Abstract
Accurate quantification of left atrium (LA) scar in patients with atrial fibrillation is essential to guide successful ablation strategies. Prior to LA scar quantification, a proper LA cavity segmentation is required to ensure exact location of scar. Both tasks can be extremely time-consuming and are subject to inter-observer disagreements when done manually. We developed and validated a deep neural network to automatically segment the LA cavity and the LA scar. The global architecture uses a multi-network sequential approach in two stages which segment the LA cavity and the LA Scar. Each stage has two steps: a region of interest Neural Network and a refined segmentation network. We analysed the performances of our network according to different parameters and applied data triaging. 200+ late gadolinium enhancement magnetic resonance images were provided by the LAScarQS 2022 Challenge. Finally, we compared our performances for scar quantification to the literature and demonstrated improved performances.
Keywords: Segmentation, Late gadolinium enhancement, Deep learning, Atrial fibrillation, Left atrium
1. Introduction
Atrial fibrillation (AF) is the most prevalent sustained heart rhythm disorder, contributing significantly to global health care costs and mortality and morbidity rates [1]. Catheter ablation may offer a cure to AF in some patients. However, AF ablation success rates are modest (50%) in patients with extensive structural remodeling (i.e., fibrosis infiltration, fat accumulation) of the left atrium (LA) [2]. Such structural remodeling is typically identified from pre-ablation late gadolinium-enhancement magnetic resonance imaging (LGE-MRI) [3]. Reconstruction of the atrial anatomy and quantification of the fibrotic substrate is clinically important for guiding catheter ablation [3]. The first step of anatomical reconstruction is the segmentation of the atrial myocardium from cardiac images. Generally, the LA endocardial walls [4] are manually segmented from the LGE-MRI to reconstruct the atrial anatomy. Location and segmentation of atrial structures such as the mitral valve, pulmonary veins, and atrial appendages are a challenge even with significant expertise. Variable left atrial anatomy and thin atrial walls, compounded with poor and inconsistent image quality, make segmentation time-consuming and challenging. Atrial segmentation is also hampered by partial volume effects as the LA is close in proximity to extra-cardiac structures [5]. Consequently, manual segmentation of the LA from LGE-MRI has high inter-operator variability even amongst experts [6]. Given the low reproducibility of existing LGE-MRI segmentation methods, a robust and fully automated LA segmentation method is critical for accurately reconstructing the atrium anatomy and identifying the structurally remodeled substrate. Recently, deep learning techniques have been applied to improve LA segmentation in LGE-MRI [7]. However, even the best networks struggle in segmenting regions with sudden changes in the LA anatomy like the pulmonary veins (PVs) [7]. Furthermore, while there has been increased development of machine learning techniques for LA segmentation in LGE-MRI, only a few methods propose segmentation of the scar [8–10]. We developed a deep learning approach to automatically segment both the LA and LA scar from LGE-MRI images. We term our network Left Atrium Scar Segmentation Network (LASSNet). LASSNet simultaneously segments the LA and LA scar and is robust to images collected by different clinical centers.
2. Methods
2.1. Training and Validation Data
The clinical data used for this work was provided by the LAScarQS 2022 Challenge [9, 11, 12]. All the data received institutional ethical approval and have been anonymized. The images and the corresponding ground truth (GT) segmentations were from four clinical centers and the images were acquired using 1.5 and 3 T MRI scanners.
LAScarQS 2022 Challenge was designed to solve two tasks: “LA Scar Quantification” and “Left Atrial Segmentation from Multi-Center LGE-MRIs”. In order to solve the first task, 60 LGE-MRIs with the corresponding GT LA cavity segmentations and GT LA scars segmentations were accessible for training. 10 LGE-MRIs without the GT were used to validate the model through an online submission of the predictions. To solve the second task, 130 LGE-MRIs with the GT left atrial cavity segmentations were made accessible for training. 20 LGE-MRIs without the corresponding GT were provided for validation of the model through an online submission of the predictions.
2.2. Data Inspection and Pre-processing
Segmentation performances of neural networks (NNs) are highly dependent on the quality of the input image data and ground truth annotation (segmentation) [13]. Therefore, data inspection was performed by a cardiac MRI expert in our team to identify image datasets of lower quality, which might have a detrimental effect on the training of LASSNet for both tasks (represented in Table 1).
Table 1.
Analysis of the training data for Task 1 (LA Scar Quantification) and Task 2 (Left Atrial Segmentation from Multi-Center LGE-MRIs).
| Analysis of the training data for Task 1 | |
| Issue | Dataset index |
| Scans with poor fat suppression can create artifacts | 28, 35, 49 |
| Scans acquired too early after contrast injection result in poor contrast between blood pool and enhancement and therefore a low accuracy of scar detection | 5, 12, 37, 49, 57, 59 |
| Scans with poor image quality for accurate scar detection (severe blurring, very noisy, etc.) | 7, 8, 19, 45, 47, 49, 50, 54 |
| Analysis of the training data for Task 2 | |
| Issue | Dataset index |
| Scans with poor fat suppression | 70, 78, 85, 109, 111, 123 |
| Scans with poor image quality | 33, 39, 75, 129 |
| Datasets with severe errors in LA cavity segmentation | 19, 24, 45, 64, 74, 95, 100, 101, 112, 126, 130 |
| Scans with partial coverage of left atrium appendage (LAA) and left superior pulmonary vein (LSPV) | 97, 100, 129 |
| Duplicate post-ablation scans of the same patient | 51, 60 |
Figure 1 shows examples of the various quality LGE scans provided for LA scar segmentation. The expert indicated that significant discrepancies in LA cavity GT segmentations were observed in regions of pulmonary veins (PVs), mitral valve, LA floor and roof, and LA appendage. Furthermore, one of the right PVs was not segmented for patients with three right PVs. Poor training data represented 25% of the dataset for Task 1 and 18.46% for Task 2. We evaluated the performance of LASSNet with and without the low quality training data in Sect. 3.1.
Fig. 1.
