Abstract
A non-redundant database of nuclear, protein-encoding, genomic DNA sequences highlighting nuclear pre-mRNA introns was constructed using information contained in the SWISS-PROT and GenBank sequence databases. This Intron DataBase (IDB) contains information about (i) introns (including nucleotide sequence, location, phase, length, GC content and consensus-sequence rule violations), (ii) exons (including nucleotide sequence, length and GC content), (iii) protein coding regions (including amino acid sequence and length), and (iv) descriptive information about the source gene and organism (including gene designations and species taxonomy). The Intron Evolution DataBase (IEDB) provides a statistical analysis of the exon and intron sequences catalogued in IDB as well as data concerning intron penetration (relative number of coding regions with introns), density (number of introns per kb of total coding sequence DNA), distribution, and consensus sequences for each species present in IDB. This supplement is provided to furnish insights into the phylogenetic distribution and evolution of introns. Both databases are extensively cross-referenced to the SWISS-PROT and GenBank databases. IDB currently contains information on over 63 000 genes and 154 000 introns; IEDB summarizes information on over 2800 species. IDB and IEDB will be updated twice a year and are available via the internet (http://nutmeg.bio.indiana.edu/intron/index.html ).
INTRODUCTION
GenBank (1), the main repository of nucleotide sequence information, contains more than 1.5 × 109 nucleotides in 2.2 × 106 entries representing over 40 000 distinct species and doubles in size every 15 months. As a result of this information explosion, data subsets that may be used to address specific problems in bioinformatics or molecular evolution are becoming increasingly difficult to extract. Numerous databases have evolved to meet such specialized needs (2) and usually embody increased levels of annotation or derivative analyses. Indeed many distinctive attributes of gene structure or function such as promoters (3), transcriptional regulatory regions (4) or signal sequences (5) are now represented in single-theme databases. Other gene subsequences, such as nuclear pre-mRNA introns, are poorly annotated in GenBank and as a result are more difficult to extract for derivative database construction.
Over the years many authors have published analyses of intron/exon structures and some have even developed relevant databases (6–11). However, as the number of sequences in GenBank grows larger and genomic redundancy (i.e., number of members in a particular gene family) increases, such efforts will become increasingly difficult and ultimately will require specialized knowledge of relational database structure and programming techniques. To address this problem, we present a suite of relational databases designed to serve as a comprehensive source of exon and intron sequence information as well as an analytical tool to facilitate phylogenetic-based statistical analysis of exons and introns.
DATABASE CONSTRUCTION
The SWISS-PROT (12) flat file databases (SPROT, TrEMBL and TrEMBL.NEW) were downloaded from the ExPASy server (ftp.expasy.ch) and all nuclear protein genes were extracted and merged into a local relational database. GenBank (1) cross-references were extracted for each species from individual SWISS-PROT entries, downloaded from NCBI (http://www. ncbi.nlm.nih.gov ) and analyzed such that the most recent, complete DNA sequences were used to build a second, relational database which contained coding region, exon and intron sequences. These data were derived from analysis of the relevant join statements in the GenBank entry feature table as identified by the Protein IDentification number (PID) contained in the SWISS-PROT cross reference. In addition to sequence information, accession numbers (including cross references to SWISS-PROT and GenBank), gene designations and descriptions, species names and taxonomic information, and derivative quantities such as sequence position, length and GC content were also recorded (see Fig. 1 for a complete description of the structure and graphical user interface of this database). Comprehensive error checking was built into these data generation algorithms such that any sequence ambiguities (including coding sequence initiation, termination and reading frame anomalies, and intron splice site-consensus rule violations) were logged and checked. Sequences that contained input artifactual errors were excluded from the database. Partial sequences were also identified and categorized as 5′ or 3′ deletions. This approach to database generation has an advantage over methodologies such as that employed by Long et al. (9) because sequence identities are minimized (i.e., the SWISS-PROT progenitor database is designed to be non-redundant) without resorting to extensive BLAST analyses to eliminate duplicate entries. In the few cases where duplicate SWISS-PROT entries existed (most commonly in the TrEMBL or TrEMBL.NEW databases), keyword and string matching algorithms were employed on curated descriptive annotations to yield a single entry for subsequent analysis. Unlike the database developed by Long et al. (9), genes which do not contain introns are included in the IDB database; this allows for measurement of parameters such as intron penetration (percentage of genes with introns) and intron density (number of introns/kb coding sequence) within particular species or gene families. Finally, since the IDB contains both intron containing and non-intron containing sequences, it can also be used as a non-redundant database for all genomic sequences from eukaryotic protein-encoding genes.
