Abstract
Purpose
To develop a deep learning approach that enables ultra-low-dose, 1% of the standard clinical dosage (3 MBq/kg), ultrafast whole-body PET reconstruction in cancer imaging.
Materials and Methods
In this Health Insurance Portability and Accountability Act–compliant study, serial fluorine 18–labeled fluorodeoxyglucose PET/MRI scans of pediatric patients with lymphoma were retrospectively collected from two cross-continental medical centers between July 2015 and March 2020. Global similarity between baseline and follow-up scans was used to develop Masked-LMCTrans, a longitudinal multimodality coattentional convolutional neural network (CNN) transformer that provides interaction and joint reasoning between serial PET/MRI scans from the same patient. Image quality of the reconstructed ultra-low-dose PET was evaluated in comparison with a simulated standard 1% PET image. The performance of Masked-LMCTrans was compared with that of CNNs with pure convolution operations (classic U-Net family), and the effect of different CNN encoders on feature representation was assessed. Statistical differences in the structural similarity index measure (SSIM), peak signal-to-noise ratio (PSNR), and visual information fidelity (VIF) were assessed by two-sample testing with the Wilcoxon signed rank t test.
Results
The study included 21 patients (mean age, 15 years ± 7 [SD]; 12 female) in the primary cohort and 10 patients (mean age, 13 years ± 4; six female) in the external test cohort. Masked-LMCTrans–reconstructed follow-up PET images demonstrated significantly less noise and more detailed structure compared with simulated 1% extremely ultra-low-dose PET images. SSIM, PSNR, and VIF were significantly higher for Masked-LMCTrans–reconstructed PET (P < .001), with improvements of 15.8%, 23.4%, and 186%, respectively.
Conclusion
Masked-LMCTrans achieved high image quality reconstruction of 1% low-dose whole-body PET images.
Keywords: Pediatrics, PET, Convolutional Neural Network (CNN), Dose Reduction
Supplemental material is available for this article.
© RSNA, 2023
Keywords: Pediatrics, PET, Convolutional Neural Network (CNN), Dose Reduction
Summary
The authors developed a longitudinal multimodality coattentional convolutional neural network transformer (Masked-LMCTrans) for ultra-low-dose (1%) and ultrafast reconstruction of whole-body PET/MRI in pediatric patients with lymphoma.
Key Points
■ Masked-LMCTrans–reconstructed PET images demonstrated less noise and more detailed structure compared with simulated 1% extremely ultra-low-dose PET, demonstrating significant improvements (P < .001) of 15.8% in the structural similarity index measure, 23.4% in peak signal-to-noise ratio, and 186% in visual information fidelity.
■ Masked-LMCTrans enables longitudinal radiologic image reconstruction by jointly reasoning multiserial images, offering a timely new perspective and pathway for general dosage reductions within and beyond PET.
Introduction
PET images are obtained by injecting patients with a radiopharmaceutical, such as fluorine 18 (18F) fluorodeoxyglucose (FDG), and thus inherently require radiation exposure (1,2). According to the “as low as reasonably achievable” (ie, ALARA) concept, decreasing the injected radiotracer dose and reducing image data acquisition times are highly desirable advancements in children with cancer. Because image quality is proportional to the number of coincidence events in the PET detector as a result of radiopharmaceutical positron annihilation (3), both actions degrade diagnostic image quality and have been limited in clinical practice.
To address this challenge, convolutional neural networks (CNNs) have been developed, which can augment high-quality PET images from ultrafast or ultra-low-dose input images (3,4). Previous studies demonstrating the capabilities of CNNs to enhance PET images have two major limitations: (a) They focus on a single anatomic region and (b) reduction in radiotracer dose or acquisition time is limited. Most previous works are confined to brain PET reconstruction (5–9), whereas whole-body reconstruction is a much more challenging task. Whole-body PET images have higher intrapatient uptake variation (notably, 18F-FDG radiotracer concentration is much higher in the brain and bladder than elsewhere), which can introduce difficulty in reconstructing images (10). In addition, the limited performance of classic CNN-based algorithms confines the relative radiotracer dose reduction for whole-body PET images to half (4,11), a quarter (3), or 6.25% (12) of the clinical standard dose. To the best of our knowledge, no studies have investigated 1% low-dose whole-body PET reconstruction.
The goal of this work is to develop a deep learning approach that allows for 1% extremely ultra-low-dose whole-body PET reconstruction. To this end, we used the information from longitudinal PET/MRI scans, a variable that has been largely ignored in previous studies. Indeed, a single patient usually undergoes multiple serial PET examinations over time for monitoring of disease progression (13). Tumor-bearing regions may be the only components in the field of view to vary substantially between examinations (14). Here, we argue that the similarity between baseline and follow-up PET/MRI scans for the same patient can be learned by the proposed model and leveraged to reduce 18F-FDG injection dose and scanning time. We proposed Masked-LMCTrans, a longitudinal multimodality coattentional CNN transformer, for 1% low-dose PET reconstruction. Masked-LMCTrans consists of two parallel streams for baseline and follow-up scan processing that communicate through the coattentional transformer layers. The coattentional transformer layers provide effective information exchange between the serial scans by using the global self-attention mechanism. This joint model also accommodates the unique processing needs of each input modality—PET and MRI—by taking advantage of the precise localization from the CNN encoder. This provides a novel approach to reducing dosage that in turn opens doors for reconstruction of longitudinal medical imaging studies.
