Highlights
-
•
1-Mortality due to ISAv exposure was significantly higher at 10 °C compared to 20 °C, despite higher acute mortality at the higher temperature.
-
•
2-Variation in family-based susceptibility to ISAv exposure ranged from 32 to 95% survival, and was linked to temperature.
-
•
3-Viral prevalence disappeared quicker at 20 °C compared to 10 °C.
-
•
4- Innate antiviral response was higher at 10 °C as demonstrated by genes such as mx1, isg15, and vprn, as well as the Th2 response, demonstrated by il10 and il4/13a.
-
•
5- The Th1 and antigen presenting cells response was initially greater at 20 °C, as seen in il12rb2 and nkl, which may therefore play a role in viral clearance.
Keywords: Temperature, Salmon, ISAv, Anti-viral, Immune response, Vaccination
Abstract
Ocean temperatures continue to rise annually due to the ever-growing consequences of global climate change. These temperature changes can have an impact on the immunological robustness of cultured fish, especially cold-water species such as Atlantic salmon. The salmon farming industry already loses hundreds of millions of dollars each year to infectious and non-infectious diseases. One particularly important and WOAH reportable disease is infectious salmon anemia caused by the orthomyxovirus ISAv. Considering the changing environment, it is necessary to find ways to mitigate the effect of diseases on the industry. For this study, 20 Atlantic salmon families were housed in each of 38 different tanks at the AVC, with half of the fish being kept at 10 °C and half being kept at 20 °C. Donor Atlantic salmon IP- injected with a highly virulent ISAv isolate (HPR4; TCID50 of 1 × 105/mL) were added to each tank as the source of co-habitation infection. Both temperatures were sampled at onset of mortality in co-habited fish and at resolution of mortality. Family background and temperature significantly impacted ISAv load, as assessed by qPCR, time to mortality and overall mortality. Mortality was more acute at 20 °C, but overall mortality was higher at 10 °C. Based on percent mortality calculated over the course of the study, different families demonstrated different levels of survival. The three families that demonstrated the highest percent mortality, and the three families with the lowest percent mortality were then assessed for their antiviral responses using relative gene expression. Genes significantly upregulated between the unexposed fish and ISAv exposed fish included mx1, il4/13a, il12rb2, and trim25, and these were further impacted by temperature. Understanding how ISAv resistance is impacted by temperature can help identify seasonal risks of ISAv outbreaks as well as ideal responses to be targeted through immunopotentiation.
1. Introduction
Infectious salmon anemia (ISA) is a viral disease which can result in millions of dollars in financial losses each year for commercial producers in the Atlantic salmon (Salmo salar) industry [1]. Fish that have been infected with the virus may show lesions such as hemorrhaging on the abdomen or viscera, exophthalmos, or an enlarged liver [1]. The first incidence of this disease reported in farmed salmon in Canada was in 1996 in New Brunswick [1]. Since then ISA has been reported on salmon farms in other Canadian provinces; Prince Edward Island, Nova Scotia, and Newfoundland and Labrador. Increased mortality with unknown cause may be an indication of ISA, with a mortality rate ranging from 0.5 to 1% daily, and an overall death rate of 90% [2]. The disease has been shown to transfer horizontally from fish to fish and from one farm site to the other through water, and boats travelling between sites and cages also playing a role in increasing outbreaks [3]. Due to the easy transmission and pathogenicity of ISA, detection of the disease often requires the entire farm population to be culled [1]. Currently there is no cure for ISA, however, there are vaccines that are regularly administered. Unfortunately, there is little scientific data on the efficacy rates of these vaccines at aquaculture farm sites [4].
Over the next 80 years, ocean temperatures are expected to rise 1–4 °C [5]. This has the potential to have significant impacts on the aquaculture industry, in particular sea cage aquaculture conducted in temperate coastal zones. Atlantic salmon aquaculture in Canada is one of these industries. It is therefore important for the industry to take a proactive approach to these potential impacts on production. As with other ectotherms, salmonid core temperatures are determined by their environment. Predicted coastal zone temperature increases and subsequent decreased oxygen capacity could push Atlantic salmon outside of their physiologically optimal environment. For instance, it has been shown that Atlantic salmon kept at higher temperatures will release a higher amount of cortisol when exposed to a stressor [6]. The increase in temperature from climate change may also alter pathogen virulence and/or temporal and spatial prevalence. In the case of ISAv, the lack of efficacious prophylaxis or intervention strategies heighten these risks making it a potential focus for inclusion in a selective breeding program. By examining the genetic and genomic components of resistance to infectious disease and how they are impacted by temperature, genetic markers can be identified and incorporated into a breeding program especially in the event that increased resistance results in fewer infected fish after exposure to ISAv in the environment. Recent work by Gervais et al. [7] has shown that ISAv resistance shows moderate heritability with a polygenic architecture in European Atlantic salmon, and mild up-regulation of the interferon pathway (mx1, mx3, irf7, trim25) which may be important markers of the host response to this virus.
We hypothesized that there would be variability in susceptibility to ISAv across different families and this variation would be linked to antiviral responses. Our second hypothesis was that high temperature (20 °C) would initiate a stronger/quicker immune response to the virus in Atlantic salmon resulting in lower mortality. To address these hypotheses, the current study examined 20 families from a Saint John River Atlantic salmon breeding program, and their natural resistance and vaccine induced resistance to ISAv, under two temperature regimes (10 and 20 °C). Co-habitation exposures were completed and 38 individual replicate tanks were assessed for survival, viral presence and immune response to infection.
2. Materials and methods
2.1. Fish maintenance and husbandry
Atlantic salmon smolts (ca.2200; Fig. 1) for this study were obtained from 20 families (100 fish per family) from a larger breeding program produced and reared at the Huntsman Marine Science center (Huntsman) in St. Andrews, New Brunswick. The families selected for this study were minimally genetically linked with contribution from 16 sires and 19 dams from three different year classes within the breeding program and having variable performance for other traits of commercial interest. All fish were anesthetized with tricaine methane sulphonate (MS-222; 150 mg/L) prior to insertion of a passive integrated transponder (PIT) tag at approximately 16.5 g mean weight. Fish were recovered in a separate bath then stocked into a single 7.5 m3 tank. Fish were cohabited in this tank (< 30 kg/m3) supplied by a partial recirculating freshwater system with ambient light and water temperature (5–12 °C depending on ambient/ seasonal temperature). When the smolts reached >50 g mean weight, all fish were anesthetized (MS-222; 150 mg/L) and half were i.p. vaccinated (100μl/fish) with Micro Forte VII (multivalent vaccine including inactivated ISA), as well as Renogen (Elanco Animal Health), while the other half of the fish from each family were given a placebo, phosphate buffered saline (PBS) i.p. injection (100 μl/fish). At approximately 1074 dd post vaccination (ddpv), fish were acclimated to increasing salinity over 2–3 weeks by the addition of seawater and maintained at 22–24 ppt (9–12 °C) until shipping. After 1074 ddpv, fish were sampled for a pre-exposure Time 0 baseline (Refer to 2.1.2). Three weeks post sampling (1253 ddpv), the fish were transported to the AQCL-2 Aquatic Facility, Atlantic Veterinary College (AVC), Charlottetown, Prince Edward Island. Upon arrival, fish were randomly distributed into 38 tanks (300 L) across 4 biofiltration units, one/room (∼9 tanks/room), with each tank containing a minimum of one vaccinated family representative, and one PBS family representative from each of the 20 families. The biomass (ca. 44 fish/tank) was approximately equal across all 38 tanks at 25 kg/m3. All fish were maintained in a seawater (33 ± 2 ppt) (Instant Ocean ®) recirculation system at 11.0 ± 1.0 °C, with a photoperiod of 14 h light: 10 h dark and allowed to acclimate to their tank for at least 3 weeks. Oxygen levels were maintained between 95 and 100% saturation and ammonia was maintained below 0.5 mg/L, nitrite below 0.5 mg/L, and nitrate below 15 mg/L.
Fig. 1.
Study design schematic. (A) Population of fish originating from Huntsman Marine Science center (20 families with 100 fish each), were (B) PIT tagged at ∼16.5 g; (C) Half of the fish in each family were i.p. injected with MicroForte vII & Renogen vaccine. The other half of the fish were given a PBS i.p. injection. (D) After 1074° days post vaccination, fish were sampled (E) for pre-exposure Time 0 (6 vaccinated and 6 PBS injected fish from each family; head kidney and serum); (F) Three weeks later fish were transported to the Atlantic Veterinary College, and (G) Randomly distributed into 38 tanks upon arrival; (H) 228 smolts were distributed across 6 tanks to be used as donor fish. (H) Donor fish were injected with ISAV-HPR4 isolate. (I) One-week later blood samples were taken of donor fish, and 6 donor fish were added to each cohabitant tank. One week post donor addition, temperature adjustments of 0.5 °C were made until half of the tanks reached 10 °C and half reached 20 °C; (J) Once mortalities were observed in non-donor cohabitant fish in each room (2 rooms per temperature), (K) 4 tanks were sampled from each room as a measure of Time 1; (L) After one week with no mortalities, Time 2 sampling occurred.
Fish were fed EWOS transfer feed at a pellet size of 3.5 mm at 1% of their body weight daily. They were hand fed twice daily during the weekdays and once daily on weekends and holidays. Feeding behavior was documented on a scale of 1–4 (1 = 100% feeding, 2 = 75% feeding, 3 = 50% feeding 4=<25% feeding) visually assessing whether fish were actively feeding both at the surface upon introduction of the feed, and the general activity level as they pursued the feed. Swimming behavior was assessed on a scale of 1–4 based on the normality of the swimming patterns with comments on use of the water column and abnormalities, e.g. presence of visible gasping and/or crowding around the air stone or water inflow. Additional recorded comments included presence or absence of fecal casts, loss of equilibrium, hyperactivity, lethargy, flashing, and change in appearance. All tanks were purged before and after feedings to remove fecal waste and uneaten feed. Fish care and husbandry practices were in accordance with the guidelines of the Canadian Council of Animal Care (Protocol # 20–017) and was approved by the University of Prince Edward Island Animal Care Committee.
