Abstract
BACKGROUND:
Estimates of overall patient health are essential to inform treatment decisions for patients diagnosed with cancer. The authors applied XWAS methods, herein referred to as “laboratory-wide association study (LWAS)”, to evaluate associations between routinely collected laboratory tests and survival in veterans with prostate cancer.
METHODS:
The authors identified 133,878 patients who were diagnosed with prostate cancer between 2000 and 2013 in the Veterans Health Administration using any laboratory tests collected within 6 months of diagnosis (3,345,083 results). Using the LWAS framework, the false-discovery rate was used to test the association between multiple laboratory tests and survival, and these results were validated using training, testing, and validation cohorts.
RESULTS:
A total of 31 laboratory tests associated with survival met stringent LWAS criteria. LWAS confirmed markers of prostate cancer biology (prostate-specific antigen: hazard ratio [HR], 1.07 [95% confidence interval (95% CI), 1.06–1.08]; and alkaline phosphatase: HR, 1.22 [95% CI, 1.20–1.24]) as well laboratory tests of general health (eg, serum albumin: HR, 0.78 [95% CI, 0.76–0.80]; and creatinine: HR, 1.05 [95% CI, 1.03–1.07]) and inflammation (leukocyte count: HR, 1.23 [95% CI, 1.98–1.26]; and erythrocyte sedimentation rate: HR, 1.33 [95% CI, 1.09–1.61]). In addition, the authors derived and validated separate models for patients with localized and advanced disease, identifying 28 laboratory markers and 15 laboratory markers, respectively, in each cohort.
CONCLUSIONS:
The authors identified routinely collected laboratory data associated with survival for patients with prostate cancer using LWAS methodologies, including markers of prostate cancer biology, overall health, and inflammation. Broadening consideration of determinants of survival beyond those related to cancer itself could help to inform the design of clinical trials and aid in shared decision making.
LAY SUMMARY:
This article examined routine laboratory tests associated with survival among veterans with prostate cancer.
Using laboratory-wide association studies, the authors identified 31 laboratory tests associated with survival that can be used to inform the design of clinical trials and aid patients in shared decision making.
Keywords: biomarkers, laboratory markers, life expectancy, methods, prostate cancer
INTRODUCTION
“Precision oncology” and “personalized medicine” are terms often applied to analyses of a tumor (eg, molecular and genomic biomarkers) that provide insight into the cancer’s behavior and may allow for targeted treatments. Ideally, a sophisticated understanding of each patient’s overall health and estimated longevity also could inform patient-centered treatment decisions. For example, an indolent cancer may be clinically significant only for patients with a long life expectancy.1 Although understanding a patient’s baseline health and competing risks of mortality is an essential component of cancer-specific screening and treatment strategies,2 determining health and life expectancy from electronic health records remains a challenge.
Prostate cancer accounts for nearly 20% of new cancer diagnoses in American men.3 A multitude of predictive models have been developed to estimate life expectancy4–6 and prostate cancer mortality risk, often incorporating biomarkers such as prostate-specific antigen (PSA); other novel urinary, serology-based, and pathological biomarkers specific to prostate cancer continue to be developed.7 However, to our knowledge, to date no approaches have incorporated routinely collected laboratory tests to refine survival models or to guide clinical decision making.
Laboratory values serve as ideal biomarkers due to their accessibility and low cost. In the Dialogue on Reverse Engineering and Assessment Methods (DREAM) challenge, select laboratory test results were found to be the most important contributors to the prediction of survival for patients with metastatic castration-resistant prostate cancer who were enrolled in clinical trials.8 However, the literature reporting associations among common laboratory tests and survival remains fragmented, often reporting a single laboratory test in a small patient cohort without validation. As a result, to our knowledge, the relative strength of associations across multiple candidate laboratory tests remains unknown. Moreover, there is concern that initially reported biomarker effect sizes are inflated and that many candidate biomarkers fail to reproduce in subsequent studies using real-world data.9,10 As such, routinely collected laboratory values have yet to be incorporated into commonly used predictive models.
To systematically evaluate laboratory values associated with survival among patients with prostate cancer, we developed a laboratory-wide association study (LWAS) approach by adapting genome-wide association study (GWAS) methods.11,12 GWAS is a popular framework with which to analyze genetic factors that correlate with disease on a genome-wide scale. GWAS studies first appeared in the literature in 2005 and became more widespread after a landmark publication in 2007.13 The LWAS framework incorporates multiple GWAS methods to facilitate reproducibility, including: 1) using the false discovery rate (FDR) to simultaneously test the association between multiple laboratory values and survival; 2) requiring concordant results in training and testing sets; and, finally, 3) requiring each test to be validated in separate cohorts. We conducted the current study using data from the Veterans Health Administration (VHA), the largest integrated health care system in the United States that cares for a large population of men with prostate cancer.14 Our goal was to test whether routinely collected laboratory values were associated with survival in men with prostate cancer of all stages.
MATERIALS AND METHODS
We obtained institutional review board approval and research permissions for VHA data access.
Patient Cohort
We identified a cohort of veterans diagnosed with incident prostate cancer between 2000 and 2013 within the VHA electronic health record using the national Veterans Affairs Corporate Data Warehouse.15 These data allow for patient identification through a unique scrambled social security number allowing linkages to determine all VHA inpatient and outpatient services as well as laboratory results.
