Abstract
Background:
It is well established that biallelic mutations in TMPRSS3 cause hearing loss. Currently, there is controversy regarding the audiological outcomes after cochlear implantation (CI) for TMPRSS3-associated hearing loss. This controversy creates confusion amongst healthcare providers regarding the best treatment options for individuals with TMPRSS3-related hearing loss.
Methods:
A literature review was performed to identify all published cases of patients with TMPRSS3-associated hearing loss who received a CI. CI outcomes of this cohort were compared to published adult CI cohorts using post-operative consonant-nucleus-consonant (CNC) word performance. TMPRSS3 expression in mouse cochlea and human auditory nerves (ANs) was determined by using hybridization chain reaction (HCR) and single-cell RNA-sequencing analysis.
Results:
In aggregate, 27 patients (30 total CI ears) with TMPRSS3-associated hearing loss treated with CI, and 85% of patients reported favorable outcomes. Post-operative CNC word scores in patients with TMPRSS3-associated hearing loss were not significantly different than those seen in adult CI cohorts (8 studies). Robust Tmprss3 expression occurs throughout the mouse organ of Corti, the spindle and root cells of the lateral wall, and faint staining within <5% of the ANs, representing type II spiral ganglion neurons. Adult human ANs express negligible levels of TMPRSS3.
Conclusion:
The clinical features after CI and physiologic expression of TMPRSS3 suggest against a major role of TMPRSS3 in auditory neurons.
Keywords: Cochlear implant, TMPRSS3, Neurons, Genetic, Hearing loss, Speech recognition
Introduction
Transmembrane serine proteases are a large family of proteins and indispensable regulators within many tissues, including the inner ear[1]. Broadly, they are classified based upon how they are anchored to the cellular membrane. Type II transmembrane serine proteases are anchored by their N-terminus to the cellular membrane and contain a group A scavenger receptor domain and an active serine protease domain[1 2]. TMPRSS3 (Transmembrane Protease, Serine 3) is a type II transmembrane serine protease required in the mammalian auditory system for proper hearing[3 4].
TMPRSS3 is essential for cochlear hair cell survival[5–7]. Defects or ablation of TMPRSS3 results in hearing loss in both mice and humans. Mice carrying a protein-truncating TMPRSS3 mutation exhibit normal cochlear and vestibular hair cell (HC) development followed by rapid HC degeneration over 48 hours starting at postnatal day 12 (P12)[5]. In humans, bi-allelic pathogenic variants of TMPRSS3 are the most common causative hearing loss gene in adults undergoing traditional and hybrid cochlear implantation (CI)[8]. Additionally, TMPRSS3 is the 5th most common gene causally associated with deafness in a multiethnic cohort of human congenital deafness[9]. Furthermore, select missense mutations (e.g. A306T, A138E, A426T) are linked to post-lingual onset high-frequency hearing loss[10]. Despite this robust and dramatic phenotype, the function of TMPRSS3 and the pathomechanism(s) that underlie TMPRSS3-related deafness remains elusive.
Cochlear implantation is the gold-standard treatment for inherited severe-to-profound sensorineural hearing loss (SNHL)[11 12]. Implants function by directly stimulating spiral ganglion neurons (SGNs), permitting sound detection and speech recognition while circumventing inner ear organs such as cochlear hair cells[6 11–13]. Currently, the post-operative CI outcomes in patients with TMPRSS3-associated hearing loss remain controversial in the published literature[6 10 13]. TMPRSS3-related damage to the SGN has been proposed as one possible explanation for poor CI outcomes in these patients[13], evidenced by TMPRSS3 immunostaining within SGNs in the murine cochlea[14]. However, recent single-cell RNA sequencing (scRNA-seq) studies have demonstrated that Tmprss3 is not expressed in type I SGNs, but only in type II SGNs—which do not transmit auditory signals and make up only 4% of the SGN population[15 16]. This disconnect between biological data and patient outcomes prompted us to critically review published reports regarding TMPRSS3 patient CI outcomes and perform in-depth expression studies of TMPRSS3 in the human and mouse auditory system.
Here we show that patients with TMPRSS3-related hearing loss have overall good post-operative performance and a TMPRSS3 genetic diagnosis should not be a reason to restrict cochlear implantation. In addition, using two different methods, we show that TMPRSS3 is not expressed in the sound transducing neurons of the cochlea and is primarily localized to the sensory epithelium portion of the cochlea. Finally, we call into question the correlation between negative CI performance and TMPRSS3-related hearing loss.
Materials and Methods
TMPRSS3-associated cochlear implant outcomes
Two independent searches of PubMed for articles published on TMPRSS3-associated hearing loss treated with CI were conducted. Search terms included the combination of “TMPRSS3” with the major subheading topics “hearing loss” and “cochlear implant”. All articles reporting CI outcomes for TMPRSS3-associated hearing loss were identified. Articles that contained subjective (favorable or unfavorable) or objective (speech scores) outcome measures were reviewed. For the adult CI cohort analysis, articles that contained more than 10 patients and had post-operative CNC word scores were reviewed. Nine articles fit these criteria and were included in this study. Demographics, age of SNHL onset, phenotypic presentation, documented TMPRSS3 mutations, patient age at time of CI, and audiologic outcomes were recorded. Individuals with hearing loss prior to two years of age were classified as having prelingual hearing loss whereas those with onset two years of age or older were designated as having post-lingual hearing loss. Objective outcomes included pre-operative and post-operative pure tone averages (PTA), consonant-nucleus-consonant (CNC) word recognition scores (WRS), and sentence scores (SS). Subjective outcomes, when reported in published articles, were classified as either favorable or poor: post-operative implant-aided CNC WRS scores less than 30% were considered poor outcomes, and scores 50% or greater were considered favorable. Change in PTA and CNC WRS scores were also calculated.
Statistical analysis
Objective data were evaluated by measures of central tendency using Microsoft Excel, 2016. The mean age difference between favorable and poor post-lingual CI performers was evaluated using a t-test using SPSS Statistics, 2020. CI outcomes in patients with TMPRSS3-associated hearing loss were compared to post-operative outcomes of eight representative CI studies in the general hearing loss population. Logistic regression was performed to determine the effects of possible predictor variables on favorable CI outcomes in the TMPRSS3 population. Linear regression was similarly performed to evaluate possible predictor effects on average change (delta) dCNC and dPTA. Mean post-operative CNC WRS analysis of variance (ANOVA) of the two populations was performed using SPSS.
Animal handling
Tmprss3-mutant mice (Tmprss3Y260X) have been described previously[5]. All mice were housed in the animal facility of Indiana University School of Medicine. All procedures were carried out in accordance with the approval of the Indiana University Institutional Animal Care and Use Committee protocol. Genotyping was performed as previously described[3].
Human tissue harvest
Human auditory nerves were harvested during vestibular schwannoma operation after obtaining patient consent (IRB approval 200634852). During the tumor removal, the auditory nerve was dissected away from the schwannoma and facial nerve within the internal auditory canal (IAC). The auditory nerve between the modiolus of the cochlea and porus of the IAC was harvested. Immediately after harvest, the tissues were flash-frozen in liquid nitrogen and stored at −80°C before RNA extraction for qRT-PCR. For HCR experiments, the auditory nerves were immediately fixed in 10% formalin for 24 hours prior to embedding.
Plasmid Construct
The coding sequence of human TMPRSS3 isoform 1 from transcript variant A (NM_024022.4) with added 5’ EcoRI and 3’ KpnI restriction sites was generated as gBlocks™ Gene Fragments (Integrated DNA Technologies, Corralville, IA). The construct p3XFLAG-CMV 7.1_TMPRSS3 was generated from the backbone of p3XFLAG-CMV 7.1_syn6 (Addgene, 50012). The Syntaxin-6 gene was replaced by TMPRSS3 gBlocks with EcoRI and KpnI restriction enzymes.
Cell culture
HEK293 cells (ATCC, Manassas, VA; CRL-1573), plated on 1% gelatin-coated cover slide in 6 well plates and were transfected with p3XFLAG-CMV 7.1_TMPRSS3 plasmid using Lipofectamine 3000 Reagent (Thermo Fisher, Waltham, MA; L3000015) for 24 hours as described by the manufacturer. Cells were washed once with ice-cold PBS and fixed with 4% paraformaldehyde for 20 mins at room temperature.
