Abstract
Objectives
We analyzed the expression of inflammatory and antiviral genes in the nasopharynx of SARS-CoV-2 infected patients and their association with the severity of COVID-19 pneumonia.
Methods
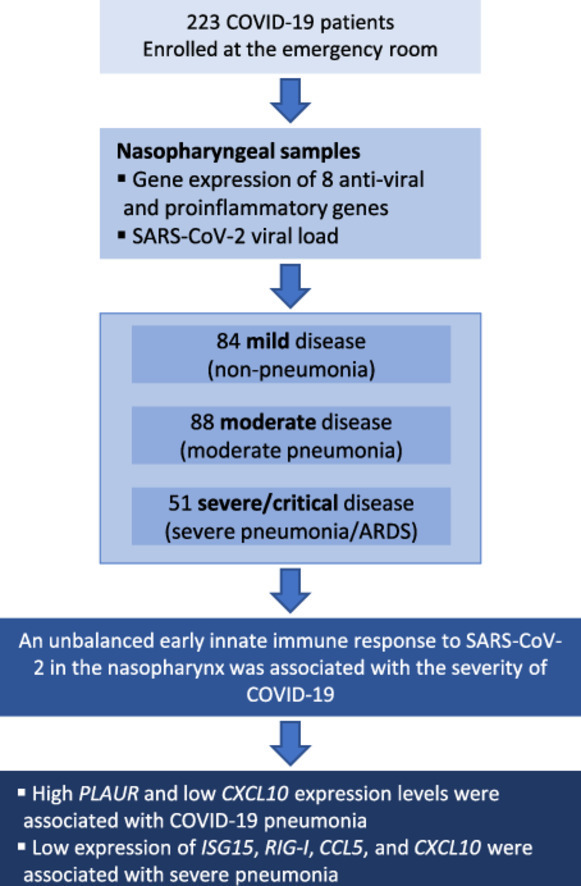
We conducted a cross-sectional study on 223 SARS-CoV-2 infected patients. Clinical data were collected from medical records, and nasopharyngeal samples were collected in the first 24 hours after admission to the emergency room. The gene expression of eight proinflammatory/antiviral genes (plasminogen activator urokinase receptor [PLAUR], interleukin [IL]-6, IL-8, interferon [IFN]-β, IFN-stimulated gene 15 [ISG15], retinoic acid-inducible gene I [RIG-I], C-C motif ligand 5 [CCL5], and chemokine C-X-C motif ligand 10 [CXCL10]) were quantified by real-time polymerase chain reaction. Outcome variables were: (i) pneumonia; (ii) severe pneumonia or acute respiratory distress syndrome. Statistical analysis was performed using multivariate logistic regression analyses.
Results
We enrolled 84 mild, 88 moderate, and 51 severe/critical cases. High expression of PLAUR (adjusted odds ratio [aOR] = 1.25; P = 0.032, risk factor) and low expression of CXCL10 (aOR = 0.89; P = 0.048, protective factor) were associated with pneumonia. Furthermore, lower values of ISG15 (aOR = 0.88, P = 0.021), RIG-I (aOR = 0.87, P = 0.034), CCL5 (aOR = 0.73, P <0.001), and CXCL10 (aOR = 0.84, P = 0.002) were risk factors for severe pneumonia/acute respiratory distress syndrome.
Conclusion
An unbalanced early innate immune response to SARS-CoV-2 in the nasopharynx, characterized by high expression of PLAUR and low expression of antiviral genes (ISG15 and RIG-I), and chemokines (CCL5 and CXCL10), was associated with COVID-19 severity.
Keywords: SARS-CoV-2, COVID-19, Pneumonia, Gene expression, Mucosal nasopharynx, Immune response
Graphical abstract
Introduction
COVID-19 follows SARS-CoV-2 infection and has caused high rates of morbidity and mortality worldwide [1]. The clinical presentation varies from mild illness to pneumonia, severe pneumonia that requires oxygen support, and critical disease with severe complications such as acute respiratory distress syndrome (ARDS) and septic shock [1]. However, these clinical presentations could be influenced by surveillance strategies, the use of therapeutics and other interventions, vaccination, and evolving variants [1].
COVID-19 has been associated with the onset of a dysregulated inflammatory response secondary to poorly controlled viral replication that produces immunopathogenic damage and contributes to the severity of the disease and the risk of death [2]. To improve our understanding of the causes of severe COVID-19 and to assist in the effective prevention and treatment of COVID-19, it is essential to discriminate protective host mechanisms that promote viral clearance and reduce disease from those that lead to severe and fatal outcomes. Most studies have reported anti-SARS-CoV-2 host responses in peripheral blood, but the systemic immune response can differ substantially from the local immune response in infected tissues, such as the upper respiratory tract (URT) [3].
