Fig. 4. Functional analysis of putative client-contacting mtHsp60 residues.
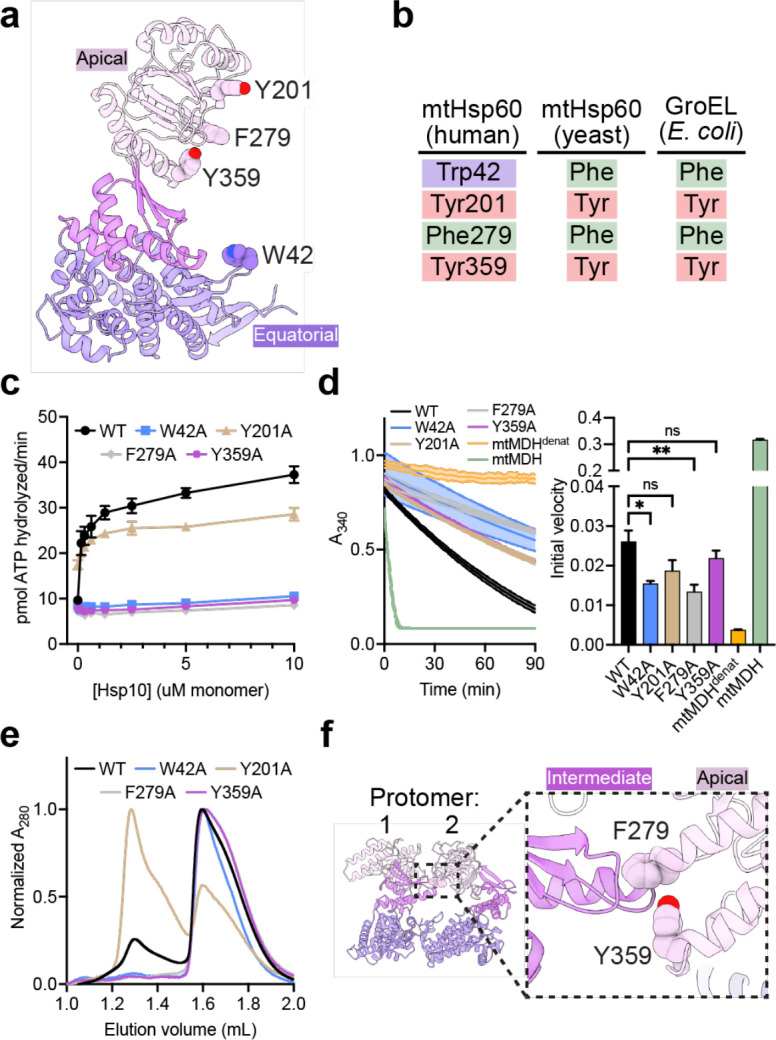
(a) Protomer of mtHsp60 from mtHsp60ATP-mtHsp10, showing residues mutated. (b) Conservation of residues in (a) among human and yeast mtHsp60 and GroEL. (c) Steady-state ATPase activity of mtHsp60 mutants vs concentration of mtHsp10. A representative experiment of three biological replicates is shown. Error bars represent standard deviation. (d) Enzymatic activity of chemically-denatured human mtMDH refolded by mtHsp60 mutants (left panel, representative of three biological replicates). Dashed lines represent standard deviation of technical triplicates. Initial velocities of absorbance curves from three biological replicates are shown at right. Error bars represent standard error of the mean. *p < 0.05, **p < 0.005, ns = not significant. (e) Analytical size exclusion chromatography traces of mtHsp60 mutants, showing complete monomerization of W42A, F279A, and Y359A mutants. (f) Model of two apo-mtHsp60 protomers, showing apical domain residues F279 and Y359 contacting the intermediate domain of an adjacent protomer.
