Abstract
We conducted a genome-wide association study (GWAS) in a multiethnic cohort of 920 at-risk infants for retinopathy of prematurity (ROP), a major cause of childhood blindness, identifying 2 loci at genome-wide significance level (p<5×10–8) and 7 at suggestive significance (p<5×10–6) for ROP ≥ stage 3. The most significant locus, rs2058019, reached genome-wide significance within the full multiethnic cohort (p=4.96×10–9); Hispanic and Caucasian infants driving the association. The lead single nucleotide polymorphism (SNP) falls in an intronic region within the Glioma-associated oncogene family zinc finger 3 (GLI3) gene. Relevance for GLI3 and other top-associated genes to human ocular disease was substantiated through in-silico extension analyses, genetic risk score analysis and expression profiling in human donor eye tissues. Thus, we report the largest ROP GWAS to date, identifying a novel locus at GLI3 with relevance to retinal biology supporting genetic susceptibilities for ROP risk with possible variability by race and ethnicity.
Introduction
Retinopathy of prematurity (ROP) is a retinal vascular disease that affects premature infants and is a leading cause of childhood blindness worldwide1–3. Birth weight (BW) less than 1500 grams, prematurity less than 32 weeks gestational age (GA), and post-natal oxygen exposure confer independent ROP risk4,5. However, this understanding of risk and pathogenesis is incomplete as only approximately 50–65% of infants with these risk factors will develop ROP disease4,6,7. For example, phenotypic extremes that do not conform to current risk profiles exist; preterm infants born at large birth weights and/or older gestational ages may develop treatment-warranted ROP whereas those within the risk profiles of GA and BW do not always develop treatment-warranted ROP8. The current stratification also does not address prevention; although post-natal oxygen exposure is the most modifiable risk, limiting oxygen increases infant mortality9–11. As a result, current screening lacks specificity and interventions are unable to modify preclinical ROP risk, instead targeting ROP once retinal disease is present and there is significant risk of blindness6,12. Improved understanding of risk will allow better detection and treatment of infants with greatest risk for severe ROP and may provide novel insights into disease patho-mechanisms in order to prevent disease.
ROP risk is multifactorial and although genetic risk is not fully elucidated, there is a growing body of evidence supporting a genetic basis for ROP, including twin studies13–15. Case control and whole exome candidate approaches have identified significant risk associations between single nucleotide polymorphisms (SNP) in brain-derived neurotrophic factor (BDNF; rs7934165 and rs2049046), thrombospondin type-1 domain-containing protein 4 (THSD4), TNF -308G/A polymorphism and angiotensin 1 converting enzyme insertion deletion (ACE ID) polymorphism16–19 (Supplementary Table 1). By contrast, recent candidate work demonstrated significant protective associations between the BDNF SNP rs7929344 and ROP and severe ROP risk associations between SNPs within VEGFA, NOS3 and EPAS120. While no SNP has been shown to reach genome-wide significance, meta-analysis has substantiated some associations18 while also beginning to identify pathobiology underlying observed differences in ROP risk relative to race; an example being recent work demonstrating association of the VEGFA+405G>C polymorphism association with ROP risk in Caucasian populations21. This observation aligns well with clinical observations showing racial differences in risk, including greater ROP severity and vision loss in white as compared to black preterm infants with equivalent BW and GA ROP risk22,23. Further, clinical progression has been shown to vary in Hispanic compared to white non-Hispanic populations24.
Taken together, our current understanding of ROP risk is incomplete, which impairs our ability to predict infants at greatest disease risk or identify pathobiology allowing for ROP prevention. While ROP risk is multifactorial, evidence supports genetic contributions to risk and protection. Herein we present a GWAS analysis based within the i-ROP consortium, including collaborators from 14 academic institutions throughout the world, demonstrating a novel ROP risk association with the SNP GLI3 rs2058019 reaching genome-wide significance (p=4.90E-09) within our multi-ethnic cohort. We further demonstrate broader applicability for this SNP and GLI3 genetic variation with other forms of retinal disease characterized by pre-retinal neovascularization, namely diabetic retinopathy. Finally, we identify additional SNPs with suggestive-level significance association with severe ROP disease, demonstrate cross-significance with previously identified ROP-associated SNPs, and further demonstrate relevance for GLI3 and these top SNPs within the human eye, finding expression in both human donor neurosensory retina as well as retinal pigment epithelium (RPE).
Results
iROP GWAS Multi-ethnic Cohort Characteristics:
The iROP cohort consists of 2187 preterm infants born at gestational ages less than 32 weeks and birth weights smaller than 1250 grams, placing them at risk to develop ROP. The iROP database was mined for infants with both biologic and phenotypic data for subsequent genotype-phenotype analysis. To identify genetic susceptibility regions for ROP, we conducted a genome-wide association study (GWAS) using the Illumina Infinium Global Screening Array with information from these infants. As published previously, phenotype was rigorously determined by a team of ophthalmologists and image graders with clinical expertise in ROP16,25,26. This subset of iROP patients represents multiple ethnic and racial groups, including 44.5% Hispanic, 35.4% White, 12.1% African American and 8% unidentified race individuals as noted in Table 1. Further, all ROP phenotypes are represented, including 197 with ROP Stages 3, 4, or 5. (Table 1). The distribution of ROP disease, ethnicity and race was similar between the infants with and without genetic data (Supplementary Table 2).
Table 1:
iROP Infants Included in GWAS Analysis
| Race/Ethnicity | No ROP | Stage 1 | Stage 2 | Stage 3 | Stage 4 | Stage 5 | Total |
|---|---|---|---|---|---|---|---|
| Caucasian | 129 | 57 | 72 | 58 | 10 | 0 | 326 |
| African American | 60 | 17 | 20 | 13 | 0 | 1 | 111 |
| Hispanic | 128 | 52 | 127 | 65 | 23 | 15 | 410 |
| Other | 34 | 11 | 16 | 11 | 0 | 1 | 73 |
| Total % | 351 (38.2%) | 137 (14.9%) | 235 (25.5%) | 147 (16%) | 33 (3.6%) | 17 (1.8%) | 920 |
A number of SNPs demonstrate significant associations with severe ROP.
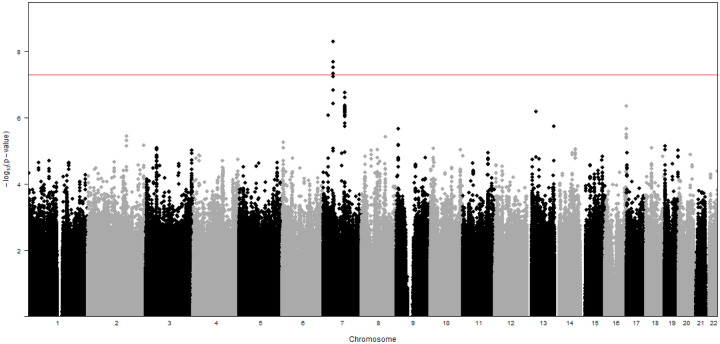
GWAS analysis was performed on DNA extracted from all 920 cohort infants to identify genetic susceptibility regions for ROP severity, defined as ROP stage 3 or greater. As depicted in Figure 1, which visualizes all p values of SNP-associations for the 22 autosomal chromosomes plotted relative to chromosome location and significance, a number of SNP associations were identified. The Y axis represents the −log transformed p value (red line representing GWAS significance level) and X axis is in genomic ordered by chromosomes and positions. SNPs reaching genome-wide significance, defined as p≤ 5×10−8, are visualized above the red line.
Figure 1. Manhattan plot of genome-wide association with ROP severity (stage 3 or greater):
Data were analyzed for all populations combined. The red line on indicates genome-wide significance (p≤ 5×10−8)
GWAS identifies ROP-associated SNPs with genome-wide significance or suggestive significance correlating with both novel and established ROP risk genes
As noted in Table 2, we identified 10 loci which demonstrate genome-wide significance (p≤ 5×10−8) or suggestive significance (p≤ 5×10−6) of association with ROP severity, defined by stage 3 disease or greater, both with and without correction for birth weight (BW) and gestational age (GA). The position, effect allele and frequency, odds ratio, p value and nearest gene is delineated in Table 2. SNPs demonstrating the lowest p value in each locus/region were selected and the bioinformatically determined nearest gene was used. Under the additive genetic model, the ROP risk corresponds to the number of copies of the effect allele when the odds ratio is greater than 1. As noted, SNPs rs2058019 (GLI3 gene) and rs11563856 (CLDN12 gene) both reach genome wide significance, although only rs2058019 remains significant at this level when controlling for the potentially confounding effects of BW and GA. In two of the loci meeting suggestive significance, the nearest gene has a prior association with ROP disease in either humans or animal models as noted by bolded text. Finally, we performed an in silico analysis to determine if each listed SNP had a described expression quantitative trait loci (eQTL) association in the EyeGEx27 database. We identified a number of established regulatory associations for our top SNPs as detailed in Table 2 within the retina, further supporting relevance to the ocular microenvironment.
