Abstract
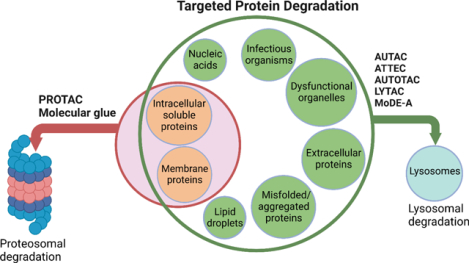
Among the scope of targeted protein degradation (TPD), Proteolysis Targeting Chimeras (PROTACs), leveraging the ubiquitin-proteasome system, have been extensively studied. However, they are limited to degrading soluble and membrane proteins, excluding the aggregated and extracellular proteins and dysfunctional organelles. As an alternative protein degradation pathway, lysosomes serve as a feasible tool to access these untouched proteins/organelles by proteosomes. Here, we focus on reviewing the emerging lysosome mediated TPD, such as AUTAC, ATTEC, AUTOTAC, LYTAC, and MoDE-A. Intracellular targets, such as soluble and aggregated proteins and organelles, can be degraded via the autophagy-lysosome pathway. Extracellular targets, such as membrane proteins, and secreted extracellular proteins can be degraded via the endosome-lysosome pathway. In addition, we summarize the mechanism and regulation of autophagy, available methods/assays monitoring the autophagy process, and the recently developed chemical probes for autophagy pathways.
Graphical Abstract
INTRODUCTION
Protein homeostasis is a coordinated and complex web of building blocks that maintains the cellular concentrations, folding, interactions, and localization of proteins essential for cellular functions. One vital role protein homeostasis plays is the clearance of unwanted misfolded proteins or proteins that fail to fold due to mutations.1 It has been demonstrated that the Ubiquitin-proteosome system (UPS) and lysosomal system are the two principal and complementary approaches for protein degradation.2,3 The UPS pathway degrades intracellular, soluble, and short-lived proteins.4,5 On the other hand, the lysosomal system can degrade many fully folded, long-lived, aggregated proteins, extracellular proteins, nucleic acids, lipids, damaged organelles and infectious organisms such as bacteria and viruses.6 In the past decade, proteolysis-targeting chimeras (PROTACs), leveraging the proteasome pathway, have made a significant progress since they emerged in 2001.7–9 Recently, several PROTACs have entered clinical trials, which demonstrated their potential in medicinal chemistry and chemical biology.10 However, the UPS related small molecule-based PROTACs approach has some limitations since it can only degrade intracellular, soluble, and short-lived proteins, limiting their applications in many diseases caused by extracellular proteins and protein aggregates, such as in Huntington’s and Alzheimer’s disease. Therefore, lysosomal degradation of biomolecules can greatly enrich the toolbox of targeted protein degradation (TPD) and expand their applications in human diseases. Lysosomes are ubiquitous acidic organelles that can degrade proteins, nucleic acids and other biomaterials. One of their cellular functions is to degrade and recycle the intracellular and extracellular materials using acidic hydrolases.11,12 There are two major lysosomal-based degradation pathways: the degradation of cytoplasmic proteins and damaged organelles by lysosome through autophagy called autophagy-lysosomal pathway and the degradation of extracellular proteins by lysosome through endocytosis called endosome-lysosomal pathway.
Autophagy-lysosomal pathway
Autophagy is a conserved catabolic process for the turnover and recycling of cytoplasmic components, such as proteins and organelles, which are then trafficked into lysosomes.13–15 Autophagy has a crucial role in maintaining cells’ homeostasis and energy balance. Any disruption in the autophagy process causes various diseases such as cancers, neurodegenerative disorders (NDDs), immune disorders, and metabolic diseases.16 Autophagy has multifaceted roles in cancers, depending on the tumor genotypes and therapeutic agents. On one hand, it controls tumor growth by removing cancer-causing cells and organelles. On the other hand, it protects tumor cells from therapy-induced death and helps to promote tumor growth.17,18 Additionally, the autophagic removal of aggregated proteins and organelles from the heart is beneficial since autophagic death of unwanted cardiac cells may lead to heart failure.19 Autophagy can also be used as a defense mechanism against intracellular pathogens.20
Autophagy is divided into three types, namely microautophagy,21,22 chaperon-mediated autophagy (CMA)23,24, and macroautophagy25. Microautophagy is the direct engulfment of cytoplasmic components by lysosomes, whereas chaperone-mediated autophagy degrades specific proteins containing a KFERQ-like motif recognized by the molecular chaperon HSPA8/HSC70, which directs the protein to the lysosomal surface protein LAMP2A for lysosomal engulfment. Macroautophagy (hereafter autophagy) involves the formation of a double-membrane vesicle called an autophagosome. The targeted, degraded material is trapped inside the autophagosome, which is later delivered into the lysosome through membrane fusion between the autophagosome and lysosome.26 The mechanism of the autophagic process is highly regulated by autophagy-related proteins. More than 40 autophagy-related genes and their encoding proteins have been identified in yeast, and most of them are also present in mammals, which indicates that autophagy is an evolutionarily conserved process.27
Mechanism and regulation of autophagy
The regulation of autophagy is a complex mechanism involving several autophagy-related (Atg) proteins. The autophagy mechanism consists of two ubiquitin-like conjugation systems resulting in the modified complexes of autophagy regulators LC3-II and Atg12-Atg5-Atg16L, respectively.28 The autophagy machinery can be divided into three stages: a) induction of autophagy b) nucleation, elongation, and maturation of autophagosome; and c) fusion with lysosome and degradation (Figure 1).
Figure 1:
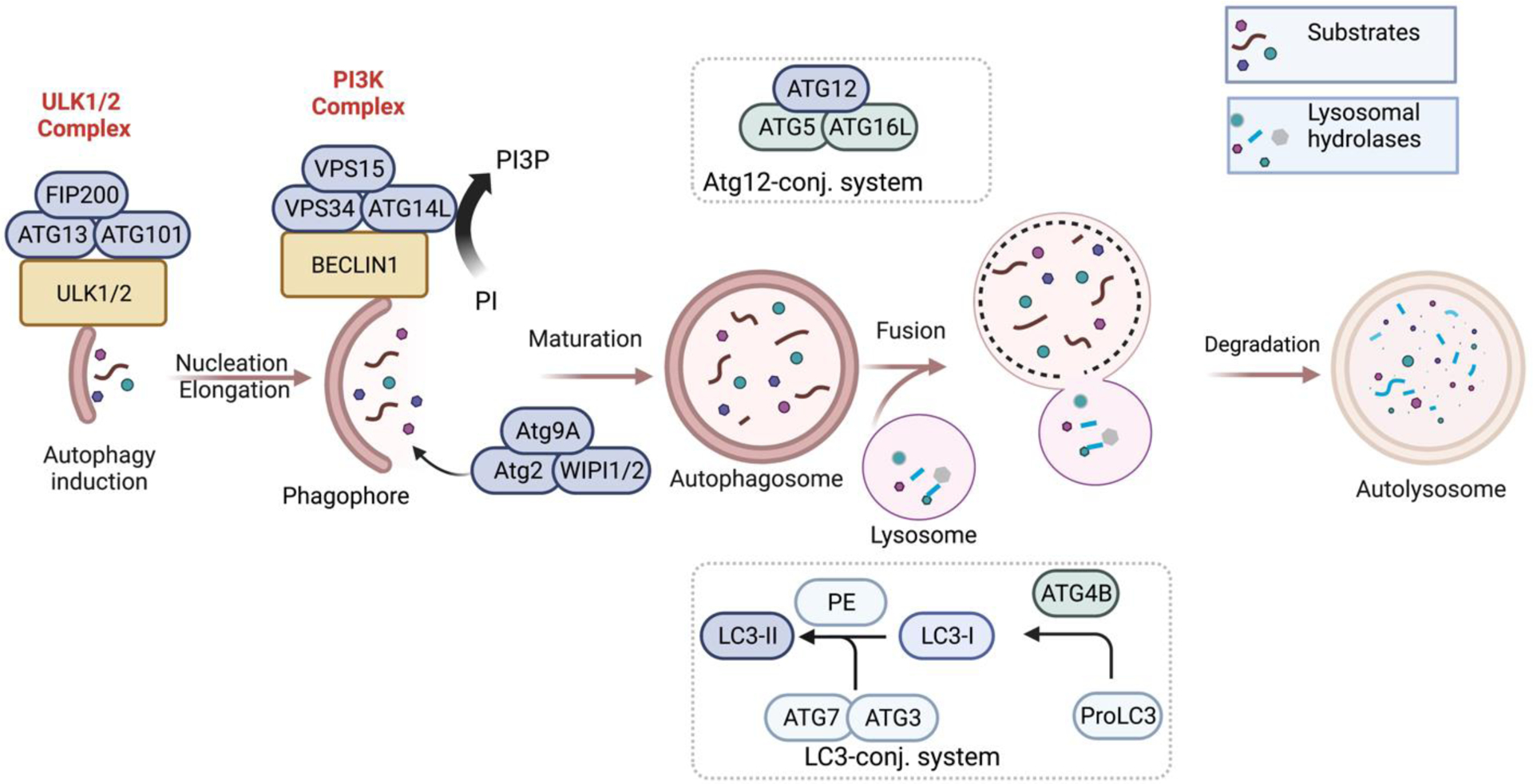
Schematic representation of autophagy.
Autophagy occurs at a low level under basal conditions and is triggered by cellular stress such as nutrient starvation, oxidative stress, and mTORC1 inhibition.29–31 The initiation of autophagosome formation occurs after the activation of ULK1/2 (unc-51 like kinase) proteins which can form ULK1/2 complexes with other proteins such as Atg13, Atg101, and FIP200.32 Autophagy nucleation is facilitated by forming the class lll phosphatidylinositol 3-kinase (PI3K) complex containing VPS34, VPS15, Beclin 1, and Atg14L.33,34 The PI3K complex also helps to phosphorylate phosphatidylinositol (PI) to phosphatidylinositol-3-phosphate (PI3P). PI3P is needed for the correct localization of Atg proteins Atg18 (WIPI1/2) and Atg2, which help to recruit Atg9 protein to the autophagosome during its nucleation step.27,35 Atg9 vesicles are the source of autophagosome membranes in nucleation step by coalescence with the ULK1 complex.36 The elongation or maturation step involves two ubiquitin-like conjugated systems, LC3-II (Atg 8 in yeast) and Atg12-Atg5-Atg16L.37 ATG4B cleaves proLC3 isoforms to form LC3-I, where the glycine residue will be the C-terminus.38 LC3-I is activated by Atg 7 (E1-like) and conjugated with Atg 3 (E2-like), followed by covalent binding to phosphatidylethanolamine (PE). This results in the lipidated LC3 (i.e., LC-3-I-PE or LC3-II). LC3-II is covalently bound to the membrane of the autophagosome.
GABARAP is another Atg 8 homologue in mammals, which is less explored than LC3.37 Atg4B also plays a role to cleave LC3-II to LC3-I referred as delipidation or deconjugation, which helps to recycle LC3.39,40 Similarly, Atg12 is activated and conjugated by Atg7 (E1-like) and Atg10 (E2-like) proteins, respectively, followed by association with Atg5 and Atg16L, resulting in an E3-like complex Atg12-Atg5-Atg16L. The Atg12-Atg5-Atg16L complex, which helps the PE-conjugation, dissociates after autophagosome formation.38,41 Atg conjugation systems are not essential for the autophagosome formation, although they facilitate and normally occur during autophagosome formation. Most importantly, Atg3 from the conjugation system is required for the opening and efficient degradation of the autophagosomal inner membrane after fusing with lysosomes.42 LC3-II acts as a binding platform for autophagy receptors (for example, p62, NBR1).43 The receptor proteins help traffic the double-membrane autophagosome to the lysosome. With the help of Rab-SNARE proteins, the fusion of lysosome and autophagosome results in autolysosomes44,45 which release their inner components as well as inner membrane into the lysosome hydrolase for their degradation.26,27,35,46,47
Selectivity in autophagy
Although autophagy was considered non-selective initially, recent studies have revealed that certain types of macroautophagy are selective.48 Most studied selective autophagy is controlled by the cargo receptor proteins, which can recognize cargos and bind to the LC3 proteins located on the isolation membrane/phagophore.49 Selective autophagy of cell organelles has been classified based on the type of organelles that act as cargos, such as mitochondria (mitophagy), protein aggregates (aggrephagy), pathogens (xenophagy), ribosomes (ribophagy), lysosome (lysophagy), liposome (lipophagy), endoplasmic reticulum (ER-phagy or reticulophagy), and ferritin (ferritinophagy).50–57 The general mechanistic pathway for selective autophagy in mammals are regulated by autophagy receptors such as p62, NBR1, NDP52, TAX1BP1, OPTN58,59 and CCT2.60 During the selective autophagy processes, autophagy receptors undergo post-translational and structural modifications, such as ubiquitylation, phosphorylation, acetylation, and oligomerization.43 In the ubiquitin-dependent cargo selection, misfolded proteins are tagged with a polyubiquitin chain which is recognized and bound to receptors through their ubiquitin-binding domain (UBD) and ultimately delivered into the autophagosome. In contrast, the ubiquitin-independent pathway involves the recognition of the specific cargos such as proteins, lipids, or sugar-based signals by the specialized autophagy receptors.61,62 For example, Dowdle et al. discovered that the selective autophagy of ferritin (i.e., ferritinophagy) is mediated by the ubiquitin-independent receptor NCOA4.57,63
p62/SQSTM1 as a receptor protein for selective autophagy
p62/SQSTM1 is a specific autophagy receptor protein found in the metazoans.64 p62 plays a crucial role in the selective autophagic degradation of ubiquitinated protein aggregates and contains several conserved domains to bind various substrates.65,66 The p62 domains facilitate the degradation of ubiquitinated cargo by binding to the UBD, undergo self-oligomerization through the PB1 domain, and then delivering the protein aggregates to the autophagosome by interacting with the autophagosome membrane protein LC3 via LC3 interacting region (LIR) (Figure 2).67–70
Figure 2:
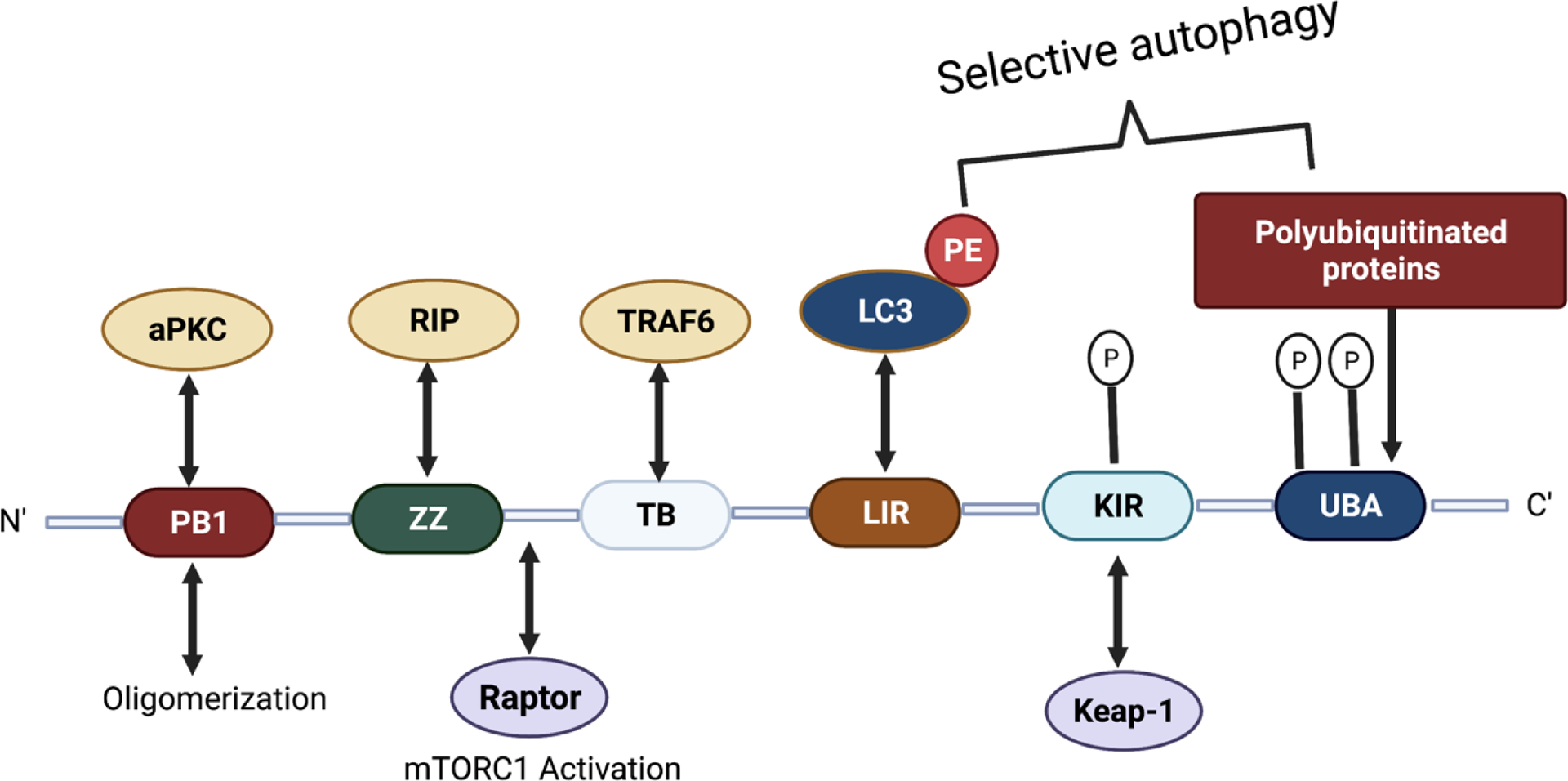
Schematic representation of p62 structure and functional domains.
