arg-rich motif (“RGRAR”) for WhiB3-dependent transcriptional activation (Fig. 7A). In contrast, deletion of the C-terminal WhiB3 (WhiB3TR) shows a negligible effect on the expression of MSMEG_4728 when compared to the wildtype. This observation indicates that the C-terminal region is not essential for the WhiB3-dependent transcriptional regulation, reconciling with the high variation of this region in the WhiB3 subclass and the nonspecific DNA binding in the complex structural analysis.
Figure 7.
Identification of the essential DNA binding motif for the WhiB3-dependent transcription activation in Msm.A and B, are the RT-qPCR analyses of relative mRNA levels of MSMEG_4728 and whiB3, respectively, in the Msm wildtype (wt), the whiB3 deletion mutant (ΔwhiB3) alone or complemented with the wildtype whiB3 (cWhiB3), the whiB3-3RtoA mutant (cWhiB3-3RtoA) or the C-terminal truncated whiB3 (cWhiB3TR). The mRNA level in each sample was normalized to the level σA, and the fold of changes were calculated relative to wt. Data are representative of three biological replicates. Statistical significance is determined by Student’s t test and displayed as ∗p < 0.05 in the comparisons as indicated. The error bars represent mean ± SD. Msm, Mycobacterium smegmatis; RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
Discussion
This study provides an atomic view of how WhiB3 interacts with σA4 and DNA. Structural comparison of the WhiB3:σA4 complex with the σA4-bound WhiB1 and WhiB7 reveals that all the three Wbl proteins share a similar molecular interface with σA4, and the subclass-specific structural features underlie the structural basis for DNA binding by WhiB3. Complemented by molecular and biochemical approaches, we uncover a conserved Arg-rich DNA binding motif near the N-terminal helix hw2 in the WhiB3 subclass and determine the importance of the C-terminal basic residues for DNA binding and transcription activation by Mtb WhiB3. It is important to note that some WhiB3 subclass members, such as those in Streptomyces hygroscopicus (WP_041665551), Geodermatophilus obscurus (WP_012950425), and Beutenbergia cavernae (WP_015883541), do not have any basic residues corresponding to the putative C-terminal DNA binding motif (aa 91–102) of Mtb WhiB3, underlying the significance of the newly identified Arg-rich motif for DNA binding by the WhiB3 subclass proteins. Furthermore, the identification of the conserved Arg-rich DNA-binding motif in WhiB3 provides a plausible explanation for the difference in DNA-binding activities between WhiB3:σA4 and WhiB1:σA4 and sheds light on the DNA binding preference of WhiB3.
It is well known that the −35 hexamer is much less conserved in mycobacterial promoters than in E. coli (37, 38, 39). Consistently, σA4 does not bind to any DNA targets used in the assays without the DNA binding motif of a Wbl protein in the EMSAs (Fig. 6) (25, 26). In this context, the Wbl proteins, which exist exclusively in Actinobacteria but not in E. coli, may be employed by mycobacteria to serve as “guide dogs” to compensate for the low DNA affinity and specificity of σA4 in the target-specific transcriptional regulation in response to environmental cues. Intriguingly, the mode of DNA binding by WhiB3 and WhiB7 are fundamentally different despite the similarity (i.e., the “RGR” sequence) in their DNA binding motif. The central “RGR” motif of WhiB7 AT-hook lies in and grips the minor groove of the A-track sequence like a hand. The mode of WhiB7 AT-hook interaction with DNA provides the structural basis for how WhiB7 AT-hook binding opens the minor groove and reversely bends the A-track DNA to facilitate σA4 binding to the −35 hexamer (26). Lacking a canonical AT-hook, WhiB3 R38 is the only DNA sequence discriminator and interacts with DNA via the guanidino group like finger touch, which explains the low DNA sequence specificity in the EMSAs and why WhiB3 cannot interact with the A-track sequence in the PwhiB7 promoter like WhiB7. The observation that σA4 is displaced from the consensus −35 element upon binding to WhiB3 hints that WhiB3 either strongly disfavors the A-track sequence or favors the GC nucleotides.
