Abstract
Stimulated Raman scattering (SRS) offers the ability to image metabolic dynamics with high signal-to-noise ratio. However, its spatial resolution is limited by the numerical aperture of the imaging objective and the scattering cross-section of molecules. To achieve super-resolved SRS imaging, we developed a deconvolution algorithm, adaptive moment estimation (Adam) optimization-based pointillism deconvolution (A-PoD) and demonstrated a spatial resolution of lower than 59 nm on the membrane of a single lipid droplet (LD). We applied A-PoD to spatially correlated multiphoton fluorescence imaging and deuterium oxide (D2O)-probed SRS (DO-SRS) imaging from diverse samples to compare nanoscopic distributions of proteins and lipids in cells and subcellular organelles. We successfully differentiated newly synthesized lipids in LDs using A-PoD-coupled DO-SRS. The A-PoD-enhanced DO-SRS imaging method was also applied to reveal metabolic changes in brain samples from Drosophila on different diets. This new approach allows us to quantitatively measure the nanoscopic colocalization of biomolecules and metabolic dynamics in organelles.
Raman imaging is a vibrational spectroscopy technique measuring the scattered light corresponding to the vibration of molecules. When incident light alters the polarizability of a molecule, the wavelength of the scattered signal is changed by the resulting vibrational modes. Although Raman scattering imaging reveals structural information of a molecule based on the wavelength change of this scattering signal, the signal of spontaneous Raman scattering is weak, making it difficult to achieve high-speed imaging. In 2008, SRS was demonstrated. To selectively accelerate the matching molecular vibrational transition by up to 108 times, SRS uses two synchronized pulsed lasers, the pump beam and the Stokes beam, to produce a beating field at their different frequencies. SRS microscopy offers greatly amplified signal intensities relative to conventional Raman microscopy and has been widely applied to bioimaging ever since1–3. About 10 years later, DO-SRS imaging was reported. DO-SRS allows bioorthogonal chemical imaging of metabolic dynamics with a greatly enhanced signal-to-noise ratio (SNR)3. Briefly, the enzymatic incorporation of deuterium (D) atoms from D2O into newly synthesized biomolecules generates a new chemical bond called the carbon–deuterium (C–D) bond in macromolecules. The shifted spectra of the C–D forms of lipids, proteins and DNA can then be observed and distinguished with macromolecular specificity using spectral unmixing. However, the spatial resolution of SRS imaging still needs improvement. Recently, several super-resolution SRS techniques have been developed4–14. Nonetheless, it is still challenging to achieve super-resolved Raman imaging without manipulating the samples and to preserve the temporal resolution without any labeling or additional physical or chemical treatment.
Image deconvolution is a computational strategy that removes distortion15. Distortion in optical microscopy results in an image blurred by light diffraction, and this blurring is expressed as a point spread function (PSF). A PSF model can be combined with deconvolution to enhance resolution of microscopic images. One traditional approach is to consider images of high-density emitters as a continuous single object and deconvolve the image. However, this approach was less accurate or produced problems such as ringing artifacts, false signals in empty spaces. Another approach to computationally improve resolution is localization of single emitters. Several deconvolution methods have been developed to achieve super-resolved images by localization of single-fluorescence emitters16–18. These methods successfully enhanced the temporal resolution of localization microscopy. However, these methods cannot localize high-density emitters in a single-frame image taken with low-sensitivity sensors and thus cannot be applied to SRS microscopy data.
To overcome these limitations, Martinez et al.19 developed a deconvolution method to fit the measured data by superposition of virtual point sources (SUPPOSe). This method takes a diffraction-limited image and approximates a super-resolution image by placing a limited number of virtual emitters on the image and optimizing the position of each emitter. It sets the total number of virtual emitters, and each emitter has the same unit intensity. The fixed total intensity prevents virtual emitters from deviating away from the optimized position. Because of this characteristic, the artifact of residual images, or ringing artifact, can be removed, as shown in Extended Data Fig. 1a,b. Additionally, due to the fixed unit intensity, each pixel can have only multiples of the unit intensity. Finally, the characteristics, fixation of total intensity and unit intensity lead to extremely high sparsity of resulting images. High sparsity keeps virtual emitters positioning on the most probable region and prevents the emitters going out from the region. In addition, because each virtual emitter represents a bundle of single emitters, this approach overcomes the limit of aforementioned sparse deconvolution methods. We sought to apply SUPPOSe directly to our SRS microscopy data; however, we found major drawbacks including low precision of the signal’s spatial location and slow processing speed.
We thus developed A-PoD, a method based on SUPPOSe that uses an Adam solver instead of a genetic algorithm for the optimization process of finding the positions of each emitter. The gradient descent algorithm Adam removes the randomness in the genetic algorithm and enables us to both enhance spatial precision and shorten the data-processing time. We applied A-PoD to SRS imaging and generated a series of super-resolved images of mammalian cells and tissues as well as Drosophila brain tissues. These images displayed nanoscopic distributions of protein and lipid in biological samples. We further measured the shapes and sizes of individual LDs in Drosophila brain samples and examined the effects of high-glucose diet on brain lipid metabolism and the size distribution of LDs. Our A-PoD algorithm achieves super-resolution images with higher spatial precision than existing deconvolution methods and at a markedly enhanced speed for image processing.
Results
Main concept of A-PoD
We converted SRS images into super-resolution images using a procedure illustrated in Fig. 1a. First, a specific number of virtual emitters proportional to the overall brightness of the image are placed on an image (X), and a blurred image (S) is created through convolution of X and the PSF. When the position of each virtual emitter is adjusted such that the difference between the blurred image S and the measured image (Y) is minimized, X becomes the image with the most optimal distribution of virtual emitters. We used a modified Adam solver for the optimization in A-PoD (see Methods for details). Using simulation image data, we compared A-PoD with the Richardson–Lucy method, a widely used deconvolution algorithm. A-PoD outperformed the Richardson–Lucy algorithm in DeconvolutionLab2 (even with 100 iterations; Extended Data Fig. 1).
Fig. 1 |. Deconvolution of SRS images using A-PoD.

a, Schematic of superresolution SRS image processing. b, Three-dimensional deconvolution result of LDs (2,850 cm−1) in a live cell. Following deconvolution, the membrane of an individual LD was clearly visualized in the intensity profile in the lower panel. AU, arbitrary units. c, Three-dimensional-rendering results of the SRS image before (left) and after (right) deconvolution. After deconvolution, the shapes of ~1-μm-sized LDs were clearly visible.
As a proof of concept, A-PoD was applied to SRS imaging of lipids (at ~2,850 cm−1) in a live cell. Application of A-PoD greatly improved spatial resolution (Fig. 1b). The increased spatial resolution clearly revealed individual LDs inside the cell, allowing us to distinguish the outer layer and the internal space of the LD. Due to the strong SNR of the SRS setup, the –CH2 vibration signal of the lipid membrane is detectable. The measured image of –CH2 vibration was deconvolved using A-PoD. As a result of this process, the 3D sizes of individual LDs can also be clearly visualized from the sharpened image (Fig. 1c).
SPIDER18 is a sparse deconvolution algorithm that can generate super-resolved images from localization microscopy data. To assess the precision of A-PoD as a super-resolution deconvolution method, we compared the localization results with those obtained using SPIDER. We used a raw mitochondrial image stack from the previous SPIDER publication18 that was composed of 100 frames. Each image frame contained information of scattered blinking emitters. The image stack was processed with the SPIDER program. The widefield image was generated by averaging the stack and was deconvolved using A-PoD. Image processing using A-PoD revealed a mitochondrial structure similar to that obtained using SPIDER (Extended Data Fig. 2a(ii,iv)). Cross-section signal-intensity profiles of images showed that the thickness of the mitochondrial membrane measured by the two methods was almost the same. These results demonstrate that A-PoD can reconstruct a super-resolved image from a single-frame widefield image. To test the processing speed of A-PoD, we deconvolved the mitochondrial image with limited virtual emitter numbers of 105. As iteration number increased, the similarity between the ground-truth image and the deconvolution image was increased (Extended Data Fig. 2b). Using A-PoD, the entire process was completed in 2 s, and the similarity was higher than that using the genetic algorithm (in 96 min with 5 × 106 iterations). By further increasing the iteration number, the genetic algorithm could improve the similarity but with a much longer processing time.
STORM image analysis
To evaluate the spatial precision of deconvolution, we compared A-PoD results with those from DAOSTORM20, a widely used algorithm to localize emitters in dense localization microscopy images, for example, stochastic optical reconstruction microscopy (STORM) images (Extended Data Fig. 3). For this comparison, an image of cultured neurons in which spectrin was labeled using a fluorescent antibody (mouse anti-β II spectrin antibody conjugated with Alexa 647) was analyzed using the two algorithms. The original STORM image stack was composed of 16,500 frames, and two regions of interest (ROI) having different emitter densities were analyzed (Extended Data Fig. 3b,c). One of the selected ROI contained a low emitter density. From the entire image stack of the selected ROI, an ‘epifluorescence’-like image was calculated. The image was deconvolved using A-PoD. Due to the low density of emitters, individual molecules in the image frame could be localized using DAOSTORM. Analysis using either A-PoD or DAOSTORM revealed the periodic structure of the membrane-associated periodic skeleton (MPS) in neurons. The intensity profile and the auto-correlation curves (Extended Data Fig. 3b(iii,iv)) showed the periodicity quantitatively, and the periodicity obtained using A-PoD was close to that obtained using DAOSTORM with less than 20% of difference. The precision might be affected by SNR, pixel size and low-frequency background of the image. From another ROI containing a high emitter density (shown in the green boxed area in Extended Data Fig. 3a(i)), we analyzed a single frame from the image stack using A-PoD (Extended Data Fig. 3c(i,ii)). Interestingly, the periodic structure of the MPS becomes more clearly visible than the other ROI with lower emitter density (Extended Data Fig. 3c(iii,iv)). Therefore, A-PoD can be used to analyze images with similar performance as that of DAOSTORM at low emitter density, indicating that A-PoD can be applied to processing images with a wider range of emitter densities.
Standard sample measurement
To quantitatively determine the resolution of A-PoD-coupled SRS imaging, we first analyzed images of standard polystyrene beads with known sizes (100 nm and 1 μm, respectively). The measured image was reproduced through convolution of the PSF and the virtual image. For an accurate prediction of the PSF, we evaluated the results of using a simulated PSF and an experimental PSF. After measurement of 100-nm beads, the experimental PSF was calculated using A-PoD (Extended Data Fig. 4a). The simulated PSF was generated by multiplication of the PSFs of the pump beam (PSFpump) and the Stokes beam (PSFStokes). These two PSFs have similar full width half maximum (FWHM) values with about 3% difference (Extended Data Fig. 4b). Using the simulated PSF, another 100-nm bead image was deconvolved, and the FWHM of the bead was decreased from 608.5 nm to 101.0 nm (Fig. 2a). To compare the two PSFs, a single-LD image was deconvolved (Extended Data Fig. 4b). Spatial resolutions of the results were evaluated based on the membrane thickness measurement and decorrelation results. The thicknesses measured were 59 nm and 76 nm by using the experimental PSF and the simulated PSF, respectively. Decorrelation21 results demonstrated the spatial resolutions of 54 nm and 57 nm in an experimental PSF result and a simulated PSF result, respectively. We next analyzed the image of 1-μm beads using the PSFsim. After deconvolution, the lateral size of the bead was expressed close to 1 μm, but the axial size of the bead was approximately 2.5 times larger. As the focal volume of a Gaussian beam has a longer shape along the vertical axis22, the axial resolution is worse than the lateral resolution. Additionally, we observed a cone-shaped afterimage appearing along the optical axis. This is because the direction and intensity of scattering are affected by the size and material of an object, and this scattering behavior is reflected in the wavefront shape of light23,24. The wavefront of light is distorted by scattering and diffraction. It is difficult to predict using an ideal PSF model. Therefore, the distortion near the bead was not removed by deconvolution. However, this can be mitigated by the combination of adaptive optics and a deep learning method that learns PSF changes around an object25,26.
Fig. 2 |. Deconvolution results of SRS images.