Table 2.
Association variants identified for all ROP samples with p <5×10−6.
| Chr | Pos | Effect Allele | AF | rsq | OR | p-value | p-value adj for BW & GA | gene | eQTL p-value |
|---|---|---|---|---|---|---|---|---|---|
| 7 | 42226712 | T | 0.06 | 0.96 | 1.27 | 5.25E-08 | 4.96E-09* | GLI3 | NA |
| 7 | 90141855 | G | 0.17 | 0.91 | 1.16 | 5.84E-08 | 1.71E-07 | CLDN12 | >0.05 |
| 16 | 90048889 | T | 0.22 | 0.81 | 1.13 | 9.67E-07 | 4.50E-07 | DEF8. TUBB3 | 2.7E-04 |
| 13 | 36559030 | A | 0.14 | 0.68 | 1.14 | 7.90E-06 | 6.50E-07 | DCLK1 | 0.02 |
| 7 | 21465569 | T | 0.06 | 0.89 | 1.21 | 3.51E-05 | 8.38E-07 | SP4 | 0.01 |
| 13 | 111811973 | G | 0.09 | 0.87 | 1.17 | 8.70E-06 | 1.83E-06 | ARHGEF7 | >0.05 |
| 9 | 8800078 | G | 0.36 | 0.93 | 1.10 | 1.40E-06 | 2.14E-06 | PTPRD | 7.7E-03 |
| 2 | 162937083 | A | 0.12 | 0.96 | 1.14 | 1.13E-06 | 3.56E-06 | DPP4 | >0.05 |
| 8 | 99105726 | C | 0.39 | 0.84 | 1.09 | 6.04E-05 | 3.77E-06 | ERICH5. RPL30. RIDA | 1.90E-21 |
| 6 | 3980472 | A | 0.08 | 0.93 | 1.17 | 9.37E-04 | 5.52E-06 | PRPF4B | >0.05 |
rs2058019 contributes to genetic susceptibility for ROP in diverse populations
Given established differences in clinical ROP risk based on race and ethnicity22,23, we analyzed the association of the lead GLI3 SNP, rs2058019, stratified by these variables. As seen in Table 3, the association between rs2058019 and ROP disease is most significant when considering all patients, although odds ratios and allele frequencies between cases and controls demonstrate that the degree and direction of association remain consistent for Caucasian and Hispanic subjects. Interestingly, the African American subjects do not appear to be contributing to this association. This latter finding does not reach significance in this sample and thus requires validation within a larger population. As noted in Supplemental Table 3, we did not find suggestive evidence for significant differences is MAF or direction of effect within racial or ethnic populations for other top SNPs.
Table 3:
Lead SNP rs2058019 demonstrates association with severe ROP disease in Hispanic and Caucasian populations but not in African American populations
| Effect Allele | AF: Case | AF: Control | AF: All | OR | P |
|---|---|---|---|---|---|
| T | 0.22 | 0.08 | 0.11 | 1.26 | 2.95E-06 |
| T | 0.04 | 0.01 | 0.01 | 1.61 | 4.0E-04 |
| T | 0 | 0.02 | 0.01 | 0.87 | 0.49 |
| T | 0.14 | 0.04 | 0.06 | 1.27 | 4.96E-9 |
HA: Hispanic Americans; CA: Caucasian Americans; AA: African Americans; All: all subjects combined.
AF: allele frequency of effect allele.
Multiple significant SNPs are identified regionally related to GLI3
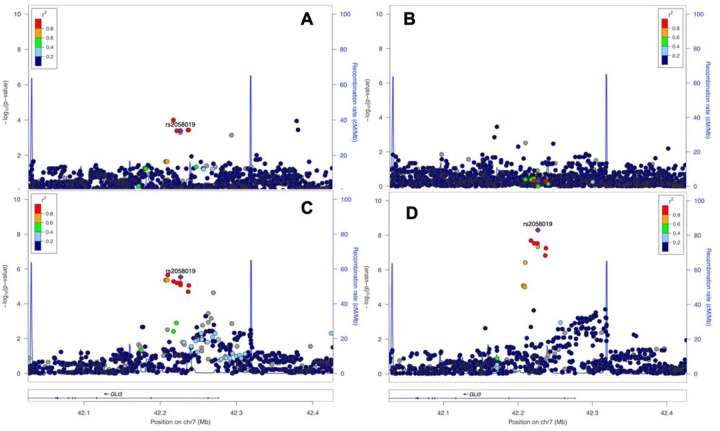
To determine the regional significance for the GLI3 pan-locus, we plotted all SNP-associations within 100 Kb of the GLI3 gene for all patients as well as for racial and ethnic subsets. As pictured in Figure 2, a regional plot with the Y axis depicting the −log transformed p value and the X axis showing chromosomal position, we illustrate the GLI3 gene regional association for all ethnic and racial groups with ROP severity. The lead SNP rs2058019 was identified as a top-associated SNP within the pan-locus for the Caucasian and Hispanic groups and both the best and sentinel SNP when all patients were considered together. Significant regional SNPs for each group are delineated fully in Supplemental Table 4. Also pictured, we determined the linkage disequilibrium (LD) for each SNP, defined by the r2 using the included color scheme. LD was determined between SNPs and genes involved/close to SNPs identified.
Figure 2. Regional association plots of top genome-wide significant SNP rs2058019 (GLI3 gene) among each ethnic group:
A: Caucasian (CA); B: African American (AA), C: Hispanic (HA), and D: all combined samples (ALL).
GWAS identifies ROP-associated SNPs with genome-wide significance or suggestive significance correlating with both novel and established ROP risk genes
As noted in Table 2, we identified 10 loci which demonstrate genome-wide significance (p≤ 5×10−8) or suggestive significance (p≤ 5×10−6) of association with ROP severity, defined by stage 3 disease or greater, both with and without correction for birth weight (BW) and gestational age (GA). The position, effect allele and frequency, odds ratio, p value and nearest gene is delineated in Table 2. SNPs demonstrating the lowest p value in each locus/region were selected and the bioinformatically determined nearest gene was used. Under the additive genetic model, the ROP risk corresponds to the number of copies of the effect allele when the odds ratio is greater than 1. As noted, SNPs rs2058019 (GLI3 gene) and rs11563856 (CLDN12 gene) both reach genome wide significance, although only rs2058019 remains significant at this level when controlling for the potentially confounding effects of BW and GA. In two of the loci meeting suggestive significance, the nearest gene has a prior association with ROP disease in either humans or animal models as noted by bolded text. Finally, we performed an in silico analysis to determine if each listed SNP had a described expression quantitative trait loci (eQTL) association in the EyeGEx27 database. We identified a number of established regulatory associations for our top SNPs as detailed in Table 2 within the retina, further supporting relevance to the ocular microenvironment.
rs2058019 contributes to genetic susceptibility for ROP in diverse populations
Given established differences in clinical ROP risk based on race and ethnicity22,23, we analyzed the association of the lead GLI3 SNP, rs2058019, stratified by these variables. As seen in Table 3, the association between rs2058019 and ROP disease is most significant when considering all patients, although odds ratios and allele frequencies between cases and controls demonstrate that the degree and direction of association remain consistent for Caucasian and Hispanic subjects. Interestingly, the African American subjects do not appear to be contributing to this association. This latter finding does not reach significance in this sample and thus requires validation within a larger population. As noted in Supplemental Table 3, we did not find suggestive evidence for significant differences is MAF or direction of effect within racial or ethnic populations for other top SNPs.
Multiple significant SNPs are identified regionally related to GLI3
To determine the regional significance for the GLI3 pan-locus, we plotted all SNP-associations within 100 Kb of the GLI3 gene for all patients as well as for racial and ethnic subsets. As pictured in Figure 2, a regional plot with the Y axis depicting the −log transformed p value and the X axis showing chromosomal position, we illustrate the GLI3 gene regional association for all ethnic and racial groups with ROP severity. The lead SNP rs2058019 was identified as a top-associated SNP within the pan-locus for the Caucasian and Hispanic groups and both the best and sentinel SNP when all patients were considered together. Significant regional SNPs for each group are delineated fully in Supplemental Table 4. Also pictured, we determined the linkage disequilibrium (LD) for each SNP, defined by the r2 using the included color scheme. LD was determined between SNPs and genes involved/close to SNPs identified.
Top-identified SNP associations vary by race and ethnicity
To further determine race and ethnicity-based differences in SNP associations with ROP severity, we performed our GWAS analysis separately for each racial and ethnic group. As pictured in Supplemental Figure 2, which visually represents SNP associations for each population, racial and ethnic differences are present. As seen in Table 4, we find several SNP associations which are racially or ethnically specific and reach genome-wide or suggestive level significance. Notably, rs9978278 (CLDN14 gene) and rs74048122 reach genome-wide significance, which remains significant after correction for BW and GA. Although the number of observations is decreased when analyzing by race and ethnicity, these findings suggest possible differences in genetic risk for ROP by population ancestry.