Some proteolytic systems are based on recognizing N-terminal residues (N-recognins) as essential components for their degradation (N-degrons), which is called the N-end rule pathway.71,72 Cha-Molstad et al. studied the mechanism of the p62 binding with ER-residing proteins BiP for the autophagy of misfolded cytosolic proteins tagged with ubiquitin (Figure 3), which is evident that p62 is a critical molecule in the crosstalk between UPS and autophagy.73 In this study, they observed that p62 follows the N-end rule pathway to the autophagosome biogenesis.66
Figure 3:
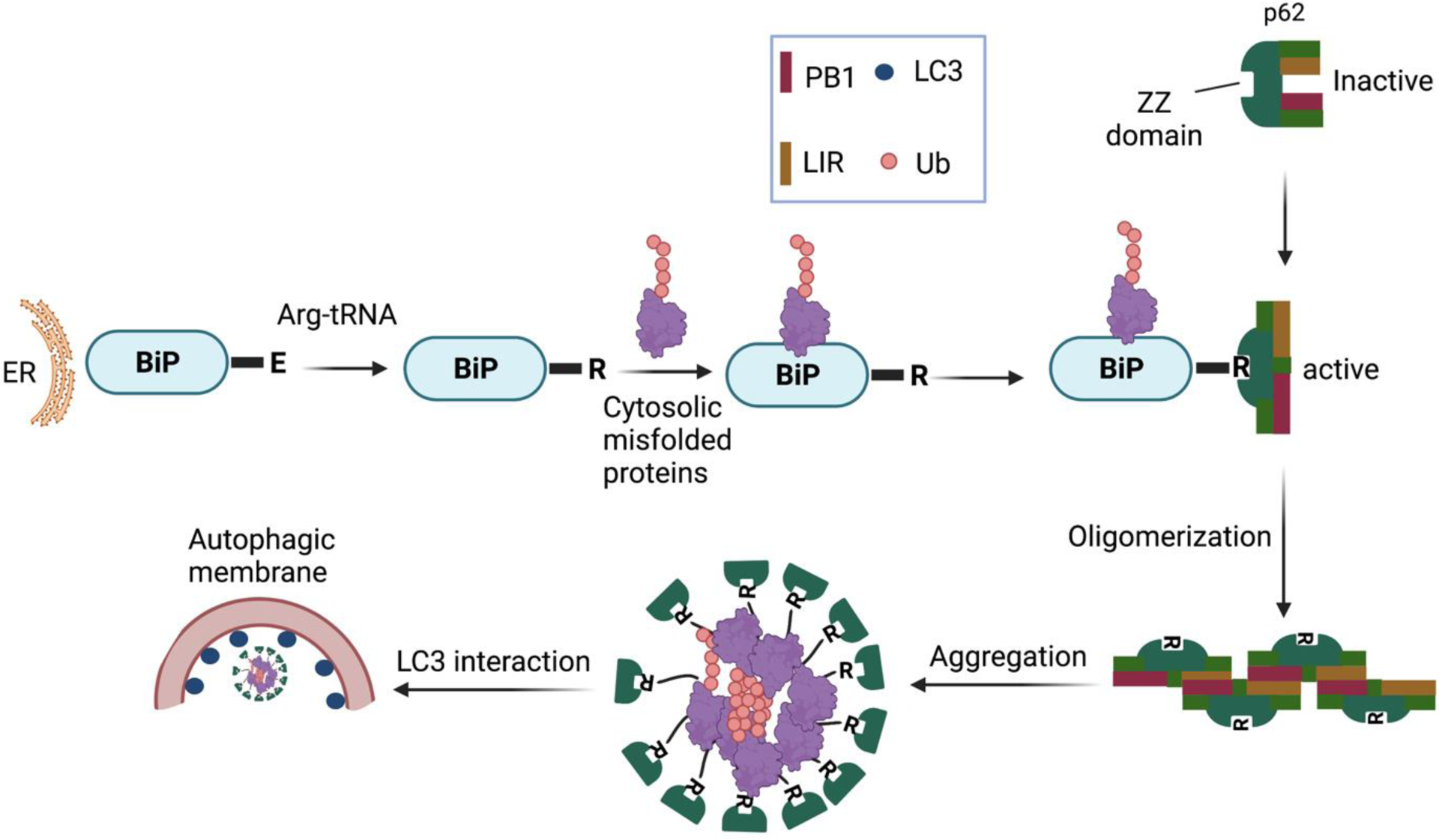
A model illustrating the role of the N-end rule pathway in N-terminal arginylation of ER-residing proteins for the regulation of autophagy through p62 binding.73,74
The ZZ domain of p62 also plays a role in inducing autophagy by selectively modulating the N-recognin site and binding to N-terminal degrons, including N-terminal arginine (Nt-R) (Figure 3). The binding of the arginylated substrates (Nt-R) to the ZZ domain of p62 facilitates disulfide bond-linked p62 self-aggregation and its interaction with LC3, which ultimately leads to the delivery of p62 and its cargo into the autophagosome.66,67 Cha-Molstad et al. developed two small molecule ligands (XIE62–1004 and XIE2008) binding to the ZZ domain of p62, which were also able to induce autophagy through p62 self-aggregation and LC3 interaction (Figure 4).74
Figure 4:
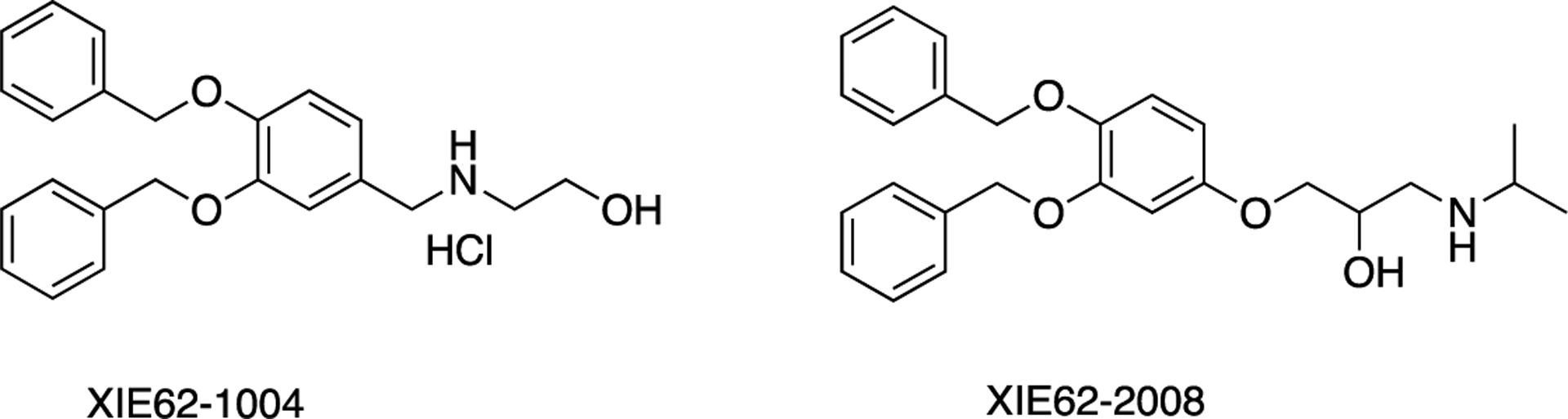
Structures of XIE62–1004 and XIE2008.
Autophagy assays
Autophagy has been extensively studied in the biomedical field. However, one of the significant challenges in the field is the limited number of methods to accurately measure autophagic activity in cells. Historically, an increase in the number of autophagosomes measured by electron microscopy has been one of the methods to measure autophagy, but autophagy is a highly dynamic process involving several steps.75 Measuring only the increase in the number of autophagosomes can be misleading since it can either mean the induction of autophagy by starvation or other stress factors or the reduction of lysosomal degradation of the autophagosomes due to lysosomal dysfunction. Measuring the autophagic flux has become a suitable alternative for autophagic activity in cells.76 Autophagy flux refers to the amount of degradation of cytoplasmic materials per unit time. In the last few years, many assays have been developed which can measure autophagic flux with excellent reliability. These assays are based on cellular expressions of specific proteins involved in autophagy, such as microtubule associated protein LC3 and autophagy receptor protein p62.76–78
1). Monitoring autophagy flux using LC3
The following three LC3-based methods have been widely used to measure the autophagic flux.
(i). LC3 turnover assay:
LC3, a mammalian homologue of yeast Atg8, has been used as a marker of autophagosome formation. As discussed before, ATG4 processes LC3 to become LC3-I which is subsequently conjugated to phosphatidylethanolamine (PE) to become LC3-II. LC3-II is found in autophagosomes and can indicate the formation of the same. LC3 turnover has been used to measure autophagic flux. Western blot has been used to visualize both LC3-I and LC3-II. The experiments are done in the presence and absence of lysosome inhibitors, and the comparison of LC3-II amounts in samples is semi-quantitatively used to determine the autophagic flux. When flux is high, the difference in LC-II between the samples will be high and vice-versa.25 Commonly used lysosomal inhibitors are lysosomal protease inhibitors, such as E64d and pepstatin A, and bafilomycin A1, and lysosomotropic reagents, such as chloroquine.79 The main advantage of this method is that it measures endogenous autophagic flux without transfection. However, the method has many drawbacks. Firstly, lysosomal inhibition can interfere with mTOR activity, resulting in further acceleration of autophagic activity. Secondly, care must be taken in selecting the type and concentration of the lysosomal inhibitor. For example, 100 nM of bafilomycin A1 blocks the fusion of autophagosomes with lysosomes affecting the measurement of autophagic flux.80
(ii). RFP-GFP-LC3 (tfLC3) reporter assay
In 2007, Kimura et al. reported an autophagy probe that analyzed the dynamics of autophagosomes using fluorescence.81 Prior to discovering an autophagy probe, scientists used GFP-LC3 as a marker probe to demonstrate the formation of autophagosomes. However, it was evident that autophagosome formation did not correlate directly to autophagy flux.82 The new probe was a novel marker protein, mRFP-GFP-LC3 tandem-tagged fluorescent protein (tfLC3), which emits both green and red fluorescence so that autophagosomes appear yellow. Once it is trafficked to autolysosome, the GFP loses fluorescence quickly due to low pH, but RFP maintains its fluorescence. The appearance of yellow or red fluorescence indicates the presence of more autophagosomes or autolysosomes. The tfLC3 probe offers many advantages over the earlier systems. For example, it can measure autophagic flux without using lysosomal inhibitors, and it can be used for selective substrates. The disadvantage to using the probe is that the system depends on transfection, and there is high background due to RFP accumulation in lysosomes.
