Abstract
Leukocyte infiltration accelerates brain injury following intracerebral hemorrhage (ICH). Yet, the involvement of T lymphocytes in this process has not been fully elucidated. Here, we report that CD4+ T cells accumulate in the perihematomal regions in the brains of patients with ICH and ICH mouse models. T cells activation in the ICH brain is concurrent with the course of perihematomal edema (PHE) development, and depletion of CD4+ T cells reduced PHE volumes and improved neurological deficits in ICH mice. Single-cell transcriptomic analysis revealed that brain-infiltrating T cells exhibited enhanced proinflammatory and proapoptotic signatures. Consequently, CD4+ T cells disrupt the blood-brain barrier integrity and promote PHE progression through interleukin-17 release; furthermore, the TRAIL-expressing CD4+ T cells engage DR5 to trigger endothelial death. Recognition of T cell contribution to ICH-induced neural injury is instrumental for designing immunomodulatory therapies for this dreadful disease.
CD4+ T cells are recruited into brain and worsen hemorrhagic stroke injury involving pro-inflammatory and pro-apoptotic action.
INTRODUCTION
The prognosis of intracerebral hemorrhage (ICH) is devastating (1). Evidence from preclinical and clinical studies suggests the detrimental role of lymphocyte-driven inflammatory response in ICH pathophysiology (2–4). These orchestrated pathways of neuroinflammation are thought to contribute toward the evolution of brain edema and secondary injury following ICH, which deteriorates patient neurological outcomes (3, 5). However, the detailed mechanisms involving lymphocyte-driven neuroinflammatory processes remain only partially understood, and no targeted treatment for post-ICH brain inflammation is currently available in clinical practice.
Blood components exuded from the hematoma and damage-associated molecular patterns released by injured neural cells activates an acute immune response following ICH (5, 6). Besides activated intrinsic brain cells such as microglia and astrocytes, lymphocytes assemble in the regions proximal to the hematoma in the acute and post-acute phase of ICH. Lymphocyte subsets—CD4+ T, CD8+ T, B, and natural killer cells—are observed within a day after ictus and peak around 3 days after (7–9).
T cells, which represent a major lymphocyte population, are among the brain-infiltrating immune cells observed in the acute stage of experimental ICH; however, their actions are comparatively less known compared to innate immune cells (7, 8). In their adaptive modality, CD4+ T cells are generally activated by T cell receptor recognition of cognate antigens presented by major histocompatibility complex II of antigen-presenting cells in the presence of costimulatory signals. Upon activation, CD4+ T cells proliferate and produce a variety of cytokines/chemokines (10, 11). Alternatively, in the absence of antigen recognition, T cells can respond to danger signals and produce cytokines (12–15), suggesting their potential involvement in the genesis or progression of the neuroinflammatory cascade in the acute stage of ICH. To dissect the roles of T cells in acute ICH brain injury, we used brain tissue sections of ICH patients and mouse models to query the potential mechanisms underlying T cell actions in ICH injury. Our study findings provide a rationale for further investigations toward the development of immunotherapies targeting T lymphocytes to modulate the development and progression of secondary brain tissue damage in ICH.
RESULTS
CD4+ T cells accumulate in perihematomal regions in ICH patients and mouse models
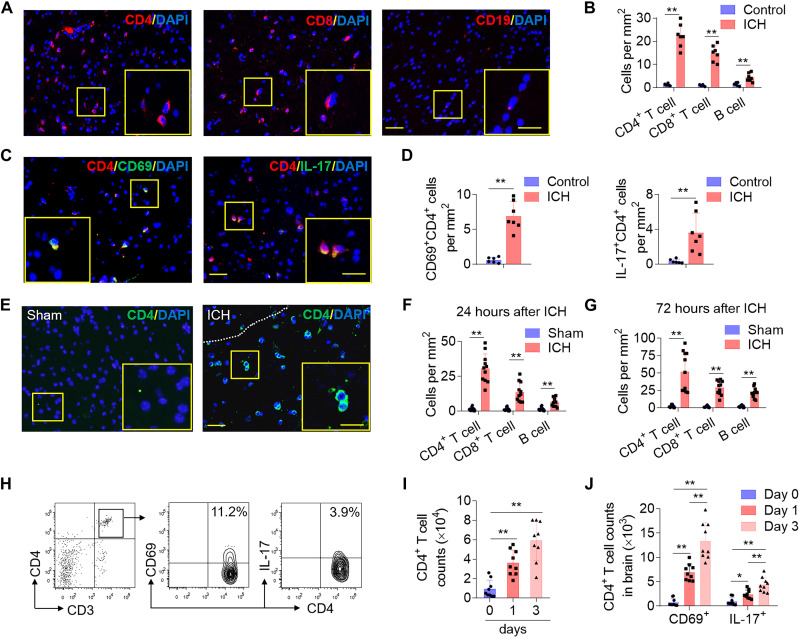
To uncover the role of T cells in the pathophysiology of ICH, we first identified T cells presence in brain tissue obtained from the perihematomal region from patients with ICH within 72-hour onset who underwent craniectomy to remove hematoma. Brain tissues were stained for cell type markers (CD4, CD8, and CD19) and phenotypic markers [CD69 and interleukin-17 (IL-17)]. An increased average count of lymphocytes was detected in ICH tissues compared to the location-matched control brain tissues (Fig. 1, A and B). In ICH tissues, average CD4+ counts outnumbered CD8+ and B cells; CD4+ cells were observed as early as 24 hours after ICH in individual brain samples (Fig. 1, A and B). In addition, T cells in the ICH group displayed a marked expression of CD69 and IL-17 compared to cells in the control group (Fig. 1, C and D), indicating a proinflammatory state of the CD4+ T cell in patient-derived tissue. The accumulation of CD4+ T cells in perihematomal regions suggests active CD4+ T cell participation in the early immune response to ICH (Fig. 1, A to D).
Fig. 1. CD4+ T cells accumulate in perihematomal areas following ICH.
Perihematomal tissues were obtained from brain basal ganglia of patients with ICH within 72-hour onset who underwent urgent evacuation of hematoma. Brain tissues for control cases were obtained from individuals that passed from non-neurologic diseases and were without history of neurological or neuropsychiatric conditions, and selected tissue sections were region-matched with ICH tissues. (A and B) Immunostaining (A) and quantification (B) of lymphocyte subsets (T and B cells) in brain sections from patients with ICH in the perihematomal area after ICH and control subjects without neurological disorders. (C and D) Immunostaining (C) and quantification (D) shows brain-infiltrating CD4+ T cells expression of the activation marker CD69 and IL-17 in perihematomal edema (PHE) after ICH. Scale bars, 40 μm and (inset) 20 μm. Quantification was averaged as positive cells per mm2. In (B) and (D), patients with ICH: n = 7; controls: n = 6. Mann-Whitney test. Means ± SD, *P < 0.05 and **P < 0.01. (E to J) ICH was induced by injection of autologous blood in C57BL/6 mice. Brain tissue was harvested at 24 or 72 hours after ICH. (E) Immunostaining of ICH brain slices show infiltrating CD4+ T cells. White dashed lines outline the hematoma area. (F and G) Quantification of lymphocyte subsets in the perihematomal region of ICH brain 24 hours (F) and 72 hours (G) after ICH induction. Scale bars, 40 μm and (inset) 20 μm. Quantification was averaged as positive cells per mm2. n = 11 mice per group. Two-tailed unpaired Student’s t test. (H to J) Flow cytometry plots and quantification show CD4+ T cell counts and the expression of CD69 and IL-17 in brain-infiltrating CD4+ T cells from days 0 to 3 after ICH. n = 9 mice per group. In (I) and (J), one-way analysis of variance (ANOVA) followed by Tukey post hoc test. Means ± SD, *P < 0.05 and **P < 0.01. DAPI, 4′, 6-diamidino-2-phenylindole.
An established animal model induced by autologous blood injection was used to experimentally confirm the observations in ICH patient-derived brain tissue. Infiltrating lymphocytes were counted in the perihematomal region of murine brains at 24 and 72 hours after ICH induction. Correspondingly, an increased accumulation of CD4+ T cells in the regions surrounding the hematoma at both 24 and 72 hours after ICH induction was observed (Fig. 1, E to G). We further noted the close proximity of these infiltrating CD4+ T cells with CD31+ endothelial cells, indicating their perivascular accumulation (fig. S1). Furthermore, CD4+ T cells increasingly expressed CD69 and IL-17 in a time-dependent manner, detected as early as 24 hours following experimental ICH (Fig. 1, H to J, and fig. S2). To ensure that observed CD4+ T cells were not derived from the injected autologous blood, blood from ubiquitin C promoter–green fluorescent protein (UBC-GFP) mice (universal cellular GFP expression) was injected into wild-type mice. Detection of GFP signal following ICH induction showed that less than 10% of CD4+ T cells in the perihematomal area were GFP+, indicating that most of these cells were actively recruited from the circulation (fig. S3).
CD4+ T cells are significantly activated in the ICH brain
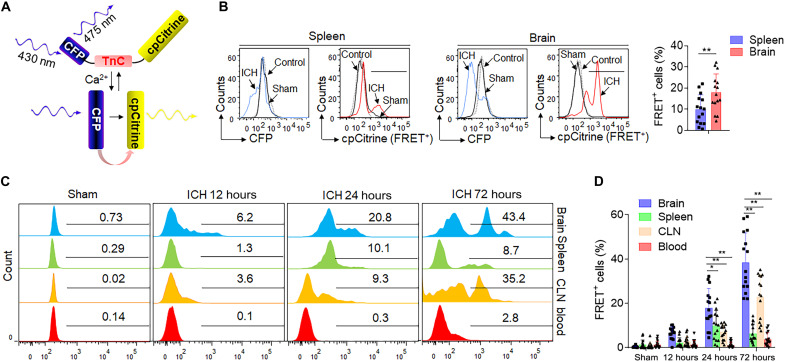
To characterize the temporal and spatial trends of T cell activation following ICH (16), we leveraged a fluorescence resonance energy transfer (FRET)–based genetically encoded calcium indicator, TN-XXL, to visualize T cell activation (Fig. 2A) (17, 18); we tested the compartment-specific T cell activation modalities in ICH mice (Fig. 2B). TN-XXL is composed of calcium-sensitive double C-terminal lobe of troponin C (TnC)–linked dual parts: cyan fluorescent protein (CFP) and cpCitrine. Upon cell activation, increased cytoplasm free calcium binds to the TnC, which then undergoes a reversible conformational change and transfers energy from the CFP to the cpCitrine, resulting in a signature drop in CFP fluorescence and concurrent increase in cpCitrine fluorescence (FRET+). The ratiometric change in fluorescent intensities reflects the cellular activation status. CD4+ T cells isolated from wild-type mice were retrovirally transduced with TN-XXL and intravenously transferred into Rag2−/− mice, followed by autologous blood ICH induction. Notably, FRET+CD4+ T cells were primarily observed in the injured brain and detected as early as 12 hours after ICH (Fig. 2, C and D). FRET+CD4+ T cells in the brain were increasingly activated in a time-dependent manner compared to cells in peripheral compartments including the spleen, cervical lymph nodes (CLNs), and blood (Fig. 2D). We also observed an increase of activated CD4+ T cells in the CLNs, but not as prominently as in the brain after ICH (Fig. 2D). These results show that CD4+ T cells are significantly activated in the hemorrhagic brain and in a time-dependent manner following ICH.