HCR Immunofluorescence
Human auditory nerves were fixed in 10% formalin for 24 hours. Then, fixed samples were cryoprotected in 30% sucrose in PBS. After embedding in TFM™ Tissue Freezing Medium (General Data Company, TFM-5), the auditory nerve was frozen and stored at −30°C. Sections of 6 μm were cut on a cryostat and air-dried for 2 hours.
Mouse cochleae were dissected from P11 mice anesthetized with isoflurane. The mice were perfused with 1X PBS through left ventricle. The cochleae were dissected and fixed in 4% paraformaldehyde overnight at room temperature and decalcified overnight at room temperature in solution containing 120 mM EDTA buffered in 1X PBS. Fixed cochleae were cryoprotected in 30% sucrose in PBS. After embedding in TFM™ Tissue Freezing Medium (General Data Company, TFM-5), cochleae were frozen and stored at −30°C. Midmodiolar cross sections of 6 μm were cut on a cryostat and air-dried for 2 hr.
The human auditory nerve sections and TMPRSS3-transfected HEK cells were stained with human TMPRSS3 HCR RNA-FISH probes while the mouse cochleae sections were stained with mouse Tmprss3 and mouse MBP HCR RNA-FISH probes (Molecular Instruments, Los Angeles, CA) following the manufacture’s protocol wth incubations at 37°C (Corning LSE mini 6800). Twenty probes were designed for human TMPRSS3 transcript variant A (NM_024022.4), mouse Tmprss3 transcript variant 1 (NM_001163776), and mouse MBP transcript variant 1 (NM_001025251), which recognizes 5’-UTR, 3’-UTR, and coding sequence (CDS). Sections were mounted with ProLong™ Gold Antifade Mountant with DAPI (ThermoFisher, P36931) and visualized on Leica DMi8 microscopy.
Real-time quantitative RT-PCR
Total RNA was extracted from human auditory nerve, HEK cells and TMPRSS3-transfected HEK cells with PureLink® RNA Mini Kit (Invitrogen) and cDNA was synthesized from 50 ng total RNA with SuperScript IV VILO Master Mix (Invitrogen). qRT-PCR was performed using SYBR-Green and primers: TMPRSS3 sense: CAT CCA GGT GGG TCT AGT TTC; TMPRSS33 antisense: AGC CTC TTT GGC TTG TAC TT; MBP sense: GGC AAG GTA CCC TGG CTA AA; MBP antisense: GGG TGG TGT GAG TCC TTG TA; GAPDH sense: CAT CAC TGC CAC CCA GAA GAC TG; GAPDH antisense: ATG CCA GTG AGC TTC CCG TTC AG. The amplification and detection were performed on QuantStudio 5 Real-Time PCR System (Invitrogen). Relative expression for TMPRSS3 and MBP were adjusted to GAPDH levels.
Immunofluorescence
The cells on the slide or tissue sections were blocked with 10% goat serum with 0.1% Tween-20 in PBS for 30 mins at room temperature (RT). Following this, sections were treated with primary antibodies in 3% goat serum with 0.1% Tween-20 in PBS at room temperature for 1 hr. The antibody we used were mouse anti-TUJ1 (BioLegend, 801202, 1:100). After washing with PBS, sections were incubated with secondary antibodies or Alexa Fluor™ 568 Phalloidin (Thermo Fisher, A12380, 1:200) in 3% goat serum with 0.1% Tween-20 in PBS at RT for 1 hr. Finally, sections were washed and mounted with ProLong™ Gold Antifade Mountant with DAPI (ThermoFisher, P36931). Staining was visualized on Leica DMi8 microscopy.
TMPRSS3 hearing loss causing variants
Eighty-six TMPRSS3 deafness-causing variants were curated from the published literature and the Deafness Variation Database (https://deafnessvariationdatabase.org/)[17] or via a Pubmed Search (July 2021). Variants were classified as Pathogenic, Likely Pathogenic or VUS according to the hearing loss specific ACMG[18].
Results
Variants throughout the TMPRSS3 gene cause hearing loss.
Of 86 variants curated in this study, 62 were classified as pathogenic/likely pathogenic according to American College of Medical Genetics (ACMG) criteria while the remaining 24 are considered variants of uncertain significance (VUS). The 62 pathogenic/likely pathogenic TMPRSS3 mutations map across all protein domains which are associated with hearing loss (Figure 1A, Supp Table 1). Of these, 35 (~56.5%) are missense or non-protein truncating (NPT) variants (Figure 1A–C). The remaining protein truncating (PT) mutations include 10 (~16%) nonsense variants, 8 (~13%) frameshift indels and 9 (14.5%) splice-altering variants (Figure 1A–C).
Figure 1: Human TMPRSS3 Gene Mutations Associated with Hearing Loss.
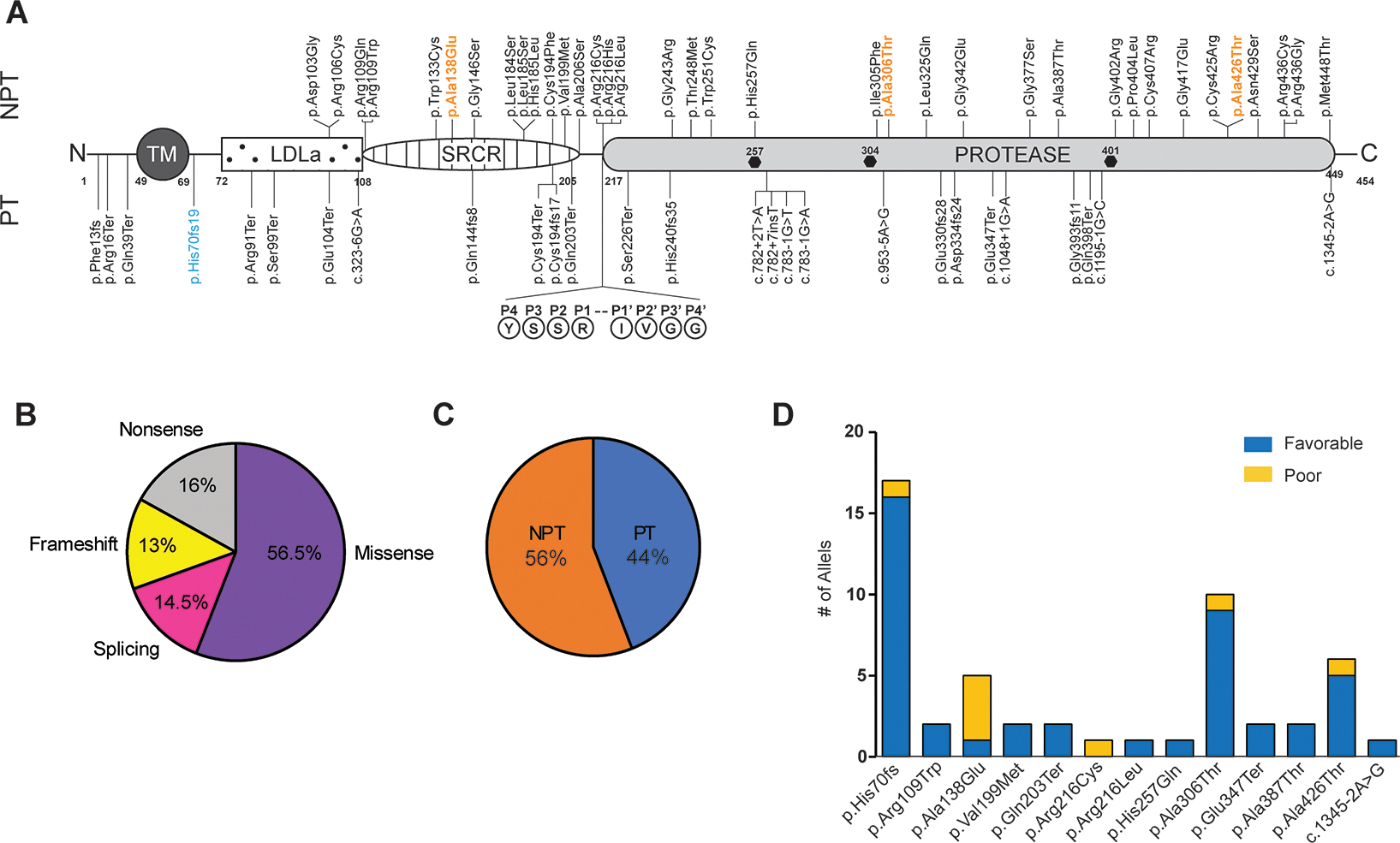
(A) Schematic of TMPRSS3 protein and pathogenic/likely pathogenic reported deafness causing variants. Frequent variants causing congenital or postlingual hearing loss denoted in blue and orange, respectively. The TMPRSS3 autocleavage site (arrowhead) at R216 is shown and hexagons indicate the conserved serine protease catalytic triad (H-D-S). TM, transmembrane domain; LDLa, low-density lipoprotein receptor domain; SRCR, scavenger receptor cysteine-rich domain, PT, protein truncating; NPT, non-protein truncating. The amino acid number for the start and end of protein domains are noted.