(iii). GFP-LC3-RFP(LC3ΔG) reporter assay
In 2018, Kaizuk et al. developed GFP-LC3-RFP(LC3ΔG), a second-generation tandem-tagged fluorescent assay single molecular probe.83 In this probe, the GFP-LC3 is conjugated to the N-terminal of RFP-LC3ΔG (ΔG means the RFP-LC3 lacks the C-terminal glycine). In cells, the probe is hydrolyzed by ATG4 family proteases generating equimolar amounts of GFP-LC3 and RFP-LC3ΔG. GFP-LC3 is conjugated to PE, localizes in autophagosome, and is subsequently degraded. Whereas the RFP-LC3ΔG is not degraded due to the lack of the C-terminal glycine. As a result, the RFP-LC3ΔG acts as an internal control and stays in the cytosol. The GFP/RFP signal ratio inversely correlates to autophagic activity. The authors applied this probe to measure autophagic flux in cells, in addition to mice and zebrafish. The probe was also used to do a high throughput screening of 1054 approved drugs to find novel autophagy inducers and inhibitors. Another advantage of this probe is that it can measure the basal autophagic activity among different tissues since the basal autophagic activity is generally too low to be measured by other probes. A significant limitation of this probe is that during transfection, a substantial proportion of clones express GFP-LC3ΔG. Thus, the authors recommend isolating clones expressing GFP-LC3-RFP-LC3ΔG after transfection.
(2). Monitoring autophagy flux using p62
p62, also called sequestosome 1 in humans, binds directly to LC3 and polyubiquitinated substrates. It becomes incorporated into the autophagosome and is itself degraded into autolysosome, thus acts as a marker of autophagic flux.76,78 The subcellular localization and level of endogenous p62 can be measured by western blotting and immunostaining.78 p62 is a multifunctional scaffold protein so it is important to validate autophagy measurement of p62 level with other available assays.
Luciferase assay-based method is also developed for the measurement of both LC3 and p62 quantification. Farakas et al84 have developed the luciferase assay-based method for LC3 measurement in autophagic flux which was later extended to p62 measurement by Min et al.77 In this method, the ratio of luciferase activity with p62 and its UBA domain deletion mutant has been used to determine the autophagic flux. Similarly, Bresciani et al.85 have developed TR-FRET assays for LC3B and p62. These assays can be used as high-throughput screening tools to identify the autophagy regulators. In this method, they have used Tb labeled LC3-II donor and D2 labelled LC3-II acceptor antibodies or Tb labelled donor p62 and Alexa-647 labeled acceptor p62 antibodies. Autophagy up-regulator induces the close proximity of LC3-II antibodies resulting in the signal for accumulation of the autophagosome vesicle.85
There are some other assay methods monitoring the autophagy markers such as ULK1, PtdIns3K, Atg9, Atg12-Atg5, Atg14, Atg16L1, WIPI family, BECN1, and STX17 (SNARE protein). Autophagic components other than LC3-family can be monitored to define specific steps of the process.76
Chemical probes for autophagy pathways:
Since tumor cells can activate autophagy to compensate for the energy shortage during cellular stress, several autophagy inhibitors targeting the Atg-related proteins have been developed to control this tumor growth mechanism by autophagy.86,87 There are some studies on the repurposed drugs for autophagy regulation, such as chloroquine and hydroxychloroquine (clinically approved anti-malarial drugs) as autophagy inhibitors.88 Some of the recently discovered chemical probes for autophagy are listed below (Table 1).
Table 1:
Some of the recently discovered chemical probes for the autophagy pathways.
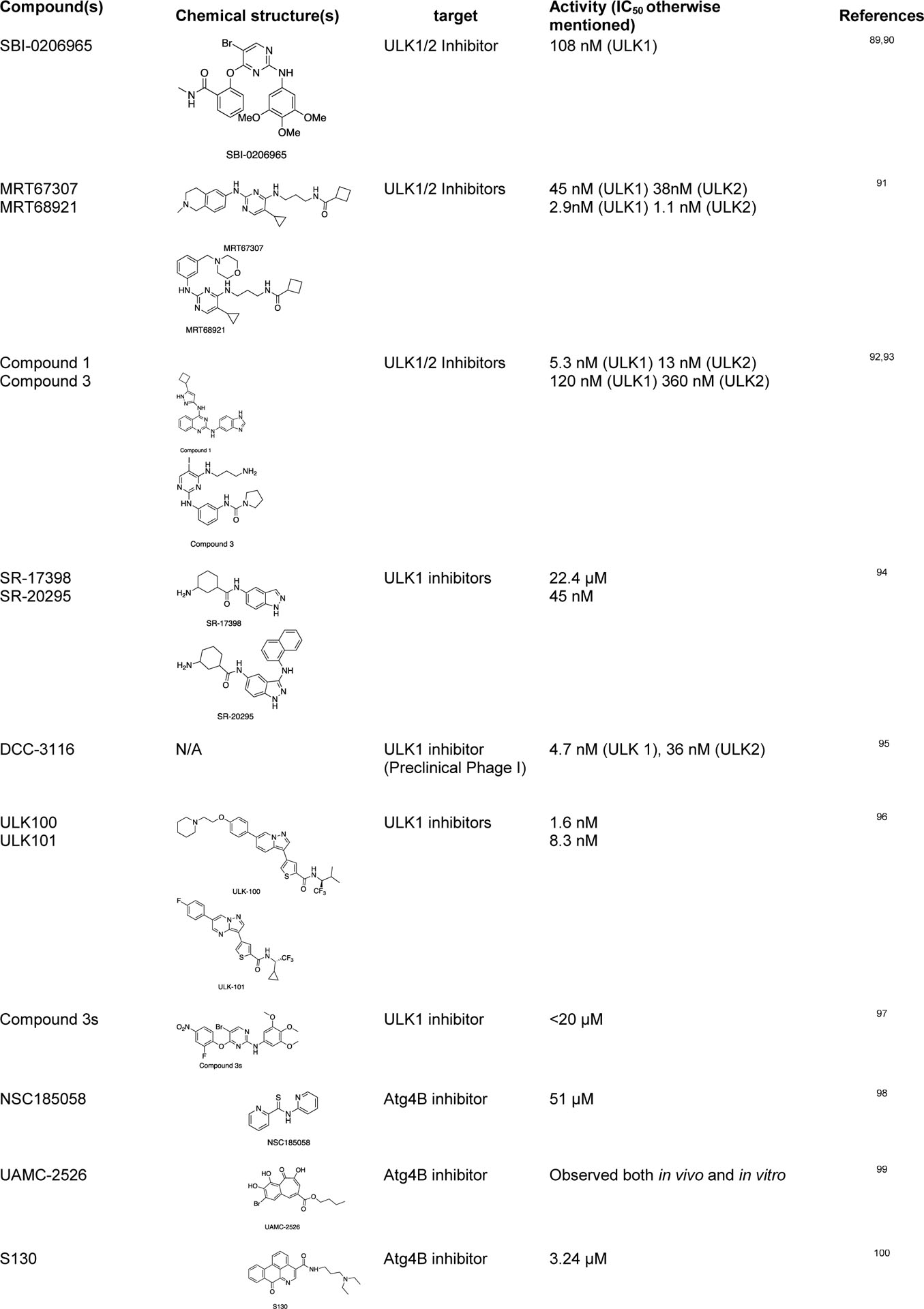
|
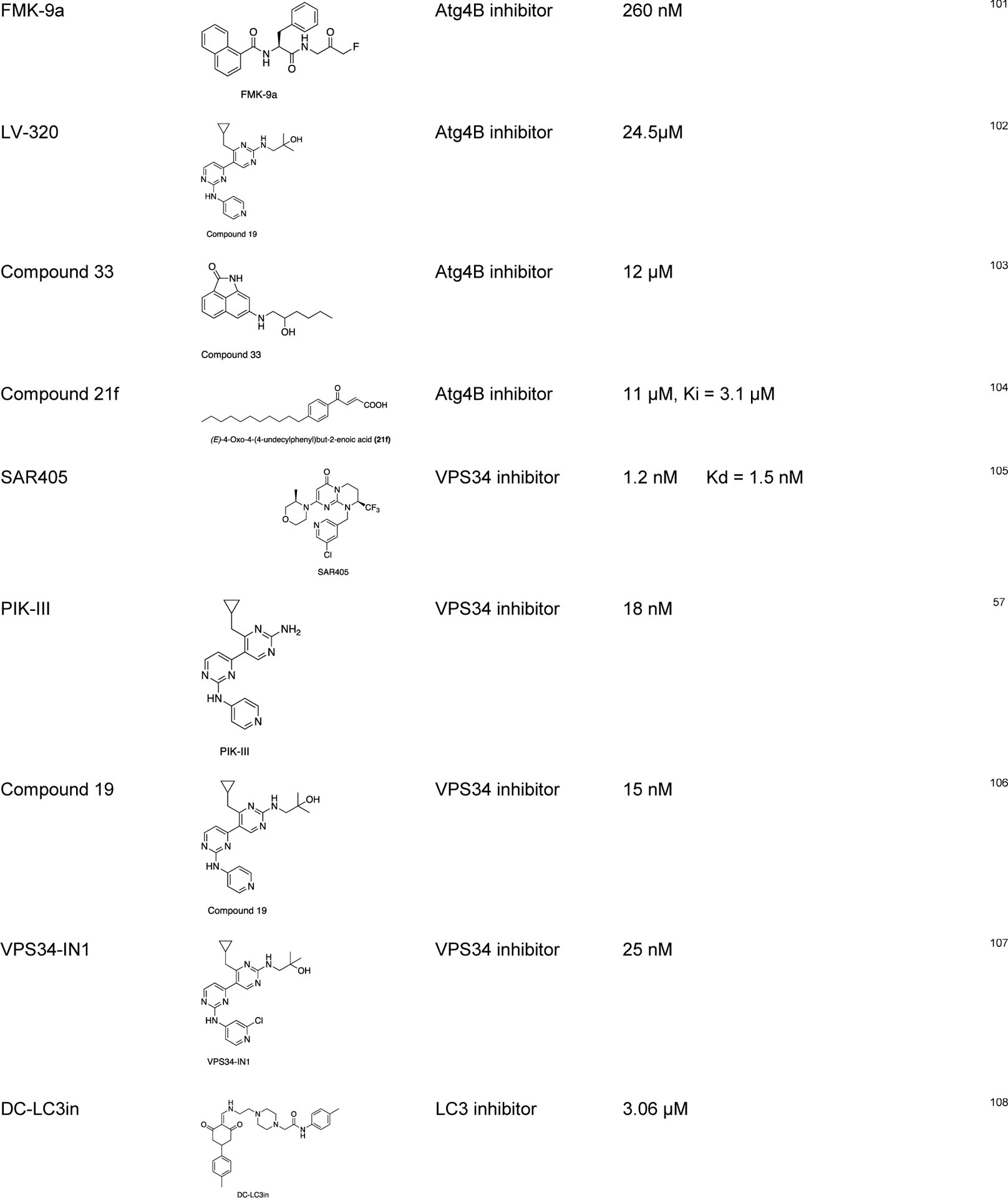
|
Endosome-lysosomal Pathway
Endocytosis is a cellular process of internalization of various components such as transmembrane proteins, receptors, receptor ligands, extracellular proteins and other biomolecules by the invagination of the plasma membranes and the formation of vesicles and vacuoles. Several steps are involved in the formation of endosomes and are classified as early endosomes, recycling endosomes, and late endosomes.109,110 The detail mechanisms of the endosome formation have been described in some other reviews.109,111 Briefly, endocytosis starts with the invagination of cargo proteins by the plasma membranes mediated by cytosolic proteins and multi-subunit complexes such as Rab proteins to form early endosome.110 Early endosomes serve as major sorting stations to send back the recycling materials into recycle endosomes or degradation materials to lysosomes by converting into late endosomes. The fusion of late endosomes with lysosomes allows the degradation of the enclosed components by the lysosomal hydrolases.109,110 Since, the UPS and autophagy based targeted degradation are only capable for degrading intercellular proteins, whereas the endosome-lysosomal pathway, described later in this review, has been applied for the degradation of extracellular proteins.112–115
Progress in the lysosome-based targeted protein degradation
Targeted protein degradation has been widely explored through PROTACs as therapeutics or chemical probes. However, this technology still has some limitations. Some other emerging technologies based on autophagy mechanisms such as autophagy-targeting chimera (AUTAC), autophagosome-tethering compounds (ATTEC), lysosome targeting chimera (LYTAC), and AUTOphagy-TArgeting Chimera (AUTOTAC) are under development. These recently acquired chemical biology platforms may overcome the PROTACs limitations. In this contribution, we summarize current lysosome-based degraders for different target proteins (Figure 5, Table 2).
Figure 5:
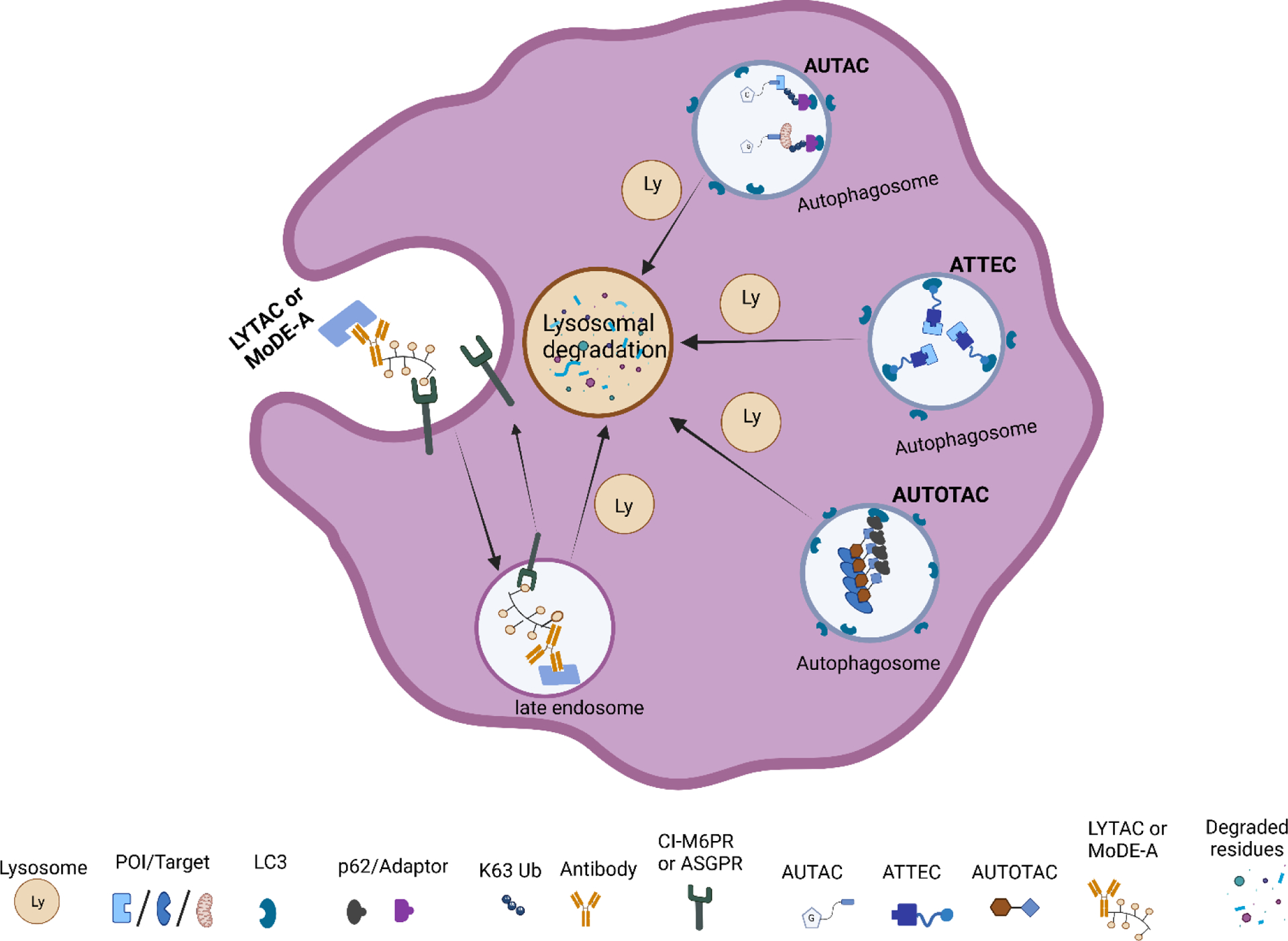
Graphical illustrations of lysosomal-based degradation technologies.