Viral preparations of Infectious Salmon Anemia Virus (ISAv), isolate ISV-HPR4 (Research and Productivity Council, Fredericton, New Brunswick; RPC/NB 04–085–1; Ritchie et al. [8]) were obtained from the Department of Fisheries and Oceans in Moncton, New Brunswick (Dr. N. Gagné). A TCID50 of 1 × 105/ml was confirmed on an Atlantic salmon kidney (ASK) cell-line monolayer in vitro using the Spearmean-Kärber method. Atlantic salmon smolts (n = 228) from the same breeding program and year class that had been kept in a separate but similar saltwater recirculating system (n = 6 tanks) were used as donors. All donor fish were anaesthetized (MS-222;150 mg/L) prior to injecting with aliquoted ISAv viral preparation and recovered in a separate tank then returned to their respective tanks. One week post injection, all donors were similarly anesthetized and a blood sample (500 μl) was taken for later assessment and to enhance progression of anemia. Six donor fish were then added to each of the 38 study tanks.
One week after the addition of donors, tanks underwent temperature adjustments of 0.5 °C per day until each of the 9 tanks in two rooms reached either 10 ± 1.0 °C, or 20 ± 1.0 °C. Additionally, oxygen levels of fish tanks maintained at 20 ± 1.0 °C, were kept at ∼80% saturation unlike 95–100% saturation in the colder temperature group.
All tanks were checked for morbidity or mortality at a minimum of 3 times daily. Any moribund fish were euthanized using an overdose of MS-222(250 mg/L). All euthanized fish and mortalities were weighed, measured, and examined (externally/internally) for clinical signs of disease.
2.2. Sampling
All fish were fasted for 24 h prior to sampling.
Time 0: A total of 6 vaccinated fish, and 6 PBS injected fish from each family were sampled at 1074° days post vaccination (ddpv). This was done to provide a baseline for all measured parameters, and to confirm the absence of viral infection.
Time 1: Once mortalities (i.e. > 2 tanks with mortalities) were observed in non-donor co-habitant fish in each room/biofiltration unit, all of the fish in each of four tanks in each room (2 rooms per temperature) were sampled. Time 1 sampling occurred at 288–308 dd post exposure (ddpe) at 20 °C and 302–312 ddpe at 10 °C. Entire tanks were sampled to prevent any bias/stress by removing uneven numbers of fish per family per group.
Time 2: Final sampling occurred after one week with no mortalities. This occurred at 667–727 ddpe at 20 °C, and at 640–661 ddpe at 10 °C. At this time, the fish from each of the remaining tanks at both temperatures were sampled.
Samples of head kidney tissue were maintained on dry ice until storage at −80 °C following sampling for later molecular analysis of immune gene responses and for determination of viral load. For each fish sampled; weight, length, and sex were recorded and condition factor (CF) was calculated. All sampled fish were also examined for any abnormalities, including ascites, external or internal hemorrhaging, enlarged spleens or kidneys, congested liver, exophthalmia, skin lesions, cataracts, and spleen or kidney nodules.
2.3. RNA isolation
Total RNA was isolated from head kidney samples (ca. 50–100 mg per fish) using an adaptation of a Tri-Reagent and metal beads method [9]. RNA was analyzed for sample quality and integrity using ThermoFisher's Nanodrop 300 followed by 1.5% Agarose gels electrophoresis. To remove any remaining contaminating DNA, each RNA sample (2–6 µg) then went through a TURBO® DNase treatment.
All samples (adjusted so the same amount of DNase treated RNA is added for each sample; 2 µg) from each time point underwent reverse transcription to create cDNA (Total reaction volume of 40 µl). This was completed using iSCRIPT™ Reverse Transcription Supermix manufactured by BioRad. A negative-RT control was also made with each lot of samples using iScript™ No-RT Control Supermix. All samples were then placed in the Eppendorf Master Cycler (EppMC) and cDNA was created using the following steps; priming for 5 min at 25 °C, reverse transcription for 20 min at 46 °C, and RT inactivation for 1 min at 95 °C. The program then held the samples at 4 °C. Once the program was complete, all samples were centrifuged and 40 µl of nuclease free water was added to each tube for a 1:2 dilution. These samples were then stored at −20 °C.
RT-qPCR (using 1 µl of diluted cDNA per reaction) was run using SsoAdvanced Universal SYBR® Green Supermix to generate a standard curve for the reference genes Elongation factor 1-alpha (ef1ab), 40S ribosomal protein S20 (rps20), and eukaryotic translation initiation factor 3 subunit E (eif3ea). The plate setup for these was 95 °C for 3 min, 95 °C for 15 s, then 60 °C for 30 s, for 40 cycles. Two different types of qPCR reactions (described in Section 2.1.6 below) were then used along with the Luminex MagPix system to establish relative expression of targets.
To test ISAv load, TaqMan™ Gene Expression Master Mix from Applied Biosystems was used (again using µl of diluted cDNA per reaction). This assay targeted segment 8 of the ISAv genome (AIVI4A5) following the protocol of Snow et al. [10] modified as below. The protocol consisted of; pre-incubation at 50 °C for 2 min for one cycle, denaturation at 95 °C for 10 min for 1 cycle, and amplification at 95 °C for 15 s then 60 °C for 1 min. This was completed for 42 cycles.
2.4. Luminex MagPix-sample selection
The Luminex MagPix system was used to run a subset of samples from all time points. There were 138 samples selected in total. Both ISAv Cq and host rps20 (to ensure lack of ISAv amplification was not due to sample quality) Cq values were analyzed for each sample. A weak Pearson correlation was observed between ISAv Cq and host rps20 Cq suggesting that the ISAv Cq values were not impacted by host RNA degradation. Time 1 samples were then sorted into 1 of 4 categories based on their ISAv Cq values. Those categories were; 〈32 Cq, 32< Cq <37, 37< Cq< 42, and Cq 〉 40/ No amplification. All the samples from the categories <32 Cq and 32< Cq <37 were chosen for analysis, as these were considered positive for ISAv. Altogether, there were 101 samples chosen from Time 1. In addition, 3 vaccinated and 3 unvaccinated size-matched fish samples were taken from each of the 6 families from Time 0. Any samples with >30 Cq for rps20 were not chosen for MagPix analysis. 150 ng of Dnase-treated RNA was then used for each sample to compare anti-viral gene expression using this technique.
2.5. Luminex MagPix protocol
Anti-viral markers for assessing immunological response to vaccination and infection in Atlantic salmon were assembled on a QuantiGene 20-plex Panel (Invitrogen, custom) and included: irf7 (NM_001136548), mx1 (NM_001139918), cd8a (NM_001123583), cd86 (XM_014215239), il4/13a (NM_001204895), isg15 (NM_001123640), il12rb2 (NM_001165330), gal-9 (XM_014215012), trim25 (XM_014207260), nkl (NM_001141110), crp (NM_001140668), il10 (XM_014186180), saa5 (NM_001146565), vprn (NM_001140939), rig-1(NM_001163699), trim16 (XM_014212487), c3 (XM_014186867) along with three reference markers rps20 (NM_001140843), ef1ab (NM_001123629) and eif3s6 (NM_001141695). The Invitrogen™ QuantiGene™ Plex Gene Expression Assay Kit was used according to the manufacturer's instructions (Thermo Fisher Scientific). Briefly, the lysis mixture was warmed at 37 °C for 30 min, followed by gentle swirling, while the target-specific probe set and blocking reagent were thawed at room temperature and briefly vortexed to mix. DNase-treated RNA, as previously described (Section 2.1.3), was thawed on ice and diluted to 150 ng in 20 µL of nuclease-free water (NFW). The Working Bead Mix was prepared for 2 to 64-plex reactions on a full 96-well plate by combining NFW, Lysis Mixture, Blocking Reagent, Capture Beads that were vortexed for 30 s before adding, and the Probe Set. The working bead mix was vortexed for 10 sec, then immediately transferred to a 25 ml reagent reservoir and 80 µL was dispensed into each well of the hybridization plate using a multi-channel pipette. The 20 µL RNA sample was pipetted into its respective well on the hybridization plate with the Working Bead Mix, so that the final volume in each well was 100 µL. Each sample was run in duplicate, with two wells serving as background controls with 80 µL of the Working Bead Mix mixed with 20 µL of NFW. To compensate for inter-run variability between plates, two wells were designated as the interplate calibrator (IPC) where the same sample was run on all plates. The hybridization plate was sealed using a pressure seal that was applied firmly with a soft-rubber roller to prevent evaporation, and then placed in an Incubating Microplate Shaker (VWR) for 18 h at 54 °C and 600 rpm.
Following incubation of the hybridization plate, the Pre-amplifier, Amplifier, and Label Probe Solutions were warmed from 4 °C to 37 °C for 30 min, then mixed well by inversion and kept at room temperature until required. The Streptavidin-conjugated R-Phycoerythrin (SAPE) Diluent was removed from 4 °C storage and brought to room temperature before use. For each plate 1X QuantiGene™ Plex Wash Buffer was prepared by mixing 0.6 mL Wash Buffer Component 1 and 10 mL of Wash Buffer Component 2 to 189 mL NFW. The hybridization plate was removed from the shaking incubator and centrifuged (Eppendorf 5804R) at 240 x g for 1 min at room temperature. The pressure seal was removed and samples were mixed and transferred to the Magnetic Separation Plate using a multichannel pipette.
The Magnetic Separation Plate was then securely locked onto the Magnetic Plate Separator (Luminex) and beads were left to accumulate at the bottom of each well for 2 min before the liquid was decanted with a manual pipette. With the magnetic separation plate locked onto the magnetic separator, 100 µL of the 1X wash buffer was added to each sample well and allowed to incubate for an additional 30 s before the liquid was decanted. This was repeated two more times before 100 µL of Pre-Amplifier Solution was added to each well. The plate was sealed and removed from the magnetic separator, then shaken on a Mini Shaker (VWR) at 800 rpm for 1 min at room temperature to resuspend beads, then transferred to the Incubating Microplate Shaker pre-set to 50 °C and 600 rpm for 1 h.