Fig. 2. CD4+ T cells are significantly and time-dependently activated in the brain after ICH.
CD4+ T cells were fluorescence-activated cell sorter (FACS) sorted from C57BL/6 mice and retrovirally transduced with TN-XXL plasmid. A total of 1 × 107 TN-XXL–expressing CD4+ T cells were then injected into Rag2−/− mice 24 hours before ICH. Twenty-four hours after ICH, FRET of TN-XXL–expressing CD4+ T cells in the brain, spleen, CLNs, and blood was detected using flow cytometry. (A) Schematic representation of the calcium indicator TN-XXL, containing donor fluorophore CFP, cpCitrine acceptor, and calcium-sensitive domain TnC, before and after binding of calcium. (B) After binding to free calcium, TnC undergoes a conformational change that leads to energy transfer from the donor to the acceptor fluorophore, resulting in a drop in CFP fluorescence and an increase in cpCitrine fluorescence with CD4+ T cells activation after ICH injury. Flow cytometry gating strategy showing negative control (mock transduced), sham operation, and ICH surgery groups in the spleen and brain separately. Bar graphs shows the FACS-based FRET measurements in TN-XXL–expressing CD4+ T cells within the spleen and brain at 24 hours after ICH. n = 15 per group. Two-tailed unpaired Student’s t test. (C and D) Flow cytometry histogram and measurements of FRET level in TN-XXL–expressing CD4+ T cells within indicated organs from 0 to 72 hours after ICH. n = 15 per group. Two-way ANOVA followed by Tukey post hoc test. Mean ± SD. *P < 0.05 and **P < 0.01.
ICH induces distinguishing transcriptomic features of T cells in the brain versus periphery
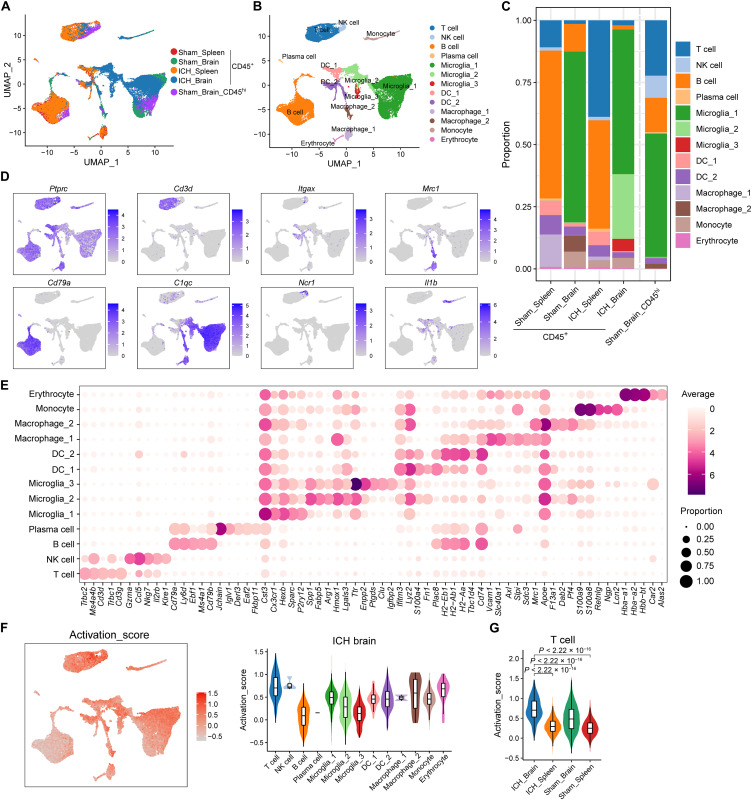
To further depict the landscape of the composition and functional states of brain-infiltrating immune cells following ICH, single-cell RNA sequencing (scRNA-seq) was performed to identify the molecular characteristics of CD45+ cells in the brain versus those in peripheral compartments of ICH mice (Fig. 3). CD45+ cells were sorted from mouse splenocytes or brain at day 3 after ICH or sham procedures and were subjected to unbiased scRNA-seq. Besides, the CD45hi cells were also isolated from sham brain (Sham brain_ CD45hi) to obtain enough T cells for subclustering and data processing. After stringent quality control, a total of 41,385 individual cells, with an average of 2372 genes per cell, were obtained (Fig. 3, A and B, and fig. S4). Thirteen cell clusters were defined on the basis of their distinct molecular features (Fig. 3, C to E). In the ICH brain, the T cell subset exhibited a higher activation score compared to the activation of other CD45+ clusters (Fig. 3F). Moreover, the T cell cluster in ICH brain exhibited a notable higher activation score compared to sham brain and spleen groups (Fig. 3G). The distinct difference in activation scores alludes to the conspicuous activation state of the T cells within the ICH brain.
Fig. 3. Single-cell analysis of CD45+ cells isolated from the brain and spleen of ICH mice.
CD45+ cells were isolated from splenocytes or brain tissues of C57BL/6 mice 72 hours after autologous blood injection induced ICH using FACS. CD45+ cells splenic (Sham spleen) or brain (Sham brain_CD45+) cells from sham-operated mice were used as controls for unbiasedly single-cell RNA mapping. The optimized FACS sorting was performed on CD45hi cell population to obtain enough CD4+ T cells in sham brains for subclustering and data processing (Sham brain_ CD45hi). scRNA-seq was performed using the 10X Genomics Chromium platform. (A and B) Uniform manifold approximation and projection (UMAP) plot along components 1 and 2 for all high-quality single cells from sham brain_CD45+ (n = 10487 cells), sham brain_CD45hi (n = 5589 cells), sham spleen (n = 8747 cells), ICH brain (n = 10,487 cells), and ICH spleen (n = 9147 cells), colored by group and cell type. (C) Stacked bar chart showing the constitution of 13 cell clusters in sham spleen, sham brain_CD45+, sham brain_CD45hi, ICH spleen, and ICH brain. (D) UMAP plots show selected marker genes for determining cell identity. (E) Dot plots show average (dot color) and percentage (dot size) expression of top five marker genes for each cluster. (F) UMAP plot illustrating activation feature scores (Ms4a4b, Ms4a6b, Cxcl10, Itk, Prkcq, Ccnd1, Ccnd2, Ccnd3, Hmox1, Ccl5, Dusp1, Klf4, Jun, and Junb) calculated using AddModuleScore function in Seurat (left). Violin plot with box plot inside showing the comparison of activation scores between T cells and other CD3−CD45+ clusters in ICH Brain (right). (G) Violin plot with box plot inside illustrating the comparison of T cell activation scores between indicated groups. In (A) to (G), data were pooled from 15 mice in each group.
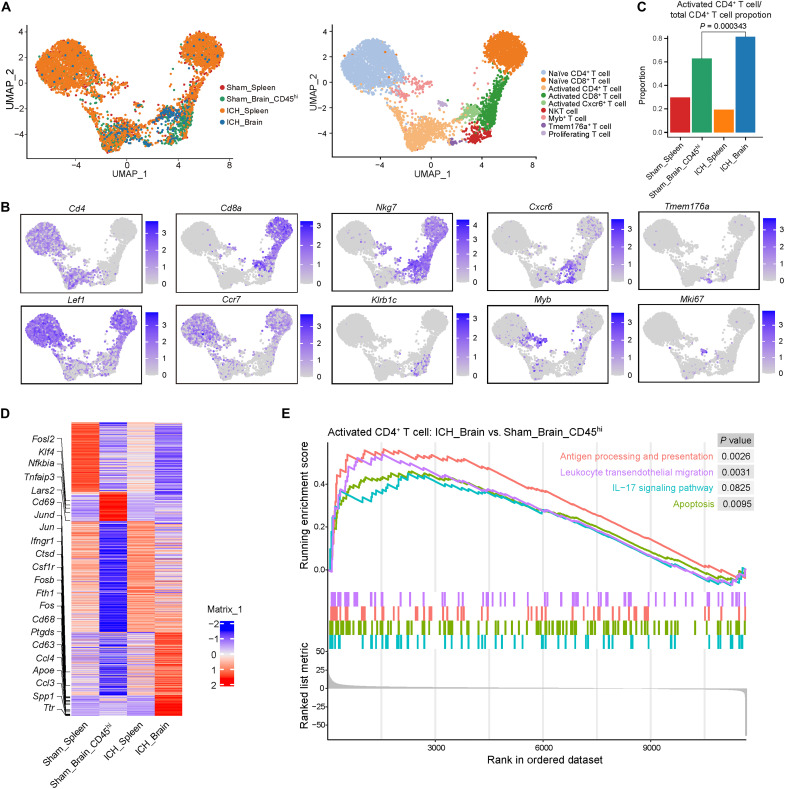
To visualize the intrinsic structure and uncover the potential functional subtypes of the overall T cell population within the context of ICH, unsupervised clustering and uniform manifold approximation and projection (UMAP) were performed for reclustered CD3+ cells from CD45+ (sham spleen, ICH spleen, and ICH brain) or CD45hi (sham brain) populations. Nine distinct T cell subclusters emerged from the sham spleen, sham brain, ICH spleen, and ICH brain, defined as naïve CD4+ T, naïve CD8+ T, activated CD4+ T, activated CD8+ T cell clusters, etc. (Fig. 4, A and B, and fig. S5). Within the ICH brain, activated CD4+ T cells were significantly increased as compared to sham brain (Fig. 4C). Compared with sham brain, ICH-brain CD4+ T cells included up-regulated genes related to inflammation (Jun, Fosb, Ifngr1, and Spp1), chemotaxis (Ccl3 and Ccl4), metabolism (ApoE and Ttr), and apoptotic-triggering death ligands (Fasl and Tnf) (Fig. 4D). Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of activated CD4+ T cells identified a pronounced up-regulation of antigen processing and presentation, apoptosis, leukocyte transendothelial migration, and IL-17 signaling pathways within the injured brain (Fig. 4E). From these high-resolution sequencing data, we can extrapolate possible targets relevant to their actions in secondary ICH injury. Yet, the salient results here find that T cells are activated in the ICH brain and display distinct signatures from peripheral T cells at day 3 after ICH.