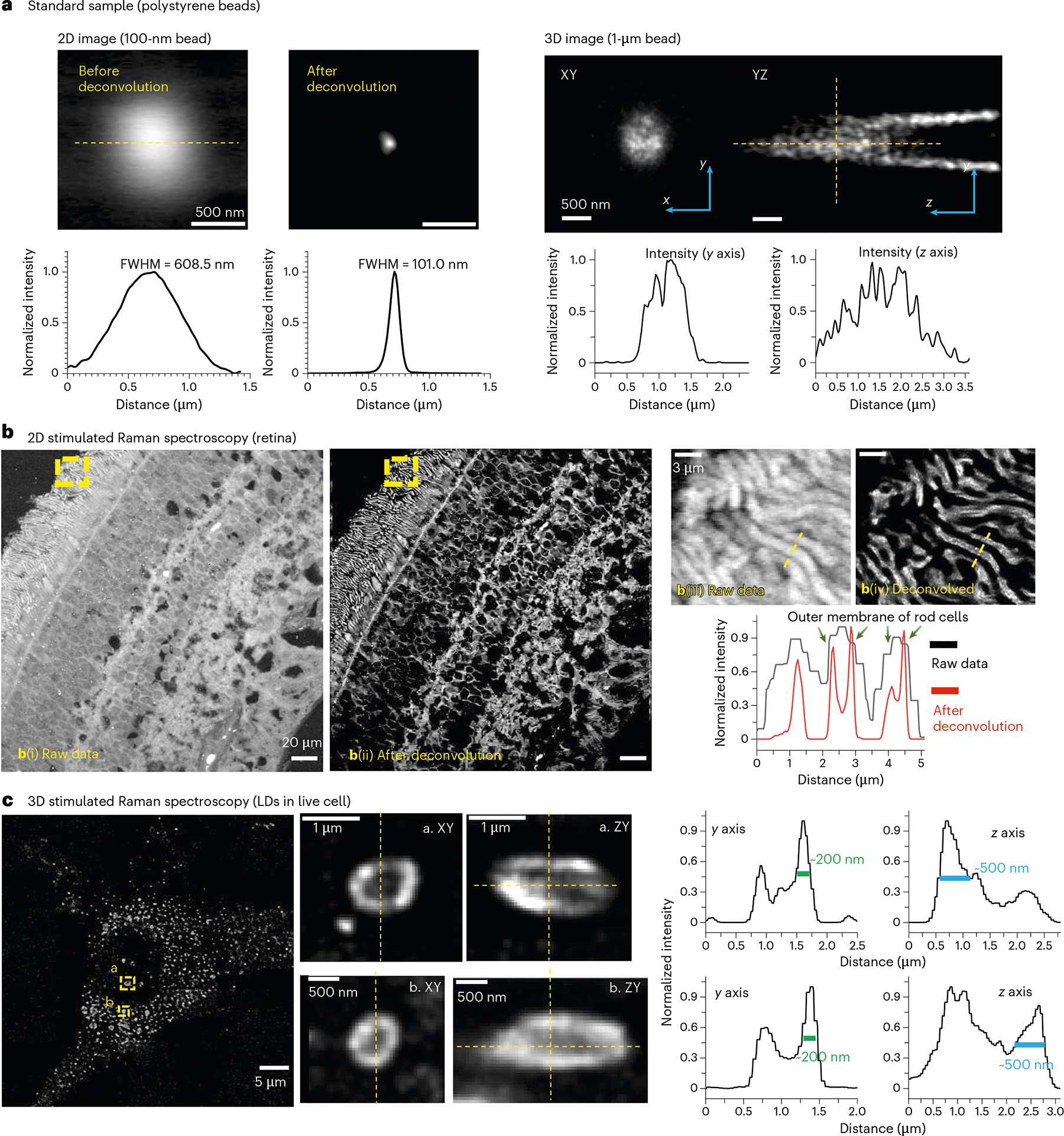
a, Images of standard beads (100 nm and 1 μm). Left, after the deconvolution of a two-dimensional (2D) image of a 100-nm bead, FWHM of the intensity profile was decreased from 608.5 nm to 101.0 nm. Right, 3D images of a 1-μm bead before and after deconvolution, together with the corresponding signal-intensity profiles. The lateral size of the bead was almost 1 μm, but the axial size was over 2.5 times bigger than the lateral size. The tail-like artifact was not removed by A-PoD. b, An SRS image of a human retinal section (at 2,930 cm−1). After deconvolution using A-PoD, contrast of the image was markedly enhanced. Deconvolution results revealed the rod outer segment cell membrane-like intensity profile. The area boxed by the dashed lines in b(i,ii) in the outer segment is enlarged and shown in b(iii,iv). c, Deconvolution result of 3D SRS images (2,850 cm−1) of LDs in a live cell. Following deconvolution, the detailed structure of LDs was more clearly visualized, including the internal score and the surface membrane.
Human retinal tissue imaging
Next, we extended A-PoD to SRS imaging of human retinal tissue samples (Fig. 2b). We focused on the outer segments of photoreceptors, which contain membranous photoreceptor disks surrounded by the cell membrane. After applying A-PoD to the SRS image, the image resolution was markedly increased, allowing for improved structural discrimination. For instance, the cell membrane could be visually distinguished in the outer segment of rod cells. The thickness of the cell membrane was about 170 nm, and the resolution of the image calculated using the decorrelation analysis method21 was approximately 100 nm. This resolution with the retinal sample image is lower than that of the standard bead image, because deconvolution accuracy depends on imaging conditions, including the intensity and sampling frequency. The bead image was measured with a sampling rate of 26 nm per pixel, but the sampling rate of the retinal image was 198 nm per pixel. Although the increase in spatial resolution was not sufficient to resolve the actual membrane thickness of ~4–5 nm, considering the wavelengths of the laser beams and the characteristics of the PSF, the resolution of ~100 nm clearly exceeded the diffraction limit. It is known that the lipid composition of the rod cell membrane is substantially different from that of the photoreceptor disks27. A-PoD-coupled SRS microscopy demonstrated a remarkable ability to distinguish these compartments.
LD imaging
LDs are organelles important for cell proliferation and survival. These ubiquitous organelles not only serve as energy stores but also play crucial roles in cell signaling and membrane trafficking. They also contain diverse spatial and chemical information that may reflect oxidative stress, metabolic flux and disease status28–38. However, it has been challenging to directly visualize LD metabolism at the organelle level, mainly due to a lack of spatial information in conventional lipidomic modalities. Using A-PoD-coupled DO-SRS imaging, we visualized the nanoscopic distribution of LDs and their metabolic activities. DO-SRS imaging (at 2,850 cm−1) clearly revealed numerous LDs in the breast cancer cell, and the size of individual LDs could be precisely measured after deconvolution (Figs. 1c and 2c and Extended Data Fig. 4b). The outer layer of each LD was visually separated from the inner space of the LD. The thickness of the LD outer layer was measured to be approximately 200 nm. This size was similar to that of the previously measured cell membrane of the rod cells. The axial thickness of the LD was approximately 500 nm, about 2.5 times larger than the lateral resolution. This difference is comparable to the resolution difference in imaging of 1-μm beads.
Next, we used a particle-analysis method to remove the background and to focus on the regions of LDs. The subcellular distribution of LDs in breast cancer cells was then analyzed (Fig. 3). We measured the distances of the detected particles from an arbitrarily chosen point near the center of the nucleus and calculated the surface area:volume (SA:V) ratio of individual LDs. The LDs were classified into three groups based on the distance and SA:V ratio using the k-mean algorithm (Fig. 3b). Group 1 had a lower SA:V ratio than the other two groups (Fig. 3d). The LDs in group 2 (Fig. 3e) were distributed more closely to the nucleus than those in group 3 (Fig. 3f). The capability of A-PoD-coupled DO-SRS to identify these different subpopulations of LDs with different SA:V ratios or subcellular distribution may facilitate future studies of dynamic interactions of LDs with other organelles (such as the endoplasmic reticulum (ER)), as previous studies suggested that nano-LDs newly detached from the ER have high SA:V ratio.
Fig. 3 |. SA:V ratio analysis.
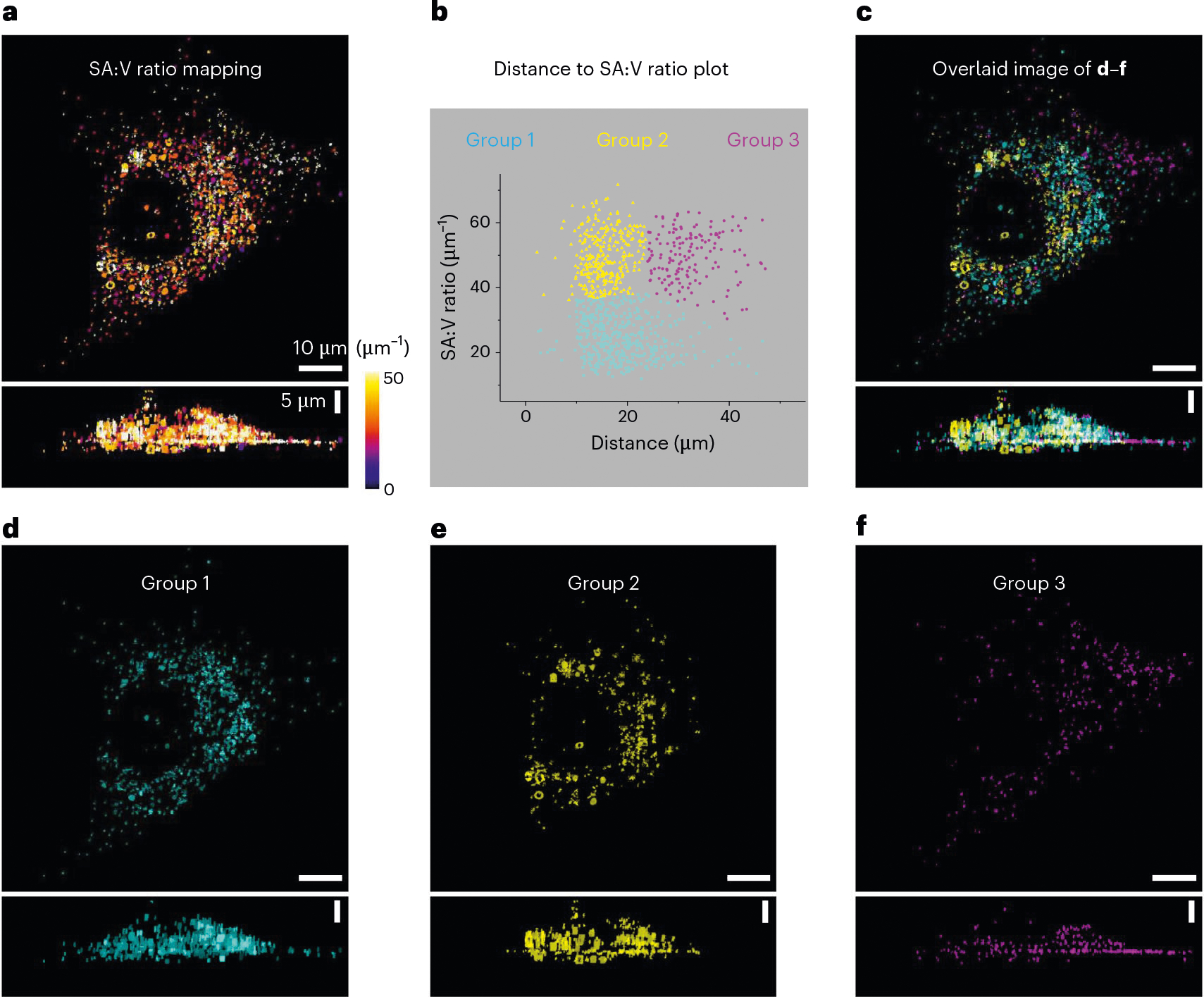
a, The SA:V ratio of LDs in the breast cancer cell image in Fig. 2c was mapped. b, k-mean clustering shows that the three groups of LDs have different SA:V ratios. c, The LD images in different groups (d–f) were overlaid. d, LDs in group 1 are widely distributed in the cell, with a low SA:V ratio. e, LDs in group 2 have a high SA:V ratio, and they are distributed closely around the nucleus. f, LDs in group 3 also have a high SA:V ratio, and they were distributed far away from the nucleus. n = 3 cells per experiment, total of six experiments.
Nanoscopic metabolic imaging with super-resolved DO-SRS
Direct visualization of LD metabolism under different conditions at the organelle level is crucial for uncovering signaling pathways and molecular mechanisms regulating lipid metabolism. Research in this area has been limited by a lack of spatial resolution in conventional lipidomic imaging modalities. We applied A-PoD-coupled DO-SRS metabolic imaging to visualizing lipid metabolism in HeLa cells cultured in the presence of D2O. The distribution of LDs in HeLa cells was imaged at 2,850 cm−1 (CH2) and 2,140 cm−1 (CD), representing the old lipids and the newly synthesized lipids, respectively (Fig. 4a). After converting the images in each channel to super-resolved ones using A-PoD, differences in the distribution of old versus new lipid signals were clearly revealed in two dimensions (Fig. 4b) and three-dimensional (3D)-rendered images (Fig. 4e). Thus, the metabolic turnover rate of subpopulations of LDs can be quantified with SA:V ratio mapping. Before deconvolution, only areas with concentrated old and new LDs were visualized. After deconvolution, the 3D shape and distribution of individual LDs were clearly visualized. Additionally, we analyzed the surface area and volume of individual LDs from cells cultured under different conditions: high tryptophan (15×,Trp) and standard control medium. The standard deviations of surface area and volume of LDs in HeLa cells cultured in high-tryptophan medium were wider than those of the control group (Fig. 4c; see Extended Data Fig. 5 for control cells). The SA:V ratio of individual LDs was mapped in deconvolved images (Fig. 4d).
Fig. 4 |. Three-dimensional super-resolution metabolic imaging of the HeLa cell.