The results from the WhiB3:σA4-βtip:PwhiB7 structure analysis described above also provide the evidence accounting for the high salt sensitivity and low specificity of WhiB3 observed in this study and the previous report (12). These observations raise the question of how WhiB3 selectively regulates target-specific gene expression. Recent work on the ortholog of Mtb WhiB2 in Streptomyces (WhiB) and Corynebacteria (WhcD) indicates that it coordinates with another regulator WhiA for DNA binding and transcriptional regulation, while WhiB does not bind to DNA alone (40, 41, 42). Likely WhiB3 may also adopt this strategy for engaging with DNA and enhancing target specificity. Our speculation is in line with the observation that the C-terminal WhiB3, which is required for enhancing WhiB3 binding to DNA in the EMSAs, does not involve in specific DNA binding in our structural analysis and is dispensable for transcription activation in Msm. It is interesting to note that the absence of the C-terminal region disrupts WhiB3 binding to DNA in the EMSAs and leads to over 7-fold increase in the whiB3 transcript level comparable to the wildtype WhiB3-complemented strain. Previous studies on WhiB1 have shown that the C-terminal Arg and Lys residues, which are also dispensable for holo-WhiB1-mediated transcriptional regulation, are required for DNA binding by apo-WhiB1 to repress several essential genes, including whiB1 itself (25, 32, 33). Further study is needed to test whether apo-WhiB3 also utilize the C-terminal domain for DNA binding and self-repression.
It is noted that the core helices of Wbl proteins involved in σA4 binding are highly conserved. However, the structural basis underlying the functional differences in the Wbl subclasses remains elusive. Our structural, biochemical, and molecular analyses of WhiB3 reported here provide crucial evidence for understanding how WhiB3 engages target DNAs differently from other characterized Wbl proteins (i.e., WhiB1 and WhiB7) via the conserved Arg-rich motif in the middle loop and regulates target-specific gene expression in coordinate with σA. A fuller view of these structural bases is critical for understanding the Wbl-dependent mechanism of pathogenesis and persistence in Mtb and building a stronger informational foundation for developing effective drugs for the treatment of the mounting threat of tuberculosis.
Experimental procedures
E. coli strains
All the E. coli strains were grown in Luria–Bertani media and at 37 °C, 200 rpm, unless otherwise specified.
Mycobacterial strains
Msm MC2 155 and the related mutant without and with a complementary gene generated in this study were grown in either Middlebrook 7H9 broth or on 7H10 agar (BD Difco) supplemented with 10% (v/v) ADS (2% dextrose, 5% bovine serum albumin and 0.85% NaCl), 0.2% (v/v) glycerol, 0.05% (v/v) Tween 80 (Sigma). When appropriate, the media were supplemented with antibiotics at following concentrations: hygromycin, 100 μg/ml for E. coli and 50 μg/ml for Msm and kanamycin, 50 μg/ml for E. coli and 25 μg/ml for Msm.
Plasmid construction for protein overexpression in E. coli
The bacterial strains and plasmids used in this study are listed in Table S1. The genes encoding Mtb WhiB3 (Rv3416, 1–102 aa) and the C-terminal domain of σA4 (Rv2703) containing the last 170 residues (aa 359–528, denoted σAC170) were amplified from Mtb H37Rv genomic DNA (a gift from Dr Midori Kato-Maeda’s group at the University of California, San Francisco) by PCR, and subsequently cloned into pET21b(+) and pCDF-1b to express tagless WhiB3 and σAC170 with a N-terminal His6-tag, respectively. The resulting plasmids, pET21-MtbWhiB3 and pCDF-1b-6HisMtbσAC170, were subsequently modified for the overexpression of the desirable proteins with either truncation or point mutation used for crystallization and biochemical assays as described in the related sections. Briefly, the pET21b-MtbWhiB3TR plasmid was modified from pET21b-MtbWhiB3 by site-directed mutagenesis for the expression of a truncated WhiB3 (WhiB3TR, containing aa 1–90) without the last ten residues in the C terminus for crystallographic work and EMSA. As previously described (26), the pET28b-6HisMtbσAC82-βtip plasmid encoding the chimera protein His6-σAC82-βtip was used for crystallographic work, and the pET28b-6HisMtbσAC112-βtip plasmid was used for the expression of the chimera protein His6-σAC112-βtip in the EMSAs.
All the plasmids were confirmed by DNA sequencing before being transformed into E. coli BL21-Gold (DE3) strain for protein expression.