Table 4:
Racial and Ethnic differences in GWAS-identified SNP associations with severe ROP disease
| chr:position | Ethnicity | Effect allele | AF: Case | AF: Control | OR | p-value | p-value adj. BW &GA P | gene |
|---|---|---|---|---|---|---|---|---|
| 4:137201813 | HA | C | 0.02 | 0.02 | Na | Na | na | none |
| CA | C | 0.14 | 0.04 | 1.39 | 4.19E-5 | 1.99E-8* | ||
| AA | C | 0.04 | 0.06 | 0.88 | 0.40 | 0.15 | ||
| All | C | 0.06 | 0.04 | Na | Na | na | ||
| 21:37894721 | HA | T | 0.06 | 0.09 | 0.96 | 0.88 | 0.48 | CLDN14 |
| CA | T | 0.12 | 0.24 | 0.94 | 5.8E-03 | 0.08 | ||
| AA | T | 0.25 | 0.03 | 1.71 | 4.04E-9* | 1.63E-8* | ||
| All | T | 0.09 | 0.13 | 0.97 | 0.16 | 0.24 | ||
| 14:43889545 | HA | G | 0.03 | 0.06 | Na | Na | na | None |
| CA | G | 0.08 | 0.07 | 0.99 | 0.55 | 0.86 | ||
| AA | G | 0.32 | 0.04 | 1.53 | 2.62E-8* | 3.14E-8* | ||
| All | G | 0.07 | 0.06 | 1.03 | 0.25 | 0.43 | ||
| 9:131003677 | HA | G | 0.06 | 0.13 | 0.93 | 0.17 | 0.14 | DNM1 |
| CA | G | 0.13 | 0.18 | 0.95 | 0.40 | 0.20 | ||
| AA | G | 0.61 | 0.19 | 1.29 | 1.74E-7 | 2.62E-8* | ||
| All | G | 0.14 | 0.17 | 0.99 | 0.86 | 0.82 |
SNPs with suggestive or greater association with ROP severity in the full iROP cohort demonstrate cross-significance within the Hispanic Diabetic Retinopathy GOLDR cohort
Our study design, which identifies SNPs associated with ROP disease stage 3 or greater, enriches our findings for genetic variation associated with pre-retinal neovascular pathobiology. Diabetic retinopathy (DR) is another form of retinal disease characterized by pre-retinal neovascularization. Therefore, to validate the significance of GLI3 genetic variation to pre-retinal neovascular disease, we performed an extension analysis of top-associated iROP SNPs, those with p ≤5×10−6 or greater significance levels, and SNPs regionally related for DR association within the Genetics of Latino Diabetic Retinopathy (GOLDR) GWAS dataset. Importantly, the GOLDR population consists of Hispanic patients with diabetic retinopathy (DR)28,29. As noted in Table 5, we found cross-significance for a number of SNPs with DR in GOLDR and with ROP severity in our iROP GWAS cohort. This included SNPs within GLI3, DCLK1, SP4, PTPRD,RPL30 and RIDA.
Table 5.
Cross-significance of top SNPs for Retinal Pathology Characterized by Pre-retinal Neovascular Disease including ROP and Diabetic Retinopathy (GOLDR)
| GOLDR | ROP | ||||||
|---|---|---|---|---|---|---|---|
| odds ratio | P value | odds ratio | P value | ||||
| GLI3 | rs74527981 | 7 | 42205730 | 2.22 | 9.3E-04 | na | Na |
| rs17172024 | 7 | 42210112 | 2.22 | 9.3E-04 | na | Na | |
| rs2058019 | 7 | 42187113 | ns | Ns | 1.27 | 4.90E-09 | |
| CLDN12 | No loci associated with DR for whole region | ||||||
| DEF8, TUBB3 | No loci associated with DR for whole region | ||||||
| DCLK1 | rs35596426 | 13 | 36452952 | 5.18 | 4.3E-04 | Na | Na |
| rs9576059 | 13 | 36452266 | 4.83 | 5.9E-04 | Na | Na | |
| rs61948265 | 13 | 35984893 | ns | Ns | 1.14 | 6.50E-07 | |
| SP4 | rs11770747 | 7 | 21552122 | 1.96 | 7.6E-04 | ns | Ns |
| rs78539005 | 7 | 21547126 | 2.51 | 8.5E-04 | 1.09 | 0.03 | |
| rs141411503 | 7 | 21425951 | ns | Ns | 1.21 | 8.38E-07 | |
| ARHGEF7 | No loci associated with DR for whole region | ||||||
| PTPRD | rs79715438 | 9 | 8809579 | 3.48 | 5.0E-04 | na | Na |
| rs75240118 | 9 | 8809921 | 3.48 | 5.0E-04 | na | Na | |
| rs80041124 | 9 | 8810631 | 3.48 | 5.0E-04 | na | Na | |
| rs76777639 | 9 | 8488087 | 3.30 | 9.6E-04 | na | Na | |
| rs529032346 | 9 | 8499029 | 3.30 | 9.6E-04 | na | Na | |
| rs9644892 | 9 | 8800078 | ns | Ns | 1.1 | 2.14E-06 | |
| DPP4 | No loci associated with DR for whole region | ||||||
| RPL30, RIDA | rs2514337 | 8 | 98113613 | 0.49 | 5.4E-04 | 0.94 | 4.4E-04 |
| rs2447504 | 8 | 98113678 | 0.49 | 5.4E-04 | 0.94 | 4.4E-04 | |
| rs2306129 | 8 | 98093498 | 0.66 | 0.04 | 1.09 | 3.77E-06 | |
| PRPF4B | No loci associated with DR for whole region | ||||||
ns: not significant.
na: association tests not applicable for SNPs with MAF less than 0.05.
ROP Genetic Risk Score (GRS) demonstrates increased significance over single SNP associations
Joint estimation of SNP effects has been suggested as a superior approach to improve SNP-based disease prediction and risk stratification, including in eye disease by our group30–35. We calculated GRSs (Score A, Score B) incorporating SNPs based on the SNP selection by LASSO method to determine if combined SNP profiles were more highly associated with ROP severity36. ROP severity was represented as an ordinal categorial variable denoting stage including none, stage 1, stage 2, etc for this analysis. Table 6 compares the association results between different GRSs. Score A consists of the top GLI3 SNPs as well as 45 other top ROP-associated SNPs and Score B consists of 255 top-ROP associated SNPs. As depicted in Table 6, each GRS was significantly associated with ROP severity. To determine the proportion of variation within the phenotype which can be explained using each risk score, and therefore the breadth of ROP phenotype that is associated with each GRS, we analyzed adjusted R-squared. As noted, score A accounted for a larger adjusted R-squared than the top SNP alone (0.63 versus 0.34), suggesting that the additional SNPs in this risk score together account for a more variance in ROP phenotype/severity than the GLI3 SNP alone. The drop in adjusted R-squared between Score A (0.63) and Score B (0.45) is likely due to the addition of noise in the model resulting from adding SNPs in this score and over-fitting. Taken together, the GRS demonstrate stronger associations with all ROP phenotypes.
Table 6.
Association tests using genetic risk score (GRS) method.
| ROP(iROP). | |||
|---|---|---|---|
| Beta | r2 | P | |
| SNP Score A (49 SNPs) | 0.14 | 0.63 | <2e-16 |
| SNP Score B (255 SNPs) | 0.05 | 0.45 | 3.8e-16 |
| GLI3 only | 0.37 | 0.34 | 3.8e-7 |
Significant retinal vascular SNPs demonstrate relevance to ROP risk
To determine the relevance of our dataset to vascular pathology more broadly, we analyzed the association of significant retinal vascular SNPs, identified in the central retinal venule equivalent (CRVE) and the central retinal arteriole equivalent (CRAE) dataset37, within our iROP dataset. Using the summary statistics from the exome chip meta-analysis for CRVE and CRAE, we tested all CRVE/CRAE loci with p ≤ 0.01 for ROP association within our iROP dataset. A total of 583 SNPs from the CRVE/CRAE dataset were available for analysis in our iROP dataset. After Bonferroni correction for multiple testing, we identified significant associations for SNPS rs13079478 (FYCO1 gene), rs33910087 (FYCO1 gene) and rs12357206 (ANK3 gene) with ROP severity as noted in Table 7.
Table 7.
Extension of CRVE/CRAE significant loci to iROP dataset
| SNP | Gene | iROP | CRVE/CRAE |
|---|---|---|---|
| P | P | ||
| rs13079478 | FYCO1 | 7.96e-6 | 3.0E-03 |
| rs33910087 | FYCO1 | 1.93e-5 | 3.0E-03 |
| rs12357206 | ANK3 | 5.07e-5 | 9.0E-05 |
summarizes SNPs (identified for CRVE/CRAE from the literature) confirmed to be significantly associated with ROP Stage.