Fig. 4. Brain CD4+ T cells bear unique molecular signatures versus peripheral T cells after ICH.
CD3+ T cells were reclustered from CD45+ cell populations isolated from the spleen or brain tissues of C57BL/6 mice at 72 hours after ICH induced by autologous blood injection, based on the scRNA-seq data. (A) UMAP plot showing T cell subtypes of CD45+ cells from sham spleen, sham brain (CD45hi), ICH spleen, and ICH brain. (B) UMAP displaying expression of maker genes for T cell subtype identity. (C) Proportion of activated CD4+ T cells within total CD4+ T cells from sham spleen, sham brain (CD45hi), ICH spleen, and ICH brain. P = 0.000343 by Fisher’s exact test. (D) Heatmap showing relative average expression of differentially expressed genes in activated CD4+ T cells across ICH brain, ICH spleen, sham brain, and sham spleen group. (E) Selected significantly enriched Kyoto Encyclopedia of Genes and Genomes pathways by gene set enrichment analysis between activated CD4+ T cells from ICH brain and those from sham brain. In (A) to (E), data were pooled from 15 mice in each group.
Brain-infiltrating CD4+ T cells promotes focal inflammation through IL-17
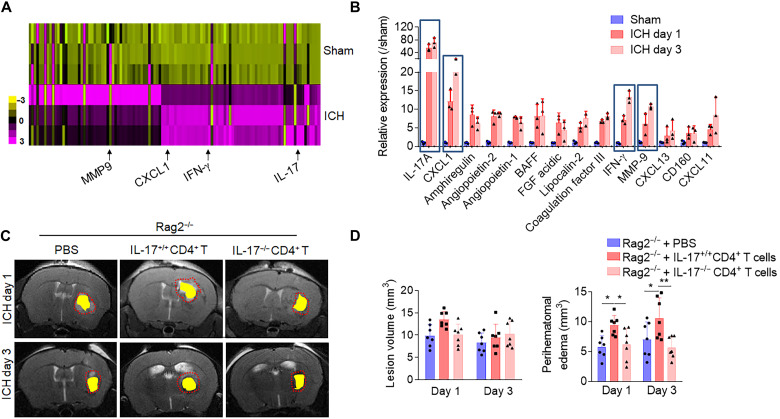
Upon uncovering the brain-specific signatures of post-ICH CD4+ T cells where the IL-17 signaling pathway was highly enriched, A Proteome Profiler Mouse XL Cytokine Array of brain-sorted CD4+ T cells, 24 and 72 hours after ICH, validated the protein expression of the enriched pathways. Compared to CD4+ T cells isolated from the sham brain, CD4+ T cells of the ICH brain registered a greater production in an array of proinflammatory cytokines, with IL-17 remaining the most expressed (Fig. 5, A and B). IL-17 has known proinflammatory functions, which initiate and drive inflammation in a tissue-dependent manner in response to injury or infection (19–21). This prompted us to interrogate the specific effects of increased IL-17 production from brain-infiltrated CD4+ cells in acute ICH injury. To this end, CD4+ T cells from IL-17−/− mice were isolated, and the IL-17−/−CD4+ T cells were intravenously transferred to Rag2−/− mice, which are devoid of mature lymphocytes. Compared to Rag2−/− mice receiving IL-17+/+ CD4+ cells, Rag2−/− mice receiving IL-17−/− CD4+ cells exhibited reduced perihematomal edema (PHE) volumes (Fig. 5, C and D), improved endothelial survival, and preservation of blood-brain barrier (BBB) integrity (fig. S6) at 1 and 3 days after ICH induction. The increased expression of inflammatory cytokines reinforces the activated status of CD4+ T cells in the ICH brain, and their increased production of IL-17 appears to be a primary cytokine-mediated method fueling perihematomal inflammation injury and worsening secondary ICH injury.
Fig. 5. Detrimental effects of brain-infiltrating CD4+ T cells in ICH involves IL-17.
(A and B) CD4+ T cells were isolated from brain of C57BL/6 mice 24 hours and 3 days after ICH induced by autologous blood injection or sham control, and cells were then lysed for Proteome Profiler Mouse XL Cytokine Array analysis. (A) Heatmap shows relative changes of cytokine and chemokine profiles in lysates of CD4+ T cells from ICH brain versus sham mice. Results were based on clustering of immunoassay measurements of the listed proteins. Purple shades represent fold change of up-regulated proteins, and yellow shades demark fold change of decreased proteins. (B) Bar graph shows highly up-regulated expression of selected cytokines/chemokines. n = 3 duplicates from 15 mice. One-way ANOVA followed by Tukey post hoc test. (C and D) CD4+ T cells were isolated from the spleen of IL-17−/− mice or their IL17+/+ littermates. IL-17−/− versus IL17+/+ CD4+ T cells were adoptively transferred into Rag2−/− mice to evaluate the role of CD4+ T cell–derived IL-17 in ICH injury. Quantification of lesion volume and PHE volume was calculated in the indicated groups of collagenase induced ICH. n = 7 mice per group. One-way ANOVA followed by Tukey post hoc test. Means ± SD. *P < 0.05 and **P < 0.01. BAFF, B cell–activating factor; FGF, fibroblast growth factor; MMP9, matrix metalloproteinase 9.
CD4+ T cells induced brain endothelial cell apoptosis involving the TRAIL-DR5 pathway
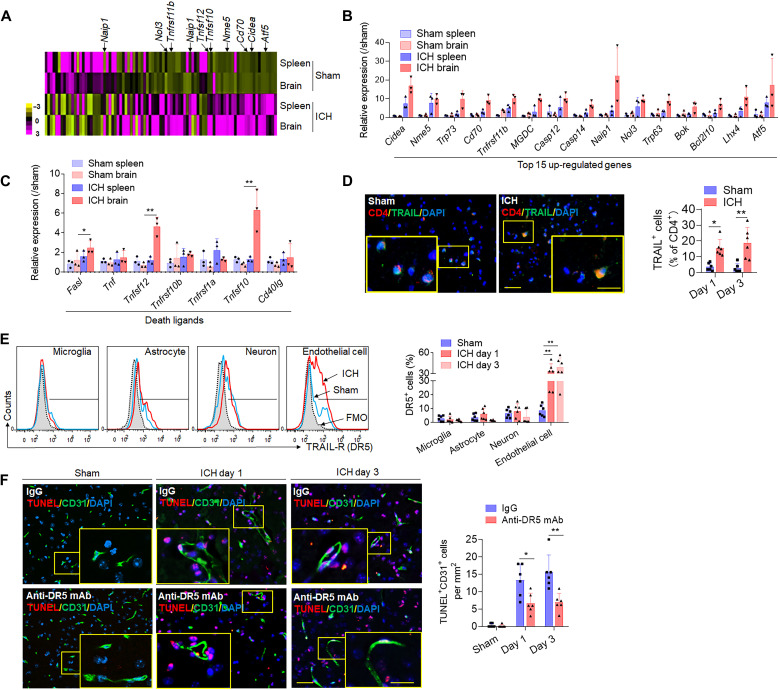
As revealed by single-cell sequencing, another potential pathway, which may underlie CD4+ T cell contribution to ICH injury, is through death ligand–mediated cell apoptosis (Fig. 4, D and E). To specifically investigate the apoptosis panel in CD4+ T subsets, RT2 Profiler PCR Array of CD4+ T lymphocytes corroborated the transcriptomic findings and identified the expression of a variety of apoptotic-related genes. At day 1 after ICH, brain-infiltrating CD4+ T cells overexpressed a variety of antiapoptotic genes (Naip1, Nol3, and Tnfrsf11b), a transcription factor for apoptotic regulation (Atf5), and the immune stimulatory factor (Cd70) (Fig. 6, A and B). Critically, brain-infiltrating CD4+ T cells were also found to up-regulate the death receptor ligands Tnfsf10, Tnfsf12, and Fasl (Fig. 6C). Immunostaining of murine ICH brain slices visualizes a notable increase in the expression of tumor necrosis factor–related apoptosis-inducing ligand (TRAIL), encoded by the Tnfsf10 gene, on CD4+ T cells (Fig. 6D). After gating for brain intrinsic cells expressing the TRAIL cognate receptor DR5, flow cytometry analysis revealed that DR5 was expressed by multiple brain intrinsic cell types; however, ICH robustly increased DR5 expression on brain endothelial cells (Fig. 6E). Considering that BBB disruption is a key pathophysiological hallmark of the ICH injury and driver of secondary injury and that endothelial cells are a critical cellular component of the neurovascular unit and participant in the evolution of brain edema, we postulate that CD4+ T cells may exert direct endothelial cell injury via TRAIL-DR5 interaction, resulting in increased BBB disruption and brain edema. We tested this notion using a TRAIL-specific antibody to neutralize the interaction. Anti-DR5 monoclonal antibody (mAb) treatment effectively blocked CD31+ endothelial cell death (Fig. 6F) and brain injury after ICH (fig. S7). These results highlight how brain-homing CD4+ T cells induce apoptosis of brain intrinsic cells through antigen-independent mechanisms, partly involving the TRAIL-DR5 pathway in ICH mice.
Fig. 6. CD4+ T cells induce apoptosis of endothelial cells through death receptor signaling.
CD4+ T cells were isolated from splenocytes or brain of mice 24 hours after autologous blood injection induced ICH or sham control. FACS-sorted CD4+ T cells were used for RT2 Profiler PCR Array analysis of 87 apoptosis related genes. (A) Heatmap shows relative changes of expression of cell apoptotic-related genes in CD4+ T cells from the brain or spleen of ICH mice versus sham mice. (B) Quantification of top 15 up-regulated apoptosis associated genes. (C) Bar graph shows genes of death receptor ligands in CD4+ T cells of ICH or sham mice. In (B) and (C), n = 3 duplicates from 15 mice. One-way ANOVA followed by Tukey post hoc test. (D) Immunostaining of death receptor ligand TRAIL in brain CD4+ T cells of ICH or sham mice. Quantification was averaged as percentage of death receptor ligand–positive cells in CD4+ T cells. Scale bars, 40 μm and (inset) 20 μm. n = 6 mice per group. Mann-Whitney test. (E) Flow cytometry plots and bar graph show expression of TRAIL receptor (DR5) in different brain cell types of sham and ICH mice, including microglia (CD45intCD11b+), astrocytes [glial fibrillary acidic protein positive (GFAP+)], neuron (NeuN+), and endothelial cells (CD31+). FMO, fluorescence minus one. n = 6 mice per group. One-way ANOVA followed by Tukey post hoc test. (F) CD31+ endothelial cell apoptosis was analyzed by TUNEL staining in sham mice and ICH mice with anti-DR5 monoclonal antibody (mAb) or IgG group. Scale bars, 40 μm and (inset) 20 μm. Quantification was averaged as positive cells per mm2 in the perihematomal region. n = 6 mice per group. Mann-Whitney test. Means ± SD. *P < 0.05 and **P < 0.01.