SARS-CoV-2 infection begins in nasal epithelial cells of the URT, where viral RNAs act as pathogen-associated molecular patterns that are recognized by pattern recognition receptors (PRR), particularly the retinoic acid-inducible gene I (RIG-I) [4]. These receptors trigger intracellular signaling pathways and lead to the activation of multiple transcription factors, such as interferon regulatory factors 1, 3, and 7 (IRF), nuclear factor-κB (NF-κB), and the AP-1 activator protein [4]. These signaling pathways induce the expression of proinflammatory cytokines and chemokines, the recruitment of inflammatory myeloid cells, and the increased expression of interferon-stimulated antiviral genes (ISGs), resulting in an innate inflammatory and antiviral immune response [4]. A rapid and balanced innate immune response of the nasopharyngeal plays a central role in the defense against SARS-CoV-2 infections and helps control virus replication and immunopathology [5]. On the contrary, a reduced innate antiviral response (e.g., a reduced level of interferon [IFN]-I and III), along with an exacerbated inflammatory response and excessive cytokine production (high expression of interleukin [IL]-6, IL-8, and tumor necrosis factor-α), has been associated with severe COVID-19 [5], [6], [7].
To gain insight into the immune response at the initial site of SARS-CoV-2 infection, we analyzed the expression of inflammatory and antiviral genes in the URT (nasopharynx) of patients infected with SARS-CoV-2 and their association with the severity of COVID-19 pneumonia.
Methods
Study design and patients
We conducted a cross-sectional study on 223 patients infected with SARS-CoV-2 who were not vaccinated against COVID-19. Samples were collected from November 2020 to March 2021 in the emergency room of the Hospital Universitario Príncipe de Asturias (HUPA). The Ethics Committee (Ref.: EXPRES-INMUNE-COVID) approved this study and authorized the patients' informed consent waiver. The STrengthening the Reporting of OBservational studies in Epidemiology checklist was followed (Supplementary Table 1). All patients were over 18 years of age and had a positive real-time polymerase chain reaction (RT-PCR) test for SARS-CoV-2. According to Nextstrain (https://nextstrain.org/sars-cov-2/), the predominant variant on November 1, 2020, was 20E (EU1) [8], accounting for 88% of sequences in Spain. This variant was replaced during the study period by the 20I variant (Alpha, V1), which became the predominant one on March 31, 2021 (85% of sequences in Spain).
Outcome variable
The severity of pneumonia in the emergency room was used to stratify patients into three groups according to the World Health Organization guideline for the clinical management of COVID-19 patients [1]: (i) mild disease (mild infection without pneumonia), (ii) moderate disease (non-severe pneumonia), and (iii) severe or critical disease (severe pneumonia or ARDS). The outcome variables were: (i) the presence of pneumonia (either moderate, severe, or critical disease); (ii) the presence of severe pneumonia/ARDS (severe or critical disease).
Data and samples
Epidemiological, clinical, and analytical data were collected from medical records. Nasopharyngeal swab samples were collected within the first 24 hours after emergency admission in an inactivating transport medium (NEST Disposable Nasopharyngeal VTM Sampler kit, Wuxi NEST Biotechnology, Wuxi, China) and stored at -80°C. This medium also prevented the degradation of SARS-CoV-2 RNA and mucosal biomarkers. The microbiological diagnosis of SARS-CoV-2 infection was performed in these nasopharyngeal swab samples using an RT-PCR assay as previously described [7].
SARS-CoV-2 RT-PCR assay
Viral RNA was extracted using ELITe Ingenius (ELITechGroup, Puteaux, France) and MagCore HF16 (RBC bioscience, Taipei, Taiwan). Real-time PCR was performed according to the usual laboratory workflow using the GeneFinder COVID-19 Plus RealAmp Kit (Osang Healthcare Co.; detected genes: E, N, and RdRP) or Viasure SARS-CoV-2 Real Time PCR Detection Kit (CerTest Biotech S.L.; detected genes: ORF1ab and N). A sample was considered positive when all SARS-CoV-2 genes included in each RT-PCR assay were amplified.
Mucosal biomarkers RT-PCR assay
Total RNA from nasopharyngeal samples was extracted using the ReliaPrepTM RNA cell Miniprep System (Promega, Fitchburg, WI, USA) and reversely transcribed with the High-Capacity complementary DNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA). The expression of the selected genes (mucosal biomarkers) was quantified in the complementary DNA by RT-PCR using TaqMan Gene Expression Assays (Applied Biosystems, Foster City, CA, USA). TaqMan probes were used for the following cellular genes: Actin-β (ACTB; Hs99999903_m1), plasminogen activator urokinase receptor (PLAUR; Hs00182181_m1), (IL-6; Hs00985639_m1), (IL-8; Hs00174103_m1), interferon-β1 (IFN-B1; Hs01077958_s1), retinoic acid-inducible gene I (RIG-I; Hs00204833_m1), IFN-stimulated gene 15 (ISG15; Hs00192713_m1), chemokine C-X-C motif ligand 10 (CXCL10; Hs00171042_m1), and chemokine C-C motif ligand 5 (CCL5; Hs00982282_m1). PCRs were performed in triplicate in 48-well plates using a StepOne RT-PCR System thermal cycler (Applied Biosystems). Differential gene expression was determined using the comparative cycle threshold (ΔΔCt) method using ACTB as endogenous control and a mixture of samples from 100 SARS-CoV-2 positive individuals as a calibrator (reference sample). Gene expression was quantified in each sample relative to the calibrator and represented as Log2 fold-change to the median of the “non-pneumonia” group. The genes analyzed were chosen based on numerous reports involving them in the innate immune response and immunopathology to respiratory viral infections, including SARS-CoV-2 [5], [6], [7],9,10]. Those genes cover classical PRR (RIG-I), antiviral genes (IFN-β), interleukins (IL-6 and IL-8), chemokines (CCL5 and CXCL10), interferon-stimulated genes (ISG15), and PLAUR (which encode for uPAR, a biomarker of COVID-19 severity) [11]. SARS-CoV-2 viral load was assessed by RT-PCR of the SARS-CoV-2 nucleocapsid gene using the SYBR-Green reaction mix (Power-Up SYBR-Green Master MIX, Applied Biosystems). The primers used were ACTB (forward: 5’-CACCAACTGGGACGACAT-3’, reverse: 5’- ACAGCCTGGATAGCAACG-3’) as endogenous control and N (forward: 5’-GGGAGCCTTGAATACACCAAAA-3’, reverse: 5’-TGTAGCACGATTGCAGCATTG-3’) as a viral marker. With the same calibrator described previously, the relative quantification was performed using the comparative method of cycle threshold (ΔΔCt).
