Abstract
SWISS-2DPAGE (http://www.expasy.ch/ch2d/ ) is an annotated two-dimensional polyacrylamide gel electrophoresis (2-DE) database established in 1993. The current release contains 24 reference maps from human and mouse biological samples, as well as from Saccharomyces cerevisiae, Escherichia coli and Dictyostelium discoideum origin. These reference maps have now 2824 identified spots, corresponding to 614 separate protein entries in the database, in addition to virtual entries for each SWISS-PROT sequence or any user-entered amino acids sequence. Last year improvements in the SWISS-2DPAGE database are as follows: three new maps have been created and several others have been updated; cross-references to newly built federated 2-DE databases have been added; new functions to access the data have been provided through the ExPASy proteomics server.
INTRODUCTION
The SWISS-2DPAGE database collects data on proteins identified on various two-dimensional polyacrylamide gel electrophoresis (2-DE) maps (1). The identification of proteins on 2-DE maps was obtained by various techniques (2), including gel comparison, microsequencing, immunoblotting, amino acid composition analysis, peptide mass fingerprinting using mass spectrometry, or a combination of some of these. The core of the SWISS-2DPAGE database consists thus in the description of the proteins identified, including mapping procedures, physiological and pathological information, experimental data (isoelectric point, molecular weight, amino acid composition, peptide masses) and bibliographical references, in addition to the 2-DE images showing the protein locations. Cross-references are provided to MEDLINE, to other 2-DE databases [ECO2DBASE (3), HSC-2DPAGE (4), YEPD (5), and the two newly built SIENA-2DPAGE (6–8) and PHCI-2DPAGE (9,10)], and to SWISS-PROT (11), which itself provides many links to other molecular databases [EMBL (12), GenBank (13), PROSITE (14), MIM (15), etc.]. The SWISS-2DPAGE database is maintained by the Swiss Institute of Bioinformatics, in collaboration with the Central Clinical Chemistry Laboratory of Geneva University Hospital.
FORMAT
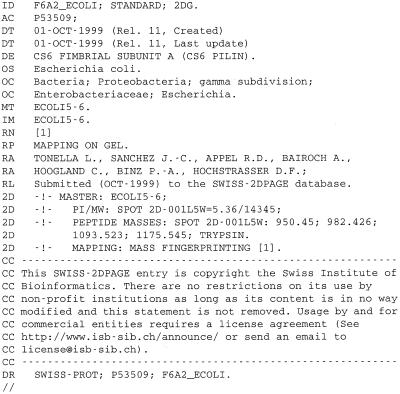
The protein entries in SWISS-2DPAGE are text files structured to be readable by human as well as by computer programs. Each entry is composed of defined lines, used to record various kinds of data (Fig. 1). For standardization purposes, the format of SWISS-2DPAGE entries is similar to the SWISS-PROT database (11) and contains specific lines dedicated to 2-DE data: (i) the master line (MT) lists the reference maps where the entry has been identified; (ii) the images line (IM) lists the 2-DE images available for the entry; (iii) the 2D lines integrate different topics including mapping procedure, spot coordinates, protein amino acid composition, peptide masses from mass spectrometry, protein expression levels and modifications. On the ExPASy Web server (see availability section) the data image associated with a protein entry displays the experimental location of the protein on the chosen map, together with a theoretical region computed from the protein sequence (Fig. 2).
Figure 1.
Example of a SWISS-2DPAGE entry.
Figure 2.