Clinical Laboratory Data and Covariates
We assembled data regarding patient demographics, tumor staging, and treatment from the Veterans Affairs Corporate Data Warehouse Oncology database. We excluded patients without complete staging information. We then stratified patient records into localized prostate cancer and locally advanced or metastatic prostate cancer risk groups (clinical classification T4, N >0, or M >0). We quantified patient comorbidities using the Deyo-Romano modification of the Charlson Comorbidity Index in the 2 years prior to the date of prostate cancer diagnosis for each patient.16 The primary treatment was defined using 4 mutually exclusive categories: 1) surgery; 2) radiotherapy; 3) androgen deprivation therapy; or 4) conservative management.17
We then obtained all laboratory data for each patient in the 6 months prior to their prostate cancer diagnosis. Laboratory tests were abstracted from the Managerial Cost Accounting System, a repository of common laboratory tests across the 170 medical centers and 1074 outpatient sites of care in the VHA. To capture routinely collected laboratory tests, we included only tests with results for at least 200 patients. We cleaned the laboratory data, and excluded extreme outlier values (eg, the lowest 1% and highest 1% of values) if these values were not biologically plausible. For example, we excluded the lowest 1% of serum creatinine measurements (<0.4 mg/dL), but not the top 1% of serum creatinine values (>5.9 mg/dL). If multiple results were available for a selected laboratory test after cleaning, we applied the average of these results as the assigned value. A total of 3,345,083 laboratory test values remained after these exclusions and consolidation, from an initial pool of 12,559,629. We normalized laboratory results using z standardization.
Survival
We evaluated survival using the VA vital status file, which compiles data from the VHA, the Centers for Medicare and Medicaid Services, the US Social Security Administration, and the National Cemetery Association.18 We censored follow-up time on December 31, 2017, for patients alive on or after this date.
LWAS Analysis
GWAS examines thousands of candidate single-nucleotide polymorphisms that may be associated with an outcome. The term “XWAS” refers to the application of statistical approaches popularized in GWAS studies to other clinical problems, particularly when there are many measured candidate markers. For example, XWAS approaches have been used to study other large-scale clinical problems19 such as environmental exposures associated with blood pressure,20 diet and nutrients associated with incident prostate cancer,21 and behavioral and health factors associated with overall survival.12
In the current study, we adapted XWAS methods to develop an LWAS approach using laboratory results available in the electronic health record to identify associations with survival for patients with prostate cancer. LWAS is a rigorous, 3-stage, hierarchical approach for evaluating the associations between individual laboratory tests and mortality. The first 2 stages use independent data sets to apply a stage-specific, model-based decision rule to a set of candidate laboratory results to determine which results proceed to the next stage. Laboratory results that do not meet criteria at any stage do not proceed to subsequent stages. Associations between outcome and laboratory results that survive to the third stage are characterized using a third independent data set (see Supporting Fig. 1).
First, we randomly split patient records into training, testing, and validation data sets, respectively, in 30%, 30%, and 40% proportions. Using the training set, we then performed Cox proportional hazards regression analysis for each laboratory value, adjusting for relevant clinical characteristics (patient age, tumor stage, treatment type, and comorbidity). We identified laboratory tests with robust associations with mortality in the training set after controlling the FDR to be ≤5% using the Benjamini-Hochberg method.22 The FDR is useful in discovery because it accounts for multiple comparisons, thereby limiting false-positive associations while providing more power than traditional Bonferroni correction approaches.23
For each laboratory test that passed the FDR threshold of ≤5%, we then fit another adjusted Cox model in the testing set (see Supporting Fig. 1). We considered laboratory tests to be significant if the associated value was <.05 in the testing set and if the direction of association was concordant in the training and testing data sets. Finally, each laboratory test then was required to be statistically significant in a third Cox model using the validation set to meet all LWAS criteria. Specifically, we reported results in terms of the standardized hazard ratio (HR, reported for a 1–standard deviation [SD] unit change in each laboratory test). For each validated clinical laboratory test, we tested the correlation among markers because future efforts might aim to select a more parsimonious panel.
We then performed this series of analyses separately for patients with localized and metastatic disease. We performed all statistical analyses within the VA Informatics and Computing Infrastructure platform using SAS Enterprise Guide 7.1 (SAS Institute Inc, Cary, North Carolina) and R statistical software (version 3.5.2; R Foundation, Vienna, Austria). We created figures using R and JMP Pro v14 (SAS Institute Inc).
RESULTS
We identified 181,009 patients who were diagnosed with prostate cancer during the study period, of whom 133,878 (74%) had had ≥1 routinely collected common outpatient laboratory tests drawn within 6 months of their diagnosis. Approximately 92.7% of patients were diagnosed with localized disease and 7.3% were diagnosed with locally advanced or metastatic disease. The mean age of the cohort was 66.6 years (±8.4 years) and the mean Charlson Comorbidity Index was 2.5 (±2.1). Table 1 provides the detailed demographic information for the current study cohort. Each patient contributed a median of 46 laboratory test results (25th, 75th percentile range, 26–72 results) in the 6 months preceding the prostate cancer diagnosis.
TABLE 1.
