Abstract
Analysis of retinal ganglion cells (RGCs) by scRNA-seq is emerging as a state-of-the-art method for studying RGC biology and subtypes, as well as for studying the mechanisms of neuroprotection and axon regeneration in the central nervous system (CNS). Rbpms has been established as a pan-RGC marker, and Spp1 has been established as an αRGC type and macrophage marker. Here, we analyzed by scRNA-seq retinal microglia and macrophages, and found Rbpms+ subpopulations of retinal microglia/macrophages, which pose a potential pitfall in scRNA-seq studies involving RGCs. We performed comparative analysis of cellular identity of the presumed RGC cells isolated in recent scRNA-seq studies, and found that Rbpms+ microglia/macrophages confounded identification of RGCs. We also showed using immunohistological analysis that, Rbpms protein localizes to stress granules in a subpopulation of retinal microglia after optic nerve injury, which was further supported by bioinformatics analysis identifying stress granule-associated genes enriched in the Rbpms+ microglia/macrophages. Our findings suggest that the identification of Rbpms+ RGCs by immunostaining after optic nerve injury should exclude cells in which Rbpms signal is restricted to a subcellular granule, and include only those cells in which the Rbpms signal is labeling cell soma diffusely. Finally, we provide solutions for circumventing this potential pitfall of Rbpm-expressing microglia/macrophages in scRNA-seq studies, by including in RGC and αRGC selection criteria other pan-RGC and αRGC markers.
Graphical Abstract
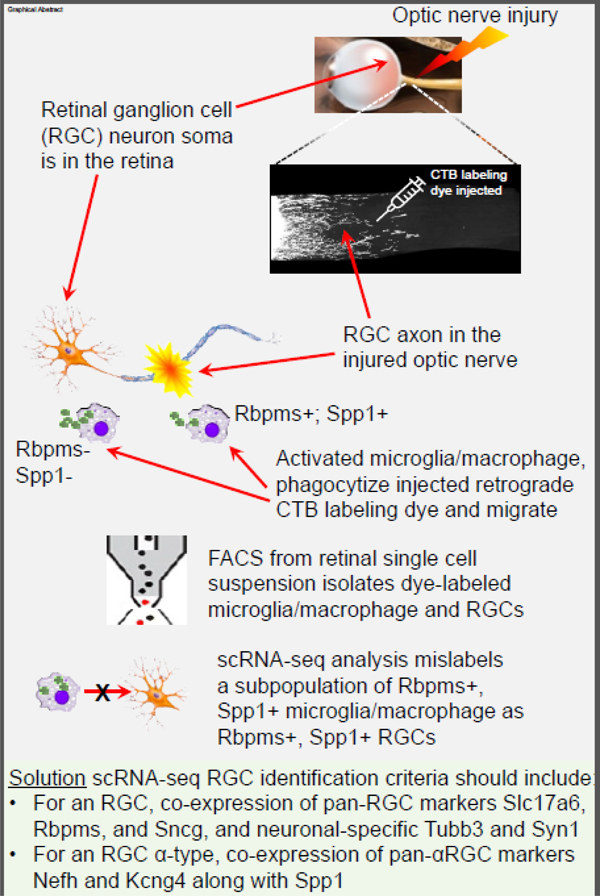
INTRODUCTION
The Pten tumor suppressor gene is one of the most potent gene-regulators of CNS axon regeneration. Pten suppresses regeneration through inhibition of the mTOR pathway, and Pten knockout (KO) and knockdown (KD) have been shown to promote various extents of axon regeneration, after optic nerve crush (ONC) injury in mice, in the α and intrinsically photosensitive (ip) subsets of retinal ganglion cells (RGCs)1–4. In order to understand the effect of Pten inhibition on the transcriptomes of RGCs that respond by regenerating axons of any length, recent studies by Li et al.5 and by Jacobi et al.6 isolated the RGCs with Pten KO that regenerated axons at least short-distance (1.5 mm) and profiled them by scRNA-seq. In contrast, we developed a new method for isolating specifically the rare subset of RGCs which respond to Pten inhibition by regenerating axons long-distance (~3 mm past the injury site or beyond) and also profiled them by scRNA-seq7. Here, we performed comparative analysis of cellular identity of the presumed RGC cells isolated in these studies, along with scRNA-seq profiled retinal and optic nerve microglia/macrophages, and reveal the existence of an Rbpms+ subpopulation of microglia/macrophages, which could confound identification of RGCs in scRNA-seq studies.
RESULTS
We report that, the pan-RGC marker Rbpms8,9 is also expressed in a subset of microglia and macrophages (which infiltrate the CNS after injury). Therefore, Rbpms alone is insufficient for identifying RGCs in scRNA-seq analysis. For example, in the study by Li et al.5, Pten KO-treated RGCs were selected from within (fluorescently pre-labeled and FACS-sorted) scRNA-seq-profiled retinal cells based on the expression of at least one RGC marker (either Rbpms, Thy1, Slc17a6, or Pou4f1–3), rather than co-expression of multiple RGC markers. As injury-activated resident microglia and peripheral macrophages phagocytize fluorescent dye and migrate (including into the retina), the reliance on single RGC marker expression rather than co-expression of multiple markers resulted in ~94% of the cells selected as presumed RGCs to be identified as microglia/macrophages by retrospect analyses (see below), which is consistent with top enriched genes reported in that study5 being microglia/macrophage markers and the top enriched gene-ontology terms being microglia/macrophage-related (e.g., leukocyte migration, lymphocyte mediated immunity). Thus, in order to determine RGC cell type identity by scRNA-seq, several of the well-established pan-RGC markers (e.g., Slc17a6 and Sncg) and neuronal markers (e.g., Tubb3 and Syn1) need to be co-expressed in a cell.