Materials and Methods
PET/MRI Cohort
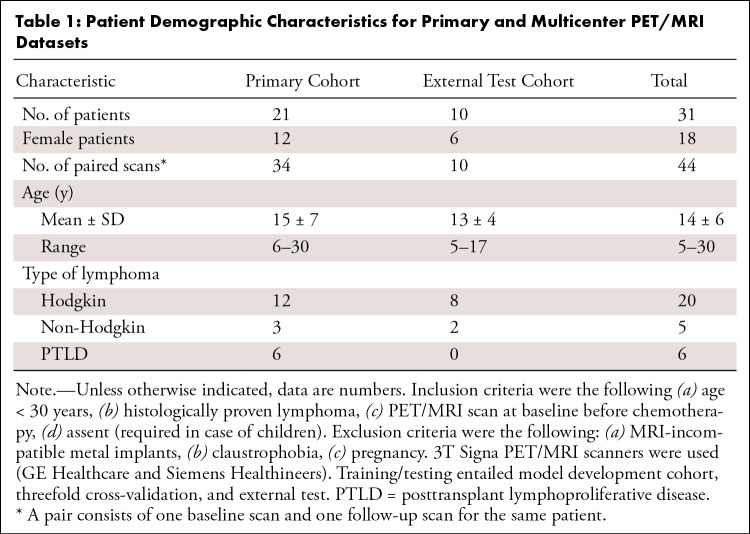
This retrospective, Health Insurance Portability and Accountability Act–compliant study included two whole-body PET/MRI cohorts (July 2015–March 2020): (a) a primary cohort and (b) a cross-continental validation cohort to assess the generalizability of our findings. We herein focus specifically on pediatric patients with lymphoma because these patients have an overall excellent prognosis, have a documented sensitivity to radiation effects, and would live long enough to experience long-term effects from radiation exposure (15). Notably, the proposed model for augmentation of ultra-low-dose PET imaging data is not confined to this scheme and can be generalized across diseases and populations given enough training data. Both participating medical centers obtained approval from their institutional review boards. In addition, the primary medical center obtained institutional review board approval to collect de-identified images in a centralized image registry, along with relevant clinical information (patient age, sex, and tumor type). Written informed consent was obtained from all adult patients and parents of pediatric patients. Children were also asked to give their assent.
For the two cohorts, we collected both pretreatment whole-body PET/MRI scans and posttreatment follow-up scans for each patient, resulting in 34 paired scans for the primary cohort (21 patients and 42 scans; a pair consists of one baseline scan and one follow-up scan) and 10 paired scans for the external test cohort (10 patients and 20 scans). Full-dose (3 MBq/kg) PET data were acquired in list mode, which helps detect coincidence events across the entire duration of the PET bed time (3 minutes 30 seconds). The reduced-dosage PET images were generated by unlisting the PET list-mode data and reconstructed based on the 1% of used counts, collected from the first 2 seconds of the scan, to simulate the ultra-low-dose scheme.
Masked-LMCTrans Approach
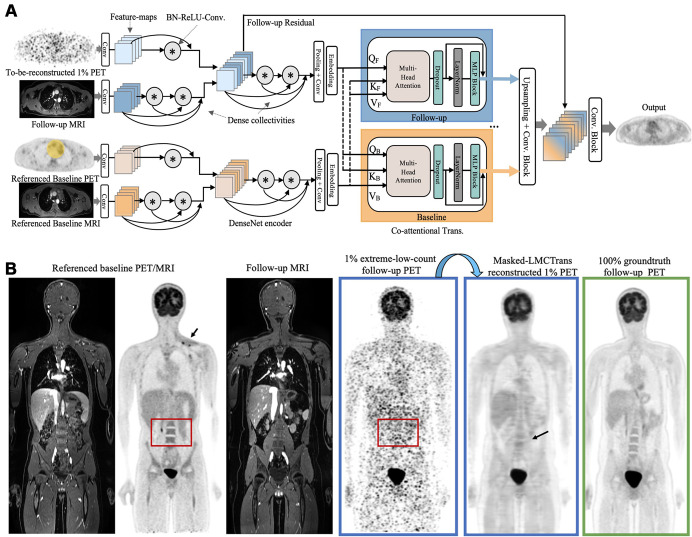
To reconstruct 1% extremely ultra-low-dose follow-up PET images by using both the acquired MRI and the referenced baseline PET/MRI scan, we had to effectively integrate four multiserial multimodality inputs. Inspired by ViLBERT's success in modeling visual-linguistic representations (16), we introduced the coattentional transformer block to provide interaction between longitudinal medical scans. Our model, known as longitudinal multimodality coattentional CNN transformer (LMCTrans), is illustrated in Figure 1A. Following the classic approach (12), the three-dimensional whole-body volume was inferenced in a section-by-section fashion and the predicted two-dimensional sections were stacked together to reconstruct the final three-dimensional prediction. To avoid introduction of erroneous upstaging for resolved tumors with low metabolic activity on the posttreatment follow-up PET scans, we masked out the tumor area of the baseline PET and thereby named the approach Masked-LMCTrans. Below we detail the essential building blocks of Masked-LMCTrans. Appendix S1 describes the loss function and training details.