Extension of associations for severe ROP SNPs identified in a candidate gene study replicate significance of the RELN gene
To independently replicate previously identified ROP-associated SNPs, we evaluated candidate SNPs found to be significantly associated with severe ROP by Harnett et al.17 within our iROP GWAS dataset. As noted in Table 8, rs10251365 in the gene RELN is significantly associated with ROP severity in both cohorts (p=0.009). The p values of SNP-associations with ROP severity based on stage using our data are included for comparisons and confirmation.
Table 8.
Association of SNPs identified with severe ROP.
| SNP | MaF | gene | P for severe ROP (Hamett, 2014) | P for ROP ≥ Stage 3 (iROP) |
|---|---|---|---|---|
| rs9332681 | Na | F5 | 0.99 | Na |
| rs379489 | 0.287 | CFH | 3.8E-03 | 0.I4 |
| rs395544 | 0.287 | CFH | 5.4E-03 | 0.14 |
| rs11587174 | 0.002 | FI3B | 0.87 | na |
| rs1467199 | 0.211 | STATI | 0.82 | 0.81 |
| rs34417936 | 0.014 | IL | 0.66 | Na |
| rs2299386 | 0.380 | RELN | 1.6E-03 | 0.73 |
| rs10251365 | 0.332 | RELN | 1.0E-03 | 0.0092* |
| rs168879811 | 0.309 | NRGI | 4.0E-04 | 0.28 |
| rs168879814 | 0.311 | NRGI | 3.0E-04 | 0.25 |
| rs2353512 | 0.002 | BDNF-AS | 0.77 | Na |
| rs7127507 | 0.295 | BDNF | 2.0E-04 | 0.70 |
| rs2049046 | 0.479 | BDNF | 3.00E-05 | 0.41 |
| rs7934165 | 0.471 | BDNF | 2.00E-05 | 0.53 |
| rs12281784 | 0.101 | LRP4 | 0.54 | 0.14 |
| rs9989002 | 0.211 | IGFI | 2.2E-03 | 0.81 |
| rs11620315 | 0.149 | FLTI | 0.45 | 0.76 |
| rs1319859 | 0.424 | IGFIR | 0.12 | 0.04 |
| rs884636 | 0.040 | PCPEPIL | 0.10 | na |
| rs7204874 | Na | NSMC EIjIL4 R | 1.4E-03 | na |
| rs2057768 | 0.287 | IL4R | 7.0E-04 | 0.65 |
| rs1551005 | Na | TTR | 0.96 | na |
MAF: minor allele frequency.
na: SNP not available in ROP dataset and/or with MAF less than 0.05.
GLI3 and other top associated genes are expressed in human retinal and retinal pigment epithelial tissues (RPE).
To determine the relevance of GLI3 genetic variation to retinal pathophysiology, we sought to determine the expression patterns of GLI3 within human retinal and RPE tissues. Human donor macular or peripheral retinal and RPE tissues from control eyes (n=10), average age 72 years, were analyzed using RNA-sequencing. Expression was analyzed using DESeq238 as we have previously published39. Analyses were done for genes relative to our top-associated ROP SNPs as listed in Table 2. All top genes except PRPF4B were expressed in human donor macular tissues; the tissue type with greatest expression and fold difference between tissue types (neurosensory retina or RPE) is listed in Table 9. As noted, GLI3 was expressed in both the neurosensory retina and RPE; macular RPE expression was significantly greater by 2.54-fold (p=3.3 × 10 −15, after correction for multiple testing using Benjamini-Hockberg) than neurosensory retinal expression (Table 9). We also analyzed differential GLI3 expression between macular and peripheral RPE tissues to determine the region with greatest expression. Interestingly, we found that GLI3 expression was 1.72 fold higher in the peripheral RPE as compared to the macular RPE (p<0.01) which is notable given the presence of ROP most commonly within the peripheral retina.
Table 9:
Top gene RNA-sequencing-based expression in human donor macular retinal or RPE tissue isolated using the Utah Protocol:
| hg19 Position | Gene Name | Tissue Type with Higher Expression | Fold Change between RPE and Retina | P-value |
|---|---|---|---|---|
| chr16:89985572–90002500 | TIJBB3 | Neurosensory Retma | 35.201 | 1.03E-130 |
| chr8:99037078–99058697 | RPL30 | RPE | 2.974 | 5.69259E-61 |
| chr13:36345477–36705443 | DCLK1 | Neurosensory Retina | 10.303 | 6.27566E-29 |
| chr7:1031 12230–103629963 | RELN | Neurosensory Retina | 10.599 | 2,41595E-28 |
| chr7:21467651–21554440 | SP4 | Neurosensory Retina | 2.467 | 2,09322E-23 |
| chr16:90014332–9003446S | DEFS | Neurosensory Retina | 1.709 | 1.10S65E-20 |
| chr7:90013034–90142716 | CLDN12 | Neurosensory Retina | 2.050 | 7.47808E-16 |
| chr7:42000547–42277469 | GL13 | RPE | 2.454 | 3.30197E-15 |
| chr9:B314245–10612723 | PTPRD | Neurosensory Retina | 3.962 | 5.21447E-13 |
| chr2:162848750–162931052 | DPP4 | RPE | 2.725 | 2.96267E-OS |
| chr13:111766158–111768025 | ARIIGEF7 | Neurosensory Retina | 2.276 | 3,91738E-05 |
Differential expression between macular retinal and RPE/Choroid was calculated and p-values corrected for multiple testing using Benjamini-Hockberg. The absolute fold change is represented between the tissue with higher expression versus the tissue with lower expression.
Discussion
Herein, we report the first successful GWAS for ROP which identifies a novel ROP risk association for the GLI3 SNP rs2058019, reaching genome-wide significance within a multiethnic population. This is the first reported genome-wide significant SNP association for ROP, which shows an association trend in the same direction for both Hispanic and Caucasian infants and therefore, may demonstrate disease applicability across populations. We further identify significance of GLI3 variation for preretinal neovascular disease and established ROP genetic risk associations using extension analysis within an independent replication cohort consisting of Hispanic individuals with diabetic retinopathy and severe ROP candidate dataset17 respectively. Finally, we demonstrate relevance of our findings to human retinal and RPE tissues using expression profiling within donor eyes. Owing to the multiethnic composition of our cohort, our analysis also is the first to demonstrate racial and ethnic differences in GWAS-determined SNP associations. While our lead SNP, rs2058019, demonstrates a less significant association within the African American populations, as noted in Table 4, we identify several SNPs with significant association within only the African American population compared with the Hispanic or non-Hispanic Caucasian populations. This is consistent with, and indeed may speak to, observed clinical differences in ROP incidence and severity, including greater disease development and severity in Caucasian populations22,23. However, while suggestive of disparate genetic risk for ROP disease within different racial and ethnic populations, these results require replication in a larger population to clarify ethnic and racial differences.
GLI3 has not previously been associated with ROP disease, though it has a defined role within ocular development. Specifically, the dual role for Gli3 as both a transcriptional activator and repressor of canonical sonic hedgehog (Shh) signaling, has been found to control differentiation of the RPE and rod photoreceptor layer during Xenopus as well as Medaka fish eye development40,41. Further, abnormal expression of the Hedgehog signaling pathway has been shown in ROP within the oxygen induced retinopathy (OIR) murine model42. More broadly Gli3 has been shown to regulate both the innate and adaptive immune response, with described roles in fetal CD4 and CD8 thymocyte and T-cell development43,44. Certainly, pre-retinal neovascular disease, including within ROP and diabetic retinopathy, has been associated with aberrant inflammation45–47. Thus, there is a feasible relationship between GLI3 and ROP disease incidence and severity.
The lead SNP, rs2058019, is present within an intronic region and has not previously been associated with disease. Thus, the functional significance is not clear, although aberrancy in GLI3 is associated with human disease, including polydactly syndromes48 and malignancy49–52. Functionally, both aberrant GLI3 repressor and activator roles have been associated with underlying patho-mechanisms, resulting in imbalance within epithelial to mesenchymal transition50,52,53, AKT/ERK1 activation54, p53 function55 and autophagy56. Significant differences in GLI3 expression between control and diseased tissues is also thought to underlie pathobiology as described in AML57 and colon cancer55, in some cases regulated by differential methylation53,57. Thus, there is significant precedent for aberrancy of GLI3 DNA structure, expression, or function in human disease. Future work will seek to determine the functional significance of this SNP association to ROP disease development in preterm infants. Our work demonstrating GLI3 expression within adult human donor eyes within both neurosensory retina and RPE/choroid tissues with a higher gradient within the RPE/choroid, supports the premise that GLI3 has relevance to human pre-retinal neovascular disease.