Statistical analysis
Statistical analysis was performed using Stata IC 17 (StataCorp, Texas, USA) and GraphPad Prism 8.0 (GraphPad Software, Inc., San Diego, CA, USA). The significance level was established at 0.05 (two-tailed).
The comparison between groups was performed using the Mann-Whitney U-test for continuous variables and Pearson´s chi-square test (χ2) or Fisher's exact test for categorical variables. Binary logistic analyses were used to evaluate the association between mucosal biomarkers and clinical outcome, providing the odds ratio (OR) and their 95% confidence intervals (CIs). Multivariate regression analyses were adjusted by the most significant covariables (age, gender, comorbidities (chronic heart disease, hypertension, chronic obstructive pulmonary disease, asthma, chronic kidney disease, obesity, diabetes, and dyslipidemia), and time from COVID-19 symptoms to sample collection), selected by a stepwise forward selection method (P in <0.05 and P out <0.10). The values of the mucosal biomarkers were log2-transformed (base-2 logarithms). The outcome variables and gene expression had no missing data. The clinical and epidemiological variables included in the adjusted regression models had less than 1% missing data and were not imputed.
We also performed hierarchical clustering heatmaps of group averages of gene expression using MetaboAnalyst 5.0 (https://www.metaboanalyst.ca/).
Results
Patient characteristics
The characteristics of patients with COVID-19 are shown in Table 1 . The patients were 61% men, and the median age was 60.5 years, which increased with the severity of COVID-19 pneumonia (P <0.001). No patient had received any vaccine against COVID-19 when the nasopharyngeal samples were obtained or during hospital admission. The main comorbidities were chronic heart disease, hypertension, chronic obstructive pulmonary disease, chronic kidney disease, obesity, diabetes, and dyslipidemia, which were much more frequent in patients with severe pneumonia/ARDS (P <0.05). Among peripheral blood biomarkers, ferritin, creatinine, lactate dehydrogenase, C-reactive protein, and glucose increased significantly with the severity of COVID-19 pneumonia (P <0.001).
Table 1.
Summary of characteristics of COVID-19 patients according to the pneumonia severity.
| Characteristic | Mild infection | Moderate pneumonia | Severe pneumonia/ acute respiratory distress syndrome | P-value |
|---|---|---|---|---|
| No. patients | 84 | 88 | 51 | |
| Age (years) | 50.2 (31.8-65.5) | 65.6 (54.4-75.5) | 72 (62.9-82.3) | <0.001 |
| Gender (male) | 43 (51.2%) | 56 (63.6%) | 38 (74.5%) | 0.023 |
| Comorbidities | ||||
| Chronic heart disease | 4 (4.8%) | 19 (21.6%) | 15 (29.4%) | <0.001 |
| Hypertension | 19 (22.6%) | 45 (51.1%) | 31 (60.8%) | <0.001 |
| Chronic obstructive pulmonary disease | 6 (7.1%) | 9 (10.2%) | 11 (21.6%) | 0.035 |
