Abstract
Tardigrades are microscopic invertebrates, which are capable of withstanding extreme environmental conditions, including high levels of radiation. A Tardigrade protein, Dsup (Damage Suppressor), protects the Tardigrade’s DNA during harsh environmental stress and X-rays. When expressed in cancer cells, Dsup protects DNA from single and double-strand breaks (DSBs) induced by radiation, increases survival of irradiated cells, and protects DNA from reactive oxygen species. These unusual properties of Dsup suggested that understanding how the protein functions may help in the design of small molecules that could protect humans during radiotherapy or space travel. Here, we investigated if Dsup is protective in cortical neurons cultured from rat embryos. We discovered that, in cortical neurons, the codon-optimized Dsup localizes to the nucleus and, surprisingly, promotes neurotoxicity leading to neurodegeneration. Unexpectedly, we found that Dsup expression results in the formation of DNA DSBs in cultured neurons. With electron microscopy, we discovered that Dsup promotes chromatin condensation. Unlike Dsup’s protective properties in cancerous cells, in neurons, Dsup promotes neurotoxicity, induces DNA damage, and rearranges chromatin. Taken together, neurons are sensitive to Dsup, and Dsup is a doubtful surrogate for DNA protection in neuronal cells.
Introduction
Tardigrades (also known as water bears or moss piglets) are microscopic organisms that can withstand extreme environmental conditions including desiccation, extreme temperatures, and cosmic radiation1–3. Recently, it was demonstrated that the DNA-binding protein Dsup (Damage Suppressor), a protein from the tardigrade Ramazzottius varieornatus, protects Tardigrade’s DNA during severe environmental stress and X-ray radiation4. When expressed in human embryonic kidney 293 cells (HEK 293), intriguingly, Dsup binds to chromatin, mitigates X-ray-induced DNA damage, and enhances cellular radiotolerance4–6. The expression of Dsup in tobacco plants enhances their tolerance to UV and X-ray radiation7. A Dsup ortholog from the tardigrade Hypsibius exemplaris binds to nucleosomes, rather than to pure DNA, to protect DNA from damage by reactive oxygen species (ROS)8. A conserved domain in the Dsup proteins that mediates nucleosome binding and protection against ROS is homologous to the nucleosome-binding domain of vertebrate high mobility group nucleosome-binding (HMGN) proteins8. It was suggested that Dsup or Dsup-like factors could be potentially introduced into more sensitive organisms to protect them from DNA damage9–11. Whether Dsup is protective for all mammalian cell types was not, however, investigated.
In this study, we investigated if the DNA-binding protein Dsup12 from the tardigrade Ramazzottius varieornatus protects neuronal DNA from damage as it does in the Tardigrade, mammalian cancerous cells, and in plants. Surprisingly, we discovered that, in cultured primary murine neurons, Dsup is neurotoxic and promotes neurodegeneration. The expression of Dsup in neurons results in chromatin rearrangement and induces DNA DSBs, whereas it does not do so in cultured cancerous cells (HEK 293), as expected. Therefore, Dsup might be a valuable new tool to dissect cell type specific mechanisms of DNA damage and repair.
Results
Expressing Dsup in primary neurons
As the sequence of the Dsup was published4, we synthesized the Dsup gene optimized for the expression in mammalian cells. To track Dsup within the cells, we tagged Dsup with a red fluorescent protein, mApple or with a blue fluorescent protein, BFP, to Dsup’s C-terminus (Figure 1a). Since HEK 293 cells were used in Hashimoto et al. to assess Dsup’s localization4, we first confirmed that the cloned Dsup is indeed nuclear in HEK 293 cells. HEK 293 cells were transfected with Dsup-mApple along with a marker of cellular morphology and viability, a green fluorescent protein, Wasabi. Cultured cells were then fixed and stained with the nuclear dye DAPI. In HEK 293 cells, Dsup-mApple was exclusively nuclear (Figure 1b).
Figure 1.