Note: p.Gly393fs11 is an approximation-based description of the variant in Scott et al. (B) Pie chart of the percentage of human TMPRSS3-associated hearing loss variant types. (C) Pie chart of the percentage of human TMPRSS3-associated hearing loss variants PT vs. NPT. (D) In cochlear implant recipients, the relationship between TMPRSS3 variants and the number of patients with post-operative performance defined as poor or favorable.
Patients with TMPRSS3-variants have excellent speech recognition outcomes with cochlear implants.
A total of nine articles describing 27 patients (30 ears) who received CI for TMPRSS3-associated hearing loss were identified (Table 1)[19–21]. After a review of the variants identified in individuals with reported CI outcomes, one individual reported in Miyagawa et al. 2015 was removed due to the reported variant c.212T>C (p.Phe71Ser), being present in >1% of control alleles in gnomAD[22]. Of the 19 patients with gender recorded, 13 (68%) were female, and 6 (32%) were male. Of 21 patients with data available, 14 (67%) were white, and 7 (33%) were Asian (Table 1). Post-lingual hearing loss occurred in 80% of patients.
Table 1.
Outcomes of cochlear implantation with or without acoustic stimulation in patients with DFNB8 or DFNB10 phenotypes of TMPRSS3 mutations based on literature review.
| Case | Reference | Ethnicity | Age of Onset (years) | Sex | Genotype | Allele Combination | Age at CI | Preop PTA [34] | Preop Word Score (%) | Preop Sentence Score (%) | Postop PTA [34] | Postop Word Score (%) | Postop Sentence Score (%) | Outcome | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Allele 1 | Allele 2 | ||||||||||||||
| 1 2 |
Battelino, 2016
Battelino, 2016 |
Slovenian Slovenian |
<2 <2 |
|
c.208delC (p.His70fs) c.208delC (p.His70fs) |
c.208delC (p.His70fs) c.208delC (p.His70fs) |
PTxPT PTxPT |
11 mo 30 mo |
80–110 95–110 |
25 45 |
Favorable Favorable |
||||
| 3 | Battelino, 2016 | Slovenian | <2 | c.208delC (p.His70fs) | c.208delC (p.His70fs) | PTxPT | 13 mo | 80–100 | 25 | Favorable | |||||
| 4 | Battelino, 2016 | Slovenian | <2 | c.208delC (p.His70fs) | c.208delC (p.His70fs) | PTxPT | 11 mo | 70–85 | 25 | Favorable | |||||
| 5 | Chung, 2014 | Korean | 6 | F | c.325C>T (p.Arg109Trp) | c.916G>A (p.Ala306Thr) | NPTxNPT | 11 yo | 70–80 | 88.5 | Favorable | ||||
| 6 | Chung, 2014 | Korean | 3 | M | c.325C>T (p.Arg109Trp) | c.916G>A (p.Ala306Thr) | NPTxNPT | 5 yo | 90–100 | 88.5 | Favorable | ||||
| 7 | Eppsteiner, 2012 | Caucasian | 5 | M | c.413C>A (p.Ala138Glu) | c.646C>T (p.Arg216Cys) | NPTxNPT | 47 yo | 93 | 28 | 47 | Poor | |||
| 8 | Eppsteiner, 2012 | Caucasian | 6 | F | c.916G>A (p.Ala306Thr) | c.413C>A (p.Ala138Glu) | NPTxNPT | 32 yo | 98 | 21 | 25 | Poor | |||
| 9 10 |
Miyagawa, 2013 (L) Miyagawa, 2013 (R) |
Japanese | 25 | F | c.607C>T (p.Gln203Ter) | c.1159G>A (p.Ala387Thr) | PTxNPT | 38 yo 38 yo |
90 90 |
30 |
90 90 |
|
Favorable Favorable |
||
| 11 | Miyagawa, 2015 | Japanese | 33 | M | c.647G>T (p.Arg216Leu) | c.771C>G (p.His257Gln) | NPTxNPT | 45 yo* | 93.8 | 30 | 90 | Favorable | |||
| 12 | Miyagawa, 2015 | Japanese | 6 | F | c.607C>T (p.Gln203Ter) | c.1159G>A (p.Ala387Thr) | PTxNPT | 38 yo* | 106.3 | 18 | 30 | 67.5 | Favorable | ||
| 13 | Shearer, 2018 *** | c.208delC (p.His70fs) | c.413C>A (p.Ala138Glu) | PTxNPT | 64 yo | 2 | Poor | ||||||||
| 14 | Shearer, 2018 *** | c.413C>A (p.Ala138Glu) | c.1276G>A (p.Ala426Thr) | NPTxNPT | 53 yo | 29 | Poor | ||||||||
| 15 | Shearer, 2018 *** | c.1345–2A>G | c.1276G>A (p.Ala426Thr) | PTxNPT | 38 yo | 83 | Favorable | ||||||||
| 16 | Weegerink, 2011 | Dutch | <2 | M | c.208delC (p.His70fs) | c.916G>A (p.Ala306Thr) | PTxNPT | 91** | Favorable | ||||||
| 17 | Weegerink, 2011 | Dutch | <2 | F | c.595G>A (p.Val199Met) | c.916G>A (p.Ala306Thr) | NPTxNPT | 80** | Favorable | ||||||
| 18 | Weegerink, 2011 | Dutch | 17 | F | c.208delC (p.His70fs) | c.1276G>A (p.Ala426Thr) | PTxNPT | 20** | 75 | Favorable | |||||
| 19 | Weegerink, 2011 | Dutch | 12 | M | c.208delC (p.His70fs) | c.1276G>A (p.Ala426Thr) | PTxNPT | 5** | 60 | Favorable | |||||
| 20 | Weegerink, 2011 | Dutch | 7 | F | c.208delC (p.His70fs) | c.1276G>A (p.Ala426Thr) | PTxNPT | 0** | 58 | Favorable | |||||
| 21 | Weegerink, 2011 | Dutch | 17 | M | c.208delC (p.His70fs) | c.1276G>A (p.Ala426Thr) | PTxNPT | 0** | 62 | Favorable | |||||
| 22 | Weegerink, 2011 | Dutch | 15 | F | c.413C>A (p.Ala138Glu) | c.595G>A (p.Val199Met) | NPTxNPT | 2.5** | 68 | Favorable | |||||
| 23 | Song, 2020 | Korean | 6 | F | c.916G>A (p.Ala306Thr) | c.1039G>T (p.Glu347Ter) | PTxNPT | 8 yo | 70 | 20 | 50 | 100 | 86 | Favorable | |
| 24 | Song, 2020 | Korean | 5 | F | c.916G>A (p.Ala306Thr) | c.1039G>T (p.Glu347Ter) | PTxNPT | 6 yo | Favorable | ||||||
| 25 26 |
Holder, 2021 (R) Holder, 2021 (L) |
3 | F | c.208delC (p.His70fs) | c.916G>A (p.Ala306Thr) | PTxNPT | 4.5 yo* 4.8 yo* |
83 83 |
44 32 |
20 24 |
68 75 |
Favorable Favorable |
|||
| 27 28 |
Holder, 2021 (R) Holder, 2021 (L) |
3 | F | c.208delC (p.His70fs) | c.916G>A (p.Ala306Thr) | PTxNPT | 4.8 yo* 3.9 yo* |
67.5 73.3 |
23.3 25 |
80 80 |
Favorable Favorable |
||||
| 29 30 |
Holder, 2021 (R) Holder, 2021 (L) |
2 | F | c.208delC (p.His70fs) | c.916G>A (p.Ala306Thr) | PTxNPT | 36 yo* 36 yo* |
44 42 |
25 22.5 |
82 92 |
Favorable Favorable |
||||
| AVERAGE (St.Dev) | 18.9 (20.7) | 82.4 (18.2) | 17.2 (15.2) | 26.7 (6.4) | 65.5 (26.3) | 67 (29.4) | |||||||||
CI = Cochlear Implantation, PTA = Pure Tone Average.