In addition to GLI3, genes associated with top identified SNPs within our dataset, as demonstrated in Table 2, have shown associations with ROP in other work. ARHGEF7, demonstrates significant differential expression within rodent models of pre-retinal neovascularization and is also different between early and late stages of the disease within the OIR model of ROP58. There is also precedent for DPP4 function within retinal vascular homeostasis which is perturbed in murine models of both ROP and diabetic retinopathy. Within the OIR model, DPP4-inhibition increased retinal vascularity and leakage while in the murine diabetic retinopathy model, DPP4-inhibition increased retinal vascular leakage59. These murine findings are substantiated in human literature, which demonstrates increased progression of proliferative diabetic retinopathy in patients taking DPP4-inhibitors60. The inverse was also found to be true, namely that topical administration of dipeptidyl peptidase prevents vascular leakage in a diabetic mouse model61. Finally, while CLDN12 has not been found to play a role in pre-retinal neovascular disease or ROP specifically, the Claudin family of proteins have precedent in neovascular ocular disease62,63. This is particularly interesting given the identification of another Claudin family member, CLDN14, with a significant SNP association for ROP severity in African Americans (Table 4).
Genes associated with our top-identified SNPs also support findings from case control candidate-approach SNP-chip studies. As noted in Table 8, extension of associations for the top 10 SNPs identified for severe ROP by Hartnett et al.17 demonstrates independent replication for rs10251365 in the RELN gene (p=0.009). While additional cross-significance was not identified, the study design, GWAS versus candidate approach, and outcome measures, “severe compared to mild ROP” versus ROP severity by stage, differ between these studies. As noted above, we also identified the greatest association for these top SNPs within a multiethnic population which is distinct from the roughly 70% Black race population examined by Hartnett et al. Thus, the lack of additional overlap may also be informed by racial and ethnic differences between studied populations or may reflect other aspects of ROP risk, such as timing of ROP presentation as well as susceptibility of disease progression given external stresses that interact with race and ethnicity. As noted, we identify unique genetic ROP risk for African American versus Caucasian or Hispanic populations. While African American unique SNPs were identified in a smaller proportion of our cohort, and therefore based in fewer observations, these findings warrant further examination within larger populations. Future meta-analysis may allow for better harmonization and additional independent validation. Taken together, our identified SNP associations demonstrate relevance to described ROP patho-mechanisms, while also identifying novel associations for further study.
The overall relevance of genetic variation to ROP risk has been debated64. Our work substantiates genetic ROP risk which is further highlighted by our genetic risk score assessment. This analysis methodology has been used in numerous disease contexts, including by our group34,65, to assess the magnitude of genetic risk for disease outcome measures. In our analysis, ROP disease severity was more significantly associated with polygenic SNP subsets than with the most significant individual GWAS SNPs. This suggests the existence of more extensive genetic variation contributing to ROP and thus, the overall importance of this continued approach.
Finally, the relevance of our data to vascular pathobiology more broadly is evidenced in our extension analyses within independent replication datasets from other forms of pre-retinal neovascular (DR/GOLDR) and retinal vascular (CVRE/CRAE) pathobiology. We identify SNP associations that “bridge” these disease contexts and thus may speak to co-occurring mechanisms of disease. Among these, SNP-associations at the GLI3, DCLK1, SP4, PTPRD,RPL30 and RIDA genes were replicated in an independent Hispanic diabetic retinopathy (DR) cohort and thus shared within the context of pre-retinal neovascular disease which is present in both ROP ≥ stage 3 and proliferative diabetic retinopathy. While DCLK166 and PTPRD67 have been previously associated with DR in animal and human studies respectively, the remaining associations are novel within the context of pre-retinal neovascular disease and represent areas of future study. Similarly, our identification of SNPs within FYCO1 and ANK3 as shared between vascular pathobiology in the CVRE/CRAE dataset and our iROP cohort, represents novel associations. Both genes have associations within the eye and retina, though most significantly with anterior segment pathology including infantile cataract and coloboma68,69.
Taken together, we report the largest ROP GWAS to date, which identifies a novel locus at GLI3 on chromosome 7 demonstrating genome wide level significance, the first ROP associated variant to do so. We further identify 7 additional loci with suggestive level significance for ROP severity. We demonstrate relevance for GLI3 and other top-associated genes to findings from prior studies and human ocular disease through in-silico extension analyses, genetic risk score analysis and expression profiling in human donor eye tissues. Significance varied by race, with greater association measured within Caucasian and Hispanic compared with African American infants. This may underlie clinically observed differences in ROP development which are also represented in our cohort which demonstrates African American ROP prevalence of 43% compared to 52% and 67% for Caucasian and Hispanic populations respectively. Replication of this study using a larger and more diverse population would allow for validation of these findings, particularly with regard to differences in genetic ROP risk relative to ethnicity and race70–72. In summary, we report the largest ROP GWAS to date, identifying a novel locus at GLI3 with relevance to retinal biology supporting genetic susceptibilities for ROP risk with possible variability by race and ethnicity.
Methods
Recruited Study Cohort: The Imaging and Informatics for ROP (iROP) study is a multicenter, prospective, ROP cohort study collecting clinical, biologic, and imaging data from preterm infants born with GA and BW risk for ROP as previously published by our group16,25. For all enrolled infants, retinal images were obtained using a wide-angle fundus camera [RetCam; Natus Medical Incorporated, Pleasanton, CA]; biologic specimens included blood or saliva and was obtained with specific consent. Informed consent was provided to the parent/guardian under approved IRB protocols at each study site which conformed to the tenets of the Declaration of Helsinki. When consent was given for biologic sample collection blood was collected in a purple top microtainer tube. White blood cells within the buffy coat were isolated using a standard protocol and DNA extracted using a Qiagen Allprep system for subsequent genetic analysis. All images were deidentified in accordance with HIPPA privacy rules. ROP phenotype was determined for each eye by consensus from at least 3 trained graders with ROP expertise employing image-based diagnoses and the clinical exam diagnosis at each study center as previously described26. ROP severity was graded based on the worst stage present for either eye. The iROP database was reviewed to identify infants with both biologic and phenotypic data availability which included 920 infants. Patients without detailed demographic, ROP screening, clinical, or imaging data were excluded.
Genotype, quality control and imputation
Genotyping was performed using genotyping arrays from Illumina Infinium Global Screening Array (GSA, Santa Clara, CA, USA). Principal component analysis (PCA) was performed using SMARTPCA implemented in EIGENSOFT73. Standard quality control procedure was performed for all genotype samples/SNPs. For example, samples with genotyping rate less than 0.92, gender mismatches, PCA outliers defined as greater than 3 standard deviations of top principal component variables (PCs), SNPs with genotyping rate less than 0.95 and Hardy-Weinberg Equilibrium (HWE) p value less than 10–6, were excluded for analysis. Imputation was conducted by Minimac using the Michigan Imputation Server74 with the reference panel from the 1000 Genomes Project Phase 3 Haplotypes75. Autosomal SNPs with the minor allele frequency (MAF) greater than 0.05 and imputation quality (rsq) greater than 0.3 were analysed.
GOLDR cohort (Genetics of Latino Diabetic Retinopathy, GOLDR)
The GOLDR study is a family-based study assessing diabetes and diabetic complications in families (siblings and/or parents) of a proband, defined as having type 2 diabetes and either known DR or a diabetes duration of ≥10 years28. Participants are all Latinos of Mexican or Central American origin, recruited, and studied between 2007 and 2012 at the Lundquist Institute (formerly called the Los Angeles BioMedical Research Institute) at Harbor-UCLA Medical Center (HUMC). In total, there were 612 participants with type 2 diabetes from 216 families, with sizes ranging from 1 to 8 members per family in the study. These samples were genotyped with the Illumina Cardio-Metabochip and imputed with 1000 Genome using Michigan Imputation Server.
Statistical Methods
Genome-wide association tests for single SNPs
Under the additive genetic model, we performed GWAS tests with allele dosage for all genotyped and imputed SNPs stratified by each ethnic group, and also combined, using Efficient Mixed Model Association eXpedited (EMMAX) method76 implemented in EPACTS (https://genome.sph.umich.edu/wiki/EPACTS). Outcome measure for association was ROP severity defined by stage 3 or higher. Birth weight, gestational age, gender, top 3 PCs, and the relatedness by the genomic relationship matrix estimated using the genomic data, were included as covariates. Associations with p < 5×10−8 were considered GWAS significant. To further investigate the relationship between birth weight/gestational age and ROP association signals, we also performed the association tests with and without adjustment for birth weight/gestational age. In addition, GWAS analyses stratified by each ethnic or racial group were also performed in the same way.
Retinal expression quantitative trait loci (eQTL) Analysis:
To investigate the relationship between association signals and gene expression, we evaluated whether top ROP SNPs identified from GWAS are related to eQTL in the genotype-tissue specific expression databases, EyeGEx27, by far the largest reference tissue bank for gene expression and regulation in the human retina.