Demographics of the Cohort of 133,878 Veterans Who Were Diagnosed With Prostate Cancer and Were Receiving Care in the VHA From 2004 Through 2013
| Patient Characteristics | Entire Cohort N = 133,878 | Those With Localized Disease N = 126,151 | Those With Locally Advanced and/or Metastatic Disease N = 7727 |
|---|---|---|---|
|
| |||
| Age (mean ± SD), y | 66.6 ± 8.4 | 66.2 ± 8.1 | 72.4 ± 10.8 |
| Charlson Comorbidity Index (mean ± SD) | 2.5 ± 2.1 | 2.4 ± 2.0 | 3.9 ± 3.1 |
| Race and/or ethnicity, % | |||
| White | 61.6 | 61.7 | 60.0 |
| Black | 26.3 | 26.3 | 24.9 |
| Other | 7.3 | 7.3 | 6.9 |
| Missing data | 4.8 | 4.6 | 7.1 |
| Prostate disease stage, % | |||
| Localized | 94.2 | ||
| Locally advanced/metastatic | 5.8 | ||
| Initial treatment, % | |||
| Surgery | 20.7 | 62.4 | 9.7 |
| Radiotherapy | 31.9 | 17.7 | 35.7 |
| Androgen deprivation therapy | 34.6 | 16.4 | 32.8 |
| Conservative management | 12.8 | 3.5 | 21.8 |
Abbreviations: SD, standard deviation; VHA, Veterans Health Administration.
We have reported the results of the LWAS approach for each laboratory test in Figure 1. When analyzing the entire cohort, 38 laboratory tests met the initial FDR threshold of ≤5% in the training set. A total of 34 tests retained significance and concordant direction of the association in the testing set, and 31 of these laboratory tests met significance thresholds in the validation set (Fig. 1A). The Pearson correlation coefficient among all laboratory tests, as well as among the laboratory tests that met LWAS criteria, are shown in Supporting Figures 2 and 3.
Figure 1.

Routinely collected laboratory tests found to be associated with survival across the entire cohort of 133,878 patients with prostate cancer. (A) Sankey diagram indicating whether each laboratory (Lab) test met each laboratory-wide association study (LWAS) criterion. The 31 laboratory tests that met all LWAS criteria are shown in brown whereas the laboratory tests that failed are shown in blue. (B) For each laboratory test, the standardized effect size (hazard ratio [HR]) is represented on the x-axis, whereas the strength of the association (the P value represented in −log10 scale) is shown on the y-axis. Each laboratory test that met the LWAS criteria and demonstrated a potentially clinically significant effect size (standardized HR <0.9 or >1) was labeled. ALT indicates alanine aminotransferase; AST, aspartate aminotransferase; BNP, brain natriuretic peptide; BUN, blood urea nitrogen; CEA, carcinoembryonic antigen; Cr, creatinine; CRP, C-reactive protein; ESR, erythrocyte sedimentation rate; FDR, false discovery rate; GFR, glomerular filtration rate; GGT, γ-glutamyl transferase; Hb, hemoglobin; HbA1c, hemoglobin A1c; HCT, hematocrit; HDL, high-density lipoprotein; HS, high sensitivity; INR, international normalized ratio; LDL, low-density lipoprotein; pCO2, partial pressure of carbon dioxide; pO2, partial pressure of oxygen; POC, point of care; Pro-BNP, pro-brain natriuretic peptide; PSA, prostate-specific antigen; PTT, partial thromboplastin time; SaO2, oxygen saturation; TSH, thyroid-stimulating hormone; Vit, vitamin; WBC, white blood count.
The relative HR and the strength of the association in the final validation set are shown in Figure 1B and Figure 2. The 31 laboratory tests that met LWAS criteria included general health markers. The laboratory tests with the largest standardized effect sizes included pro-brain natriuretic peptide and γ-glutamyl transferase, which were most strongly and directly associated with mortality (HR, 1.43 and HR, 1.23, respectively); higher serum albumin (HR, 0.78) and hemoglobin (HR, 0.81) were found to be inversely associated with mortality. Additional tests identified by LWAS also included known prostate cancer biomarkers associated with mortality (PSA: HR, 1.07; and alkaline phosphatase: HR, 1.18). The distribution of the HRs for all covariates in the final adjusted models are shown in Supporting Figure 4.
Figure 2.