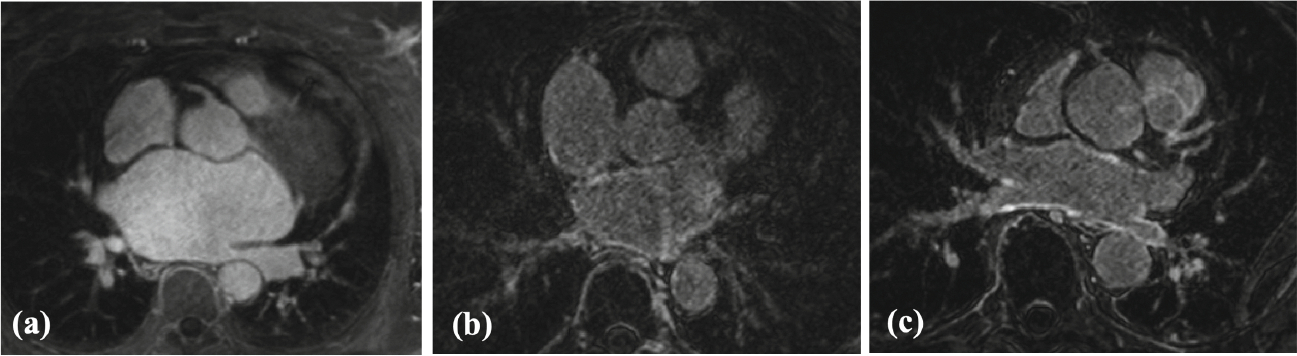
Representative examples of various quality LGE scans for LA scar segmentation: (a) poor quality (very low contrast between scar and blood, sub-optimal inversion time (TI) value, bad signal-to-noise ratio (SNR)); (b) fair quality (good scar-blood contrast, optimal TI, low SNR, artifact in LA); (c) good quality (high scar-blood contrast, optimal TI, good SNR).
Then before feeding the image volumes into the NN, the images were pre-processed to obtain the same voxel size and dimensions. Images and corresponding GT segmentations were pre-processed and normalized before training the NN. Image augmentation was applied to the normalized images to increase the amount of data available for training and encourage generalizable performance.
During the scar GT inspection, mislabeled scar voxels were noticed: deep inside LA cavity (Fig. 2(a)), far outside LA wall (Fig. 2(b)) and enhanced voxels of anatomical structures adjacent to LA (Fig. 2(c)). Therefore, we constrained the scar GT voxels to be located into LA wall region of interest (ROI) masks.
Fig. 2.
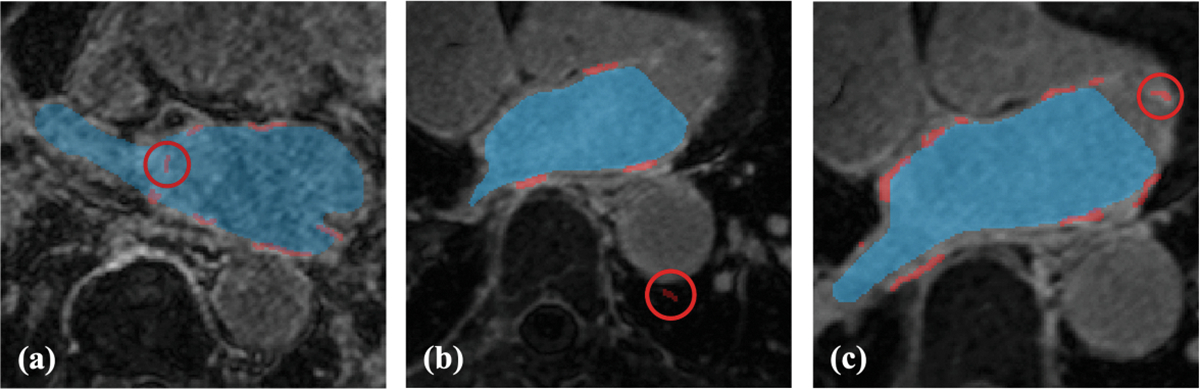
Representative examples of various mislabeled scar voxels (red circles): (a) scar voxels deep inside LA cavity; (b) scar voxels far outside LA wall ROI; (c) enhanced voxels of the mitral valve were mislabeled as LA scar.
2.3. The Neural Network Architecture
The global architecture is presented in Fig. 3. It is composed of four NN models. LASSNet uses a multi-network sequential approach in two stages to segment the LA cavity and LA Scar. Each stage consists of two steps: (1) A ROI NN that first detects the anatomical location of the LA (or LA Scar) to be segmented, followed by (2) a LA (or LA Scar) segmentation (LA(S)SEG) NN that generates a refined LA cavity segmentation for the first stage or a refined LA Scar segmentation for the second stage.
Fig. 3. LASSNet architecture.
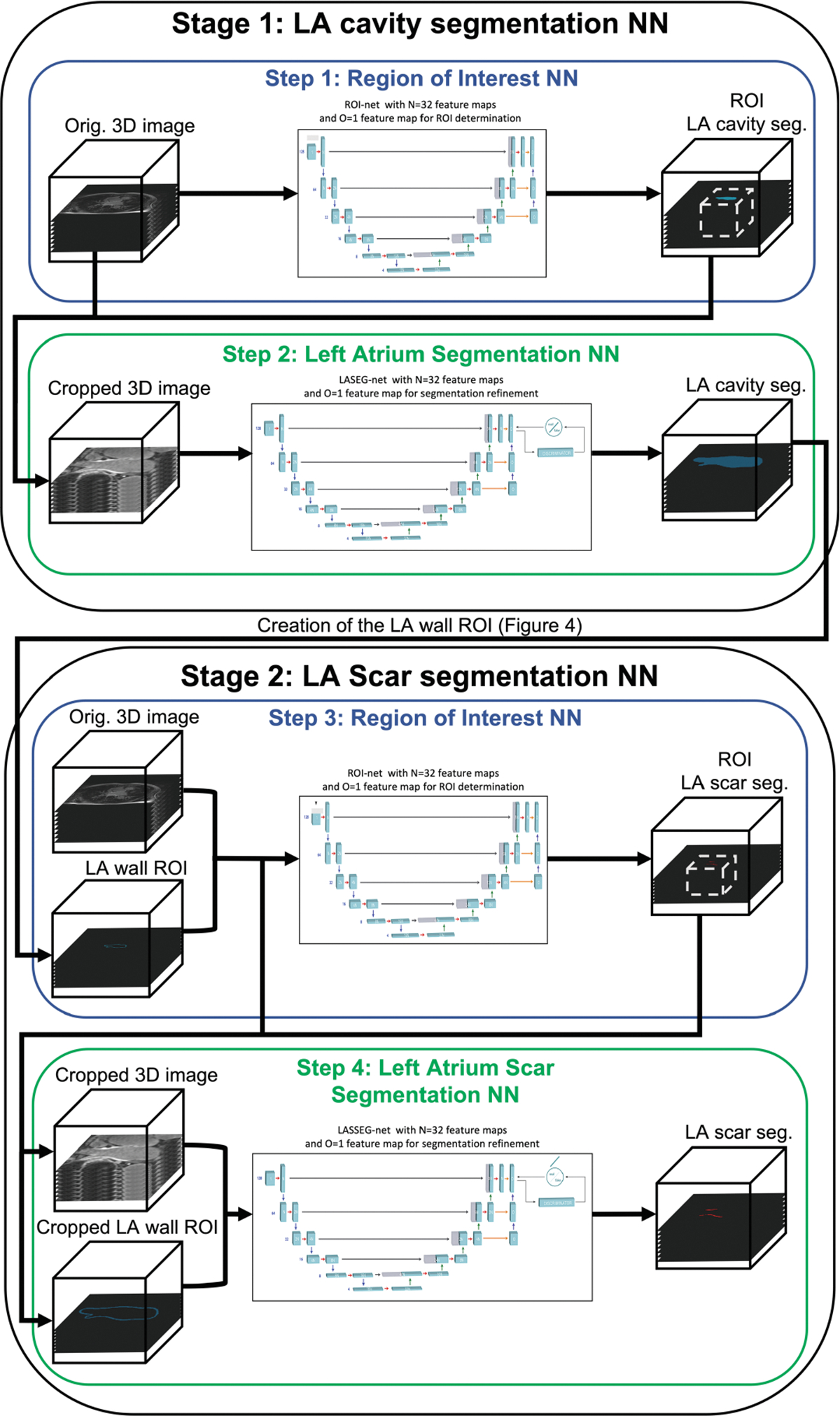