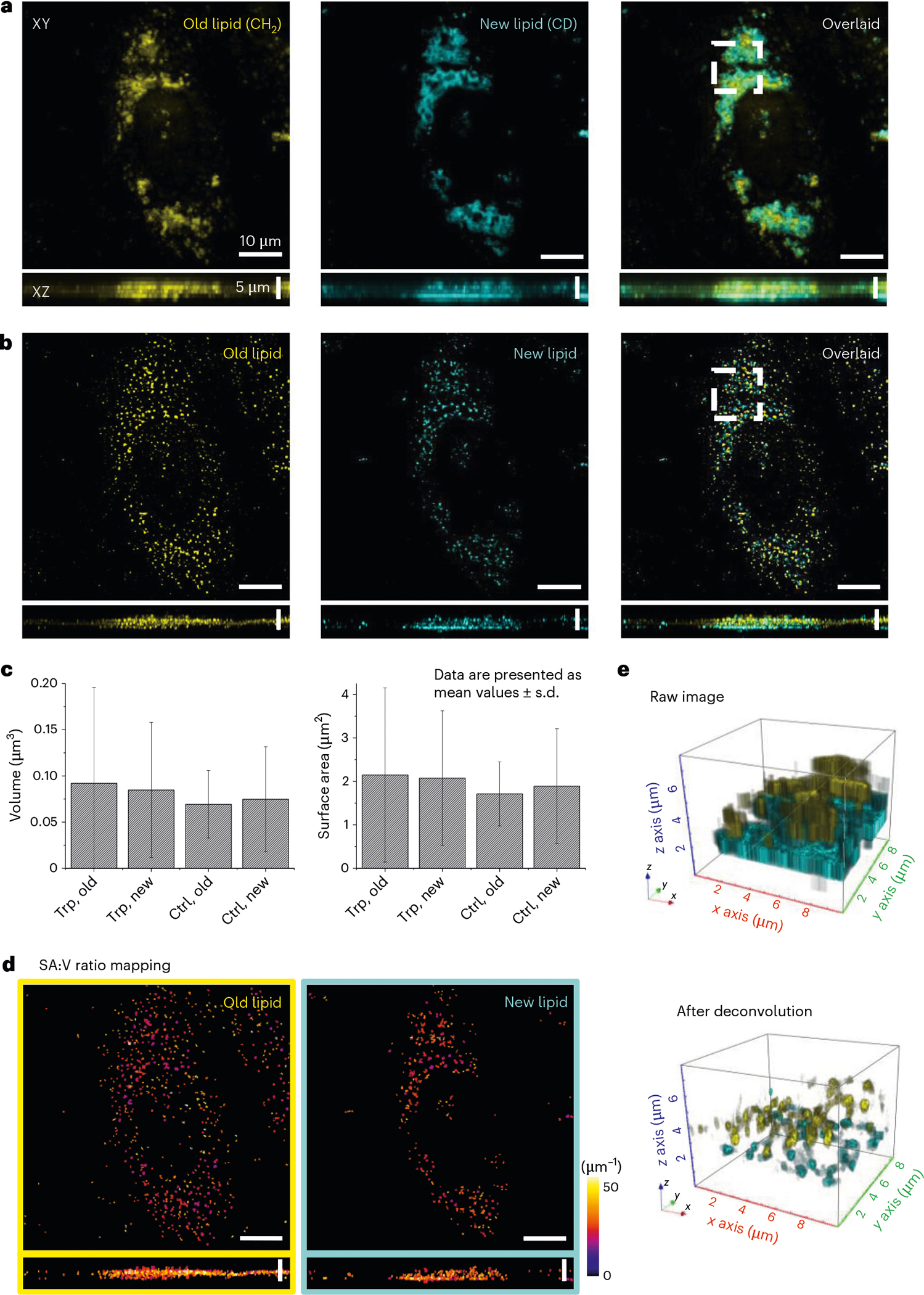
a, DO-SRS images of LDs in CH2 and CD channels. The CH2 channel represents the distribution of old LDs (left), and the CD vibration image shows the distribution of newly synthesized LDs (middle). To compare the two images (left and middle), the images were overlaid (right). b, DO-SRS images were deconvolved using A-PoD, and the results clearly separate the signals of two different types of LDs, old versus newly synthesized (left, middle and right). c, Averaged volume and surface area of each LD in the two different culture conditions were plotted. The LDs in the cell cultured with excessive tryptophan (Trp) have wider distribution than those in the control group (Ctrl) (number of LDs: 264 (new LDs in Trp), 108 (new LDs in Trp), 653 (old LDs in control) and 736 (new LDs in control)). Images of the control cell are presented in Extended Data Fig. 4. Bar plots are presented as mean values ± s.d. d, The SA:V ratio of individual LDs was mapped. Using the color code, the SA:V ratio was visualized. e, The 3D-rendering images of the white dashed boxed regions in a (right) and b (right) show the resolution difference before and after deconvolution (top and bottom). n = 45 (five cells in each ROI, three ROIs per experiment and three experiments).
It has been proposed that LDs play a critical role in neuroblast cell division and brain development. One major hurdle for understanding functional roles of LDs under physiological or pathological conditions is the limited imaging methods for direct observation of LD metabolic activity changes under physiological or pathological conditions. We applied A-PoD-enhanced DO-SRS imaging to directly visualize metabolic changes in Drosophila larval brains collected from animals on different diets. The DO-SRS image of the entire brain lobe collected at 2,850 cm−1 showed a large amount of lipids in larvae fed with the standard control diet (Fig. 5). To determine the subcellular location of the lipids, magnified images were taken from the central brain region. These images clearly revealed lipids inside LDs (small dot-like structures). Using A-PoD, we were able to acquire the profile of individual LDs and compare the size distribution of LDs in brain samples of flies fed a standard diet with that of those fed with a high-glucose diet (3× glucose) (Fig. 5a–d; also see histograms in Extended Data Fig. 6a and images before overlay in Extended Data Fig. 7). Size analysis showed that LDs in the ~0.2–0.3-μm2 range were predominant in the control group, whereas LDs in the high-glucose group showed a wider range of size distribution, with many small LDs in the ~0.1–0.2-μm2 range. To better visualize the subcellular distribution of LDs of different sizes in situ, color-coded images were generated to show the distribution of small (~0.05–0.2 μm2), medium (~0.2–0.3 μm2) and large (~0.3–0.45 μm2) LDs, respectively (Extended Data Fig. 6a). Considering the small difference in the two histograms and the pixel size (163 nm) in the raw images, it is worth noting that this A-PoD-enhanced SRS approach can measure LD sizes of a wide range (~0.05–0.45 μm2).
Fig. 5 |. Super-resolution metabolic imaging of Drosophila brain samples.
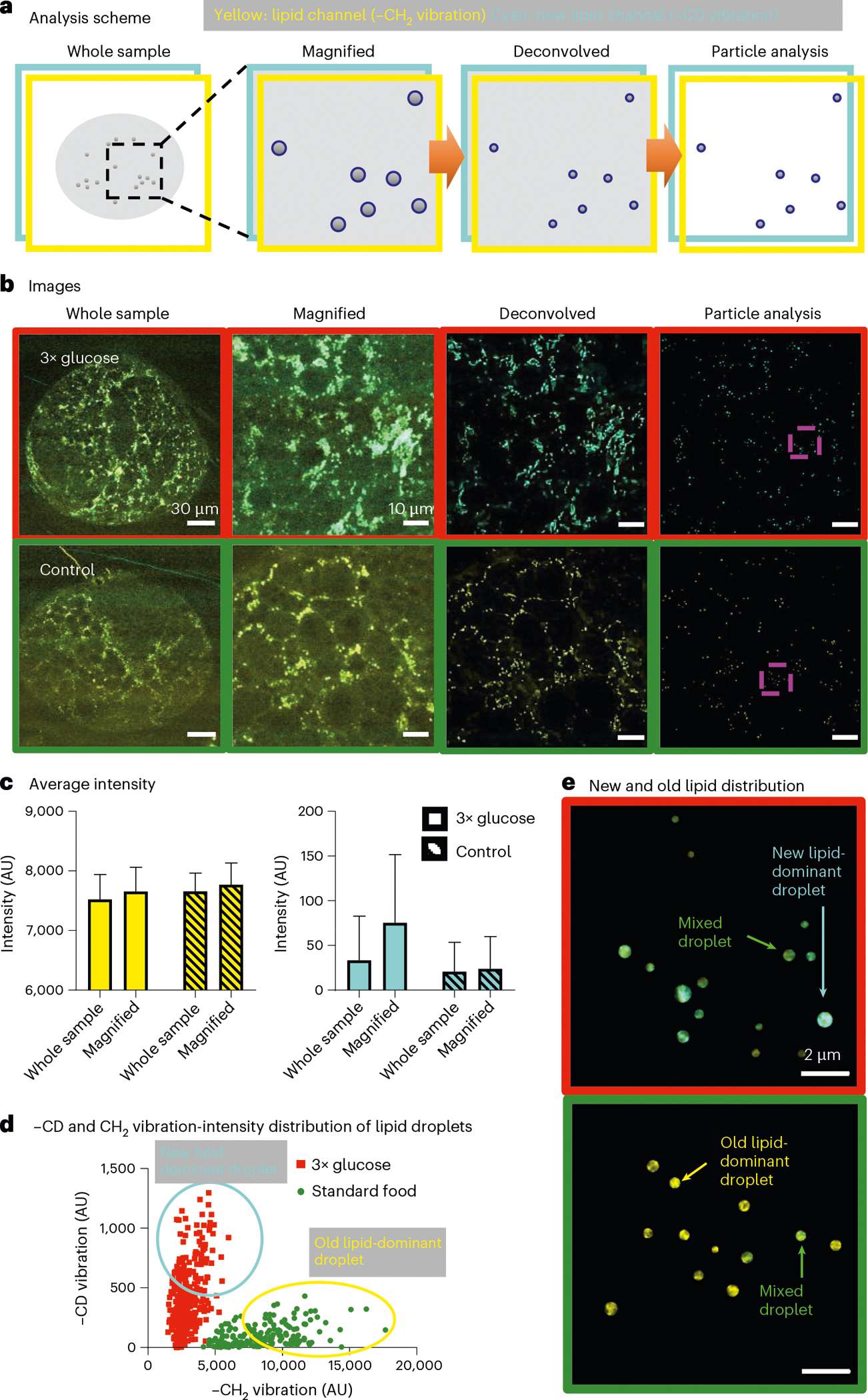
a, Schematic of the analysis method. The whole-sample image represents the overall lipid distribution. The image was magnified to compare the signal distribution of old and new lipids. The nanoscopic distribution of lipids was revealed after deconvolution. The particle-analysis method enables us to remove background and analyze individual LDs. b, Brain samples from flies on two different diets were measured using DO-SRS microscopy. The sample in the 3× glucose group (red boxed images, yellow, –CH2 signal; cyan, –CD signal) and the control group (green boxed images, yellow, –CH2 signal; cyan, –CD signal) were analyzed. The images before the overlay are displayed in Extended Data Fig. 7. c, The average signal intensity of the images in two groups. The average signal intensity of old lipid in the control group was slightly higher than that of the 3× glucose group. The new lipid signal in the 3× glucose group was much higher than that in the control group. The new lipid signal difference was clearer in the magnified image. Bar plots are presented as mean values ± s.d. of each image intensity. d, The scatterplot shows the distribution of the new lipid:old lipid (CD/CH2) signal ratio of individual LDs. Under the two different dietary conditions, the LDs have a clearly distinguishable CD/CH2 signal ratio. The averaged turnover rate in the 3× glucose group is over ten times larger than the rate of the control group (number of LDs: 179 (standard food), 299 (3× glucose)). e, Using particle analysis, we can visualize the nanoscopic distribution of newly synthesized lipids in individual LDs. Areas boxed by pink dashed lines in the images in b are enlarged and shown. n = 3 brains per group.
Combined with D2O labeling, lipid metabolic activities in the brain samples were measured. By measuring the LD size and turnover rates (Extended Data Fig. 6b), we quantified the correlation between size and metabolic activity. The correlation coefficients were 0.44 in control flies and 0.40 in the high-glucose group, with no significant differences detected. Both groups showed a positive correlation between LD size and metabolic activity, suggesting that larger LDs have higher metabolic activity. This result is consistent with our studies on Drosophila fat body metabolic activity39–41. Importantly, quantitative analyses of the CD/CH2 ratio showed that the average lipid-turnover rate in the high-glucose group was about ten times higher than that in the control group, suggesting that more newly synthesized lipids had accumulated in flies on the high-glucose diet (Fig. 5d and Extended Data Fig. 6b). A-PoD-coupled DO-SRS combined with particle analysis further enabled us to map distinct subpopulations of LDs: new lipid-dominant, old lipid-dominant and mixed LDs (Fig. 5e). Further studies are necessary to determine molecular mechanisms by which a high-glucose diet modulates lipid-turnover rates.
Nanoscopic colocalization of macromolecules and fluorophores
Applying A-PoD to spatially correlated multiphoton fluorescence (MPF) imaging and SRS imaging, we next examined nanoscopic spatial distributions of proteins and lipids in mitochondria of live cells (Fig. 6). We imaged HEK293 cells with the mitochondria stably labeled with mitochondria-red fluorescent protein (Mito-RFP). The fluorescence signals of Mito-RFP were measured using MPF. At the same time, SRS images of 2,930 cm−1 (CH3 protein, in cyan) and 2,850 cm−1 (CH2 lipid, in yellow) were also measured. The SRS images in the two different Raman shifts were unmixed into the protein channel and the lipid channel using an existing protocol (Fig. 6b)3,42. The images of these three measured channels were then converted into super-resolved images using A-PoD (Fig. 6c). Before deconvolution, there was a substantial overlap of different types of signals (white areas in Fig. 6d, left). After deconvolution, the white area was reduced, and the different distribution of each component was clearly revealed (Fig. 6d, middle). This is consistent with the fact that SRS signals for protein and lipid panels are not mitochondrion-specific proteins or lipids. On the other hand, most Mito-RFP signals (in magenta) were overlapping with lipid signals (in yellow), consistent with the fact that Mito-RFP marked the mitochondrial membrane. Furthermore, in the signal-intensity profile of the cross-section, the influence of the blurry background signal was reduced after deconvolution, and the position of each component was accurately expressed (Fig. 6d, right). These data showed that applying A-PoD to multiplexed MPF–SRS imaging substantially enhanced the resolution.