To demonstrate this potential pitfall of relying only on expression of a single RGC marker rather than co-expression of several markers in scRNA-seq studies, we performed comparative analysis of the cells with Pten KO/KD labeled as RGCs in the studies by Li et al.5, Jacobi et al.6, and ourselves7. We also included in the analysis scRNA-seq-profiled microglia/macrophages, which we isolated after ONC from retinas and optic nerves, and separated based on the expression of Rbpms (Fig. 1A). In total, 6.6% of microglia/macrophage cells were Rbpms+. Rbpms detection was not a consequence of a piece of broken RGC sticking to microglia/macrophage as we applied doublet filters and did not find other RGC/neuronal markers expressed in either subpopulation of microglia/macrophages (Fig. 1A-H). The range of Rbpms expression in the Rbpms+ microglia/macrophages was equivalent to levels observed in uninjured and injured RGCs (Fig. 1A). Rbpms was expressed in the RGC-labeled cells from studies by Li et al.5, Jacobi et al.6, and ourselves7 (Fig. 1A). However, the other RGC-specific markers, Slc17a6, Sncg, and Brn3a, and neuronal-specific marker Syn1, were not enriched in the RGC-labeled cells in Li et al.5, but were enriched in cells from studies by Jacobi et al.6 and ourselves7 (Fig. 1A-H). We also examined expression of established ipRGC type markers, melanopsin (Opn4) and Tbr2 (Eomes)9–13. Both Li et al.5 and Jacobi et al.6 cells were negative for these markers, whereas Opn4 and to a lesser extent Eomes were enriched in non-αRGC cells we isolated ourselves (which regenerated axons long-distance)7 (Fig. 1E-F; see in more detail below). Neuronal-specific marker Tubb3 (which is also RGC-specific within the retina9) was expressed in cells from our study7 and in uninjured and injured (untreated) RGCs, but not enriched in cells from the other studies5,6 (Fig. 1G). Only 17 cells, which represent 5.7% of all RGC-labeled cells that regenerated axons in the study by Li et al.5, expressed the appropriate RGC markers.
Figure 1. Analysis of pan-RGC, pan-neuronal, ipRGC, and microglia/macrophage gene markers in scRNA-seq datasets from different conditions and cell types.
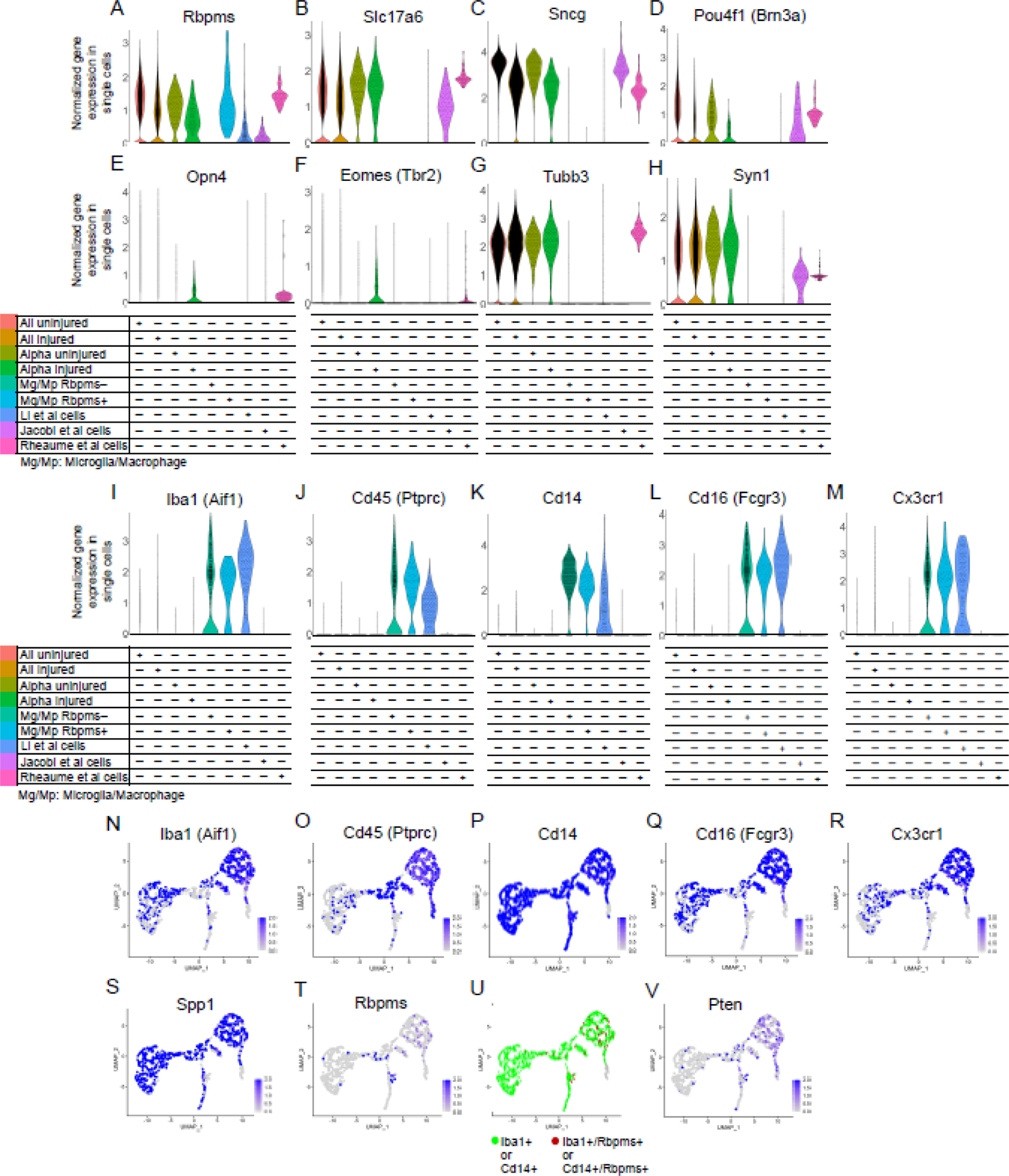
(A-H) Pan-RGC gene markers (A-D) are enriched in Tran et al. (2019) uninjured and injured RGCs, including in the αRGC subset, but with an exception for Rbpms (A) are not expressed in the microglia/macrophages (Mg/Mp). IpRGC markers (E-F) are not enriched in Li et al. (2022) or Jacobi et al. (2022) cells, but are expressed in non-αRGCs isolated by ourselves (Rheaume et al., 2023). Neuronal-specific (G-H) gene markers are also enriched in axon-regenerating RGCs isolated by Jacobi et al. (2022) and ourselves (Rheaume et al., 2023) but are absent from the majority of presumed RGC cells isolated by Li et al. (2022), with an exception for Rbpms (A) which is also enriched in the Rbpms+ subpopulation of microglia/macrophages. Tubb3 (G) is enriched in uninjured and injured untreated RGCs (Tran et al., 2019), and in axon-regenerating RGCs isolated by ourselves (Rheaume et al., 2023).
(I-M) Established microglia/macrophage markers, Iba1 (I), Cd45 (J), Cd14 (K), Cd16 (L), and Cx3cr1 (M), are enriched in all microglia/macrophages (regardless of Rbpms expression) and in the presumed RGC cells isolated by Li et al. (2022), but not enriched in isolated RGCs from any other condition/group.
(N-S) UMAPs of scRNA-seq-profiled retinal microglia/macrophages after optic nerve injury show that, established microglia/macrophage markers, Iba1 (N), Cd45 (O), Cd14 (P), Cd16 (Q), Cx3cr1 (R), and Spp1 (S), between them are expressed in all (and co-expressed in most of) the microglia/macrophages we isolated.
