Abstract
The Prostate Expression Database (PEDB) is an online resource designed to access and analyze gene expression information derived from the human prostate. PEDB archives >55 000 expressed sequence tags (ESTs) from 43 cDNA libraries in a curated relational database that provides detailed library information including tissue source, library construction methods, sequence diversity and sequence abundance. The differential expression of each EST species can be viewed across all libraries using a Virtual Expression Analysis Tool (VEAT), a graphical user interface written in Java for intra- and inter-library species comparisons. Recent enhancements to PEDB include: (i) the functional categorization of annotated EST assemblies using a classification scheme developed at The Institute for Genome Research; (ii) catalogs of expressed genes in specific prostate tissue sources designated as transcriptomes; and (iii) the addition of prostate proteome information derived from two-dimensional electrophoreses and mass spectrometry of prostate cancer cell lines. PEDB may be accessed via the WWW at http://www.mbt.washington.edu/PEDB/
INTRODUCTION
The Prostate Expression Database (PEDB) was established to provide a rapid and convenient means of accessing data characterizing the subset of the human genome that is used or expressed in the human prostate. With current technology, it is most straightforward to define this repertoire of genes at the level of transcripts, comprehensively termed the transcriptome, though ultimately the analysis of protein level and function, comprehensively termed the proteome, provides a more accurate assessment of gene expression activity. The PEDB is a curated resource comprised of expressed sequence tags (ESTs) produced from cDNA libraries representing a wide spectrum of normal, benign and malignant prostate disease states. Detailed library information including tissue source, library construction methods, sequence diversity and abundance are maintained in a relational database management system (RDBMS). Prostate ESTs are assembled into distinct species groups using the multiple alignment program CAP2 (1), and annotated with information from GenBank (2), dbEST (3) and the Unigene (4) public sequence databases. The primary user work sites involve: (i) database queries with nucleotide sequence information using the BLAST algorithm (5) or word searches to find homologous sequences in PEDB that could be useful in extending and further defining the user’s sequence; and (ii) virtual expression analysis using a graphical user interface to perform intra- and inter-library sequence abundance comparisons. The PEDB also provides links to other relevant WWW resources involving prostate disease, cancer biology and genomics. A detailed description of the database development, data inventory and utilities is available online (Table 1) (http://www.mbt.washington.edu/PEDB/overview/ ).
Table 1. Table of contents for PEDB overview (http://www.mbt.washington.edu/PEDB/overview/ ).
| 1. | Introduction to PEDB | |
| 2. | PEDB construction schema and dataflow | |
| 3. | PEDB utilities | |
| A | BLAST queries and work-search | |
| B | Virtual analysis expression tool (VEAT) | |
| 4. | PEDB enhancements 1999 | |
| A | Functional categorization | |
| B | Prostate transcriptome | |
| C | Prostate proteome | |
| 5. | PEDB references and resources |
PEDB DATA
The foundation of gene expression information contained in PEDB consists of archives of ESTs derived from a variety of prostate cDNA libraries. These ESTs are either obtained from publicly available sequence repositories such as GenBank, the database of ESTs (dbEST), and The Institute for Genome Research (TIGR), or from in-house EST sequencing projects. The methods of sequence processing and curation involves a pipeline of sequence submission, sequence masking, sequence clustering and cluster annotation that we have previously described in detail (6). The CAP2 multiple alignment program is used to assemble ESTs into clusters using a variant of the Smith–Waterman algorithm. A consensus sequence is provided for each cluster which is then used for BLAST queries against the Unigene, GenBank and dbEST databases to provide cluster annotation and to further facilitate the assembly process.
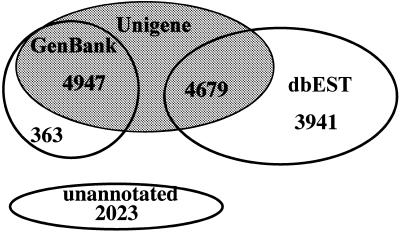
The most recent build of PEDB ESTs was assembled starting with 55 000 prostate ESTs. Portions of EST sequences with homology to cloning vector, Escherichia coli genomic DNA, and human repetitive DNA sequences were masked and ESTs with >100 bp of high quality sequence were admitted to the assembly process. A total of 49 816 ESTs were assembled using CAP2 to produce 21 114 clusters. Each cluster was annotated by searching the Unigene, GenBank and dbEST databases with the CAP2 generated cluster consensus sequences using BLASTN. Clusters annotating to the same database sequence were joined to further reduce the number of distinct clusters to 15 953 (Fig. 1).