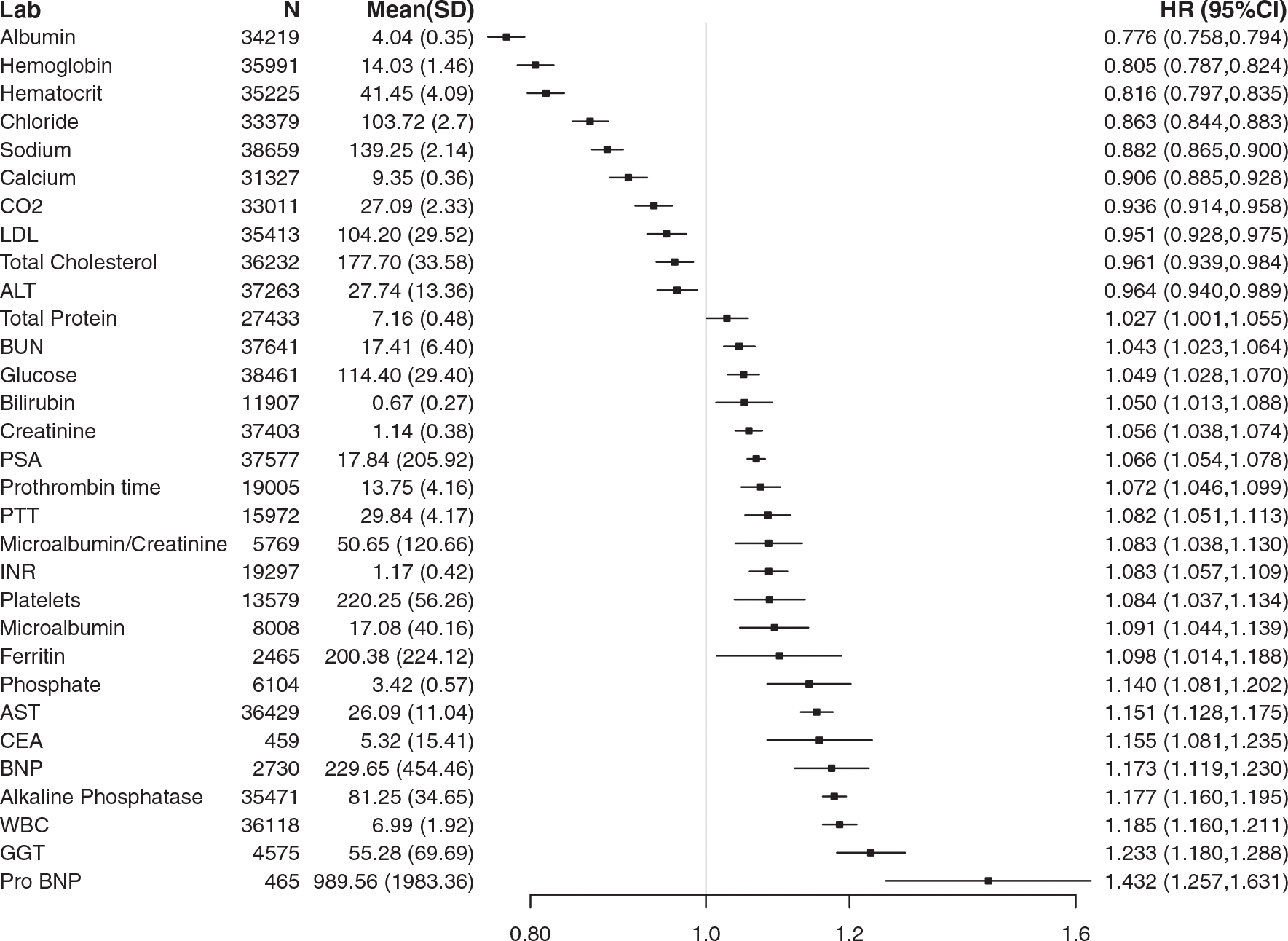
Thirty-one laboratory (Lab) tests that met laboratory-wide association study (LWAS) criteria for the entire cohort of 133,878 patients with prostate cancer. Each standardized hazard ratio (HR) is reported for a 1-standard deviation change in the laboratory test result after adjusting for patient age, comorbidity, prostate cancer stage of disease, and treatment type. The corresponding means and standard deviations of the laboratory test results are shown for reference. ALT indicates alanine aminotransferase; AST, aspartate aminotransferase; BNP, brain natriuretic peptide; BUN, blood urea nitrogen; CEA, carcinoembryonic antigen; CO2, carbon dioxide; GGT, γ-glutamyl transferase; INR, international normalized ratio; LDL, low-density lipoprotein; Pro-BNP, pro-brain natriuretic peptide; PSA, prostate-specific antigen; PTT, partial thromboplastin time.
Analyses in the Subcohort of Patients With Localized Disease
We then analyzed the cohort of patients with localized prostate cancer (124,089 patients) separately (Figs. 3 and 4). A total of 28 laboratory tests met LWAS criteria. The erythrocyte sedimentation rate (HR, 1.33), leukocyte count (HR, 1.23), and alkaline phosphatase (HR, 1.22) were found to be directly associated with mortality, whereas serum albumin (HR, 0.78) and hemoglobin (HR, 0.80) were found to be inversely associated with mortality.
Figure 3.

Routinely collected laboratory (Lab) tests found to be associated with survival in 124,089 patients with localized prostate cancer. (A) Sankey diagram indicating whether each laboratory test met each laboratory-wide association study (LWAS) criterion. The 28 laboratory tests that met all LWAS criteria are shown in brown whereas the laboratory tests that failed are shown in blue. (B) For each laboratory test, the standardized effect size (hazard ratio [HR]) is represented on the x-axis whereas the strength of the association (the P value represented in −log10 scale) is shown on the y-axis. Each laboratory test that met the LWAS criteria and demonstrated a potentially clinically significant effect size (standardized HR <0.9 or >1) was labeled. ALT indicates alanine aminotransferase; AST, aspartate aminotransferase; BNP, brain natriuretic peptide; BUN, blood urea nitrogen; CEA, carcinoembryonic antigen; Cr, creatinine; CRP, C-reactive protein; ESR, erythrocyte sedimentation rate; FDR, false discovery rate; GFR, glomerular filtration rate; GGT, γ-glutamyl transferase; Hb, hemoglobin; HbA1c, hemoglobin A1c; HCT, hematocrit; HDL, high-density lipoprotein; HS, high sensitivity; INR, international normalized ratio; LDL, low-density lipoprotein; pCO2, partial pressure of carbon dioxide; pO2, partial pressure of oxygen; POC, point of care; Pro-BNP, pro-brain natriuretic peptide; PSA, prostate-specific antigen; PTT, partial thromboplastin time; SaO2, oxygen saturation; TSH, thyroid-stimulating hormone; Vit, vitamin; WBC, white blood count.