(T-V) UMAPs of scRNA-seq-profiled retinal microglia/macrophages after optic nerve injury show that, Rbpms is expressed in a subpopulation of these cells (T), which co-express Iba1 and/or Cd14 microglia/macrophage markers (U), and nearly half of them do not express Pten (V).
We then analyzed the expression of established microglia/macrophage markers, Iba1, Cd45, Cd14, Cd16, and Cx3cr1. We found that these markers were enriched in microglia/macrophages (regardless of whether they expressed Rbpms) and in the presumed RGC cells from the study by Li et al.5, but not in any of the RGC conditions, even after Pten KO/KD in studies by Jacobi et al.6 and ourselves7 (Fig. 1I-M). For reference, UMAPs show that between them, these microglia/macrophage markers were expressed in all (and co-expressed in most of) the microglia/macrophages we isolated (Fig. 1N-R), along with Spp1 marker of macrophages14–16 (Fig. 1S). After ONC, injury-activated microglia and infiltrating peripheral macrophages migrate within the optic nerve and into the retina. They can phagocytize the fluorescent tracer dye injected into the injured optic nerve, and some of them migrate into the retina within a day (thus, they are FACS-sorted as positive for the labeling dye). We also show that, all Rbpms+ microglia/macrophages we identified are positive for established microglia/macrophages markers Iba1 and/or Cd14 (Fig. 1T-U) and nearly half of them do not express Pten (Fig. 1V). Taken together, these data demonstrate that, it is insufficient to rely on expression of only one pan-RGC marker to identify RGCs within a mixed population of scRNA-seq-profiled retinal cell types, as this can lead to inclusion of microglia/macrophages. Co-expression of pan-RGC markers (including Slc17a6 and Sncg) is necessary, and the inclusion of neuronal-specific markers (e.g., Tubb3 and Syn1) is preferred, in order to identify RGCs by scRNA-seq.
Because Li et al.5 found expression of certain αRGC markers in their cells, we analyzed whether they are expressed in microglia/macrophages as well. The αRGC marker genes17, Nefh (which encodes Neurofilament-H containing a non-phosphorylated epitope recognized by an SMI-32 antibody)2,10, Kcng42,18, and Etv110,19, were not enriched in cells from the study by Li et al.5, with only a handful of cells expressing it within the 5.7% of cells that were indeed RGCs (Fig. 2A-C). However, Nefh and Kcng4 were enriched in the Jacobi et al.6 RGCs. Furthermore, although Kcng4 and Etv1 were downregulated in αRGCs after ONC, Kcng4 downregulation was prevented by Pten KO pre-treatment in Jacobi et al.6 cells (Fig. 2B-C). Other αRGC markers, Spp12,10,11 and subtype C42-specific Fes11, however, were enriched in the Li et al.5 cells, but they were also enriched in the macrophages/microglia (Fig. 2D-E). Spp1 was also enriched in Jacobi et al.6 RGCs. As cells we isolated ourselves7 were non-αRGC subtypes (which regenerated axons long-distance in response to Pten KD), they did not express these αRGC markers except for low levels of Spp1, consistent with prior reports that Pten inhibition leads to Spp1 upregulation in some cell types20–22. Spp1 expression was also substantially higher in the Li et al.5 cells compared to Jacobi et al. and our cells (Fig. 2D), consistent with that Spp1 is an established marker of macrophages14–16 (Fig. 1S). Another αRGC subtype-specific marker, Kit (for C41)11, was not expressed except for in a few cells isolated by Li et al.5, and the remaining αRGC subtype-specific markers, Il1rapl2 (for C43) and Tpbg (for C45)11, were not enriched in the Li et al.5 cells. Thus, when using these markers for αRGCs in a mixed population of scRNA-seq-profiled cells, it is important to first evaluate for co-expression of pan-RGC markers.
Figure 2. Analysis of αRGC gene markers and neuroprotective genes in, and global transcriptome correlation analysis of, scRNA-seq datasets from different conditions and cell types.
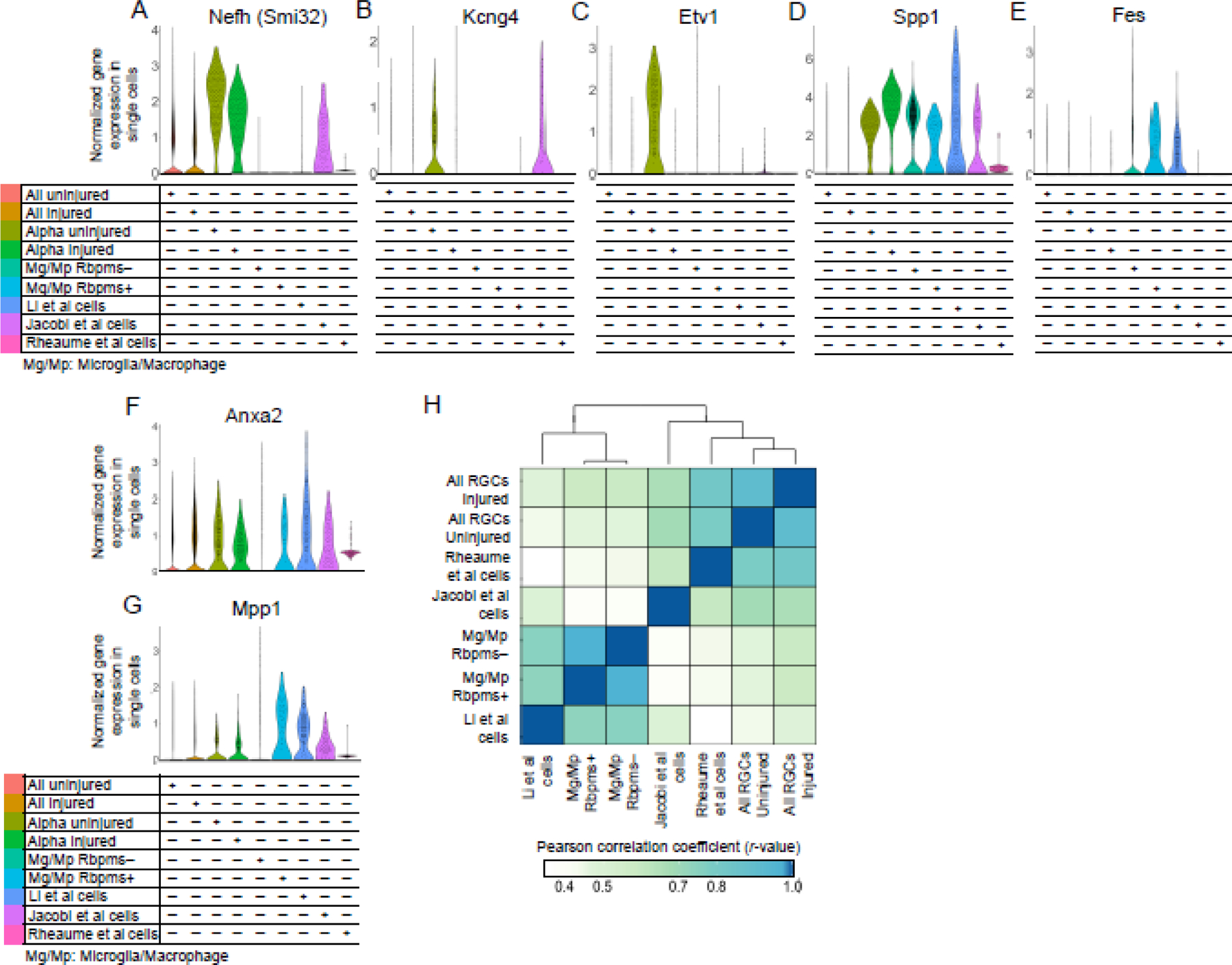