Patients who underwent electric acoustic stimulation (both cochlear implantation and hearing aid use).
Phoneme score rather than monosyllable word recognition score. PT, Protein truncating; NPT, nonprotein truncating.
Variants listed in Table 1 of Shearer 2018 are shifted up by 1 line. Grey shadding denotes poor outcomes.
The age of implantation was widely variable with a mean (standard deviation) age of 19.8 (±20.2) years (Table 1). Favorable hearing outcome after implantation was reported by 85% of patients. The mean PTA improved from 82.4 (±18.2) dB to 26.7 (±6.4) dB with an average change in PTA (delta-PTA or dPTA), defined as post-op PTA minus pre-op PTA, of −55.7 (±17.9). Multivariable linear regression found no significant difference between dPTA and age (0.954), although higher pre-operative PTA (worse hearing) was associated with a greater dPTA (b= −.284, p<0.001). Average dCNC, defined as post-op CNC score minus pre-op score, was 48.3% (±14.7%) (Table 1) with an average of 17.2% (±15.2%) pre-operatively and 65.5% (±26.3%) after implantation. Multivariable linear regression found no significant association between dCNC and age (p=0.973) or pre-operative CNC (p=0.124). Poor post-lingual performers were significantly older than good performers [t(17.4) = 4.7, P=0.018]. Multivariable logistic regression found no significant association between age and subjective performance (p=0.413).
TMPRSS3 mutations contributing to hearing loss occurred on 13 different alleles in our cohort (Figure 1D). Mutations were recorded for each of the two alleles present in each individual (Table 1). The most common allele was the frameshift variant c.208delC (p.His70fs) which accounts for ~1/3 of total alleles (17/52) present in this cohort. Other common variants included c.916G>A (p.Ala306Thr), c.1276G>A (p.Ala426Thr), and c.413C>A (p.Ala138Glu), which comprised 10, 6, and 4 alleles, respectively. The remaining 10 variants accounted for 2 or 1 alleles each (Figure 1D). Favorable and poor CI performance related to each variant was determined (Figure 1D).
The most common allelic combination was protein truncating (PT) by non-protein truncating (NPT), accounting for 14 cases (13 favorable outcomes vs 1 poor outcome). NPTxNPT accounted for the next highest allelic combination with 8 cases (5 favorable outcomes vs 3 poor outcomes), and 4 cases had PTxPT (4 favorable outcomes vs 0 poor outcomes). Statistical analysis (Chi-Square test) could not be performed because several conditions had fewer than 5 events.
Four patients reported subjectively poor outcomes. Three of the four patients carried the c.413C>A (p.Ala138Glu) allele (Table 1 and Figure 1D), although the second allele was different in each case (p.Arg216Cys, p.Ala306Thr, and p.Ala426Thr). The remaining patient also carried the c.1276G>A (p.Ala426Thr) allele along with the c.208delC (p.His70fs) allele. The c.1276G>A (p.Ala426Thr) allele was observed in 5 patients with favorable outcomes, while the c.413C>A (p.Ala138Glu) allele was observed in one favorable outcome in combination with the c.595G>A (p.Val199Met) allele. Alleles p.His70fs and p.Ala306Thr were also seen in patients with favorable outcomes (Table 1). Of the four poor performers, two (both with p.Ala138Glu allele) had experienced hearing loss for greater than 25 years while the remaining two (one with p.Ala138Glu allele, both with p.Ala426Thr allele) did not have age at onset recorded, but were in their 50s and 60s, respectively, at time of implantation. The p.Ala138Glu and p.Ala426Thr mutations typically results in post-lingual progressive high-frequency sensorineural hearing loss prior to age 15[10]. Therefore, it is likely that both of these patients had a significant duration of hearing loss prior to implantation, which is associated with poor CI performance[23]. Both mutations are, however, typically associated with less severe hearing loss than other TMPRSS3 mutations[10], such as p.Val199Met and p.Ala306Thr, which were not present in as high of a proportion of poor CI performers. Additionally, one patient (Case 8) who carries both the p.Ala306Thr and the p.Ala138Glu variants, required revision CI surgery due to hardware infection.
CI outcomes in patients with TMPRSS3-mediated hearing loss are similar to post-lingual adults
Nine large studies of more than 10 patients reported post-lingual adult CI outcomes for 565 individuals and these patients exhibited a mean post-operative CNC word score of 51.2% (22.2%) (Table 2)[24–31]. Of the seven studies with pre- and post-operative CNC scores recorded, the average improvement in CNC score (defined as final score minus initial score) was 40.4%. An ANOVA test found no significant difference in post-operative CNC WRS between the TMPRSS3 and general hearing loss cohorts [66.2% (25.8) vs. 50.1% (12.5); F(1,6) = 1.97, P=0.21].
Table 2.
Cochlear implant outcomes in TMPRSS3-associated hearing loss (bold) and the general hearing loss population.
| Study, year | Subjects (n) | Preop CNC WRS - % [14] | Postop CNC WRS - % [14] |
|---|---|---|---|
| This Study | 30 | 17.4 (15.3) | 66.2 (25.8) |
| Kelsall et al, 2021 | 100 | 14.6 (11.6) | 65.2 (18.8) |
| Sullivan et al, 2019 | 60 | 8.4 (19) | 33 |
| Pillsbury III et al, 2018 | 73 | 30.4 (13.4) | 66.9 (18.5) |
| Cusumano et al, 2017 | 26 | 6.6 (10.1) | 42.1 (33.2) |
| O’Connell et al, 2016 | 220 | n/a | 46.6 (23.0) |
| Kamakura et al, 2016 | 17 | n/a | 40.3 (19.4) |
| Buchman et al, 2014 | 13 | 0 | 60 |
| Zwolan et al, 2001 | 56 | 2.25 (3.95) | 42.2 (23.5) |
TMPRSS3 expression is minimal in auditory neurons
To determine the spatial and quantitative expression of Tmprss3, we performed hybridization chain reaction (HCR) on mouse cochlea at P11. We observe robust expression of Tmprss3 in cells throughout the organ of Corti including Myo7A+ inner and outer hair cells (Figure 2A–D). Tmprss3 is also detected in the spindle and root cells of the lateral wall, but no expression is detected in the stria vascularis (Figure 2A–C). Very faint expression is detected in <5% of the SGNs, but not in the peripheral neuronal processes near the habenula perforate (Figure 2E&F). Based upon the location and low quantity of these SGN cells, they most likely represent type II SGNs.
Figure 2: Tmprss3 is robustly expressed in mouse otic sensory epithelium and not in type I SGNs.