| Asthma | 4 (4.8%) | 7 (7.9%) | 2 (3.9%) | 0.539 |
| Chronic kidney disease | 1 (1.2%) | 9 (10.2%) | 7 (13.7%) | 0.014 |
| Liver cirrhosis | 4 (4.8%) | 3 (3.4%) | 5 (9.8%) | 0.260 |
| Cancer | 0 (0.0%) | 2 (2.3%) | 1 (1.9%) | 0.394 |
| Obesity | 8 (9.5%) | 38 (43.2%) | 30 (58.8%) | <0.001 |
| Diabetes | 5 (5.9%) | 18 (20.4%) | 14 (27.4%) | 0.002 |
| Dyslipidemia | 14 (16.7%) | 29 (32.9%) | 23 (45.1%) | 0.001 |
| Smoker | 10 (11.9%) | 7 (7.9%) | 3 (5.9%) | 0.451 |
| Charlson comorbidity index | 0 (0-2.5) | 3 (1-4.5) | 4 (2-6) | <0.001 |
| Time from COVID-19 symptoms to sample collection (days) | 5.7 (2.8-7.8) | 7.1 (3.6-7.4) | 7.4 (4.6-8.9) | 0.179 |
| Oxygen saturation (%) | 0.97 (0.95-0.98) | 0.94 (0.92-0.96) | 0.85 (0.8-0.87) | <0.001 |
| Baseline laboratory findings | ||||
| Hematocrit (%) | 42.9 (40.4-46.7) | 42.9 (39.8-45.3) | 43.3 (39.9-46.2) | 0.950 |
| White blood cells (x 103 cells/µl) | 5.8 (5-7.2) | 5.6 (4.5-7.9) | 7.8 (5.6-10.3) | 0.007 |
| Lymphocytes (x 103 cells/µl) | 1.3 (1-1.7) | 1 (0.8-1.4) | 0.75 (0.5-1.2) | <0.001 |
| Neutrophils (x 103 cells/µl) | 4 (3-5) | 3.9 (2.9-5.8) | 6.2 (3.9-8.9) | <0.001 |
| Thrombocytes (x 109 cells/l) | 188 (161-257) | 185 (127.5-231) | 175 (146-247) | 0.429 |
| International normalized ratio | 0.99 (0.94-1.1) | 0.98 (0.94-1) | 1.1 (0.97-1.2) | 0.009 |
| Glucose (mg/dl) | 105 (95-114) | 113 (103-137) | 132 (117-164) | <0.001 |
| Creatinine (mg/dl) | 0.8 (0.7-1) | 0.9 (0.8-1.2) | 1.1 (0.8-1.5) | <0.001 |
| Estimated glomerular filtration rate (ml/min) | 87.3 (76-100.7) | 79.9 (58.9-90.9) | 67.4 (44-85.9) | <0.001 |
| Albumin (g/dl) | 4.4 (4-4.5) | 4 (3.8-4.3) | 3.7 (3.5-4) | <0.001 |
| Alanine aminotransferase (IU/l) | 23 (18-36) | 32 (19-56) | 29 (19-59) | 0.089 |
| Lactate dehydrogenase (IU/l) | 219 (197-280) | 309 (249.5-388.5) | 433 (328-522) | <0.001 |
| Ferritin (ug/l) | 209 (99-522) | 413 (198-914) | 672 (372-1343) | <0.001 |
| C-reactive Protein (mg/l) | 13.9 (3.4-38.1) | 56.1 (26.8-108.5) | 123.3 (77.7-200.8) | <0.001 |
Statistics: Values are expressed as the median and interquartile range for continuous variables and absolute count (percentage) for categorical variables. Comparisons between groups were performed using the Mann-Whitney U-test for continuous variables and the chi-square test (χ2) or Fisher's exact test for categorical variables. Statistically significant differences are shown in bold.
Nasopharyngeal biomarkers and pneumonia
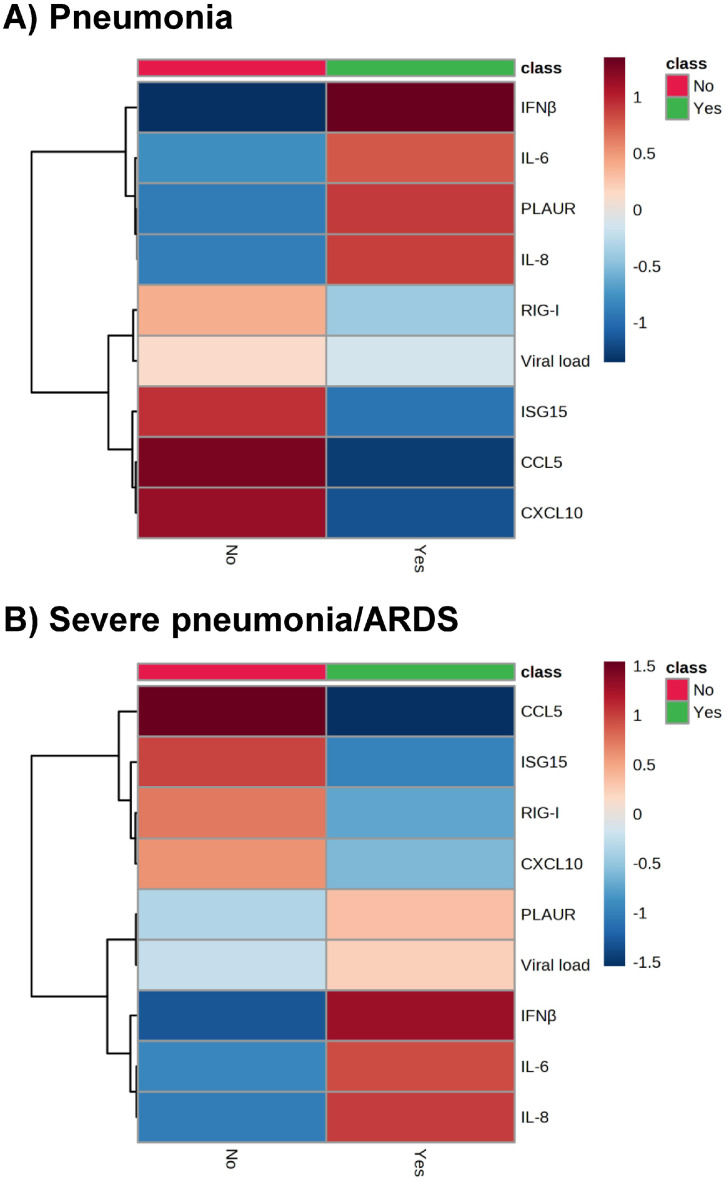
Patients with pneumonia had higher values of PLAUR, IL-8, and IFN-β than those without pneumonia, while the CCL5 and CXCL10 values were lower in patients with pneumonia (P <0.05; Figure 1 ). The hierarchical clustering heatmap (Figure 2 a) showed that high values of IFN-β, IL-6, PLAUR, and IL-8; and low values of RIG-I, viral load, ISG15, CCL5, and CXCL10 were characteristic of patients with pneumonia.