Following the 1 h Pre-Amplifier incubation, the Magnetic Separation Plate was removed from the shaking incubator and inserted into the Handheld Magnetic Plate Separator. The plate seal was removed, and the washing procedure was repeated as above, making sure to give time between the three washes for the beads to accumulate at the bottom of each well. Following decanting of the last wash, 100 µL of the Amplifier Solution was added to each well and the plate was sealed and removed from the Handheld Magnetic Plate Separator and processed as previously described. This process was then repeated with the addition of 100 µL of the Label Probe Solution (3 x wash, add 100 µL solution to each well, shake 1 min, incubate at 50 °C and 600 rpm for 1 h).
During the 1 h Label Probe hybridization, SAPE Working Reagent was made by diluting 36 µL of SAPE with 12 mL of SAPE Diluent and protected from light until required. After the 1 h incubation, the plate was washed as described above and 100 µL of SAPE Working Reagent was added to each well. The plate was sealed and covered with aluminum foil to protect from light, then placed on the Mini Shaker at room temperature, shaking for 1 min at 800 rpm followed by shaking for 30 min at 600 rpm.
Following the incubation with SAPE, the plate secured onto the magnetic separator and washed three times as described above, using the SAPE Wash Buffer. Following decanting of the last wash, 130 µL of SAPE Wash Buffer was added to each well. The plate was then sealed and removed from the magnetic separator, where it was wrapped in aluminum foil to protect from light. The wrapped plate was placed on the Mini Shaker at room temperature for 3 min at 800 rpm to resuspend beads. The plate was immediately loaded into the pre-warmed and calibrated Magpix® (Luminex) and read using the Luminex xPONENT® MAGPIX Version 4.2 Build 1324 software
2.6. Reverse transcription quantitative polymerase chain reaction (RT-qPCR)
Four genes (mx1, il4/13a, il12rb2, and trim25) of interest were selected for further analysis of the final time point in the study (Time 2). The standard curves generated for the reference genes ef1ab, rps20, eif3s6 for ISAv load analysis were also used for these samples (Table 1).
Table 1.
RT-qPCR primers used for Time 2 samples.
| Gene Target | Name | Reference | Accession # | Primers (5′−3′) |
Amplicon size (bp) | Efficiency (%) |
|---|---|---|---|---|---|---|
| Myxovirus (influenza virus) resistance 1 | mx1 | Workenhe et al., 2009 |
NM_001123693.1 |
F- TGCAGCTGGGAAGCAAACT R- CAACGTTTGGCTGATCAGATTC |
123 | 95.4 |
| Interleukin 4/interleukin-13 | il-4/13a | Purcell, S.L. | AB574337 | F-CTGGGACCACCACAAAATGC R- GTGGCATTTTTCACGGAGGC |
73 | 94.5 |
| Interleukin-12 β | il-12β | Skugor et al., 2NEN-1b |
F-TCTACCTACACGACATTGTCCAG R- ATCCATCACCTGGCACTTCATCC |
62 | 93.1 | |
| Tripartite Motif Containing 25 | trim25 | Workenhe et al., 2009 |
F-ATAGGACCCTGCCTTCACTG R- CTGGAGACTGGAGCACACTG |
64 | N/A | |
| Eukaryotic Elongation Factor-1Alpha paralog B | ef1ab | Olsvik et al., 2005 |
F-TGCCCCTCCAGGATGTCTAC R- CACGGCCCACAGGTACTG |
59 | 98.0 | |
| S20 Ribosomal protein S20 | rps20 | Olsvik et al., 2005 |
F-GCACTTATCCGTGGAGCTA R- TGGTGATGCGCAGAGTCTTG |
85 | 101.0 | |
| Eukaryotic translation initiation factor 3 subunit 6 | eif3s6 | Skugor et al., 2008 |
F-GTCGCCGTACCAGCAGGTGATT R- CGTGGGCCATCTTCTTCTCGA |
92 | 97.3 |
To generate standard curves, a pool of the cDNA from all Time 2 samples was created. To achieve a wide dynamic range standard curve a modified pool, with samples of lower Cq, was made (n = 16 samples) that represented all treatment groups and temperatures, with a Cq value between 24.0–27.2.
The ef1ab and eif3ea assays were run at a 1:5 dilution. Each of the Time 2 samples were then run on two 384-well plates. For each well in the 384-well plate, 10μl of Master Mix was added, with the mix including the SsoAdvanced Universal™ SYBR® Green. 2 µl of cDNA from each sample was added to three triplicate wells. The Master Mix and the cDNA was added to each well using the Aurora plate loading robot (Aurora- VERSA 10 Automated Liquid Handling).
A temperature optimization plate was run to determine the best annealing temperature to use for each gene. An optimal temperature of 61.6 °C was used for trim25, mx1 and il4/il13a, and for il12rb2 the optimal annealing temperature was 54.8 °C. The plate setup was 95 °C for 3 min, 95 °C for 15 s, the chosen annealing temperature for 30 s, 65 °C for 5 s, for 40 cycles. The mx1 assay was run at a 1:15 dilution. Expression for il4/13a and il12rb2 was very low, so a PCR product was first made to spike into the cDNA pool so that a standard curve could be generated. Time 2 samples for il4/13a and il12rb2 were run at a 1:2 dilution. Due to the absence of trim25 in most samples analysis of this gene for Time 2 was discontinued.
2.7. Data analysis
Survival data from each family was analyzed using a nonparametric distribution analysis with right censoring and Kaplan-Meier survival estimates in Stata (Version 12) software (reporting the Log-Rank p-value). An α=0.05 was used. It was also used to determine which families should be further analyzed. The three families with the lowest survival and the three families with the highest survival across both temperatures were chosen for further analysis.
2.7.1. MagPix
Due to variability in the eif3ea over time, it was not carried forward for relative expression calculations but, instead, mean normalized expression was calculated using just ef1ab and rps20. Average mean fluorescence intensity (MFI) was calculated between the two reference genes. To calculate the gene expression for each gene, for each individual sample, the average net MFI was divided by the average MFI determined for the reference genes. In this manner, the gene expression was calculated for each of the 17 genes of interest (GOI) for each sample. All data was combined into one spreadsheet and imported into RStudio (version 3.1.) for statistical analysis. In RStudio, a multi-variable ANOVA table was generated for each of the individual variables, ‘Family’, ‘Time’, ‘Temperature’, ‘Vaccination status’. An ANOVA table was also generated for the relationship between ‘Family’ and each of the other three variables. A Tukey post hoc test was used to conduct the pairwise comparison if it was found to be significant in the ANOVA tables. Boxplots were generated in RStudio.
2.7.2. qPCR
Bio-Rad CFX Maestro was used to assess ef1ab, eif3s6 and rps20 reference gene stability (GeNorm). Analysis of all three reference genes together showed instability. Out of the three genes tested, ef1ab and rps20 were the most stable together. The mean Cq and standard deviation (STDEV) values were calculated for both rps20 (26.27 ± 1.51 Cq) and ef1ab (28.69 ± 2.34 Cq), however M value >1.0 and stability were still unacceptable. As rps20 had the smallest STDEV, as well as the tightest Cq range, it was chosen as the reference gene for data normalization. The data was then analyzed through RStudio and graphs were generated in the same way that data was analyzed for MagPix.
3. Results
3.1. Temperatures of the tanks
To avoid any effect of stress on the fish due to sudden changes in water temperature, all tanks were slowly adjusted from 11 to 12 °C, to their intended study temperature. The temperature in half of the tanks (19) was decreased to 10.2 °C ± 0.11 °C, while the temperature in the other half of the tanks (19) was increased to 19.0 °C ± 0.37 °C (Suppl. Fig. 1). There were no mortalities or abnormal behavior associated with the temperature change.
3.2. Mortalities following exposure to ISAv
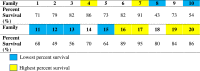
Overall percent survival at the end of the study was calculated by using the cumulative mortality and the total number of fish from each family before donor fish were added to cohabitant tanks. This was calculated for each family at both temperature points. The six families with highest percent survival at 10 °C, were 7, 9, 16, 17, 19, and 20 (yellow highlight; Table 2a) and the 6 families with the lowest percent survival were 1, 8, 11, 12, 15, and 18 (blue highlight; Table 2a). The six families with the highest percent survival at 20 °C were 17, 7, 16, 20, 4, and 19 whereas the 6 families with the lowest percent survival were 8, 12, 10, 13, 15, and 11 (Table 2b).
Table 2a.
Percent survival (%) at 10 °C for Atlantic salmon families 1–20 challenged with ISAv.
 |
Table 2b.
Percent survival (%) at 20 °C for Atlantic salmon families 1–20 challenged with ISAv.
 |
Overall, percent mortality was 57% at 10 °C and 35% at 20 °C. In order to select the families most likely to show contrasting gene expressions that could result in clear survival differences in fish infected with ISAv, six total families were chosen for follow up analysis. Families were chosen based on a combination of the percent survival and the number of samples available at both time points. These six families were also in the top six for survival (7, 16, 17) or bottom six for survival (8, 12, 15) at both 10 °C and 20 °C (Table 2c).
Table 2c.
Cumulative percent survival (%) at 10 °C and 20 °C combined for families 1–20.
 |
Peak mortality occurred more quickly at 20 °C (Fig. 2). Survival by family ranged from 32 – 85% and 43 to 95% at 10 °C and 20 °C, respectively (Tables 2a and 2b; Suppl. Fig 2). The differential survival rates in the six families selected as most resistant or susceptible at 10 °C and 20 °C are depicted in Fig. 3a and 3b. There was no significant difference between vaccinated and unvaccinated fish regardless of family at 10 °C (p = 0.9314; Suppl. Fig. 2), or at 20 °C (p = 0.6772; Supple. Fig. 2).