Step 1: LGE-MRI images are passed into ROI NN which identifies the region where the left atrium is located. The image volume is then cropped to the ROI and resampled. Step 2: The image is inputted into the Left Atrium Segmentation Network (LASEG NN) to perform the LA cavity segmentation. LASEG NN includes a discriminator for adversarial training. Step 3: LGE-MRI images and LA wall ROI masks are passed into ROI NN which identifies the region where the left atrium scar is located. The image volume and the LA wall ROI are then cropped to the identified ROI and resampled. Step 4: Cropped LGE-MRI images and LA wall ROI masks are then inputted into the Left Atrium Scar Segmentation Network (LASSEG NN) to refine the left atrium scar segmentation. LASSEG NN includes a discriminator for adversarial training.
Steps 1 and 3: Region Of Interest Neural Networks (ROI NNs):
The objective of the ROI NNs is to identify the region where the LA (or LA scar) is located to reduce the number of background voxels due to non-atrial structures. The ROI NNs are a variation of the 3D U-Net [14]. The Stage 1 NN was modified to accept an image volume and a single binary label GT segmentation of the LA. The Stage 2 ROI NN was modified to accept both an image volume and a single binary label LA wall ROI image volume as well as a single binary label GT segmentation of the LA scar. The LA wall ROI image volume was created from the non-overlapping region of the dilated and eroded segmentation output of the Stage 1 NN in order to obtain a LA wall where post-ablation scar should exist [15, 16]. We evaluate the performance of LASSNet with different ROI wall thicknesses in Sect. 3.1. The process to create the LA wall ROI is represented on Fig. 4.
Fig. 4.
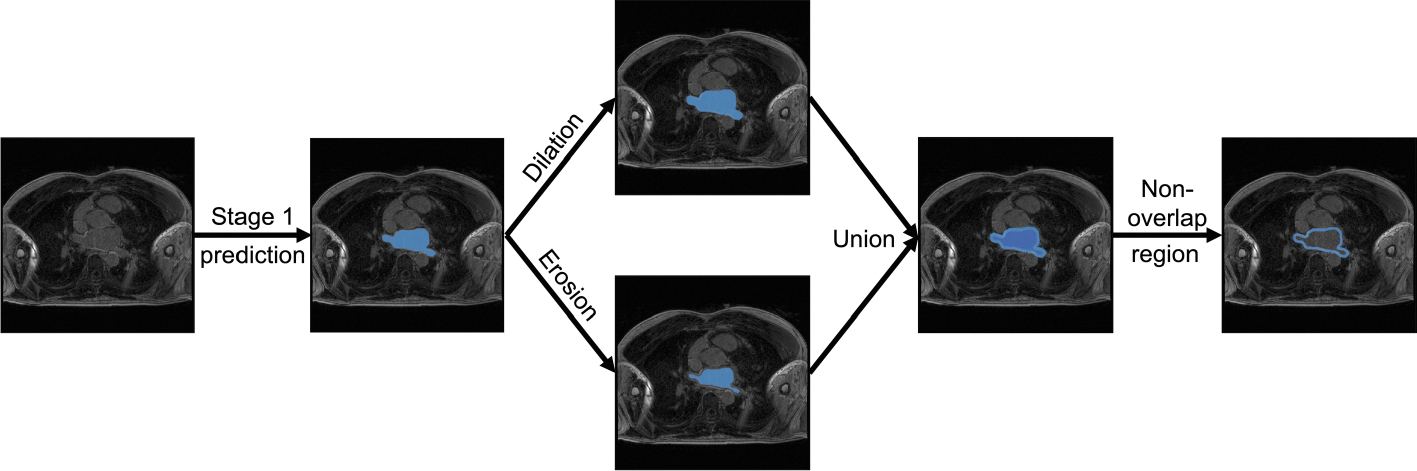
Process to create the LA wall ROI where the scar should subsist. We use the LA cavity segmentation (output of the Stage 1 NN) to create a dilated and an eroded mask. Both masks are unionized and the non-overlapping is kept as the LA wall ROI.
The U-Nets are fully convolutional NNs which is the most used approach for biomedical image segmentation [30]. The architecture of the U-Nets is presented in Fig. 5; it consists of an encoder which down-samples the input image through a series of convolutions and max-pooling operations down to a bottleneck layer, followed by a decoder that up-samples the bottleneck representation back to the original image resolution. We implemented instance normalization instead of traditional batch normalization [31] to normalize our image volumes across spatial locations.
Fig. 5.