General procedures for protein expression, purification, and analysis
Overexpression and purification of the proteins of interest from E. coli BL21-Gold (DE3) for structural and biochemical studies were done as described in our study of WhiB7 (26). Samples after each step of purification were analyzed by SDS-PAGE and by UV-Visible (UV-Vis) spectroscopy. Unless otherwise specified, the final purified proteins in 50 mM Tris-HCl, pH 8.0, 100 mM NaCl, and 0.2 mM tris(2-carboxyethyl)phosphine were stored in liquid nitrogen until use. UV-Vis spectra of the purified proteins were recorded using an HP 8452a diode array UV-Vis spectrophotometer (Agilent Technologies Inc). The absorption at 410 nm, characteristic of proteins containing [4Fe-4S]2+ clusters (43, 44), was used to estimate the occupancy of the Fe–S cluster in the protein samples containing a Wbl protein. Protein concentrations were estimated either by the Pierce Bradford Assay Kit (Thermo Fisher Scientific) or UV-Vis absorption spectroscopy.
Pull-down assays
To verify the role of the residues in WhiB3 and σA4 for the complex formation, the two plasmids encoding a tagless Mtb WhiB3 (wildtype or mutant) and a His6-σAC170 (wildtype or mutant), respectively, were transformed into E. coli BL21-Gold (DE3) for protein coexpression and affinity purification by Ni-NTA Sepharose resin as previously described (26). The purified protein samples were analyzed by UV-Vis spectroscopy and SDS-PAGE.
DNA-binding assays
Nondenaturing gel EMSAs were used to test the binding of Mtb WhiB3 (wildtype and mutants) in complex with His6-σAC112-βtip to the promoter of the pks3 gene (Ppks3) as previously described (25, 26). The Ppks3 dsDNA containing 316 bp upstream of the start codon of the pks3 gene, the same promoter region used in the previous EMSA (12), was amplified from Mtb H37Rv genomic DNA using the biotin-labeled primer pairs (forward primer: biotin-labeled 5′-AACGGATTTCGGGGCCTTTTGCGTCTGCT-3′; reverse primer: 5′-TTACCAACACATTCGGGCTCAGGAT-3′). In each of the 10-μl binding reactions, 0.2 nM biotin-labeled Ppks3 dsDNA was incubated with σAC112-βtip-bound Mtb WhiB3 (wildtype or mutant) in the presence of 25 mM Tris pH 8.0, 5 mM MgCl2, 0.1 mg/ml BSA, 1 mM dithiothreitol, and 20 mM KCl. After incubation at room temperature for 20 min, the samples were analyzed on 6% native polyacrylamide gels and UV crosslinked to Hybond-N+ nylon membrane (GE Healthcare Life Sciences) after gel transfer. The biotin-labeled DNA was detected using the LightShift Chemiluminescent EMSA Kit (Thermo Scientific, Inc) according to the manufacturer’s instructions.
Creation of a whiB3 deletion mutant in Msm
The whiB3 (MSMEG_1597) deletion mutant (ΔwhiB3) in Msm was generated using homologous recombination–based in-frame unmarked deletion as previously described (45). The resulting ΔwhiB3 strain was verified by PCR and further confirmed by DNA sequencing.
Creation of the whiB3-complemented strains
For the complementation test in the ΔwhiB3 strain, the DNA fragment encoding MsmWhiB3 (wildtype or mutant) was cloned into the integration plasmid pKW08-Lx-Int (integrated plasmid) for expression of tagless MtbWhiB3 from the native whiB3 promoter (46). The resulting plasmids, pKW08-Int-PwhiB3-msmWhiB3, pKW08-Int-PwhiB3-msmWhiB3TR, and pKW08-Int-PwhiB3-msmWhiB3-3RtoA, are listed in Table S1. Each of these plasmids was transformed into the ΔwhiB3 strain to determine whether it can complement WhiB3-dependent transcription by RT-qPCR.