Figure 1.
Summary of gene expression in the upper respiratory tract of patients infected with SARS-CoV-2 according to the severity of COVID-19 pneumonia. Statistics: Differences between groups were assessed using the Mann-Whitney U-test. Abbreviations: ARDS, acute respiratory distress syndrome; CCL5, chemokine C-C motif ligand 5; CXCL10, chemokine C-X-C motif ligand 10; IFN, interferon; IL, interleukin; ISG15, IFN-stimulated gene 15; PLAUR, plasminogen activator, urokinase receptor; RIG-I, retinoic acid-inducible gene I.
Figure 2.
Hierarchical clustering heatmaps of groups show averages of gene expression in the upper respiratory tract of patients infected with SARS-CoV-2 according to the severity of COVID-19 pneumonia (a, Pneumonia; b, Severe pneumonia/ARDS). Statistics: Hierarchical clustering heatmaps were performed using MetaboAnalyst 5.0 (https://www.metaboanalyst.ca/). Settings used were normalization by a pooled sample from a group (group PQN), automatic scaling, Euclidean distance measure, and the Ward clustering method. Abbreviations: ARDS, acute respiratory distress syndrome; CXCL5, chemokine C-C motif ligand 5; CXCL10, chemokine C-X-C motif ligand 10; IFN, interferon; IL, interleukin; ISG15, IFN-stimulated gene 15; PLAUR, plasminogen activator, urokinase receptor; RIG-I, retinoic acid-inducible gene I.
We performed logistic regressions adjusted for the most significant covariates for each of the genes studied (Table 2 ). After stepwise selection, significant covariates that remained in the adjusted logistic regression model were obesity and age (see Supplementary Table 2). Only high PLAUR values (adjusted OR [aOR] = 1.25; P = 0.032, risk factor) and low CXCL10 values (aOR = 0.89; P = 0.048, protective factor) were associated with pneumonia.
Table 2.
Summary of adjusted associations between mucosal biomarkers (log2 values) in the upper respiratory tract and severity of COVID-19 pneumonia in the emergency department.
| Biomarkers | Pneumonia |
Severe pneumonia/acute respiratory distress syndrome |
||
|---|---|---|---|---|
| aOR (95% CI) | P-value | aOR (95% CI) | P-value | |
| Inflammation genes | ||||
| PLAUR | 1.25 (1.02; 1.53) | 0.032 | 1.19 (1.01; 1.41) | 0.427 |
| IL-6 | 1.02 (0.91; 1.14) | 0.718 | 1.07 (0.98; 1.18) | 0.055 |
| IL-8 | 1.11 (0.97; 1.28) | 0.126 | 1.16 (1.03; 1.32) | 0.062 |
| Antiviral genes | ||||
| ISG15 | 0.94 (0.80; 1.11) | 0.456 | 0.88 (0.76; 1.01) | 0.021 |
| RIG-I | 0.94 (0.76; 1.15) | 0.527 | 0.87 (0.73; 1.03) | 0.034 |
| IFN-β | 1.13 (0.96; 1.34) | 0.139 | 1.15 (0.99; 1.34) | 0.411 |
| Chemokine genes | ||||
| CCL5 | 0.89 (0.71; 1.13) | 0.351 | 0.73 (0.61; 0.89) | <0.001 |
| CXCL10 | 0.89 (0.79; 1.00) | 0.048 | 0.84 (0.76; 0.94) | 0.002 |
| SARS-CoV-2 | ||||
| SARS-CoV-2 Viral load | 0.98 (0.92; 1.04) | 0.470 | 0.99 (0.94; 1.04) | 0.678 |
Statistics: The values of the mucosal biomarkers were log2-transformed (base-2 logarithms). The association analysis was performed by binary logistic regression adjusted by the most significant covariables. Covariates that remained in the logistic regression model after the forward stepwise selection method were obesity and age. Statistically significant differences are shown in bold.
Abbreviations: aOR, adjusted odds ratio; CCL5, Chemokine (C-C motif) ligand 5; CI, confidence interval; CXCL10, C-X-C motif chemokine ligand 10; IFN, interferon; IL, interleukin; ISG15, IFN-stimulated gene 15; PLAUR, plasminogen activator, urokinase receptor; RIG-I, retinoic acid-inducible gene I.