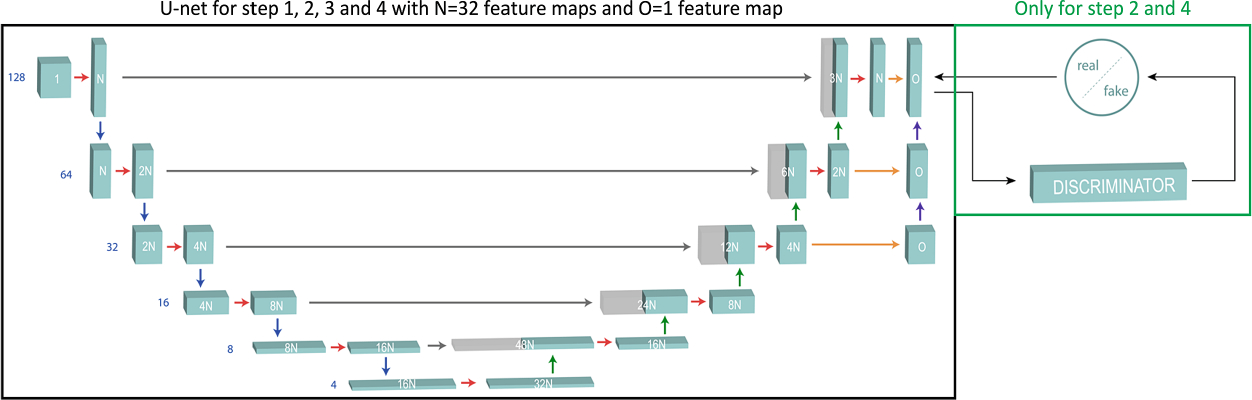
U-Net model used for each step in LASSNet architecture.
Predictions generated by the ROI NNs are activated by a sigmoid function to generate probabilities that a voxel is either ROI (voxel value 1) or background (voxel value 0). The predicted atrial (or atrial scar) ROI is then used to determine an appropriate bounding box by identifying the smallest rectangular prism containing all predicted ROI voxels. The prism is then padded with a 10-voxel buffer along all three dimensions. The original image is then cropped to the padded bounding box and converted to a volumetric array as input to the LA(S)SEG NN.
Steps 2 and 4: Left Atrium (Scar) Segmentation Neural Networks (LA(S)SEG NNs)
The LA(S)SEG NNs are conditional generative adversarial network (GAN) combinations of a generator and a discriminator, and follow the PatchGan [17] framework. The generator of the LA(S)SEG NNs follows the same variant of the 3D U-Net used for the ROI NNs and generates the predicted segmentations. The architecture is presented in Fig. 5. The step 2 NN was modified to accept a cropped image volume and a cropped single binary label GT segmentation of the LA. The step 4 NN was modified to accept both a cropped image volume and a cropped single binary label LA wall ROI image volume as well as a single binary label GT segmentation of the LA scar. The discriminator is a deep convolutional NN that performs image classification. It accepts the GT segmentations and the predicted segmentation output from the generator as inputs and predicts the likelihood of the predicted segmentation being real (GT) or fake (generated). The discriminator is penalized if it misclassifies a predicted segmentation as GT or vice versa. The discriminator is then used to train the generator. Since the generator’s output is connected directly to the discriminator’s input, through backpropagation, the discriminator provides feedback to the generator so it can generate more realistic segmentation predictions. The goal of the generator is to encourage the discriminator to misclassify the predicted segmentations as the GT [18].
2.4. Implementation
Overall, 130 image volumes (LGE-MRI with LA cavity GT segmentations) were used in the training of Stage 1 and 60 image volumes (LGE-MRI with LA cavity and LA scar GT segmentations) were used in the training of Stage 2. To avoid overfitting, early stopping was implemented. Step 1 NNs ran on average for 205 epochs (≈ 12 hours computation time), step 2 NNs for 148 epochs (≈ 12 hours computation time), step 3 NNs for 175 epochs (≈ 6 hours computation time) and step 4 NNs for 146 epochs (≈ 6 hours computation time). All networks used the Adam optimizer [19] with an adaptive learning rate (LR) starting at 10−3. The NNs were trained using Keras [20] and Tensorflow [21] on an Nvidia K80 GPU (24GB GDDR5). The loss function is based on the Sørensen-Dice Coefficient (Dice). The Dice measures the overlap between two areas (2D) or volumes (3D). Dice values range from [0, 1], with a Dice of 0 indicating that there is no overlap and a Dice of 1 indicating a perfect match.
| (1) |
where and are the 3D image volume ground truth and the predicted 3D image volume.
3. Experimental Results
The LAScarQS 2022 Challenge was divided in three different phases: training phase, validation phase and test phase. We present the results obtained from the validation phase in the following section. To evaluate the performances of the LA segmentation network (Stage 1) of LASSNet, the Sørensen-Dice Coefficient (Dice) [22], the Average Surface Distance (ASD) [23, 24] and Hausdorff distance (HD) [25] were used for each image volume as metrics. In addition to those metrics that focuses on the LA cavity; the accuracy, the specificity, the sensitivity, the Sørensen-Dice Coefficient and the Generalized Dice score (GDice) [26, 27] were used to evaluate the performances of the LA scar segmentation (Stage 2).
3.1. Segmentation Performances and Discussion
We present a two stages deep learning (DL) approach to automatically segment both LA and LA scar from LGE-MRI. We show that LASSNet provides a continuous and realistic scar pattern and promising results for scar quantification.
First, we trained LASSNet with a ROI wall thickness of 2.5 mm and analyzed the performances depending on whether or not data quality selection was applied in Table 2. The LA cavity segmentation varied in Dice from 0.8659 to 0.8892, ASD from 2.59 to 2.179 mm and HD from 30.66 to 26.27 mm when changing the training data of Stage 1. Dice, ASD and HD were all improved when increasing the dataset from 46 to 130 scans; 46 scans is not enough data for LASSNet to learn to segment the LA cavity correctly. When data quality selection was applied to the 130 scans dataset by removing the 24 worst quality scans, it further increased all Stage 1 performances, showing how important it is to have a high quality training dataset.
Table 2. LASSNet average segmentation performances depending on the data quality selection.
Left atrium cavity Dice coefficient (LAcav Dice), Left atrium cavity average surface distance (LAcav ASD), Left atrium cavity Hausdorff distance (LAcav HD), Left atrium scar accuracy (LAscar Acc), Left atrium scar specificity (LAscar Spe), Left atrium scar sensitivity (LAscar Sen), Left atrium scar Dice coefficient (LAscar Dice), Left atrium scar generalized Dice coefficient (LAscar GDice) are shown for 4 LASSNet trainings with the Dice as loss function and a ROI wall thickness of 2.5 mm and depending on whether data quality selection was applied (green if applied and red if not): LASSNet1 both stages were trained on the Task 1 selected data (46 LGE scans), LASSNet2 Stage 1 was trained on the Task 2 full data (130 LGE scans) and Stage 2 was trained on the Task 1 full data (60 LGE scans), LASSNet3 Stage 1 was trained on the Task 2 selected data (106 LGE scans) and Stage 2 was trained on the Task 1 selected data (46 LGE scans), LASSNet4 Stage 1 was trained on the Task 2 selected data (106 LGE scans) and Stage 2 was trained on the Task 1 full data (60 LGE scans). Acc, Spe and Sen are expressed in percentage terms, Dice and GDice are adimensional, and HD and ASD are in millimeters. The best performing model is written in bold.