RT-qPCR
Msm cells were cultured in 50 ml 7H9 broth supplemented with 50 ug/ml hygromycin, if applicable, until A600nm reached ∼0.7 to 0.8. The cells were then harvested, and RNA extraction was done using RNeasy Mini kit (QIAGEN) according to the manufacturer’s instructions. RNA was then treated with gDNA wipeout buffer supplied in the QuantiTect Reverse Transcription Kit (Qiagen). Complementary DNA was obtained by reverse transcription using the QuantiTect Reverse Transcription Kit from 1 μg of each of the RNA samples. Quantification PCR (qPCR) was performed using CFX Connect Real-Time PCR Detection System and SsoAdvanced Universal SYBR Green Supermix (Bio-Rad). All RT-qPCR reactions were performed in biological triplicates. The mRNA level of the sigA gene was used as the reference. The extent of expressional changes was calculated using the 2-ΔΔCt method and scaled to the Msm wildtype strain. The results were analyzed using the Origin software. The primers used for RT-qPCR were:
sigA: forward primer, 5′- GTGTGGGACGAGGAAGAGTC-3′
reverse primer, 5′- ACCTCTTCTTCGGCGTTGAG-3′
whiB3: forward primer, 5′-CAACTGCGACACATTTCCTTCGCAC-3′
reverse primer, 5′- GAATCCGAGCGTGAGCTTCTGC -3′
MSMEG_4728: forward primer, 5′- ACCGTTCCGGTGTGGAACAT-3′
reverse primer, 5′- CGGTGAACTCGAAACGGCTG-3′.
These primers were designed to be in the coding regions of the transcripts.
Crystallization
Initial crystallization screens of the WhiB3TR:σAC82-βtip complex were carried out at 18 °C in a Coy anaerobic chamber using the sitting-drop vapor diffusion method, followed by optimization of the crystallization hits. High-quality crystals were obtained by mixing 1 μl WhiB3TR:σA4-βtip at 80 mg/ml with an equal volume of the reservoir solution containing 10 to 20 mM nickel chloride and 0.8 to 1.0 M lithium sulfate.
For the crystallization of the WhiB3:σAC82-βtip:PwhiB7 complex, 16-bp synthetic PwhiB7 duplex DNA with a 5′ G/C overhang at each end (5′-GAAAATCGGTTGTGGT-3′/5′-TTTTAGCCAACACCAC-3′, Sigma-Aldrich) was used for cocrystallization with the WhiB3:σAC82-βtip complex. The WhiB3:σAC82-βtip complex was first mixed in a 1:1 M ratio with 100 μM PwhiB7 and subsequently concentrated to 40 mg/ml before crystallization. The best crystals were obtained by mixing 1 μl of the protein–DNA complex with the reservoir solution containing 0.1 M calcium acetate, 0.1 M sodium cacodylate, pH 6.5, 10%∼13% polyethylene glycol 8000. All the crystals were briefly soaked in the reservoir solution supplemented with 20% glycerol for cryoprotection before flash-cooling in liquid nitrogen.
X-ray crystallographic data collection, structural determination, and analysis
X-ray diffraction data were collected at the beamlines 9-2 and 12-2 of the Stanford Synchrotron Radiation Lightsource from single crystals maintained at 100 K using a 6M Pixel Array Detector. The diffraction data were collected at Se K-edge (12,658 eV) and used for the final structural refinement for all three structures. The SAD data collected from a single crystal of WhiB3TR:σAC82-βtip at the Fe K-edge absorption peak (7200 eV) were used for experimental phasing. The diffraction data were indexed, integrated, and scaled using HKL2000 (47). Model building and structure refinement were performed in COOT and Phenix (48, 49). The data collection and refinement statistics are summarized in Table 1.
Two forms of the WhiB3TR:σAC82-βtip crystals in space groups P43212 and R3, respectively, were observed in the same crystallization drop. The phases for the P43212 form were solved by SAD using Phenix.Autosol, with a figure of merit of 0.58. Model building and structure refinement were carried out using COOT and Phenix (48, 49). The final model in the P43212 form was refined to 1.35 Å with one trimer of the WhiB3TR:σAC82-βtip complexes per asymmetric unit. The R3 crystal structure was solved by molecular replacement using a single copy of WhiB3TR:σA4-βtip from the P43212 crystal structure as a search model and refined to 1.5 Å in the final model. The trimer axis in the R3 crystal structure coincides with the crystallographic threefold axis, resulting in one WhiB3:σA4-βtip complex per asymmetric unit. The WhiB3TR:σA4-βtip structures in the two crystal forms are essentially identical except for the N-terminal residues (aa 6–11) of WhiB3.
The phases for the WhiB3: σA4-βtip:PwhiB7 structure were determined by molecular replacement using the WhiB3:σA4-βtipʹ structure in the R3 form as the search model. Phenix.autobuid was then used to build the first partial DNA model of PwhiB7. The assignment of the rest nucleotides was done manually, followed by multiple cycles of refinements using COOT and PHENIX (48, 49).