Figure 4.

Twenty-eight laboratory (Lab) tests that met laboratory-wide association study (LWAS) criteria for the entire cohort of 124,089 patients with localized prostate cancer. Each standardized hazard ratio (HR) is reported for a 1-standard deviation change in the laboratory test result after adjusting for patient age, comorbidity, prostate cancer stage of disease, and treatment type. The corresponding means and standard deviations of the laboratory test results are shown for reference. AST indicates aspartate aminotransferase; BNP, brain natriuretic peptide; BUN, blood urea nitrogen; ESR, erythrocyte sedimentation rate; GGT, γ-glutamyl transferase; HDL, high-density lipoprotein; INR, international normalized ratio; LDL, low-density lipoprotein; Pro-BNP, pro-brain natriuretic peptide; PSA, prostate-specific antigen; PTT, partial thromboplastin time; WBC, white blood count.
Analyses in the Subcohort With Locally Advanced and Metastatic Disease
Among the 9789 patients diagnosed with metastatic prostate cancer, we found 15 laboratory tests associated with survival that met LWAS criteria (Figs. 5 and 6). The laboratory tests with the largest standardized effect size directly associated with mortality were alkaline phosphatase (HR, 1.33), ferritin (HR, 1.23), and platelet count (HR, 1.22). In contrast, serum albumin (HR, 0.75) and hemoglobin (HR, 0.75) were found to be inversely associated with mortality in this subcohort.
Figure 5.

Routinely collected laboratory (Lab) tests found to be associated with survival in 9789 patients with locally advanced and/or metastatic prostate cancer. (A) Sankey diagram indicating whether each laboratory test met each laboratory-wide association study (LWAS) criterion. The 15 laboratory tests that met all LWAS criteria are shown in brown whereas the laboratory tests that failed are shown in blue. (B) For each laboratory test, the effect size (hazard ratio [HR]) is represented on the x-axis, whereas the strength of the association (the P value represented in −log10 scale) is shown on the y-axis. Each laboratory test that met the LWAS criteria and demonstrated a potentially clinically significant effect size (standardized HR <0.9 or >1) was labeled. ALT indicates alanine aminotransferase; AST, aspartate aminotransferase; BNP, brain natriuretic peptide; BUN, blood urea nitrogen; CEA, carcinoembryonic antigen; Cr, creatinine; CRP, C-reactive protein; ESR, erythrocyte sedimentation rate; FDR, false discovery rate; GFR, glomerular filtration rate; GGT, γ-glutamyl transferase; Hb, hemoglobin; HbA1c, hemoglobin A1c; HCT, hematocrit; HDL, high-density lipoprotein; HS, high sensitivity; INR, international normalized ratio; LDL, low-density lipoprotein; pCO2, partial pressure of carbon dioxide; pO2, partial pressure of oxygen; POC, point of care; Pro-BNP, pro-brain natriuretic peptide; PSA, prostate-specific antigen; PTT, partial thromboplastin time; SaO2, oxygen saturation; TSH, thyroid-stimulating hormone; Vit, vitamin; WBC, white blood count.
Figure 6.

Fifteen laboratory (Lab) tests that met laboratory-wide association study (LWAS) criteria for the entire cohort of 9789 patients with locally advanced and/or metastatic prostate cancer. Each hazard ratio (HR) is reported for a 1-standard deviation change in the laboratory test result after adjusting for patient age, comorbidity, prostate cancer stage of disease, and treatment type. The corresponding means and standard deviations of the laboratory test results are shown for reference. AST, aspartate aminotransferase; BUN, blood urea nitrogen; CO2, carbon dioxide; PSA, prostate-specific antigen; WBC, white blood count.
DISCUSSION
Identifying laboratory measures of overall health may aid in the challenging task of estimating life expectancy among men with prostate cancer, among whom overdiagnosis is common and in whom competing risks of mortality dominate prostate cancer–specific mortality risk.24 We demonstrated the use of LWAS, using GWAS methods, to identify routinely collected laboratory test results associated with all-cause mortality among patients diagnosed with prostate cancer. The LWAS approach applies a rigorous statistical analysis to confirm these associations on a large scale using real-world data. We identified 31 laboratory tests that passed LWAS criteria and were found to be associated with mortality in a large, national cohort.
The patterns we identified in the subcohorts with localized or metastatic disease generally were similar. The findings of the current study suggested that laboratory proxies of the inflammatory state are associated with mortality in patients with prostate cancer. Systemic inflammation increasingly has been shown to be associated with the diagnosis and progression of many cancers, including urologic malignancies.25 Prior studies have reported associations between C-reactive protein26 and the erythrocyte sedimentation rate27 and mortality in patients with localized prostate cancer. These LWAS findings have confirmed prior reports demonstrating that markers of the systemic inflammatory response are associated with mortality among patients diagnosed with cancer.28,29 To the best of our knowledge, the question of whether these findings more broadly reflect the overall health of a patient, the systemic effects of cancer before a formal diagnosis of prostate cancer, or a susceptibility to cancer progression is unknown.