Genetic risk score (GRS) of ROP:
Three genetic risk scores were tested. For the “GLI3” risk score was composed of the genotype for the most significant SNP, rs2058019. In order to reduce overfitting of the genetic risk score to ROP stage due to the large number of SNPs relative to the number of subjects, we fit a linear regression model with a LASSO penalty using the R package “glmnet” with 451,565 directly genotyped SNPs77,78. The outcome variable was ROP Stage as an ordinal categorial variable (None, Stage 1, Stage 2 and Stage 3 and above). The glmnet software performs 10-fold cross-validation of the mean-squared error with a series of 100 penalties selected by a coordinate-descent; two models result, SNP score B was composed of 255 SNPs selected at the minimum penalty, and SNP score A was composed of 49 SNPs at 1 standard error (1 s.e.) from the mean-squared error. Selecting SNPs at 1 s.e. is an additional way to reduce overfitting. SNP score B and SNP score A were the sum of the number of risk alleles across all the SNPs for that score. Scores were tested in a linear model with sex, birthweight, gestation age, and the first two principal components of the SNP data. Principal components were calculated using a singular value decomposition.
Extension of the association signals for diabetic retinopathy in GOLDR:
Since many infants are Hispanic origin, we further performed the SNP association tests in an independent Hispanic cohort (GOLDR). Using diabetic retinopathy (DR) patients as cases (72 cases vs 491 controls), the association tests were performed for the top significant GWAS and suggestive ROP association genes/loci identified above (10 loci) in GOLDR cohort with the logistic regression. The significance threshold for the extension analysis was defined by Bonferroni correction with the number of genes/loci tested (0.05/10=0.005).
Extension of association signals for CRVE/CRAE loci:
Further extension analysis was performed to cross-check the overlapping SNP-associations with CRVE and/or CRAE. Using the summary statistics from the exome chip meta-analysis for CRVE and CRAE37, we evaluated association signals for all available CRVE/CRAE loci with p values<0.01 (583 SNPs in total) in both ROP dataset. The significance threshold for the extension analysis was defined by Bonferroni correction with the number of SNPs tested (0.05/583=9×10−5).
Extension of the association signals for severe ROP identified by Hartnett et al. candidate gene study:
In order to replicate the ROP association signals by other studies, we tested the significant SNPs identified by Harnett et al17 associated with severe ROP. Out of 22 top severe ROP signals, we evaluated 15 SNP-associations available in our analysis. The significance threshold for the extension analysis was defined by Bonferroni correction with the number of SNPs tested (p=0.05/15=0.0033).
Donor Eye Tissue Repository: Methods for human donor eye collection were previously described in detail according to a standardized protocol79. In brief, in collaboration with the Utah Lions Eye Bank, donor eyes were procured within a 6-hour post-mortem interval, defined as death-to-preservation time. Both eyes of the donor underwent post-mortem phenotyping with ocular imaging, including spectral domain optical coherence tomography (SD-OCT), and color fundus photography as published. Retinal pigment epithelium/choroid was immediately dissected from the overlying retina, and macula separated from periphery using an 8mm macular punch. For both peripheral and macular tissues, RPE/choroid was separated from the overlying retinal tissue using microdissection; tissue planes were optimized to minimize retinal contamination of RPE/choroid samples using a subsequent 6mm RPE/choroid tissue punch. Isolated macular and peripheral RPE/choroid samples were preserved in RNAlater (Ambion, ThermoFisher, Waltham, MA, USA), and stored at −20°C for 24 hours then transferred to −80°C. Phenotype analysis was performed as described79 by a team of 4 retinal specialists and ophthalmologists at the University of Utah School of Medicine, Moran Eye Center and the Massachusetts Eye and Ear Infirmary Retina Service. Agreement of all 4 specialists upon independent review of the color fundus and OCT imaging was deemed diagnostic; discrepancies were resolved by collaboration between a minimum of three specialists to ensure a robust and rigorous phenotypic analysis. One eye was per donor was biochemically analyzed. Institutional approval, and the consent of patients to donate their eyes and for research purposes was obtained from the University of Utah and conformed to the tenets of the Declaration of Helsinki. All tissue was deidentified in accordance with HIPPA privacy rules.
Nucleic acid extraction and RNA-Sequencing:
Transcriptional profiling of macular and peripheral retina and RPE/choroid tissues from 10 unrelated control eye tissue donors, average age 72 years, was performed using RNA-sequencing. DNA and RNA were extracted from peripheral and macular neurosensory retinal and RPE/choroid tissues, prepared as described above, using the Qiagen All-prep DNA/RNA mini kit (cat #80204) per the manufacturer’s protocol. Quality of RNA samples was assessed with an RNA Nano Chip (Agilent). Total RNA was poly-A selected and cDNA libraries were constructed using the Illumina TruSeq Stranded mRNA Sample Preparation Kit (cat# RS-122-2101, RS-122-2102) according to the manufacturer’s protocol. Sequencing libraries (18 pM) were chemically denatured and applied to an Illumina TruSeq v3 single read flow cell using an Illumina cBot. Hybridized molecules were clonally amplified and annealed to sequencing primers with reagents from an Illumina TruSeq SR Cluster Kit v3-cBot-HS (GD-401-3001). Following transfer of the flowcell to an Illumina HiSeq instrument (HCS v2.0.12 and RTA v1.17.21.3), a 50-cycle single read sequence run was performed using TruSeq SBS v3 sequencing reagents (FC-401-3002)
Primary Analysis of RNA Sequencing Data:
Each of the 50bp, poly-A selected, non-stranded, Illumina HiSeq fastq datasets were processed as follows: Reads were aligned using NovoCraft’s novoalign 2.08.03 software (http://www.novocraft.com/) with default settings plus the -o SAM -r All 50 options to output multiple repeat matches. The genome index contained human hg19 chromosomes, phiX (an internal control), and all known and theoretical splice junctions based on Ensembl transcript annotations. Additional details for this aspect of the protocol are described elsewhere (http://useq.sourceforge.net/usageRNASeq.html).Next, raw novoalignments were processed using the open source USeq SamTranscriptiomeParser (http://useq.sourceforge.net) to remove alignments with an alignment score greater than 90 (~ 3 mismatches), convert splice junction coordinates to genomic, and randomly select one alignment to represent reads that map equally well to multiple locations. Relative read coverage tracks were generated using the USeq Sam2USeq utility (http://useq.sourceforge.net/cmdLnMenus.html#Sam2USeq) for each sample and sample type (Normal Retina, Neovascular AMD Retina, Intermediate AMD Retina, Normal RPE, Neovascular AMD RPE, and Intermediate AMD RPE). These data tracks are directly comparable in genome browsers and good tools to visualize differential expression and splicing. Estimates of sample quality were determined by running the Picard CollectRnaSeqMetrics application (http://broadinstitute.github.io/picard/) on each sample. These QC metrics were then merged into one spreadsheet to identify potential outliers. Agilent Bioanalyzer RIN and library input concentration columns were similarly added for QC purposes (http://www.genomics.agilent.com).
Acknowledgements
Supported by the National Institutes of Health, National Eye Institute (NIH/NEI) Contract RO1EY019474, R01 EY031331, R21 EY031883, and P30 EY10572 the National Center for Advancing Translational Sciences, CTSI grant UL1TR001881, and the National Institute of Diabetes and Digestive and Kidney Disease Diabetes Research Center (DRC) grant DK063491 to the Southern California Diabetes Endocrinology Research Center. unrestricted departmental funding and a Career Development Award (Dr Campbell) from Research to Prevent Blindness (New York, NY), and with support from the US Agency for International Development, the Seva Foundation, and Helen Keller International. Infrastructure for the CHARGE Consortium is supported in part by the National Heart, Lung, and Blood Institute (NHLBI) grant R01HL105756. LAO: 1K08EY031800-03; (PI). This work was supported by National Institutes of Health Core Grant (EY014800), and an Unrestricted Grant from Research to Prevent Blindness, New York, NY, to the Department of Ophthalmology & Visual Sciences, University of Utah. MEH: R01EY015130 and R01EY107011 (PI). R21: R21EY033579 (co-PI). MMD: Ira G. Ross and Elizabeth Olmsted Ross Endowed Chair. MMD receives research grant support from Genentech (San Francisco, CA). This paper was supported in part by the Genetics of Latinos Diabetic Retinopathy (GOLDR) Study grant EY14684.
Competing interests
Dr. Campbell receives research support from Genentech (San Francisco, CA). Dr. Campbell was a consultant to Boston AI Lab (Boston, MA). Dr. Chan is on the Scientific Advisory Board for Phoenix Technology Group (Pleasanton, CA), a consultant for Alcon (Ft Worth, TX). Dr. Chiang was previously a consultant for Novartis (Basel, Switzerland), and was previously an equity owner of InTeleretina, LLC (Honolulu, HI). Drs. Campbell and Chan are equity owners of Siloam Vision.
Footnotes
Supplementary Files
Contributor Information
Jerome Rotter, Lundquist Institute.
