Abstract
ProDom contains all protein domain families automatically generated from the SWISS-PROT and TrEMBL sequence databases (http://www.toulouse. inra.fr/prodom.html ). ProDom-CG results from a similar domain analysis as applied to completed genomes (http://www.toulouse.inra.fr/prodomCG.html ). Recent improvements to the ProDom database and its server include: scaling up to include sequences from TrEMBL, addition of Pfam-A entries to the set of expert validated families, assignment of stable accession numbers, consistency indicators for domain families, domain arrangements of sub-families and links to Pfam-A.
INTRODUCTION
ProDom is a database of protein domain families obtained by an automated analysis of available protein sequence data (1,2). It is useful for analysing the domain arrangements of complex protein families and helps to analyse homology relationships in modular proteins. The clustering of homologous domains provides a rational way of organising protein sequence data. An interactive graphical interface was designed to allow for easy navigation between schematic domain arrangements, multiple alignments, phylogenetic trees, SWISS-PROT entries (3), PROSITE patterns (4), Pfam-A families (5) and 3-D structures in the PDB (6). Alignments and trees can be reduced or developed to facilitate the analysis of sequence relationships within large domain families (7). New sequences can be searched against ProDom and aligned with existing domain families, and modelled on the basis of homologous domains in the PDB.
Recently, we scaled up the process to include TrEMBL sequences in the source database. We have also added Pfam-A families to the set of expert validated families used in the ProDom construction procedure. Other recent improvements in ProDom make it easier to keep track of a protein family across successive releases.
BUILDING ProDom
Since version 35, the automated process that builds ProDom has been complemented by the result of an expertise. For some domain families, experts were asked to correct domain boundaries. To increase the number of these expert-validated families, we used the curated part of Pfam (5): the seed alignments of 1403 Pfam-A families were added to the list of 21 ProDom expert-validated multiple alignments and used to build new ProDom families with the PSI-BLAST program (8). Other families are built with an automated process based on a recursive use of PSI-BLAST as described previously (2,9). This process can be applied to any set of protein sequences, provided there are enough sequences available to detect domain boundaries. Since version 99.1, the ProDom source database is SWISS-PROT and its TrEMBL supplement (3). A set of available complete genomes is also used to build ProDom-CG; release 20 was built by automatic clustering of protein domains from 20 complete genomes available on April 8, 1999: four archaea, 14 bacteria and two eukaryotes.
ProDom STATISTICS
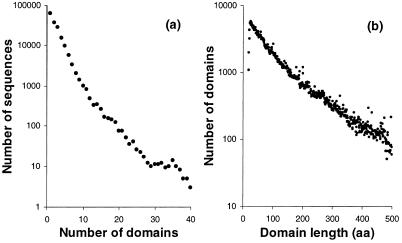
ProDom, version 99.2, contains 157 167 families (Table 1). ProDom covers >95% of the residues in the source database. The inclusion of TrEMBL represents a 2.4-fold increase in the source database. The ProDom building process scaled up with no major difficulties and with stable results. The average number of domains per sequence remains stable, close to three domains per sequence, with an exponential distribution (Fig. 1a). Surprisingly, domain lengths in ProDom also show an exponential distribution (Fig. 1b), contrary to the expectation of a more balanced distribution centred on the mean. Thus short ‘domains’ are over-represented in ProDom in its current state, which may be due to numerous sequence ends and inter-domain linkers which are generated as a result of the automated process. Fifty-six percent of all ProDom sequence residues are found in families containing 10 or more members. There are 6264 ProDom entries linked to 1462 Pfam-A entries (v 4.0), 5787 linked to 1056 PROSITE entries (v15) and 2378 linked to PDB.