(A-E) Pan-αRGC markers Nefh (A), Kcng4 (B), and Spp1 (D), are enriched in Tran et al. (2019) αRGC subset and in axon-regenerating RGCs isolated by Jacobi et al. (2022). Spp1 (D) is enriched in the presumed RGC cells isolated by Li et al. (2022) more than in any other condition, but is also substantially enriched in the microglia/macrophages, regardless of Rbpms expression. Kcng4 (B) and Etv1 (C) αRGC markers are downregulated after ONC injury in Tran et al. (2019) αRGC subsets, but Kcng4 downregulation is prevented by Pten KO in axon-regenerating RGCs isolated by Jacobi et al. (2022). Fes (E) was enriched only in the presumed RGC cells isolated by Li et al. (2022) and in the microglia/macrophages. No αRGC markers were meaningfully enriched in long-distance axon-regenerating RGCs isolated by ourselves (Rheaume et al., 2023), except for low levels of Spp1 (which is consistent with reports that Pten inhibition leads to Spp1 upregulation).
(F-G) Neuroprotective genes found in the screen by Li et al. (2022), Anxa2 (F) and Mpp1 (G), are robustly expressed in the Rbpms+ microglia/macrophages, as well as in RGCs from every other condition/group.
(H) Analysis of the global transcriptomes clustered and correlated presumed RGC cells from the study by Li et al. (2022) more strongly with the microglia/macrophage datasets than with other RGC datasets, whereas Jacobi et al. (2022) cells and Rheaume et al. (2023) cells clustered and correlated more strongly with RGC datasets than with microglia/macrophage datasets.
We also found that the neuroprotective genes, Anxa2 and Mpp1, found by Li et al.5, are robustly expressed in the Rbpms+ microglia/macrophages (Fig. 2F-G), and along with Rbpms are also present in macroglia previously analyzed by bulkRNA-seq (see mouse microglia dataset at https://www.brainrnaseq.org)23. We further confirmed that the presumed RGC cells from the study by Li et al.5 are microglia/macrophages, by analyzing global transcriptomes (and not only the canonical cell type markers) and finding that they correlated and clustered more strongly with our microglia/macrophage dataset than with any RGC dataset (Fig. 2H). Analysis of global transcriptomes also confirmed that Rbpms+ microglia/macrophages are near identical (by correlation and associated clustering) to the Rbpms− microglia/macrophages (Fig. 2H). Thus, inadvertently, Li et al.5 screened microglia/macrophage genes (instead of the genes enriched in the RGCs that regenerated axons after Pten KO), and found several neuroprotective hits, such as Anxa2 and Mpp1, which are expressed in both Rbpms+ microglia/macrophage and RGCs (Fig. 2F-G).
To test whether Rbpms is translated in microglia/macrophages which express Rbpms mRNA, we co-immunostained the retinas after ONC for Rbpms, Iba1, and Tubb3 (anti-Tuj1 antibody). All Iba1+ cells were Tuj1-, whereas Rbpms+/Tuj1+ RGCs were present in the ganglion cell layer (GCL), as expected. Amongst the Iba1+ cells, we found a subpopulation of cells containing subcellular Rbpms+ structures which morphologically resembled stress granules, in contrast to the diffusely distributed Rbpms signal in Tuj1+ RGCs in the same retinas (Fig. 3A-I). Thus, subcellular distribution/localization of Rbpms differs between the injured retinal microglia and RGCs, consistent with that Rbpms localizes to cytoplasmic stress granules in other cell types24,25 and stress granules form in microglia activated in neurodegenerative diseases26–28. Furthermore, genes ontology (GO) analysis of genes that are significantly enriched in Rbpms+ relative to Rbpms− microglia/macrophages identified “response to stress” amongst the topmost associated biological processes (Fig. 3J). Moreover, the genes enriched in the Rbpms+ microglia/macrophages included stress granule-associated genes (Hspa1b, Hspa1a, Hspa5, Hsph1, Dnajb1, Egr1, Fos, Dusp1, Ppp1r15a29) along with G3bp1, which has been previously established to colocalize with Rbpms in stress granules24 (Fig. 3K). Taken together, immunohistological and bioinformatics analyses suggests that, Rbpms protein localizes to stress granules in a subpopulation of retinal microglia/macrophages after optic nerve injury. Therefore, histological quantification of Rbpms+ RGCs by immunostaining after optic nerve injury should exclude cells in which Rbpms signal is restricted to subcellular granules and include only those cells in which the Rbpms signal is labeling cell soma diffusely.
Figure 3. Rbpms protein localizes to stress granules in a subpopulation of retinal microglia/macrophages which are enriched for stress response genes after optic nerve injury.