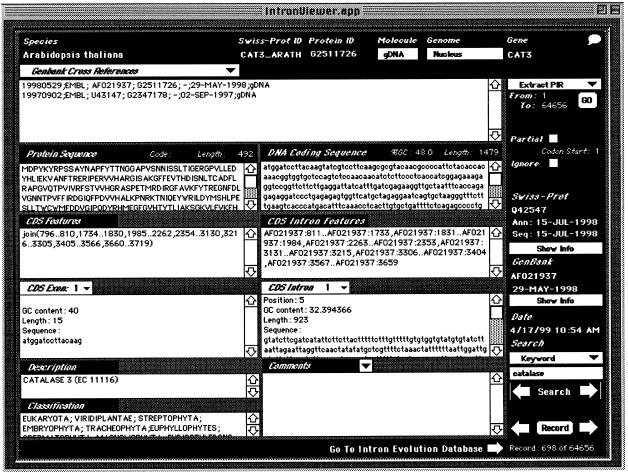
Figure 1.
Structure and Macintosh™ graphical user interface of the IDB. From left, top to bottom: fields are shown that identify the species, molecule, genome, and gene from which a particular nucleotide sequence is derived. SWISS-PROT Identification codes and the protein PID are also noted. Directly below the species and molecular identifiers is a list of all known database cross references followed by the coding region amino acid and DNA sequences. GC content and the length of particular molecules are noted where appropriate. The GenBank coding sequence join statement and derivative intron locations used to extract individual exon and intron sequences are available via pull-down menus. Position, phase, GC content, and sequence length are noted where appropriate. A description of the gene and the species classification taken from the GenBank Taxonomy database are also provided. The right panel contains additional information concerning the particular cross-references from SWISS-PROT and GenBank (including all accession numbers and database update dates) that were used to generate the database entry. The user may select from various navigational modes that allow examination of the database sequentially, by user-defined record, or by a search of genus, species, or protein description. User-defined ranges of records may be exported as text files for incorporation into other databases.
The data contained in the Intron DataBase (IDB) was summarized on a species by species basis to yield the Intron Evolution DataBase (IEDB). One version (identified in Table 1 as the redundant database) used all available data present in IDB; a second was produced using the Pfam database (13) to eliminate all but one representative sequence from each protein family within a species (identified in Table 1 as the non-redundant database). Use of the curated Pfam database that is generated from hidden Markov model profiles would be expected to eliminate sequence redundancy within potentially divergent protein families more efficiently than the uniformly applied 20% similarity criterion used in the FASTA alignments advocated by Long et al. (9) for this purpose. Appropriate use of individual IEDB databases thus ensures minimization of protein family bias (e.g., elimination of paralogous sequences) and maintenance of orthologous genes for the assessment of species-specific attributes. Statistical measures of central tendency and distribution for a variety of coding sequence, exon and intron attributes such as position, length and GC content were included for each species in the IEDB (see Fig. 2 for a complete description of the structure and graphical user interface of this database). Statistical outliers (defined as the most extreme 1% of a particular dataset for each species) were cross-referenced to IDB sequence entries, which facilitates inspection of potentially interesting data subsets. Finally, measurements of intron penetration, density, distribution, consensus patterns and mutations were included for each species in IEDB.
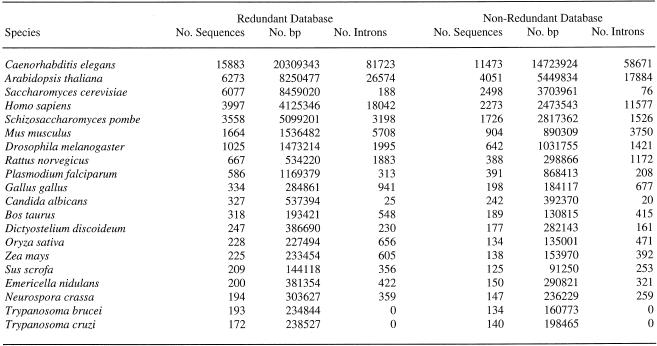
Table 1. Top 20 species in the IEDB.
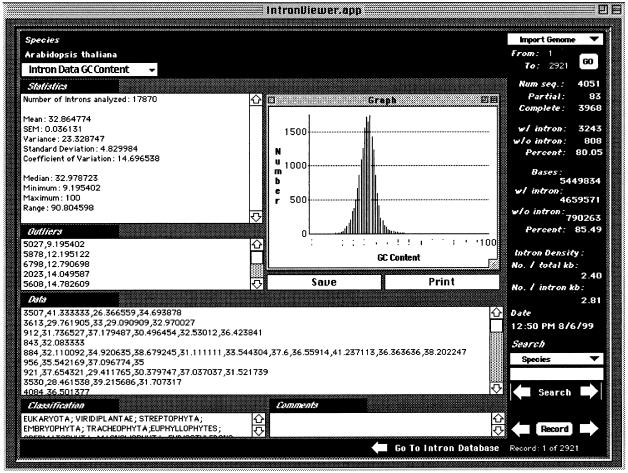
Figure 2.