Figure 1:
Proposed Masked-LMCTrans (multimodality coattentional convolutional neural network transformer) for 1% extremely ultra-low-dose PET/MRI reconstruction. (A) Framework of Masked-LMCTrans. The referenced baseline PET (with tumor area masked out as covered in the yellow mask) and MRI, along with the follow-up 1% PET/MRI scans, are fed into the model as combined inputs. The DenseNet feature encoder encodes PET and MRI separately before aggregation, with batch normalization, rectified linear unit, and 3 × 3 convolution (BN-ReLU-Conv) composite operations and dense collectivities. The coattentional transformer block fuses the information from the baseline and the follow-up (as indicated by the feature maps colored in orange and blue, respectively; the fused feature maps in the latter layers are mixed colored). The fusion is performed through baseline and follow-up information exchange by query, key, and value (denoted as Q, K, V). In this manner, Masked-LMCTrans reconstructs a 1% follow-up PET image, making use of the longitudinal similarity. (B) Representative posttreatment fluorine 18 fluorodeoxyglucose PET/MRI scan in a 14-year-old male patient with Hodgkin lymphoma. The contrast and structural details are significantly improved on Masked-LMCTrans–reconstructed PET as opposed to the simulated 1% PET. The red bounding box shows the spine anatomic structure, which is completely missing in the simulated 1% PET but successfully reconstructed by Masked-LMCTrans, with the help of the referenced baseline PET. The small tumor around the left supraclavicular region (arrow) in the baseline PET was resolved after treatment and was not shown on the reconstructed PET.
DenseNet encoder for feature extraction.—We applied DenseNet (17) block as our encoder of choice because it demonstrates strong representational power with convolution operations and dense collectivities (Fig 1A). The lth layer receives the “collective knowledge”—the feature maps from all preceding layers, x0, x1, …, xl-1—as inputs:
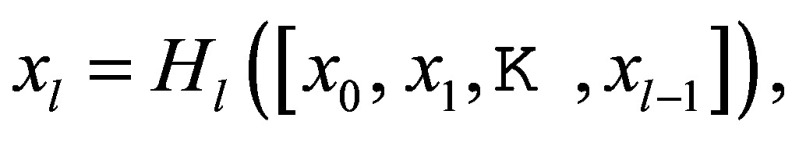 |
where [x0, x1, …,xl-1] refers to the concatenation of the feature maps produced in layer 0, 1, …,l-1. Hl denotes the composite function of three operations: batch normalization, rectified linear unit, and 3 × 3 convolutions (BN-ReLU-Conv). Such attributes introduce diversified features with all complexity levels and richer patterns that benefit the final PET reconstruction. Moreover, PET and MRI need different levels of feature encoding due to their inherent complexity and initial abstract nature of their input representations. Therefore, we set PET feature encoder simpler than MRI (one vs two BN-ReLU-Conv [Fig 1A]) before fusing them, as PET features are themselves already the output of the PET reconstruction network. We further examined the representational power of different CNN encoders.
Coattentional transformer layer for longitudinal fusion.—Although initial feature extractions are essential, equally important is how the baseline and follow-up PET/MRI scans relate to one another (eg, how to make use of the higher-quality baseline PET to benefit the reconstruction of the extremely ultra-low-dose follow-up PET). Thus, we used the coattentional transformer layer to capture the complex long-range temporal dependency. As opposed to CNNs, in which the receptive fields are gradually expanded through a series of convolution operations, the self-attention operations inherited in the transformer allow full coverage of the entire input space through token (ie, region) matching–in other words, each token is “matched” with all tokens within the input. Three vectors are calculated for each token: query, key, and value. Token matching computes a dot product score between the query (the token in consideration) and the key (the token being matched with) to weigh the value (of the token being matched with). This score determines how much focus to place on other regions of the input as we encode the region at this certain position. In this way, global self-attention, which refers to the dependencies between regions even when they are distant, is obtained. The coattentional transformer layer in Masked-LMCTrans computes query, key, and value matrices, as in a standard transformer block, given intermediate feature representations of baseline and follow-up PET/MRI scans. However, the keys and values from each sequence are passed as inputs to the other sequence's multiheaded attention block. Consequently, the attention block produces attention-pooled features for each sequence conditioned on the other, in effect performing baseline conditioned follow-up attention in the baseline stream and follow-up conditioned baseline attention in the follow-up stream. Thus, when the model processes each region in the to-be-reconstructed follow-up PET/MRI, the module allows it to look at other positions in the referenced baseline PET/MRI for clues that may improve encoding for this region. As a result, the correlation and similarity between baseline and follow-up streams is completely encapsulated.
Masked-LMCTrans.—Chemotherapy leads to a decrease in size and metabolic activity of lymphomas. On interim scans, 2–4 weeks after start of chemotherapy, a metabolic tumor response is defined by a decline in tumor metabolic activity at or below the metabolic activity of the liver as an internal standard of reference. Stable disease is defined as unchanged metabolic activity, and progressive disease is defined as increased metabolic activity or development of new tumor nodules. Acknowledging that variance of this nature should not be introduced in the follow-up PET reconstructions, we elected to mask out the prominent tumor regions in the baseline (pretreatment) PET scans before feeding them into the proposed model (Fig 1A inputs).
Computational Assessment
For evaluation, we adopted three quantitative metrics to measure the quality of the reconstructed PET images: structural similarity index measure (SSIM), peak signal-to-noise ratio (PSNR), and visual information fidelity (VIF). SSIM is the most widely used metric in imaging reconstruction. It is a combination of luminance, contrast, and structural comparison functions (18,19). Specifically, the SSIM score was derived by comparing the artificial intelligence (AI)–reconstructed PET and true standard-dose PET sequences and quantifying the similarity on a scale of 0 (no similarity) to 1 (perfect similarity). PSNR is most commonly used to measure the reconstruction quality of a lossy transformation (20). The higher the PSNR, the better the degraded image has been reconstructed to resemble the original image. SSIM and PSNR mainly focus on pixelwise similarity; thus, VIF was introduced. VIF uses natural statistics models to evaluate psychovisual features of the human visual system (21). The code for calculating the performance was written with Python. Standardized uptake values are the most widely used metric in clinical FDG PET oncologic imaging in assessing tumor glucose metabolism. The maximum standardized uptake value of the liver was measured by placing a three-dimensional volume of interest over the liver.