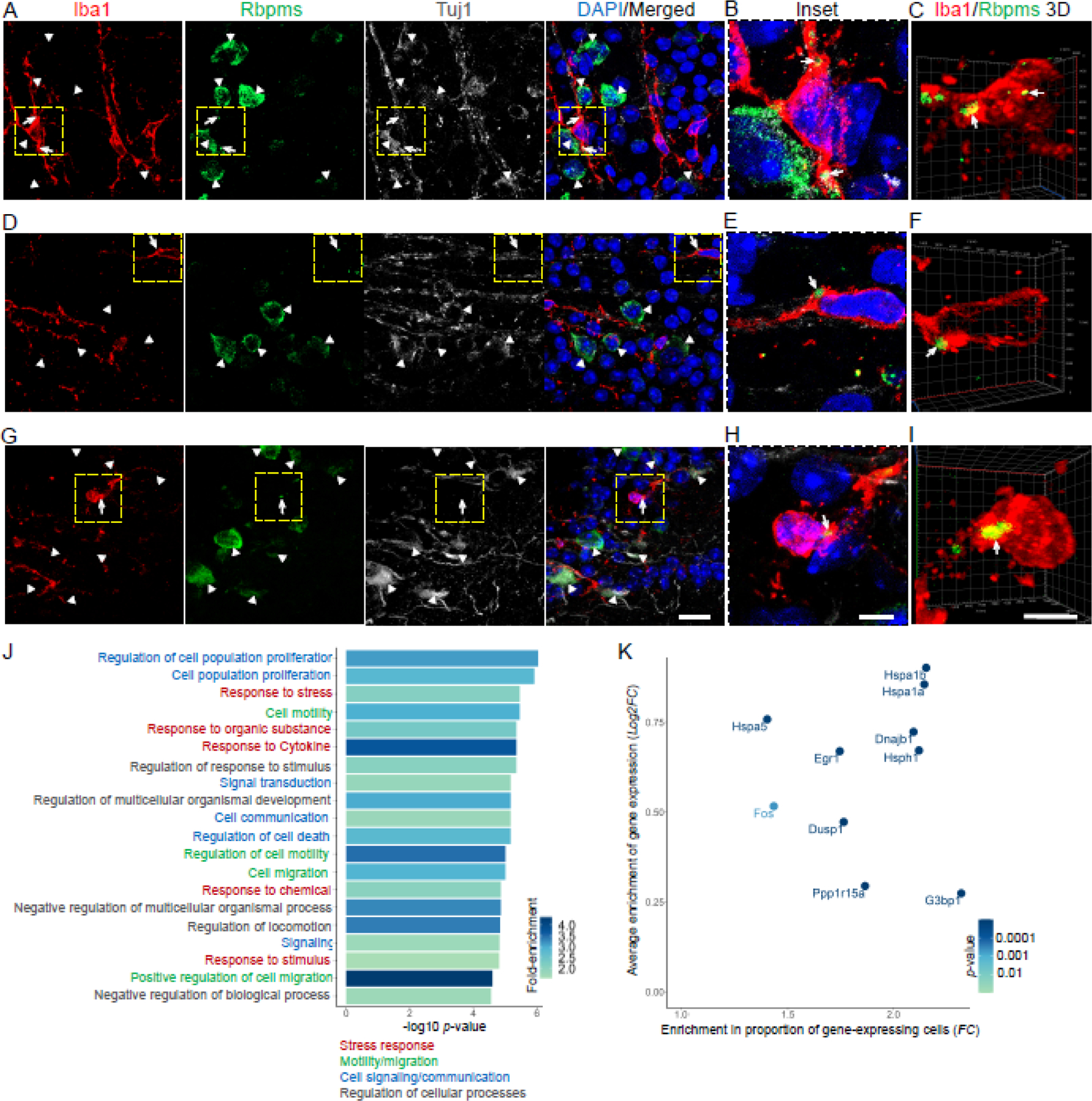
(A-I) Confocal images of the retinal flatmounts (at 2 weeks after ONC) co-immunostained for Rbpms, Iba1, Tuj1, and counterstained for DAPI (to label nuclei), show examples of Rbpms signal localization to granules in Iba1-labled microglia/macrophages, as compared to diffuse cytoplasmic Rbpms signal in Tuj1-labled RGCs, in the injured retinas’ GCL. White arrowheads indicate Rbpms+/Tuj1+ RGCs. White arrows indicate Rbpms+ granules within the Iba1+ microglia/macrophages. Dashed yellow-line outlined regions in the main panels (A,D,G) are shown in insets (B,E,H) and in 3D renderings (C,F,I) at higher magnification for better visualization of the Rbpms+ granules’ subcellular localization within the Iba1+ microglia/macrophages. Main panels scale bar: 20 µm; insets and 3D panels scale bars: 5 µm.
(J) Gene ontology (GO) analysis of genes enriched in Rbpms+ relative to Rbpms− microglia/macrophages identified GO biological processes (GO:BPs; 20 topmost significant by adjusted p-value are shown) that included multiple cellular stress response-related terms. Further functional annotation and semantic similarity clustering of GO:Terms (using the EnrichPlot R package) identified 4 overarching enriched pathways, which included “stress response”. GO:BPs are color-coded according to overarching enriched pathways to which they belong, as marked. Color scale bar indicates GO:BP terms’ fold-change (FC) enrichment (see Methods).
(K) Enrichment of stress granule-associated genes in the Rbpms+ relative to Rbpms− microglia/macrophages is demonstrated by higher average expression of these genes (y-axis; log2 FC) and by higher proportion of Rbpms+ cells as compared to Rbpms− cells expressing these genes (x-axis; FC). Color scale bar indicates significance of average expression enrichment (adjusted p-value by Wilcoxon rank-sum test; see Methods).
DISCUSSION
We revealed herein the existence of an Rbpms+ subpopulation of retinal microglia/macrophages, some of which also migrate into the retina from the optic nerve after activation by ONC injury. Rbpms, Anxa2, and Mpp1 are also expressed in microglia from other CNS regions, which were previously analyzed by bulkRNA-seq (see mouse microglia dataset at https://www.brainrnaseq.org)23. Rbpms is an established pan-RGC marker8,9, and Spp1 is an established αRGC type and macrophage marker2,10,11,14–16. Therefore, Rbpms+ and Spp1+ microglia/macrophages within retinal suspension could confound the identification of RGCs by scRNA-seq analysis, as occurred in a study by Li et al.5. However, serendipitous discovery of the neuroprotective potential of Anxa2 and Mpp1 genes by Li et al.5 study has significant implications for developing neuroprotective therapies, regardless of that microglia/macrophages analysis assisted in their identification.
We demonstrate that in order to circumvent the potential pitfall of mistaking Rbpms+ microglia/macrophages for RGCs in scRNA-seq studies, detection of co-expression of pan-RGC markers (including Rbpms, Slc17a6, and Sncg) is necessary, the inclusion of neuronal-specific markers (e.g., Tubb3 and Syn1) is preferred (Tubb3 is also RGC-specific within the retina), and the inclusion of Brn3A-C, which between them cover all RGCs9,30,31, should be considered. Similarly, in order to circumvent the potential pitfall of mistaking Spp1+ microglia/macrophages for αRGCs in scRNA-seq studies, it is important to first evaluate for co-expression of pan-RGC markers other than just Rbpms, and the inclusion of other pan-αRGC markers (e.g., Nefh and Kcng4) is preferred, with the caveat that Kcng4 is downregulated after ONC injury unless pre-treated (with Pten KO in this case). Similar considerations need to be made when relying on αRGC subtype C42 marker Fes, as it is also expressed in a subpopulation of Rbpms+ microglia/macrophages. Furthermore, because our immunohistological and bioinformatics analyses demonstrated Rbpms protein localization to stress granules in a subpopulation of retinal microglia after optic nerve injury, histological quantification of Rbpms+ RGCs by immunostaining after optic nerve injury should exclude cells in which Rbpms signal is restricted to a subcellular granule, and include only those cells in which the Rbpms signal is labeling cell soma diffusely (as even if there is Rbpms in the injured RGCs’ stress granules it would be masked by the diffuse Rbpms signal throughout RGC soma).