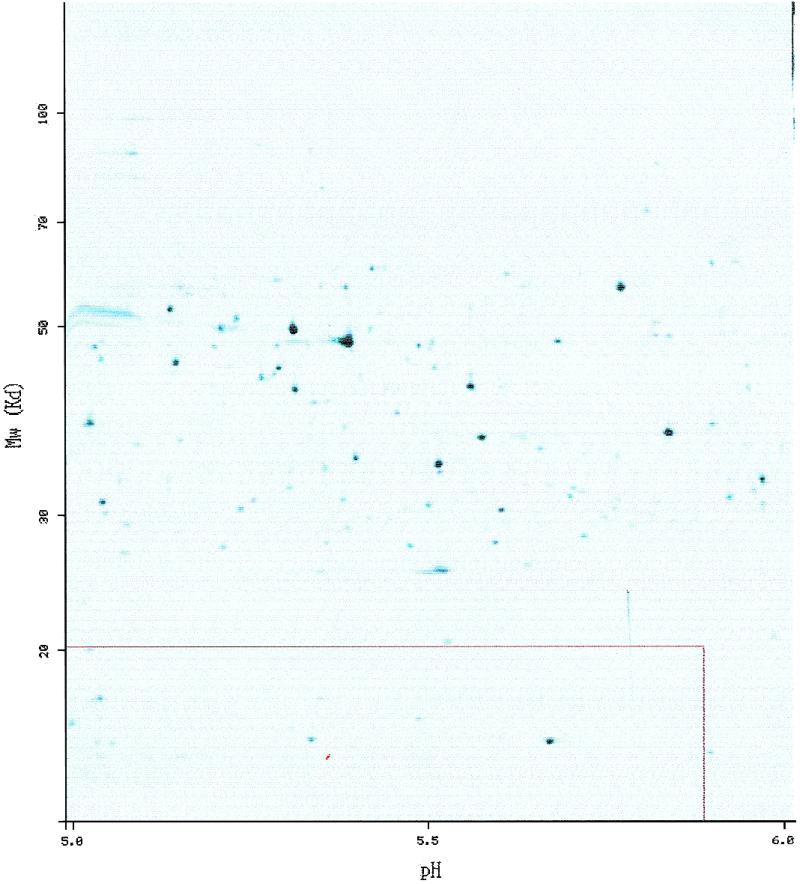
The protein from Figure 1 shown on the SWISS-2DPAGE E.coli reference map for pI range from 5 to 6 (ECOLI5–6). The protein spot itself is highlighted in red, and the red rectangle around shows the theoretical region where one would expect to see the protein, computed from the SWISS-PROT protein sequence.
CURRENT CONTENT
Release 11 of SWISS-2DPAGE (October 1999) contains twenty-four reference maps. Fourteen are from human cells or tissues (kidney, liver, lymphoma, platelet cells, red blood cells, colorectal epithelia cells), body fluids (cerebrospinal fluid, plasma), culture cells (colorectal adenocarcinoma cells, erythroleukemia cell line, hepatoblastoma carcinoma derived cells, hepatoblastoma carcinoma derived cell line secreted proteins, macrophage like cell line, promyelocytic leukemia derived cells); four are from mouse cells or tissues (liver, gastrocnemius muscle, pancreatic islet cells, epididymal fat pad); the other maps are from Saccharomyces cerevisiae, Escherichia coli (obtained for four pI ranges: 3.5–10, 4–5, 4.5–5.5, 5–6) and Dictyostelium discoideum origin. Table 1 gives detailed descriptions for each of these maps, including creation and release dates, number of detected spots, number of identified spots, and number of distinct protein entries. On these maps a total of 2824 spots have been identified by gel matching (51%), immunoblotting (24%), co-migration (3%), microsequencing (17%), amino acids composition (6%) and mass spectrometry (15%). Total exceeds 100% as some of the spots have been identified by several methods. These identifications correspond currently to 614 different protein entries from human, mouse, yeast, E.coli and D.discoideum origin (Fig. 3). In addition, there are as many virtual entries as protein sequences in the SWISS-PROT database or as any user-entered amino acids sequence.
Table 1. Content of SWISS-2DPAGE.