Xiaohui Li, The Institute for Translational Genomics and Population Sciences.
Leah A. Owen, University of Utah, Moran Eye Center
Kent Taylor, The Institute for Translational Genomics and Population Sciences.
Susan Ostmo, Oregon Health & Science University.
Yii-Der Ida Chen, Lundquist Institute for Biomedical Innovation at Harbor-UCLA Medical Center.
Aaron Coyner, Oregon Health & Science University.
Kemal Sonmez, Casey Eye Institute Oregon Health & Science University.
M.Elizabeth hartnett, University of Utah.
Xiuqing Guo, The Institute for Translational Genomics and Population Sciences.
Eli Ipp, THE LUNDQUIST INSTITUTE.
Kathryn Roll, THE LUNDQUIST INSTITUTE.
Pauline Genter, THE LUNDQUIST INSTITUTE.
R.V. Paul Chan, University of Illinois at Chicago.
Margaret DeAngelis, SUNY-University at Buffalo.
Michael Chiang, National Eye Institute.
J Peter Campbell, OHSU.
References
- 1.Gilbert C. & Foster A. Childhood blindness in the context of VISION 2020--the right to sight. Bull. World Health Organ. 79, 227–232 (2001). [PMC free article] [PubMed] [Google Scholar]
- 2.Blencowe H., Lawn J. E., Vazquez T., Fielder A. & Gilbert C. Preterm-associated visual impairment and estimates of retinopathy of prematurity at regional and global levels for 2010. Pediatr. Res. 74 Suppl 1, 35–49 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gilbert C. Retinopathy of prematurity: a global perspective of the epidemics, population of babies at risk and implications for control. Early Hum. Dev. 84, 77–82 (2008). [DOI] [PubMed] [Google Scholar]
- 4.Owen L. A., Morrison M. A., Hoffman R. O., Yoder B. A. & DeAngelis M. M. Retinopathy of prematurity: A comprehensive risk analysis for prevention and prediction of disease. PloS One 12, e0171467 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Slidsborg C. et al. Neonatal Risk Factors for Treatment-Demanding Retinopathy of Prematurity: A Danish National Study. Ophthalmology 123, 796–803 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Cryotherapy for Retinopathy of Prematurity Cooperative Group. Multicenter Trial of Cryotherapy for Retinopathy of Prematurity: ophthalmological outcomes at 10 years. Arch. Ophthalmol. Chic. Ill 1960 119, 1110–1118 (2001). [DOI] [PubMed] [Google Scholar]
- 7.Schaffer D. B. et al. Prognostic factors in the natural course of retinopathy of prematurity. The Cryotherapy for Retinopathy of Prematurity Cooperative Group. Ophthalmology 100, 230–237 (1993). [DOI] [PubMed] [Google Scholar]
- 8.Kim S. J. et al. Identification of candidate genes and pathways in retinopathy of prematurity by whole exome sequencing of preterm infants enriched in phenotypic extremes. Sci. Rep. 11, 4966 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Owen L. A. & Hartnett M. E. Current Concepts of Oxygen Management in Retinopathy of Prematurity. J. Ophthalmic Vis. Res. 9, 94–100 (2014). [PMC free article] [PubMed] [Google Scholar]
- 10.SUPPORT Study Group of the Eunice Kennedy Shriver NICHD Neonatal Research Network et al. Target ranges of oxygen saturation in extremely preterm infants. N. Engl. J. Med. 362, 1959–1969 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schmidt B. et al. Effects of targeting higher vs lower arterial oxygen saturations on death or disability in extremely preterm infants: a randomized clinical trial. JAMA 309, 2111–2120 (2013). [DOI] [PubMed] [Google Scholar]
- 12.Good W. V. FINAL RESULTS OF THE EARLY TREATMENT FOR RETINOPATHY OF PREMATURITY (ETROP) RANDOMIZED TRIAL. Trans. Am. Ophthalmol. Soc. 102, 233–250 (2004). [PMC free article] [PubMed] [Google Scholar]
- 13.Flitcroft D. I., Nolan L., O’Connor J., Lanigan B. & O’Keefe M. Comparison of Retinopathy of Prematurity (ROP) in mono– and dizygotic twins: Evidence for genetic and intrauterine environmental factors in ROP aetiology. Invest. Ophthalmol. Vis. Sci. 45, 4020–4020 (2004). [Google Scholar]
- 14.Woo S. J. et al. A co-twin study of the relative effect of birth weight and gestational age on retinopathy of prematurity. Eye Lond. Engl. 25, 1478–1483 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yau G. S. K. et al. Differences in risk factors for retinopathy of prematurity development in paired twins: a Chinese population study. ScientificWorldJournal 2014, 212183 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim S. J. et al. Identification of candidate genes and pathways in retinopathy of prematurity by whole exome sequencing of preterm infants enriched in phenotypic extremes. Sci. Rep. 11, 4966 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hartnett M. E. et al. Genetic Variants Associated With Severe Retinopathy of Prematurity in Extremely Low Birth Weight Infants. Invest. Ophthalmol. Vis. Sci. 55, 6194–6203 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gohari M. et al. Association of eNOS and ACE Polymorphisms with Retinopathy of Prematurity: A Systematic Review and Meta-Analysis. Fetal Pediatr. Pathol. 39, 334–345 (2020). [DOI] [PubMed] [Google Scholar]
- 19.Lei X.-J., Zhao Y.-X. & Qiao T. Influence of polymorphisms in VEGF, ACE, TNF and GST genes on the susceptibility to retinopathy of prematurity among Chinese infants. Int. J. Ophthalmol. 11, 1451–1457 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ilguy S. et al. The relationship of retinopathy of prematurity with brain-derivated neurotrophic factor, vascular endotelial growth factor-A, endothelial PAD domain protein 1 and nitric oxide synthase 3 gene polymorphisms. Ophthalmic Genet. 42, 725–731 (2021). [DOI] [PubMed] [Google Scholar]
- 21.Gohari M. et al. An Updated and Comprehensive Meta-Analysis of Association between VEGA −634G > C, −460T > C, +405G > C and +936C > T Polymorphisms and Retinopathy of Prematurity Risk. Fetal Pediatr. Pathol. 40, 233–249 (2021). [DOI] [PubMed] [Google Scholar]
- 22.Saunders R. A. et al. Racial variation in retinopathy of prematurity. The Cryotherapy for Retinopathy of Prematurity Cooperative Group. Arch. Ophthalmol. Chic. Ill 1960 115, 604–608 (1997). [DOI] [PubMed] [Google Scholar]
- 23.Port A. D., Chan R. V. P., Ostmo S., Choi D. & Chiang M. F. Risk Factors for Retinopathy of Prematurity: Insights from Outlier Infants. Graefes Arch. Clin. Exp. Ophthalmol. Albrecht Von Graefes Arch. Klin. Exp. Ophthalmol. 252, 1669–1677 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eliason K. J., Dane Osborn J., Amsel E. & Richards S. C. Incidence, progression, and duration of retinopathy of prematurity in Hispanic and white non-Hispanic infants. J. AAPOS Off. Publ. Am. Assoc. Pediatr. Ophthalmol. Strabismus 11, 447–451 (2007). [DOI] [PubMed] [Google Scholar]
- 25.Brown J. M. et al. Automated Diagnosis of Plus Disease in Retinopathy of Prematurity Using Deep Convolutional Neural Networks. JAMA Ophthalmol. 136, 803–810 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ryan M. C. et al. Development and Evaluation of Reference Standards for Image-based Telemedicine Diagnosis and Clinical Research Studies in Ophthalmology. AMIA Annu. Symp. Proc. AMIA Symp. 2014, 1902–1910 (2014). [PMC free article] [PubMed] [Google Scholar]
- 27.Ratnapriya R. et al. Retinal transcriptome and eQTL analyses identify genes associated with age-related macular degeneration. Nat. Genet. 51, 606–610 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuo J. Z. et al. Association of fasting insulin and C peptide with diabetic retinopathy in Latinos with type 2 diabetes. BMJ Open Diabetes Res. Care 2, e000027 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gao X. et al. Native American Ancestry Is Associated With Severe Diabetic Retinopathy in Latinos. Invest. Ophthalmol. Vis. Sci. 55, 6041–6045 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Literature-Based Genetic Risk Scores for Coronary Heart Disease | Circulation: Cardiovascular Genetics. https://www.ahajournals.org/doi/full/10.1161/CIRCGENETICS.111.960708. [DOI] [PubMed]
- 31.Dareng E. O. et al. Polygenic risk modeling for prediction of epithelial ovarian cancer risk. Eur. J. Hum. Genet. EJHG 30, 349–362 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao Y., Tao Z. & Chen X. Identification of a three-m6A related gene risk score model as a potential prognostic biomarker in clear cell renal cell carcinoma. PeerJ 8, e8827 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang T. et al. Detecting genetic risk factors for Alzheimer’s disease in whole genome sequence data via Lasso screening. in 2015 IEEE 12th International Symposium on Biomedical Imaging (ISBI) 985–989 (2015). doi: 10.1109/ISBI.2015.7164036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McKean-Cowdin R. et al. Genetic Susceptibility to Diabetic Retinopathy Using a Genetic Risk Score in Multiethnic Cohorts. Invest. Ophthalmol. Vis. Sci. 61, 3849 (2020). [Google Scholar]