Fig. 6 |. Multiplexed super-resolution MPF-SRS imaging of mitochondria.
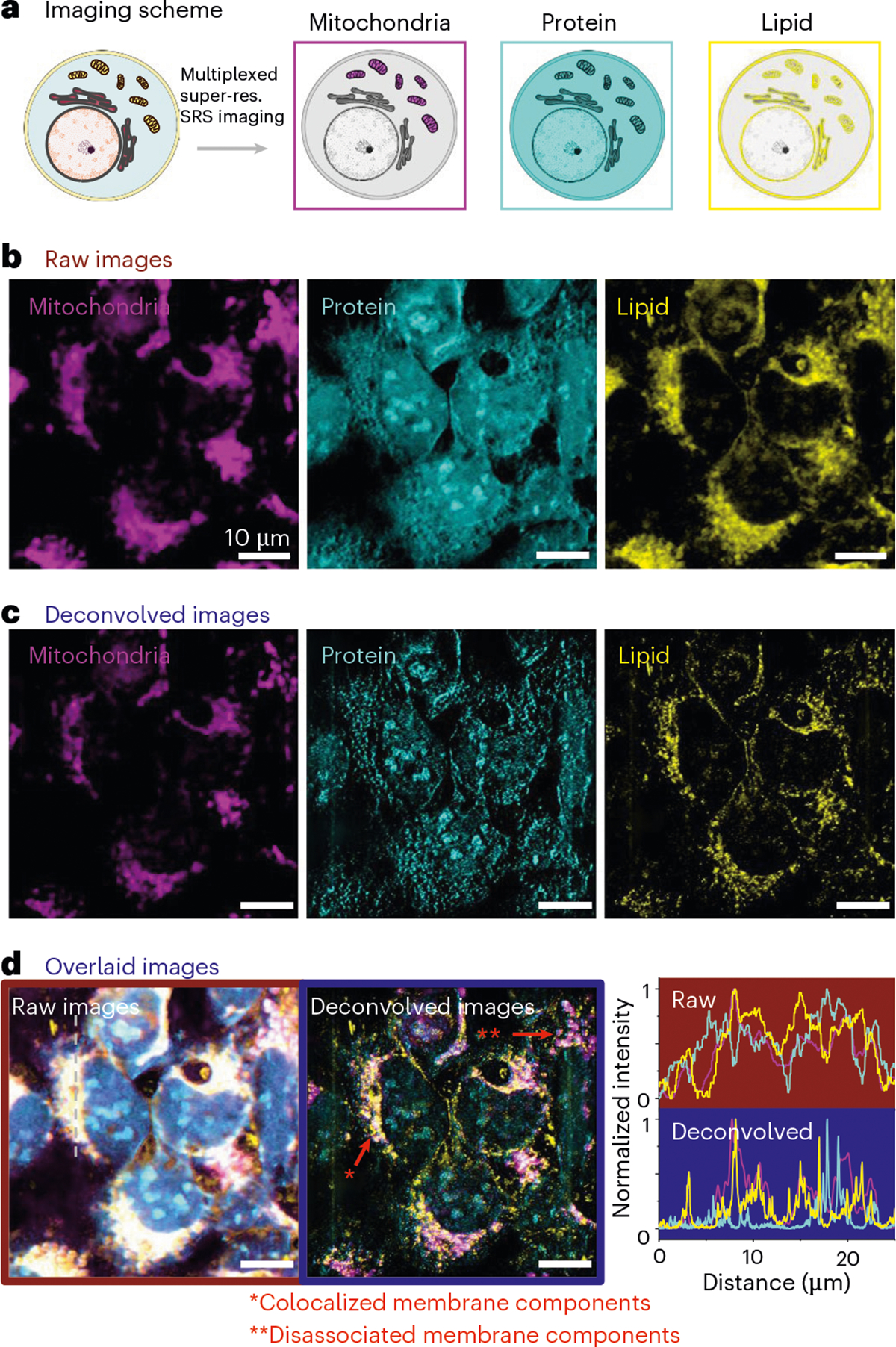
a, The multiplexed imaging scheme. Images were taken in the MPF channel (mitochondria) and two SRS channels (protein, lipid) simultaneously and then deconvolved using A-PoD. Super-res., super-resolution. b, Mitochondria in HEK293 cells were labeled with Mito-RFP (magenta) and imaged using MPF and SRS microscopy. The SRS images in 2,930 cm−1 and 2,850 cm−1 were unmixed to protein (cyan) and lipid (yellow) channels, respectively. c, The multiplexed images of mitochondria were deconvolved and converted to super-resolution images (magenta, Mito-RFP; cyan, protein; yellow, lipid). d, The superimposed images show the resolution difference before and after deconvolution. Left, superimposed image before deconvolution. Middle, after deconvolution, the white area where the three components were overlapping was much reduced. Right, top and bottom, normalized signal-intensity profiles before and after deconvolution. The three components show distinct spatial distribution after deconvolution as shown in the signal-intensity profiles. n = 15 (five ROIs per sample were imaged in three samples).
Discussion
In this study, we have developed the A-PoD algorithm and integrated it with SRS, DO-SRS and MPF–SRS imaging methods. A-PoD substantially enhances the spatial resolution of images at a high processing speed and spatial accuracy when an appropriate PSF is defined, regardless of the imaging modality. A-PoD can be applied not only to widefield fluorescence microscopy19 but also to various other microscopy techniques. The super-resolution A-PoD-coupled SRS microscopy introduced here also has broad applications including deep-tissue imaging, hyperspectral imaging and multiplex imaging43–46.
We first characterized A-PoD as a sparse deconvolution method by analyzing the simulated data. The capability of A-PoD to generate super-resolved images was evaluated by comparison with localization microscopy data (Extended Data Fig. 2a). Although the genetic algorithm in SUPPOSe works well for optimizing variables in an integer domain (for example, the address of specific pixels), it contains randomness in the process. Gradient values of the function need to be calculated in every optimization step, which is time consuming. Of note, by changing the genetic algorithm to A-PoD, the image-deconvolution process was shortened from a few hours to a few seconds (Extended Data Fig. 2b). Compared with the Richardson–Lucy algorithm (Extended Data Figs. 1c and 8), the most widely used deconvolution method, A-PoD offers much richer information at a high resolution.
For analysis of STORM imaging data (Extended Data Fig. 3), A-PoD demonstrated potential as an image-processing tool for localization microscopy. Generally, to achieve a super-resolved image using STORM, we need to keep a low concentration of emitters. By contrast, for A-PoD, strong signals are desirable to achieve higher resolution. Due to this unique characteristic of A-PoD, we could clearly visualize the periodic structure of the MPS in neurons from a single image in the bright ROI. This finding implies that A-PoD substantially improves the temporal resolution of localization microscopy, allowing us to extract image features from a single frame to a few frames rather than analyzing tens of thousands of image frames. Depending on the imaging rate of the image stack, it would be possible to take a super-resolved image in a few-microsecond range when enough emitter density is secured.
Using A-PoD-coupled SRS microscopy, we successfully examined the distributions of proteins and lipids in cultured cells and tissue samples at the nanoscopic level. The nanoscopic distribution of LDs in cancer cells and the membranous outer segments of rod cells in the retinal tissue were clearly resolved. Furthermore, integration of A-PoD into our DO-SRS platform enabled us to examine different distributions of newly synthesized lipids versus the pre-existing lipids in live cells and tissues. This combination provides a powerful tool for direct visualization of lipid metabolic changes not only in cells but also in brain tissues (Figs. 4 and 5).
Using cultured HeLa cells and breast cancer cells, we demonstrated the power of A-PoD-coupled SRS imaging in examining subcellular organelles, such as LDs and mitochondria (Figs. 3, 4 and 6). We mapped the SA:V ratio of individual LDs. Because the accuracy of the measured surface area and volume depends on the spatial resolution of images, A-PoD is a valuable tool for analyzing the exact values of these parameters. Using A-PoD-coupled DO-SRS, we examined the spatial distribution of distinct subpopulations of LDs, those predominantly containing newly synthesized lipids, those mostly containing old lipids and LDs containing mixed lipids. Mapping the old and new lipid domains in individual LDs (Fig. 5e) provides useful information in studying lipid metabolism at the nanoscale. Future experiments are necessary for understanding the pathophysiological roles of LD heterogeneity. Nevertheless, our A-PoD-based DO-SRS imaging system provides a robust method for studying molecular heterogeneity in living organisms.
Analyses of the LD size distribution and lipid-turnover rate in Drosophila brain samples indicate that the subpopulation of LDs with higher turnover rate increased in the brain in flies on a high-glucose diet and that the average lipid-turnover rate in the high-glucose group was much higher than that in the control group. These analyses suggest that smaller LDs, which are usually newly born LDs connected to the ER47, may have lower de novo lipid-synthesis ability. They may obtain lipid content directly from the ER lumen. This is consistent with a previous study48 reporting that enzymes (such as DGAT2 and CCT1) mediating de novo lipid synthesis were mainly localized in larger mature LDs detached from the ER. Here, our A-PoD-enhanced super-resolution DO-SRS imaging has revealed the metabolic diversity of LDs, which had not been detected by other methods. Previous studies reported that ER stress was induced by high glucose49 and that ER stress increased LD number50,51. Our A-PoD-based super-resolution DO-SRS imaging system provides an effective tool for future studies on dynamic changes in LDs, functional roles of LDs and underlying mechanisms under various physiological and pathological conditions.
To define nanoscopic distribution of different molecules, we can use A-PoD in multiplex SRS imaging. We prepared HEK293 cells stably expressing Mito-RFP and examined subcellular distribution of mitochondria, proteins and lipids using A-PoD-based MPF and SRS imaging. As expected, most Mito-RFP signals overlap with CH2 lipid (membrane) signals (Fig. 6d). By comparing the spatial localization of different components, we can clearly define colocalized and non-overlapping components. Furthermore, A-PoD-coupled multiplex SRS can be applied to imaging other biomolecules such as nucleic acids, etc.
To improve the spatial resolution, we used image deconvolution for SRS images, which is not trivial as SRS uses two laser beams. For the deconvolution algorithm, an accurate PSF model is needed, which can be either measured experimentally or simulated. Here, we tested both experimental and simulated PSFs and compared the results from them. For the simulated PSF, we chose it to be the product of the PSF of the pump beam and the Stokes beam. Simulated PSF has several advantages: first, it can be applied to existing images of which the experimental PSF might not be obtained; second, simulated PSF shows broad applications in other imaging modalities in addition to SRS.
The potential application of A-PoD in new localization microscopy methods was demonstrated in the analyses of STORM images. For existing localization microscopy methods, the amount of emitter signals has to be precisely adjusted. To make this adjustment, one needs to take numerous different frames to reconstruct a single super-resolved image. However, A-PoD can maximize the temporal resolution by overcoming the limitation of emitter density. Therefore, this program allows us to take not only a super-resolution SRS image but also a super-resolution fluorescence image at a high speed.
A-PoD has a wide range of applications. In this study, we presented the results combining A-PoD with STORM, DO-SRS and multiplex MPF–SRS. It is also applicable to other imaging techniques. First of all, if we have enough information about the blurring kernel, spatial resolution of every old image can be improved. For example, for atomic force microscopy, the PSF model of optical microscopy cannot be applied because the morphology of the sample is measured by force between the tip end and the sample. However, the tip convolution effect blurs images due to the shape of the tip end. An attempt to deconvolute assuming the shape of the tip end was made 2 decades ago52, but it has not yet improved the quality of the atomic force microscopy image dramatically when A-PoD can be applied as a solution. In addition, the resolution of super-resolved images can be further improved by using A-PoD. Structured illumination microscopy (SIM), one of the super-resolution imaging techniques, is an example. Studies on the super-resolved Raman imaging technique using SIM were published recently53,54. Although SIM improves spatial resolution over twofold by reducing the size of the PSF, the resolution of SIM images can be further increased using A-PoD, because deconvolution is also possible based on the reduced PSF. This approach has been applied using a different deconvolution program, Sparse-SIM55. Finally, A-PoD can be applied to astronomy56, which is a research field in which deconvolution is widely used. In fact, the Richardson–Lucy algorithm was originally published for astronomy studies57. In sum, the results shown in this study represent the beginning of many different applications of A-PoD from the nanoscale to the astronomic scale.
Online content
Any methods, additional references, Nature Portfolio reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at https://doi.org/10.1038/s41592-023-01779-1.
Methods
Image preprocessing
The image of the 1-μm bead was interpolated two times along the optical axis direction, and the retina image was interpolated six times in all directions. The 3D live-cell images were interpolated ten times along the optical axis direction. The measured DO-SRS images were resampled before deconvolution. For all resampling processes, the Fourier interpolation code about f-SOFI was used58. To increase the SNR, the PURE denoise filter was used ten times to reduce noise in imaging the standard bead; and the automatic correction of the sCMOS-related noise algorithm was used for the retina image59,60.