Table 2:
Summary of recently discovered lysosome-based degraders.
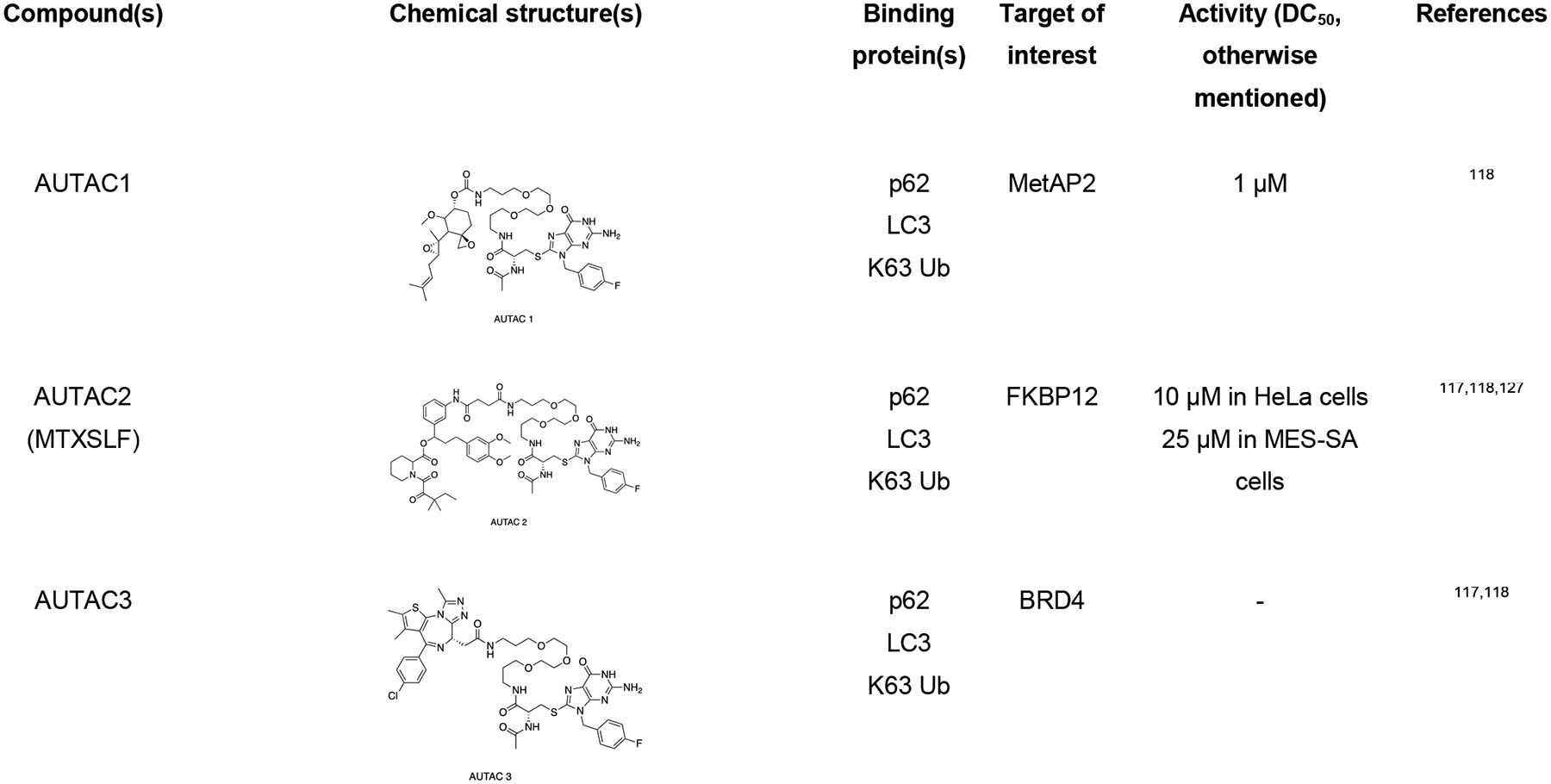
|
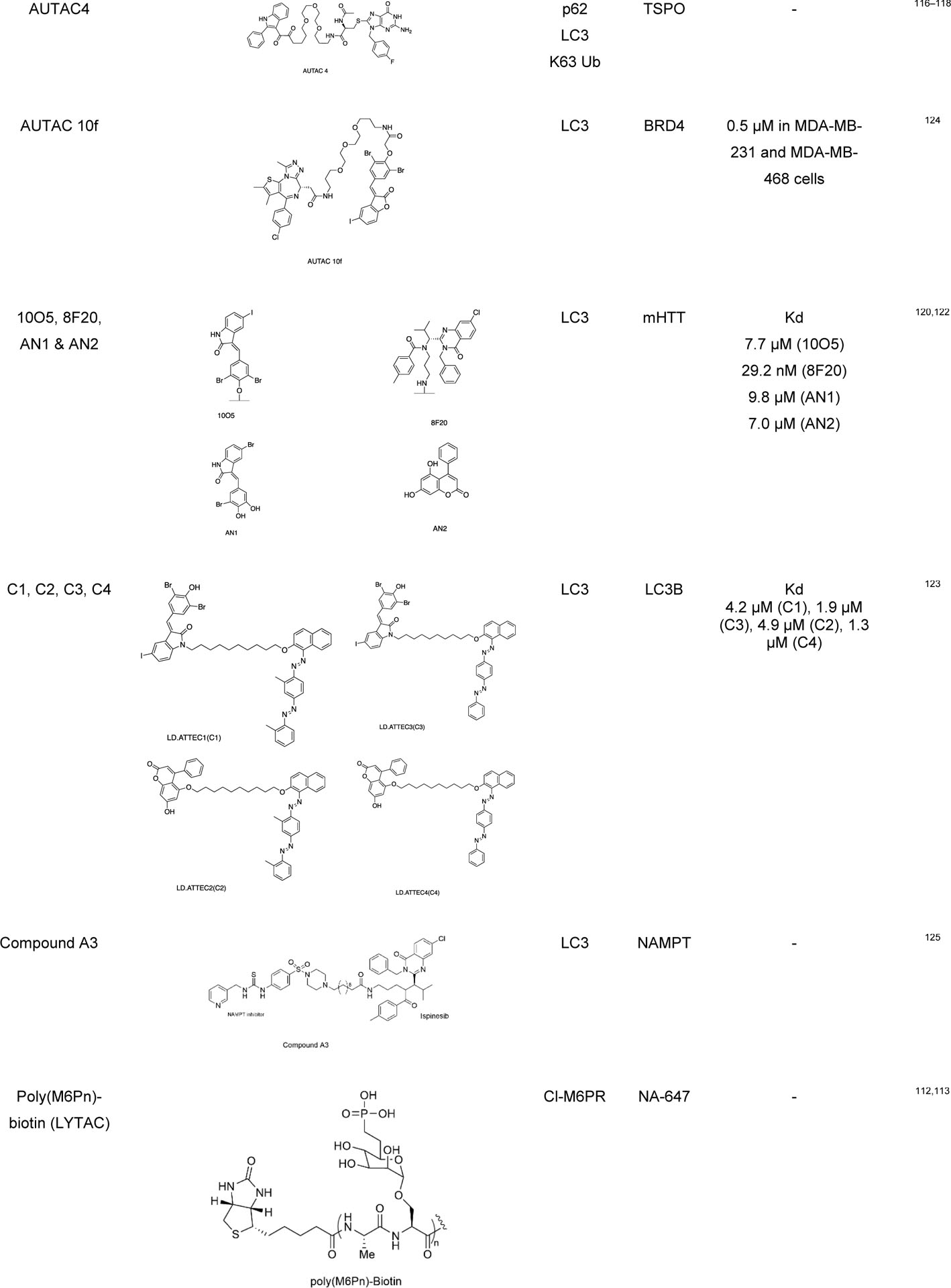
|
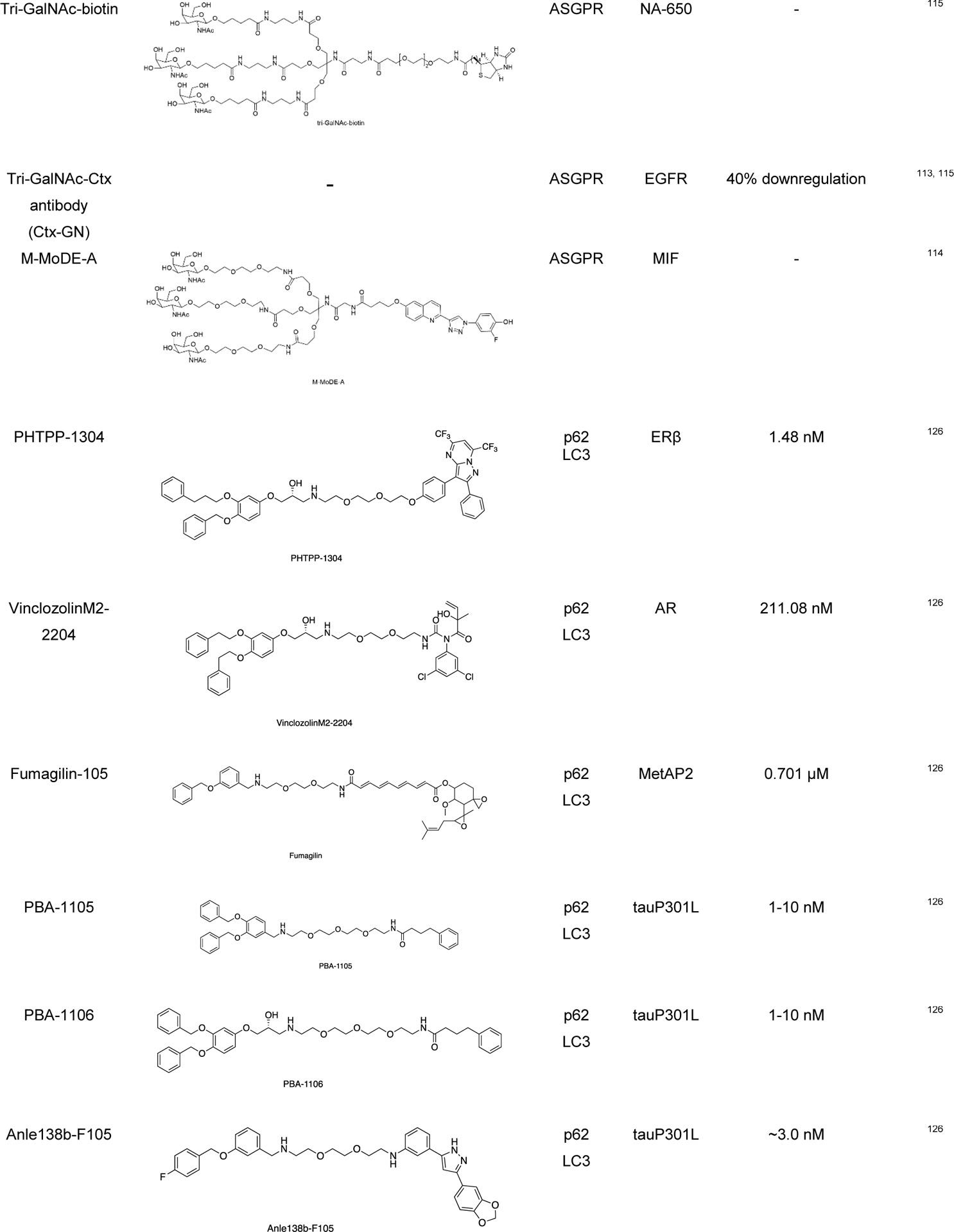
|
AUTACs
The Arimoto group developed autophagy-targeting chimera (AUTAC) molecules to remove targeted cytosolic proteins or mitochondria in xenophagy.76,116–118 The S-guanylation of group A Streptococcus (GAS) bacteria by 8-nitro-cGMP promotes the K63-linked polyubiquitination, which ultimately signals the selective transportation to autophagosome followed by degradation.119 The Arimoto group took advantage of GAS bacteria’s selective process to develop a chimeric molecule capable of targeted protein degradation. AUTAC molecule contains a p-fluorobenzylguanylation tag (FBnG unit) and a target-specific binder linked to polyethylene glycol (PEG). Arimoto et al. developed AUTAC1 containing a methionine aminopeptidase 2 (MetAP2) protein binder fumagillin that successfully degraded the MetAP2 at 1 μM. Similarly, AUTAC2 was designed to degrade FK506-binding protein (FKBP12), whose non-covalent synthetic ligand of FKBP (SLF) has been used to degrade with 10 μM concentration through the AUTAC system. AUTAC3 was also designed to target nuclear protein Brd4 with its binder JQ1, but degradation was not as effective as cytosolic proteins MetAP2 and FKBP12. Most importantly, AUTAC4 was designed for selective degradation of mitochondria. AUTAC4 contains a phenylindole moiety that binds to mitochondrial translocator proteins (TSPO) located on the outer mitochondrial membrane (OMM) and can selectively remove the dysfunctional mitochondria. In summary, one end of AUTAC molecule can selectively bind to the protein of interest (POIs), and the other end, which contains S-guanine moiety, helps to induce K63 polyubiquitination. The autophagy receptors such as p62 recognize K63 polyubiquitinated protein cargoes and traffic them to the autophagosomes for subsequent degradation. The major limitation for AUTAC is that the mechanism K63 polyubiquitination induced by S-guanylation is still unknown.