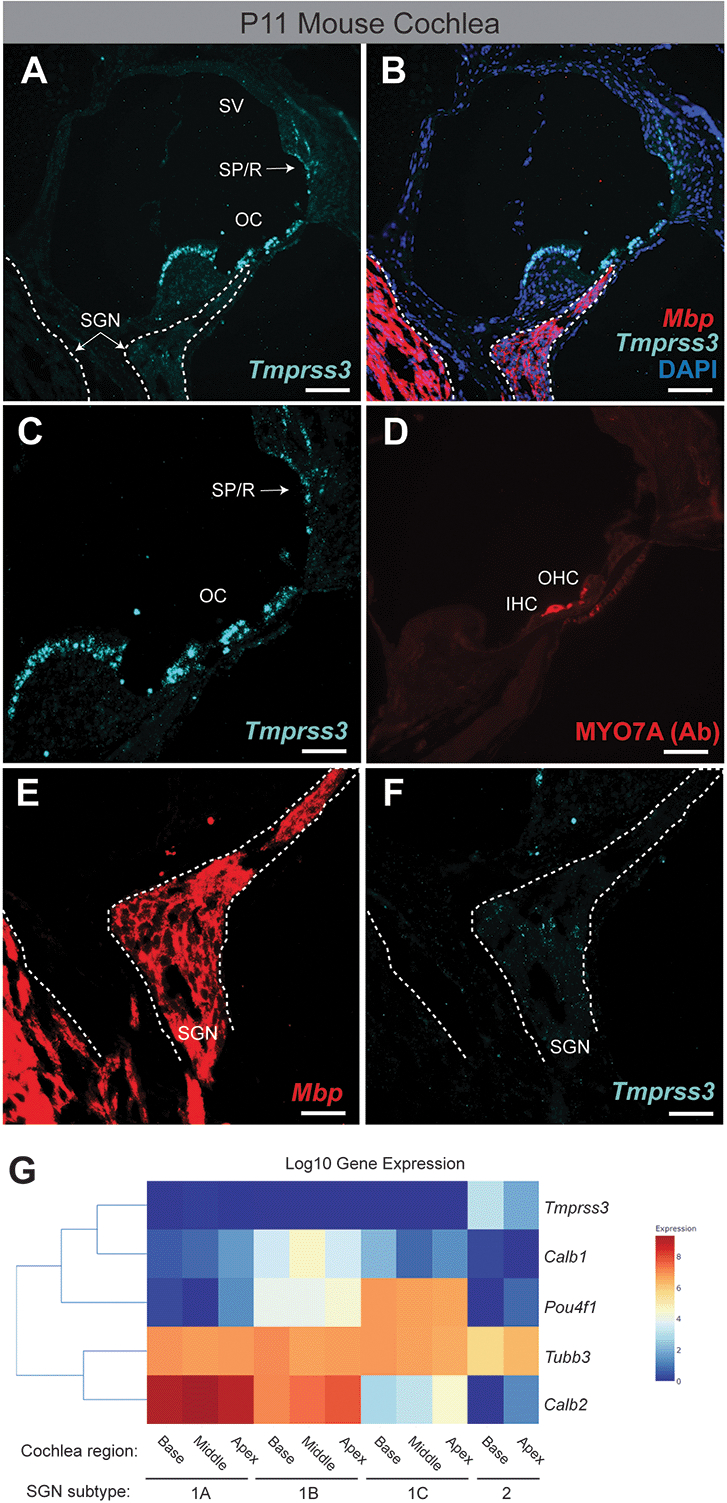
(A) Hybridization chain reaction (HCR) labeling of Tmprss3 in the P11 mouse cochlea. The SGN is outlined with a dotted line. (B) HCR labeling of Tmprss3 and Mbp to label neuron of the SGN. (C) Higher magnification insets of the otic epithelium. (D) Antibody labeling of the Myo7A+ inner and outer hair cells. (E&F) Higher magnification of the SGN showing minimal expression of Tmprss3 near the location of Type II SGNs. (G) Single-cell RNA-sequencing gene expression data for mouse SGN subtypes using gEAR (http://umgear.org). Expression data from the P25P27, mouse, scRNA-seq, cochlear sensory neurons dataset[16]. Tmprss3 is only expressed in Type II SGNs. OC, organ of Corti; SGN, spiral ganglion neurons; SP/R, spindle and root cells; SV, stria vascularis; Ab, antibody. Scale bars: 25 um (A, B); 10 um (C-F).
Next, we analyzed the expression of Tmprss3 in the publicly available Gene Expression Analysis Resource (http://umgear.org)[32]. We compared the expression of Tmprss3 across SGN subtypes (Type Ia, Ib, Ic and Type II) across three single-cell RNA-sequencing datasets from SGNs (Supplemental Figure)[15 16 33]. Consistent with our HCR data, we observed no expression of Tmprss3 in all three subtypes (A, B, C) of Type I SGNs across all three datasets (Figure 2 and Supplemental Figure). The neuronal marker TuJ1 (Tubb3 gene) was expressed across all subtypes of SGNs and all datasets (Figure 2 and Supplemental Figure). One dataset demonstrated expression of Tmprss3 in Type II SGNs in P25 – P27 mice, but expression in Type II SGNs was not seen in 9-week-old mice (Figure 2 and Supplemental Figure).
Next, we harvested human auditory nerves (HAN) from three patients undergoing translabyrinthine craniotomy for vestibular schwannoma surgery. As expected, these patients had preoperative hearing loss with speech reception thresholds (SRT) between 30–45 dB and word discrimination scores of 0% (patients 1 & 2) and 84% (patient 3) (Figure 3A). To confirm the specificity of our human HCR probe and for qRT-PCR, we cloned the human TMPRSS3 gene into an expression vector and expressed TMPRSS3 in HEK293 cells. Using quantitative RT-PCR on HAN, control HEK cells and transfected HEK293 cells we observed robust expression of TMPRSS3 in the transfected cells and only minimal expression of TMPRSS3 in the HAN (Figure 3B). As expected, we observed robust expression of myelin basic protein (MBP) in HAN and not in HEK cells (Figure 3C). Next, we performed TMPRSS3 HCR and confirmed the specificity of the probe using TMPRSS3 transfected HEK293 cells (Figure 3D&E). We did not detect TMPRSS3 expression within the HAN of either patients 2 or 3 (Figure 3F&G). We confirmed the presence of auditory neurons using TuJ1 immunostaining (Figure 3H–I).
Figure 3: TMPRSS3 expression is minimal in human auditory neurons (HAN).
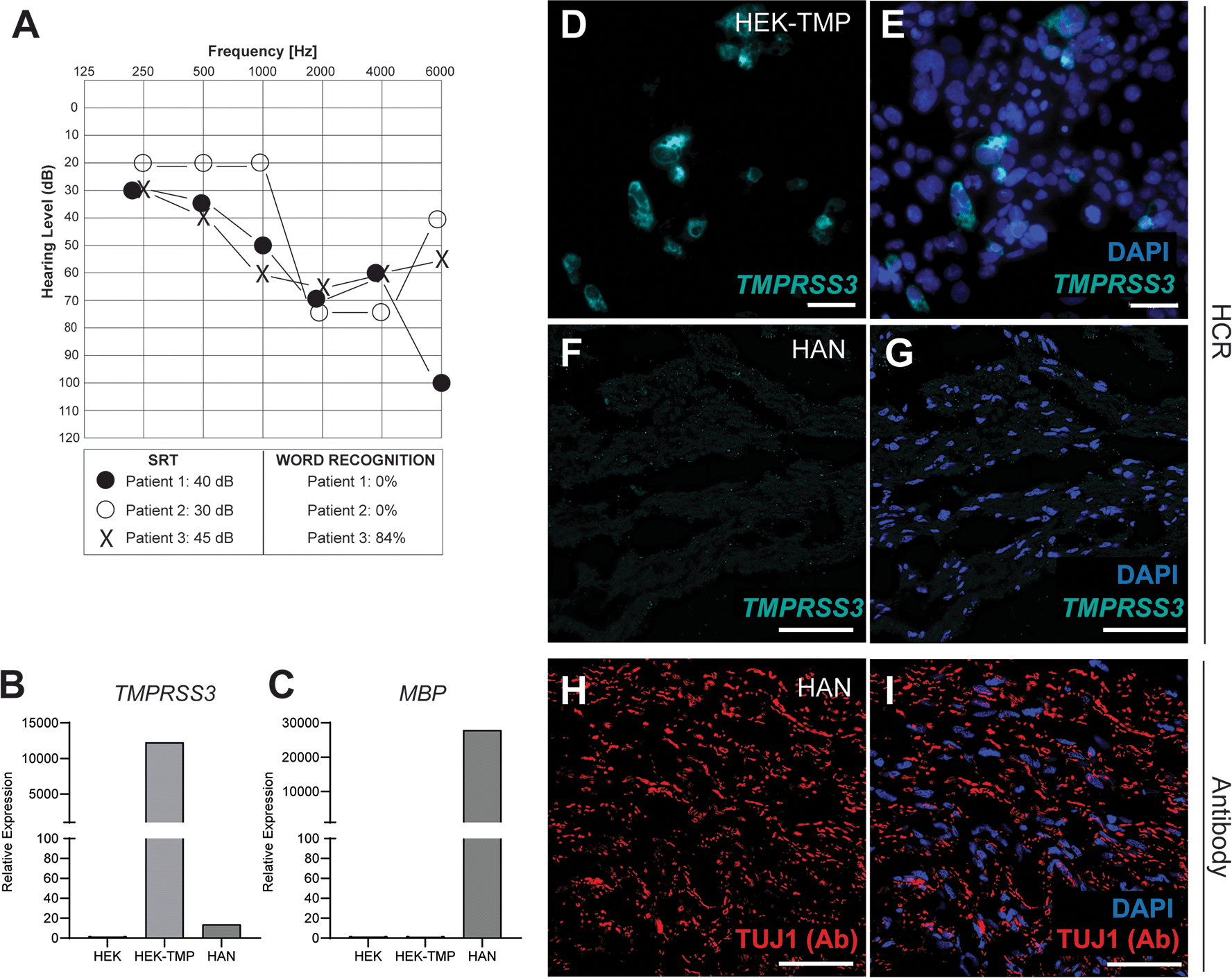
(A) Audiogram from three patients whose auditory nerves were analyzed. Circles denote right ear and X denotes the left ear. (B&C) qRT-PCR analysis of TMPRSS3 and MBP of patient #3 HAN shows minimal expression of TMPRSS3. (D&E) HCR labeling of human TMPRSS3 in TMPRSS3-tranfected HEK293 cells. (F&G) HCR of HAN with TMPRSS3 shows minimal expression of TMPRSS3. (H&I) Antibody (Ab) labeling of HAN with neuron-specific TUJ1. SRT, Speech reception threshold. Scale bar, 10μm (D, E), 20 μm (F - I).