A-PoD algorithm
The A-PoD algorithm described in the paper was newly implemented for SRS analysis. We adopted the Adam solver as the optimization method and used a gradient algorithm instead of a genetic algorithm. The optimization method was changed to a gradient descent algorithm from a genetic algorithm. The optimization method used is the Adam solver61. Because the variables of A-PoD are positions of each virtual emitter, the numbers are set to the address value of the pixel. Therefore, all these numbers have integer values, and, for this, the gradient equation of the Adam solver was modified as follows.
Here, is an objective function for deconvolution of 3D images.
PSFs for deconvolution processes were simulated using the PSF generator in the ImageJ plugin according to the physical conditions of each measurement14,15. To efficiently process a 3D image, the image was deconvolved by dividing the image into several pieces as used in the SPIDER algorithm18. A-PoD was implemented using Tensorflow 1.15 and Python 3.6. The number of virtual emitters used was manually controlled under the condition that the image contrast improved. All calculations were performed on a Xeon W-2145 CPU with 64 GB of RAM and a NVIDIA Quadro P4000 GPU.
Lipid droplet analysis
After deconvolution, individual LDs were counted with the 3D object counter in ImageJ. Based on the information of position, volume, surface area and mean distance, we prepared the plots in Figs. 3 and 6 and Extended Data Figs. 5 and 6. After detection of individual LDs, we mapped the SA:V ratio using home-built Matlab code.
Standard beads
A colloid suspension of polystyrene beads 100 nm in diameter with a solid content of 1.0% (wt) (Thermo Scientific) was used in the following experiments. To tailor the suspension for the CAPA experiments, the colloidal solution was further diluted tenfold to a concentration of 0.1% (wt) (9.33 × 1,010 parts per ml) using deionized water.
Retinal section preparation
Human retinal tissue sections were obtained from a donor (age 83) (San Diego Eye Bank, CA, USA) with appropriate consent from the San Diego Eye Bank and following a protocol approved by the University of California, San Diego Human Research Protection Program. The donor had had no history of eye disease, diabetes or any neurological diseases. Following fixation, the retina was processed for cryostat sections (12 μm) and stored at –80 °C.
Frozen sections were defrosted (10 min, room temperature) and washed with 1× PBS three times, for 10 min each time, and then sandwiched between a 170-nm coverslip and a glass slide with PBS solution. The coverslips were sealed with nail polish.
MCF-7 breast cancer cells
MCF-7 cells were cultured in DMEM growth medium supplemented with 10 mg l−1 insulin (Sigma-Aldrich), 1% (vol/vol) penicillin–streptomycin mix (Fisher Scientific) and 5% (vol/vol) heat-inactivated FBS on a #1 thickness coverglass (GG-12-Laminin, Neuvitro) for 48 h. Cells were fixed with 4% (vol/vol) paraformaldehyde (PFA) solution for 15 min and then mounted on 1-mm-thick glass slides.
HEK293 cells
HEK293 cells were stably transfected with a plasmid for expressing monomeric red fluorescent protein containing a mitochondrial targeting sequence (Mito-RFP)62. Cells were cultured on coverglasses in 24-well cell culture dishes at 37 °C (5% CO2) in DMEM supplemented with 10% FBS (Atlanta Biological) and 1% penicillin–streptomycin (Fisher Scientific). Cells were fixed with 4% PFA in PBS. Following washes with PBS, the coverglasses were mounted in PBS before imaging.
HeLa cells
HeLa cells were cultured in DMEM, supplemented with 10% FBS and 1% penicillin–streptomycin (Fisher Scientific), and incubated with 5% CO2 at 37 °C. After passing at 80% confluence, cells were seeded at a concentration of 2 × 105 cells per ml onto a coverglass in a 24-well plate. DMEM with 0.5% FBS and 1% penicillin–streptomycin was used to synchronize the cells for 8 h. The medium was then changed to 50% (vol/vol) heavy water (D2O) and treatment medium as described below.
For the excess aromatic amino acid condition, phenylalanine and tryptophan were increased as two separate test conditions at a 15× concentration. L-phenylalanine powder (SLCF3873, Sigma-Aldrich) and L-tryptophan powder (SLCF2559, Sigma-Aldrich) were added to DMEM for the excess groups. Cells were then cultured for 36 h. Next, the cells were gently rinsed with 1× PBS with calcium and magnesium ions at 37 °C (Fisher Scientific, 14040216) and fixed in 4% methanol-free PFA solution (VWR, 15713-S) for 15 min. The coverglass was finally mounted on the cleaned 1-mm-thick glass microscope slides with 120-μm spacers filled with 1× PBS for imaging and spectroscopy. These samples were stored at 4 °C when not in use.
Drosophila
The w1118 parent flies were raised in vials containing standard food (Bloomington cornmeal–yeast–sugar recipe) at 25 °C in an environment with controlled light (12–12-h light–dark cycle) and humidity (>70%) for several generations. Embryos from the young females (~7 d old) were collected in a 4-h window to synchronize larval development. Two groups of 10–15 first instar larvae were placed in vials containing 20% D2O-labeled standard food (100 g yeast, 50 g sucrose, 5 g agar per liter) and 3× high-glucose food (100 g yeast, 150 g sucrose, 5 g agar per liter), respectively. The larvae were allowed to develop until the wandering third-instar stage, and then brains were dissected in PBS and fixed in 4% formaldehyde for 21 min at room temperature. After fixation, brains were washed four times with PBS in glass wells and were then sandwiched between a coverglass and the slide with PBS solution. To prevent tissue drying, nail polish was used to seal the surrounding coverglass.
STORM imaging
Mouse hippocampal neuronal culture and immunostaining were performed as described previously63. STORM imaging64 was performed on a custom inverted microscope (Applied Scientific Imaging) with a 60× Nikon objective (MRD01605). A custom Lumencor CELESTA system was used to illuminate the sample. A laser line (~1 W, 640 nm) was used to image a hippocampal neuron immunostained using anti-β II spectrin antibody conjugated to the Alexa 647 dye conjugated to the anti-spectrin antibody, and a laser line (~200 mW, 405 nm) was used to stimulate the cycling of the dyes. The Teledyne Kinetix camera was used for imaging at 50 Hz.
The other imaging conditions and the parameters for the DAOSTORM fitting and processing were set as described previously63.
The neuron culture was performed as described previously65.
Stimulated Raman scattering microscopy
A custom-built upright laser-scanning microscope (Olympus) with a 25× water objective (XLPLN, WMP2, 1.05 NA, Olympus) was applied for near-IR throughput. A synchronized pulsed pump beam (tunable wavelength, 720–990 nm; pulse width, 5–6 ps; repetition rate, 80 MHz) and a Stokes beam (wavelength at 1,032 nm; pulse width, 6 ps; repetition rate, 80 MHz) were supplied by a picoEmerald system (Applied Physics & Electronics) and coupled into the microscope. The pump and Stokes beams were collected in transmission by a high-NA oil condenser (1.4 NA). A high-O.D. shortpass filter (950 nm, Thorlabs) was used that would completely block the Stokes beam and transmit the pump beam only onto an Si photodiode for detecting the stimulated Raman loss signal. The output current from the photodiode was terminated, filtered and demodulated by a lock-in amplifier at 20 MHz. The demodulated signal was fed into the FV3000 software module FV-OSR (Olympus) to form an image during laser scanning. All images obtained were 512 × 512 pixels, with a dwell time of 80 μs and imaging speed of ~23 s per image.
Fluorescence microscopy
MPF microscopy was integrated with the DIY SRS microscopy together for imaging the same ROI with different modalities (DO-SRS signals and fluorescence signals). The Mitored signal was imaged with 800-nm ultrafast laser-scanning two-photon fluorescence excitation and detected by PMT with a 610-nm bandpass filter in front of it.
Extended Data
Extended Data Fig. 1 |. Comparison of A-PoD with Richardson-Lucy method using simulation data.
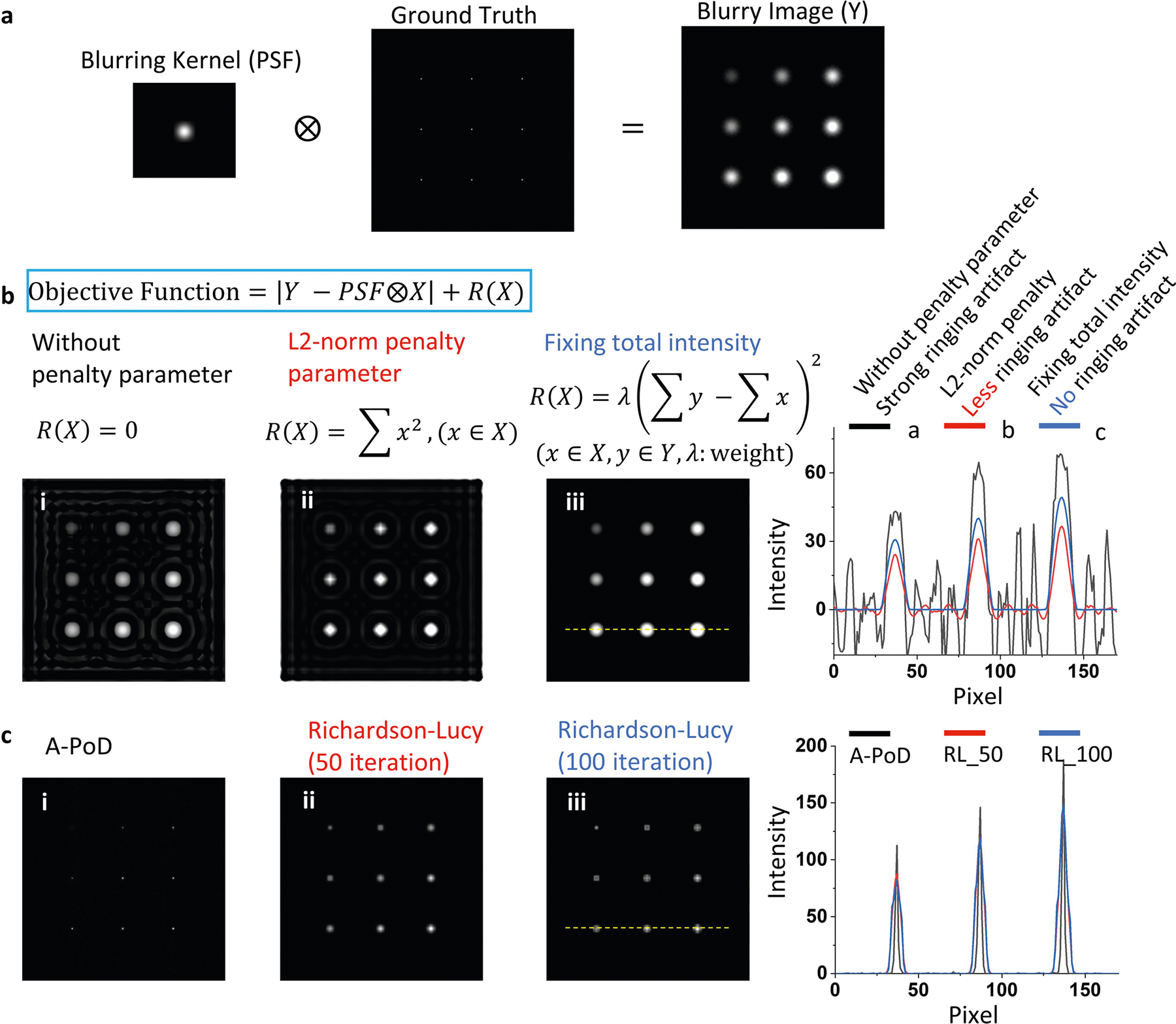
a. To compare different deconvolution methods, we generated an artificial image composed of single pixel sized 9 dots. The dots in the image have different intensity values. By convolution with an artificial PSF, a blurry image (Y) was generated. The image (Y) was deconvolved using a penalized regression method. b. When we minimize the objective function in panel b, the images, X results. Depending on the penalty parameter, R(X), X has various forms. The optimization result without any penalty parameter has strong ringing artifact as shown in panel b(i), and the result with L2-norm penalty parameter has reduced ringing artifact as shown in panel b(ii). By limiting summation of total intensity, we can reduce the ringing artifact as shown in panel b(iii). The penalty parameter limiting the total intensity as a fixed value makes the values in empty space to zeros. Accordingly, one of the main characteristics of A-PoD, the fixed total intensity of X, can increase sparsity of resulting images. c. Comparison of A-PoD with Richardson-Lucy method. When we apply another characteristic of A-PoD, quantization of intensity value, together, the resulting image of A-PoD has higher resolution than that obtained using Richardson-Lucy method. The signal intensity profile shows the difference in resolutions. The dots in the A-PoD image have narrower width than Richardon-Lucy images. The calculation time of A-PoD was 1.9 s, and Deconvolutionlab2 using Richardson-Lucy algorithm calculated the image for 1.1 s (50 iteration) and 2.2 s (100 iteration).
Extended Data Fig. 2 |. Precision and speed of A-PoD in comparison with SPIDER.