Structure and Macintosh™ graphical user interface of the IEDB. From left, top to bottom: the IEDB summarizes all information in the IDB for a particular species. Information on number, GC content, length, and distribution of both introns and exons; intron phase; and splice-site consensus sequences and mutations may be obtained using the pull-down menu. A full range of statistics are included for each of these quantities including mean, standard error of the mean (SEM), variance, standard deviation, coefficient of variation, medium, minimum, maximum, and range. Distribution column graphs are available for most data types; the most extreme 1% of the data are identified as outliers. A Chi-squared test of the relative frequencies of phase 0, 1 and 2 introns and a Kolmogorov-Smirnov analysis of intron position distributions within the species’ genes are also provided. The right panel summarizes the number and type of sequences analyzed for a particular species, the intron penetration (relative number of coding regions with introns), and the intron density (defined as either the number of introns per kb of total coding sequence DNA or the number of introns per kb of intron-containing coding sequence DNA. The user may navigate this database in a fashion akin to the IDB and may export information or create spreadsheet-compatible reports as necessary. All data points are cross-referenced with the IDB.
CONTENTS OF THE CURRENT RELEASE
The most recent version of the IDB and IEDB databases is based on information obtained from releases 37 and 111 of the SWISS-PROT and GenBank databases, respectively, with updates to March 31, 1999. A total of 313 465 proteins from 13 616 species were analyzed for inclusion in IDB. Of these, 78 350 proteins were obtained from the SPROT division, 178 468 from the TrEMBL division and 56 647 from the TrEMBL.NEW division of the SWISS-PROT database.
The current release of IDB and IEDB comprises over 63 000 genes and approximately 154 000 introns from more than 2800 species. Table 1 summarizes the number of sequences and introns obtained from the top 20 most sequenced species in the database.
FUTURE PROSPECTS
It is our intent to provide two releases of IDB and IEDB per year, coinciding with major releases of the SWISS-PROT database. Announcements concerning the availability of these releases will be made through the appropriate bionet newsgroups.
Currently, only introns from the coding regions of protein-encoding nuclear genes are included in the databases; it is our hope to extend coverage to introns located in the 5′ and 3′ untranslated regions of these genes in a future release. The feasibility of including group I and group II introns is also being investigated.
AVAILABILITY AND CITATION
The IDB and IEDB databases are available for download through the internet (http://nutmeg.bio.indiana.edu/intron/index.html ) in either a text-based flat-file format that may be imported into a variety of database programs or a relational format readable by included applications developed in house for the Macintosh™ computer. A cgi-based search engine is currently under development for browsing the databases over the internet.
No inclusion of IDB or IEDB into other databases is allowed without the explicit permission of the authors. All rights are reserved. Users are asked to cite this publication when reporting results based on the use of either database.
Acknowledgments
ACKNOWLEDGEMENT
This work is supported by the National Science Foundation (MCD-9318858).
REFERENCES
- 1.Benson D.A., Boguski,M., Lipman,D.J., Ostell,J., Ouellete,B.F., Rapp,B.A. and Wheeler,D.L. (1999) Nucleic Acids Res., 27, 12–17. Updated article in this issue: Nucleic Acids Res. (2000), 28, 15–18.9847132 [Google Scholar]
- 2.Burks C. (1999) Nucleic Acids Res., 27, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Perier R.C., Junier,T., Bonnard,C. and Bucher,P. (1999) Nucleic Acids Res., 27, 307–309. Updated article in this issue: Nucleic Acids Res. (2000), 28, 302–303.9847211 [Google Scholar]
- 4.Kolchanov N.A., Ananko,E.A., Podkolodnaya,O.A., Ignatieva,E.V., Stepanenko,I.L., Kel-Margoulis,O.V., Kel,A.E., Merkulova,T.I., Goryachkovskaya,T.N., Busygina,T.V., Kolpakov,F.A., Podkolodny,N.L., Naumochkin,A.N. and Romashchenko,A.G. (1999) Nucleic Acids Res., 27, 303–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ponting C.P., Schultz,J., Milpetz,F. and Bork,P. (1999) Nucleic Acids Res., 27, 229–232. Updated article in this issue: Nucleic Acids Res. (2000), 28, 231–234.9847187 [Google Scholar]
- 6.Hawkins J.D. (1988) Nucleic Acids Res., 16, 9893–9908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dorit R.L. and Gilbert,W. (1991) Curr. Opin. Genet. Dev., 1, 464–469. [DOI] [PubMed] [Google Scholar]
- 8.Mount S.M., Burks,C., Hertz,G., Stormo,G.D., White,O. and Fields,C. (1992) Nucleic Acids Res., 20, 4255–4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Long M., Rosenberg,C. and Gilbert,W. (1995) Proc. Natl Acad. Sci. USA, 92, 12495–12499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fedorov A., Fedorova,L., Starshenko,V., Filatov,V. and Grigorev,E. (1998) J. Mol. Evol., 46, 263–271. [DOI] [PubMed] [Google Scholar]
- 11.Deutsch M. and Long,M. (1999) Nucleic Acids Res., 27, 3219–3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bairoch A. and Apweiler,R. (1999) Nucleic Acids Res., 27, 49–54. Updated article in this issue: Nucleic Acids Res. (2000), 28, 45–48.9847139 [Google Scholar]
- 13.Bateman A., Birney,E., Durbin,R., Eddy,S.R., Finn,R.D. and Sonnhammer,E.L.L. (1999) Nucleic Acids Res., 27, 260–262. Updated article in this issue: Nucleic Acids Res. (2000), 28, 263–266.9847196 [Google Scholar]