| Map | Creation | Last modification | Detected spots | Identified spots | No. of entries |
|---|---|---|---|---|---|
| HUMAN | |||||
| CEC | Jan-98 | Jan-98 | 3077 | 56 | 40 |
| CSF | Aug-93 | Nov-98 | 1664 | 309 | 30 |
| DLD1 | Jan-99 | Jan-99 | 3440 | 108 | 67 |
| ELC | Aug-93 | Jun-97 | 2144 | 35 | 19 |
| HEPG2 | Aug-93 | Sep-97 | 2862 | 98 | 45 |
| HEPG2SP | Aug-93 | Jan-96 | 1734 | 155 | 25 |
| HL60 | Sep-97 | Sep-97 | 3164 | 26 | 17 |
| KIDNEY | Sep-97 | Sep-97 | 2896 | 42 | 27 |
| LIVER | Aug-93 | Sep-97 | 2413 | 138 | 68 |
| LYMPHOMA | Aug-93 | Jun-97 | 1890 | 60 | 33 |
| PLASMA | Aug-93 | Sep-97 | 1966 | 626 | 70 |
| PLATELET | Aug-93 | Jun-99 | 2193 | 41 | 18 |
| RBC | Aug-93 | Sep-97 | 1800 | 190 | 33 |
| U937 | Aug-93 | Jun-97 | 895 | 42 | 32 |
| DICTYSLUG | Sep-97 | Sep-97 | 3164 | 25 | 16 |
| ECOLI | |||||
| 3.5–10 | Aug-95 | Jan-98 | 2364 | 206 | 180 |
| 4–5 | Jun-99 | Oct-99 | 449 | 18 | 12 |
| 4.5–5.5 | Oct-98 | Jun-99 | 1025 | 86 | 59 |
| 5–6 | Oct-99 | Oct-99 | 542 | 24 | 21 |
| YEAST | Feb-95 | Jun-97 | 1940 | 101 | 62 |
| MOUSE | |||||
| EFP | Sep-98 | Sep-98 | 3540 | 87 | 52 |
| ISLETS | Sep-98 | Jun-99 | 2528 | 7 | 2 |
| LIVER | Sep-98 | Oct-99 | 2836 | 201 | 107 |
| MUSCLE | Sep-98 | Oct-99 | 2650 | 143 | 72 |
Abbreviations are as follows: CEC, colorectal epithelia cells; CSF, cerebrospinal fluid; DLD1, colorectal adenocarcinoma cells; ELC, erythroleukemia cell line; HEPG2, hepatoblastoma carcinoma derived cells; HEPG2SP, hepatoblastoma carcinoma derived cell line secreted proteins; HL60, promyelocytic leukemia derived cells; RBC, red blood cells; U937, macrophage like cell line; DICTYSLUG, D.discoideum; ECOLI, E.coli, with pI range from 3.5 to 10 (3.5–10), from 4 to 5 (4–5), from 4.5 to 5.5 (4.5–5.5) and from 5 to 6 (5–6); YEAST, S.cerevisiae; EFP, epididymal fat pad; ISLETS, pancreatic islet cells; MUSCLE, gastrocnemius muscle.
Figure 3.

Species distribution of the SWISS-2DPAGE entries. Abbreviations as follows: E.COLI, E.coli; YEAST, S.cerevisiae; DICTYSLUG, D.discoideum.
AVAILABILITY
The SWISS-2DPAGE database is freely available for non-profit organizations. It is submitted to an annual subscription fee for commercial entities (for details, see URL http://www. expasy.ch/announce/ ). In both cases, users can get a local copy of SWISS-2DPAGE using anonymous ftp from the ExPASy FTP server (ftp://ftp.expasy.ch/databases/swiss-2dpage/ ). In addition, SWISS-2DPAGE entries can be reached through a 2-D gel analysis software such as Melanie 3 (http://www.expasy. ch/melanie/ ) (16).
However, the most efficient and user-friendly way to inter-actively browse in SWISS-2DPAGE is through the ExPASy proteomics server (http://www.expasy.ch/ ). The SWISS-2DPAGE top page provides several textual and graphical queries and displays results with active links to other databases. A SWISS-2DPAGE entry may be reached by keyword search (protein name, accession number, description, authors or full text) or by selecting a spot on one of the 2-DE maps. New features have been added to improve interactions in accessing the SWISS-2DPAGE data. First, all searching functions in the database are now accessible from the top page and results page of each keyword search function. This feature (http://www.expasy.ch/cgi-bin/ch2d-search-de ) has been designed to facilitate the navigation between the different ways to query the database. Second, for both textual and graphical queries, the result now displays SWISS-2DPAGE entries using a ‘nice view’ tool. This tool provides users with an easily readable alternative to the original text file representation, by collecting in blocks related information such as general information, origin of the protein, references, etc. Moreover the 2D part combines all 2-DE data in a cooperating view (eg., http://www.expasy.ch/cgi-bin/nice2dpage.pl?P00938 ). For each reference maps where the protein shown has been identified, this integrated view includes a map icon in front of the detailed 2-DE information. Direct links have also been added from amino acid composition and peptide masse data to the concerned section in the user manual (http://www.expasy.ch/ch2d/manch2d.html ) describing data format and protocols. Finally, an additional tool allows users to retrieve in a table all the protein entries identified on a given reference map, with all the 2-DE attributes and references (http:// www.expasy.ch/cgi-bin/get-ch2d-table.pl ).