Microglia and macrophages are involved in retinal degenerative diseases that affect RGCs (e.g., glaucoma, traumatic optic neuropathy, ischemic optic neuropathy) and other retinal cell types32–36. The application of scRNA-seq technologies for studying the cellular and molecular mechanisms of retinal degeneration resulting from diseases or injuries is becoming more widely used in the field. Therefore, the use of proper marker combinations we report, and consideration of the Rbpms+ subpopulation of microglia/macrophages we reveal, will assist future scRNA-seq studies of retinal degeneration and RGC biology. Furthermore, because the global transcriptomes of Rbpms+ and Rbpms− microglia/macrophages are overall similar, it remains to be investigated further whether the Rbpms+ subpopulation has unique functions. As stress granules are involved in other neurodegenerative diseases26–28, our finding of Rbpms localization to stress granules in a subpopulation of microglia/macrophages after injury suggests that they may be involved in retinal degeneration as well.
MATERIALS AND METHODS
Animal use and surgeries.
All animal studies were performed at the University of Connecticut Health Center with approval of the Institutional Animal Care and Use Committee and of the Institutional Biosafety Committee, and performed in accordance with the ARVO Statement for the Use of Animals in Ophthalmic and Visual Research. Mice were housed in the animal facility with a 12-h light/12-h dark cycle (lights on from 7:00 AM to 7:00 PM) and a maximum of five adult mice per cage. The study used wild-type 129S1/SvImJ mice (The Jackson Laboratory). Optic nerve surgeries were carried out on mice of both sexes 8–12 weeks of age (average body weight 20–26 g) under general anesthesia, as described previously3,37–39. Animals were sacrificed 2 weeks after ONC for histological analysis.
Immunohistological processing and analysis.
Standard histological procedures were used, as described previously3,37–40. Briefly, anesthetized mice were transcardially perfused with isotonic saline followed by 4% paraformaldehyde (PFA) at 2 weeks after ONC, the eyes were dissected, postfixed 2 hours, flattened retinas were resected, after making 4 symmetrical slits embedded in OCT Tissue Tek Medium (Sakura Finetek), frozen, and cryosectioned at 14 µm horizontality (capturing the ganglion cell layer of the whole retina), and then immunostained and mounted on coated glass slides for imaging, as we described previously41. For immunostaining, the tissues were blocked with appropriate sera, incubated overnight at 4 °C with primary antibodies, Rbpms (1:500; guinea pig, 1832-RBPMS, PhosphoSolutions), Iba1 (1:600; rabbit, 019–19741, Wako), βIII-Tubulin/Tuj1 (1:500; mouse, MMS-435P, BioLegend), and counterstained with DAPI (1:5000; Thermo Fisher Scientific) to label nuclei, then washed 3 times, incubated with appropriate fluorescent dye-conjugated secondary antibodies (1:500; all were IgG H+L; Alexa Fluor, Thermo Fisher Scientific) overnight at 4 °C, washed 3 times again, and mounted for imaging. Z-stacked images were acquired at 0.2 µm intervals using a confocal microscope (63x Oil; LSM800, Zeiss) and processed using ZEN software (Zeiss).
ScRNA-seq and data availability.
The cells identified as RGCs, which regenerated axons in response to Pten inhibition, were processed for scRNA-seq by plate-based SmartSeq2 in studies by Li et al.5 and Jacobi et al.6 and ourselves7. ScRNA-seq counts-matrix were assembled following alignment to the mouse genome and transcriptome using the Hisat2-Cufflinks pipeline. Counts-matrix for Li et al.5 (GSE206626) and Jacobi et al.6 (GSE202155) were obtained from Gene Expression Omnibus. RGCs were selected per criteria specified in the respective paper’s methods sections. In scRNA-seq dataset from Li et al.5, 297 cells were initially identified as presumed RGCs (that regenerated axons), but only 17 cells were left after exclusion of macrophages/microglia in retrospect analysis reported herein (the other group of RGC-labeled cells in Li et al.5 study that survived but did not regenerate axons had just 8 RGCs and the remaining cells were microglia/macrophages when the appropriate markers were used). In the scRNA-seq dataset from Jacobi et al.6, 120 cells were identified as RGCs. In the scRNA-seq dataset we generated, 101 cells were identified as RGCs7 (GSE210137). 10x Genomics 3’-droplet based scRNA-seq dataset of adult uninjured and injured (2 weeks after ONC) RGCs, which included αRGCs (average of α subtypes C41, C42, C43, and C45), was obtained from Tran et al.11 (GSE137400). We generated 10x Genomics 3’-droplet based scRNA-seq dataset of macrophage/microglia from adult (10 weeks old) uninjured and injured (2 weeks after ONC) retinas/optic nerves, using the methods we described previously10,41. After demultiplexing and aligning to the mouse genome and transcriptome using the CellRanger pipeline, 1168 macrophage/microglia passed quality control (including filters of a minimum of 200 genes, 6500 maximum genes). Batches were merged, normalized and scaled using Seurat and batch correction/integration of data sets was achieved using the R package harmony42. Comparative analysis of scRNA-seq datasets by Violin plots was performed using the R package Seurat v4.3.0. Correlation matrix was generated using base R software and heatmap with dendrogram was generated using the R package Superheat43,44. The macrophage/microglia scRNA-seq data we generated, and the integrated data-frame from different cell types and conditions we assembled, are available through the NCBI GEO accession GSE228986. Seurat’s FindMarkers function (with Wilcoxon rank sum test, and default parameters)45 was used to identify significantly differentially expressed genes (DEGs; , ) in Rbpms+ microglia/macrophage relative to the Rbpms− population. Gene ontology (GO) enrichment analysis was performed using the R package gprofiler246, with significantly upregulated DEGs as input and all genes expressed > 0.001 in the macrophage/microglia dataset as background. GO term enrichment plots and functional annotation and semantic clustering of GO terms was performed using the treeplot function from the Enrichplot R package along with the ggplot2 R package47,48.
Highlights.
Pan-retinal ganglion cell (RGC) marker Rbpms found in a subpopulation of retinal microglia/macrophages can confound identification of RGCs by scRNA-seq.