Depletion CD4+ T cells reduced secondary ICH injury
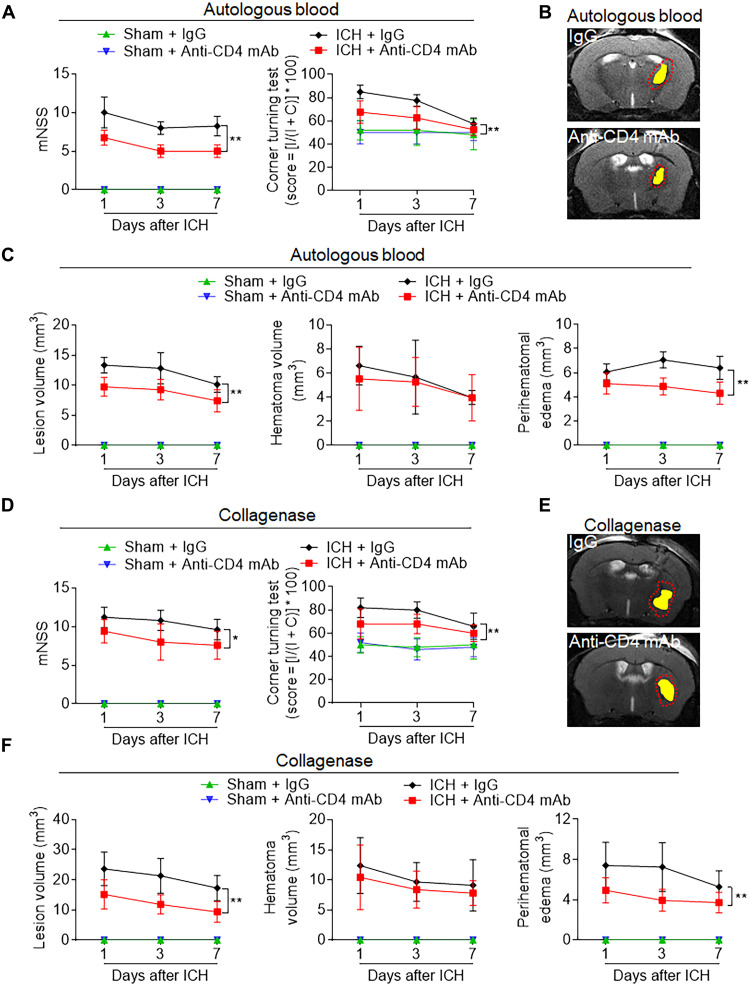
To ascertain their contribution to secondary brain injury in ICH, CD4+ T cells were depleted with an anti-CD4 mAb before ICH induction in mice. Flow cytometry quantification verified the efficacy of CD4+ T cell depletion, without influencing other cell populations in peripheral blood of control mice (fig. S8). Ablation of CD4+ T cells in both autologous blood and collagenase models of hemorrhagic stroke demonstrated T cell participation in the processes of secondary brain injury. CD4+ depleted animals recorded reductions in PHE volume and improvement of functional outcomes at days 1, 3, and 7, compared to mice receiving IgG control, in both ICH models (Fig. 7). Depletion of CD4+ cells before model induction ameliorated parameters of secondary injury, functionally supporting the detrimental capacity of CD4+ T cells in acute ICH injury.
Fig. 7. Depletion of CD4+ T cells ameliorates neurodeficits and ICH injury in mice.
C57BL/6 mice received intraperitoneal injection of 50 μg of anti-CD4 mAb or lgG control 24 hours before ICH induction with injection of autologous blood or collagenase. (A) Behavioral tests including modified neurological scale score (mNSS) and corner turning test were performed in indicated groups of autologous blood–induced ICH mice. (B) Multimodal 7-T magnetic resonance imaging (MRI) was used to visualize lesion (T2) and hematoma (SWI) in autologous blood–induced ICH mice. PHE volume was calculated by subtracting the hematoma volume from lesion volume. Representative MRI for hematoma and PHE in mice receiving anti-CD4 mAb or lgG following ICH induction for 24 hours. (C) Quantification of lesion volume, hematoma, and PHE volume in the indicated groups of autologous blood–induced ICH mice. (D) Behavioral tests in indicated groups of collagenase induced ICH mice. (E) Representative MRI for hematoma and PHE in mice receiving anti-CD4 mAb or lgG following ICH induction for 24 hours. Yellow shadows represent hematoma, and red lines delineate the lesion area. (F) Quantification of lesion volume and PHE volume in the indicated groups of collagenase-induced ICH mice. In (A) to (F), n = 7 mice per group. Two-way ANOVA followed by Tukey post hoc test. Means ± SD. *P < 0.05 and **P < 0.01.
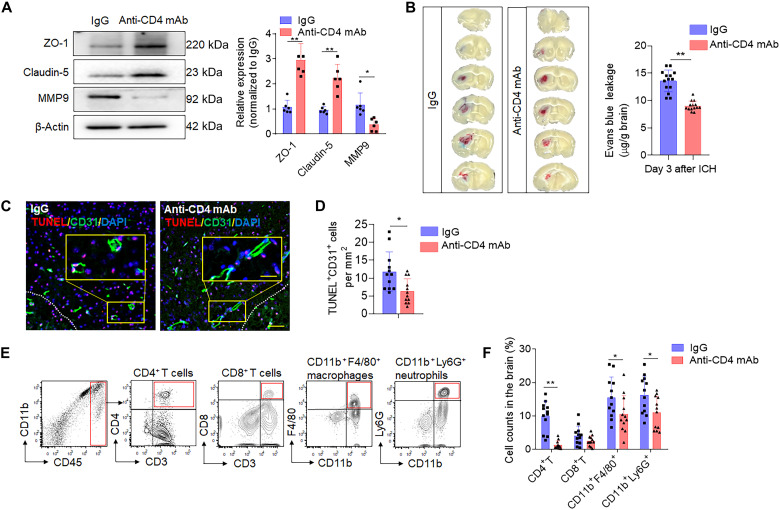
BBB permeability and neuroinflammation are key contributors toward PHE expansion. To further elucidate their roles, we looked into CD4+ T cell influence on BBB integrity and local inflammation following ICH. CD4+ depletion before ICH preserved the expression of major tight junction proteins, claudin-5 and ZO-1, at 3 days after ICH (Fig. 8A). In addition, CD4+ T cell depletion reduced matrix metalloproteinase 9 (MMP9) protein in the ICH brain at day 3 (Fig. 8A); MMP9 is an immunemodulatory molecule that damages the matrices of the BBB following stroke and is considered a surrogate marker of BBB disruption (22, 23). In CD4+ T cell–depleted animals, we observed reduced leakage of Evans blue (EB) into the brain parenchyma (Fig. 8B) and a reduction brain CD31+ endothelial cell death (Fig. 8, C and D). Last, we evaluated whether brain-infiltrated CD4+ T lymphocytes affect the focal inflammatory milieu by quantifying leukocyte trafficking into the ICH brain of CD4+ T cell–depleted mice. Compared to immunoglobulin G (IgG)–treated control mice, counts of brain-infiltrating neutrophils and monocytes were notably reduced in CD4+ T cell–depleted mice at day 3 after ICH induction (Fig. 8, E and F, and fig. S9). These findings inform how infiltrating CD4+ T cells engage in critical processes known to drive secondary injury following cerebral hemorrhage; considering CD4+ ablation in ICH preserved BBB integrity, attenuated local inflammation, reduced lesion volume, and improved sensorimotor function in animal models. These broad responses may point to the multifaceted involvement of CD4+ T cells in secondary ICH injury. Together, these results suggest that CD4+ T cells are acutely recruited to the ICH brain where they exacerbate BBB disruption and focal inflammation to contribute to the secondary brain injury.
Fig. 8. Depletion of CD4+ T cells reduces BBB permeability and leucocyte infiltration in ICH mice.
C57BL/6 mice received intraperitoneal injection of 50 μg of anti-CD4 mAb or lgG control 24 hours before ICH induction with injection of autologous blood. (A) Brain tissue was collected at day 3 after ICH. Western blot shows tight junction protein level (claudin-5 and ZO-1) and MMP9 expression in ICH mice receiving anti-CD4 mAb or lgG control. Right: Quantification of ZO-1, claudin-5, and MMP9 in brain homogenates of ICH mice. n = 6 mice per group. Mann-Whitney test. (B) Concentrations of evans blue were measured at day 3 after ICH, with anti-CD4 mAb or IgG injection. n = 14 per group. Two-tailed unpaired Student’s t test. (C and D) Immunostaining (C) and quantification (D) of TUNEL in CD31+ endothelial cells of ICH mice received IgG control or anti-CD4 mAb separately. White dashed lines outline the hematoma area. Scale bars, 40 μm and (inset) 20 μm. n = 12 mice per group. Two-tailed unpaired Student’s t test. (E and F) Single-cell suspensions from brain tissue were collected at day 3 after ICH. (E) Gating strategy of brain-infiltrating neutrophils (CD11b+CD45hiLy6G+), monocytes/macrophages (CD11b+CD45hiF4/80+), CD4+ T cells (CD45hiCD3+CD4+), and CD8+ T cells (CD45hiCD3+CD8+) was shown. (F) Counts of the immune cell populations in ICH brains from mice receiving anti-CD4 mAb or IgG. n = 12 mice per group. Two-tailed unpaired Student’s t test. Means ± SD. *P < 0.05 and **P < 0.01.
DISCUSSION
This study provides the first thorough evidence that CD4+ T cells accelerate PHE development in ICH. As documented here, activated CD4+ T cells accumulate in the perihematomal region and drive local inflammation via the production of IL-17 and engagement of endothelial cell death receptors, resulting in diminishing BBB integrity and augmented brain edema (fig. S10). Monoclonal antibody depletion of CD4+ T cells before ICH induction limits PHE and lesion volume, attenuates local brain inflammation, maintains key tight junction proteins, and improves functional outcomes through the acute time points after ICH. The varied responses elicited by the absence of CD4+ in ICH pathophysiology not only indicates their expansive effector functions but may also illustrate the complex interaction and possible redundant pathways encompassed in the pathophysiology of ICH secondary injury.
PHE is a cause of progressive tissue injury after ICH, and the extent of brain edema surrounding the hematoma directly correlates with poor outcomes in patients with ICH (24). PHE represents an attractive therapeutic target in part because of the extended time window during which it occurs; however, it has thus far not been successfully therapeutically modulated with available drugs, as shown by clinical trials. For example, in MISTIE III trial (Minimally Invasive Surgery Plus Alteplase for Intracerebral Hemorrhage Evacuation), Shah et al. reported that a minimally invasive procedure for hematoma clearance failed to show superiority to standard medical care (25). Inflammation contributes to the development and progression of PHE and thus subsequent neurological deterioration (26, 27). In targeting ICH-induced neuroinflammation, numerous studies have explored the potential of multiple sclerosis disease-modifying drugs in the setting of stroke (28). Two clinical trials involving natalizumab showed that it does not confer any benefits, but four human studies with fingolimod have showcased its potential in improving recovery prospects (29, 30). Therefore, there is an unmet need for the development of alternative immune-modulatory approaches in alleviating ICH injury.