Statistical Analysis
Masked-LMCTrans was written in Python 3, with model training and testing performed using the PyTorch package (version 1.10). All statistical analyses were performed using R software (R Project for Statistical Computing). To assess the significance of the difference between two-sample tests, we used the Wilcoxon signed rank t test as implemented in R software. We used a predefined P value of less than .05 to indicate statistical significance. Performance results for the primary and external test cohorts are reported as mean, SD, and first (25%) and third (75%) quartiles of the data.
Data Availability
All of the code of the algorithm, the models, and the de-identified data can be made available from the authors upon reasonable request.
Results
Patient Characteristics
The primary cohort included 21 patients (mean age, 15 years ± 7 [SD]; 12 female, nine male). The external test cohort included 10 patients (mean age, 13 years ± 4; six female, four male) (Table 1).
Table 1:
Patient Demographic Characteristics for Primary and Multicenter PET/MRI Datasets
Comparison of AI-reconstructed and Ultra-Low-Dose PET Image Quality
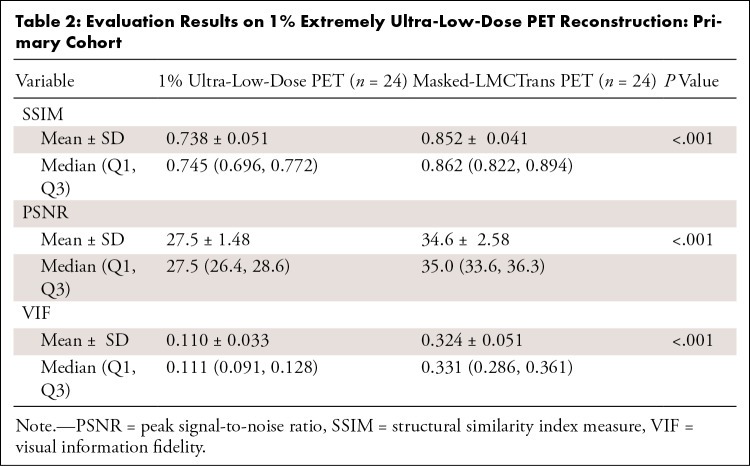
Masked-LMCTrans–reconstructed PET images demonstrated significantly less noise and more detailed structure compared with the simulated 1% extremely ultra-low-dose PET (Fig 1B, Table 2). Pairwise t tests (P < .001) indicated an improvement of 15.8% in SSIM, 23.4% in PSNR, and 186% in VIF.
Table 2:
Evaluation Results on 1% Extremely Ultra-Low-Dose PET Reconstruction: Primary Cohort
Benefit of Longitudinal PET/MRI Reconstruction
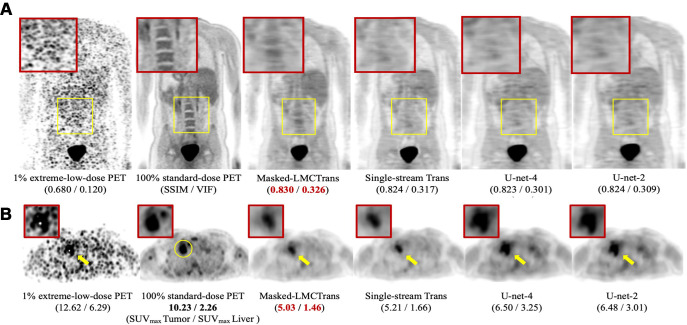
Compared with the single-stream model, Masked-LMCTrans–reconstructed PET better captured structural details because of its access to high-quality baseline PET/MRI reference data and capacity to grasp similarities between longitudinal scans (Figs 2, 3). On all of the 24 paired PET/MRI testing scans, Masked-LMCTrans consistently outperformed the single-stream baseline in SSIM and VIF (P < .05 for all patients).
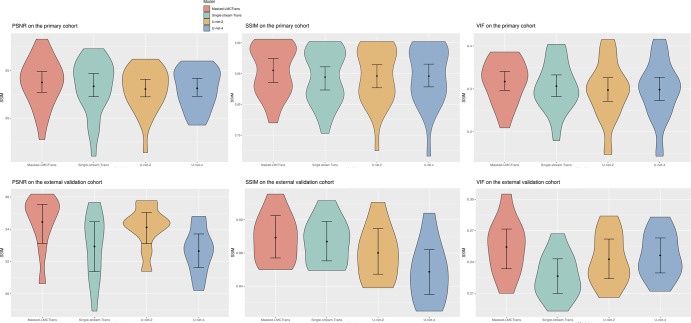
Figure 2:
Qualitative results of Masked-LMCTrans (multimodality coattentional convolutional neural network transformer) with comparison models: single-stream transformer, U-Net-4, and U-Net-2. (A) Representative posttreatment fluorine 18 (18F) fluorodeoxyglucose (FDG) PET/MRI scan in a 14-year-old male patient with Hodgkin lymphoma from the primary cohort. The spine structure is completely missed by the other models but recovered by Masked-LMCTrans, although not perfectly, whereas the 1% PET is extremely noisy. (B) Representative posttreatment 18F-FDG PET/MRI scan in a 19-year-old female patient with Hodgkin lymphoma from the primary cohort. Tumor-to-liver maximum standardized uptake value (SUVmax tumor/SUVmax liver) contrast is preserved in all reconstructions. The reconstructions for the unresolved tumor (as the yellow circle indicates in 100% standard-dose PET) from models of the transformer family (Masked-LMCTrans and single-stream transformer) resemble that of 100% PET in terms of structural fidelity. SSIM = structural similarity index measure, VIF = visual information fidelity.