Rbpms protein localizes to stress granules in retinal microglia/macrophages after optic nerve injury.
Pan-RGC markers that are necessary for reliable RGC identification by scRNA-seq specified.
ACKNOWLEDGMENTS.
This work was supported by grants from The University of Connecticut School of Medicine, Start-Up Funds (to E.F.T.), and the National Institutes of Health (NIH) (Grant R01-EY029739, to E.F.T.). Portions of this research were conducted at the High Performance Computing Facility, University of Connecticut.
ABBREVIATIONS
- RGC
retinal ganglion cell
- ipRGC
intrinsically photosensitive retinal ganglion cell
- αRGC
α retinal ganglion cell
- CNS
central nervous system
- Mg/Mp
microglia/macrophage
- scRNA-seq
single cell RNA sequencing
- ONC
optic nerve crush
- KO
knockout
- KD
knockdown
- FACS
fluorescence-activated cell sorting
- GCL
ganglion cell layer
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- 1.Park KK, Liu K, Hu Y, et al. Promoting axon regeneration in the adult CNS by modulation of the PTEN/mTOR pathway. Science Nov 7 2008;322(5903):963–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Duan X, Qiao M, Bei F, Kim IJ, He Z, Sanes JR. Subtype-Specific Regeneration of Retinal Ganglion Cells following Axotomy: Effects of Osteopontin and mTOR Signaling. Neuron Mar 2015;85(6):1244–56. doi: 10.1016/j.neuron.2015.02.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim J, Sajid MS, Trakhtenberg EF. The extent of extra-axonal tissue damage determines the levels of CSPG upregulation and the success of experimental axon regeneration in the CNS. Sci Rep Jun 2018;8(1):9839. doi: 10.1038/s41598-018-28209-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yungher BJ, Luo X, Salgueiro Y, Blackmore MG, Park KK. Viral vector-based improvement of optic nerve regeneration: characterization of individual axons’ growth patterns and synaptogenesis in a visual target. Gene Ther Oct 2015;22(10):811–21. doi: 10.1038/gt.2015.51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li L, Fang F, Feng X, et al. Single-cell transcriptome analysis of regenerating RGCs reveals potent glaucoma neural repair genes. Neuron 08 17 2022;110(16):2646–2663.e6. doi: 10.1016/j.neuron.2022.06.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jacobi A, Tran NM, Yan W, et al. Overlapping transcriptional programs promote survival and axonal regeneration of injured retinal ganglion cells. Neuron 08 17 2022;110(16):2625–2645.e7. doi: 10.1016/j.neuron.2022.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rheaume BA, Xing J, Lukomska A, et al. PTEN inhibition dedifferentiates long-distance axon-regenerating intrinsically photosensitive retinal ganglion cells and upregulates mitochondria-associated DYNLT1A and LARS2. Development 2023;In Press [DOI] [PMC free article] [PubMed]
- 8.Rodriguez AR, de Sevilla Müller LP, Brecha NC. The RNA binding protein RBPMS is a selective marker of ganglion cells in the mammalian retina. J Comp Neurol Apr 2014;522(6):1411–43. doi: 10.1002/cne.23521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nadal-Nicolás FM, Galindo-Romero C, Lucas-Ruiz F, et al. Pan-retinal ganglion cell markers in mice, rats, and rhesus macaques. Zool Res Jan 18 2023;44(1):226–248. doi: 10.24272/j.issn.2095-8137.2022.308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rheaume BA, Jereen A, Bolisetty M, et al. Single cell transcriptome profiling of retinal ganglion cells identifies cellular subtypes. Nat Commun Jul 2018;9(1):2759. doi: 10.1038/s41467-018-05134-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tran NM, Shekhar K, Whitney IE, et al. Single-Cell Profiles of Retinal Ganglion Cells Differing in Resilience to Injury Reveal Neuroprotective Genes. Neuron 12 2019;104(6):1039–1055.e12. doi: 10.1016/j.neuron.2019.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pérez de Sevilla Müller L, Sargoy A, Rodriguez AR, Brecha NC Melanopsin ganglion cells are the most resistant retinal ganglion cell type to axonal injury in the rat retina. PLoS One 2014;9(3):e93274. doi: 10.1371/journal.pone.0093274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen CK, Kiyama T, Weber N, et al. Characterization of Tbr2-expressing retinal ganglion cells. J Comp Neurol Oct 2021;529(15):3513–3532. doi: 10.1002/cne.25208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Y, Du W, Chen Z, Xiang C. Upregulation of PD-L1 by SPP1 mediates macrophage polarization and facilitates immune escape in lung adenocarcinoma. Exp Cell Res Oct 15 2017;359(2):449–457. doi: 10.1016/j.yexcr.2017.08.028 [DOI] [PubMed] [Google Scholar]
- 15.Dong B, Wu C, Huang L, Qi Y. Macrophage-Related SPP1 as a Potential Biomarker for Early Lymph Node Metastasis in Lung Adenocarcinoma. Front Cell Dev Biol 2021;9:739358. doi: 10.3389/fcell.2021.739358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morse C, Tabib T, Sembrat J, et al. Proliferating SPP1/MERTK-expressing macrophages in idiopathic pulmonary fibrosis. Eur Respir J Aug 2019;54(2)doi: 10.1183/13993003.02441-2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gallego-Ortega A, Norte-Muñoz M, Di Pierdomenico J, et al. Alpha retinal ganglion cells in pigmented mice retina: number and distribution. Front Neuroanat 2022;16:1054849. doi: 10.3389/fnana.2022.1054849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krieger B, Qiao M, Rousso DL, Sanes JR, Meister M. Four alpha ganglion cell types in mouse retina: Function, structure, and molecular signatures. PLoS One 2017;12(7):e0180091. doi: 10.1371/journal.pone.0180091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martersteck EM, Hirokawa KE, Evarts M, et al. Diverse Central Projection Patterns of Retinal Ganglion Cells. Cell Rep Feb 2017;18(8):2058–2072. doi: 10.1016/j.celrep.2017.01.075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim HJ, Ryu J, Woo HM, et al. Patterns of gene expression associated with Pten deficiency in the developing inner ear. PLoS One 2014;9(6):e97544. doi: 10.1371/journal.pone.0097544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sato W, Horie Y, Kataoka E, et al. Hepatic gene expression in hepatocyte-specific Pten deficient mice showing steatohepatitis without ethanol challenge. Hepatol Res Apr 2006;34(4):256–65. doi: 10.1016/j.hepres.2006.01.003 [DOI] [PubMed] [Google Scholar]