- 35.Bailey J. N. C. et al. The Application of Genetic Risk Scores in Age-Related Macular Degeneration: A Review. J. Clin. Med. 5, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hahn G. et al. A Smoothed Version of the Lassosum Penalty for Fitting Integrated Risk Models Using Summary Statistics or Individual-Level Data. Genes 13, 112 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jensen R. A. et al. Novel Genetic Loci Associated with Retinal Microvascular Diameter. Circ. Cardiovasc. Genet. 9, 45–54 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Love M. I., Huber W. & Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim S. et al. Generation, transcriptome profiling, and functional validation of cone-rich human retinal organoids. Proc. Natl. Acad. Sci. U. S. A. 116, 10824–10833 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sun J. et al. Zic5 stabilizes Gli3 via a non-transcriptional mechanism during retinal development. Cell Rep. 38, 110312 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reinhardt R. et al. Sox2, Tlx, Gli3, and Her9 converge on Rx2 to define retinal stem cells in vivo. EMBO J. 34, 1572–1588 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu M., Chen X., Liu H. & Di Y. Expression and significance of the Hedgehog signal transduction pathway in oxygen-induced retinal neovascularization in mice. Drug Des. Devel. Ther. 12, 1337–1346 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Matissek S. J. & Elsawa S. F. GLI3: a mediator of genetic diseases, development and cancer. Cell Commun. Signal. 18, 54 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hager-Theodorides A. L., Dessens J. T., Outram S. V. & Crompton T. The transcription factor Gli3 regulates differentiation of fetal CD4- CD8- double-negative thymocytes. Blood 106, 1296–1304 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Al-Kharashi A. S. Role of oxidative stress, inflammation, hypoxia and angiogenesis in the development of diabetic retinopathy. Saudi J. Ophthalmol. 32, 318–323 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bulka C. M. et al. Placental CpG Methylation of Inflammation, Angiogenic, and Neurotrophic Genes and Retinopathy of Prematurity. Invest. Ophthalmol. Vis. Sci. 60, 2888–2894 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Holm M. et al. Systemic Inflammation-Associated Proteins and Retinopathy of Prematurity in Infants Born Before the 28th Week of Gestation. Invest. Ophthalmol. Vis. Sci. 58, 6419–6428 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xiang Y. et al. A Novel Nonsense GLI3 Variant Is Associated With Polydactyly and Syndactyly in a Family by Blocking the Sonic Hedgehog Signaling Pathway. Front. Genet. 11, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang G. et al. The Microarray Gene Profiling Analysis of Glioblastoma Cancer Cells Reveals Genes Affected by FAK Inhibitor Y15 and Combination of Y15 and Temozolomide. Anticancer Agents Med. Chem. 14, 9–17 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rodrigues M. F. S. D. et al. GLI3 knockdown decreases stemness, cell proliferation and invasion in oral squamous cell carcinoma. Int. J. Oncol. 53, 2458–2472 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Han W., Ibarra G., Gupta M., Yin Y. & Elsawa S. F. Elevated GLI3 expression in germinal center diffuse large B cell lymphoma. Leuk. Lymphoma 59, 2743–2745 (2018). [DOI] [PubMed] [Google Scholar]
- 52.Li J., Qiu M., An Y., Huang J. & Gong C. miR-7–5p acts as a tumor suppressor in bladder cancer by regulating the hedgehog pathway factor Gli3. Biochem. Biophys. Res. Commun. 503, 2101–2107 (2018). [DOI] [PubMed] [Google Scholar]
- 53.Fu L. et al. Set7 mediated Gli3 methylation plays a positive role in the activation of Sonic Hedgehog pathway in mammals. eLife 5, e15690 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Karin M. The regulation of AP-1 activity by mitogen-activated protein kinases. J. Biol. Chem. 270, 16483–16486 (1995). [DOI] [PubMed] [Google Scholar]
- 55.Hn K., Sc O., Js K. & Ya Y. Abrogation of Gli3 expression suppresses the growth of colon cancer cells via activation of p53. Exp. Cell Res. 318, (2012). [DOI] [PubMed] [Google Scholar]
- 56.Lo Ré A. E. et al. Novel AKT1-GLI3-VMP1 pathway mediates KRAS oncogene-induced autophagy in cancer cells. J. Biol. Chem. 287, 25325–25334 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chaudhry P., Singh M., Triche T. J., Guzman M. & Merchant A. A. GLI3 repressor determines Hedgehog pathway activation and is required for response to SMO antagonist glasdegib in AML. Blood 129, 3465–3475 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Recchia F. M., Xu L., Penn J. S., Boone B. & Dexheimer P. J. Identification of Genes and Pathways Involved in Retinal Neovascularization by Microarray Analysis of Two Animal Models of Retinal Angiogenesis. Invest. Ophthalmol. Vis. Sci. 51, 1098–1105 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee C.-S. et al. Dipeptidyl Peptidase-4 Inhibitor Increases Vascular Leakage in Retina through VE-cadherin Phosphorylation. Sci. Rep. 6, 29393 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kang E. Y.-C. et al. Association between Add-On Dipeptidyl Peptidase-4 Inhibitor Therapy and Diabetic Retinopathy Progression. J. Clin. Med. 10, 2871 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hernández C. et al. Topical administration of DPP-IV inhibitors prevents retinal neurodegeneration in experimental diabetes. Diabetologia 60, 2285–2298 (2017). [DOI] [PubMed] [Google Scholar]
- 62.Gonçalves A., Ambrósio A. F. & Fernandes R. Regulation of claudins in blood-tissue barriers under physiological and pathological states. Tissue Barriers 1, e24782 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Differential expression of claudins in retinas during normal development and the angiogenesis of oxygen-induced retinopathy - PubMed. https://pubmed.ncbi.nlm.nih.gov/21862644/. [DOI] [PubMed]
- 64.Swan R. et al. The Genetics of Retinopathy of Prematurity: A Model for Neovascular Retinal Disease. Ophthalmol. Retina 2, 949–962 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dornbos P. et al. A combined polygenic score of 21,293 rare and 22 common variants improves diabetes diagnosis based on hemoglobin A1C levels. Nat. Genet. 54, 1609–1614 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yang H., Son G. W., Park H. R., Lee S. E. & Park Y. S. Effect of Korean Red Ginseng treatment on the gene expression profile of diabetic rat retina. J. Ginseng Res. 40, 1–8 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Peng D. et al. CDKAL1 rs7756992 is associated with diabetic retinopathy in a Chinese population with type 2 diabetes. Sci. Rep. 7, 8812 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Owen N. et al. Identification of 4 novel human ocular coloboma genes ANK3, BMPR1B, PDGFRA, and CDH4 through evolutionary conserved vertebrate gene analysis. Genet. Med. 24, 1073–1084 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chen J. et al. Mutations in FYCO1 Cause Autosomal-Recessive Congenital Cataracts. Am. J. Hum. Genet. 88, 827–838 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Visscher P. M. et al. 10 Years of GWAS Discovery: Biology, Function, and Translation. Am. J. Hum. Genet. 101, 5–22 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cantor R. M., Lange K. & Sinsheimer J. S. Prioritizing GWAS Results: A Review of Statistical Methods and Recommendations for Their Application. Am. J. Hum. Genet. 86, 6–22 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Barr J. L. et al. Heritable Risk and Protective Genetic Components of Glaucoma Medication Non-Adherence. Int. J. Mol. Sci. 24, 5636 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Price A. L. et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 38, 904–909 (2006). [DOI] [PubMed] [Google Scholar]
- 74.Das S. et al. Next-generation genotype imputation service and methods. Nat. Genet. 48, 1284–1287 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.L C. et al. The international Genome sample resource (IGSR): A worldwide collection of genome variation incorporating the 1000 Genomes Project data. Nucleic Acids Res. 45, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kang H. M. et al. Variance component model to account for sample structure in genome-wide association studies. Nat. Genet. 42, 348–354 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Friedman J., Hastie T. & Tibshirani R. Regularization Paths for Generalized Linear Models via Coordinate Descent. J. Stat. Softw. 33, 1–22 (2010). [PMC free article] [PubMed] [Google Scholar]
- 78.Tibshirani R. et al. Strong rules for discarding predictors in lasso-type problems. J. R. Stat. Soc. Ser. B Stat. Methodol. 74, 245–266 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Owen L. A. et al. The Utah Protocol for Postmortem Eye Phenotyping and Molecular Biochemical Analysis. Invest. Ophthalmol. Vis. Sci. 60, 1204–1212 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]