Data visualization
Sequence alignments were performed using Clustal Omega and ESpript online server (https://espript.ibcp.fr), and the sequence logo was generated using WebLogo (50, 51, 52). The representative WhiB3 subclass sequences used for the alignments are modified from the study by Chandra et al. (53) and listed in the legend of Fig. S1. 3D structure figures were prepared with the PyMol Molecular Graphics System v2.3 (https://pymol.org/2/). The molecular interface between WhiB3 and σA4-βtipʹ was estimated using the online macromolecular interface tool PISA (54).
Data availability
Atomic coordinates and structure factors have been deposited in the RCSB Protein Data Bank (PDB) under the accession codes 8CWT and 8CWR for the WhiB3:σA4-βtip complex in the P43212 form and the R3 form, respectively, and 8CYF for the WhiB3:σA4-βtip:PwhiB7 complex, respectively.
Supporting information
This article contains supporting information (25, 26, 50, 54).
Conflict of interest
The authors declare no conflict of interest with the contents of this article.
Acknowledgments
We thank the staff at beamlines 9-2 and 12-2 of SSRL for the assistance during the X-ray diffraction data collection and Dr M. Kato-Maeda for the Mtb H37Rv genomic DNA.
Author contributions
L-M. Z. project administration; L-M. Z. and T. W. methodology; T. W., V. K., M. H., C. J., S. L., and A. S. investigation; X. S. supervision; T. W and L-M. Z. writing-original draft; .T. W., V. K., M. H., C. J., S. L., X. S., L-M. Z., and A. S. writing–review and editing.
Funding and additional information
This work was supported in part by National Institutes of Health (R35 GM138157-01 to L-M. Z.) and by National Science Foundation CAREER Award (CLP 1846908 to L-M. Z.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health and the National Science Foundation. Use of the Stanford Synchrotron Radiation Lightsource, SLAC National Accelerator Laboratory, is supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences under Contract No. DE-AC02-76SF00515. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research, and by the National Institutes of Health, National Institute of General Medical Sciences (including P41GM103393).
Reviewed by members of the JBC Editorial Board. Edited by Patrick Sung
Footnotes
Present addresses for: Shanren Li, College of Life Sciences, Fujian Normal University, Fuzhou, Fujian, 350117, China; Andrew Schacht, Division of Chemistry and Chemical Engineering, California Institute of Technology, Pasadena, CA, 91125, USA.
Supporting information
References
- 1.Pai M., Kasaeva T., Swaminathan S. Covid-19's devastating effect on tuberculosis care - a path to recovery. N. Engl. J. Med. 2022;386:1490–1493. doi: 10.1056/NEJMp2118145. [DOI] [PubMed] [Google Scholar]
- 2.Pawlowski A., Jansson M., Skold M., Rottenberg M.E., Kallenius G. Tuberculosis and HIV co-infection. PLoS Pathog. 2012;8 doi: 10.1371/journal.ppat.1002464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davis N.K., Chater K.F. The Streptomyces coelicolor whiB gene encodes a small transcription factor-like protein dispensable for growth but essential for sporulation. Mol. Gen. Genet. 1992;232:351–358. doi: 10.1007/BF00266237. [DOI] [PubMed] [Google Scholar]
- 4.Soliveri J.A., Gomez J., Bishai W.R., Chater K.F. Multiple paralogous genes related to the Streptomyces coelicolor developmental regulatory gene whiB are present in Streptomyces and other actinomycetes. Microbiology. 2000;146:333–343. doi: 10.1099/00221287-146-2-333. [DOI] [PubMed] [Google Scholar]
- 5.Gomez J.E., Bishai W.R. whmD is an essential mycobacterial gene required for proper septation and cell division. Proc. Natl. Acad. Sci. U. S. A. 2000;97:8554–8559. doi: 10.1073/pnas.140225297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morris R.P., Nguyen L., Gatfield J., Visconti K., Nguyen K., Schnappinger D., et al. Ancestral antibiotic resistance in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U. S. A. 2005;102:12200–12205. doi: 10.1073/pnas.0505446102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen Z., Hu Y., Cumming B.M., Lu P., Feng L., Deng J., et al. Mycobacterial WhiB6 differentially regulates ESX-1 and the Dos regulon to modulate granuloma formation and virulence in Zebrafish. Cell Rep. 2016;16:2512–2524. doi: 10.1016/j.celrep.2016.07.080. [DOI] [PubMed] [Google Scholar]