Discussion
Determining factors that impact CI performance is critical for identifying CI candidates and managing patient expectations. CI performance is linked to both age of implantation and hearing loss etiology[23 34 35]. As genomic sequencing has become more feasible, increasing efforts are being made to determine which genetic causes may predict good or poor outcomes after implantation—and, consequently, how these patients should be counseled pre-operatively. Much of this interest has centered on TMPRSS3 and the still poorly-understood mechanism by which mutations to this gene adversely affect hearing and, possibly, CI outcomes. In this study, however, we find that patients with TMPRSS3-related hearing loss do not have worse CI outcomes compared to the general population. Furthermore, we demonstrate that Tmprss3 is highly expressed in otic sensory epithelia with limited SGN expression.
Poor CI outcomes in TMPRSS3-associated hearing loss were originally attributed to TMPRSS3 expression in SGNs[13]. However, these data were obtained with mouse tissue using an antibody that was not validated for specificity with methods such as Tmprss3-knockout tissue or western blot analysis[14]. Our compiled CI outcomes data here indicate that TMPRSS3 does not play a significant physiologic role in the neuronal aspects of hearing. Our data using validated and highly specific probes for Tmprss3 in mouse and TMPRSS3 in human tissues corroborates published scRNA-seq datasets from SGNs (Supplementary Figure 1)[15 16 33], the sensory epithelia[36 37] and the lateral wall[38].
Post-implantation outcomes in TMPRSS3 patients compared to the general population[24–27 39]. These outcomes compare similarly to a large systematic review of adult CI outcomes of 2798 patients across 46 studies and showed CNC scores improved from 8.3% (12.4) to 54.0% (22.5)[40]. The absence of significantly different outcomes between the TMPRSS3 cohort and the general population also suggest that TMPRSS3 may have no clinically relevant biological role in the auditory nerve.
Duration of deafness is one of the most important factors to predict speech perception after cochlear implantation in postligually deaf patients[23]. This may be due to progressive degeneration of SGNs. Mouse models mimicking human deafness with degeneration of sensory hair cells uniformly exhibit progressive neuronal death, a secondary effect to the primary lesion[41–43]. Loss of TMPRSS3 in mouse does not alter the total number of sensory hair cells or total number of SGNs during development[3], yet we and others have shown that loss of TMPRSS3 in mouse results in rapid HC degeneration at P12 leading to profound deafness[3 7]. Similar to other genetic causes of hearing loss in which the pathology affects the hair cell or the supporting cells of the sensory epithilium, Tmprss3-mutant mice also exhibt a delayed-onset progressive SGN degeneration started after P90[3]. The selected TMPRSS3 patients who exhibited poor performance were all implanted at a significantly older age—and, in at least two cases, with prolonged duration of hearing loss, which most likely affected the overall number and health of residual SGNs. These factors likely played a substantial role in their poor outcome[44].
In order to assess possible genotype-phenotype correlations among various TMPRSS3 allelic combinations, we compared hearing outcomes in patients with PTxPT, NPTxPT or NPTxNPT combinations. No difference in performance between these groups.
Taken together, patients with TMPRSS3-associated SNHL have comparable CI outcomes relative to the general population. Duration of deafness appears to be the key factor predictive of poor performance. While the cause of deafness in patients harboring TMPRSS3 mutations remains poorly understood, our results suggest that TMPRSS3 mutations largely affect cochlear hair cells and not the SGN, which would explain the positive CI outcomes in these patients.
Supplementary Material
Supplemental Figure 1 Single-cell RNA-sequencing gene expression data for mouse SGN subtypes using gEAR (http://umgear.org) from 3 different data sets.
Key messages.
WHAT IS ALREADY KNOWN ON THIS TOPIC
Bi-allelic variants in TMPRSS3 result in congenital and post-lingual sensorineural hearing loss. Previous reports suggest TMPRSS3 is expressed in spiral ganglion neurons (SGNs) and portend for poor cochlear implantation (CI) outcomes in patients with TMPRSS3-related hearing loss
WHAT THIS STUDY ADDS
We find that CI outcomes in patients with TMPRSS3-related hearing loss are similar to other cohorts of CI patients. Tmprss3 is not expressed in acoustic stimuli-transmitting type I SNGs of mice or humans.
HOW THIS STUDY MIGHT AFFECT RESEARCH, PRACTICE AND/OR POLICY
This study shows that patients with bi-allelic TMPRSS3-related hearing loss have excellent CI performance and argues against the physiologic role of TMPRSS3 in SGNs.
Acknowledgements:
The authors are grateful to all patients for consenting to biological sampling and data collection.
Funding:
This work was supported by the National Institutes of Health (K08-DC016034 to R.F.N.), the Triological Society and American College of Surgeons (Clinician Scientist Development Award to R.F.N.) and NIGMS T32 Training in Genetics Fellowship (T32 GM007748 to K.T.B.).
Footnotes
Declarations
Competing interests: The authors have no competing interests and declare that the research was conducted in the absence of any potential conflict of interest.
Ethics approval statement: The care and use of animals were approved by the Indiana University School of Medicine’s Institutional Animal Care and Use Committee and by the the Indiana University School of Medicine IRB (IRB approval 200634852).