Figure 3:
Model performance comparison on the primary cohort and external test cohort. The violin plots show the quantitative metrics in peak signal-to-noise ratio (PSNR), visual information fidelity (VIF), and structural similarity index measure (SSIM), along with 95% CIs. Masked-LMCTrans refers to multimodality coattentional convolutional neural network transformer, and single-stream trans refers to single-stream transformer. P values between Masked-LMCTrans and the other three models are less than .05 for SSIM and VIF metrics over both the primary and external cohorts, indicating statistically significant differences.
Advantage of Coattentional Transformer Layer
Despite the addition of baseline PET/MRI, U-Net-4 had similar values to U-Net 2 in SSIM, VIF, and PSNR in the primary cohort (Fig 2). Masked-LMCTrans—with the help of coattentional transformer layers—significantly outperformed U-Net models (Fig 3).
Generalizability in an Independent External Test Cohort
Image quality was significantly improved on Masked-LMCTrans–reconstructed PET as opposed to the original 1% extremely ultra-low-dose PET images by 5.15 dB in PSNR, 4.50% in SSIM, and 0.167 in VIF, demonstrating good model generalization across different institutions (Fig S1, Table S1). In addition, Masked-LMCTrans consistently outperformed the single-stream transformer baseline and U-Net models in the SSIM, demonstrating the model's generalizability and robustness to variations in scanner hardware and software.
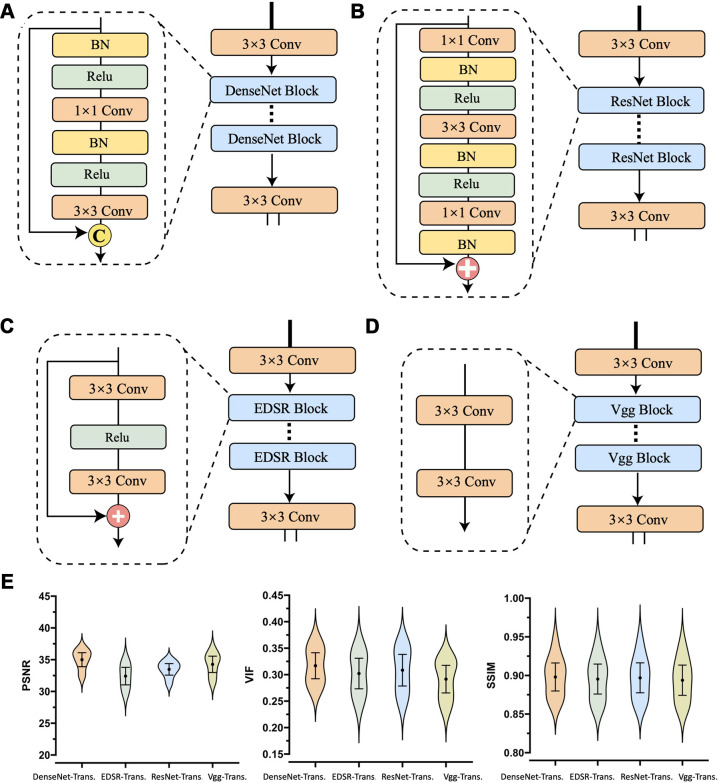
Assessment of Four CNN Feature Encoders
When moving from EDSR (enhanced deep super-resolution network; the worst performer) to DenseNet (the best performer) with 32 growth rate (the number of new feature maps added by each layer within a dense block) and three blocks, we observed a 2.5-dB increase in PSNR, indicating the superiority of DenseNet in modeling PET/MRI modalities (Fig 4).
Figure 4:
Convolutional neural network (CNN) encoder architectures and performance comparisons. The representational power of the four most commonly used CNN feature extractors were examined: DenseNet, ResNet, EDSR, and VGG. The CNN-encoder part of Masked-LMCTrans (multimodality coattentional convolutional neural network transformer) was replaced with the four structures, respectively, and the resultant models were named as “Encoder-Trans.” (A–D) The architecture and operation composition for DenseNet, ResNet, EDSR, and VGG encoder, respectively. (E) Violin plots show the model performances with 95% CIs. P values between DenseNet encoder and the other three encoders are less than .05 for peak signal-to-noise ratio (PSNR) and VIF (visual information fidelity) metrics, indicating statistical significance. BN = batch normalization, conv = convolution, EDSR = enhanced deep super-resolution network, ReLU = rectified linear unit, SSIM = structural similarity index measure, VGG = Visual Geometry Group.
Discussion
Our data showed that a longitudinal multimodality coattentional CNN transformer for PET reconstruction (Masked-LMCTrans) enabled high image quality reconstruction of 1% low-dose whole-body PET images. Our approach is unique in that it uses paired baseline PET/MRI scans to aid in the reconstruction of extremely ultra-low-dose follow-up PET scans along with the high-resolution structural MRI. Masked-LMCTrans accommodates the unique processing needs of each input modality by taking advantage of the strong representational power from the DenseNet encoder. Meanwhile, this joint model integrates complex long-term dependency for the longitudinal images by using the global self-attention mechanism introduced by the coattentional transformer block. Masked-LMCTrans effectively captures the dynamic information from longitudinal PET/MRI scans to reduce the image noise and recover the structural details of tumors and the liver. As a result, target tumors could be detected in the AI-reconstructed 1% extreme-low-dose PET images. From here, we can apply this algorithm for reconstruction of higher count images to determine which minimal 18F-FDG dose will lead to clinically acceptable image quality.
The model may enable reduced radiation dose and/or faster PET imaging, which could further affect patient care by offering safer scans and higher patient throughput.