Table 1. Comparison of ProDom versions.
| ProDom version | 36 | 99.1 | 99.2 |
|---|---|---|---|
| Release date | 08/27/1998 | 01/13/1999 | 07/23/1999 |
| Source database (non fragmentary sequences) | SP36 | SP36 + TrEMBL + TrEMBL updates 10/17/1998 | SP37 + TrEMBL+ TrEMBL updates 04/26/1999 |
| Number of sequences | 66 756 | 158 880 | 170 648 |
| Number of residues | 25 356 087 | 61 545 344 | 66 703 627 |
| ProDom domain families | 57 976 | 149 606 | 157 167 |
| Sequence coverage | 95.9% | 95.4% | 95.6% |
| Mean number of domains per sequence | 2.7 | 3.3 | 2.8 |
| Mean domain length (residues) | 136 | 112 | 132 |
| ProDom domain families with at least two members | 17 777 | 44 345 | 43 965 |
| Sequence coverage | 77.4% | 76.1% | 76.6% |
| Mean domain length (residues) | 141 | 111 | 138 |
Figure 1.
(a) Distribution of the number of domains per sequence in ProDom 99.2. (b) Distribution of the length in amino acids of individual domains in ProDom 99.2.
RECENT ProDom IMPROVEMENTS
Accession numbers
Each ProDom entry now has a unique and stable accession number (AC) that will provide access to the same domain family across successive releases. These numbers are formed with the letters PD followed by exactly six digits (e.g. PD002243). As ProDom is built anew every time, domain families are not exactly conserved from one release to the next. We have derived a tool that links families in release n to families in release n–1. For each family in release n, it searches for overlaps with families of release n–1; it sorts the hits in decreasing order using the absolute and relative numbers of sub-sequences involved in the overlap; AC numbers are assigned by selecting the first available number in the list, or, if none is left, a new AC is assigned.
Consistency indicator
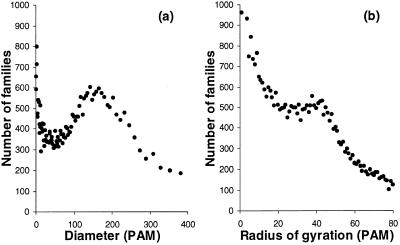
As ProDom families are computed by an automated process, the sequence homogeneity can vary considerably between families. Some families may include hundreds of nearly identical or alternatively very diverged sequences. We have introduced two indicators that measure the consistency of a family: the diameter and the radius of gyration. The diameter is the maximal distance between two domains of the family. The radius of gyration is the weighted root mean square of the distance between each domain and the family consensus sequence. To help the selection of a sequence that represents the family well, we also indicate which sequence lies closest to the consensus. Among the 43 965 ProDom families containing at least two sequences, 24% had a diameter <10 PAM and 90% <240 PAM; 30% had a radius <10 PAM and 90% <71 PAM. The diameter distribution (Fig. 2) presents two modes, indicating that there are two classes of families in ProDom. In the first class, domains are overly similar, indicating sequence redundancy in the source database. In the second class, families are truly complex and include more diverged homologous domains.
Figure 2.
Distribution of the diameter (a) and radius of gyration (b) of ProDom families (release 99.2). Note that the first points of both distributions lie outside the scale: 5411 families are completely redundant with a diameter of 0 PAM.
Graphical representation
As described earlier (2), the ProDom Web server provides a graphical representation of protein domain arrangements. Each protein is shown on a single line with schematic boxes hypertext-linked to corresponding ProDom entries. Each domain family has a unique representation that is linked to the ProDom accession number, which ensures its stability between successive releases. The graphical representation of the domain arrangements for all proteins sharing a homologous domain can be large and difficult to comprehend. As ProDom families can be divided into sub-families following a phylogenetic tree, it is now possible to display the domain arrangements for all proteins from the same sub-family. For example, ProDom domain PD000612 includes 94 sequences of cytochrome b5 and heme-binding domains of homologous oxidoreductases: the user can readily display the protein domain arrangements specifically for the 42 nitrate reductases or for the three sulfite oxidases (see example of ProDom WWW server usage in Supplementary Material).