REFERENCES
- 1.Szabo R, Bugge TH. Membrane-anchored serine proteases in vertebrate cell and developmental biology. Annu Rev Cell Dev Biol 2011;27:213–35 doi: 10.1146/annurev-cellbio-092910-154247[published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barre O, Dufour A, Eckhard U, Kappelhoff R, Beliveau F, Leduc R, Overall CM. Cleavage specificity analysis of six type II transmembrane serine proteases (TTSPs) using PICS with proteome-derived peptide libraries. PloS one 2014;9(9):e105984 doi: 10.1371/journal.pone.0105984[published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fasquelle L, Scott HS, Lenoir M, Wang J, Rebillard G, Gaboyard S, Venteo S, Francois F, Mausset-Bonnefont AL, Antonarakis SE, Neidhart E, Chabbert C, Puel JL, Guipponi M, Delprat B. Tmprss3, a transmembrane serine protease deficient in human DFNB8/10 deafness, is critical for cochlear hair cell survival at the onset of hearing. Journal of Biological Chemistry 2011;286(19):17383–97 doi: 10.1074/jbc.M110.190652[published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scott HS, Kudoh J, Wattenhofer M, Shibuya K, Berry A, Chrast R, Guipponi M, Wang J, Kawasaki K, Asakawa S, Minoshima S, Younus F, Mehdi SQ, Radhakrishna U, Papasavvas MP, Gehrig C, Rossier C, Korostishevsky M, Gal A, Shimizu N, Bonne-Tamir B, Antonarakis SE. Insertion of beta-satellite repeats identifies a transmembrane protease causing both congenital and childhood onset autosomal recessive deafness. Nat Genet 2001;27(1):59–63 doi: 10.1038/83768[published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 5.Fasquelle L, Scott HS, Lenoir M, Wang J, Rebillard G, Gaboyard S, Venteo S, François F, Mausset-Bonnefont AL, Antonarakis SE, Neidhart E, Chabbert C, Puel JL, Guipponi M, Delprat B. Tmprss3, a transmembrane serine protease deficient in human DFNB8/10 deafness, is critical for cochlear hair cell survival at the onset of hearing. J Biol Chem 2011;286(19):17383–97 doi: 10.1074/jbc.M110.190652[published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Battelino S, Klancar G, Kovac J, Battelino T, Trebusak Podkrajsek K. TMPRSS3 mutations in autosomal recessive nonsyndromic hearing loss. Eur Arch Otorhinolaryngol 2016;273(5):1151–4 doi: 10.1007/s00405-015-3671-0[published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 7.Tang PC, Alex AL, Nie J, Lee J, Roth AA, Booth KT, Koehler KR, Hashino E, Nelson RF. Defective Tmprss3-Associated Hair Cell Degeneration in Inner Ear Organoids. Stem Cell Reports 2019;13(1):147–62 doi: 10.1016/j.stemcr.2019.05.014[published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Seligman KL, Shearer AE, Frees K, Nishimura C, Kolbe D, Dunn C, Hansen MR, Gantz BJ, Smith RJH. Genetic Causes of Hearing Loss in a Large Cohort of Cochlear Implant Recipients. Otolaryngol Head Neck Surg 2022;166(4):734–37 doi: 10.1177/01945998211021308[published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bademci G, Foster J 2nd, Mahdieh N, Bonyadi M, Duman D, Cengiz FB, Menendez I, Diaz-Horta O, Shirkavand A, Zeinali S, Subasioglu A, Tokgoz-Yilmaz S, Huesca-Hernandez F, de la Luz Arenas-Sordo M, Dominguez-Aburto J, Hernandez-Zamora E, Montenegro P, Paredes R, Moreta G, Vinueza R, Villegas F, Mendoza-Benitez S, Guo S, Bozan N, Tos T, Incesulu A, Sennaroglu G, Blanton SH, Ozturkmen-Akay H, Yildirim-Baylan M, Tekin M. Comprehensive analysis via exome sequencing uncovers genetic etiology in autosomal recessive nonsyndromic deafness in a large multiethnic cohort. Genet Med 2016;18(4):364–71 doi: 10.1038/gim.2015.89[published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weegerink NJ, Schraders M, Oostrik J, Huygen PL, Strom TM, Granneman S, Pennings RJ, Venselaar H, Hoefsloot LH, Elting M, Cremers CW, Admiraal RJ, Kremer H, Kunst HP. Genotype-phenotype correlation in DFNB8/10 families with TMPRSS3 mutations. J Assoc Res Otolaryngol 2011;12(6):753–66 doi: 10.1007/s10162-011-0282-3[published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nishio SY, Usami SI. Outcomes of cochlear implantation for the patients with specific genetic etiologies: a systematic literature review. Acta Otolaryngol 2017;137(7):730–42 doi: 10.1080/00016489.2016.1276303[published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 12.Shearer AE, Tejani VD, Brown CJ, Abbas PJ, Hansen MR, Gantz BJ, Smith RJH. In Vivo Electrocochleography in Hybrid Cochlear Implant Users Implicates TMPRSS3 in Spiral Ganglion Function. Sci Rep 2018;8(1):14165 doi: 10.1038/s41598-018-32630-9[published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eppsteiner RW, Shearer AE, Hildebrand MS, Deluca AP, Ji H, Dunn CC, Black-Ziegelbein EA, Casavant TL, Braun TA, Scheetz TE, Scherer SE, Hansen MR, Gantz BJ, Smith RJ. Prediction of cochlear implant performance by genetic mutation: the spiral ganglion hypothesis. Hear Res 2012;292(1–2):51–8 doi: 10.1016/j.heares.2012.08.007[published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guipponi M, Toh MY, Tan J, Park D, Hanson K, Ballana E, Kwong D, Cannon PZ, Wu Q, Gout A, Delorenzi M, Speed TP, Smith RJ, Dahl HH, Petersen M, Teasdale RD, Estivill X, Park WJ, Scott HS. An integrated genetic and functional analysis of the role of type II transmembrane serine proteases (TMPRSSs) in hearing loss. Hum Mutat 2008;29(1):130–41 doi: 10.1002/humu.20617[published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 15.Sun S, Babola T, Pregernig G, So KS, Nguyen M, Su SM, Palermo AT, Bergles DE, Burns JC, Müller U. Hair Cell Mechanotransduction Regulates Spontaneous Activity and Spiral Ganglion Subtype Specification in the Auditory System. Cell 2018;174(5):1247–63.e15 doi: 10.1016/j.cell.2018.07.008[published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shrestha BR, Chia C, Wu L, Kujawa SG, Liberman MC, Goodrich LV. Sensory Neuron Diversity in the Inner Ear Is Shaped by Activity. Cell 2018;174(5):1229–46.e17 doi: 10.1016/j.cell.2018.07.007[published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Azaiez H, Booth KT, Ephraim SS, Crone B, Black-Ziegelbein EA, Marini RJ, Shearer AE, SloanHeggen CM, Kolbe D, Casavant T, Schnieders MJ, Nishimura C, Braun T, Smith RJH. Genomic Landscape and Mutational Signatures of Deafness-Associated Genes. Am J Hum Genet 2018;103(4):484–97 doi: 10.1016/j.ajhg.2018.08.006[published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oza AM, DiStefano MT, Hemphill SE, Cushman BJ, Grant AR, Siegert RK, Shen J, Chapin A, Boczek NJ, Schimmenti LA, Murry JB, Hasadsri L, Nara K, Kenna M, Booth KT, Azaiez H, Griffith A, Avraham KB, Kremer H, Rehm HL, Amr SS, Abou Tayoun AN, ClinGen Hearing Loss Clinical Domain Working G. Expert specification of the ACMG/AMP variant interpretation guidelines for genetic hearing loss. Hum Mutat 2018;39(11):1593–613 doi: 10.1002/humu.23630[published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miyagawa M, Nishio SY, Ikeda T, Fukushima K, Usami S. Massively parallel DNA sequencing successfully identifies new causative mutations in deafness genes in patients with cochlear implantation and EAS. PLoS One 2013;8(10):e75793 doi: 10.1371/journal.pone.0075793[published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chung J, Park SM, Chang SO, Chung T, Lee KY, Kim AR, Park JH, Kim V, Park WY, Oh SH, Kim D, Park WJ, Choi BY. A novel mutation of TMPRSS3 related to milder auditory phenotype in Korean postlingual deafness: a possible future implication for a personalized auditory rehabilitation. J Mol Med (Berl) 2014;92(6):651–63 doi: 10.1007/s00109-014-1128-3[published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 21.Song MH, Jung J, Rim JH, Choi HJ, Lee HJ, Noh B, Lee JS, Gee HY, Choi JY. Genetic Inheritance of Late-Onset, Down-Sloping Hearing Loss and Its Implications for Auditory Rehabilitation. Ear Hear 2020;41(1):114–24 doi: 10.1097/aud.0000000000000734[published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 22.Miyagawa M, Nishio SY, Sakurai Y, Hattori M, Tsukada