After brain injury, peripheral immune cells are activated and infiltrated the injured brain where they propagate acute neuroinflammation and contribute to secondary brain injury (8, 31, 32). CD4+ T cells have been suggested to be the predominant infiltrating leukocyte populations in the acute stage in experimental ICH (8). Emerging literatures support a role for meningeal vessels as a route for immune cell migration and post-stroke inflammation, as well (33, 34). Studies, depleting lymphocyte infiltration to the brain or limiting their influx, show that T cells absence generally alleviates acute neuroinflammation and injury after ICH (35). Given their broad effector actions, several studies detailed T cell action in ICH related to inflammation (36), BBB injury (37), neurotoxicity (38), and edema formulation (39). Despite these reported studies, the fine kinetics and specific T cell activation in ICH brain were previously unknown. In this study, we detail their activation status by leveraging the TN-XXL approach, which tracks their differential activation in vivo. This approach revealed CD4+ T cells to be substantially and time-dependently activated in the hemorrhagic brain, suggesting that the microenvironment of the ICH brain primes the infiltrating T cells.
scRNA-seq of CD4+ T cell identified specific proinflammatory and proapoptotic signatures unique to the ICH brain versus their profiles in peripheral compartments. Of inflammatory genes altered in brain CD4+ T cells following ICH, the IL-17 pathway stood out in its degree of enrichment. One recent study by Yang et al. (40) showed prominent accumulation of T helper 1 (TH1) and TH17 cell subsets of CD4+ T cells at 14 days after ICH and suggested that modulating CD4+ T cell differentiation may alleviate post-ICH outcomes. Our data show an early increase in CD4+ T cells and the IL-17+CD4+ T cell (TH17) subset in patients with ICH, which is consistent and enhances the results from the aforementioned study. From this present data, we deduce that CD4+ T cells augment acute local inflammation in the ICH brain and worsen secondary injury primarily through IL-17 production.
We further demonstrate that CD4+ T cells can induce apoptosis of intrinsic brain cells by expressing death receptor ligands, and their interaction with cells bearing cognate death receptors was restricted after receptor blockade. We highlight a series of apoptotic-related genes and death ligands up-regulated in brain CD4+ T cells following ICH, particularly their surface expression of TRAIL. The unique role of TRAIL in the nervous system is revealed by the widespread expression of its receptor DR5 on various brain cell types, including neurons, astrocytes, and endothelial cells. Notably, DR5 was found to be significantly up-regulated in endothelial cells following ICH. The detrimental role of TRAIL-DR5 pathway in BBB injury was attributed with reduced endothelial cell apoptosis in the ICH brain by DR5 mAb pretreatment. These findings expose an innovative mechanism by which cells in the ICH brain can be damaged by invading T cells through the engagement of cell death receptors.
This study has several limitations. First, restricted by the source of control samples, we adopted postmortem tissue sections for control, as previously published (9). Second, female mice were not used. In addition, several treatments such as T cell depletion and antibody neutralization were given before ICH. The effect of treatment after injury is warranted in future preclinical studies. On the basis of single-cell sequencing data, the impact of other activated T cell subsets on ICH brain injury warrants study. Moreover, our data demonstrates that CD4+ T cells are also activated in the CLN after ICH, supplementing a recent study that brain-to-CLN pathway may be involved in acute brain injury (41). Further studies could determine the dynamic activation of T cells within the ICH brain and CLN, as well as their specific features in the respective two compartments.
As a major cellular component of the adaptive immune system, T cells are usually activated by specific antigens presented on the surface of antigen-presentng cells (42), a process elapsing from days to weeks until maturity. Yet, the early T cells activation we observe in acute ICH injury indicates the involvement of antigen-independent mechanisms. This study identifies how T lymphocytes respond and participate in ICH pathology. Together, our results broaden the understanding of T lymphocyte–mediated neuroinflammation in relation to PHE expansion after ICH, providing insights toward targeted intervention for ICH.
MATERIALS AND METHODS
Human brain tissues
Collection of human samples was performed according to protocols approved by the Institutional Review Boards of Tianjin Medical University General Hospital (Tianjin, China) and Beijing Tiantan Hospital (Beijing, China). For immunostaining of human brain tissues, six cases of control postmortem and seven ICH human brain sections were included. All seven patients had primary ICH at the basal ganglia. Fresh brain tissues of perihematomal regions were collected from patients with ICH undergoing hematoma evacuation within 72 hours after onset. Brain tissues for control cases were obtained from deceased individuals with non-neurologic diseases and without history of neurological or neuropsychiatric conditions. The selected tissue sections were region-matched with ICH tissues. Lymphocytes populations were counted at 12 different view fields for each brain slide and then quantified by averaging positive cells per mm2. Image analysis was performed using ImageJ software (National Institutes of Health, MD, USA). Quantification was performed in a double-blinded manner. The average age between ICH and control subjects did not differ significantly [ICH: 54.7 ± 9.52 years old (y/o); control: 56.5 ± 8.55 y/o; means ± SEM; P > 0.05; unpaired t test].
Mice
All animal experiments were performed in accordance with the ARRIVE (Animal Research: Reporting In Vivo Experiments) guidelines and obtained approved protocols from respective Institutional Animal Care and Use Committee (IACUC). All studies were performed with adult 3- to 4-month-old male mice. C57BL/6 (RRID: MGI:2159769), Rag2−/− (RRID: NSRRC_0035), UCB-GFP (RRID:IMSR_JAX:004353), and IL-17−/− mice (RRID:IMSR_JAX:034140) and IL17+/+ littermates were purchased from the Jackson Laboratory (Bar Harbor, Maine). All mutant mice were backcrossed to the C57BL/6 background for 12 generations. Animals were housed under pathogen-free conditions, with maximum of five animals per cage, standardized light-dark cycle conditions, and free access to food and water.
Induction of murine ICH model
ICH in mice was induced by intra-striatal injection of autologous blood or bacterial collagenase as previously described (4, 43). Briefly, mice were anesthetized with ketamine/xylazine mixture by intraperitoneal injection and positioned prone in a stereotactic head frame. A ~1-mm-diameter hole was drilled on the right side of the skull (2.3 mm lateral to midline, 0.5 mm anterior to bregma). In the autologous blood model, 30 μl of nonheparinized blood was withdrawn from angular vein and infused via infusion pump to the coordinates; 5 μl was initially injected at a rate of 1 μl/min at a depth of 3.0 mm, and the remaining 25 μl was injected at an identical rate at 3.7 mm depth. The needle was left for 15 min to prevent backflow and then gently withdrawn. In the collagenase model, 0.0375 U of bacterial collagenase (type IV, Sigma-Aldrich, St. Louis, MO) in 0.5 μl of saline was administered at the same coordinates at rate of 0.5 μl/min. An identical surgical procedure with equal volume of sterile saline injection for sham-operated groups. Cranial burr hole was sealed with bone wax, and incision was sutured. Body temperature was maintained at 37.0° ± 0.5°C throughout the procedures. The total mortality rate of mice subjected to ICH was ~4.8%.
Evaluation of neurological deficit
Neurological outcome of ICH mice was evaluated by modified neurological scale score (mNSS) and corner test as reported previously (4, 44). The mNSS consisted of motor, sensory, reflex, and balance assessments with the highest possible score being 18. The rating scale was as follows: A score of 13 to 18 indicates severe injury, 7 to 12 indicates moderate injury, and 1 to 6 indicates mild injury. Following surgery, each mouse was assessed on a scale from 0 to 18 after recovery from the ICH surgical procedure. Mice with a score of <6 or above a score of 13 at 24 hours after ICH (before treatment) were not included in the study.
Magnetic resonance imaging
Brain lesion size and PHE in ICH mice were evaluated with 7-T small animal magnetic resonance imaging scanner (Bruker, Billerica, MA). T2-weighted images of the brain were acquired with fat-suppressed rapid acquisition with relaxation enhancement sequence (repetition time, 4000 ms; echo time, 60 ms; slice thickness, 0.5 mm) to visualize lesion volume. Susceptibility-weighted images (SWIs; repetition time, 21.0 ms; echo time, 8.0 ms; 0.3-mm thickness) visualized hematoma volume. T2 and SWI images were manually traced and calculated by summing the volume by the distance between sections via ImageJ (National Institutes of Health), PHE volume was calculated as difference of lesion and hematoma volumes (44). Magnetic resonance imaging data were analyzed by two experimenters blinded to experimental conditions.
Administration of monoclonal antibodies
Ultra-LEAF–purified anti-CD4 mAb (100470, GK1.5, BioLegend, San Diego, CA) was intraperitoneally injected to deplete CD4+ T cells, with a dose of 50 μg at 24 hours before ICH induction and every 5 days until the end of experiments. IgG antibody (Rat IgG2b, 400671, BioLegend) was used as control for anti-CD4 mAb. Anti-DR5 mAb (119909, MD5-1, BioLegend) was administered intraperitoneally at a dosage of 100 μg at 24 hours before ICH induction. Purified hamster IgG isotype antibody (400902, HTK888, BioLegend) was used as a control for anti-DR5 mAb.
Immunostaining
Immunostaining procedure of brain slices was conducted as previously described (4). Primary antibodies were incubated at 4°C overnight at vendor concentration. After washing with cold PBS, slices were incubated with fluorescence-conjugated secondary antibodies at room temperature for 1 hour. Slides were washed and mounted with a mounting medium containing 4′, 6-diamidino-2-phenylindole (DAPI; ab104139, Abcam, Cambridge, UK). Primary antibodies used were as follows: anti-human CD4 (EPR6855, ab133616, Abcam), anti-human CD8a (D8A8Y, 85336, Cell Signaling Technology), anti-human CD69 (EPR21814, ab233396, Abcam), anti-human IL-17 (4K5F6, ab189377, Abcam), anti-human IL-4 (PA5-25165, Invitrogen), anti-human interferon-γ (IFN-γ) (15365-1-AP, Proteintech), anti-human Foxp3 (12632, Cell Signaling Technology), anti-mouse CD4 (GK1.5, MAB554-100, R&D Systems), anti-mouse CD8 (EPR20305, ab209775, Abcam), anti-human/mouse CD19 (6OMP31, 14-0194-82, eBioscience), anti-human/mouse TRAIL-R2 (DR5, PA5-19895, Invitrogen), and anti-mouse CD31 (D8V9E, 77699, Cell Signaling Technology). Secondary antibodies used were as follows: Alexa Fluor 594 anti-rat IgG (ab150160, Abcam), Alexa Fluor 594 anti-rabbit IgG (ab150080, Abcam), Alexa Fluor 488 anti-rabbit IgG (ab150077, Abcam), Alexa Fluor 488 anti-mouse IgG (ab150113, Abcam), and Alexa Fluor 488 Anti-rat IgG (ab150157, Abcam). TUNEL (terminal deoxynucleotidyl transferase–mediated deoxyuridine triphosphate nick end labeling) staining used the in situ 5-bromo-2´-deoxyuridine–Red DNA Fragmentation (TUNEL) Assay Kit (ab66110, Abcam). All Images were acquired on a fluorescent microscope (Olympus, model BX-61, Center Valley, PA, USA). Image analysis was performed using ImageJ software (National Institutes of Health, MD, USA).