ATTECs
Lu and coworkers developed a new approach for targeted protein degradation called autophagosome-tethering compounds (ATTEC).120–123 ATTECs are bifunctional chimeric compounds that tether the POI to a specific protein degradation machinery (PDM) component, such as LC3. These compounds are found to be allele selective for a specific protein. They also designed a small molecule microarray screening for compounds that interact with LC3 and disease-causing protein mutant Huntington (HTT) with an expanded polyglutamine (PolyQ) stretch that is found in Huntington’s disease.122 From the microarray screening, the group identified four allele selective compounds (10O5, 8F20 or Ispinesib, AN1, and AN2) that can interact only with mHTT but not with wild-type (WT) HTT. These compounds also reduced some other polyQ expansion proteins such as ATXN3. Once the ATTEC molecule tether expanded polyQ proteins to autophagosome through LC3 domain, it is directed for subsequent degradation.
Recently, Lu group has applied their ATTEC strategy to degrade non-protein biomolecules, such as lipid droplets (LDs), and developed a new class of molecules called LD-ATTEC.123 Since lipid droplets are composed of lipids, PROTACs and AUTACs cannot target or degrade the LDs. LD-ATTECs were designed as bifunctional molecules that link the selective LD detecting probe (i.e., Sudan dyes) and LC3-binding molecules. These molecules constitute a ternary complex between triacylglycerol and LC3, which induces the proximity of LDs and autophagosomes in cells leading to autophagic degradation. During this mechanism, only the LDs induced by oleic acid in fibroblasts and endogenous LDs in differentiated adipocytes were degraded, leaving other lipid-containing membranes unaffected. Also, the global autophagy was not influenced by this selective LD degradation. The successful degradation of stored fats in the cell with LD-ATTEC opens a novel approach against the diseases caused by the accumulation of lipid droplets, such as obesity, cardiovascular diseases, or fatty liver disease. Although ATTEC technology is potentially effective in many cell types, the binding site in LC3 is not yet known.
Pei and coworkers have demonstrated an LC3 targeting autophagy chimeric molecule that can successfully degrade the BRD4 protein through the autophagy pathway. They have used a reversible BET bromodomain inhibitor JQ1 as a warhead for BRD4 and GW5074 for LC3 and linker to get the potent AUTAC molecule that downregulates the level of BRD4.124 Recently, Sheng group have applied the ATTEC principle to develop the first generation of autophagic degrader of nicotinamide phosphoribosyl transferase (NAMPT). The NAMPT ATTEC were synthesized by connecting NAMPT inhibitor and LC3 binding warhead Ispinesib through a flexible linker.125 Mechanistic studies confirmed that the NAMPT degradation occurs via autophagy-lysosomal pathway.
AUTOTACs
Ji et al. developed another autophagy based chemical tools called AUTOphagy-TArgeting Chimera (AUTOTAC).126 It is a bifunctional molecule which contains an autophagy targeting ligand (ATL) or p62 binding ligand and a target binding ligand (TBL). Autophagy targeting ligands bind to the ZZ domain of the autophagy cargo receptor p62, which can activate them for self-oligomerization for the autophagosome biogenesis through LC3 interactions. p62 binding ligands follows the N-end rule pathways. PHTPP-1304, Vinclozolin M2 and Fumagillin have been used as target binding ligands (TBLs) for estrogen receptor beta (ERβ), androgen receptors (AR) and methionine aminopeptidase-2 (MetAP2), respectively. The resulting AUTOTACs were able to degrade the respective target proteins in nanomolar range, which was not possible with ATL or TBL alone. AUTOTAC is applicable for the autophagic clearance of a wide range of intracellular target proteins. It can target not only monomeric proteins but also aggregated oligomeric proteins with sustained efficacy. Another feature of AUTOTAC is that unlike PROTACs, its potency is not critically dependent on the linker length, rendering straightforward AUTOTAC design.
LYTACs and MoDE-As
The Bertozzi group developed lysosomal targeting chimeras (LYTACs) for the degradation of the extracellular secreted proteins and plasma membrane-associated proteins.112,113 LYTACs consist of a target binding moiety (small molecule or antibody) linked to a glycan (polypeptide) ligand that can bind to the lysosome targeting receptor such as the cation-independent mannose-6-phosphate receptor (CI-M6PR) or asialoglycoprotein receptor (ASGPR) for liver-specific lysosomal degradation. LYTAC contains a glycan tag to help recognize the extracellular and membrane bound protein of interest (i.e., EGFR, CD71, PD-L1) for lysosomal degradation. They used neutravidin (NA), a fluorescently labeled protein that is stable under endosome and lysosome condition, to measure its uptake when combined with the LYTAC molecule. In parallel, the Tang group also developed a trivalent-N-acetylgalactosamine (tri-GalNAc) to target LYTACs to ASGPR on hepatocytes.115 They conjugated the ASGPR ligand (tri-GalNAc) to biotin and antibodies generating a new class of degraders, which were able to internalize and degrade neutravidin and EGFR. Capitalizing a similar concept, the Spiegel group developed MoDE-As (molecular degraders of extracellular proteins through the asialoglycoprotein receptor (ASGPR)), a small molecule version of ASGPR targeted LYTACs.114 MoDE-As molecules were able to recruit and induce the degradation α-DNP antibody and cytokine MIF protein.
The degradation of ASGPR was dependent on ASGPR internalization through the clathrin-mediated endocytosis-lysosomal system. The capability of lysosomes to maintain homeostasis stability was not affected in cells treated with LYTACs, suggesting that this modality may be safe at the cellular level. The success of LYTAC degraders results from endogenous kinetics of protein trafficking and turnover for the targeted protein. However, LYTACs cannot be applied to intracellular targets due to the nature of the degraders. As small molecules, MoDE-As might achieve deeper tissue penetration compared with antibody-based LYTACs.
Tissue Selectivity of Lysosome-Mediated Targeted Protein Degradation
For different lysosome-mediated TPD technologies, different key partner proteins in the lysosomal pathway are involved. Based the quantitative proteomics study in human tissues128, we examined the tissue distribution levels of p62 (gene: SQSTM1), LC3 (genes: MAP1LC3A and MAP1LC3B), ASGPR (genes: ASGR1 and ASGR2), and CI-M6PR (gene: IGF2R) based on their tissue specificity (TS) scores (Figure 6). It is interesting to note that p62 and ASGPR are highly enriched in skeletal muscle and liver, respectively. In brain, while LC3 is enriched, CI-M6PR is deficient compared to other tissues. Future work can take advantage of the differential expression of the lysosome pathway related proteins to achieve tissue selectivity.
Figure 6:
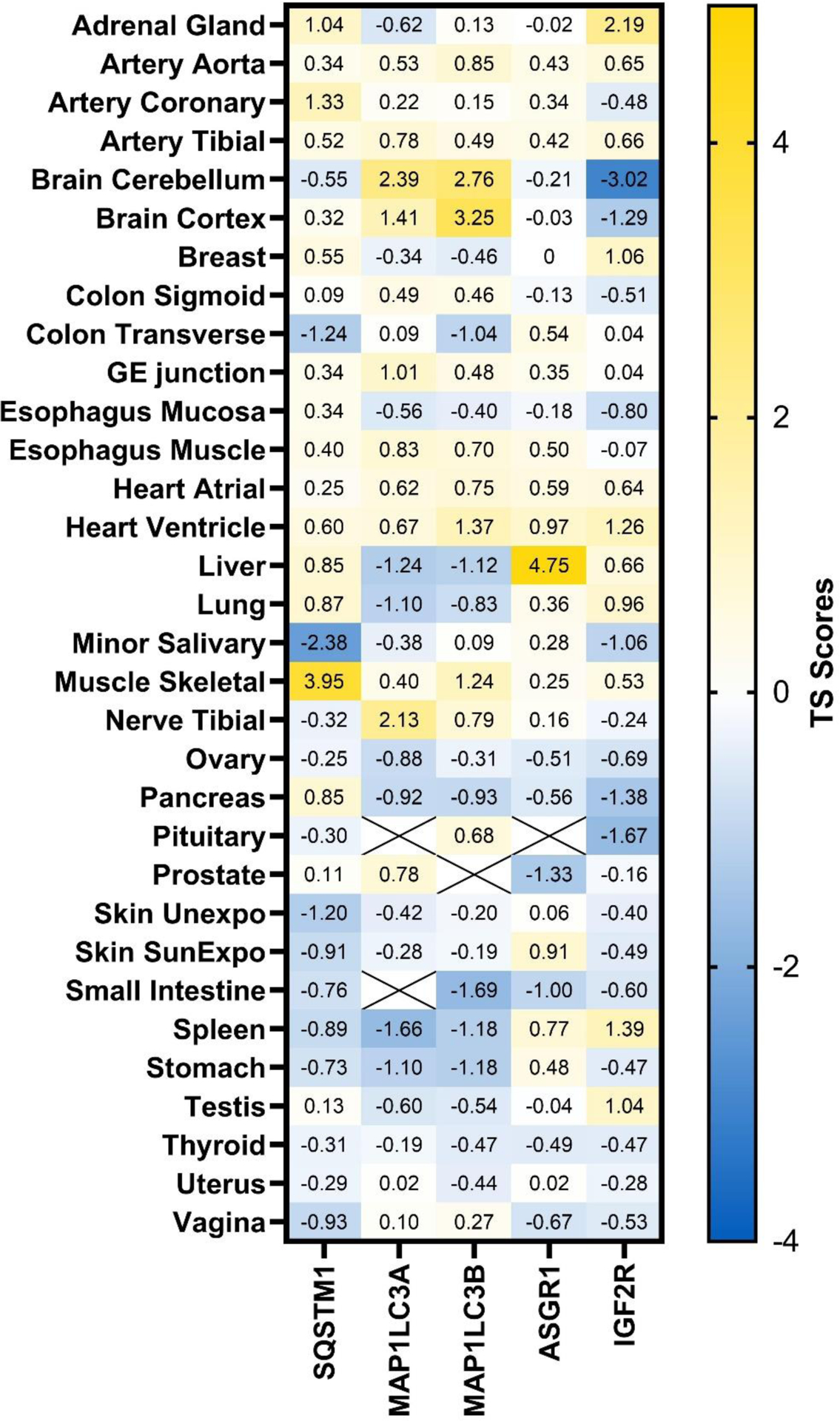
Tissue distribution of lysosome pathway related proteins. Protein abundance was measured using quantitative proteomics.128 Tissue selectivity (TS) scores were calculated and plotted. TS score >2.5 is considered highly enriched in certain tissues. SQSTM1: sequestosome 1 or p62; MAP1LC3A: microtubule associated protein 1 light chain 3 α; MAP1LC3B: microtubule associated protein 1 light chain 3 β; ASGR1: asialoglycoprotein receptor 1; ASGR2: asialoglycoprotein receptor 2; IGF2R: insulin like growth factor 2 receptor or CI-M6PR. The TS score for ASGR2 is unavailable.
Conclusion and Future Perspectives
The regulatory cycle of proteins has a crucial role in the fate of a living cell. Unwanted proteins need to be recycled through degradation into their constituent amino acids. Targeted protein degradation is one of the recently developed therapeutic approaches which is used to degrade disease-causing proteins. PROTACs are the most studied TPD technologies based on the UPS pathway, but these technologies are limited only to soluble and membrane proteins. There are some PROTAC molecules reported for the degradation of protein aggregates, for examples; tau,129 huntingtin,130 and α-synuclein proteins.131 Ubiquitination-dependent PROTAC cannot be applied to degrade insoluble and complex protein aggregates and defective cell organelles such as mitochondria. Therefore, advancements lysosomal-based degradation are inevitable. When studying autophagy-mediated lysosomal degradation, multiple assays are available. However, it is highly recommended not to use any single assay to measure autophagy and to validate the results using various assays depending on the experimental design.
Some newly explored TPD approaches such as AUTAC, ATTEC, AUTOTAC, and LYTAC are based on the lysosomal pathways. These novel approaches can selectively recognize and traffic the proteins/organelles to lysosomes for degradation. The lysosome-based degradation of the target proteins can overcome the limitations of proteasome-mediated degradation. However, complete understanding and broad application of these approaches are still in its infancy. AUTAC and ATTEC are used to degrade intracellular proteins or biomolecules, but their mechanism is not completely understood. The critical unanswered questions with these techniques are how S-guanylation of AUTAC induces K63 -polyubiquitination. ATTEC molecules are more selective for the targets, but their binding mechanism to LC3 is still unknown. The Kim group discovered that AUTOTAC can activate the autophagy cargo receptor (i.e., p62), which is self-polymerized, sequestered, and delivered into phagophore for autophagic degradation.126 To further improve our understanding of these Autophagy Lysosome System (ALS) mediated degraders, assays commonly used in PROTAC studies should be adopted, such as proteomics profiling to evaluate the degradation specificity, target engagement in cells, protein degradation kinetics and in vivo PKPD modeling.126 Comparing PROTACs with the ALS mediated degraders, we observed that the ALS mediated degraders tend to have μM or sub-μM potencies, while PROTACs can usually achieve single digit nM or sub-nM potencies. This may be due to the difference of degradation kinetics between the UPS and the ALS. To differentiate from PROTACs, the development of ALS-based degraders should primarily focus on targets that cannot be degraded by PROTACs, such as misfolded proteins or aggregates and dysfunctional organelles. Similarly, LYTACs and MoDE-As are primarily designed for extracellular protein degradation. These modalities need to find their niche applications that can differentiate from neutralizing antibodies, which are proven clinical modalities. Nonetheless, the lysosomal-based degradation approach will greatly expand the TPD toolkit, not only as chemical biology tools but also as therapeutic modalities. Lysosomal-based degradation techniques might not be able to degrade nuclear proteins efficiently, as the lysosomal system mostly operate in the cytoplasm. Moreover, the degradation efficiency of these methods may vary among different cell and tissue types (Figure 6), which could be taken advantage of to achieve tissue selectivity. In summary, as an alternative platform, lysosome-mediated targeted protein degradation will further expand the TPD field and provide exciting opportunities for therapeutic development.