PUTTING ProDom TO USE IN GENOME PROJECTS
ProDom is widely used to analyse protein domain relationships in genomic sequences. For instance ProDom was used systematically by Marcotte et al. (10) in order to infer protein–protein interactions on the basis of ‘Rosetta Stone’ sequence combinations. Another example of a systematic use of ProDom concerns structural genomics. Several projects have recently emerged aiming at a systematic study of the protein structure universe (see for instance http://www.nih.gov/nigms/news/meetings/structural_genomics_targets.html ). These projects require a comprehensive protein family classification scheme in order to adequately sample the protein structure space. We have contributed to such a scheme in the framework of the Protein Structure Initiative (http://www.genome3d.org ). Target proteins for structure determination were selected for 2587 ProDom families on the following criteria: (i) no 3-D structure was available; (ii) they contain at least two members (true family); (iii) they contain at least one protein with only one domain, shorter than 500 amino acids (ProDom domain span is correct); (iv) the two most distant sequences in the family share at least 10% identity (family is homogeneous). Proposed targets are single domain proteins, preferably human. The choice of single domain proteins obviates the need to engineer specific domains and should make expression and purification easier to achieve.
Another concerted effort is the InterPro project aiming at integrating resources for protein families (http://www.ebi.ac. uk/interpro ). We selected 2883 ProDom families that appear to be good candidates for new families to be documented in InterPro. They were selected on the following criteria: (i) they are not referenced in PROSITE 15.0; (ii) they contain at least two members; (iii) they contain at least one single-domain protein from SWISS-PROT, shorter than 500 amino acids; (iv) the similarity between the most distant sequences in the family lies between 10 and 90% identity (family is homogeneous, yet not overly redundant). These criteria ensure that domain boundaries are well defined for each new family.
AVAILABILITY
Available via anonymous FTP site: ftp://ftp.toulouse.inra.fr/pub/prodom
or WWW server: http://www.toulouse.inra.fr/prodom.html
http://www.toulouse.inra.fr/prodomCG.html
SUPPLEMENTARY MATERIAL
See Supplementary Material available at NAR Online.
REFERENCES
- 1.Sonnhammer E.L.L. and Kahn,D. (1994) Protein Sci., 3, 482–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Corpet F., Gouzy,J. and Kahn,D. (1999) Nucleic Acids Res., 27, 263–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bairoch A. and Apweiler,R. (1999) Nucleic Acids Res., 27, 49–54. Updated article in this issue: Nucleic Acids Res. (2000), 28, 45–48.9847139 [Google Scholar]
- 4.Hofmann K., Bucher,P., Falquet,L. and Bairoch,A. (1999) Nucleic Acids Res., 27, 215–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bateman A., Birney,E., Durbin,R., Eddy,S.R., Finn,R.D. and Sonnhammer,E.L.L. (1999) Nucleic Acids Res., 27, 260–262. Updated article in this issue: Nucleic Acids Res. (2000), 28, 263–266.9847196 [Google Scholar]
- 6.Abola E.E., Bernstein,F.C., Bryant,S.H., Koetzle,T.F. and Weng,J. (1987) In Allen,F.H., Bergerhoff,G. and Sievers,R. (eds), Crystallographic Databases-Information Content, Software Systems, Scientific Applications. Data Commission of the International Union of Crystallography, Bonn/Cambridge/Chester, pp. 107–132.
- 7.Corpet F., Gouzy,J. and Kahn,D. (2000) Bioinformatics, in press. [Google Scholar]
- 8.Altschul S.F., Madden,T.L., Schäffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,J.L. (1997) Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gouzy J., Corpet,F. and Kahn,D. (1999) Computers Chem., 23, 333–340. [DOI] [PubMed] [Google Scholar]
- 10.Marcotte E.M., Pellegrini,M, Ng,H.L., Rice,D.W., Yeates,T.O. and Eisenberg,D. (1999) Science, 285, 751–753. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.