| NNs | LAcav Dice | LAcav ASD | LAcav HD | LAscar Acc | LAscar Spe | LAscar Sen | LAscar Dice | LAscar GDice |
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| LASSNet1 | 0.8659 | 2.59 | 30.66 | 0.9999246 | 0.9999627 | 0.576 | 0.559 | 0.8953 |
| Stage 1 | ± 0.0751 | ± 1.31 | ± 8.01 | ±2.23×10−5 | ±1.05×10−5 | ± 0.137 | ± 0.160 | ± 0.0229 |
|
| ||||||||
| LASSNet2 | 0.8737 | 2.44 | 27.14 | 0.9999258 | 0,9999603 | 0.621 | 0.586 | 0.8987 |
| Stage 1 & 2 | ± 0.0648 | ± 1.33 | ± 8.77 | ±2.39×10−5 | ±1.38×10−5 | ± 0.116 | ± 0.134 | ± 0.0237 |
|
| ||||||||
| LASSNet3 | 0.8892 | 2.179 | 26.27 | 0.9999231 | 0.9999596 | 0.588 | 0.563 | 0.8942 |
| Stage 1 & 2 | ± 0.0432 | ± 0.950 | ± 11.24 | ±2.65×10−5 | ±1.32×10−5 | ± 0.136 | ± 0.151 | ± 0.0281 |
|
| ||||||||
| LASSNet4 | 0.8892 | 2.179 | 26.27 | 0.9999273 | 0.9999616 | 0.624 | 0.591 | 0.9004 |
| Stage 1 & 2 | ± 0.0432 | ± 0.950 | ± 11.24 | ±2.38×10−5 | ±1.22×10−5 | ± 0.124 | ± 0.137 | ± 0.0241 |
For Stage 2, the LA scar segmentation averaged the same accuracy and specificity (0.99993 and 0.99996) throughout the different LASSNet trainings. The sensitivity varied from 0.576 to 0.624, scar Dice from 0.559 to 0.591 and GDice from 0.8942 to 0.9004. Since fewer scans were available, using the full dataset achieved better results than when quality selection was used. Therefore, it is preferable to train the model over the 60 scans available with scar GT than over the 46 higher quality scans only.
Then we analyzed how the Stage 2 segmentation performance changed depending on the LA wall ROI thickness (2.5 mm or 5 mm). Since Stage 1 was identical for each LASSNet, all models obtained the same LA cavity segmentation performances: 0.8892 for Dice, 2.179 mm for ASD and 26.27 mm for HD. The LA scar segmentation averages the same accuracy and specificity (0.99992 and 0.99996). Sensitivity varied between 0.588 and 0.680, Dice between 0.563 and 0.588 and GDice from 0.8942 to 0.8948. We obtain very similar performance for the GDice but better results for the Dice and sensitivity when trained on a thicker LA wall ROI; LASSNet segments more scar and overlaps more with the GT. Figure 7 provides visualizations for the scar segmentation results of LASSNet trained on 5 mm ROI wall thickness and 2.5 mm ROI wall thickness compared to the ground truth. Both LASSNet predicted scar distributions agree better with typical ablation locations than the scar GT. Figure 6 shows a representative example of the two different thicknesses used for training (5 mm and 2.5 mm) and illustrates the area each LA wall ROI covers. It shows that the definition of the LA wall ROI used for this study includes too much blood pool assuming perfect LA cavity segmentation. Typical LA wall thicknesses range from 0.5 to 3.5 mm [16]. The LA wall ROI of 5 mm thickness generates better results because the majority of LA wall voxels are included in the LA wall ROI. On the other hand, for the LA wall ROI of 2.5 mm, only part of LA wall is included and some scar voxels are situated outside the LA wall ROI. To prevent this, a more realistic definition of LA wall thickness can be used to retrain the NN. The 2.5 mm LA wall ROI was created through an erosion and dilation process with the identical kernel size. To get a more realistic definition of the LA wall ROI and keep a thickness of 2.5 mm, we need to apply a smaller kernel size for the erosion and a larger kernel size for the dilation.
Fig. 7.
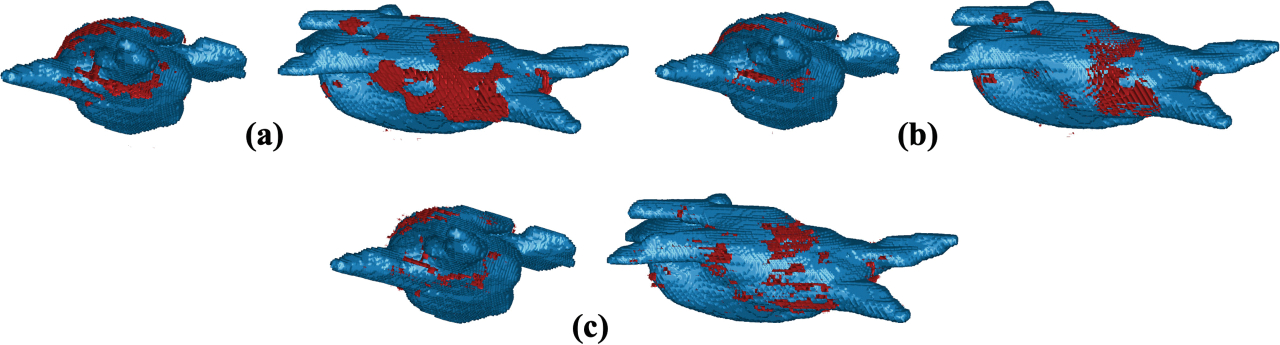
Comparison of the LA scar prediction depending on the ROI wall thickness to the ground truth on Test 3: (a) LA scar prediction of LASSNet trained on 5 mm ROI wall thickness; (b) LA scar prediction of LASSNet trained on 2.5 mm ROI wall thickness; (c) LA scar ground truth.
Fig. 6.