Acknowledgements
The research was supported in part by National Institute of Health (R01-GM115622, R01-CA250503, and R01-CA268518 to J.W.), Cancer Prevention & Research Institute of Texas (CPRIT, RP220480 to J.W.), and the Michael E. DeBakey, M.D., Professorship in Pharmacology (to J.W.). Figures with illustrations are created with BioRender.com. We also appreciate the anonymous reviewers’ help to significantly improve this manuscript.
Footnotes
Conflicts of interest
J.W. is the co-founder of CoActigon Inc. and Chemical Biology Probes LLC.
References:
- (1).Jackson MP; Hewitt EW Cellular Proteostasis: Degradation of Misfolded Proteins by Lysosomes. Essays Biochem. 2016, 60 (2), 173–180. 10.1042/EBC20160005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Ciechanover A Intracellular Protein Degradation: From a Vague Idea, through the Lysosome and the Ubiquitin–Proteasome System, and onto Human Diseases and Drug Targeting (Nobel Lecture). Angew. Chem. Int. Ed 2005, 44 (37), 5944–5967. 10.1002/anie.200501428. [DOI] [PubMed] [Google Scholar]
- (3).Dikic I Proteasomal and Autophagic Degradation Systems. Annu. Rev. Biochem 2017, 86 (1), 193–224. 10.1146/annurev-biochem-061516-044908. [DOI] [PubMed] [Google Scholar]
- (4).Rousseau A; Bertolotti A Regulation of Proteasome Assembly and Activity in Health and Disease. Nat. Rev. Mol. Cell Biol 2018, 19 (11), 697–712. 10.1038/s41580-018-0040-z. [DOI] [PubMed] [Google Scholar]
- (5).Varshavsky A Regulated Protein Degradation. Trends Biochem. Sci 2005, 30 (6), 283–286. 10.1016/j.tibs.2005.04.005. [DOI] [PubMed] [Google Scholar]
- (6).Lilienbaum A Relationship between the Proteasomal System and Autophagy. Int. J. Biochem. Mol. Biol 2013, 4 (1), 1–26. [PMC free article] [PubMed] [Google Scholar]
- (7).Hughes SJ; Testa A; Thompson N; Churcher I The Rise and Rise of Protein Degradation: Opportunities and Challenges Ahead. Drug Discov. Today 2021, 26 (12), 2889–2897. 10.1016/j.drudis.2021.08.006. [DOI] [PubMed] [Google Scholar]
- (8).Bond MJ; Crews CM Proteolysis Targeting Chimeras (PROTACs) Come of Age: Entering the Third Decade of Targeted Protein Degradation. RSC Chem. Biol 2021, 2 (3), 725–742. 10.1039/D1CB00011J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Burslem GM; Crews CM Proteolysis-Targeting Chimeras as Therapeutics and Tools for Biological Discovery. Cell 2020, 181 (1), 102–114. 10.1016/j.cell.2019.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Békés M; Langley DR; Crews CM PROTAC Targeted Protein Degraders: The Past Is Prologue. Nat. Rev. Drug Discov 2022, 21 (3), 181–200. 10.1038/s41573-021-00371-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Wang F; Gómez-Sintes R; Boya P Lysosomal Membrane Permeabilization and Cell Death. Traffic 2018, 19 (12), 918–931. 10.1111/tra.12613. [DOI] [PubMed] [Google Scholar]
- (12).Pei J; Wang G; Feng L; Zhang J; Jiang T; Sun Q; Ouyang L Targeting Lysosomal Degradation Pathways: New Strategies and Techniques for Drug Discovery. J. Med. Chem 2021, 64 (7), 3493–3507. 10.1021/acs.jmedchem.0c01689. [DOI] [PubMed] [Google Scholar]
- (13).He C; Klionsky DJ Regulation Mechanisms and Signaling Pathways of Autophagy. Annu. Rev. Genet 2009, 43 (1), 67–93. 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Yang Z; Klionsky DJ Eaten Alive: A History of Macroautophagy. Nat. Cell Biol 2010, 12 (9), 814–822. 10.1038/ncb0910-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Eskelinen E-L; Saftig P Autophagy: A Lysosomal Degradation Pathway with a Central Role in Health and Disease. Biochim. Biophys. Acta BBA - Mol. Cell Res 2009, 1793 (4), 664–673. 10.1016/j.bbamcr.2008.07.014. [DOI] [PubMed] [Google Scholar]
- (16).Baek K-H; Park J; Shin I Autophagy-Regulating Small Molecules and Their Therapeutic Applications. Chem. Soc. Rev 2012, 41 (8), 3245. 10.1039/c2cs15328a. [DOI] [PubMed] [Google Scholar]
- (17).Yang ZJ; Chee CE; Huang S; Sinicrope FA The Role of Autophagy in Cancer: Therapeutic Implications. Mol. Cancer Ther 2011, 10 (9), 1533–1541. 10.1158/1535-7163.MCT-11-0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Levy JMM; Towers CG; Thorburn A Targeting Autophagy in Cancer. Nat. Rev. Cancer 2017, 17 (9), 528–542. 10.1038/nrc.2017.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).De Meyer GRY; Martinet W Autophagy in the Cardiovascular System. Biochim. Biophys. Acta BBA - Mol. Cell Res 2009, 1793 (9), 1485–1495. 10.1016/j.bbamcr.2008.12.011. [DOI] [PubMed] [Google Scholar]
- (20).Wileman T Autophagy as a Defence against Intracellular Pathogens. Essays Biochem. 2013, 55, 153–163. 10.1042/bse0550153. [DOI] [PubMed] [Google Scholar]
- (21).Mizushima N; Levine B; Cuervo AM; Klionsky DJ Autophagy Fights Disease through Cellular Self-Digestion. Nature 2008, 451 (7182), 1069–1075. 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Schuck S Microautophagy – Distinct Molecular Mechanisms Handle Cargoes of Many Sizes. J. Cell Sci 2020, 133 (17), jcs246322. 10.1242/jcs.246322. [DOI] [PubMed] [Google Scholar]
- (23).Kaushik S; Cuervo AM Chaperone-Mediated Autophagy: A Unique Way to Enter the Lysosome World. Trends Cell Biol. 2012, 22 (8), 407–417. 10.1016/j.tcb.2012.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Stricher F; Macri C; Ruff M; Muller S HSPA8/HSC70 Chaperone Protein: Structure, Function, and Chemical Targeting. Autophagy 2013, 9 (12), 1937–1954. 10.4161/auto.26448. [DOI] [PubMed] [Google Scholar]
- (25).Mizushima N; Yoshimori T How to Interpret LC3 Immunoblotting. Autophagy 2007, 3 (6), 542–545. 10.4161/auto.4600. [DOI] [PubMed] [Google Scholar]
- (26).Yorimitsu T; Klionsky DJ Autophagy: Molecular Machinery for Self-Eating. Cell Death Differ. 2005, 12 (S2), 1542–1552. 10.1038/sj.cdd.4401765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Li X; He S; Ma B Autophagy and Autophagy-Related Proteins in Cancer. Mol. Cancer 2020, 19 (1), 12. 10.1186/s12943-020-1138-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Harada K; Kotani T; Kirisako H; Sakoh-Nakatogawa M; Oikawa Y; Kimura Y; Hirano H; Yamamoto H; Ohsumi Y; Nakatogawa H Two Distinct Mechanisms Target the Autophagy-Related E3 Complex to the Pre-Autophagosomal Structure. eLife 2019, 8, e43088. 10.7554/eLife.43088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Kroemer G; Mariño G; Levine B Autophagy and the Integrated Stress Response. Mol. Cell 2010, 40 (2), 280–293. 10.1016/j.molcel.2010.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Andersson A-M; Andersson B; Lorell C; Raffetseder J; Larsson M; Blomgran R Autophagy Induction Targeting MTORC1 Enhances Mycobacterium Tuberculosis Replication in HIV Co-Infected Human Macrophages. Sci. Rep 2016, 6 (1), 28171. 10.1038/srep28171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Kim YC; Guan K-L MTOR: A Pharmacologic Target for Autophagy Regulation. J. Clin. Invest 2015, 125 (1), 25–32. 10.1172/JCI73939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Hosokawa N; Sasaki T; Iemura S; Natsume T; Hara T; Mizushima N Atg101, a Novel Mammalian Autophagy Protein Interacting with Atg13. Autophagy 2009, 5 (7), 973–979. 10.4161/auto.5.7.9296. [DOI] [PubMed] [Google Scholar]
- (33).Lindmo K; Brech A; Finley KD; Gaumer S; Contamine D; Rusten TE; Stenmark H The PI 3-Kinase Regulator Vps15 Is Required for Autophagic Clearance of Protein Aggregates. Autophagy 2008, 4 (4), 500–506. 10.4161/auto.5829. [DOI] [PubMed] [Google Scholar]
- (34).Russell RC; Tian Y; Yuan H; Park HW; Chang Y-Y; Kim J; Kim H; Neufeld TP; Dillin A; Guan K-L ULK1 Induces Autophagy by Phosphorylating Beclin-1 and Activating VPS34 Lipid Kinase. Nat. Cell Biol 2013, 15 (7), 741–750. 10.1038/ncb2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Yin Z; Pascual C; Klionsky DJ Autophagy: Machinery and Regulation. Microb. Cell 3 (12), 588–596. 10.15698/mic2016.12.546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Karanasios E; Walker SA; Okkenhaug H; Manifava M; Hummel E; Zimmermann H; Ahmed Q; Domart M-C; Collinson L; Ktistakis NT Autophagy Initiation by ULK Complex Assembly on ER Tubulovesicular Regions Marked by ATG9 Vesicles. Nat. Commun 2016, 7 (1), 12420. 10.1038/ncomms12420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Tanida I; Ueno T; Kominami E LC3 Conjugation System in Mammalian Autophagy. Int. J. Biochem. Cell Biol 2004, 36 (12), 2503–2518. 10.1016/j.biocel.2004.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Fan S; Yue L; Wan W; Zhang Y; Zhang B; Otomo C; Li Q; Lin T; Hu J; Xu P; Zhu M; Tao H; Chen Z; Li L; Ding H; Yao Z; Lu J; Wen Y; Zhang N; Tan M; Chen K; Xie Y; Otomo T; Zhou B; Jiang H; Dang Y; Luo C Inhibition of Autophagy by a Small Molecule through Covalent Modification of the LC3 Protein. Angew. Chem. Int. Ed 2021, 60 (50), 26105–26114. 10.1002/anie.202109464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Nakatogawa H; Ishii J; Asai E; Ohsumi Y Atg4 Recycles Inappropriately Lipidated Atg8 to Promote Autophagosome Biogenesis. Autophagy 2012, 8 (2), 177–186. 10.4161/auto.8.2.18373. [DOI] [PubMed] [Google Scholar]
- (40).Maruyama T; Noda NN Autophagy-Regulating Protease Atg4: Structure, Function, Regulation and Inhibition. J. Antibiot. (Tokyo) 2018, 71 (1), 72–78. 10.1038/ja.2017.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Hanada T; Noda NN; Satomi Y; Ichimura Y; Fujioka Y; Takao T; Inagaki F; Ohsumi Y The Atg12-Atg5 Conjugate Has a Novel E3-like Activity for Protein Lipidation in Autophagy. J. Biol. Chem 2007, 282 (52), 37298–37302. 10.1074/jbc.C700195200. [DOI] [PubMed] [Google Scholar]
- (42).Tsuboyama K; Koyama-Honda I; Sakamaki Y; Koike M; Morishita H; Mizushima N The ATG Conjugation Systems Are Important for Degradation of the Inner Autophagosomal Membrane. Science 2016, 354 (6315), 1036–1041. 10.1126/science.aaf6136. [DOI] [PubMed] [Google Scholar]
- (43).Gubas A; Dikic I A Guide to the Regulation of Selective Autophagy Receptors. FEBS J. 2022, 289 (1), 75–89. 10.1111/febs.15824. [DOI] [PubMed] [Google Scholar]
- (44).Wang Y; Li L; Hou C; Lai Y; Long J; Liu J; Zhong Q; Diao J SNARE-Mediated Membrane Fusion in Autophagy. Semin. Cell Dev. Biol 2016, 60, 97–104. 10.1016/j.semcdb.2016.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Nakamura S; Yoshimori T New Insights into Autophagosome–Lysosome Fusion. J. Cell Sci 2017, jcs.196352. 10.1242/jcs.196352. [DOI] [PubMed] [Google Scholar]
- (46).Das G; Shravage BV; Baehrecke EH Regulation and Function of Autophagy during Cell Survival and Cell Death. Cold Spring Harb. Perspect. Biol 2012, 4 (6), a008813. 10.1101/cshperspect.a008813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Badadani M Autophagy Mechanism, Regulation, Functions, and Disorders. ISRN Cell Biol. 2012, 2012, 1–11. 10.5402/2012/927064. [DOI] [Google Scholar]
- (48).Faruk MO; Ichimura Y; Komatsu M Selective Autophagy. Cancer Sci. 2021, 112 (10), 3972–3978. 10.1111/cas.15112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Zaffagnini G; Martens S Mechanisms of Selective Autophagy. J. Mol. Biol 2016, 428 (9), 1714–1724. 10.1016/j.jmb.2016.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Lemasters JJ Selective Mitochondrial Autophagy, or Mitophagy, as a Targeted Defense Against Oxidative Stress, Mitochondrial Dysfunction, and Aging. Rejuvenation Res. 2005, 8 (1), 3–5. 10.1089/rej.2005.8.3. [DOI] [PubMed] [Google Scholar]