Acknowledgments
ACKNOWLEDGEMENTS
The authors thank M. Cawthorne for providing mouse sample, R. Joubert-Carron for HL60 sample, M. Raymond for colorectal sample, C. Sarto for kidney sample, J. Yan for Dictyostelium sample; and W. Bienvenut, I. Demalte, S. Frutiger, G. Hughes, S. Jaccoud, S. Paesano, C. Pasquali, F. Ravier, G. Rosselat, V. Rouge for their technical assistance.
REFERENCES
- 1.Appel R.D, Sanchez,J.-C., Bairoch,A., Golaz,O., Miu,M., Vargas,R. and Hochstrasser,D.F. (1993) Electrophoresis, 14, 1232–1238. [DOI] [PubMed] [Google Scholar]
- 2.Wilkins M.R. and Gooley,A.A. (1997) In Wilkins,M.R., Williams,K.L., Appel,R.D. and Hochstrasser,D.F. (eds.), Proteome Research: New Frontiers in FunctionalGenomics. Springer Verlag, pp. 35–64.9286698 [Google Scholar]
- 3.VanBogelen R.A., Abshire,K.Z., Moldover,B., Olson,E.R. and Neidhardt,F.C. (1997) Electrophoresis, 18, 1243–1251. [DOI] [PubMed] [Google Scholar]
- 4.Dunn M.J., Corbett,J.M. and Wheeler,C.H. (1997) Electrophoresis, 18, 2795–2802. [DOI] [PubMed] [Google Scholar]
- 5.Payne W.E. and Garrels,J.I. (1997) Nucleic Acids Res., 25, 57–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bini L., Sanchez-Campillo,M., Santucci,A., Magi,B., Marzocchi,B., Comanducci,M., Christiansen,G., Birkelund,S., Cevenini,R., Vretou,E., Ratti,G. and Pallini V. (1996) Electrophoresis, 17, 185–190. [DOI] [PubMed] [Google Scholar]
- 7.Bini L., Heid,H., Liberatori,S., Geier,G., Pallini,V. and Zwilling,R. (1997) Electrophoresis, 18, 557–562. [DOI] [PubMed] [Google Scholar]
- 8.Bini L., Magi,B., Marzocchi,B., Arcuri,F., Tripodi,S., Cintorino,M., Sanchez,J.-C., Frutiger,S., Hughes,G.J., Pallini,V., Hochstrasser,D.F. and Tosi,P. (1997) Electrophoresis, 18, 2832–2841. [DOI] [PubMed] [Google Scholar]
- 9.Shaw A.C., Larsen,M.R., Roepstorff,P., Holm,A., Christiansen,G. and Birkelund,S. (1999) Electrophoresis, 20, 977–983. [DOI] [PubMed] [Google Scholar]
- 10.Shaw A.C, Larsen,M.R., Roepstorff,P., Justesen,J., Christiansen,G. and Birkelund,S. (1999) Electrophoresis, 20, 984–993. [DOI] [PubMed] [Google Scholar]
- 11.Bairoch A. and Apweiler,R. (1999) Nucleic Acids Res., 27, 49–54. Updated article in this issue: Nucleic Acids Res. (2000), 28, 45–48.9847139 [Google Scholar]
- 12.Stoesser G., Tuli,M.A., Lopez,R. and Sterk,P. (1999) Nucleic Acids Res., 27, 18–24. Updated article in this issue: Nucleic Acids Res. (2000), 28, 19–23.9847133 [Google Scholar]
- 13.Benson D.A., Boguski,M.S., Lipman,D.J., Ostell,J., Ouellette B.F.F., Rapp,B.A. and Wheeler,D.L. (1999) Nucleic Acids Res., 27, 12–17. Updated article in this issue: Nucleic Acids Res. (2000), 28, 15–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hofmann K., Bucher,P., Falquet,L. and Bairoch,A. (1999) Nucleic Acids Res., 27, 215–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pearson P., Francomano,C., Foster,P., Bocchini,C., Li,P. and McKusick,V.A. (1994) Nucleic Acids Res., 22, 3470–3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Appel R.D., Palagi,P.M., Walther,D., Vargas,J.R., Sanchez,J.-C., Ravier,F., Pasquali,C. and Hochstrasser,D.F. (1997) Electrophoresis, 18, 2724–2734. [DOI] [PubMed] [Google Scholar]