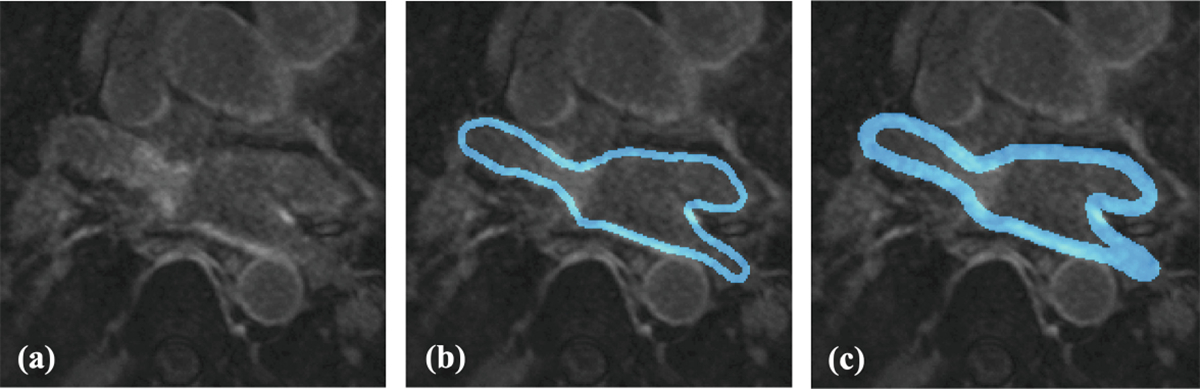
Representative example of the two different thicknesses used for training: (a) LGE-MRI; (b) 2.5 mm LA wall ROI thickness; (c) 5 mm LA wall ROI thickness.
Table 3 shows how the best GDice performing LASSNet behaves for every scan in the testing dataset. Three scans (Test 1, 5 and 8) failed to achieve a LA cavity Dice above 0.86. This is because Stage 1 creates artifacts for some scans when segmenting the LA cavity. Those artifacts are easily noticeable and quick to correct manually. When excluded, the LA cavity Dice changes to 0.9129 which correspond to the values obtained in the literature. For scar segmentation, Test 5 and 7 get very low Dice scores (under 0.5) because both have low contrast between scar (enhancement) and blood pool. GT and LASSNet predictions are non-accurate for such scans; they are shown on Fig. 8 and Fig. 9. Figure 10 illustrates LASSNet results with the best scar Dice and scar GDice scores for testing dataset (Test 9). LASSNet segments more realistic continuous fully connected scar on the posterior LA wall compared to the GT. Figure 7 also shows more continuous scar on the posterior LA wall and around the right superior pulmonary vein compare to the GT. Figure 10 demonstrates obvious inconsistencies through slice direction in scar GT. Multiple scans in the testing set have similar GT scar appearance with obvious scar discontinuities in slice direction. Such post-ablation LA scar distributions are not realistic and are caused by scar segmentation methods without scar contiguity constraint in slice direction can be easily seen in coronal or sagittal view of scar GT segmentations. Figure 10(b) shows a comparison between scar GT and LASSNet prediction in coronal view.
Table 3. Detailed segmentation performances of LASSNet 4 on the testing set.
Left atrium scar accuracy (LAscar Acc), Left atrium scar specificity (LAscar Spe), Left atrium scar sensitivity (LAscar Sen), Left atrium scar Dice coefficient (LAscar Dice), Left atrium scar generalized Dice score (LAscar GDice) are shown for each LGE scan from the testing set. Acc, Spe and Sen are expressed in percentage terms, Dice and GDice are adimensional, and HD and ASD are in millimeters. Worst scar Dice is written in red and the best is written in green.
| NNs | LAcav Dice | LAcav ASD | LAcav HD | LAscar Acc | LAscar Spe | LAscar Sen | LAscar Dice | LAscar GDice |
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| Test0 | 0.9135 | 1.871 | 28.16 | 0.9998893 | 0.9999653 | 0.573 | 0.648 | 0.8665 |
| Test1 | 0.8005 | 3.925 | 36.52 | 0.9999533 | 0.9999693 | 0.704 | 0.620 | 0.9121 |
| Test2 | 0.8931 | 1.755 | 15.33 | 0.9999294 | 0.9999622 | 0.686 | 0.671 | 0.8858 |
| Test3 | 0.9382 | 1.150 | 16.91 | 0.9998901 | 0.9999459 | 0.643 | 0.647 | 0.8968 |
| Test4 | 0.8871 | 2.697 | 29.02 | 0.9999296 | 0.9999574 | 0.681 | 0.628 | 0.8997 |
| Test5 | 0.8530 | 2.310 | 18.87 | 0.9999459 | 0.9999580 | 0.622 | 0.425 | 0.9110 |
| Test6 | 0.9176 | 1.472 | 26.12 | 0.9999270 | 0.9999529 | 0.701 | 0.625 | 0.8807 |
| Test7 | 0.9316 | 1.543 | 16.00 | 0.9999362 | 0.9999663 | 0.288 | 0.277 | 0.9353 |
| Test8 | 0.8487 | 3.608 | 51.75 | 0.9999137 | 0.9999494 | 0.665 | 0.621 | 0.8776 |
| Test9 | 0.9089 | 1.460 | 24.04 | 0.9999587 | 0.9999889 | 0.673 | 0.751 | 0.9388 |
Fig. 8.
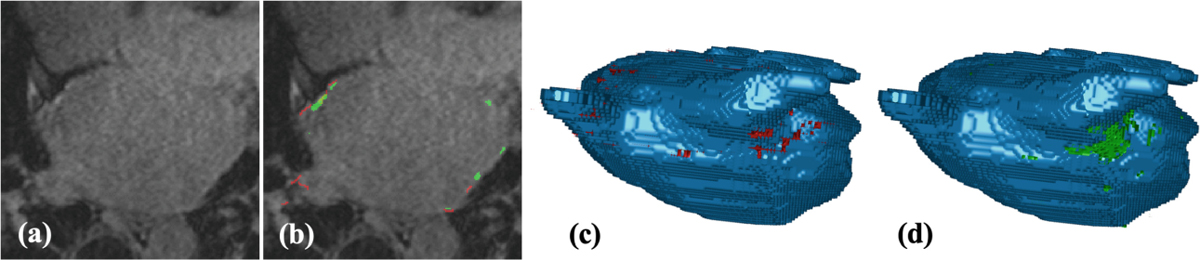
LASSNet result with the worst Dice score and scar detection sensitivity for testing dataset (Test 7 - scan with low contrast between scar and blood pool): (a) LGE-MRI; (b) Scar GT (red) and scar prediction (green); (c) 3D view of the LA cavity with scar GT (red); (d) 3D view of the LA cavity with scar prediction (green).
Fig. 9.