Cell isolation and passive transfer of CD4+ T cells
CD4+ T cells were sorted from pooled splenocytes of wild type, IL-17−/− mouse strains, or IL17+/+ littermates. Spleen tissues were separated and placed on a premoistened 70-μm cell strainer and then gently homogenized with the end of a 1-ml syringe plunger. The strainer was washed with 10 ml of erythrocyte lysis buffer (349202, BD FACS, San Jose, CA, USA), and the eluted cells were incubated for 5 min at room temperature and washed with 40 ml of phosphate-buffered saline (PBS). Cell suspensions were simultaneously incubated for 30 min with alloohycocyanin (APC)-conjugated anti-CD4 (RM4-4, 116013), and phycoerythrin (PE)–conjugated anti-CD3 (17A2, 100205). Then, CD4+ T cells were purified via two rounds of cell sorting selection (CD4+CD3+) with the high-speed sort of FACS Aria III flow cytometer. Purity of sorted CD4+ T cells was verified by flow cytometry before transfer. Highly purified (>99%) CD4+ T cells (1 × 107) were injected into tail vein of Rag2−/− recipient mice.
Cytokine array
Cytokine secretion profile of ICH brain CD4+ T cells was evaluated by Proteome Profiler Mouse XL Cytokine Array. CD4+ cells were sorted from the whole brain of ICH or sham mice. Cytokine levels in these samples were detected using the Mouse XL Cytokines Array Kit (R&D Systems Inc. Minneapolis, MN) according to the manufacturer’s instructions.
Flow cytometry
Preparation of single-cell suspensions for flow cytometry from the spleen, brain, CLNs, and blood were prepared as previously reported (4, 45). Briefly, mice were euthanized by lethal anesthesia via isoflurane, and then the brain and spleen were removed. Brain tissues were minced and incubated with collagenase IV and deoxyribonuclease at 37°C for 30 min. After removing the myelin debris by centrifugation in 30% Percoll, single cells were suspended in 1% bovine serum albumin. Spleen tissues were minced and strained through a 70-μm cell strainer; red blood cells were removed using a lysis buffer. For CLN tissue, the ventral neck area of mice was dissected, and bilateral CLNs were removed before or after ICH. The lymph node tissues were minced and strained through a 70-μm cell strainer. Thereafter, single-cell suspensions were placed in conical centrifuge tubes (106 cells per tube) and stained with fluorescently conjugated antibodies. For intracellular staining, cells were fixed in fixation buffer for 20 min after surface marker staining. After washing twice in permeabilization buffer, cells were incubated with antibodies in the staining buffer for 45 min. All flow cytometry antibodies were sourced from BioLegend, unless otherwise indicated. Antibodies used were as follows: CD45-APC-Cy7 (30-F11, 103116), CD4-APC (RM4-4, 116013), CD4-PE (GK1.5, 100408), CD3–fluorescein isothiocyanate (FITC) (17A2, 100204), CD3-PE (17A2, 100205), CD8a-BV421 (53-6.7, 100737), CD69-BV421 (H1.2F3, 104527), IL-17A–PE-Cy7 (TC11-18H10.1, 506921), F4/80-FITC (BM8, 123017), Ly6G-APC (1A8, 127613), CD11b-PE-Cy7 (17A2, 101216), TRAIL-R2-FITC (MD5-1, A15750, Invitrogen), NeuN-AF647 (EPR12763, ab190565, Abcam), CD31-BV421 (390, 102423), glial fibrillary acidic protein–PE (1B4, 561483, BD Bioscience), IL-4–APC (11B11, 504106), IFN-γ–BV605 (XMG1.2, 505839), CD25-BV421 (PC61, 102034), and Foxp3-PE (MF-14, 126404). Samples were run on FACSAria (BD Biosciences) and analyzed using FACS Diva and FlowJo X software (Informer Technologies, Ashland, OR).
Single-cell RNA sequencing
CD45+ single cells were sorted by flow cytometry for sequencing. The optimized fluorescence-activated cell sorter (FACS) sorting were performed on CD45hi cell population to obtain enough CD4+ T cells in sham brains for subclustering and data processing (Sham brain_2 in Fig. 3A).10X Genomics platform of Tulane University Single Cell Sequencing Core and Huada Gene (Shenzhen, PRC) prepared the barcode libraries with the Single Cell 3′ Reagent Kit v2 (10X Genomics) along the manufacturer’s instructions. RNA-seq performed at a sequencing depth of approximately 50,000 reads per cell.
scRNA-seq data processing
Raw files (FASTQ format) were processed with Cell Ranger software (v5.0.1), which performed mapping to the mm10 transcriptome, filtering, barcode counting, and unique molecular identifier (UMI) counting and lastly generated feature-by-barcode UMI matrix. Scrublet (46) software was used to remove potential doublets for individual samples. Seurat (v3.1.5) was used for downstream analysis. Low-quality cells, <500 genes expressed or with <2500 UMIs, were excluded at the initial quality control step. Cells >15% mitochondrial UMIs were removed. Library-size normalization was performed on the UMI-collapsed gene expression values for each cell barcode by scaling the total number of transcripts and multiplying by 10,000. Data were then log-transformed for further downstream analysis.
Two thousand genes with highest variance were initially identified using the FindVariableGenes function. Batch effects were corrected by using the fastMNN function of batchelor R package (47), a fast version of the mutual nearest neighbors (MNNs) method, and the first 20 components of MNN was considered to reduce the dimensionality. Cells were clustered with Seurat’s FindClusters function. For visualization, we applied run UMAP to cluster cells on scatter plots. Marker genes were defined as genes with an adjusted P value < 0.05 tested with a nonparametric Wilcoxon rank-sum test (table S1). KEGG pathway enrichment analysis was performed by gene set enrichment analysis function in clusterProfiler (v3.14.3) (48). Activation feature scores were calculated by using AddModuleScore function in Seurat with an array of cell activated feature genes based on published literatures (Ms4a4b, Ms4a6b, Cxcl10, Itk, Prkcq, Ccnd1, Ccnd2, Ccnd3, Hmox1, Ccl5, Dusp1, Klf4, Jun, and Junb) (49–57).
Apoptosis-related gene evaluation
Apoptosis-related genes in FACS-sorted CD4+ T cells from the spleen or brain of ICH or sham mice were evaluated by the 87 gene RT2 Profiler PCR Array Kit (330231, QIAGEN). mRNAs were extracted from isolated cells with TRIzol reagent (Invitrogen). The expression of apoptosis-related genes was determined according to the manufacturer’s protocol.
In vivo detection and T cell activation via TN-XXL
TN-XXL pcDNA3 was a gift from Oliver Griesbeck (Addgene plasmid # 45797; http://n2t.net/addgene:45797; RRID:Addgene_45797). FACS-isolated CD4+ T cells from wild-type mice spleen were transfected with TN-XXL as reported (18, 58). Briefly, isolated cells were suspended in retrovirus supernatant supplemented with polybrene (8 μg/ml; Sigma-Aldrich) and IL-2 (10 ng/ml). Transfection was achieved by centrifuge at 450 relative centrifugal force for 90 min at room temperature. Cells were washed and then intravenously transferred to recipient mice. Mock transduced (vector without TN-XXL) CD4+ T cells were used as a control. TN-XXL–expressing or mock-transduced CD4+ T cells were then injected into Rag2−/− mice. The FRET of TN-XXL–expressing CD4+ T cells in the brain and peripheral tissues was detected using flow cytometry. TN-XXL consists of CFP as FRET donor and cpCitrine as FRET acceptor. These proteins are linked by the calcium-sensitive double C-terminal lobe of TnC. After binding to free calcium, TnC undergoes a conformational change that leads to energy transfer from the donor to the acceptor fluorophore, resulting in a drop in CFP fluorescence and an increase in cpCitrine fluorescence. FACSCanto system was used for intracellular calcium measurements, with a 405-nm laser for excitation of CFP and 465/30 nm and 530/30 nm bandpass filters for CFP and FRET emission, respectively, along with a 488-nm laser with a 530/30-nm bandpass filter for excitation and emission of cpCitrine.
Western blot
Mice treated with anti-CD4 mAb or IgG control were euthanized days 1 and 3 after ICH. After pericardiac perfusion with cold PBS, ipsilateral brain were harvested and homogenized in radioimmunoprecipitation assay lysis buffer (Thermo Fisher Scientific, Waltham, MA) with 1 mM phenyl-methanesulfonyl fluoride (Thermo Fisher Scientific, Waltham, MA) and phosphatase inhibitor cocktail (sc-45065, Santa Cruz Biotechnology, Dallas, TX). The supernatant from centrifugation-collected total proteins was separated by 10% SDS–polyacrylamide gel electrophoresis and transferred onto a nitrocellulose membrane (Amersham Biosciences, Piscataway, NJ). Immunoblot analysis for primary antibody incubation of ZO-1 (a0659, ABclonal), claudin 5 (29767-1-AP, Proteintech), and MMP9 (RM1020, ab283575, Abcam) at 4°C overnight. β-Actin (3700, Cell Signaling Technology) was internal control. Membrane was incubated with horseradish peroxidase–conjugated anti-rabbit or anti-mouse secondary antibody (Thermo Fisher Scientific, Waltham, MA) for 1 hour at room temperature. Chemiluminescent intensity was measured with ImageQuant LAS 4000 (GE Healthcare Life Sciences, Uppsala, Sweden).
EB analysis
To determine BBB permeability, EB dye (Sigma-Aldrich, St. Louis, MO) was used for each group of mice. Mice were intravenously injected with EB dye (2% in saline, 4 ml/kg) 4 hours before euthanization. The ipsilateral hemisphere was weighed and then homogenized into a tube with 2 ml of formamide (Sigma-Aldrich, St. Louis, MO). After incubation in water bath at 60°C for 72 hours, supernatants were collected and the optical density at 600 nm was measured by a microplate reader (Thermo Fisher Scientific, Varioskan Flash, USA). The concentration of EB was calculated with the following formula: EB content in brain tissue (μg/g wet brain) = EB concentration × formamide (ml)/wet weight (g).