Fig. 2.
Daily cumulative mortalities of Atlantic salmon (Salmo salar) maintained at either 10 °C or 20 °C, where day 0 represents addition of donor infected Atlantic salmon to each of the tanks. Atlantic salmon were infected with infectious salmon anemia virus. T1 and T2 indicate time points in which Atlantic salmon were assessed for weight and length.
Fig. 3.
Cumulative daily percent survival at 10 °C (a) and 20 °C (b) for best and worst surviving families (7, 8, 12, 15, 16, 17) of Atlantic salmon infected with ISAv.
3.3. Condition factor and weight of sampled fish
Due to the lack of differences observed in survival based on vaccination, both PBS and vaccinated fish were grouped together for CF and weight analysis. The mean weight of fish sampled (n>400) at 10 °C was 227.7 ± 100.2 g and mean fork length was 27.7 ± 13.0 cm with a condition factor of 1.08 ± 0.17. The fish sampled (n>400) at 20 °C had a mean weight of 221.5 ± 95.2 g and mean fork length of 27.7 ± 4.3 cm with a condition factor of 1.03 ± 0.17. A comparison of condition factor for the fish at 10 °C and 20 °C indicated no significant difference (p = 0.454). When comparing the condition factors between families (10 °C and 20 °C combined) there was no significant difference between condition factor at Time 1 (p = 0.2518) or at Time 2 (p = 0.2069).
When comparing weights between families (10 °C and 20 °C combined), family differences were observed over time (Fig. 4). For Time 1, family 12 had a significantly higher weight than family 7 (p = 0.0001), family 8 (p = 0.004), and family 16 (p = 0.001; Fig. 3). Family 15 had a significantly higher weight than family 7 (p = 0.019) and family 17 (p = 0.0001). When comparing weights for families in Time 2, family 12 had a significantly higher weight than family 7 (p = 0.037), family 16 (p = 0.046), and family 17 (p = 0.0001). Family 15 had a significantly higher weight than family 7 (p = 0.006), family 8 (p = 0.038), and family 17 (p = 0.0001). Between Time 1 and Time 2, family 7 (p = 0.0483) and family 15 (p = 0.0282) had a significant increase in weight (Fig. 4).
Fig. 4.
Weight of sampled Atlantic salmon (Salmo salar) at Time 1 and Time 2 at both 10 °C and 20 °C for families 7, 8, 12, 15, 16, 17 exposed to Infectious salmon anemia (ANOVA, p ≤ 0.05; n = 6–10 fish per family per temperature at each time point).
3.4. ISAv load in head kidney from sampled fish
Differences between temperatures were recorded for ISAv load (Cq values) in the 6 families (7, 8, 12, 15, 16, 17) analyzed using qPCR (Table 3a, Table 3b). At Time 1, there was no amplification (i.e., not positive for ISAv) for 30.5% of the fish at 10 °C and 57.0% at 20 °C. At Time 2, there was no amplification in 41.8% of the fish at 10 °C and 80.8% at 20 °C. Samples with a Cq value below 37 were considered positive for the terms of our study. For Time 1, there were 30.7% positive samples at 10 °C and 35.4% positive at 20 °C. For Time 2, there were 20.0% positive at 10 °C and 0% at 20 °C. These 6 families represented the entire exposure group, as when comparing all 20 families at the two temperatures, there were 18.9% positive at 10 °C and 0.01% (1/115) at 20 °C.
Table 3a.
ISAv load as assessed by qPCR in head kidney from fish sampled at Time 1. Highest relative percent survival families highlighted in yellow.
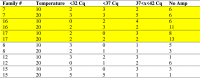 |
Table 3b.
ISAv load as assessed by qPCR in head kidney from fish sampled at Time 2. Highest relative percent survival families highlighted in yellow.
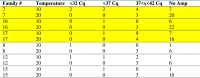 |
3.5. Immune gene expression and vaccination status
There was no impact of vaccination status on gene regulation for 14 of the 17 genes of interest investigated (il10, il12rb2, isg15, mx1, vprn, nkl, saa5, rig1, il4/13a, trim25, crp, c3, irf76, cd8a, cd86). Unvaccinated fish at Time 1 had significantly higher expression of nkl compared to the unvaccinated fish at Time 0 (p = 0.0066, data not shown) and trim16 expression was significantly higher in vaccinated compared to unvaccinated fish at Time 0 (p = 0.0036, data not shown). Galectin-9 was not impacted by sampling time, but significantly affected by the vaccination status of the fish (at Time 0 and Time 1 combined; Fig. 5), where unvaccinated fish had a significantly higher expression (p = 0.0029). No other differences with respect to vaccination status and gene expression were observed.
Fig. 5.
Mean normalized relative quantity (MNRQ) of Galectin 9 (GAL9) in Atlantic salmon (Salmo salar) smolts (head kidney) between vaccinated and unvaccinated fish at both Time 0 and Time 1. Letters indicate significant difference in Galectin 9 gene expression regardless of time (ANOVA, p ≤ 0.05; n = 33–36 per group at each time point.
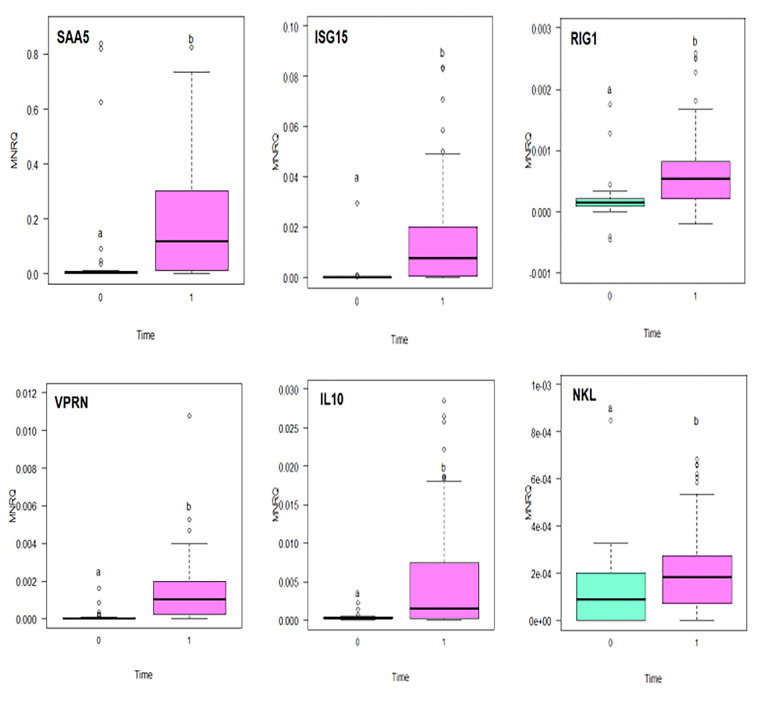
3.6. Immune gene expression and time
Ten genes of interest were found to have significantly different expression between Time 0 and Time 1. Combining data from all the 6 groups (7, 8, 12, 15, 16, 17), six genes [saa5 (p = 0.011), isg15 (p = 0.055), rig1 (p = 2.42×10−05), vprn (p = 2.4 × 10−0.6), il10 (p = 0.00018), and nkl (p = 0.014)] were found to be significantly upregulated at Time 1 compared to Time 0 (Fig. 6). Similarly, when all families were combined, expression of il4/13a was significantly upregulated between Time 0 and Time 1 (p<0.001) (Fig. 7a). When the time points were separated into families, a significant difference was observed in all 6 families between Time 0 and Time 1 for family 17 and family 8 (p = 0.0047), family 17 and 15 (p = 0.014), and family 17 and 7 for il4/13a (p = 0.05) (Fig.7b).
Fig. 6.
Mean normalized relative quantity (MNRQ) of Serum amyloid A protein (SAA5), Interferon stimulated gene 15 (ISG15), Retinoic acid inducible gene I (RIG-I),VPRN, Interleukin-10 (IL-10), NK-lysin (NKL) in Atlantic salmon (Salmo salar) smolts (head kidney) for families 7, 8, 12, 15, 16, 17 exposed to Infectious Salmon Anemia (ISAv) at Time 0 and Time 1 sampling points. Letters indicate significant difference between the two time points in gene expression (ANOVA, p ≤ 0.05; n = 8–12 fish per family per temperature at each time point).
Fig. 7.
Mean normalized relative quantity (MNRQ) of interleukin-4/13A (il4/13a) in Atlantic salmon (Salmo salar) smolts (head kidney) for fish exposed to Infectious Salmon Anemia (ISAv) at both 10 °C and 20 °C for families 7, 8, 12, 15, 16, 17 separated into Time 0 and Time 1(a) and by family (b). Letters indicate significant difference in gene expression between the two time points or family (p < 0.001; n = 8–12 fish per family per temperature at each time point).