- 8.Burian J., Ramon-Garcia S., Howes C.G., Thompson C.J. WhiB7, a transcriptional activator that coordinates physiology with intrinsic drug resistance in Mycobacterium tuberculosis. Expert Rev. Anti Infect. Ther. 2012;10:1037–1047. doi: 10.1586/eri.12.90. [DOI] [PubMed] [Google Scholar]
- 9.Rohde K.H., Abramovitch R.B., Russell D.G. Mycobacterium tuberculosis invasion of macrophages: linking bacterial gene expression to environmental cues. Cell Host Microbe. 2007;2:352–364. doi: 10.1016/j.chom.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 10.Mehta M., Rajmani R.S., Singh A. Mycobacterium tuberculosis WhiB3 responds to vacuolar pH-induced changes in mycothiol redox potential to modulate phagosomal maturation and virulence. J. Biol. Chem. 2016;291:2888–2903. doi: 10.1074/jbc.M115.684597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Singh A., Guidry L., Narasimhulu K.V., Mai D., Trombley J., Redding K.E., et al. Mycobacterium tuberculosis WhiB3 responds to O2 and nitric oxide via its [4Fe-4S] cluster and is essential for nutrient starvation survival. Proc. Natl. Acad. Sci. U. S. A. 2007;104:11562–11567. doi: 10.1073/pnas.0700490104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Singh A., Crossman D.K., Mai D., Guidry L., Voskuil M.I., Renfrow M.B., et al. Mycobacterium tuberculosis WhiB3 maintains redox homeostasis by regulating virulence lipid anabolism to modulate macrophage response. PLoS Pathog. 2009;5 doi: 10.1371/journal.ppat.1000545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saini V., Farhana A., Steyn A.J. Mycobacterium tuberculosis WhiB3: a novel iron-sulfur cluster protein that regulates redox homeostasis and virulence. Antioxid. Redox Signal. 2012;16:687–697. doi: 10.1089/ars.2011.4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Larsson C., Luna B., Ammerman N.C., Maiga M., Agarwal N., Bishai W.R. Gene expression of Mycobacterium tuberculosis putative transcription factors whiB1-7 in redox environments. PLoS One. 2012;7 doi: 10.1371/journal.pone.0037516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saini V., Cumming B.M., Guidry L., Lamprecht D.A., Adamson J.H., Reddy V.P., et al. Ergothioneine maintains redox and bioenergetic homeostasis essential for drug susceptibility and virulence of Mycobacterium tuberculosis. Cell Rep. 2016;14:572–585. doi: 10.1016/j.celrep.2015.12.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cumming B.M., Rahman M.A., Lamprecht D.A., Rohde K.H., Saini V., Adamson J.H., et al. Mycobacterium tuberculosis arrests host cycle at the G1/S transition to establish long term infection. PLoS Pathog. 2017;13 doi: 10.1371/journal.ppat.1006389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mehta M., Singh A. Mycobacterium tuberculosis WhiB3 maintains redox homeostasis and survival in response to reactive oxygen and nitrogen species. Free Radic. Biol. Med. 2019;131:50–58. doi: 10.1016/j.freeradbiomed.2018.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Steyn A.J., Collins D.M., Hondalus M.K., Jacobs W.R., Jr., Kawakami R.P., Bloom B.R. Mycobacterium tuberculosis WhiB3 interacts with RpoV to affect host survival but is dispensable for in vivo growth. Proc. Natl. Acad. Sci. U. S. A. 2002;99:3147–3152. doi: 10.1073/pnas.052705399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Molle V., Palframan W.J., Findlay K.C., Buttner M.J. WhiD and WhiB, homologous proteins required for different stages of sporulation in Streptomyces coelicolor A3(2) J. Bacteriol. 2000;182:1286–1295. doi: 10.1128/jb.182.5.1286-1295.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jakimowicz P., Cheesman M.R., Bishai W.R., Chater K.F., Thomson A.J., Buttner M.J. Evidence that the Streptomyces developmental protein WhiD, a member of the WhiB family, binds a [4Fe-4S] cluster. J. Biol. Chem. 2005;280:8309–8315. doi: 10.1074/jbc.M412622200. [DOI] [PubMed] [Google Scholar]