- (51).Lamark T; Johansen T Aggrephagy: Selective Disposal of Protein Aggregates by Macroautophagy. Int. J. Cell Biol 2012, 2012, 1–21. 10.1155/2012/736905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Bauckman KA; Owusu-Boaitey N; Mysorekar IU Selective Autophagy: Xenophagy. Methods 2015, 75, 120–127. 10.1016/j.ymeth.2014.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Wyant GA; Abu-Remaileh M; Frenkel EM; Laqtom NN; Dharamdasani V; Lewis CA; Chan SH; Heinze I; Ori A; Sabatini DM NUFIP1 Is a Ribosome Receptor for Starvation-Induced Ribophagy. Science 2018, 360 (6390), 751–758. 10.1126/science.aar2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Koerver L; Papadopoulos C; Liu B; Kravic B; Rota G; Brecht L; Veenendaal T; Polajnar M; Bluemke A; Ehrmann M; Klumperman J; Jäättelä M; Behrends C; Meyer H The Ubiquitin‐conjugating Enzyme UBE 2 QL 1 Coordinates Lysophagy in Response to Endolysosomal Damage. EMBO Rep. 2019, 20 (10). 10.15252/embr.201948014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Shin DW Lipophagy: Molecular Mechanisms and Implications in Metabolic Disorders. Mol. Cells 2020, 43 (8), 686–693. 10.14348/molcells.2020.0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Cebollero E; Reggiori F; Kraft C Reticulophagy and Ribophagy: Regulated Degradation of Protein Production Factories. Int. J. Cell Biol 2012, 2012, e182834. 10.1155/2012/182834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Dowdle WE; Nyfeler B; Nagel J; Elling RA; Liu S; Triantafellow E; Menon S; Wang Z; Honda A; Pardee G; Cantwell J; Luu C; Cornella-Taracido I; Harrington E; Fekkes P; Lei H; Fang Q; Digan ME; Burdick D; Powers AF; Helliwell SB; D’Aquin S; Bastien J; Wang H; Wiederschain D; Kuerth J; Bergman P; Schwalb D; Thomas J; Ugwonali S; Harbinski F; Tallarico J; Wilson CJ; Myer VE; Porter JA; Bussiere DE; Finan PM; Labow MA; Mao X; Hamann LG; Manning BD; Valdez RA; Nicholson T; Schirle M; Knapp MS; Keaney EP; Murphy LO Selective VPS34 Inhibitor Blocks Autophagy and Uncovers a Role for NCOA4 in Ferritin Degradation and Iron Homeostasis in Vivo. Nat. Cell Biol 2014, 16 (11), 1069–1079. 10.1038/ncb3053. [DOI] [PubMed] [Google Scholar]
- (58).Johansen T; Lamark T Selective Autophagy: ATG8 Family Proteins, LIR Motifs and Cargo Receptors. J. Mol. Biol 2020, 432 (1), 80–103. 10.1016/j.jmb.2019.07.016. [DOI] [PubMed] [Google Scholar]
- (59).Kirkin V; Rogov VV A Diversity of Selective Autophagy Receptors Determines the Specificity of the Autophagy Pathway. Mol. Cell 2019, 76 (2), 268–285. 10.1016/j.molcel.2019.09.005. [DOI] [PubMed] [Google Scholar]
- (60).Ma X; Lu C; Chen Y; Li S; Ma N; Tao X; Li Y; Wang J; Zhou M; Yan Y-B; Li P; Heydari K; Deng H; Zhang M; Yi C; Ge L CCT2 Is an Aggrephagy Receptor for Clearance of Solid Protein Aggregates. Cell 2022, 185 (8), 1325–1345.e22. 10.1016/j.cell.2022.03.005. [DOI] [PubMed] [Google Scholar]
- (61).Matsumoto G; Wada K; Okuno M; Kurosawa M; Nukina N Serine 403 Phosphorylation of P62/SQSTM1 Regulates Selective Autophagic Clearance of Ubiquitinated Proteins. Mol. Cell 2011, 44 (2), 279–289. 10.1016/j.molcel.2011.07.039. [DOI] [PubMed] [Google Scholar]
- (62).Khaminets A; Behl C; Dikic I Ubiquitin-Dependent And Independent Signals In Selective Autophagy. Trends Cell Biol. 2016, 26 (1), 6–16. 10.1016/j.tcb.2015.08.010. [DOI] [PubMed] [Google Scholar]
- (63).Mancias JD; Wang X; Gygi SP; Harper JW; Kimmelman AC Quantitative Proteomics Identifies NCOA4 as the Cargo Receptor Mediating Ferritinophagy. Nature 2014, 509 (7498), 105–109. 10.1038/nature13148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Katsuragi Y; Ichimura Y; Komatsu M P62/SQSTM1 Functions as a Signaling Hub and an Autophagy Adaptor. FEBS J. 2015, 282 (24), 4672–4678. 10.1111/febs.13540. [DOI] [PubMed] [Google Scholar]
- (65).Pankiv S; Clausen TH; Lamark T; Brech A; Bruun J-A; Outzen H; Øvervatn A; Bjørkøy G; Johansen T P62/SQSTM1 Binds Directly to Atg8/LC3 to Facilitate Degradation of Ubiquitinated Protein Aggregates by Autophagy. J. Biol. Chem 2007, 282 (33), 24131–24145. 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- (66).Cha-Molstad H; Yu JE; Feng Z; Lee SH; Kim JG; Yang P; Han B; Sung KW; Yoo YD; Hwang J; McGuire T; Shim SM; Song HD; Ganipisetti S; Wang N; Jang JM; Lee MJ; Kim SJ; Lee KH; Hong JT; Ciechanover A; Mook-Jung I; Kim KP; Xie X-Q; Kwon YT; Kim BY P62/SQSTM1/Sequestosome-1 Is an N-Recognin of the N-End Rule Pathway Which Modulates Autophagosome Biogenesis. Nat. Commun 2017, 8 (1), 102. 10.1038/s41467-017-00085-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Zhang Y; Mun SR; Linares JF; Ahn J; Towers CG; Ji CH; Fitzwalter BE; Holden MR; Mi W; Shi X; Moscat J; Thorburn A; Diaz-Meco MT; Kwon YT; Kutateladze TG ZZ-Dependent Regulation of P62/SQSTM1 in Autophagy. Nat. Commun 2018, 9 (1), 4373. 10.1038/s41467-018-06878-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).Wurzer B; Zaffagnini G; Fracchiolla D; Turco E; Abert C; Romanov J; Martens S Oligomerization of P62 Allows for Selection of Ubiquitinated Cargo and Isolation Membrane during Selective Autophagy. eLife 2015, 4, e08941. 10.7554/eLife.08941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69).Itakura E; Mizushima N P62 Targeting to the Autophagosome Formation Site Requires Self-Oligomerization but Not LC3 Binding. J. Cell Biol 2011, 192 (1), 17–27. 10.1083/jcb.201009067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Kumsta C; Chang JT; Lee R; Tan EP; Yang Y; Loureiro R; Choy EH; Lim SHY; Saez I; Springhorn A; Hoppe T; Vilchez D; Hansen M The Autophagy Receptor P62/SQST-1 Promotes Proteostasis and Longevity in C. Elegans by Inducing Autophagy. Nat. Commun 2019, 10 (1), 5648. 10.1038/s41467-019-13540-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Tasaki T; Sriram SM; Park KS; Kwon YT The N-End Rule Pathway. Annu. Rev. Biochem 2012, 81 (1), 261–289. 10.1146/annurev-biochem-051710-093308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72).Varshavsky A The N-End Rule Pathway of Protein Degradation. Genes Cells 1997, 2 (1), 13–28. 10.1046/j.1365-2443.1997.1020301.x. [DOI] [PubMed] [Google Scholar]
- (73).Cha-Molstad H; Sung KS; Hwang J; Kim KA; Yu JE; Yoo YD; Jang JM; Han DH; Molstad M; Kim JG; Lee YJ; Zakrzewska A; Kim S-H; Kim ST; Kim SY; Lee HG; Soung NK; Ahn JS; Ciechanover A; Kim BY; Kwon YT Amino-Terminal Arginylation Targets Endoplasmic Reticulum Chaperone BiP for Autophagy through P62 Binding. Nat. Cell Biol 2015, 17 (7), 917–929. 10.1038/ncb3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (74).Cha-Molstad H; Yu JE; Feng Z; Lee SH; Kim JG; Yang P; Han B; Sung KW; Yoo YD; Hwang J; McGuire T; Shim SM; Song HD; Ganipisetti S; Wang N; Jang JM; Lee MJ; Kim SJ; Lee KH; Hong JT; Ciechanover A; Mook-Jung I; Kim KP; Xie X-Q; Kwon YT; Kim BY P62/SQSTM1/Sequestosome-1 Is an N-Recognin of the N-End Rule Pathway Which Modulates Autophagosome Biogenesis. Nat. Commun 2017, 8 (1), 102. 10.1038/s41467-017-00085-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (75).Autophagosome and Phagosome / Edited by Vojo Deretic; Deretic V, Ed.; Methods in molecular biology; Humana Press: Totowa, NJ, 2008. [DOI] [PubMed] [Google Scholar]
- (76).Klionsky DJ; Abdelmohsen K; Abe A; Abedin MJ Guidelines for the Use and Interpretation of Assays for Monitoring Autophagy (3rd Edition). Autophagy 2016, 12 (1), 1–222. 10.1080/15548627.2015.1100356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (77).Min Z; Ting Y; Mingtao G; Xiaofei T; Dong Y; Chenguang Z; Wei D Monitoring Autophagic Flux Using P62/SQSTM1 Based Luciferase Reporters in Glioma Cells. Exp. Cell Res 2018, 363 (1), 84–94. 10.1016/j.yexcr.2017.12.027. [DOI] [PubMed] [Google Scholar]
- (78).Bjørkøy G; Lamark T; Pankiv S; Øvervatn A; Brech A; Johansen T Chapter 12 Monitoring Autophagic Degradation of P62/SQSTM1. In Methods in Enzymology; Elsevier, 2009; Vol. 452, pp 181–197. 10.1016/S0076-6879(08)03612-4. [DOI] [PubMed] [Google Scholar]
- (79).Mizushima N; Murphy LO Autophagy Assays for Biological Discovery and Therapeutic Development. Trends Biochem. Sci 2020, 45 (12), 1080–1093. 10.1016/j.tibs.2020.07.006. [DOI] [PubMed] [Google Scholar]
- (80).Klionsky DJ; Elazar Z; Seglen PO; Rubinsztein DC Does Bafilomycin A 1 Block the Fusion of Autophagosomes with Lysosomes? Autophagy 2008, 4 (7), 849–850. 10.4161/auto.6845. [DOI] [PubMed] [Google Scholar]
- (81).Kimura S; Noda T; Yoshimori T Dissection of the Autophagosome Maturation Process by a Novel Reporter Protein, Tandem Fluorescent-Tagged LC3. Autophagy 2007, 3 (5), 452–460. 10.4161/auto.4451. [DOI] [PubMed] [Google Scholar]
- (82).Ni H-M; Bockus A; Wozniak AL; Jones K; Weinman S; Yin X-M; Ding W-X Dissecting the Dynamic Turnover of GFP-LC3 in the Autolysosome. Autophagy 2011, 7 (2), 188–204. 10.4161/auto.7.2.14181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (83).Kaizuka T; Morishita H; Hama Y; Tsukamoto S; Matsui T; Toyota Y; Kodama A; Ishihara T; Mizushima T; Mizushima N An Autophagic Flux Probe That Releases an Internal Control. Mol. Cell 2016, 64 (4), 835–849. 10.1016/j.molcel.2016.09.037. [DOI] [PubMed] [Google Scholar]
- (84).Farkas T; Høyer-Hansen M; Jäättelä M Identification of Novel Autophagy Regulators by a Luciferase-Based Assay for the Kinetics of Autophagic Flux. Autophagy 2009, 5 (7), 1018–1025. 10.4161/auto.5.7.9443. [DOI] [PubMed] [Google Scholar]
- (85).Bresciani A; Spiezia MC; Boggio R; Cariulo C; Nordheim A; Altobelli R; Kuhlbrodt K; Dominguez C; Munoz-Sanjuan I; Wityak J; Fodale V; Marchionini DM; Weiss A Quantifying Autophagy Using Novel LC3B and P62 TR-FRET Assays. PLOS ONE 2018, 13 (3), e0194423. 10.1371/journal.pone.0194423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (86).Yang ZJ; Chee CE; Huang S; Sinicrope FA The Role of Autophagy in Cancer: Therapeutic Implications. Mol. Cancer Ther 2011, 10 (9), 1533–1541. 10.1158/1535-7163.MCT-11-0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (87).Targeting Autophagy in Cancer Therapy; Yang J-M, Ed.; Current Cancer Research; Springer International Publishing: Cham, 2016. 10.1007/978-3-319-42740-9. [DOI] [Google Scholar]
- (88).Bu F; Zhang J; Shuai W; Liu J; Sun Q; Ouyang L Repurposing Drugs in Autophagy for the Treatment of Cancer: From Bench to Bedside. Drug Discov. Today 2021, S1359644621004943. 10.1016/j.drudis.2021.11.013. [DOI] [PubMed] [Google Scholar]
- (89).Egan DF; Chun MGH; Vamos M; Zou H; Rong J; Miller CJ; Lou HJ; Raveendra-Panickar D; Yang C-C; Sheffler DJ; Teriete P; Asara JM; Turk BE; Cosford NDP; Shaw RJ Small Molecule Inhibition of the Autophagy Kinase ULK1 and Identification of ULK1 Substrates. Mol. Cell 2015, 59 (2), 285–297. 10.1016/j.molcel.2015.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (90).Tang F; Hu P; Yang Z; Xue C; Gong J; Sun S; Shi L; Zhang S; Li Z; Yang C; Zhang J; Xie C SBI0206965, a Novel Inhibitor of Ulk1, Suppresses Non-Small Cell Lung Cancer Cell Growth by Modulating Both Autophagy and Apoptosis Pathways. Oncol. Rep 2017, 37 (6), 3449–3458. 10.3892/or.2017.5635. [DOI] [PubMed] [Google Scholar]
- (91).Petherick KJ; Conway OJL; Mpamhanga C; Osborne SA; Kamal A; Saxty B; Ganley IG Pharmacological Inhibition of ULK1 Kinase Blocks Mammalian Target of Rapamycin (MTOR)-Dependent Autophagy *. J. Biol. Chem 2015, 290 (18), 11376–11383. 10.1074/jbc.C114.627778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (92).Lazarus MB; Shokat KM Discovery and Structure of a New Inhibitor Scaffold of the Autophagy Initiating Kinase ULK1. Bioorg. Med. Chem 2015, 23 (17), 5483–5488. 10.1016/j.bmc.2015.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (93).Lazarus MB; Novotny CJ; Shokat KM Structure of the Human Autophagy Initiating Kinase ULK1 in Complex with Potent Inhibitors. ACS Chem. Biol 2015, 10 (1), 257–261. 10.1021/cb500835z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (94).Wood SD; Grant W; Adrados I; Choi JY; Alburger JM; Duckett DR; Roush WR In Silico HTS and Structure Based Optimization of Indazole-Derived ULK1 Inhibitors. ACS Med. Chem. Lett 2017, 8 (12), 1258–1263. 10.1021/acsmedchemlett.7b00344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (95).Smith BD; Vogeti L; Gupta A; Singh J; Al-Ani G; Bulfer SL; Caldwell TM; Timson MJ; Vogeti S; Ahn YM; Al-Hashimi H; Crawley CK; Heiniger CL; Leary CB; Proto JT; Shen Q; Telikepalli H; Yates K; Lu W-P; Flynn DL Abstract B129: Preclinical Studies with DCC-3116, an ULK Kinase Inhibitor Designed to Inhibit Autophagy as a Potential Strategy to Address Mutant RAS Cancers. Mol. Cancer Ther 2019, 18 (12_Supplement), B129. 10.1158/1535-7163.TARG-19-B129. [DOI] [Google Scholar]