We also noted the prognostic power of basic hematologic laboratory studies (eg, hemoglobin, hematocrit, platelets, and leukocyte count) in patients with localized or advanced disease. Previous studies have highlighted lower disease-specific survival in patients with lower serum albumin among patients with localized30 and metastatic prostate cancer.31,32 These may be measures of patient health (including the state of inflammation and nutritional status), and are consistent with prior multivariable models derived from a secondary analysis of clinical trial data that found that hemoglobin, alkaline phosphatase, aspartate aminotransferase, and albumin were significantly associated with survival in patients with metastatic disease.33,34 These data may reinforce the concept that patients with a diagnosis of prostate cancer are more likely to die with prostate cancer rather than of prostate cancer. Alternatively, measures of nutrition and inflammation (eg, albumin and hemoglobin) also have been associated with response to treatment among patients with advanced prostate cancer, such as response to chemotherapy and radioligand therapy.33,34
LWAS identified PSA and alkaline phosphatase as being directly related to morality; these tests are well established markers of prostate cancer. It is worth noting that among patients diagnosed with prostate cancer, a 1-SD change in PSA levels was found to be only modestly associated with survival; many other laboratory tests demonstrated standardized HRs of higher magnitude. Alkaline phosphatase is a marker of bone turnover and therefore is known to be linked with bone metastasis volume and overall survival in patients with metastatic prostate cancer.35,36 It is interesting to note that alkaline phosphatase also was associated with survival in patients with localized disease.
These laboratory test and survival associations hold considerable clinical value. An increasing number of models, particularly in populations of patients with metastatic disease, are attempting to use laboratory values to stratify clinical trial participants and refine study design. For example, the DREAM challenge demonstrates the ability of baseline clinical data, including laboratory tests, to optimize predictive models for patients with metastatic, castration-resistant prostate cancer.8 The inclusion of routinely collected laboratory data would strengthen countless models, such as the PREDICT model, that do not yet include laboratory data to stratify risk among patients with prostate cancer and improve model estimates.37
The current study has notable strengths. Herein, we have introduced the LWAS approach, which provides a method with which to systematically identify multiple laboratory tests associated with an outcome using a stringent framework designed to maximize reproducibility. We demonstrated how laboratory tests can appear to be promising markers but fail to retain significance in validation data sets. LWAS appears to be less likely to identify false-positive findings compared with conventional reports of a single laboratory test in a single patient cohort. The LWAS framework also provides some insight into the relative importance of each laboratory test by using standardized laboratory test results. Second, the current study leveraged data from the VHA electronic health record. Specifically, the VHA provides a large, national cohort, distributed across 170 hospitals, with high-quality laboratory and survival data. The large cohort size, and capture of routine clinical laboratory tests, allowed for the validation of these findings in randomly selected cohorts.
There also were several limitations to the current study. First, an LWAS approach using standardized estimated effect sizes assumes a linear relation between laboratory test results and mortality. It is possible that some laboratory tests that do not meet LWAS criteria would prove to be associated with mortality if they were analyzed using nonlinear models or when analyzed as dichotomous variables using clinical cutoff values. It is interesting to note that the approach used in the current study did not assume relevant clinical cutoff values, which can vary by patient age and health status.38 Moreover, using dichotomized results from continuous laboratory test results risks the loss of power and precision,39–42 and causes concern for the misclassification of patients around the selected “high” or “low” cutoff values.43 Similarly, we did not evaluate the associations observed using combinations of tests that might reflect organ dysfunction (eg, alanine aminotransferase plus bilirubin) or the ratio of specific tests (eg, the neutrophil-to-lymphocyte ratio). Second, although the LWAS approach used in the current study standardized laboratory test results to compare the relative effects of individual laboratory tests (ie, the HR for a 1-SD change in the laboratory result), patients did not necessarily undergo a complete panel of laboratory tests. Third, although we selected the most common laboratory tests used in clinical care, there are available laboratory studies that were not included in the current analysis. Thus, we might have missed an association between an infrequently used laboratory test and mortality. Fourth, we tested for associations among laboratory tests and mortality, but other outcomes may be of interest to clinicians and patients with prostate cancer. We chose all-cause mortality because it is of paramount importance and the data were reliable. Prostate cancer progression and cancer-specific survival, as well as other important outcomes, are much more difficult to capture from large populations and administrative data, as are parameters of health status such as functional capacity or health-related quality of life. Last, although the VHA cares for a large, diverse, and nationally distributed population, the VHA population does differ from the general population and therefore the findings of the current study may not be fully generalizable outside of this system. However, nonveteran patients may be more similar to patients seen in the community than patients selected for clinical trials, who have been shown not to reflect the general population.44
Conclusions
The results of the current study have demonstrated the usefulness of LWAS to identify routinely collected laboratory tests associated with survival among men with prostate cancer. In the current study, LWAS identified 31 routinely collected laboratory tests associated with survival. Proinflammatory markers were found consistently and directly to be associated with mortality in patients with localized and locally advanced and/or metastatic disease. In this era of personalized medicine, the addition of laboratory data to survival models and clinical trial design has the potential to individualize and refine approaches to the management of patients with prostate cancer.