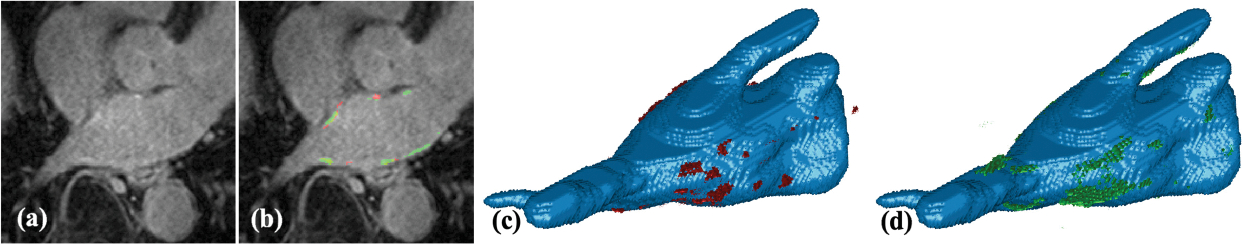
LASSNet result for LGE scan with low contrast between scar (enhancement) and blood pool (Test 5): (a) LGE-MRI, (b) Scar GT (red) and scar prediction (green), (c) 3D view of the LA cavity with corresponding scar GT (red) and (d) 3D view of the LA cavity with corresponding scar prediction (green).
Fig. 10.

LASSNet result with the best Dice and GDice scores for Testing dataset (Test 9): (a) LGE-MRI with scar GT (red) and scar prediction (green) in the axial view; (b) LGE-MRI with scar GT (red) and scar prediction (green) in the coronal view; (c) 3D view of the LA cavity with scar GT (red); (d) 3D view of the LA cavity with scar prediction (green).
Table 4 further demonstrates the performances of LASSNet by comparing it with the previously published methods. For the segmentations of LA and LA scars, we compared the segmentation performances of LASSNet to the JAS-GAN model of Jun Chen et al. [8], AtrialJSQnet NN of Lei Li et al. [9], and a multiview two-task (MVTT) method proposed by Guang Yang et al. [10]. Each method used different datasets or additional cardiac magnetic resonance (CMR) scans. Jun Chen et al. and Lei Li et al. results come from the MICCAI 2018 Atrial Segmentation Challenge dataset which provided 100 scans with labels of LA wall and LA endocardium. Guang Yang et al. used their own dataset which consisted of 190 scans. Table 4 shows that LASSNet achieves the highest GDice among those LA scar segmentation methods. LASSNet also achieves the best LA scar Dice when Test 5 and Test 7 (scans with poor contrast between blood pool and scar) are excluded. The LA cavity Dice score is slightly lower, compare to the other methods’ performances and could be further improve.
Table 4. Comparison of combined LA and LA scar average segmentation NNs performances.
Left atrium cavity Dice coefficient (LAcav Dice), Left atrium scar sensitivity (LAscar Sen), Left atrium scar Dice coefficient (LAscar Dice), Left atrium scar generalized Dice score (LAscar GDice) are shown for 5 NNs: LASSNet4, LASSNet4BIS without Test 5&7, Jun Chen et al. [8] Lei Li et al. [9] and Guang Yang et al. [10]. Sen is expressed in percentage terms, Dice and GDice are adimensional. The best scores are written in bold. Note: As the cited sources use different data sets or additional cardiac magnetic resonance scans, an exact comparison with our results cannot be made. The benchmarks are thus provided solely as information.
| Method | LAcav Dice | LAScar Sen | LAscar Dice | LAscar GDice |
|---|---|---|---|---|
| LASSNet4 | 0.8892 ±0.0432 | 0.624±0.124 | 0.591±0.137 | 0.9004 ±0.0241 |
| LASSNet4BIS | 0.8885 ±0.0443 | 0.6647 ±0.0422 | 0.6515 ±0.0438 | 0.8947 ±0.0241 |
| Jun Chen [8] | 0.913 ±0.027 | - | 0.621 ±0.0110 | - |
| Lei Li [9] | 0.913 ±0.032 | - | 0.543±0.097 | 0.872 ±0.024 |
| Guang Yang [10] | 0.931 ±0.018 | 0.8677 ±0.0464 | - | 0.8659 ±0.0560 |
Accurate and reproducible segmentation of the atrial anatomy and quantification of the fibrotic substrate is clinically important for guiding catheter ablation in AF patients. However, this clinically essential information is only available to clinicians in a few research centers because manual segmentation of LA and LA scar is time-consuming (30–60 mins), challenging and requires very specific expertise. We have developed deep neural network LASSNet to drastically speedup and simplify segmentation of LA anatomy and LA scar from atrial LGE-MRI. Once the model is trained, it takes only a couple of minutes to predict the segmentations. The network was validated on LGE scans acquired at 4 clinical centers using 1.5 and 3 T MRI scanners. LASSNet demonstrated excellent performance in LA and LA scar segmentation making it a viable tool for use in wide clinical practice.
4. Conclusion
In this work, we proposed a deep learning approach to automatically segment both the LA cavity and LA scar which proved to be robust to atrial LGE-MRI collected by different clinical centers. Although, the LA cavity segmentation could be further improved, LASSNet achieves superior scar segmentation performances over previously published methods and shows promising results for scar segmentation with realistic scar pattern in agreement with typical ablation locations. Limitations to this work are the limited number of adequate quality LGE-MRI scans in training and validation datasets, inconsistencies in LA cavity GT and scar GT segmentations, and LGE-MRI scans from only 4 clinical centers. The LASSNet framework could easily be applied to other segmentation tasks that demand a two stages refined segmentation task.