We would like to motivate the exploration of longitudinal radiologic image reconstruction that may enable a new pathway for general dosage reduction in modalities beyond PET. To date, few efforts have been reported in this direction (22,23). The inability of the conventional methods to allow for jointly reasoning multiserial images hinders current exploration in this direction, even though it is common clinical practice for patients to undergo multiple serial scans. A framework that integrates complex global longitudinal dependency from sequential data with the precise localization from each input stream is crucial for jointly reasoning multiserial images. We anticipate that the proposed Masked-LMCTrans may serve as a common foundation for a wide array of radiology imaging reconstruction tasks, including those for CT and contrast-enhanced MRI.
Our study had some limitations. Lymphoma typically has robust baseline metabolic activity in pediatric patients. Further work is needed to assess whether the proposed approach can also perform well with low-grade, more subtle hypermetabolic lesions. In addition, we follow existing lesions that have been diagnosed on the baseline scan. It is necessary to see whether the model performs equally well in other tissues besides soft tissue and lymph nodes, such as bone, as well as in new lesions on the interim scan. Further evaluation of image quality and lesion detection by experienced radiologists is necessary to assess the clinical utility of the diagnostic images. Furthermore, the innate data demand of deep learning models and the lack of large-scale PET/MRI images represent a major bottleneck for further scale-up and transition of our research outcomes to practical use. Future prospective studies should validate our observations in larger patient populations, perhaps through a multicenter approach. Another limitation was that our study used simulated low-dose PET images; evidence of AI-enabled 1% low-dose reconstruction in true injected low-dose cases is still needed. Future prospective studies must show whether PET/MRI images reconstructed from digital ultra-low-dose PET images captured prospectively provide similar results. Finally, PET detector hardware and postprocessing techniques are constantly evolving. The comparative performance of standard PET scans enhanced with Masked-LMCTrans versus novel PET detector designs, such as the Explorer whole-body PET imaging technology (24), remains an area for future work. The recently developed photon-counting detector CT (25,26) uses new energy-resolving x-ray detectors that lead to higher contrast-to-noise ratio and optimized imaging. Large field-of-view PET scanners (27,28) have also substantially reduced the required PET data acquisition times by collecting PET information from a larger field of view than currently available with most conventional PET scanners. It is worth comparing these techniques. In addition, we could cross-train our algorithm to augment image data from these new scanners and further accelerate their image data acquisition time.
In conclusion, our approach holds promise for advancing the development of safe imaging tests, shortening examination durations, and increasing possibilities for frequent follow-up examinations.
Y.R.J.W. and L.Q. contributed equally to this work.
D.R. and H.E.D.L. are co–senior authors.
Current addresses: Department of Statistics & Actuarial Science, University of Hong Kong, Hong Kong.
Current addresses: Department of Biomedical Engineering, University of Virginia, Charlottesville, Va.
Supported by the National Cancer Institute of the U.S. National Institutes of Health (grant R01CA269231) and the Andrew McDonough B+ Foundation.
Disclosures of conflicts of interest: Y.R.J.W. Grant from the National Cancer Institute of the U.S. National Institutes of Health, grant number R01CA269231, and the Andrew McDonough B + Foundation. L.Q. No relevant relationships. N.D.S. National Institutes of Health Director's Early Independence Award DP5 (DP5OD031846), National Cancer Institute F99/K00 Predoctoral to Postdoctoral Fellow Transition Award (K00CA234954). X.L. No relevant relationships. J.W. No relevant relationships. K.E.H. No relevant relationships. A.J.T. No relevant relationships. S.G. No relevant relationships. X.X. No relevant relationships. A.P. No relevant relationships. D.R. Grant support from NCI; associate editor of Radiology: Artificial Intelligence. H.E.D.L. Grant from the National Cancer Institute of the US National Institutes of Health, grant number R01CA269231; 5R01AR054458-11, NIH/NIAMS Title: Monitoring Stem Cell Engraftment in Arthritic Joints with MR Imaging Dates: 8/01/2017-7/31/2023, Role: PI 10% effort; R01CA269231 NIH/NCI Title: Advanced Imaging Tools to Assess Cancer Therapeutics in Pediatric Patients, Dates: 2/1/2022-01/31/2027, Role: PI 10% effort; R01HD103638 NIH/NICHD Title: Theranostics for Pediatric Brain Cancer Dates: 4/15/2021-4/15/2026, Role: PI 20% effort; UG3CA268112 NIH/NCI Title: Cellular Senescence Network: New Imaging Tools for Arthritis Imaging; Dates: 10/1/2021-11/31/2026, Role: PI 10% effort; R21AR075863 NIH/NIAMS Title: Instant Stem Cell Labeling with a New Microfluidic Device Dates: 7/1/2019-6/30/2023 (one year NCE), Role: PI 5% effort; R21HD101129 Title: Imaging Chemotherapy-Induced Brain Damage in Pediatric Cancer Survivors Dates: 8/1/2020-7/31/2023 (one year