Nasopharyngeal biomarkers and severe pneumonia/acute respiratory distress syndrome
Patients with severe pneumonia/ARDS had higher expression levels of IL-6, IL-8, and IFN-β than those with non-severe pneumonia, while the values of RIG-I, ISG15, CCL5, and CXCL10 were lower in patients with severe pneumonia/ARDS (P <0.05; Figure 1). The hierarchical clustering heatmap (Figure 2b) showed that low values of CCL5, ISG15, RIG-I, and CXCL10 and high values of PLAUR, viral load, IFN-β, IL-6, and IL-8 were characteristic of patients with severe pneumonia/ARDS.
We also performed logistic regressions adjusted for the most significant covariates (Table 2), once again obesity and age being the significant covariates that remained in the adjusted logistic regression models (see Supplementary Table 2). Lower values of ISG15 (aOR = 0.88, P = 0.021), RIG-I (aOR = 0.87, P = 0.034), CCL5 (aOR = 0.73, P <0.001), and CXCL10 (aOR = 0.84, P = 0.002) were risk factors for severe pneumonia/ARDS. CCL5 was the only marker with a suitable area under the curve = 0.745.
Discussion
This study shows that increased PLAUR gene expression and decreased CXCL10 gene expression in the nasopharynx were associated with COVID-19 pneumonia, and lower gene expression of antiviral genes (ISG15 and RIG-I) and chemokines (CCL5 and CXCL10) were related to severe pneumonia/ARDS. These findings provide information about the immunopathology of SARS-CoV-2 infection. Still, they may also help assess the prognosis and improve patient management in the early COVID-19 phase when the patient is diagnosed.
The innate mucosal immune response in the URT is critical for the early control of SARS-CoV-2 infection [4], as it occurs at the initial infection site and triggers the subsequent adaptive response, thus affecting the progression of COVID-19 disease [12]. In line with this, an imbalanced host antiviral/inflammatory response contributes to the immunopathology of COVID-19 and can increase the severity of the disease [5], [6], [7].
We initially found increased expression of IL-6, IL-8, and IFN-β with increasing pneumonia severity, but they did not reach statistical significance in the multivariable analysis.
Previous studies have reported a correlation between elevated levels of proinflammatory cytokines in the blood, including IL-6 and IL-8, and the severity or unfavorable outcomes of COVID-19 [2,13]. Our data about expression levels of IL-6 and IL-8 are in concordance with these previous reports, which reported elevated IL-6 and/or IL-8 in URT samples from patients with severe COVID-19 [14,15]. IL-6 and IL-8 are components of the cytokine storm that promotes neutrophil accumulation at the site of infection, leading to tissue injury [16].
In this study, after multivariate logistic regression analysis, only PLAUR expression levels increased significantly in patients with pneumonia. PLAUR encodes uPAR, a membrane receptor that, after proteolytic cleavage, generates a soluble form (suPAR) that plays an essential role in the innate immune response by attracting proinflammatory cells to the site of infection [17]. Elevated levels of suPAR in plasma are considered a biomarker of severe prognosis in patients with COVID-19 and are indicative of prolonged underlying inflammation [11]. These data are consistent with our results, suggesting that suPAR may also be overexpressed in the nasopharynx of patients with COVID-19 pneumonia.
The univariate analysis also found that IFN-β expression levels increased with increasing pneumonia severity. Since IFN-β induces the expression of ISGs [18,19], the low expression of RIG-I, ISG15, CCL5, and CXCL10 seems contradictory. However, Sposito et al. have recently shown that the expression of protective ISGs in the upper airways after SARS-CoV-2 infections is induced mainly by IFN-λ1 and IFN-l3 and not by IFN-I [20]. Furthermore, IFN-β was up-regulated in severe patients with reduced ISG response [20].
In contrast to IFN-β expression, we found that four ISGs linked to antiviral activity (RIG-I and ISG15) and chemotaxis (CCL5 and CXCL10) were down-regulated in serious cases of COVID-19 (severe pneumonia/ARDS). On the one hand, innate antiviral activity is vital in the control of SARS-CoV-2 infection [4]. RIG-I recognizes viral RNA and triggers a signaling pathway that leads to the expression of type I IFN and ISGs, establishing an antiviral state that limits the spread of SARS-CoV-2 infection [4]. ISG15, a ubiquitin-like protein, is a prototype of ISG with broad-spectrum antiviral activity [21]. On the other hand, chemokines recruit immune cells to inflammation sites, which are involved in the immunopathology of COVID-19 disease [22]. However, chemotaxis also promotes the interaction between T cells and dendritic cells, helping to control early viral replication in the nasopharyngeal mucosa [5]. CCL5 (also known as RANTES) is a potent chemoattractant for several immune cells and, therefore, an important link between innate and adaptive immune responses [23]. Other COVID-19 studies have shown low levels of nasopharyngeal CCL5 expression in patients with severe pneumonia [23] and critical disease [7,15]. Although high plasma levels of CXCL10 have generally been found to be associated with disease progression and a worse clinical course [22], our results indicate the opposite in the nasopharynx. According to our findings, high levels of CXCL10 in the early innate nasopharyngeal response against SARS-CoV-2 have been reported to prevent the development of severe COVID-19 [24], as it plays a central role in the activation of several immune cells [22]. Therefore, its lower expression levels result in an immune response that cannot limit the spread of infection, leading to severe pneumonia [25]. Furthermore, patients with mild or moderate symptoms develop a T helper profile "TH1-TH17" with high levels of CXCL10 that protect against severe disease [14].