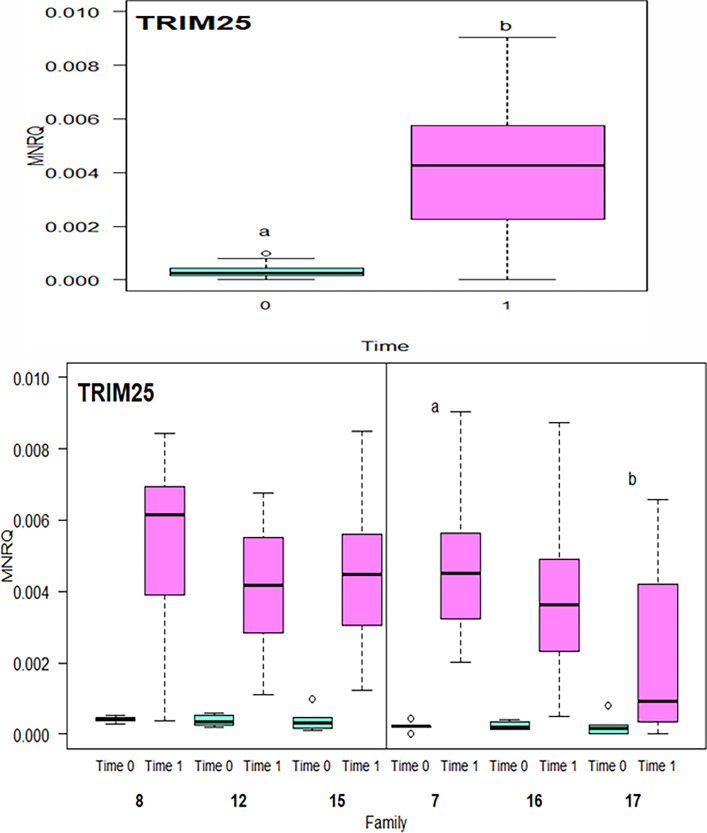
With all families combined, expression of trim25 was significantly upregulated at Time 1 compared to Time 0 (p<0.001) (Fig. 8a). When the timepoints were separated into families, there was a significant downregulation of trim25 in family 17 compared to family 7 (p = 0.006) (Fig. 8b).
Fig. 8.
Mean normalized relative quantity (MNRQ) of Tripartite Motif Containing 25 (trim25), in Atlantic salmon (Salmo salar) smolts (head kidney) for fish exposed to Infectious Salmon Anemia (ISAv) at both 10 °C and 20 °C for families 7, 8, 12, 15, 16, 17 separated into Time 0 and Time 1 (a) and by family (b). Letters indicate significant difference between the two time points in gene expression (p < 0.001; n = 8–12 fish per family per temperature at each time point).
With all families combined, expression of il12rb2 was significantly upregulated at Time 1 compared to Time 0 (p = 2.5 × 10−06) (Fig. 9a). There was a significant difference observed between families 7 and 12 (p = 0.05), and between families 7 and 16 (p = 0.038) (Fig. 9b). Similarly, with all families combined, expression of mx1 was significantly upregulated at Time 1 compared to Time 0 (p = 0.006) (Fig. 10a). There were no significant differences in expression of mx1 between families (Fig. 10b).
Fig. 9.
Mean normalized relative quantity (MNRQ) of interleukin-12 (il12) in Atlantic salmon (Salmo salar) smolts (head kidney) for fish exposed to Infectious Salmon Anemia (ISAv) at both 10 °C and 20 °C for families 7, 8, 12, 15, 16, 17 separated into Time 0 and Time 1 (a) and by family (b). Letters indicate significant difference between the two time points in gene expression (p<0.05; n = 8–12 fish per family per temperature at each time point).
Fig. 10.
Mean normalized relative quantity (MNRQ) of MX dynamin like GTPase 1(mx1) in Atlantic salmon (Salmo salar) smolts (head kidney) for fish exposed to Infectious Salmon Anemia (ISAv) at both 10 °C and 20 °C for families 7, 8, 12, 15, 16, 17 separated into Time 0 and Time 1 (a) and by family (b). Letters indicate significant difference between the two time points in gene expression (p<0.05; n = 8–12 fish per family per temperature at each time point).
3.7. Immune gene expression and temperature
At Time 1, three genes of interest were found to be significantly different in Atlantic salmon head kidney depending on water temperature. Interleukin-10 (p = 1 × 10−07) and isg15 (p = 0.030) were significantly upregulated at 10 °C compared to 20 °C (Fig. 11). Conversely, il12rb2 was significantly up regulated at 20 °C (p = 0.0020). At Time 2, there were no significant differences in expression at the different temperatures for two of the three genes of interest analysed, with the exception of il12rb2 expression in family 17 (Suppl. Fig. 3). Of 130 head kidneys analysed, only 13 showed any expression at this time point for il12rb2, and 12 of those 13 head kidneys were from fish in family 17. The remaining positive head kidney was from family 16 (Suppl. Fig. 3).
Fig. 11.
Mean normalized relative quantity (MNRQ) of Interleukin-10 (il10), interleukin-12 (il12), interferon-stimulated gene (isg15) in Atlantic salmon (Salmo salar) smolts (head kidney) for families 7, 8, 12, 15, 16, 17 at either 10 °C or 20 °C in fish exposed to Infectious Salmon Anemia (ISAv) at Time 1. Letters indicate significant difference between the two temperature points in gene expression (ANOVA, p ≤ 0.05; n = 8–12 fish per family per temperature at each time point).
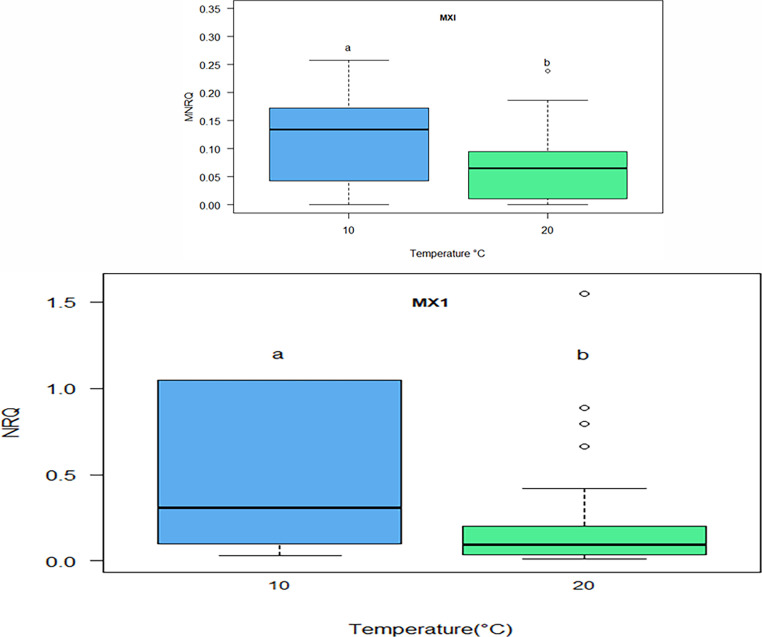
At Time 1, mx1 (p = 5.48×10−05) expression was significantly upregulated at 10 °C compared to 20 °C (Fig. 12a). At Time 2, expression of mx1 was still significantly upregulated at 10 °C compared to 20 °C (p = 2.5 × 10−06) (Fig. 12b). No significant difference in mx1 expression was found between families at Time 2 for mx1 (Suppl. Fig. 4). Other genes were only impacted by temperature at Time 1, such as viperin (vprn) which was significantly upregulated at 10 °C compared to 20 °C (p = 0.0043; Fig. 13a) and NK-lysin (nkl) which was significantly upregulated at 20 °C compared to 10 °C (p = 0.0049, Fig. 13b). In addition, il4/13a, which showed no difference in temperature at Time 1, had significantly higher expression at 10 °C compared to 20 °C, at Time 2 (p = 2.83e−10, Fig. 13c). Notably, il4/13a was also significantly upregulated in family 17 compared to family 8 (p = 0.000042), family 12 (p = 0.0001), family 15 (p = 0.00042), family 7 (p = 0.0000001), and family 16 (p = 0.0000013) (Suppl. Fig. 5).
Fig. 12.
Mean normalized relative quantity (MNRQ) of MX dynamin like GTPase 1(mx1) in Atlantic salmon (Salmo salar) smolts (head kidney) for families 7, 8, 12, 15, 16, 17 at either 10 °C or 20 °C in fish exposed to Infectious Salmon Anemia (ISAv) at Time 1 (a) and Time 2 (b). Letters indicate significant difference between the two temperature points in gene expression (p ≤ 0.05; n = 10–14 fish per family per temperature at each time point).
Fig. 13.
Mean normalized relative quantity (MNRQ) of Viperin (vprn, a), NK-lysin (nkl, b) and Interleukin-4/13A (il4/13a, c) in Atlantic salmon (Salmo salar) smolts (head kidney) for families 7, 8, 12, 15, 16, 17 at either 10 °C or 20 °C in fish exposed to Infectious Salmon Anemia (ISAv) at Time 1 (a,b) and time 2 (c). Letters indicate significant difference between the two temperatures in gene expression (ANOVA, p ≤ 0.05; n = 10–14 fish per family per temperature at each time point).
3.8. ISAv load
All samples analyzed for Time 1 and Time 2 were normalized to the measure of ISAv load to see if there was any correlation with viral load and vaccination status or the genes of interest. Again, no relationship between vaccination status and ISAv was observed, however, all genes except il12rb2 and c3 (and crp which was <0.01/NA) were positively correlated with viral load (Table 4). While mx1, gal9 and il10 showed the strongest positive correlation with viral load, the correlations were only moderately correlated (0.31, 0.30 and 0.37, respectively).
Table 4.
Pearson correlation coefficient for genes of interest with viral load in Atlantic salmon infected with ISAv.
|
Pearson Coefficient |
0.25 | 0.31 | 1.1E−01 | 0.10 | 9.7E−03 | 0.20 | −0.024 | 0.30 | 0.21 |
| Gene | NKL | CRP | IL10 | SAA5 | VPRN | RIG1 | TRIM16 | C3 | |
|---|---|---|---|---|---|---|---|---|---|
|
Pearson Coefficient |
2.0E−01 | N/A | 0.37 | 0.23 | 0.10 | 0.053 | 2.3E−01 | −0.037 |
4. Discussion
4.1. Percent survival against ISAv
Over the course of the study, Atlantic salmon maintained at 10 °C experienced a higher total mortality compared to fish at 20 °C. A prior study published by Falk et al. [11], showed that replication of ISA virus, in vitro in SHK-1 cells, was at it its maximum between 10 °C and 15 °C, and a decrease greater than 99% in viral replication was seen at 20 °C, while at 25 °C, no viral replication was seen at all. This [11], and prior unpublished work in our lab, was the reason for allowing cohabitation with exposed donors to occur prior to temperature increase. While donor fish were exposed i.p. and allowed to shed the virus for some time prior to the increase in temperature, by 288–308 ddpe (Time 1) at 20 °C only 35.4% of cohabitant fish exhibited measurable viral load in the kidney by qPCR, and zero at 667–727 ddpe (Time 2). In contrast, while Time 1 samples (302–312 dd) at 10 °C showed similar prevalence (30.7%), by Time 2 (640–661 dd) there were still 20% of samples positive for ISAv. Initiation of donor mortality began at 20 °C before 10 °C, acute mortality of cohabitants was higher at this temperature and the same prevalence of infection was observed at the first sampling point at each temperature, suggesting that the higher temperature did not prevent viral replication and/or abrogate the virulence/pathogenicity of the virus. As we did not measure ISAv in the water, we cannot directly determine whether high temperatures impact virus survival in seawater or whether the virus was more efficient at killing susceptible hosts at the higher temperature and burned out the source of infective material resulting in overall lower shedding as daily mortalities were removed. In the absence of this information, our data would suggest that either the virus replication or virulence increases at some temperature between 10 °C and 20 °C allowing for the observed mortality spike at the higher temperature, and then decreases drastically thereafter and/or the host response is also enhanced between these temperatures.