NCE), Role: PI 5% effort; U24CA264298 NIH/NCI Title: Co-clinical research for imaging tumor associated macrophages Dates: 7/1/2021-6/30/2026, Role: Co-PI 10% effort; 5P30CA124435-10 NIH/NCI Title: Stanford Cancer Institute Support Grant (PI: Steven Artandi) Dates: 6/01/2017-5/31/2027, Role: Co-I 10% effort (H. Daldrup-Link, Codirector, Cancer Imaging & Early Detection Program); Cancer Research Institute Technology Impact Award Title: Mechanoporation creates new Biomarkers for Cancer Immunotherapy Dates: 7/1/2021-6/30/2023, Role: PI 1% effort; Sarcoma Foundation of America Title: Imaging response to CD47 mAb immunotherapy in pediatric patients with osteosarcoma Dates: 6/1/2020-5/31/2023, Role: PI 1% effort; R24OD019813-01 NIH Office of the Director Title: Expanding the Utility of Severe Combined ImmunoDeficient (SCID) pig models Dates: 5/1/2015-4/30/2025 Role: External Advisory Board Member (PI Christopher Tuggle); The ReMission Alliance Against Brain Tumors Title: Imaging CAR-T cells in Glioblastoma Dates: 9/1/2020-8/30/2023, Role: PI of subaward 5% effort; MegaPro Inc. Title: Evaluation of MegaPro Nanoparticles for MRI monitoring of Cancer Immunotherapy Dates: 7/1/2021-6/30/2023 Role: PI 1% effort; R01CA263500 NIH/NCI (PI C. Mackall) Title: Developing Safe and Effective GD2-CAR T Cell Therapy for Diffuse Midline Gliomas Dates: 7/1/2021–6/30/2026, Role: Co-Investigator 2% effort; Stanford Center for Artificial Intelligence in Medicine and Imaging (AIMI) Title: Standardized Therapy Response Assessments of Pediatric Cancers Dates: 11/1/2021–10/31/2023 Role: PI 2% effort; Patent: US6009342 Patent Assignee: University of California; Daldrup-Link, H.: Immunotherapy for cancer treatment using iron oxide nanoparticles; Patent: US20130344003 Patent Assignee: Stanford University; Daldrup-Link, H.: In vivo iron labeling of stem cells and tracking these labeled stem cells after their transplantation; Patent: US9579349, issued 2/28/17 Patent Assignee: Stanford University; Nejadnik H., Lenkov O., Daldrup-Link, H.: Compositions and methods for mesenchymal and/or chondrogenic differentiation of stem cells Patent: US20140271616 A Patent Assignee: Stanford University; Falconer R., Loadman P., Gill J.; Rao, J.; Daldrup H.E.: Activatable theranostic nanoparticles; Patent: WO 2015014756. Patent Assignee: Bradford University, UK and Stanford University, USA; Li K., Nejadnik H., Daldrup-Link, H.: Dual-modality Imaging Probe for Combined Localization and Apoptosis Detection of Stem Cells; Patent: US20180036435 Patent Assignee: Stanford University; Daldrup-Link, H., Mohanty S.: AntiWarburg Nanoparticles; Patent: WO/2018/217943 Patent Assignee: Stanford University; Co-Chair, Steering Committee, The NIH Common Fund's Cellular Senescence Network (SenNet) Program; Awards Committee, World Molecular Imaging Society (WMIS); MR Imaging Committee, Society for Pediatric Radiology (SPR) Diversity & Inclusion Committee, Society for Pediatric Radiology (SPR); Oncology Imaging Committee, Society for Pediatric Radiology (SPR); ad hoc reviewer, Nanotechnology (Nano) Study Section. NIH CSR (Center for Scientific Review); Ad Hoc Reviewer, ZRG1 SBIB-Q 03 Study Section; Biomedical Imaging & Bioengineering, Center for Scientific Reviews; reviewer for research grant applications for Emerson Collective Cancer Research Fund; reviewer for research grant applications for the Swiss Cancer Research Foundation; reviewer for the Florida Department of Health's Biomedical Research Program; reviewer for the Belgian Foundation Against Cancer; receipt of equipment, materials, drugs or other services from MegaPro Biomedical; Managing Director of Monasteria Press; associate editor for Radiology: Imaging Cancer; associate chair for Diversity, Department of Radiology, Stanford School of Medicine, Co-Program Director, Mentoring to AdVance womEN in Science (MAVENS) program, Stanford School of Medicine, Advisory committee member, Justice Diversity, Equity and Inclusion (JEDI) at the Stanford Cancer Institute (SCI).
Abbreviations:
- AI
- artificial intelligence
- BN-ReLU-Conv
- batch normalization, rectified linear unit, and 3 × 3 convolutions
- CNN
- convolutional neural network
- FDG
- fluorodeoxyglucose
- PSNR
- peak signal-to-noise ratio
- SSIM
- structural similarity index measure
- VIF
- visual information fidelity
References
- 1. Huang B , Law MWM , Khong PL . Whole-body PET/CT scanning: estimation of radiation dose and cancer risk . Radiology 2009. ; 251 ( 1 ): 166 – 174 . [DOI] [PubMed] [Google Scholar]
- 2. Leide-Svegborn S . Radiation exposure of patients and personnel from a PET/CT procedure with 18F-FDG . Radiat Prot Dosimetry 2010. ; 139 ( 1-3 ): 208 – 213 . [DOI] [PubMed] [Google Scholar]
- 3. Chaudhari AS , Mittra E , Davidzon GA , et al . Low-count whole-body PET with deep learning in a multicenter and externally validated study . NPJ Digit Med 2021. ; 4 ( 1 ): 127 . [Published correction appears in NPJ Digit Med 2021;4(1):139.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Theruvath AJ , Siedek F , Yerneni K , et al . Validation of deep learning-based augmentation for reduced 18F-FDG dose for PET/MRI in children and young adults with lymphoma . Radiol Artif Intell 2021. ; 3 ( 6 ): e200232 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Xiang L , Qiao Y , Nie D , An L , Wang Q , Shen D . Deep auto-context convolutional neural networks for standard-dose PET image estimation from low-dose PET/MRI . Neurocomputing 2017. ; 267 : 406 – 416 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Xu J , Gong E , Pauly J , Zaharchuk G . 200x Low-dose PET reconstruction using deep learning . arXiv preprint arXiv:1712.04119. https://arxiv.org/abs/1712.04119. Posted December 12, 2017. Accessed May 2022. [Google Scholar]