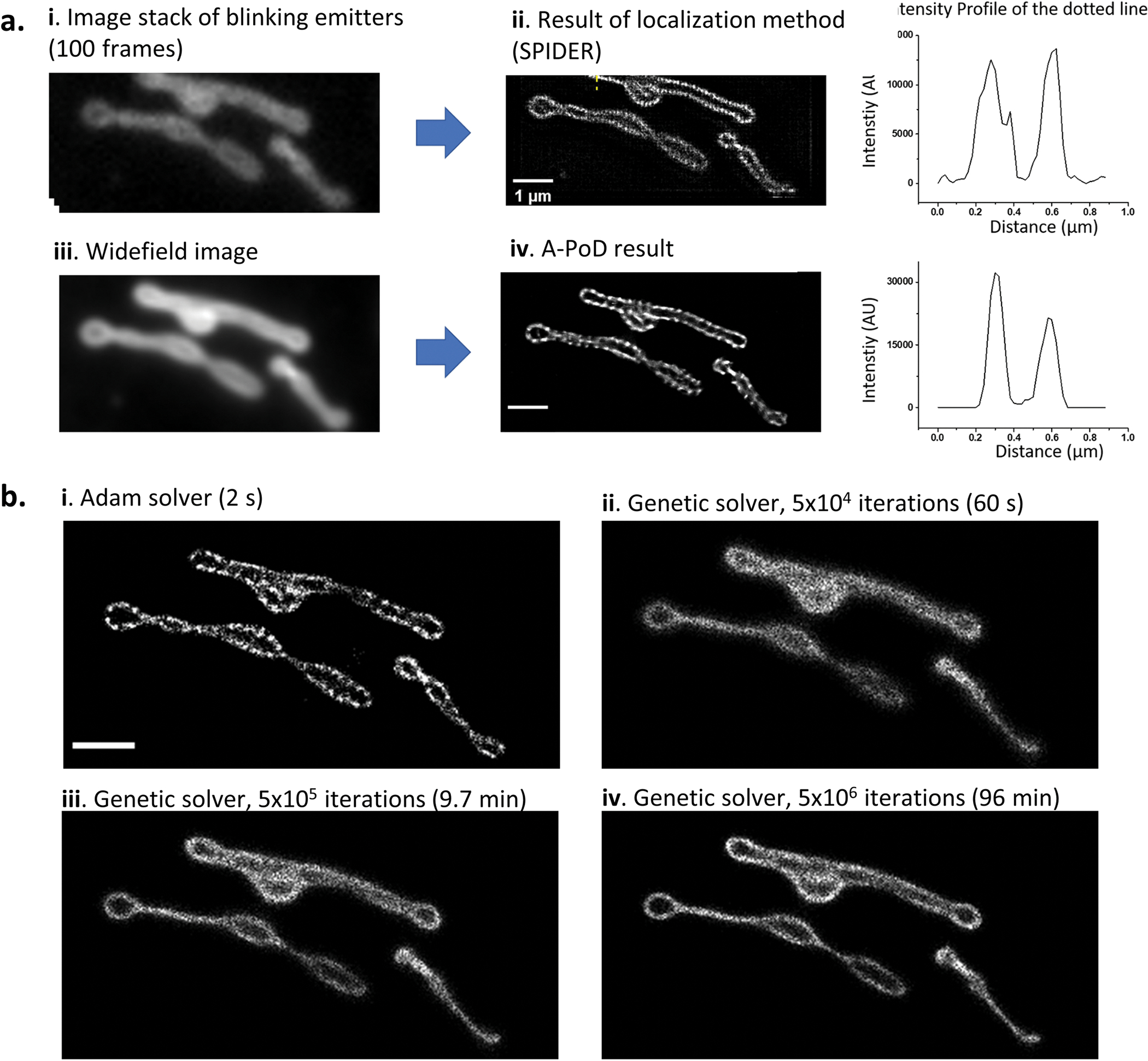
a. To compare the localization microscopy image with A-PoD result, we deconvolved a mitochondrial image. The image stack is composed of 100 frames. Each image frame contains information about blinking emitters. The emitters were localized using SPIDER deconvolution algorithm. By averaging the image stack, we generated a widefield image, and the widefield image was deconvolved using A-PoD. The intensity profiles of the cross-section in the deconvolved images show the similarity between the two results. b. Two optimization methods for the deconvolution process were compared. An image composed of 100000 virtual emitters was deconvolved using the two different optimizers. The results of Adam solver (i) finished calculation within 2 s. By increasing the iteration number, the deconvolution results using genetic solver (ii, iii, and iv with different iteration numbers) were compared with the result of Adam solver. The deconvolution result with a high iteration number shows more precise image. However, to generate an image having same quality as that obtained with the Adam solver, we need to increase the iteration number further beyond 5 × 106 more.
Extended Data Fig. 3 |. Comparison of the deconvolution results on STORM images using DAO STORM versus A-PoD.
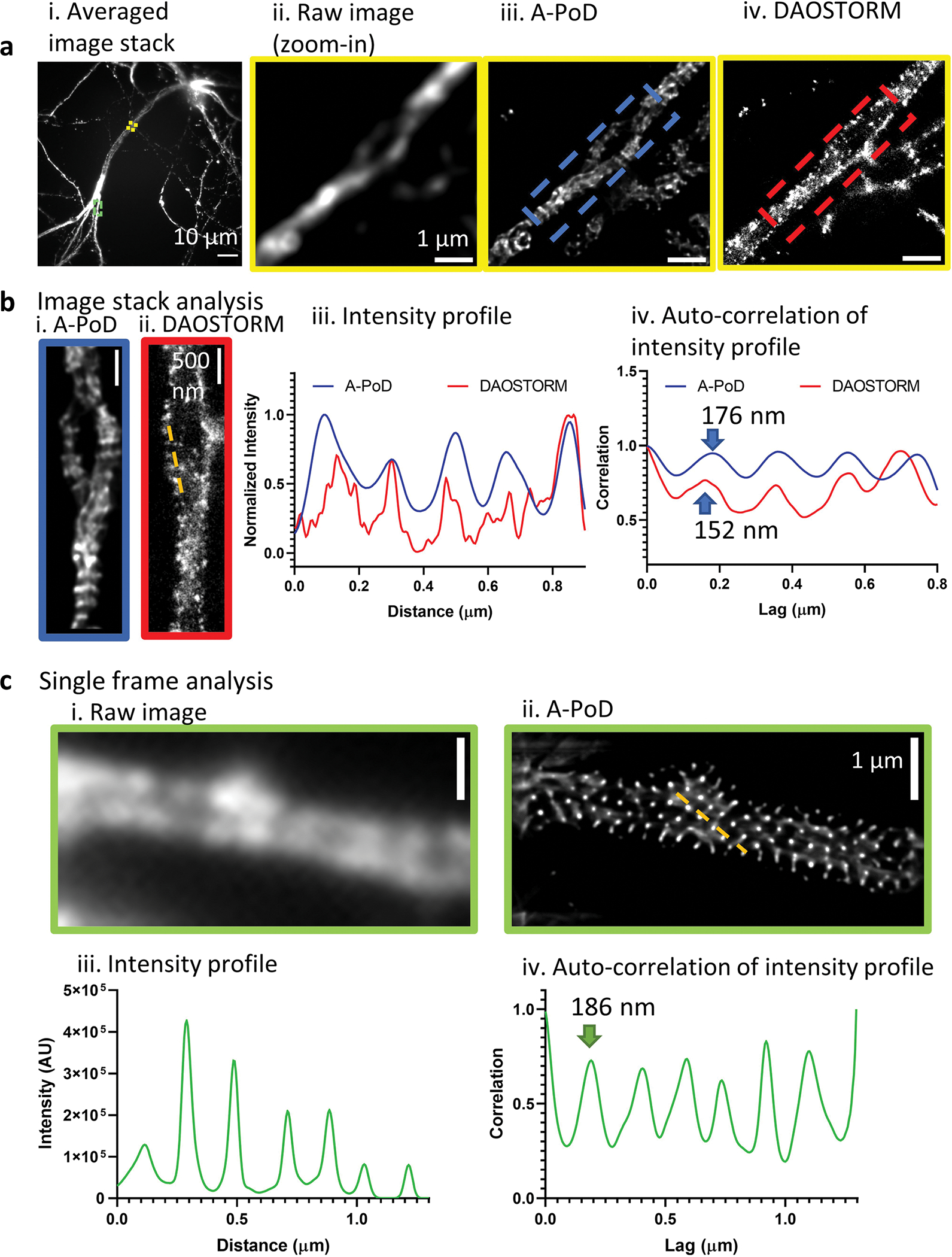
a. (i) A single ‘epifluorescence’-like image was calculated by averaging the STORM-stack. (ii) We selected an area with low emitter density (yellow rectangle region in (i)) than other areas. (iii) The averaged image stack of the chosen area was deconvolved using A-PoD. (iv) From the whole stack of the selected area, the individual single emitters were localized using DAOSTORM. b. The two areas marked by the blue and red rectangle areas in (a. i and b. ii) were selected. (iii and iv) The intensity profiles and auto-correlation data shows the periodicity of the structure of the membrane-associated periodic skeleton (MPS) in neurons. c. Another bright area with high emitter density (green rectangle area in a.i) where we cannot localize the individual molecules using DAOSTORM was selected. (i) From the image stack of the selected area, we chose a single frame. (ii) Using A-PoD, we deconvolved the chosen frame. (iii and iv) The intensity profile and the auto-correlation result show the periodicity. Due to the strong intensity, the periodic structure was clearly revealed, and the interval in the MPS is also close to the previous published result, 190 nm.
Extended Data Fig. 4 |. Comparison of two PSF models.
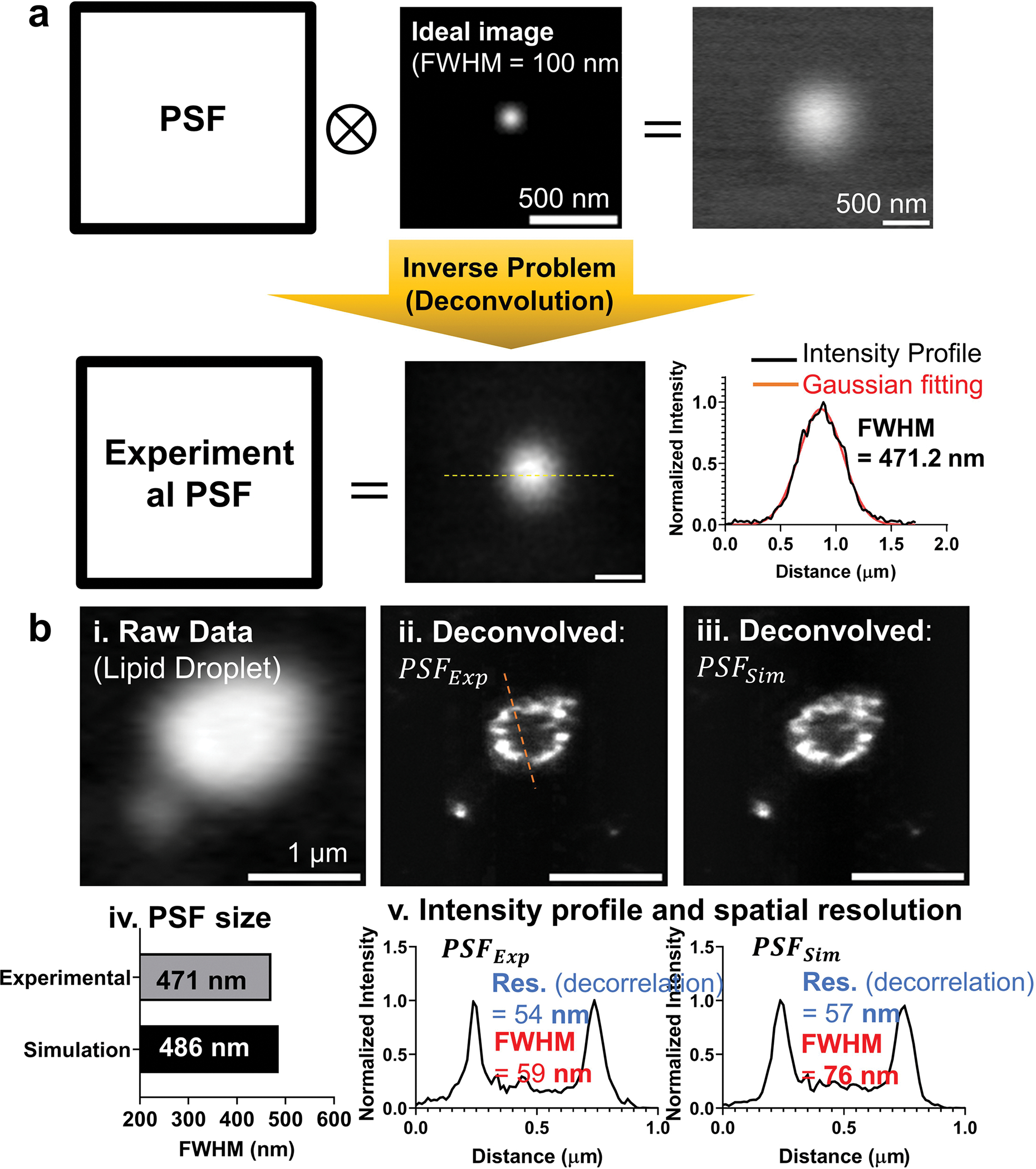
a. Experimental PSF was extracted from 100 nm bead image. As shown in a, by deconvolving the measured bead image with artificial 2D Gaussian image having 100 nm FWHM, experimental PSF was calculated. The FWHM of the experimental PSF was 471.2 nm. b. Single LD image was deconvolved using simulated PSF and experimental PSF. After deconvolution, the raw LD image (in b, i) was converted to the two images (in b, ii and iii). Two PSF has almost similar size with about 5% error (bar graphs in b, iv). From the intensity profiles of the two deconvolved images, membrane thickness was measured. The thinnest part has 59 nm and 76 nm for experimental PSF and simulated PSF, respectively. Spatial resolutions measured with the decorrelation method were 54 nm and 57 nm for experimental PSF and simulated PSF, respectively.