People with better control of SARS-CoV-2 infection usually have higher ISG expression levels and a lower risk of developing severe COVID-19 [25]. Therefore, low levels of RIG-I, ISG15, CCL5, and CXCL10 expression in the URT may reflect a weak antiviral response that cannot restrict viral replication and spread to the lower respiratory tract, leading to pneumonia. Strategies aimed to induce the production of critical antiviral proteins in the URT, or their exogenous administration, could aid in preventing the progression to severe COVID-19 and represent an interesting new avenue of investigation. Moreover, these genes may be useful markers for the severity of COVID-19 pneumonia. Profiling the expression of PLAUR, antiviral genes (ISG15 and RIG-I), and chemokines (CCL5 and CXCL10) in the nasopharynx can easily be performed from the same sample used for the SARS-CoV-2 diagnosis. This information can help improve the prognosis and management of patients in the initial infection phase.
Limitations of the study
First, the sample size was small, which could affect statistical power, making it difficult to reach statistical significance and increasing the false positive rate. Second, the design of this study was retrospective, and biases may have been introduced. Third, the quality of the RNA extracted from nasopharyngeal swabs could vary due to differences in sample collection, affecting the precision, validity, and generalizability of the results obtained in our investigation. Finally, this study was conducted during the first year of the pandemic, so we do not know the impact of Omicron variants and vaccination on our results.
Conclusion
An unbalanced early innate immune response to SARS-CoV-2 in the nasopharynx, characterized by high expression of PLAUR and low expression of antiviral genes (ISG15 and RIG-I), and chemokines (CCL5 and CXCL10), is associated with the severity of COVID-19 in unvaccinated patients against SARS-CoV-2. Additional studies are needed to determine if our findings are maintained against other SARS-CoV-2 variants and in patients vaccinated against COVID-19.
Declarations of Competing Interest
The authors have no competing interests to declare.
Acknowledgments
Funding
This work was supported by awards from the Canadian Institutes of Health Research (CIHR OV2 – 170357, [JFBM]). The study was also funded by the Centro de Investigación Biomédica en Red (CIBER) en Enfermedades Infecciosas (CB21/13/00044 [SR]) y Enfermedades Respiratorias (CB22/06/00035 [JFBM]).
Ethics approval and consent to participate
This study was approved by the Ethics Committee of the “Hospital Universitario Príncipe de Asturias” (Ref.: EXPRES-INMUNE-COVID). The Institutional Review Board of the Hospital also approved this study. The Ethics Committee authorized the informed consent waiver.
Acknowledgments
We want to acknowledge the patients in this study for their participation. This study would not have been possible without the collaboration of all medical and nursing staff and data managers who participated in the project.
Author contributions
Funding acquisition: JFBM, SR, and IM. Study concept and design: SR and IM. Patients' selection and clinical data acquisition: FPG, LCG, IHF, VGV, and JCG. Sample preparation, RNA isolation, and reverse transcription-polymerase chain reactions: CPM, MMV, AVB, and MJMG. Statistical analysis and interpretation of data: CPM, FPG, JFBM, SR, and IM. Writing – original draft preparation: CPM, FPG, SR, and IM. Writing – review & editing: JFBM. Supervision and visualization: SR and IM.
Consent for publication
Not applicable.
Availability of data and materials
The datasets used and analyzed during the current study are available from the corresponding authors upon reasoned request.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ijid.2023.06.001.
Appendix. Supplementary materials
References
- 1.World Health Organization . 2022. Clinical management of COVID-19: living guideline. Geneva, Switzerland. [PubMed] [Google Scholar]; https://www.ncbi.nlm.nih.gov/books/NBK582435/ [accessed 20 March 2023].