- 7. Schramm G , Rigie D , Vahle T , et al . Approximating anatomically-guided PET reconstruction in image space using a convolutional neural network . Neuroimage 2021. ; 224 : 117399 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chen KT , Toueg TN , Koran MEI , et al . True ultra-low-dose amyloid PET/MRI enhanced with deep learning for clinical interpretation . Eur J Nucl Med Mol Imaging 2021. ; 48 ( 8 ): 2416 – 2425 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chen KT , Schürer M , Ouyang J , et al . Generalization of deep learning models for ultra-low-count amyloid PET/MRI using transfer learning . Eur J Nucl Med Mol Imaging 2020. ; 47 ( 13 ): 2998 – 3007 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang T , Lei Y , Fu Y , et al . Machine learning in quantitative PET: a review of attenuation correction and low-count image reconstruction methods . Phys Med 2020. ; 76 : 294 – 306 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Whiteley W , Luk WK , Gregor J . DirectPET: full-size neural network PET reconstruction from sinogram data . J Med Imaging (Bellingham) 2020. ; 7 ( 3 ): 032503 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang YJ , Baratto L , Hawk KE , et al . Artificial intelligence enables whole-body positron emission tomography scans with minimal radiation exposure . Eur J Nucl Med Mol Imaging 2021. ; 48 ( 9 ): 2771 – 2781 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Unterrainer M , Eze C , Ilhan H , et al . Recent advances of PET imaging in clinical radiation oncology . Radiat Oncol 2020. ; 15 ( 1 ): 88 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ellis S , Reader AJ . Longitudinal multi-dataset PET image reconstruction . 2017 IEEE Nuclear Science Symposium and Medical Imaging Conference (NSS/MIC) 2017. ; 1 – 3 . [Google Scholar]
- 15. Brenner DJ , Doll R , Goodhead DT , et al . Cancer risks attributable to low doses of ionizing radiation: assessing what we really know . Proc Natl Acad Sci U S A 2003. ; 100 ( 24 ): 13761 – 13766 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lu J , Batra D , Parikh D , Lee S . ViLBERT: Pretraining Task-Agnostic Visiolinguistic Representations for Vision-and-Language Tasks . arXiv 1908.02265 [preprint] https://arxiv.org/abs/1908.02265. Posted August 6, 2019. Accessed May 2022. [Google Scholar]
- 17. Huang G , Liu Z , Van Der Maaten L , Weinberger KQ . Densely connected convolutional networks . Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition 2017. ; 4700 – 4708 . [Google Scholar]
- 18. Jayachandran Preetha C , Meredig H , Brugnara G , et al . Deep-learning-based synthesis of post-contrast T1-weighted MRI for tumour response assessment in neuro-oncology: a multicentre, retrospective cohort study . Lancet Digit Health 2021. ; 3 ( 12 ): e784 – e794 . [DOI] [PubMed] [Google Scholar]
- 19. Wang Z , Bovik AC , Sheikh HR , Simoncelli EP . Image quality assessment: from error visibility to structural similarity . IEEE Trans Image Process 2004. ; 13 ( 4 ): 600 – 612 . [DOI] [PubMed] [Google Scholar]
- 20. Horé A , Ziou D . Image Quality Metrics: PSNR vs SSIM . 2010 20th International Conference on Pattern Recognition 2010. ; 2366 – 2369 . [Google Scholar]
- 21. Sheikh HR , Bovik AC . Image information and visual quality . IEEE Trans Image Process 2006. ; 15 ( 2 ): 430 – 444 . [DOI] [PubMed] [Google Scholar]
- 22. Ellis S , Reader AJ . Synergistic longitudinal PET image reconstruction . 2016 IEEE Nuclear Science Symposium, Medical Imaging Conference and Room-Temperature Semiconductor Detector Workshop (NSS/MIC/RTSD) 2016. ; 1 – 3 . [Google Scholar]
- 23. Ellis S , Reader AJ . Simultaneous maximum a posteriori longitudinal PET image reconstruction . Phys Med Biol 2017. ; 62 ( 17 ): 6963 – 6979 . [DOI] [PubMed] [Google Scholar]
- 24. Badawi RD , Shi H , Hu P , et al . First human imaging studies with the EXPLORER total-body PET scanner . J Nucl Med 2019. ; 60 ( 3 ): 299 – 303 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Willemink MJ , Persson M , Pourmorteza A , Pelc NJ , Fleischmann D . Photon-counting CT: technical principles and clinical prospects . Radiology 2018. ; 289 ( 2 ): 293 – 312 . [DOI] [PubMed] [Google Scholar]
- 26. Flohr T , Petersilka M , Henning A , Ulzheimer S , Ferda J , Schmidt B . Photon-counting CT review . Phys Med 2020. ; 79 : 126 – 136 . [DOI] [PubMed] [Google Scholar]
- 27. van Sluis J , Boellaard R , Dierckx RAJO , Stormezand GN , Glaudemans AWJM , Noordzij W . Image quality and activity optimization in oncologic 18F-FDG PET using the digital biograph vision PET/CT system . J Nucl Med 2020. ; 61 ( 5 ): 764 – 771 . [DOI] [PubMed] [Google Scholar]
- 28. van Sluis J , van Snick JH , Brouwers AH , et al . EARL compliance and imaging optimisation on the Biograph Vision Quadra PET/CT using phantom and clinical data . Eur J Nucl Med Mol Imaging 2022. ; 49 ( 13 ): 4652 – 4660 . [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All of the code of the algorithm, the models, and the de-identified data can be made available from the authors upon reasonable request.