Statistical analysis
Animals were randomly assigned to treatment conditions. Randomization was based on the random number generator function in Microsoft Excel software. All results were analyzed by investigators blinded to the treatment. The exclusion criteria are described in the individual method sections. No power analysis predetermined sample sizes were applied; however, the sample sizes were similar to those reported in our prior publications (59, 60). Data are presented as the means ± SD. Statistical significance was determined by two-tailed unpaired Student’s t test for parametric data in univariate analysis, Mann-Whitney test for nonparametric data, one-way analysis of variance (ANOVA) followed by Tukey post hoc test for three or more groups, and two-way ANOVA to assess the entire time course variation, accompanied by Tukey post hoc test for multiple comparisons. Fisher’s exact test was used to determine differentially expressed genes in scRNA-seq between indicated groups. P < 0.05 was considered statistically significant. All statistical analyses were performed using Prism 8.0 software (GraphPad, San Diego, CA, USA).
Acknowledgments
Funding: This study was supported in part by the American Heart Association (grant ID: 833696) and National Science Foundation of China (82122021).
Author contributions: X.W. and W.-N.J. formulated the concept and designed the studies. S.X.S., Y.X., Y.L., W.-N.J., and K.S., M.Y. performed the experiments and analyzed the results. S.X.S., K.S., Q.L., and W.-N.J. interpreted the results and drafted the manuscript.
Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. scRNA-seq data have been deposited in the NCBI Gene Expression Omnibus (GEO) (www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE230414).
Supplementary Materials
This PDF file includes:
Figs. S1 to S10
Table S1
References and notes
- 1.Q. Bai, M. Xue, V. W. Yong, Microglia and macrophage phenotypes in intracerebral haemorrhage injury: Therapeutic opportunities. Brain 143, 1297–1314 (2020). [DOI] [PubMed] [Google Scholar]
- 2.Y. Fu, J. Hao, N. Zhang, L. Ren, N. Sun, Y. J. Li, Y. Yan, D. Huang, C. Yu, F. D. Shi, Fingolimod for the treatment of intracerebral hemorrhage: A 2-arm proof-of-concept study. JAMA Neurol. 71, 1092–1101 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Y. Fu, Q. Liu, J. Anrather, F. D. Shi, Immune interventions in stroke. Nat. Rev. Neurol. 11, 524–535 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.S. X. Shi, Y. J. Li, K. Shi, K. Wood, A. F. Ducruet, Q. Liu, IL (Interleukin)-15 Bridges Astrocyte-Microglia Crosstalk and Exacerbates Brain Injury Following Intracerebral Hemorrhage. Stroke 51, 967–974 (2020). [DOI] [PubMed] [Google Scholar]
- 5.K. Shi, D. C. Tian, Z. G. Li, A. F. Ducruet, M. T. Lawton, F. D. Shi, Global brain inflammation in stroke. Lancet Neurol. 18, 1058–1066 (2019). [DOI] [PubMed] [Google Scholar]
- 6.R. F. Keep, Y. Hua, G. Xi, Intracerebral haemorrhage: Mechanisms of injury and therapeutic targets. Lancet Neurol. 11, 720–731 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.M. C. Loftspring, J. McDole, A. Lu, J. F. Clark, A. J. Johnson, Intracerebral hemorrhage leads to infiltration of several leukocyte populations with concomitant pathophysiological changes. J. Cereb. Blood Flow Metab. 29, 137–143 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.E. Mracsko, E. Javidi, S. Y. Na, A. Kahn, A. Liesz, R. Veltkamp, Leukocyte invasion of the brain after experimental intracerebral hemorrhage in mice. Stroke 45, 2107–2114 (2014). [DOI] [PubMed] [Google Scholar]
- 9.Z. Li, M. Li, S. X. Shi, N. Yao, X. Cheng, A. Guo, Z. Zhu, X. Zhang, Q. Liu, Brain transforms natural killer cells that exacerbate brain edema after intracerebral hemorrhage. J. Exp. Med. 217, e20200213 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.T. Korn, A. Kallies, T cell responses in the central nervous system. Nat. Rev. Immunol. 17, 179–194 (2017). [DOI] [PubMed] [Google Scholar]
- 11.M. Lever, P. K. Maini, P. A. van der Merwe, O. Dushek, Phenotypic models of T cell activation. Nat. Rev. Immunol. 14, 619–629 (2014). [DOI] [PubMed] [Google Scholar]
- 12.J. T. Walsh, S. Hendrix, F. Boato, I. Smirnov, J. Zheng, J. R. Lukens, S. Gadani, D. Hechler, G. Golz, K. Rosenberger, T. Kammertons, J. Vogt, C. Vogelaar, V. Siffrin, A. Radjavi, A. Fernandez-Castaneda, A. Gaultier, R. Gold, T. D. Kanneganti, R. Nitsch, F. Zipp, J. Kipnis, MHCII-independent CD4+ T cells protect injured CNS neurons via IL-4. J. Clin. Invest. 125, 699–714 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.G. Caron, D. Duluc, I. Fremaux, P. Jeannin, C. David, H. Gascan, Y. Delneste, Direct stimulation of human T cells via TLR5 and TLR7/8: Flagellin and R-848 up-regulate proliferation and IFN-gamma production by memory CD4+ T cells. J. Immunol. 175, 1551–1557 (2005). [DOI] [PubMed] [Google Scholar]
- 14.M. Komai-Koma, L. Jones, G. S. Ogg, D. Xu, F. Y. Liew, TLR2 is expressed on activated T cells as a costimulatory receptor. Proc. Natl. Acad. Sci. U.S.A. 101, 3029–3034 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.C. Kleinschnitz, N. Schwab, P. Kraft, I. Hagedorn, A. Dreykluft, T. Schwarz, M. Austinat, B. Nieswandt, H. Wiendl, G. Stoll, Early detrimental T-cell effects in experimental cerebral ischemia are neither related to adaptive immunity nor thrombus formation. Blood 115, 3835–3842 (2010). [DOI] [PubMed] [Google Scholar]
- 16.J. Zhang, K. Shi, Z. Li, M. Li, Y. Han, L. Wang, Z. Zhang, C. Yu, F. Zhang, L. Song, J.-F. Dong, A. La Cava, K. N. Sheth, F.-D. Shi, Organ- and cell-specific immune responses are associated with the outcomes of intracerebral hemorrhage. FASEB J. 32, 220–229 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.N. I. Kyratsous, I. J. Bauer, G. Zhang, M. Pesic, I. Bartholomaus, M. Mues, P. Fang, M. Worner, S. Everts, J. W. Ellwart, J. M. Watt, B. V. L. Potter, R. Hohlfeld, H. Wekerle, N. Kawakami, Visualizing context-dependent calcium signaling in encephalitogenic T cells in vivo by two-photon microscopy. Proc. Natl. Acad. Sci. U.S.A. 114, E6381–E6389 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.M. Mues, I. Bartholomaus, T. Thestrup, O. Griesbeck, H. Wekerle, N. Kawakami, G. Krishnamoorthy, Real-time in vivo analysis of T cell activation in the central nervous system using a genetically encoded calcium indicator. Nat. Med. 19, 778–783 (2013). [DOI] [PubMed] [Google Scholar]
- 19.H. Kebir, K. Kreymborg, I. Ifergan, A. Dodelet-Devillers, R. Cayrol, M. Bernard, F. Giuliani, N. Arbour, B. Becher, A. Prat, Human TH17 lymphocytes promote blood-brain barrier disruption and central nervous system inflammation. Nat. Med. 13, 1173–1175 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.I. M. Stromnes, L. M. Cerretti, D. Liggitt, R. A. Harris, J. M. Goverman, Differential regulation of central nervous system autoimmunity by T(H)1 and T(H)17 cells. Nat. Med. 14, 337–342 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.H. Park, Z. Li, X. O. Yang, S. H. Chang, R. Nurieva, Y. H. Wang, Y. Wang, L. Hood, Z. Zhu, Q. Tian, C. Dong, A distinct lineage of CD4 T cells regulates tissue inflammation by producing interleukin 17. Nat. Immunol. 6, 1133–1141 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Y. Shigemori, Y. Katayama, T. Mori, T. Maeda, T. Kawamata, Matrix metalloproteinase-9 is associated with blood-brain barrier opening and brain edema formation after cortical contusion in rats. Acta Neurochir. Suppl. 96, 130–133 (2006). [DOI] [PubMed] [Google Scholar]
- 23.T. L. Barr, L. L. Latour, K. Y. Lee, T. J. Schaewe, M. Luby, G. S. Chang, Z. El-Zammar, S. Alam, J. M. Hallenbeck, C. S. Kidwell, S. Warach, Blood-brain barrier disruption in humans is independently associated with increased matrix metalloproteinase-9. Stroke 41, e123–e128 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.S. Urday, W. T. Kimberly, L. A. Beslow, A. O. Vortmeyer, M. H. Selim, J. Rosand, J. M. Simard, K. N. Sheth, Targeting secondary injury in intracerebral haemorrhage--perihaematomal oedema. Nat. Rev. Neurol. 11, 111–122 (2015). [DOI] [PubMed] [Google Scholar]
- 25.V. A. Shah, R. E. Thompson, G. Yenokyan, J. N. Acosta, R. Avadhani, R. Dlugash, N. McBee, Y. Li, B. M. Hansen, N. Ullman, G. Falcone, I. A. Awad, D. F. Hanley, W. C. Ziai, One-Year Outcome Trajectories and Factors Associated with Functional Recovery Among Survivors of Intracerebral and Intraventricular Hemorrhage With Initial Severe Disability. JAMA Neurol. 79, 856–868 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.X. Hu, T. M. De Silva, J. Chen, F. M. Faraci, Cerebral vascular disease and neurovascular injury in ischemic stroke. Circ. Res. 120, 449–471 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.C. Iadecola, J. Anrather, The immunology of stroke: From mechanisms to translation. Nat. Med. 17, 796–808 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.A. M. Aloizou, V. Siokas, G. Pateraki, I. Liampas, C. Bakirtzis, Z. Tsouris, G. Lazopoulos, D. Calina, A. O. Docea, A. Tsatsakis, D. P. Bogdanos, E. Dardiotis, Thinking outside the ischemia box: Advancements in the use of multiple Sclerosis drugs in ischemic stroke. J. Clin. Med. 10, 630 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.J. Elkins, R. Veltkamp, J. Montaner, S. C. Johnston, A. B. Singhal, K. Becker, M. G. Lansberg, W. Tang, I. Chang, K. Muralidharan, S. Gheuens, L. Mehta, M. S. V. Elkind, Safety and efficacy of natalizumab in patients with acute ischaemic stroke (ACTION): A randomised, placebo-controlled, double-blind phase 2 trial. Lancet Neurol. 16, 217–226 (2017). [DOI] [PubMed] [Google Scholar]