- 2.Ragab D, Salah Eldin H, Taeimah M, Khattab R, Salem R. The COVID-19 cytokine storm; what we know so far. Front Immunol. 2020;11:1446. doi: 10.3389/fimmu.2020.01446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang W, Chua BY, Selva KJ, Kedzierski L, Ashhurst TM, Haycroft ER, et al. SARS-CoV-2 infection results in immune responses in the respiratory tract and peripheral blood that suggest mechanisms of disease severity. Nat Commun. 2022;13:2774. doi: 10.1038/s41467-022-30088-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Diamond MS, Kanneganti TD. Innate immunity: the first line of defense against SARS-CoV-2. Nat Immunol. 2022;23:165–176. doi: 10.1038/s41590-021-01091-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ziegler CGK, Miao VN, Owings AH, Navia AW, Tang Y, Bromley JD, et al. Impaired local intrinsic immunity to SARS-CoV-2 infection in severe COVID-19. Cell. 2021;184:4713–4733. doi: 10.1016/j.cell.2021.07.023. .e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blanco-Melo D, Nilsson-Payant BE, Liu WC, Uhl S, Hoagland D, Møller R, et al. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020;181:1036–1045. doi: 10.1016/j.cell.2020.04.026. .e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pérez-García F, Martin-Vicente M, Rojas-García RL, Castilla-García L, Muñoz-Gomez MJ, Hervás Fernández I, et al. High SARS-CoV-2 viral load and low CCL5 expression levels in the upper respiratory tract are associated with COVID-19 severity. J Infect Dis. 2022;225:977–982. doi: 10.1093/infdis/jiab604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hodcroft EB, Zuber M, Nadeau S, Vaughan TG, Crawford KHD, Althaus CL, et al. Spread of a SARS-CoV-2 variant through Europe in the summer of 2020. Nature. 2021;595:707–712. doi: 10.1038/s41586-021-03677-y. [DOI] [PubMed] [Google Scholar]
- 9.Li A, Xu J. Innate immune response in respiratory system: a double-edged sword against virus infection. Infect Dis Immunity. 2022;2:132–134. doi: 10.1097/ID9.0000000000000031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Newton AH, Cardani A, Braciale TJ. The host immune response in respiratory virus infection: balancing virus clearance and immunopathology. Semin Immunopathol. 2016;38:471–482. doi: 10.1007/s00281-016-0558-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chalkias A, Mouzarou A, Samara E, Xanthos T, Ischaki E, Pantazopoulos I. Soluble urokinase plasminogen activator receptor: A biomarker for predicting complications and critical care admission of COVID-19 patients. Mol Diagn Ther. 2020;24:517–521. doi: 10.1007/s40291-020-00481-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gallo O, Locatello LG, Mazzoni A, Novelli L, Annunziato F. The central role of the nasal microenvironment in the transmission, modulation, and clinical progression of SARS-CoV-2 infection. Mucosal Immunol. 2021;14:305–316. doi: 10.1038/s41385-020-00359-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mangalmurti N, Hunter CA. Cytokine storms: understanding COVID-19. Immunity. 2020;53:19–25. doi: 10.1016/j.immuni.2020.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rodriguez C, de Prost N, Fourati S, Lamoureux C, Gricourt G, N'Debi M, et al. Viral genomic, metagenomic and human transcriptomic characterization and prediction of the clinical forms of COVID-19. PLoS Pathog. 2021;17 doi: 10.1371/journal.ppat.1009416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chua RL, Lukassen S, Trump S, Hennig BP, Wendisch D, Pott F, et al. COVID-19 severity correlates with airway epithelium-immune cell interactions identified by single-cell analysis. Nat Biotechnol. 2020;38:970–979. doi: 10.1038/s41587-020-0602-4. [DOI] [PubMed] [Google Scholar]
- 16.Gustine JN, Jones D. Immunopathology of hyperinflammation in COVID-19. Am J Pathol. 2021;191:4–17. doi: 10.1016/j.ajpath.2020.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mondino A, Blasi F. uPA and uPAR in fibrinolysis, immunity and pathology. Trends Immunol. 2004;25:450–455. doi: 10.1016/j.it.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 18.Kim YM, Shin EC. Type I and III interferon responses in SARS-CoV-2 infection. Exp Mol Med. 2021;53:750–760. doi: 10.1038/s12276-021-00592-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bhat SA, Shibata T, Leong M, Plummer J, Vail E, Khan Z. Comparative upper respiratory tract transcriptomic profiling reveals a potential role of early activation of interferon pathway in severe COVID-19. Viruses. 2022;14:2182. doi: 10.3390/v14102182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sposito B, Broggi A, Pandolfi L, Crotta S, Clementi N, Ferrarese R, et al. The interferon landscape along the respiratory tract impacts the severity of COVID-19. Cell. 2021;184:4953–4968. doi: 10.1016/j.cell.2021.08.016. .e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perng YC, Lenschow DJ. ISG15 in antiviral immunity and beyond. Nat Rev Microbiol. 2018;16:423–439. doi: 10.1038/s41579-018-0020-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gudowska-Sawczuk M, Mroczko B. What is currently known about the role of CXCL10 in SARS-CoV-2 infection? Int J Mol Sci. 2022;23:3673. doi: 10.3390/ijms23073673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Montalvo Villalba MC, Valdés Ramírez O, Muné Jiménez M, Arencibia Garcia A, Martinez Alfonso J, González Baéz G, et al. Interferon gamma, TGF-β1 and RANTES expression in upper airway samples from SARS-CoV-2 infected patients. Clin Immunol. 2020;220 doi: 10.1016/j.clim.2020.108576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheemarla NR, Watkins TA, Mihaylova VT, Wang B, Zhao D, Wang G, et al. Dynamic innate immune response determines susceptibility to SARS-CoV-2 infection and early replication kinetics. J Exp Med. 2021;218 doi: 10.1084/jem.20210583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ramasamy R. Innate and adaptive immune responses in the upper respiratory tract and the infectivity of SARS-CoV-2. Viruses. 2022;14:933. doi: 10.3390/v14050933. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and analyzed during the current study are available from the corresponding authors upon reasoned request.