4.2. Condition factor and weight of sampled fish
Condition factors between 10 °C and 20 °C were not found to be significantly different. A study by Ignatz et al. [12], looked at transgenic triploid female Atlantic salmon's nutrient utilization as well as growth performance. They held fish at 10.5 °C, 13.5 °C, and 16.5 °C in freshwater, until the fish reached 1500 g in weight. They found that the fish grown at the highest temperature, 16.5 °C, reached the 1500 g weight faster than at the other temperatures, however, they were the least efficient at nutrient deposition. A study by Crouse et al. [13], consisted of smolts that were housed at both 12 °C and 14 °C in freshwater recirculating systems for 8 months. At 14 °C, the fish were significantly greater in length at both the 2-month and 4-month sampling period, while the weight was only significantly heavier at the 4-month sampling period. In our work, the fish were not significantly heavier at one temperature in comparison to the other. However, the time span of the challenge was limited. Interestingly, families 7 and 12 (lowest survival rate families) did show a significantly heavier weight at both time points compared to families with the best survival rate. This unfortunately may suggest some trade-off between growth and ISAv resistance or could relate to susceptibility to ISAv being higher in the smaller fish within these families resulting in their removal through mortality. Numbers of fish within different size ranges within a family however were too few to examine this further, but could be followed up in the future. When water temperature increases, each 10 °C increment results in an approximate 10 to 20% decrease in the amount of dissolved oxygen available [14]. With this, comes a decrease in normal metabolic functions, such as reproduction, eating, movement, and growth [14]. A study by Gamperl et al. [15] exposed Atlantic salmon to a temperature range of 12 °C to 20 °C with both a normal oxygen saturation as well as ∼70% oxygen saturation. The fish with a 70% oxygen saturation grew slower than the fish at normoxic conditions. At 18 °C and 19 °C, both oxygen saturations ended up having a decreased feed consumption. In our study, a temperature of 20 °C was associated with an 80% oxygen saturation, and a temperature of 10 °C was associated with a 100% oxygen saturation, so although the results did not reveal a significant difference in condition factor, it is possible that if the oxygen saturation was dropped to 70%, the outcomes may have changed. It is possible that due to the relatively short time period of the study the fish were not housed at a long enough time at the distinct temperatures to observe any significant changes in length and weight and subsequently condition factor.
4.3. Effectiveness in vaccination
There was no significant difference in survival between vaccinated and unvaccinated fish. Lauscher et al. [16] tested an inactivated ISAv vaccine, with varying quantities of the inactivated ISAv (4, 20 and 100%). In short, the study demonstrated that the highest levels of protection, as well as the production of ISAv -specific antibodies was found in the vaccine containing the highest antigen dose. As a commercial vaccine was used in this study, inappropriate dose would be unlikely unless this occurred through faulty administration. As the vaccination team is highly experienced in administration this also seems unlikely. Orthomyxovirus vaccines in mammals often target the hemagglutinin (HE) surface protein, which loses efficacy across different strains due to the high variability within the HE [17]. Specific antibodies (IgM or IgT) to ISAv were not evaluated here (i.e. no specific test has been developed), but this could be a useful future direction to determine whether they were indeed produced and/or to determine pathogen variant specificity. This vaccine, however was examined for and, induced specific IgM antibodies to Aeromonas salmonicida [one of the multivalent components and tested using ELISA], as previously reported [18]. This induction was 16.3-fold compared to PBS injected fish prior to exposure, and 113-fold compared to PBS injected fish [time matched] following infection (time point 2; average across temp), suggesting no issue with the vaccine or administration. Across the 17 GOIs examined surprisingly the only gene that had a significant upregulation between vaccinated and unvaccinated salmon in this study was galectin-9 (gal9). Although this gene has been significantly studied in mammals, its importance in fish is not as well examined. Galectin-9 has previously been shown to be impacted by co-infection of salmon with sea lice and ISAv and has been proposed as an influenza biomarker, impacting efficacy of influenza vaccines by activating the virus-specific CD8 T-cell response, and enhancing virus-specific antibody generation [19,20]. CD8 and other T-cell related transcripts were not impacted by vaccination in this study, again supporting a lack of vaccine induced responses/protection toward virus in these fish. Global transcriptomics, currently being investigated, and/or greater sample sizes within individual families may help identify more subtle impacts of this vaccine, however recent work by Fanuzzo et al. [21] examining Micro Forte VII impact on salmon kidney transcriptome also found no induction of the anti-viral genes studied (mx1, vpn, isg15, etc.). Further characterization of the impacts this multivalent vaccine has on the host responses against individual pathogens is warranted.
4.4. Immune gene expression
Head kidneys of infected Atlantic salmon at two temperatures were examined over time for immunological gene expression using both the MagPix multiplex and qRT-PCR. Due to the family background component of the study, it was not possible to have uninfected time matched controls as this would have added an additional 2000+ animals. Therefore, Time 0 baseline (pre-infection) samples were taken from representatives of each family, both vaccinated and unvaccinated as an uninfected comparator. When comparing this baseline gene expression against Time 1 post infection, induction of 10 immunological genes associated with anti-viral immunity, acute phase, and T-cell mediated responses and cytokine signaling were all induced, regardless of temperature. As previously observed, ISAv infection induced strong responses by each of these genes [7,22]. Retinoic acid-inducible gene I (RIG-I), a cytosolic pattern recognition receptor (PRR) has been shown in various studies to play an important role in inducing type I interferons, and in antiviral defense in other species [23]. Type I interferons limit the virus from spreading to nearby cells, promote an innate immune response, including inflammatory responses, and help activate the adaptive immune system. A study by Wang et al. [24], also showed that during the late stages of viral infection, nuclear RIG-I will start to promote apoptosis. Rehwinkel and Gack [23], noted that RIG-I can detect Orthomyxoviridae (Influenza, ISAv, etc.) infections. Due to the important role that RIG-I plays in the antiviral response; it is unsurprising that levels were significantly higher post-ISA exposure (Time 1). Trim25 plays a major role in the gene induction of RIG-I, so it follows that this gene was also significantly upregulated at Time 1 compared to Time 0, just as rigI was [23].
ISAv infection under the two temperature regimes, affected expression of these genes in two ways. Several interferon pathway and T-cell signaling genes were upregulated at 10 °C. ISG15 mRNA expression was also found to be significantly higher in the 10 °C exposed fish. When interferons (IFN)- α/β become stimulated, isg15 becomes induced [25]. Røkenes et al. [25] noted that Atlantic salmon produced isg15 in response to ISAv exposure. These authors proposed that a high amount of ISG-15 may not affect viral replication directly, but create an environment which is hostile for their reproduction. Based on the results of this study, it seems therefore possible that isg15 production got ramped up at 10 °C, due to the slight increase in viral load that was also seen at the lower temperature, in an attempt to decrease viral replication and its associated damage to the host.
The mx1 gene was also found to be significantly higher at 10 °C. MX proteins have been found to stop cytoplasmic RNA viruses from replicating in multiple fish species [26]. However, Fernaández-Trujillio et al. [27] noted that even though MX proteins act as antiviral proteins, there are different MX proteins, and each one comes with its unique antiviral specificity. An MX protein that works on one virus may not work on a different isolate of the same virus [27]. In their study with gilthead seabream these authors found that MX to virus ratio was an important factor in the antiviral response. Kim and Kim [26] showed that mx1 had no effect in endothelial progenitor cells (EPC) when exposed to VHSv, proposing that maybe MX1 is not effective at inhibiting viral replication with this type of virus. And prior work has shown that ISAv is a powerful inducer of type I interferon system genes in Atlantic salmon but not inhibited by this interferon system [28].
Along with mx1, vpn (Viperin; Virus Inhibitory Protein, Endoplasmic Reticulum-associated, Interferon inducible) was also found to be lower at 20 °C. Viperin can also play a role in stopping viral replication, as well as entry, through binding to the viral proteins [29]. Viperin is a gene that can be activated by both type I and type II interferons. During the final stage of the life cycle of the human influenza virus, where the virus bud is released from the plasma membrane [30], an enzyme, Farnesyl Diphosphate Synthase (FPPS) will interact with Viperin, resulting in an inhibition of FPPS, which, in turn disturbs the lipid-raft microdomains [30]. ISAv isolates have been shown to induce vpn expression in Atlantic salmon, however, based on these results it seems possible that vpn is temperature dependent, with the lower temperature leading to higher upregulation at Time 1.