- 30.Y. Fu, N. Zhang, L. Ren, Y. Yan, N. Sun, Y. J. Li, W. Han, R. Xue, Q. Liu, J. Hao, C. Yu, F. D. Shi, Impact of an immune modulator fingolimod on acute ischemic stroke. Proc. Natl. Acad. Sci. U.S.A. 111, 18315–18320 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.M. Durocher, B. Knepp, A. Yee, G. Jickling, F. Rodriguez, K. Ng, X. Zhan, F. Hamade, E. Ferino, H. Amini, P. Carmona-Mora, H. Hull, B. P. Ander, F. R. Sharp, B. Stamova, Molecular Correlates of Hemorrhage and Edema Volumes Following Human Intracerebral Hemorrhage Implicate Inflammation, Autophagy, mRNA Splicing, and T Cell Receptor Signaling. Transl. Stroke Res. 12, 754–777 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.E. Mracsko, R. Veltkamp, Neuroinflammation after intracerebral hemorrhage. Front. Cell. Neurosci. 8, 388 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.R. Cai, C. Pan, A. Ghasemigharagoz, M. I. Todorov, B. Forstera, S. Zhao, H. S. Bhatia, A. Parra-Damas, L. Mrowka, D. Theodorou, M. Rempfler, A. L. R. Xavier, B. T. Kress, C. Benakis, H. Steinke, S. Liebscher, I. Bechmann, A. Liesz, B. Menze, M. Kerschensteiner, M. Nedergaard, A. Erturk, Panoptic imaging of transparent mice reveals whole-body neuronal projections and skull-meninges connections. Nat. Neurosci. 22, 317–327 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.J. H. Ahn, H. Cho, J. H. Kim, S. H. Kim, J. S. Ham, I. Park, S. H. Suh, S. P. Hong, J. H. Song, Y. K. Hong, Y. Jeong, S. H. Park, G. Y. Koh, Meningeal lymphatic vessels at the skull base drain cerebrospinal fluid. Nature 572, 62–66 (2019). [DOI] [PubMed] [Google Scholar]
- 35.W. B. Rolland, T. Lekic, P. R. Krafft, Y. Hasegawa, O. Altay, R. Hartman, R. Ostrowski, A. Manaenko, J. Tang, J. H. Zhang, Fingolimod reduces cerebral lymphocyte infiltration in experimental models of rodent intracerebral hemorrhage. Exp. Neurol. 241, 45–55 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.L. L. Mao, H. Yuan, W. W. Wang, Y. J. Wang, M. F. Yang, B. L. Sun, Z. Y. Zhang, X. Y. Yang, Adoptive regulatory T-cell therapy attenuates perihematomal inflammation in a mouse model of experimental intracerebral hemorrhage. Cell. Mol. Neurobiol. 37, 919–929 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.X. Zhang, W. Liu, J. Yuan, H. Zhu, Y. Yang, Z. Wen, Y. Chen, L. Li, J. Lin, H. Feng, T lymphocytes infiltration promotes blood-brain barrier injury after experimental intracerebral hemorrhage. Brain Res. 1670, 96–105 (2017). [DOI] [PubMed] [Google Scholar]
- 38.L. Gao, P. P. Li, T. Y. Shao, X. Mao, H. Qi, B. S. Wu, M. Shan, L. Ye, H. W. Cheng, Neurotoxic role of interleukin-17 in neural stem cell differentiation after intracerebral hemorrhage. Neural Regen. Res. 15, 1350–1359 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Q. Zhong, K. Zhou, Q. L. Liang, S. Lin, Y. C. Wang, X. Y. Xiong, Z. Y. Meng, T. Zhao, W. Y. Zhu, Y. R. Yang, M. F. Liao, Q. W. Gong, L. Liu, A. Xiong, J. Hao, J. Wang, Q. W. Yang, Interleukin-23 secreted by activated macrophages drives γδT cell production of interleukin-17 to aggravate secondary injury after intracerebral hemorrhage. J. Am. Heart Assoc. 5, e004340 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.H. Yang, X. Gao, W. Xiao, J. Su, Y. Li, W. Ni, Y. Gu, Minocycline Alleviates White Matter Injury following Intracerebral Hemorrhage by Regulating CD4+ T Cell Differentiation via Notch1 Signaling Pathway. Oxid. Med. Cell. Longev. 2022, 3435267 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.E. Esposito, B. J. Ahn, J. Shi, Y. Nakamura, J. H. Park, E. T. Mandeville, Z. Yu, S. J. Chan, R. Desai, A. Hayakawa, X. Ji, E. H. Lo, K. Hayakawa, Brain-to-cervical lymph node signaling after stroke. Nat. Commun. 10, 5306 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.S. L. Swain, K. K. McKinstry, T. M. Strutt, Expanding roles for CD4+ T cells in immunity to viruses. Nat. Rev. Immunol. 12, 136–148 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.M. Li, Z. Li, H. Ren, W. N. Jin, K. Wood, Q. Liu, K. N. Sheth, F. D. Shi, Colony stimulating factor 1 receptor inhibition eliminates microglia and attenuates brain injury after intracerebral hemorrhage. J. Cereb. Blood Flow Metab. 37, 2383–2395 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.S. X. Shi, K. Shi, Q. Liu, Brain injury instructs bone marrow cellular lineage destination to reduce neuroinflammation. Sci. Transl. Med. 13, (2021). [DOI] [PubMed] [Google Scholar]
- 45.Y. Gan, Q. Liu, W. Wu, J. X. Yin, X. F. Bai, R. Shen, Y. Wang, J. Chen, A. La Cava, J. Poursine-Laurent, W. Yokoyama, F. D. Shi, Ischemic neurons recruit natural killer cells that accelerate brain infarction. Proc. Natl. Acad. Sci. U.S.A. 111, 2704–2709 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.S. L. Wolock, R. Lopez, A. M. Klein, Scrublet: Computational identification of cell doublets in single-cell transcriptomic data. Cell Syst. 8, 281–291.e9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.L. Haghverdi, A. T. L. Lun, M. D. Morgan, J. C. Marioni, Batch effects in single-cell RNA-sequencing data are corrected by matching mutual nearest neighbors. Nat. Biotechnol. 36, 421–427 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.G. Yu, L. G. Wang, Y. Han, Q. Y. He, clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 16, 284–287 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Y. Sui, W. Zeng, MS4A4A Regulates Arginase 1 Induction during Macrophage Polarization and Lung Inflammation in Mice. Eur. J. Immunol. 50, 1602–1605 (2020). [DOI] [PubMed] [Google Scholar]
- 50.L. A. Hohsfield, A. R. Najafi, Y. Ghorbanian, N. Soni, E. E. Hingco, S. J. Kim, A. D. Jue, V. Swarup, M. A. Inlay, K. N. Green, Effects of long-term and brain-wide colonization of peripheral bone marrow-derived myeloid cells in the CNS. J. Neuroinflammation 17, 279 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.R. Reschke, J. Yu, B. Flood, E. F. Higgs, K. Hatogai, T. F. Gajewski, Immune cell and tumor cell-derived CXCL10 is indicative of immunotherapy response in metastatic melanoma. J. Immunother. Cancer 9, e003521 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.S. De Munck, M. Provost, M. Kurikawa, I. Omori, J. Mukohyama, J. Felix, Y. Bloch, O. Abdel-Wahab, J. F. Bazan, A. Yoshimi, S. N. Savvides, Structural basis of cytokine-mediated activation of ALK family receptors. Nature 600, 143–147 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.T. N. Sims, T. J. Soos, H. S. Xenias, B. Dubin-Thaler, J. M. Hofman, J. C. Waite, T. O. Cameron, V. K. Thomas, R. Varma, C. H. Wiggins, M. P. Sheetz, D. R. Littman, M. L. Dustin, Opposing effects of PKCtheta and WASp on symmetry breaking and relocation of the immunological synapse. Cell 129, 773–785 (2007). [DOI] [PubMed] [Google Scholar]
- 54.J. Zhang, X. Bu, H. Wang, Y. Zhu, Y. Geng, N. T. Nihira, Y. Tan, Y. Ci, F. Wu, X. Dai, J. Guo, Y. H. Huang, C. Fan, S. Ren, Y. Sun, G. J. Freeman, P. Sicinski, W. Wei, Cyclin D-CDK4 kinase destabilizes PD-L1 via cullin 3-SPOP to control cancer immune surveillance. Nature 553, 91–95 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.A. M. Mitterstiller, D. Haschka, S. Dichtl, M. Nairz, E. Demetz, H. Talasz, M. P. Soares, E. Einwallner, H. Esterbauer, F. C. Fang, S. Geley, G. Weiss, Heme oxygenase 1 controls early innate immune response of macrophages to Salmonella Typhimurium infection. Cell. Microbiol. 18, 1374–1389 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.K. L. Jeffrey, M. Camps, C. Rommel, C. R. Mackay, Targeting dual-specificity phosphatases: Manipulating MAP kinase signalling and immune responses. Nat. Rev. Drug Discov. 6, 391–403 (2007). [DOI] [PubMed] [Google Scholar]
- 57.T. Herta, A. Bhattacharyya, M. Rosolowski, C. Conrad, C. Gurtner, A. D. Gruber, P. Ahnert, B. Gutbier, D. Frey, N. Suttorp, S. Hippenstiel, J. Zahlten, Krueppel-Like Factor 4 Expression in Phagocytes Regulates Early Inflammatory Response and Disease Severity in Pneumococcal Pneumonia. Front. Immunol. 12, 726135 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.M. Mank, A. F. Santos, S. Direnberger, T. D. Mrsic-Flogel, S. B. Hofer, V. Stein, T. Hendel, D. F. Reiff, C. Levelt, A. Borst, T. Bonhoeffer, M. Hubener, O. Griesbeck, A genetically encoded calcium indicator for chronic in vivo two-photon imaging. Nat. Methods 5, 805–811 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Q. Liu, W. N. Jin, Y. Liu, K. Shi, H. Sun, F. Zhang, C. Zhang, R. J. Gonzales, K. N. Sheth, A. La Cava, F. D. Shi, Brain ischemia suppresses immunity in the periphery and brain via different neurogenic innervations. Immunity 46, 474–487 (2017). [DOI] [PubMed] [Google Scholar]
- 60.W. N. Jin, K. Shi, W. He, J. H. Sun, L. Van Kaer, F. D. Shi, Q. Liu, Neuroblast senescence in the aged brain augments natural killer cell cytotoxicity leading to impaired neurogenesis and cognition. Nat. Neurosci. 24, 61–73 (2021). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S10
Table S1