Extended Data Fig. 5 |. SRS images of a HeLa cell cultured in the standard medium.
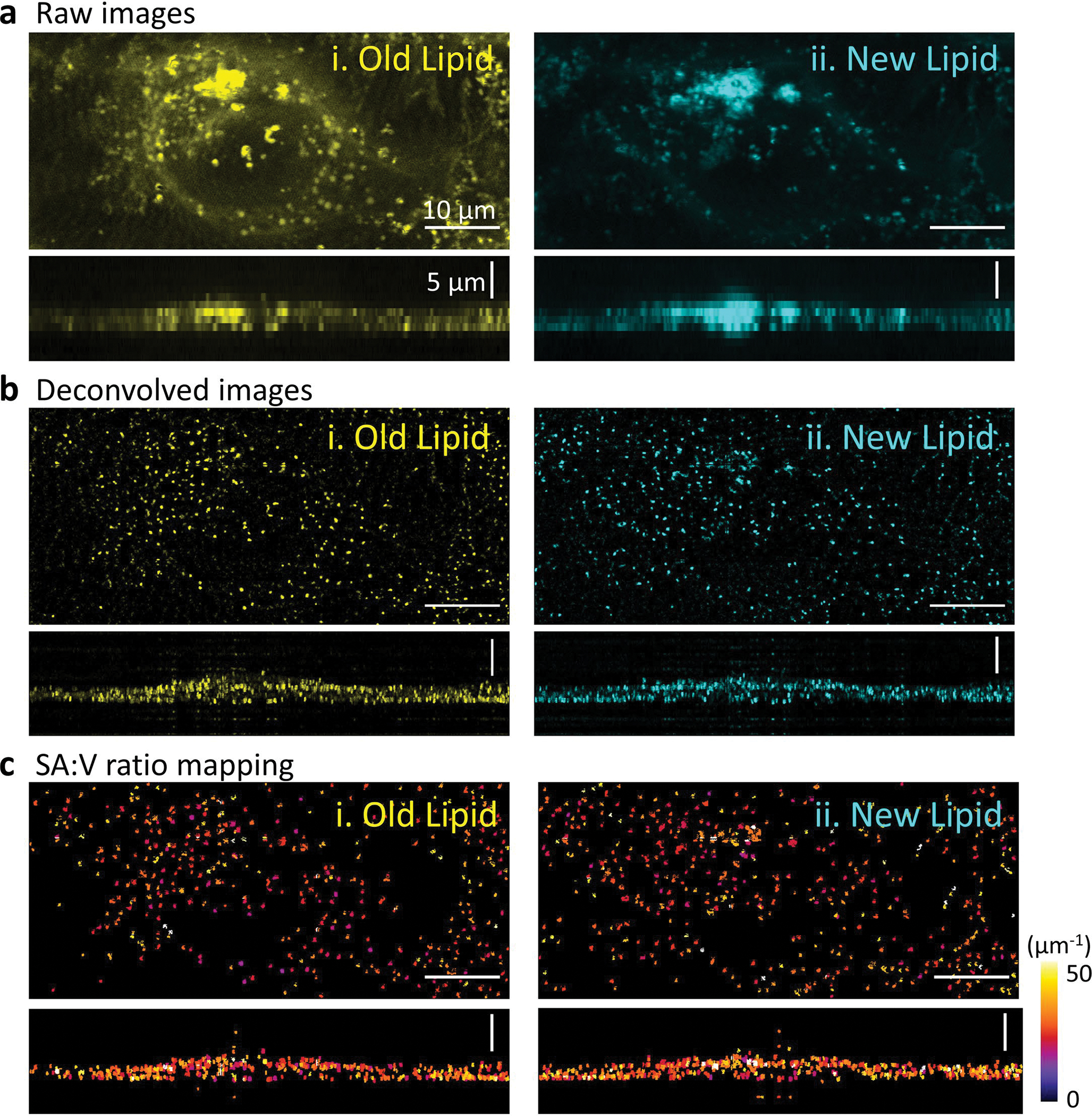
a. Raw DO-SRS images of the HeLa cell. b. Deconvolution results of the images. The images show the shape and distribution of the lipid droplets in sub-micron scale. c. After measuring the surface area and volume of individual lipid droplets, the surface area to volume ratio of individual LDs was mapped.
Extended Data Fig. 6 |. LD size and lipid turnover rate distribution.
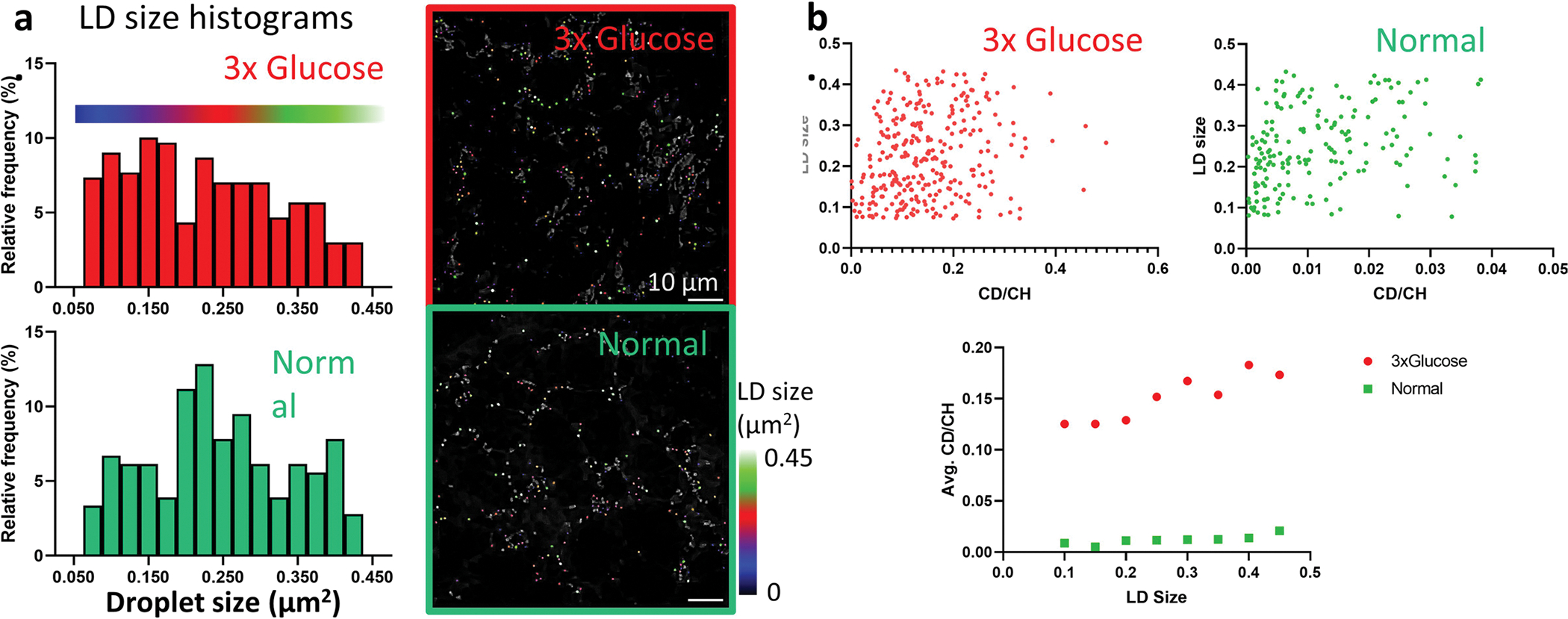
a. In flies fed on different diets, LDs have different size distribution. In high glucose group, the LD size was widely distributed, and the number of LDs in 0.1~0.2 μm2 range was higher than the other size. In control dietary condition, the control group with standard diet, the number of LDs in 0.2~0.3 μm2 range was high. LDs were labeled on the images with three colors according to the size (Blue, 0.05~0.2 μm2; Red, 0.2~0.3 μm2; Green, 0.3~0.45 μm2). b. To compare the LD size and lipid turnover rate, the two parameters of individual LDs were plotted. Under both conditions, LD size and lipid turnover rate show positive correlation. Correlation coefficient: 0.40 (3x glucose), 0.44 (control).
Extended Data Fig. 7 |. SRS images of larvae brain samples from flies fed on different diets.

a. DO-SRS images of a drosophila larvae brain in 3x glucose group. The wide range new lipid (CD) and old lipid (CH2) signal show the distribution of newly synthesized lipids and old lipids in whole sample, respectively. In the zoomed-in images, the microscopic distribution of two different lipid components is clearly shown. After deconvolution, the nanoscopic distribution and shape of lipid droplets are getting clearer. By using the particle analysis method, we can remove the background and focus on the areas of lipid droplets. b. SRS images of a drosophila larvae brain in the control group were processed with the same manner in a. These images were analyzed, and the analysis result is explained in Fig. 6.
Extended Data Fig. 8 |. Comparison between A-PoD and the Richardson-Lucy method.
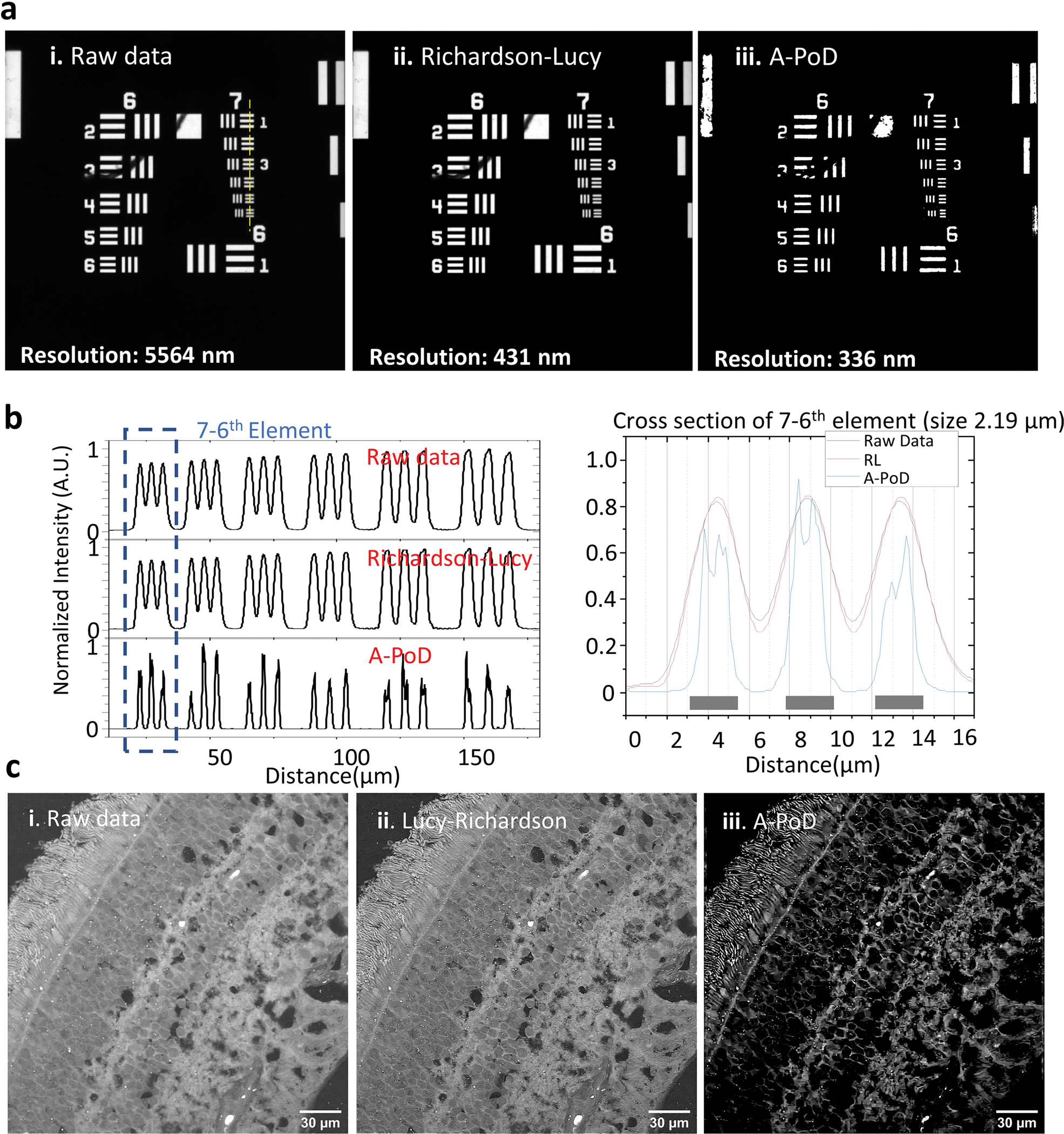
a. USAF-1951 resolution target. The fluorescence image of the resolution target in the paper65 was deconvolved using Richardson-Lucy algorithm (Deconvolutionlab2 program)20. b. Intensity profiles of the yellow dotted line in figure A show the resolution difference. A-PoD result resolved each line perfectly, but Richardson-Lucy result could not resolve them. c. Deconvolution results of retinal tissue image. The raw image (i) was deconvolved with Richardson-Lucy algorithm (ii) and A-PoD (iii). The image contrast was significantly improved when we used A-PoD for deconvolution.