- 21.Burian J., Yim G., Hsing M., Axerio-Cilies P., Cherkasov A., Spiegelman G.B., et al. The mycobacterial antibiotic resistance determinant WhiB7 acts as a transcriptional activator by binding the primary sigma factor SigA (RpoV) Nucleic Acids Res. 2013;41:10062–10076. doi: 10.1093/nar/gkt751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kudhair B.K., Hounslow A.M., Rolfe M.D., Crack J.C., Hunt D.M., Buxton R.S., et al. Structure of a Wbl protein and implications for NO sensing by M. tuberculosis. Nat. Commun. 2017;8:2280. doi: 10.1038/s41467-017-02418-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feng L., Chen Z., Wang Z., Hu Y., Chen S. Genome-wide characterization of monomeric transcriptional regulators in Mycobacterium tuberculosis. Microbiology (Reading) 2016;162:889–897. doi: 10.1099/mic.0.000257. [DOI] [PubMed] [Google Scholar]
- 24.Stewart M.Y.Y., Bush M.J., Crack J.C., Buttner M.J., Le Brun N.E. Interaction of the Streptomyces Wbl protein WhiD with the principal sigma factor σHrdB depends on the WhiD [4Fe-4S] cluster. J. Biol. Chem. 2020;295:9752–9765. doi: 10.1074/jbc.RA120.012708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wan T., Li S., Beltran D.G., Schacht A., Zhang L., Becker D.F., et al. Structural basis of non-canonical transcriptional regulation by the σA-bound iron-sulfur protein WhiB1 in M. tuberculosis. Nucleic Acids Res. 2020;48:501–516. doi: 10.1093/nar/gkz1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wan T., Horova M., Beltran D.G., Li S., Wong H.X., Zhang L.M. Structural insights into the functional divergence of WhiB-like proteins in Mycobacterium tuberculosis. Mol. Cell. 2021;81:2887–2900.e5. doi: 10.1016/j.molcel.2021.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lilic M., Darst S.A., Campbell E.A. Structural basis of transcriptional activation by the Mycobacterium tuberculosis intrinsic antibiotic-resistance transcription factor WhiB7. Mol. Cell. 2021;81:2875–2886.e5. doi: 10.1016/j.molcel.2021.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haran T.E., Mohanty U. The unique structure of A-tracts and intrinsic DNA bending. Q. Rev. Biophys. 2009;42:41–81. doi: 10.1017/S0033583509004752. [DOI] [PubMed] [Google Scholar]
- 29.You D., Xu Y., Yin B.C., Ye B.C. Nitrogen regulator GlnR Controls redox sensing and lipids anabolism by directly activating the whiB3 in Mycobacterium smegmatis. Front. Microbiol. 2019;10:74. doi: 10.3389/fmicb.2019.00074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blanco A.G., Canals A., Bernues J., Sola M., Coll M. The structure of a transcription activation subcomplex reveals how σ70 is recruited to PhoB promoters. EMBO J. 2011;30:3776–3785. doi: 10.1038/emboj.2011.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Campbell E.A., Muzzin O., Chlenov M., Sun J.L., Olson C.A., Weinman O., et al. Structure of the bacterial RNA polymerase promoter specificity sigma subunit. Mol. Cell. 2002;9:527–539. doi: 10.1016/s1097-2765(02)00470-7. [DOI] [PubMed] [Google Scholar]
- 32.Smith L.J., Stapleton M.R., Buxton R.S., Green J. Structure-function relationships of the Mycobacterium tuberculosis transcription factor WhiB1. PLoS One. 2012;7 doi: 10.1371/journal.pone.0040407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith L.J., Stapleton M.R., Fullstone G.J., Crack J.C., Thomson A.J., Le Brun N.E., et al. Mycobacterium tuberculosis WhiB1 is an essential DNA-binding protein with a nitric oxide-sensitive iron-sulfur cluster. Biochem. J. 2010;432:417–427. doi: 10.1042/BJ20101440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Solans L., Aguilo N., Samper S., Pawlik A., Frigui W., Martin C., et al. A specific polymorphism in Mycobacterium tuberculosis H37Rv causes differential ESAT-6 expression and identifies WhiB6 as a novel ESX-1 component. Infect. Immun. 