Supplementary Material
FUNDING SUPPORT
Supported in part by the VA Merit Review (I01 HX0021261 to John T. Leppert) from the US Department of Veterans Affairs Health Services Research and Development Service. Work was supported using resources and facilities at the Veterans Affairs Informatics and Computing Infrastructure (VINCI) (VA HSR RES 13–457). The contents herein do not represent the views of the US Department of Veterans Affairs or the US government.
Footnotes
CONFLICT OF INTEREST DISCLOSURES
Jaden Yang was supported by a grant from the US Department of Veterans Affairs Health Services Research and Development Service for work performed as part of the current study. Kristopher Kapphahn was supported by a grant from the US Department of Veterans Affairs Health Services Research and Development Service for work performed as part of the current study. Todd H. Wagner has received grants from the US Department of Veterans Affairs for work performed outside of the current study. Chirag J. Patel has received grants from the National Institutes of Health for work performed as part of the current study and has received personal fees and nonfinancial support from XY Health Inc, personal fees from the Centers for Disease Control and Prevention, and personal fees from Janssen Inc for work performed outside of the current study. The other authors made no disclosures.
Additional supporting information may be found in the online version of this article.
REFERENCES
- 1.Welch HG, Black WC. Overdiagnosis in cancer. J Natl Cancer Inst. 2010;102:605–613. [DOI] [PubMed] [Google Scholar]
- 2.Walter LC, Covinsky KE. Cancer screening in elderly patients: a framework for individualized decision making. JAMA. 2001;285:2750–2756. [DOI] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7–30. [DOI] [PubMed] [Google Scholar]
- 4.Kent M, Vickers AJ. A systematic literature review of life expectancy prediction tools for patients with localized prostate cancer. J Urol. 2015;193:1938–1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Daskivich TJ, Thomas IC, Luu M, et al. External validation of the Prostate Cancer Specific Comorbidity Index: a claims based tool for the prediction of life expectancy in men with prostate cancer. J Urol. 2019;202:518–524. [DOI] [PubMed] [Google Scholar]
- 6.Sohlberg EM, Thomas IC, Yang J, et al. Life expectancy estimates for patients diagnosed with prostate cancer in the Veterans Health Administration. Urol Oncol. 2020;38:734.e1–734.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cooperberg MR, Carroll PR, Dall’Era MA, et al. The State of the Science on Prostate Cancer Biomarkers: The San Francisco Consensus Statement. Eur Urol. 2019;76:268–272. [DOI] [PubMed] [Google Scholar]
- 8.Guinney J, Wang T, Laajala TD, et al. Prediction of overall survival for patients with metastatic castration-resistant prostate cancer: development of a prognostic model through a crowdsourced challenge with open clinical trial data. Lancet Oncol. 2017;18:132–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ioannidis JP, Panagiotou OA. Comparison of effect sizes associated with biomarkers reported in highly cited individual articles and in subsequent meta-analyses. JAMA. 2011;305:2200–2210. [DOI] [PubMed] [Google Scholar]
- 10.Collins GS, Reitsma JB, Altman DG, Moons KGM; members of the TRIPOD group. Transparent Reporting of a Multivariable Prediction Model for Individual Prognosis or Diagnosis (TRIPOD): the TRIPOD Statement. Eur Urol. 2015;67:1142–1151. [DOI] [PubMed] [Google Scholar]
- 11.Patel CJ, Bhattacharya J, Butte AJ. An environment-wide association study (EWAS) on type 2 diabetes mellitus. PLoS One. 2010;5:e10746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Patel CJ, Rehkopf DH, Leppert JT, et al. Systematic evaluation of environmental and behavioural factors associated with all-cause mortality in the United States National Health and Nutrition Examination Survey. Int J Epidemiol. 2013;42:1795–1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zullig LL, Sims KJ, McNeil R, et al. Cancer incidence among patients of the U.S. Veterans Affairs health care system: 2010 update. Mil Med. 2017;182:e1883–e1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fihn SD, Francis J, Clancy C, et al. Insights from advanced analytics at the Veterans Health Administration. Health Aff (Millwood). 2014;33:1203–1211. [DOI] [PubMed] [Google Scholar]
- 16.Deyo RA, Cherkin DC, Ciol MA. Adapting a clinical comorbidity index for use with ICD-9-CM administrative databases. J Clin Epidemiol. 1992;45:613–619. [DOI] [PubMed] [Google Scholar]
- 17.Barbosa PV, Thomas IC, Srinivas S, et al. Overall survival in patients with localized prostate cancer in the US Veterans Health Administration: is PIVOT generalizable? Eur Urol. 2016;70:227–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sohn MW, Arnold N, Maynard C, Hynes DM. Accuracy and completeness of mortality data in the Department of Veterans Affairs. Popul Health Metr. 2006;4:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Patel CJ, Ioannidis JP. Studying the elusive environment in large scale. JAMA. 2014;311:2173–2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McGinnis DP, Brownstein JS, Patel CJ. Environment-wide association study of blood pressure in the National Health and Nutrition Examination Survey (1999–2012). Sci Rep. 2016;6:30373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Papadimitriou N, Muller D, van den Brandt PA, et al. A nutrient-wide association study for risk of prostate cancer in the European Prospective Investigation into Cancer and Nutrition and the Netherlands Cohort Study. Eur J Nutr. 2020;59:2929–2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57:289–300. [Google Scholar]