As mentioned, macrophage/T-cell signaling genes were also impacted by temperature. Interleukin-10 displayed higher expression in the infected group at 10 °C compared to 20 °C. Interleukin-10 has been described as a Th2 cytokine, which can thus play a role in immunosuppression and is also able to inhibit synthesis of other cytokines in particular Th1 [31]. Interleukin-10 has been shown to help down-regulate inflammation to prevent uncontrolled immunopathology [32]. These authors showed mandarin fish splenocytes pre-incubated with IL-10 decreased the production of LPS-stimulated inflammatory genes il1β, il6, il8, and tnfα. Further studies in vertebrate monocyte-bacteria interactions have shown that, while pathological effects of excessive inflammatory cytokine production can be mediated by IL-10, it also has the ability to inhibit potentially protective cell-mediated immune responses in humans against Burkholderia pseudomallei and Atlantic salmon against Aeromonas salmonicida [33,34]. The crucial role IL-10 plays in homeostasis can also be hijacked by viruses, as numerous viruses produce IL-10 homologues giving them the ability to polarize antigen presenting cell (i.e. macrophage) subtypes, as discussed above in bacteria, to allow for survival and reduced adaptive immune recognition [35]. In our case, upregulation of IL-10 may have helped control the inflammation response after the initial onset of infection and protected against the more severe acute mortality observed at 20 °C, but may have led to improved survival for the virus and greater chronic mortality observed at this temperature. Similarly, il4/13a was lower at 20 °C. Interleukin-4/13A is one of three IL-4 and IL-13 cytokine paralogues [36] and also a part of the Th2 immune response which triggers B cell immunity [36]. Even though this gene was not expressed higher at 20 °C, where the salmon had better overall survival, one of the families that had the best survival, family 17, had significantly higher upregulation. Therefore, this may indicate that the gene is temperature dependant, may play a role in surviving ISAv and requires further examination.
At Time 1, il12rb2 was significantly upregulated at 20 °C compared to 10 °C, whereas at Time 2 it was significantly upregulated at 10 °C. Conversely IL-10, IL-12 and its receptors (such as the β2 receptor examined here) play a role in Th1 development and are considered proinflammatory cytokines [37]. Toll-like receptor ligation can alert the dendritic cells to produce IL-12, which can then be increased even more if pro-inflammatory signaling occurs secondary to that [37]. Interleukin-12 has multiple subunits which are all expressed and regulated differently through signaling different pathways. A study by Wang and Husain [37], showed it is possible that a competition may result in dendritic cells, B-cells, or macrophages for both secretion and pairing, when there are different subunits of IL-12. The expression of il12rb2 was one of the few genes exhibiting family-based differences over both time and temperature, and of greatest interest, similar to il4/13a and trim25, was the fact that resistant families 7, 16, 17 were the ones exhibiting significant differences in these genes.
NK-Lysin's are antimicrobial peptides (AMPs) that play an important role in the innate immune system [38]. The study by Acosta et al. [38], were able to show that nkl can induce IFN-γ. It also showed that it can recruit immune cells to help clear the infection through inflammation. Hence, nkl upregulation at 20 °C and at Time 1 is to be expected with relation to the other interferon related transcripts.
5. Conclusion
This study demonstrated that temperature plays a major role in infectious salmon anemia progression and the ability of the virus to invade the Atlantic salmon's immune system. The ability of the salmon immune system to upregulate the necessary genes to defend against the virus, and the virus's difficulty to replicate at 20 °C seem to mutually play a role in overall survival at the higher temperature. At 10 °C, a higher viral load was present in the kidney, along with a higher overall mortality. The innate antiviral response was higher at this temperature, as demonstrated by genes such as mx1, isg15, and vprn, as well as the Th2 response, demonstrated by il10 and il4/13a. The lower response seen at 20 °C may have been turned off due to quicker clearance of the virus, or it may have been inhibited due to stress on the fish initially at the higher temperature, resulting in the greater acute mortality. As fish die and are removed quicker there is less viral shedding into the tank and likely reducing the length an anti-viral response must be maintained. The Th1 and antigen presenting cells response was initially greater at 20 °C, as seen in il12rb2 and nkl, which may therefore play a role in viral clearance, as the viral load was lower at this temperature, as well as the overall mortality. Activation of NK cells is an important process in RNA viral clearance (i.e. influenza and flaviviruses) in humans, and similarly is mediated by induction of IL-12 among other cytokines [39]. It is also possible that at 20 °C, the virus itself may not be as easily transmitted due to the obstacle in replicating. Significant family-based survival differences could also be linked with some molecular markers. Similar to Gervais et al. [7], where upregulation of trim25 was associated with resistance to ISAv, trim25 differences were exhibited within the families with highest survival and one of the most resistant families (family 17) also showed significant differences with other families with respect to il4/13a and il12rb2 over time. It is evident that family-based resistance to ISAv exists, but further work is required to identify appropriate markers for selection and enhancing ISAv vaccines must also be considered.
Author contributions
Carrying out the investigation, qPCR methodology, formal analysis, validation, visualization, original draft writing, software usage was LG; Conceptualization, Funding acquisition, project administration, supervision, writing – review & editing, was carried out by MDF, AFG. Contributing to additional laboratory work, formal analysis, writing (review & editing), MagPix and qPCR methods was SLP, SKW, WCC and DM.
Supplemental Figures
Supplemental Figure 1: Temperature profile for the 38 tanks from acclimation the day Atlantic salmon (Salmo salar) arrived at the Atlantic Veterinary College to adjusted temperatures needed for the study (10 °C or 20 °C) up until Time 2 for the 20 °C tanks.
Supplemental Figure 2: Cumulative daily percent survival at 10 °C (a) and 20 °C (b) for 20 families of Atlantic salmon infected with ISAv. (c-n) Individual family survival rate sin vaccinated and PBS-injected fish at both temperatures.
Supplemental Figure 3: Expression relative to reference (NRQ) of interleukin-12 (il12) at Time 2 in 6 families of Atlantic salmon (Salmo salar) smolts (head kidney) maintained at 10 °C or 20 °C and exposed to Infectious Salmon Anemia (ISAv). Letters indicate significant difference in gene expression (ANOVA, p ≤ 0.05).
Supplemental Figure 4: Expression relative to reference (NRQ) of mx1 at Time 2 in 6 families of Atlantic salmon (Salmo salar) smolts (head kidney at either 10 °C or 20 °C exposed to Infectious Salmon Anemia (ISAv). Letters indicate significant difference between the two temperature points in gene expression (ANOVA, p ≤ 0.05).
Supplemental Figure 5: Expression relative to reference (NRQ) of Interleukin-4/13A (il4/13a) at Time 2 in 6 genetic families in Atlantic salmon (Salmo salar) smolts (head kidney) maintained at either 10 °C or 20 °C and exposed to Infectious Salmon Anemia (ISAv). Letters indicate significant difference between families in gene expression (ANOVA, p ≤ 0.05).
Declaration of Competing Interest
Dr. Garber works with Huntsman Marine Science center and is in charge of their breeding program.
Acknowledgements
This work was possible through funding from the Atlantic Canada Opportunities Agency (ACOA) – Atlantic Innovation Fund (Grant – MICCSA), as such access to any additional raw data not supplied in the supplemental material can be provided upon request; a graduate research stipend from AVC-UPEI; Innovation PEI; Huntsman Marine Science center; staff within the AVC - Aquatic Animal Facilities, Nelly Gagne, DFO, and Jonathon Perreira.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.fsirep.2023.100099.
Appendix. Supplementary materials
Data availability
Data will be made available on request.
References
- 1.Gagne N., Leblanc F. Overview of infectious salmon anemia virus (ISAV) in Atlantic Canada and the first report of an ISAV North American-HPR-0 subtype. J. Fish Dis. 2017;41 doi: 10.1111/jfd.12670. [DOI] [PubMed] [Google Scholar]
- 2.Office International des Epizooties, Manual of Diagnostic Tests for Aquatic Animals (2019) 1–16.