Figure 1.
Results of PEDB cluster annotations against the GenBank, Unigene and dbEST nucleotide sequence database. Joining clusters based upon an identical public database sequence annotation reduced the number of distinct species from 21 114 to 15 953. As the Unigene database is comprised of sequences from both GenBank and dbEST, many PEDB clusters share annotations with two databases. Unannotated PEDB sequences represent clusters that do not have significant homology to any public database sequence.
QUERIES, VISUALIZATION AND ANALYSIS TOOLS
The primary work sites of PEDB involve a BLAST interface for sequence-based queries against PEDB and the Unigene datasets. A word-search option has been added to facilitate the queries of known genes and GenBank accession numbers. A second work site is used for the generation of dynamic gene expression profiles based upon EST assembly and annotation information. Expression data are generated, viewed and manipulated using the Virtual Expression Analysis Tool (VEAT). The VEAT provides user-directed inter- and intra-library analysis of transcript abundance, diversity and differential expression.
Annotated PEDB clusters are now functionally categorized using minor modifications to a classification system available on the TIGR web site (http://www.tigr.org/ ) (7). This scheme provides an initial framework for building a transcriptome comprised of that portion of the genome expressed in the prostate. We have used this prostate transcriptome to facilitate large-scale gene expression studies by designing prostate-centric gene expression cDNA microarrays. Finally, the PEDB site has recently added a section for viewing portions of the prostate proteome in the form of two-dimensional gel electrophoresis images. This section will be expanded in the future to include spot annotations and correlations with cDNA array transcript expression data.
SUMMARY AND FUTURE DEVELOPMENT
The Prostate Expression Database (PEDB) functions as a centralized archive of gene expression information derived from the human prostate that can be utilized by investigators studying normal and neoplastic prostate development. Expression data are stored in a fashion suitable for sequence- and keyword-based queries, assessment of gene expression diversity, comparative analyses of expression and functional categorization. Current work and future enhancements to PEDB focus on completing the prostate transcriptome through the acquisition of low abundance transcripts, providing an interactive cluster or contig assembly viewer suitable for the identification of single nucleotide polymorphisms (SNPs) and alternatively spliced transcripts, and ultimately developing a corresponding database correlating protein expression information with transcript data. PEDB is accessible via the WWW at http://www.mbt.washington.edu/PEDB
Acknowledgments
ACKNOWLEDGEMENTS
We thank collaborators in the CaPCURE Genetics Consortium for helpful advice, Xiaoqiu Huang for the CAP2 program, Steve Lasky and the University of Washington Molecular Biotechnology sequencing group for sequencing support. This work was supported by the CaPCURE Foundation and grants (K08 CA75173-01A1) from the National Cancer Institute and (DAMD17-98-1-8499) from the USAMRMC (both to P.S.N.).
REFERENCES
- 1.Huang X. (1996) Genomics, 33, 21–31. [DOI] [PubMed] [Google Scholar]
- 2.Benson D.A., Boguski,M.S., Lipman,D.J., Ostell,J. and Ouellette,B.F.F. (1998) Nucleic Acids Res., 26, 1–7. Updated article in this issue: Nucleic Acids Res. (2000), 28, 15–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boguski M.S., Lowe,T.M.J. and Tolstoshev,C.M. (1993) Nature Genet., 4, 332–333. [DOI] [PubMed] [Google Scholar]
- 4.Schuler G.D. (1997) J. Mol. Med., 75, 694–698. [DOI] [PubMed] [Google Scholar]
- 5.Altschul S.F., Gish,W., Miller,W., Myers,E.W. and Lipman,D.J. (1990) J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 6.Hawkins V., Doll,D., Bumgarner,R., Smith,T., Abajian,C., Hood,L. and Nelson,P.S. (1999) Nucleic Acids Res., 27, 204–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Adams M.D., Kerlavage,A.R., Fleischman,R.D., Fuldner,R.A., Bult,C.J., Lee,N.H., Kirkness,E.F., Weinstock,K.G., Gocayne,J.D. and White,O. (1995) Nature, 377 (Suppl. 28), 3–174. [PubMed] [Google Scholar]