K, Moteki H, Kojima H, Usami S. The patients associated with TMPRSS3 mutations are good candidates for electric acoustic stimulation. Ann Otol Rhinol Laryngol 2015;124 Suppl 1:193s–204s doi: 10.1177/0003489415575056[published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 23.Bernhard N, Gauger U, Romo Ventura E, Uecker FC, Olze H, Knopke S, Hansel T, Coordes A. Duration of deafness impacts auditory performance after cochlear implantation: A meta-analysis. Laryngoscope Investig Otolaryngol 2021;6(2):291–301 doi: 10.1002/lio2.528[published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Buchman CA, Dillon MT, King ER, Adunka MC, Adunka OF, Pillsbury HC. Influence of cochlear implant insertion depth on performance: a prospective randomized trial. Otol Neurotol 2014;35(10):1773–9 doi: 10.1097/mao.0000000000000541[published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 25.Pillsbury HC 3rd, Dillon MT, Buchman CA, Staecker H, Prentiss SM, Ruckenstein MJ, Bigelow DC, Telischi FF, Martinez DM, Runge CL, Friedland DR, Blevins NH, Larky JB, Alexiades G, Kaylie DM, Roland PS, Miyamoto RT, Backous DD, Warren FM, El-Kashlan HK, Slager HK, Reyes C, Racey AI, Adunka OF. Multicenter US Clinical Trial With an Electric-Acoustic Stimulation (EAS) System in Adults: Final Outcomes. Otol Neurotol 2018;39(3):299–305 doi: 10.1097/mao.0000000000001691[published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kelsall D, Lupo J, Biever A. Longitudinal outcomes of cochlear implantation and bimodal hearing in a large group of adults: A multicenter clinical study. Am J Otolaryngol 2021;42(1):102773 doi: 10.1016/j.amjoto.2020.102773[published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 27.O’Connell BP, Cakir A, Hunter JB, Francis DO, Noble JH, Labadie RF, Zuniga G, Dawant BM, Rivas A, Wanna GB. Electrode Location and Angular Insertion Depth Are Predictors of Audiologic Outcomes in Cochlear Implantation. Otol Neurotol 2016;37(8):1016–23 doi: 10.1097/mao.0000000000001125[published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zwolan T, Kileny PR, Smith S, Mills D, Koch D, Osberger MJ. Adult cochlear implant patient performance with evolving electrode technology. Otol Neurotol 2001;22(6):844–9 doi: 10.1097/00129492-200111000-00022[published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 29.Kamakura T, Nadol JB, Jr. Correlation between word recognition score and intracochlear new bone and fibrous tissue after cochlear implantation in the human. Hear Res 2016;339:132–41 doi: 10.1016/j.heares.2016.06.015[published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cusumano C, Friedmann DR, Fang Y, Wang B, Roland JT Jr., Waltzman SB. Performance Plateau in Prelingually and Postlingually Deafened Adult Cochlear Implant Recipients. Otol Neurotol 2017;38(3):334–38 doi: 10.1097/mao.0000000000001322[published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 31.Sullivan CB, Al-Qurayshi Z, Zhu V, Liu A, Dunn C, Gantz BJ, Hansen MR. Long-term audiologic outcomes after cochlear implantation for single-sided deafness. Laryngoscope 2020;130(7):1805–11 doi: 10.1002/lary.28358[published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Orvis J, Gottfried B, Kancherla J, Adkins RS, Song Y, Dror AA, Olley D, Rose K, Chrysostomou E, Kelly MC, Milon B, Matern MS, Azaiez H, Herb B, Colantuoni C, Carter RL, Ament SA, Kelley MW, White O, Bravo HC, Mahurkar A, Hertzano R. gEAR: Gene Expression Analysis Resource portal for community-driven, multi-omic data exploration. Nat Methods 2021;18(8):843–44 doi: 10.1038/s41592-021-01200-9[published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Milon B, Shulman ED, So KS, Cederroth CR, Lipford EL, Sperber M, Sellon JB, Sarlus H, Pregernig G, Shuster B, Song Y, Mitra S, Orvis J, Margulies Z, Ogawa Y, Shults C, Depireux DA, Palermo AT, Canlon B, Burns J, Elkon R, Hertzano R. A cell-type-specific atlas of the inner ear transcriptional response to acoustic trauma. Cell Rep 2021;36(13):109758 doi: 10.1016/j.celrep.2021.109758[published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blamey P, Arndt P, Bergeron F, Bredberg G, Brimacombe J, Facer G, Larky J, Lindstrom B, Nedzelski J, Peterson A, Shipp D, Staller S, Whitford L. Factors affecting auditory performance of postlinguistically deaf adults using cochlear implants. Audiol Neurootol 1996;1(5):293–306 doi: 10.1159/000259212[published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 35.Lazard DS, Vincent C, Venail F, Van de Heyning P, Truy E, Sterkers O, Skarzynski PH, Skarzynski H, Schauwers K, O’Leary S, Mawman D, Maat B, Kleine-Punte A, Huber AM, Green K, Govaerts PJ, Fraysse B, Dowell R, Dillier N, Burke E, Beynon A, Bergeron F, Baskent D, Artieres F, Blamey PJ. Pre-, per- and postoperative factors affecting performance of postlinguistically deaf adults using cochlear implants: a new conceptual model over time. PLoS One 2012;7(11):e48739 doi: 10.1371/journal.pone.0048739[published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kolla L, Kelly MC, Mann ZF, Anaya-Rocha A, Ellis K, Lemons A, Palermo AT, So KS, Mays JC, Orvis J, Burns JC, Hertzano R, Driver EC, Kelley MW. Characterization of the development of the mouse cochlear epithelium at the single cell level. Nat Commun 2020;11(1):2389 doi: 10.1038/s41467-020-16113-y[published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kubota M, Scheibinger M, Jan TA, Heller S. Greater epithelial ridge cells are the principal organoid-forming progenitors of the mouse cochlea. Cell Rep 2021;34(3):108646 doi: 10.1016/j.celrep.2020.108646[published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Korrapati S, Taukulis I, Olszewski R, Pyle M, Gu S, Singh R, Griffiths C, Martin D, Boger E, Morell RJ, Hoa M. Single Cell and Single Nucleus RNA-Seq Reveal Cellular Heterogeneity and Homeostatic Regulatory Networks in Adult Mouse Stria Vascularis. Front Mol Neurosci 2019;12:316 doi: 10.3389/fnmol.2019.00316[published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Teagle HFB, Park LR, Brown KD, Zdanski C, Pillsbury HC. Pediatric cochlear implantation: A quarter century in review. Cochlear Implants Int 2019;20(6):288–98 doi: 10.1080/14670100.2019.1655868[published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 40.Boisvert I, Reis M, Au A, Cowan R, Dowell RC. Cochlear implantation outcomes in adults: A scoping review. PLoS One 2020;15(5):e0232421 doi: 10.1371/journal.pone.0232421[published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Giese APJ, Tang YQ, Sinha GP, Bowl MR, Goldring AC, Parker A, Freeman MJ, Brown SDM, Riazuddin S, Fettiplace R, Schafer WR, Frolenkov GI, Ahmed ZM. CIB2 interacts with TMC1 and TMC2 and is essential for mechanotransduction in auditory hair cells. Nat Commun 2017;8(1):43 doi: 10.1038/s41467-017-00061-1[published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Calabro KR, Boye SL, Choudhury S, Fajardo D, Peterson JJ, Li W, Crosson SM, Kim MJ, Ding D, Salvi R, Someya S, Boye SE. A Novel Mouse Model of MYO7A USH1B Reveals Auditory and Visual System Haploinsufficiencies. Front Neurosci 2019;13:1255 doi: 10.3389/fnins.2019.01255[published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pawlowski KS, Kikkawa YS, Wright CG, Alagramam KN. Progression of inner ear pathology in Ames waltzer mice and the role of protocadherin 15 in hair cell development. J Assoc Res Otolaryngol 2006;7(2):83–94 doi: 10.1007/s10162-005-0024-5[published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Holden LK, Finley CC, Firszt JB, Holden TA, Brenner C, Potts LG, Gotter BD, Vanderhoof SS, Mispagel K, Heydebrand G, Skinner MW. Factors affecting open-set word recognition in adults with cochlear implants. Ear Hear 2013;34(3):342–60 doi: 10.1097/AUD.0b013e3182741aa7[published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1 Single-cell RNA-sequencing gene expression data for mouse SGN subtypes using gEAR (http://umgear.org) from 3 different data sets.