- (96).Martin KR; Celano SL; Solitro AR; Gunaydin H; Scott M; O’Hagan RC; Shumway SD; Fuller P; MacKeigan JP A Potent and Selective ULK1 Inhibitor Suppresses Autophagy and Sensitizes Cancer Cells to Nutrient Stress. iScience 2018, 8, 74–84. 10.1016/j.isci.2018.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (97).Sun D; Yang Z; Zhen Y; Yang Y; Chen Y; Yuan Y; Zhang L; Zeng X; Chen L Discovery of 5-Bromo-4-Phenoxy-N-Phenylpyrimidin-2-Amine Derivatives as Novel ULK1 Inhibitors That Block Autophagy and Induce Apoptosis in Non-Small Cell Lung Cancer. Eur. J. Med. Chem 2020, 208, 112782. 10.1016/j.ejmech.2020.112782. [DOI] [PubMed] [Google Scholar]
- (98).Akin D; Wang SK; Habibzadegah-Tari P; Law B; Ostrov D; Li M; Yin X-M; Kim J-S; Horenstein N; Dunn WA A Novel ATG4B Antagonist Inhibits Autophagy and Has a Negative Impact on Osteosarcoma Tumors. Autophagy 2014, 10 (11), 2021–2035. 10.4161/auto.32229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (99).Kurdi A; Cleenewerck M; Vangestel C; Lyssens S; Declercq W; Timmermans J-P; Stroobants S; Augustyns K; De Meyer GRY; Van Der Veken P; Martinet W ATG4B Inhibitors with a Benzotropolone Core Structure Block Autophagy and Augment Efficiency of Chemotherapy in Mice. Biochem. Pharmacol 2017, 138, 150–162. 10.1016/j.bcp.2017.06.119. [DOI] [PubMed] [Google Scholar]
- (100).Fu Y; Hong L; Xu J; Zhong G; Gu Q; Gu Q; Guan Y; Zheng X; Dai Q; Luo X; Liu C; Huang Z; Yin X-M; Liu P; Li M Discovery of a Small Molecule Targeting Autophagy via ATG4B Inhibition and Cell Death of Colorectal Cancer Cells in Vitro and in Vivo. Autophagy 2019, 15 (2), 295–311. 10.1080/15548627.2018.1517073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (101).Chu J; Fu Y; Xu J; Zheng X; Gu Q; Luo X; Dai Q; Zhang S; Liu P; Hong L; Li M ATG4B Inhibitor FMK-9a Induces Autophagy Independent on Its Enzyme Inhibition. Arch. Biochem. Biophys 2018, 644, 29–36. 10.1016/j.abb.2018.03.001. [DOI] [PubMed] [Google Scholar]
- (102).Bosc D; Vezenkov L; Bortnik S; An J; Xu J; Choutka C; Hannigan AM; Kovacic S; Loo S; Clark PGK; Chen G; Guay-Ross RN; Yang K; Dragowska WH; Zhang F; Go NE; Leung A; Honson NS; Pfeifer TA; Gleave M; Bally M; Jones SJ; Gorski SM; Young RN A New Quinoline-Based Chemical Probe Inhibits the Autophagy-Related Cysteine Protease ATG4B. Sci. Rep 2018, 8 (1), 11653. 10.1038/s41598-018-29900-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (103).Quintana M; Bilbao A; Comas-Barceló J; Bujons J; Triola G Identification of Benzo[Cd]Indol-2(1H)-Ones as Novel Atg4B Inhibitors via a Structure-Based Virtual Screening and a Novel AlphaScreen Assay. Eur. J. Med. Chem 2019, 178, 648–666. 10.1016/j.ejmech.2019.05.086. [DOI] [PubMed] [Google Scholar]
- (104).Kudo Y; Endo S; Fujita M; Ota A; Kamatari YO; Tanaka Y; Ishikawa T; Ikeda H; Okada T; Toyooka N; Fujimoto N; Matsunaga T; Ikari A Discovery and Structure-Based Optimization of Novel Atg4B Inhibitors for the Treatment of Castration-Resistant Prostate Cancer. J. Med. Chem 2022, 65 (6), 4878–4892. 10.1021/acs.jmedchem.1c02113. [DOI] [PubMed] [Google Scholar]
- (105).Ronan B; Flamand O; Vescovi L; Dureuil C; Durand L; Fassy F; Bachelot M-F; Lamberton A; Mathieu M; Bertrand T; Marquette J-P; El-Ahmad Y; Filoche-Romme B; Schio L; Garcia-Echeverria C; Goulaouic H; Pasquier B A Highly Potent and Selective Vps34 Inhibitor Alters Vesicle Trafficking and Autophagy. Nat. Chem. Biol 2014, 10 (12), 1013–1019. 10.1038/nchembio.1681. [DOI] [PubMed] [Google Scholar]
- (106).Honda A; Harrington E; Cornella-Taracido I; Furet P; Knapp MS; Glick M; Triantafellow E; Dowdle WE; Wiedershain D; Maniara W; Moore C; Finan PM; Hamann LG; Firestone B; Murphy LO; Keaney EP Potent, Selective, and Orally Bioavailable Inhibitors of VPS34 Provide Chemical Tools to Modulate Autophagy in Vivo. ACS Med. Chem. Lett 2016, 7 (1), 72–76. 10.1021/acsmedchemlett.5b00335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (107).Bago R; Malik N; Munson MJ; Prescott AR; Davies P; Sommer E; Shpiro N; Ward R; Cross D; Ganley IG; Alessi DR Characterization of VPS34-IN1, a Selective Inhibitor of Vps34, Reveals That the Phosphatidylinositol 3-Phosphate-Binding SGK3 Protein Kinase Is a Downstream Target of Class III Phosphoinositide 3-Kinase. Biochem. J 2014, 463 (3), 413–427. 10.1042/BJ20140889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (108).Fan S; Yue L; Wan W; Zhang Y; Zhang B; Otomo C; Li Q; Lin T; Hu J; Xu P; Zhu M; Tao H; Chen Z; Li L; Ding H; Yao Z; Lu J; Wen Y; Zhang N; Tan M; Chen K; Xie Y; Otomo T; Zhou B; Jiang H; Dang Y; Luo C Inhibition of Autophagy by a Small Molecule through Covalent Modification of the LC3 Protein. Angew. Chem. Int. Ed 2021, 60 (50), 26105–26114. 10.1002/anie.202109464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (109).Hu Y-B; Dammer EB; Ren R-J; Wang G The Endosomal-Lysosomal System: From Acidification and Cargo Sorting to Neurodegeneration. Transl. Neurodegener 2015, 4 (1), 18. 10.1186/s40035-015-0041-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (110).Lamb CA; Dooley HC; Tooze SA Endocytosis and Autophagy: Shared Machinery for Degradation. BioEssays 2013, 35 (1), 34–45. 10.1002/bies.201200130. [DOI] [PubMed] [Google Scholar]
- (111).Huotari J; Helenius A Endosome Maturation: Endosome Maturation. EMBO J. 2011, 30 (17), 3481–3500. 10.1038/emboj.2011.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (112).Banik SM; Pedram K; Wisnovsky S; Ahn G; Riley NM; Bertozzi CR Lysosome-Targeting Chimaeras for Degradation of Extracellular Proteins. Nature 2020, 584 (7820), 291–297. 10.1038/s41586-020-2545-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (113).Ahn G; Banik SM; Miller CL; Riley NM; Cochran JR; Bertozzi CR LYTACs That Engage the Asialoglycoprotein Receptor for Targeted Protein Degradation. Nat. Chem. Biol 2021, 17 (9), 937–946. 10.1038/s41589-021-00770-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (114).Caianiello DF; Zhang M; Ray JD; Howell RA; Swartzel JC; Branham EMJ; Chirkin E; Sabbasani VR; Gong AZ; McDonald DM; Muthusamy V; Spiegel DA Bifunctional Small Molecules That Mediate the Degradation of Extracellular Proteins. Nat. Chem. Biol 2021, 17 (9), 947–953. 10.1038/s41589-021-00851-1. [DOI] [PubMed] [Google Scholar]
- (115).Zhou Y; Teng P; Montgomery NT; Li X; Tang W Development of Triantennary N-Acetylgalactosamine Conjugates as Degraders for Extracellular Proteins. ACS Cent. Sci 2021, 7 (3), 499–506. 10.1021/acscentsci.1c00146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (116).Takahashi D; Arimoto H Targeting Selective Autophagy by AUTAC Degraders. Autophagy 2020, 16 (4), 765–766. 10.1080/15548627.2020.1718362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (117).Takahashi D; Arimoto H Selective Autophagy as the Basis of Autophagy-Based Degraders. Cell Chem. Biol 2021, 28 (7), 1061–1071. 10.1016/j.chembiol.2021.05.006. [DOI] [PubMed] [Google Scholar]
- (118).Takahashi D; Moriyama J; Nakamura T; Miki E; Takahashi E; Sato A; Akaike T; Itto-Nakama K; Arimoto H AUTACs: Cargo-Specific Degraders Using Selective Autophagy. Mol. Cell 2019, 76 (5), 797–810.e10. 10.1016/j.molcel.2019.09.009. [DOI] [PubMed] [Google Scholar]
- (119).Ito C; Saito Y; Nozawa T; Fujii S; Sawa T; Inoue H; Matsunaga T; Khan S; Akashi S; Hashimoto R; Aikawa C; Takahashi E; Sagara H; Komatsu M; Tanaka K; Akaike T; Nakagawa I; Arimoto H Endogenous Nitrated Nucleotide Is a Key Mediator of Autophagy and Innate Defense against Bacteria. Mol. Cell 2013, 52 (6), 794–804. 10.1016/j.molcel.2013.10.024. [DOI] [PubMed] [Google Scholar]
- (120).Li Z; Wang C; Wang Z; Zhu C; Li J; Sha T; Ma L; Gao C; Yang Y; Sun Y; Wang J; Sun X; Lu C; Difiglia M; Mei Y; Ding C; Luo S; Dang Y; Ding Y; Fei Y; Lu B Allele-Selective Lowering of Mutant HTT Protein by HTT–LC3 Linker Compounds. Nature 2019, 575 (7781), 203–209. 10.1038/s41586-019-1722-1. [DOI] [PubMed] [Google Scholar]
- (121).Ding Y; Fei Y; Lu B Emerging New Concepts of Degrader Technologies. Trends Pharmacol. Sci 2020, 41 (7), 464–474. 10.1016/j.tips.2020.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (122).Li Z; Zhu C; Ding Y; Fei Y; Lu B ATTEC: A Potential New Approach to Target Proteinopathies. Autophagy 2020, 16 (1), 185–187. 10.1080/15548627.2019.1688556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (123).Fu Y; Chen N; Wang Z; Luo S; Ding Y; Lu B Degradation of Lipid Droplets by Chimeric Autophagy-Tethering Compounds. Cell Res. 2021, 31 (9), 965–979. 10.1038/s41422-021-00532-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (124).Pei J; Pan X; Wang A; Shuai W; Bu F; Tang P; Zhang S; Zhang Y; Wang G; Ouyang L Developing Potent LC3-Targeting AUTAC Tools for Protein Degradation with Selective Autophagy. Chem. Commun 2021, 57 (97), 13194–13197. 10.1039/D1CC04661F. [DOI] [PubMed] [Google Scholar]
- (125).Dong G; Wu Y; Cheng J; Chen L; Liu R; Ding Y; Wu S; Ma J; Sheng C Ispinesib as an Effective Warhead for the Design of Autophagosome-Tethering Chimeras: Discovery of Potent Degraders of Nicotinamide Phosphoribosyltransferase (NAMPT). J. Med. Chem 2022, 65 (11), 7619–7628. 10.1021/acs.jmedchem.1c02001. [DOI] [PubMed] [Google Scholar]
- (126).Ji CH; Kim HY; Lee MJ; Heo AJ; Park DY; Lim S; Shin S; Ganipisetti S; Yang WS; Jung CA; Kim KY; Jeong EH; Park SH; Bin Kim S; Lee SJ; Na JE; Kang JI; Chi HM; Kim HT; Kim YK; Kim BY; Kwon YT The AUTOTAC Chemical Biology Platform for Targeted Protein Degradation via the Autophagy-Lysosome System. Nat. Commun 2022, 13 (1), 904. 10.1038/s41467-022-28520-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (127).Braun PD; Barglow KT; Lin Y-M; Akompong T; Briesewitz R; Ray GT; Haldar K; Wandless TJ A Bifunctional Molecule That Displays Context-Dependent Cellular Activity. J. Am. Chem. Soc 2003, 125 (25), 7575–7580. 10.1021/ja035176q. [DOI] [PubMed] [Google Scholar]
- (128).Jiang L; Wang M; Lin S; Jian R; Li X; Chan J; Dong G; Fang H; Robinson AE; Snyder MP A Quantitative Proteome Map of the Human Body. Cell 2020, 183 (1), 269–283.e19. 10.1016/j.cell.2020.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (129).Silva MC; Ferguson FM; Cai Q; Donovan KA; Nandi G; Patnaik D; Zhang T; Huang H-T; Lucente DE; Dickerson BC; Mitchison TJ; Fischer ES; Gray NS; Haggarty SJ Targeted Degradation of Aberrant Tau in Frontotemporal Dementia Patient-Derived Neuronal Cell Models. eLife 2019, 8, e45457. 10.7554/eLife.45457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (130).Tomoshige S; Nomura S; Ohgane K; Hashimoto Y; Ishikawa M Degradation of Huntingtin Mediated by a Hybrid Molecule Composed of IAP Antagonist Linked to Phenyldiazenyl Benzothiazole Derivative. Bioorg. Med. Chem. Lett 2018, 28 (4), 707–710. 10.1016/j.bmcl.2018.01.012. [DOI] [PubMed] [Google Scholar]
- (131).Kargbo RB PROTAC Compounds Targeting α-Synuclein Protein for Treating Neurogenerative Disorders: Alzheimer’s and Parkinson’s Diseases. ACS Med. Chem. Lett 2020, 11 (6), 1086–1087. 10.1021/acsmedchemlett.0c00192. [DOI] [PMC free article] [PubMed] [Google Scholar]