- 3.Lyngstad T.M., Jansen T.M., Sindre H., Jonassen C.M., Hjortaas M.J., Johnsen S., Brun E. Epidemiological investigation of infectious salmon anaemia (ISA) outbreaks in Norway 2003-2005. Prev. Vet. Med. 2008:213–217. doi: 10.1016/j.prevetmed.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 4.Andresen A.M.S., Boudinot P., Gjøen T. Kinetics of transcriptional response against poly (I:C) and infectious salmon anemia virus (ISAV) in Atlantic salmon kidney (ASK) cell line. Dev. Comp. Immunol. 2020;110:1–16. doi: 10.1016/j.dci.2020.103716. [DOI] [PubMed] [Google Scholar]
- 5.Laffoley D., Baxter J.M. IUCN; Gland, Switzerland: 2016. Explaining Ocean Warming: Causes, Scale, Effects and Consequences. Full report; p. 456. editors. [Google Scholar]
- 6.Madaro A., Folkedal O., Maiolo S., Alvanopoulopu M., Olsen R.E. Effects of acclimation temperature on cortisol and oxygen consumption in Atlantic salmon (Salmo salar) post-smolt exposed to acute stress. Aquacult. 2018;497:331–335. [Google Scholar]
- 7.Gervais O., Barria A., Papadopoulou A., Gratacap R.L., Hillestad B., Tinch A.E., Martin S.A.M., Robledo D., Houston R.D. Exploring genetic resistance to infectious salmon anemia virus in Atlantic salmon by genome-wide association and RNA sequencing. BMC Gen. 2021;22:345. doi: 10.1186/s12864-021-07671-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ritchie R.J., McDonald J.T., Glebe B., Young-Lai W., Johnsen E., Gagné N. Comparative virulence of Infectious salmon anemia virus isolates in Atlantic Salmon, Salmo Salar L. J. Fish Dis. 2009;32:157–171. doi: 10.1111/j.1365-2761.2008.00973.x. [DOI] [PubMed] [Google Scholar]
- 9.Chomczynski P., Sacchi N. Single-step method of RNA isolation by acid guanidium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 1987;162(1):156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 10.Snow M., McBeath A.J.A., Doig J.B., Kerr R., Cunningham C.O., Nylund A., Devold M. Development, application and validation of a Taqman real-time RT-PCR assay for the detection of infectious salmon anaemia virus (ISAV) in Atlantic salmon (Salmo salar) Dev. Biol. 2006;126:133–145. [PubMed] [Google Scholar]
- 11.Falk K., Namork E., Rimstad E., Mjaaland S., Dannevig B.H. Characterization of infectious salmon anemia virus, an orthomyxo-like virus isolated from Atlantic salmon (Salmo salar L.) ASM. 1997;71(12):9016–9023. doi: 10.1128/jvi.71.12.9016-9023.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ignatz E.H., Dumas A., Benfey T.J., Hori T.S., Braden L.M., Runighan C.D., Rise M.K., Westcott J.D. Growth performance and nutrient utilization of growth hormone transgenic female triploid Atlantic salmon (Salmo salar) reared at three temperatures in a land-based freshwater recirculating aquaculture system (RAS) Aquacult. 2020:519. [Google Scholar]
- 13.Crouse C., Davidson J., Good C. C. The effects of two water temperature regimes on Atlantic salmon (Salmo salar) growth performance and maturation in freshwater recirculating aquaculture systems. Aquacult. 2022:553. [Google Scholar]
- 14.Farrell A.P., Richards J.R. Chapter 11 defining hypoxia: an integrative synthesis of the responses of fish to hypoxia. Fish Physiol. 2009;27:487–503. [Google Scholar]
- 15.Gamperl A.K., Ajiboye O.O., Zanuzzo F.S., Sandrelli R.M., Peroni E.F.C., Beemelmanns A. The impacts of increasing temperature and moderate hypoxia on the production characteristics, cardiac morphology and haematology of Atlantic salmon (Salmo salar) Aquacult. 2020:519. [Google Scholar]
- 16.Lauscher A., Krossøy B., Frost P., Grove S., König M., Bohlin J., Falk K., Austbø L., Rimstad E. Immune responses in Atlantic salmon (Salmo salar) following protective vaccination against Infectious salmon anemia (ISA) and subsequent ISA virus infection. Vaccine. 2011;29:6392–6401. doi: 10.1016/j.vaccine.2011.04.074. [DOI] [PubMed] [Google Scholar]
- 17.McMillan C.L.D., Young P.R., Watterson D., Chappell K.J. The next generation of influenza vaccines: towards a universal solution. Vaccines (Basel) 2021;9(1) doi: 10.3390/vaccines9010026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Braden L.M., Whyte S.K., Brown A.B.J., Van Iderstine C., Letendre C., Groman D., Lewis J., Purcell S.L., Hori T., Fast M.D. Vaccine-induced protection against furunculosis involves pre-emptive priming of humoral immunity in Arctic Charr. Front. Immunol. 2019;(10):4. doi: 10.3389/fimmu.2019.00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Y., Ke F., Ma J., Zhou S. A tandem-repeat galectin-9 involved in immune response of yellow catfish, Pelteobagrus fulvidraco, against Aeromonas hydrophilia. Fish Shellfish Immunol. 2016;51:152–160. doi: 10.1016/j.fsi.2016.02.018. [DOI] [PubMed] [Google Scholar]
- 20.Barker S.E., Bricknell I.R., Covello J., Purcell S., Fast M., Wolters W., Bouchard A. Sea lice, Lepeophtheirus salmonis (Krøyer 1837), infected Atlantic salmon (Salmo salar L.) are more susceptible to infectious salmon anemia virus. PLoS ONE. 2019;14(1) doi: 10.1371/journal.pone.0209178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fanuzzo F.S., Beemelmanns A., Hall J.R., Rise M.L., Gamperl A.K. The innate immune response of Atlantic salmon (Salmo salar) is not negatively affected by high temperature and moderate hypoxia. Front. Immunol. 2020 doi: 10.3389/fimmu.2020.01009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Workenhe S.T., Hori T.S., Rise M.K., Kibenge M.J.T., Kibenge F.S.B. Infectious salmon anaemia virus (ISAV) isolates induce distinct gene expression responses in the Atlantic salmon (Salmo salar) macrophage/dendritic-like cell line TO, using genomic techniques. Mol. Immunol. 2009;46:2955–2974. doi: 10.1016/j.molimm.2009.06.015. [DOI] [PubMed] [Google Scholar]
- 23.Rehwinkel J., Gack M.U. RIG-I-like receptors: their regulation and roles in RNAa sensing. Nat. Rev. Immunol. 2020;20:537–551. doi: 10.1038/s41577-020-0288-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang C., Zhour W., Liu Y., Xu Y., Zhang X., Jiang C., Minghing J., Cao X. Nuclear transloation of RIG-I promotes cellular apoptosis. J. Autoimmun. 2022:130. doi: 10.1016/j.jaut.2022.102840. [DOI] [PubMed] [Google Scholar]
- 25.Røkenes T.P., Larsen R., Robertson B. Atlantic salmon ISG15: expression and conjugation to cellular proteins in response to interferon, double-stranded RNA and virus infections. Mol. Immunol. 2006;33:950–959. doi: 10.1016/j.molimm.2006.03.016. [DOI] [PubMed] [Google Scholar]
- 26.Kim M.S., Kim K.H. Effect of CRISPR/Cas9-mediated knockout of either Mx1 or ISG15 gene in EPC cells on resistance against VHSV infection. Fish Shellfish Immunol. 2019;93:1041–1046. doi: 10.1016/j.fsi.2019.08.058. [DOI] [PubMed] [Google Scholar]
- 27.Fernández-Trujillo M.A., García-Rosado E., Alonso M.C., Castro D., Álvarez M.C., Béjar J. Mx1, Mx2 and Mx3 proteins from the gilthead seabream (Sparus aurata) show in vitro antiviral activity against RNA and DNA viruses. Mol. Immunol. 2013;56:630–636. doi: 10.1016/j.molimm.2013.06.018. [DOI] [PubMed] [Google Scholar]
- 28.Kileng O., Brundtland M.I., Robertsen B. Infectious salmon anemia virus is a powerful inducer of key genes of the type I interferon system in Atlantic salmon, but is not inhibited by interferon. Fish Shellfish Immunol. 2007;23:378–389. doi: 10.1016/j.fsi.2006.11.011. [DOI] [PubMed] [Google Scholar]
- 29.Madushani K.P., Shanaka K.A.S.N., Yang H., Lim C., Jeong T., Tharuka M.D.N., Lee J. Molecular characterization, expression profile, and antiviral activity of redlip mullet (Liza haematocheila) viperin. Comp. Biochem. Physiol. B. 2022;258 doi: 10.1016/j.cbpb.2021.110699. [DOI] [PubMed] [Google Scholar]
- 30.Wang X., Hinson E.R., Cresswell P. The interferon-inducible protein viperin inhibits influenza virus release by pertubing lipid rafts. Cell Host Microbe. 2007;2(2):96–105. doi: 10.1016/j.chom.2007.06.009. [DOI] [PubMed] [Google Scholar]
- 31.Ingerslev H.-.C., Rønneseth A., Pettersen E.F., Werfeland H.I. Differential expression of immune genes in Atlantic salmon (Salmo salar L). Challenged intraperitoneally or by cohabitation with IPNV. Scand. J. Immunol. 2009;69(20):90–98. doi: 10.1111/j.1365-3083.2008.02201.x. [DOI] [PubMed] [Google Scholar]
- 32.Huo H.J., Chen S.N., Li L., Nie P. Functional characterization of IL-10 and its receptor subunits in a perciform fish, the mandarin fish, Siniperca chuatsi. Dev. Comp. Immunol. 2019;97:64–75. doi: 10.1016/j.dci.2019.03.017. [DOI] [PubMed] [Google Scholar]
- 33.Kessler B., Rinchai D., Kewcharoenwong C., Nithichanon A., Biggart R., Hawrylowicz C.M., Bancroft G.J., Lertememongkolchai G. Interleukin 10 inhibits pro-inflammatory cytokine responses and killing of Burkholderia pseudomallei. Sci. Rep. 2017:7L42781. doi: 10.1038/srep42791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fast M.D., Tse B., Boyd J.M., Johnson S.C. Mutations in the Aeromonas salmonicida subsp. Salmonicida type III secretion system affect Atlantic salmon leucocyte activation and downstream immune responses. Fish Shellfish Immunol. 2009;27:721–728. doi: 10.1016/j.fsi.2009.09.009. [DOI] [PubMed] [Google Scholar]
- 35.Nqash A., Stuart G., Kemp R., Wise L. Parapoxvirus interleukin-10 homologues vary in their receptor binding, anti-inflammatory, and stimulatory activities. Pathogens. 2022;11(507) doi: 10.3390/pathogens11050507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sequeida A., Castillo A., Cordero N., Wong V., Montero R., Vergara C., Valenzuela B., Vargas D., Valdés N., Morales J., Tello M., Sandino A.M., Maisey K., Imarai M. The Atlantic salmon interleukin 4/13 receptor family: structure, tissue distribution and modulation of gene expression. Fish Shellfish Immunol. 2020;98:773–787. doi: 10.1016/j.fsi.2019.11.030. [DOI] [PubMed] [Google Scholar]
- 37.Wang T., Husain M. The expanding repertoire of the IL-12 cytokine family. In teleost fish: identification of three paralogues each of the p35 and p40 genes in salmonids, and comparative analysis of their expression and modulation in Atlantic salmon Salmo salar. Dev. Comp. Immunol. 2014;46:194–207. doi: 10.1016/j.dci.2014.04.008. [DOI] [PubMed] [Google Scholar]
- 38.Acosta J., Roa F., González-Chavarría I., Astuya A., Maura R., Montesino R., Muñoz C., Camacho F., Saavedra P., A Valenzuela, Sánchez O., Toledo J.R. In vitro immunomodulatory activities of peptides derived from Salmo salar NK-lysin and cathelicidin in fish cells. Fish Shellfish Immmunol. 2019;88:587–594. doi: 10.1016/j.fsi.2019.03.034. [DOI] [PubMed] [Google Scholar]
- 39.Björkström N., Strunz B., Björkström H.Ljunggren. Natural killer cells in anti-viral immunity. Nature Rev. Immunol. 2021 doi: 10.1038/s41577-021-00558-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.