- 23.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100:9440–9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Esserman LJ, Thompson IM Jr, Reid B. Overdiagnosis and overtreatment in cancer: an opportunity for improvement. JAMA. 2013;310:797–798. [DOI] [PubMed] [Google Scholar]
- 25.Sylman JL, Mitrugno A, Atallah M, et al. The predictive value of inflammation-related peripheral blood measurements in cancer staging and prognosis. Front Oncol. 2018;8:78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hall WA, Nickleach DC, Master VA, et al. The association between C-reactive protein (CRP) level and biochemical failure-free survival in patients after radiation therapy for nonmetastatic adenocarcinoma of the prostate. Cancer. 2013;119:3272–3279. [DOI] [PubMed] [Google Scholar]
- 27.Johansson JE, Sigurdsson T, Holmberg L, Bergstrom R. Erythrocyte sedimentation rate as a tumor marker in human prostatic cancer. An analysis of prognostic factors in 300 population-based consecutive cases. Cancer. 1992;70:1556–1563. [DOI] [PubMed] [Google Scholar]
- 28.Sylman JL, Boyce HB, Mitrugno A, et al. A temporal examination of platelet counts as a predictor of prognosis in lung, prostate, and colon cancer patients. Sci Rep. 2018;8:6564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shafique K, Proctor MJ, McMillan DC, Qureshi K, Leung H, Morrison DS. Systemic inflammation and survival of patients with prostate cancer: evidence from the Glasgow Inflammation Outcome Study. Prostate Cancer Prostatic Dis. 2012;15:195–201. [DOI] [PubMed] [Google Scholar]
- 30.Wang Y, Chen W, Hu C, et al. Albumin and fibrinogen combined prognostic grade predicts prognosis of patients with prostate cancer. J Cancer. 2017;8:3992–4001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arthur R, Williams R, Garmo H, et al. Serum inflammatory markers in relation to prostate cancer severity and death in the Swedish AMORIS study. Int J Cancer. 2018;142:2254–2262. [DOI] [PubMed] [Google Scholar]
- 32.Nadal R, Tsai HL, Sinibaldi VJ, et al. Prognostic factors for clinical outcomes in patients with metastatic castration resistant prostate cancer treated with sequential novel androgen receptor–directed therapies. Prostate. 2016;76:512–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ahmadzadehfar H, Schlolaut S, Fimmers R, et al. Predictors of overall survival in metastatic castration-resistant prostate cancer patients receiving [177Lu]Lu-PSMA-617 radioligand therapy. Oncotarget. 2017; 8:103108–103116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Belderbos BPS, de Wit R, Hoop EO, et al. Prognostic factors in men with metastatic castration-resistant prostate cancer treated with cabazitaxel. Oncotarget. 2017;8:106468–106474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heinrich D, Bruland O, Guise TA, Suzuki H, Sartor O. Alkaline phosphatase in metastatic castration-resistant prostate cancer: reassessment of an older biomarker. Future Oncol. 2018;14:2543–2556. [DOI] [PubMed] [Google Scholar]
- 36.Robinson D, Sandblom G, Johansson R, et al. ; Scandinavian Prostate Cancer Group (SPCG)-5. Prediction of survival of metastatic prostate cancer based on early serial measurements of prostate specific antigen and alkaline phosphatase. J Urol. 2008;179:117–122; discussion 122–123. [DOI] [PubMed] [Google Scholar]
- 37.Thurtle DR, Greenberg DC, Lee LS, Huang HH, Pharoah PD, Gnanapragasam VJ. Individual prognosis at diagnosis in nonmetastatic prostate cancer: development and external validation of the PREDICT prostate multivariable model. PLoS Med. 2019;16:e1002758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Manrai AK, Patel CJ, Ioannidis JPA. In the era of precision medicine and big data, who is normal? JAMA. 2018;319:1981–1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Collins GS, Ogundimu EO, Cook JA, Manach YL, Altman DG. Quantifying the impact of different approaches for handling continuous predictors on the performance of a prognostic model. Stat Med. 2016;35:4124–4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bennette C, Vickers A. Against quantiles: categorization of continuous variables in epidemiologic research, and its discontents. BMC Med Res Methodol. 2012;12:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Greenland S Avoiding power loss associated with categorization and ordinal scores in dose-response and trend analysis. Epidemiology. 1995;6:450–454. [DOI] [PubMed] [Google Scholar]
- 42.Naggara O, Raymond J, Guilbert F, Roy D, Weill A, Altman DG. Analysis by categorizing or dichotomizing continuous variables is inadvisable: an example from the natural history of unruptured aneurysms. AJNR Am J Neuroradiol. 2011;32:437–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Altman DG, Royston P. The cost of dichotomising continuous variables. BMJ. 2006;332:1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Al-Refaie WB, Vickers SM, Zhong W, Parsons H, Rothenberger D, Habermann EB. Cancer trials versus the real world in the United States. Ann Surg. 2011;254:438–442; discussion 442–433. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