2014;82:3446–3456. doi: 10.1128/IAI.01824-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fonfria-Subiros E., Acosta-Reyes F., Saperas N., Pous J., Subirana J.A., Campos J.L. Crystal structure of a complex of DNA with one AT-hook of HMGA1. PLoS One. 2012;7 doi: 10.1371/journal.pone.0037120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aravind L., Landsman D. AT-hook motifs identified in a wide variety of DNA-binding proteins. Nucleic Acids Res. 1998;26:4413–4421. doi: 10.1093/nar/26.19.4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cortes T., Schubert O.T., Rose G., Arnvig K.B., Comas I., Aebersold R., et al. Genome-wide mapping of transcriptional start sites defines an extensive leaderless transcriptome in Mycobacterium tuberculosis. Cell Rep. 2013;5:1121–1131. doi: 10.1016/j.celrep.2013.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shell S.S., Wang J., Lapierre P., Mir M., Chase M.R., Pyle M.M., et al. Leaderless transcripts and small proteins are common features of the mycobacterial translational landscape. PLoS Genet. 2015;11 doi: 10.1371/journal.pgen.1005641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Newton-Foot M., Gey van Pittius N.C. The complex architecture of mycobacterial promoters. Tuberculosis (Edinb.) 2013;93:60–74. doi: 10.1016/j.tube.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 40.Lee D.S., Kim P., Kim E.S., Kim Y., Lee H.S. Corynebacterium glutamicum WhcD interacts with WhiA to exert a regulatory effect on cell division genes. Antonie Van Leeuwenhoek. 2018;111:641–648. doi: 10.1007/s10482-017-0953-0. [DOI] [PubMed] [Google Scholar]
- 41.Bush M.J., Chandra G., Bibb M.J., Findlay K.C., Buttner M.J. Genome-wide chromatin immunoprecipitation sequencing analysis shows that WhiB is a transcription factor that cocontrols its regulon with WhiA to initiate developmental cell division in Streptomyces. mBio. 2016;7 doi: 10.1128/mBio.00523-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lilic M., Holmes N.A., Bush M.J., Marti A.K., Widdick D.A., Findlay K.C., et al. Structural basis of dual activation of cell division by the actinobacterial transcription factors WhiA and WhiB. Proc. Natl. Acad. Sci. U. S. A. 2023;120 doi: 10.1073/pnas.2220785120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Johnson M.K. Iron-sulfur proteins: new roles for old clusters. Curr. Opin. Chem. Biol. 1998;2:173–181. doi: 10.1016/s1367-5931(98)80058-6. [DOI] [PubMed] [Google Scholar]
- 44.Crack J.C., Smith L.J., Stapleton M.R., Peck J., Watmough N.J., Buttner M.J., et al. Mechanistic insight into the nitrosylation of the [4Fe-4S] cluster of WhiB-like proteins. J. Am. Chem. Soc. 2011;133:1112–1121. doi: 10.1021/ja109581t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mao X.J., Yan M.Y., Zhu H., Guo X.P., Sun Y.C. Efficient and simple generation of multiple unmarked gene deletions in Mycobacterium smegmatis. Sci. Rep. 2016;6 doi: 10.1038/srep22922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Williams K.J., Joyce G., Robertson B.D. Improved mycobacterial tetracycline inducible vectors. Plasmid. 2010;64:69–73. doi: 10.1016/j.plasmid.2010.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Otwinowski Z., Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 48.Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., et al. Phenix: a comprehensive python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Crooks G.E., Hon G., Chandonia J.M., Brenner S.E. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Robert X., Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014;42:W320–324. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sievers F., Higgins D.G. Clustal omega for making accurate alignments of many protein sequences. Protein Sci. 2018;27:135–145. doi: 10.1002/pro.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chandra G., Chater K.F. Developmental biology of Streptomyces from the perspective of 100 actinobacterial genome sequences. FEMS Microbiol. Rev. 2014;38:345–379. doi: 10.1111/1574-6976.12047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Krissinel E., Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic coordinates and structure factors have been deposited in the RCSB Protein Data Bank (PDB) under the accession codes 8CWT and 8CWR for the WhiB3:σA4-βtip complex in the P43212 form and the R3 form, respectively, and 8CYF for the WhiB3:σA4-βtip:PwhiB7 complex, respectively.