- 22.Lu TL, Huang YF, You LR, et al. Conditionally ablated Pten in prostate basal cells promotes basal-to-luminal differentiation and causes invasive prostate cancer in mice. Am J Pathol Mar 2013;182(3):975–91. doi: 10.1016/j.ajpath.2012.11.025 [DOI] [PubMed] [Google Scholar]
- 23.Bennett ML, Bennett FC, Liddelow SA, et al. New tools for studying microglia in the mouse and human CNS. Proc Natl Acad Sci U S A Mar 22 2016;113(12):E1738–46. doi: 10.1073/pnas.1525528113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Farazi TA, Leonhardt CS, Mukherjee N, et al. Identification of the RNA recognition element of the RBPMS family of RNA-binding proteins and their transcriptome-wide mRNA targets. RNA Jul 2014;20(7):1090–102. doi: 10.1261/rna.045005.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Furukawa MT, Sakamoto H, Inoue K. Interaction and colocalization of HERMES/RBPMS with NonO, PSF, and G3BP1 in neuronal cytoplasmic RNP granules in mouse retinal line cells. Genes Cells Apr 2015;20(4):257–66. doi: 10.1111/gtc.12224 [DOI] [PubMed] [Google Scholar]
- 26.Ghosh S, Geahlen RL. Stress Granules Modulate SYK to Cause Microglial Cell Dysfunction in Alzheimer’s Disease. EBioMedicine Nov 2015;2(11):1785–98. doi: 10.1016/j.ebiom.2015.09.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.LeBlang CJ, Medalla M, Nicoletti NW, et al. Reduction of the RNA Binding Protein TIA1 Exacerbates Neuroinflammation in Tauopathy. Front Neurosci 2020;14:285. doi: 10.3389/fnins.2020.00285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu Z, Mei F, Gan Y, et al. FAM69C functions as a kinase for eIF2α and promotes stress granule assembly. EMBO Rep Mar 16 2023:e55641. doi: 10.15252/embr.202255641 [DOI] [PMC free article] [PubMed]
- 29.Somasekharan SP, Zhang F, Saxena N, et al. G3BP1-linked mRNA partitioning supports selective protein synthesis in response to oxidative stress. Nucleic Acids Res Jul 09 2020;48(12):6855–6873. doi: 10.1093/nar/gkaa376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Badea TC, Nathans J. Morphologies of mouse retinal ganglion cells expressing transcription factors Brn3a, Brn3b, and Brn3c: analysis of wild type and mutant cells using genetically-directed sparse labeling. Vision Res Jan 2011;51(2):269–79. doi: 10.1016/j.visres.2010.08.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sajgo S, Ghinia MG, Brooks M, et al. Molecular codes for cell type specification in Brn3 retinal ganglion cells. Proc Natl Acad Sci U S A May 2017;114(20):E3974–E3983. doi: 10.1073/pnas.1618551114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Andereggen L, Trakhtenberg EF, Yin Y, Benowitz LI. Inflammation and Optic Nerve Regeneration. Neuroinflammation: New Insights into Beneficial and Detrimental Functions. Wiley; 2015:189–204.
- 33.Rashid K, Akhtar-Schaefer I, Langmann T. Microglia in Retinal Degeneration. Front Immunol 2019;10:1975. doi: 10.3389/fimmu.2019.01975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Levin LA, Patrick C, Choudry NB, Sharif NA, Goldberg JL. Neuroprotection in neurodegenerations of the brain and eye: Lessons from the past and directions for the future. Front Neurol 2022;13:964197. doi: 10.3389/fneur.2022.964197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.García-Bermúdez MY, Freude KK, Mouhammad ZA, van Wijngaarden P, Martin KK, Kolko M. Glial Cells in Glaucoma: Friends, Foes, and Potential Therapeutic Targets. Front Neurol 2021;12:624983. doi: 10.3389/fneur.2021.624983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu C, Roubeix C, Sennlaub F, Saban DR. Microglia versus Monocytes: Distinct Roles in Degenerative Diseases of the Retina. Trends Neurosci Jun 2020;43(6):433–449. doi: 10.1016/j.tins.2020.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.de Lima S, Koriyama Y, Kurimoto T, et al. Full-length axon regeneration in the adult mouse optic nerve and partial recovery of simple visual behaviors. Proc Natl Acad Sci U S A Jun 2012;109(23):9149–54. doi: 10.1073/pnas.1119449109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Trakhtenberg EF, Li Y, Feng Q, et al. Zinc chelation and Klf9 knockdown cooperatively promote axon regeneration after optic nerve injury. Exp Neurol 02 2018;300:22–29. doi: 10.1016/j.expneurol.2017.10.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lukomska A, Kim J, Rheaume BA, et al. Developmentally upregulated Transcriptional Elongation Factor A like 3 suppresses axon regeneration after optic nerve injury. Neurosci Lett Sep 21 2021:136260. doi: 10.1016/j.neulet.2021.136260 [DOI] [PMC free article] [PubMed]
- 40.Kurimoto T, Yin Y, Omura K, et al. Long-distance axon regeneration in the mature optic nerve: contributions of oncomodulin, cAMP, and pten gene deletion. J Neurosci Nov 2010;30(46):15654–63. doi: 10.1523/JNEUROSCI.4340-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xing J, Lukomska A, Rheaume BA, et al. Post-injury born oligodendrocytes incorporate into the glial scar and contribute to the inhibition of axon regeneration. Development Mar 27 2023;doi: 10.1242/dev.201311 [DOI] [PMC free article] [PubMed]
- 42.Korsunsky I, Millard N, Fan J, et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat Methods Dec 2019;16(12):1289–1296. doi: 10.1038/s41592-019-0619-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.R-Core_Team. R: A language and environment for statistical computing R Foundation for Statistical Computing. http://www.R-project.org; 2014. [Google Scholar]
- 44.Barter RL, Yu B. Superheat: An R package for creating beautiful and extendable heatmaps for visualizing complex data. Journal of Computational and Graphical Statistics 2018;doi: 10.1080/10618600.2018.1473780 [DOI] [PMC free article] [PubMed]
- 45.Stuart T, Butler A, Hoffman P, et al. Comprehensive Integration of Single-Cell Data. Cell 06 2019;177(7):1888–1902.e21. doi: 10.1016/j.cell.2019.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kolberg L, Raudvere U, Kuzmin I, Vilo J, Peterson H. gprofiler2 -- an R package for gene list functional enrichment analysis and namespace conversion toolset g:Profiler. F1000Res 2020;9doi: 10.12688/f1000research.24956.2 [DOI] [PMC free article] [PubMed]
- 47.Wu T, Hu E, Xu S, et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation (Camb) Aug 28 2021;2(3):100141. doi: 10.1016/j.xinn.2021.100141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wickham H ggplot2: Elegant Graphics for Data Analysis. Springer; 2009. [Google Scholar]