Supplementary Material
Acknowledgements
We thank K. Zhang, W. Min and J. Enderlein for helpful discussions and suggestions. Thanks to M. Shtrahman and S. Saidi for providing 1 μm bead samples. We acknowledge University of California, San Diego startup funds, NIH U54CA132378, NIH 5R01NS111039, NIH R21NS125395, NIH U54DK134301, NIH U54 HL165443 and a Hellman Fellow Award.
Footnotes
Competing interests
A provisional patent application has been filed by the UC San Diego patent office for L.S. and H.J. under the title ‘SUPER-RESOLUTION STIMULATED RAMA SCATTERING MICROSCOPY WITH A-POD’, U.S. provisional application serial no. _63/379,226_, filed 12 October 2022. All other authors declare no competing interests.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Code availability
Exemplary data and source code for A-PoD with explanations about parameters and the installation protocol are available at https://github.com/lingyanshi2020/A-PoD/.
Additional information
Extended data is available for this paper at https://doi.org/10.1038/s41592-023-01779-1.
Supplementary information The online version contains supplementary material available at https://doi.org/10.1038/s41592-023-01779-1.
Reprints and permissions information is available at www.nature.com/reprints.
Data availability
All the data supporting the findings of this study are available within the paper and its Supplementary Information.
References
- 1.Freudiger CW et al. Label-free biomedical imaging with high sensitivity by stimulated Raman scattering microscopy. Science 322, 1857–1861 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ploetz E, Laimgruber S, Berner S, Zinth W & Gilch P Femtosecond stimulated Raman microscopy. Appl. Phys. B 87, 389–393 (2007). [Google Scholar]
- 3.Shi L et al. Optical imaging of metabolic dynamics in animals. Nat. Commun. 9, 2995 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ao J et al. Switchable stimulated Raman scattering microscopy with photochromic vibrational probes. Nat. Commun. 12, 3089 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Qian C et al. Super-resolution label-free volumetric vibrational imaging. Nat. Commun. 12, 3648 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xiong H et al. Super-resolution vibrational microscopy by stimulated Raman excited fluorescence. Light Sci. Appl. 10, 87 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gong L, Zheng W, Ma Y & Huang Z Saturated stimulated-Raman-scattering microscopy for far-field superresolution vibrational imaging. Phys. Rev. Appl. 11, 034041 (2019). [Google Scholar]
- 8.Gong L & Wang H Breaking the diffraction limit by saturation in stimulated-Raman-scattering microscopy: a theoretical study. Phys. Rev. A 90, 013818 (2014). [Google Scholar]
- 9.Gong L & Wang H Suppression of stimulated Raman scattering by an electromagnetically-induced-transparency-like scheme and its application for super-resolution microscopy. Phys. Rev. A 92, 023828 (2015). [Google Scholar]
- 10.Silva WR, Graefe CT & Frontiera RR Toward label-free super-resolution microscopy. ACS Photonics 3, 79–86 (2016). [Google Scholar]
- 11.Shi L et al. Super-resolution vibrational imaging using expansion stimulated Raman scattering microscopy. Adv. Sci. 9, 2200315 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tzang O, Pevzner A, Marvel RE, Haglund RF & Cheshnovsky O Super-resolution in label-free photomodulated reflectivity. Nano Lett. 15, 1362–1367 (2015). [DOI] [PubMed] [Google Scholar]
- 13.Guilbert J et al. Label-free super-resolution chemical imaging of biomedical specimens. Preprint at bioRxiv 10.1101/2021.05.14.444185 (2021). [DOI] [Google Scholar]
- 14.Kirshner H, Aguet F, Sage D & Unser M 3-D PSF fitting for fluorescence microscopy: implementation and localization application. J. Microsc. 249, 13–25 (2013). [DOI] [PubMed] [Google Scholar]
- 15.Sage D et al. DeconvolutionLab2: an open-source software for deconvolution microscopy. Methods 115, 28–41 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Zhu L, Zhang W, Elnatan D & Huang B Faster STORM using compressed sensing. Nat. Methods 9, 721–723 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Min J et al. FALCON: fast and unbiased reconstruction of high-density super-resolution microscopy data. Sci. Rep. 4, 4577 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hugelier S et al. Sparse deconvolution of high-density super-resolution images. Sci. Rep. 6, 21413 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martínez S, Toscani M & Martinez OE Superresolution method for a single wide-field image deconvolution by superposition of point sources. J. Microsc. 275, 51–65 (2019). [DOI] [PubMed] [Google Scholar]
- 20.Holden SJ, Uphoff S & Kapanidis AN DAOSTORM: an algorithm for high-density super-resolution microscopy. Nat. Methods 8, 279–280 (2011). [DOI] [PubMed] [Google Scholar]
- 21.Descloux A, Grußmayer KS & Radenovic A Parameter-free image resolution estimation based on decorrelation analysis. Nat. Methods 16, 918–924 (2019). [DOI] [PubMed] [Google Scholar]
- 22.Shi L, Rodríguez-Contreras A & Alfano RR Gaussian beam in two-photon fluorescence imaging of rat brain microvessel. J. Biomed. Opt. 19, 126006 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chaigneau E, Wright AJ, Poland SP, Girkin JM & Silver RA Impact of wavefront distortion and scattering on 2-photon microscopy in mammalian brain tissue. Opt. Express 19, 22755–22774 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tzarouchis D & Sihvola A Light scattering by a dielectric sphere: perspectives on the Mie resonances. Appl. Sci. 8, 184 (2018). [Google Scholar]
- 25.Ji N, Milkie DE & Betzig E Adaptive optics via pupil segmentation for high-resolution imaging in biological tissues. Nat. Methods 7, 141–147 (2010). [DOI] [PubMed] [Google Scholar]
- 26.Zhang B, Zhu J, Si K & Gong W Deep learning assisted zonal adaptive aberration correction. Front. Phys. 8, 634 (2021). [Google Scholar]
- 27.Boesze-Battaglia K & Yeagle PL Rod outer segment disc membranes are capable of fusion. Invest. Ophthalmol. Vis. Sci. 33, 484–493 (1992). [PubMed] [Google Scholar]
- 28.Abramczyk H et al. The role of lipid droplets and adipocytes in cancer. Raman imaging of cell cultures: MCF10A, MCF7, and MDA-MB-231 compared to adipocytes in cancerous human breast tissue. Analyst 140, 2224–2235 (2015). [DOI] [PubMed] [Google Scholar]
- 29.Bagheri P, Hoang K, Fung AA, Hussain S & Shi L Visualizing cancer cell metabolic dynamics regulated with aromatic amino acids using DO-SRS and 2PEF microscopy. Front. Mol. Biosci. 8, 779702 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fung A et al. Imaging sub-cellular methionine and insulin interplay in triple negative breast cancer lipid droplet metabolism. Front. Oncol. 12, 858017 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jarc E & Petan T Focus: organelles: lipid droplets and the management of cellular stress. Yale J. Biol. Med. 92, 435–452 (2019). [PMC free article] [PubMed] [Google Scholar]
- 32.Li X et al. Quantitative imaging of lipid synthesis and lipolysis dynamics in Caenorhabditis elegans by stimulated Raman scattering microscopy. Anal. Chem. 91, 2279–2287 (2018). [DOI] [PubMed] [Google Scholar]
- 33.Lisec J, Jaeger C, Rashid R, Munir R & Zaidi N Cancer cell lipid class homeostasis is altered under nutrient-deprivation but stable under hypoxia. BMC Cancer 19, 501 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paar M et al. Remodeling of lipid droplets during lipolysis and growth in adipocytes. J. Biol. Chem. 287, 11164–11173 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rysman E et al. De novo lipogenesis protects cancer cells from free radicals and chemotherapeutics by promoting membrane lipid saturation. Cancer Res. 70, 8117–8126 (2010). [DOI] [PubMed] [Google Scholar]
- 36.Schott MB et al. Lipid droplet size directs lipolysis and lipophagy catabolism in hepatocytes. J. Cell Biol. 218, 3320–3335 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schug Z et al. Acetyl-CoA synthetase 2 promotes acetate utilization and maintains cell growth under metabolic stress. Cancer Cell 27, 57–71 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wolins NE et al. S3–12, adipophilin, and TIP47 package lipid in adipocytes. J. Biol. Chem. 280, 19146–19155 (2005). [DOI] [PubMed] [Google Scholar]
- 39.Li Y, Zhang W, Fung AA & Shi L DO-SRS imaging of diet regulated metabolic activities in Drosophila during aging processes. Aging Cell 21, e13586 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li Y, Zhang W, Fung AA & Shi L DO-SRS imaging of metabolic dynamics in aging Drosophila. Analyst 146, 7510–7519 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li Y et al. Direct imaging of lipid metabolic changes in Drosophila ovary during aging using DO-SRS microscopy. Front. Aging 2, 819903 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lu F-K et al. Label-free DNA imaging in vivo with stimulated Raman scattering microscopy. Proc. Natl Acad. Sci. USA 112, 11624–11629 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wei M et al. Volumetric chemical imaging by clearing-enhanced stimulated Raman scattering microscopy. Proc. Natl Acad. Sci. USA 116, 6608–6617 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bae K et al. Mapping the intratumoral heterogeneity in glioblastomas with hyperspectral stimulated Raman scattering microscopy. Anal. Chem. 93, 2377–2384 (2021). [DOI] [PubMed] [Google Scholar]
- 45.Gong L, Lin S & Huang Z Stimulated Raman scattering tomography enables label-free volumetric deep tissue imaging. Laser Photonics Rev. 15, 2100069 (2021). [Google Scholar]
- 46.Shi L et al. Highly-multiplexed volumetric mapping with Raman dye imaging and tissue clearing. Nat. Biotechnol. 40, 364–373 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wilfling F, Haas JT, Walther TC & Farese RV Jr. Lipid droplet biogenesis. Curr. Opin. Cell Biol. 29, 39–45 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wilfling F et al. Triacylglycerol synthesis enzymes mediate lipid droplet growth by relocalizing from the ER to lipid droplets. Dev. Cell 24, 384–399 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Back SH & Kaufman RJ Endoplasmic reticulum stress and type 2 diabetes. Annu. Rev. Biochem. 81, 767–793 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yamamoto K et al. Induction of liver steatosis and lipid droplet formation in ATF6α-knockout mice burdened with pharmacological endoplasmic reticulum stress. Mol. Biol. Cell 21, 2975–2986 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moncan M et al. Regulation of lipid metabolism by the unfolded protein response. J. Cell. Mol. Med. 25, 1359–1370 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tabet M & Urban KF III. Deconvolution of tip affected atomic force microscope images and comparison to Rutherford back-scattering spectrometry. J. Vac. Sci. Technol. B 15, 800–804 (1997). [Google Scholar]
- 53.Lee H et al. Super-resolved Raman microscopy using random structured light illumination: concept and feasibility. J. Chem. Phys. 155, 144202 (2021). [DOI] [PubMed] [Google Scholar]
- 54.Watanabe K et al. Structured line illumination Raman microscopy. Nat. Commun. 6, 10095 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhao W et al. Sparse deconvolution improves the resolution of live-cell super-resolution fluorescence microscopy. Nat. Biotechnol. 40, 606–617 (2021). [DOI] [PubMed] [Google Scholar]
- 56.Starck JL, Pantin E & Murtagh F Deconvolution in astronomy: a review. Publ. Astron. Soc. Pac. 114, 1051–1069 (2002). [Google Scholar]
- 57.Lucy LB An iterative technique for the rectification of observed distributions. Astron. J. 79, 745–754 (1974). [Google Scholar]
- 58.Stein SC, Huss A, Hähnel D, Gregor I & Enderlein J Fourier interpolation stochastic optical fluctuation imaging. Opt. Express 23, 16154–16163 (2015). [DOI] [PubMed] [Google Scholar]
- 59.Mandracchia B et al. Fast and accurate sCMOS noise correction for fluorescence microscopy. Nat. Commun. 11, 94 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Blu T & Luisier F The SURE-LET approach to image denoising. IEEE Trans. Image Process. 16, 2778–2786 (2007). [DOI] [PubMed] [Google Scholar]
- 61.Kingma DP & Ba J Adam: a method for stochastic optimization. Preprint at arXiv 10.48550/arXiv.1412.6980 (2014). [DOI] [Google Scholar]
- 62.Deng J et al. FUS interacts with HSP60 to promote mitochondrial damage. PLoS Genet. 11, e1005357 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bintu B et al. Super-resolution chromatin tracing reveals domains and cooperative interactions in single cells. Science 362, eaau1783 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rust MJ, Bates M & Zhuang X Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat. Methods 3, 793–796 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhou R, Han B, Xia C & Zhuang X Membrane-associated periodic skeleton is a signaling platform for RTK transactivation in neurons. Science 365, 929–934 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data supporting the findings of this study are available within the paper and its Supplementary Information.