References
- 1.Stewart S, et al. : Cost of an emerging epidemic: an economic analysis of atrial fibrillation in the UK. Heart 90(3), 286–292 (2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burstein B, Nattel S: Atrial fibrosis: mechanisms and clinical relevance in atrial fibrillation. J. Am. College Cardiol. 51(8), 802–809 (2008) [DOI] [PubMed] [Google Scholar]
- 3.Marrouche NF, et al. : Association of atrial tissue fibrosis identified by delayed enhancement MRI and atrial fibrillation catheter ablation: the DECAAF study. In: Jama 311(5), 498–506 (2014) [DOI] [PubMed] [Google Scholar]
- 4.Karim R, Mohiaddin R, Rueckert D: Left atrium segmentation for atrial fibrillation ablation. In: Medical Imaging 2008: Visualization, Image-Guided Procedures, and Modeling, vol. 6918. SPIE, pp. 941–948 (2008) [Google Scholar]
- 5.Ho SY, McCarthy KP, Faletra FF: Anatomy of the left atrium for interventional echocardiography. Eur. J. Echocardiography 12(10), i11–i15 (2011) [DOI] [PubMed] [Google Scholar]
- 6.Mohrs OK, et al. : Thrombus detection in the left atrial appendage using contrast-enhanced MRI: a pilot study. Am. J. Roentgenol. 186(1), 198–205 (2006) [DOI] [PubMed] [Google Scholar]
- 7.Xiong Z, et al. : A global benchmark of algorithms for segmenting the left atrium from late gadolinium-enhanced cardiac magnetic resonance imaging. Med. Image Anal. 67, 101832 (2021) [DOI] [PubMed] [Google Scholar]
- 8.Chen J, et al. : JAS-GAN: generative adversarial network based joint atrium and scar segmentations on unbalanced atrial targets. IEEE J. Biomed. Health Inf. 26(1), 103–114 (2022) [DOI] [PubMed] [Google Scholar]
- 9.Li L, et al. : AtrialJSQnet: a New framework for joint segmentation and quantification of left atrium and scars incorporating spatial and shape information. Med. Image Analys. 76, 102303 (2022). issn: 1361–8415 [DOI] [PubMed] [Google Scholar]
- 10.Yang G, et al. : Simultaneous left atrium anatomy and scar segmentations via deep learning in multiview information with attention. Futur. Gener. Comput. Syst. 107, 215–228 (2020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li L, et al. : Medical image analysis on left atrial LGE-MRI for atrial fibrillation studies: a review. Med. Image Anal. 77, 102360 (2022). issn: 1361–8415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li L, Zimmer VA, Schnabel JA, Zhuang X: AtrialGeneral: domain generalization for left atrial segmentation of multi-center LGE MRIs. In: de Bruijne M, Cattin PC, Cotin S, Padoy N, Speidel S, Zheng Y, Essert C (eds.) MICCAI 2021. LNCS, vol. 12906, pp. 557–566. Springer, Cham: (2021). 10.1007/978-3-030-87231-1_54 [DOI] [Google Scholar]
- 13.Li L, Zimmer VA, Schnabel JA, Zhuang X: AtrialGeneral: domain generalization for left atrial segmentation of multi-center LGE MRIs. In: de Bruijne M, Cattin PC, Cotin S, Padoy N, Speidel S, Zheng Y, Essert C (eds.) MICCAI 2021. LNCS, vol. 12906, pp. 557–566. Springer, Cham: (2021). 10.1007/978-3-030-87231-1_54 [DOI] [Google Scholar]
- 14.Ronneberger O, Fischer P, Brox T: U-Net: convolutional networks for biomedical image segmentation. In: Navab N, Hornegger J, Wells WM, Frangi AF (eds.) MICCAI 2015. LNCS, vol. 9351, pp. 234–241. Springer, Cham: (2015). 10.1007/978-3-319-24574-4_28 [DOI] [Google Scholar]
- 15.Karim R, et al. : Evaluation of current algorithms for segmentation of scar tissue from late gadolinium enhancement cardiovascular magnetic resonance of the left atrium: an open-access grand challenge. J. Cardiovascular Magnetic Resonance 15(1), 1–17 (2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roy B, et al. : Left atrial wall thickness variability measured by CT scans in patients undergoing pulmonary vein isolation. J. Cardiovascular Electrophysiology 22(11), 1232–1236 (2011) [DOI] [PubMed] [Google Scholar]
- 17.Chen C, et al. : Deep learning for cardiac image segmentation: a review. Front. Cardiovascular Med. 7, 25 (2020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Isola P, et al. : Image-to-image translation with conditional adversarial networks. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 1125–1134 (2017) [Google Scholar]
- 19.Kingma DP, Ba J: Adam: a method for stochastic optimization. In: arXiv preprint arXiv:1412.6980 (2014) [Google Scholar]
- 20.Chollet F, et al. : Keras (2015). https://keras.io
- 21.Abadi M, et al. : TensorFlow: Large-Scale Machine Learning on Heterogeneous Systems. Software available from tensorflow.org (2015). https://www.tensorflow.org/
- 22.Dice LR: Measures of the amount of ecologic association between species. Ecology 26(3), 297–302 (1945) [Google Scholar]
- 23.Teguh DN, et al. : Clinical validation of atlas-based auto-segmentation of multiple target volumes and normal tissue (swallowing/mastication) structures in the head and neck. Int. J. Radiation Oncology* Biology* Phys 81(4), 950–957 (2011) [DOI] [PubMed] [Google Scholar]
- 24.Kiser KJ, et al. : Novel autosegmentation spatial similarity metrics capture the time required to correct segmentations better than traditional metrics in a thoracic cavity segmentation workflow. J. Digital Imaging 34(3), 541–553 (2021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Birsan T, Tiba D: One hundred years since the introduction of the set distance by Dimitrie Pompeiu. In: IFIP Conference on System Modeling and Optimization, pp. 35–39. Springer (2005) [Google Scholar]
- 26.Sudre CH, Li W, Vercauteren T, Ourselin S, Jorge Cardoso M: Generalised dice overlap as a deep learning loss function for highly unbalanced segmentations. In: Cardoso MJ, Arbel T, Carneiro G, Syeda-Mahmood T, Tavares JMRS, Moradi M, Bradley A, Greenspan H, Papa JP, Madabhushi A, Nascimento JC, Cardoso JS, Belagiannis V, Lu Z (eds.) DLMIA/ML-CDS −2017. LNCS, vol. 10553, pp. 240–248. Springer, Cham: (2017). 10.1007/978-3-319-67558-9_28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Crum WR, Camara O, Hill DLG: Generalized overlap measures for evaluation and validation in medical image analysis. IEEE Trans. Med. Imaging 25(11, 1451–1461 (2006) [DOI] [PubMed] [Google Scholar]
- 28.Badger TJ, et al. : Evaluation of left atrial lesions after initial and repeat atrial fibrillation ablation: lessons learned from delayed-enhancement MRI in repeat ablation procedures. Circulation: Arrhythmia Electrophysiology 3(3), 249–259 (2010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McGann CJ, et al. : New magnetic resonance imaging-based method for defining the extent of left atrial wall injury after the ablation of atrial fibrillation. J. Am. Coll. Cardiol. 52(15), 1263–1271 (2008) [DOI] [PubMed] [Google Scholar]
- 30.Taghanaki Asgari, Saeid, et al. : Deep semantic segmentation of natural and medical images: a review. Artif. Intell. Rev. 54(1), 137–178 (2021) [Google Scholar]
- 31.Ulyanov D, Vedaldi A, Lempitsky V: Instance normalization: the missing ingredient for fast stylization. arXiv preprint arXiv:1607.08022 (2016) [Google Scholar]