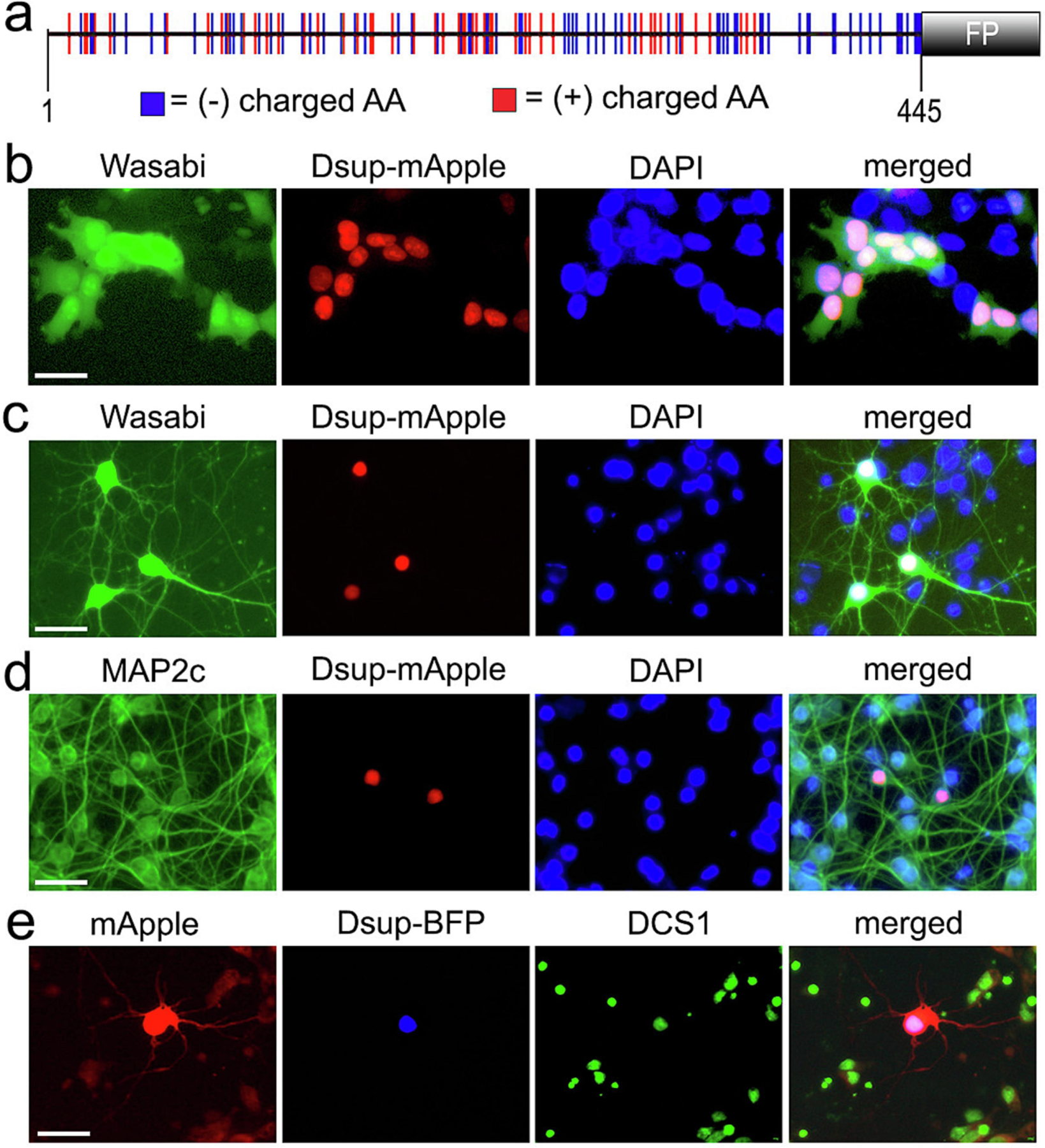
Expression Dsup in primary neurons. (a) A scheme of the tardigrade Ramazzottius varieornatus Dsup protein, optimized for the expression in mammalian cells, fused to a fluorescent protein (mApple or BFP). The positions of acidic and basic amino acid residues within the 445 amino acid protein indicate that the Dsup protein is highly charged. (b) HEK 293 cells were co-transfected with Dsup-mApple and Wasabi. Cells were fixed 24 hours after transfection, stained the DAPI dye, and imaged. Note that the Dsup construct is nuclear. Scale bar, 10 µm. (c) Primary cortical neurons were co-transfected with Dsup-mApple and Wasabi. 24 hours after transfection, live cells were stained the DAPI dye and imaged. Note that Dsup-mApple is localized to the nucleus. Scale bar, 20 µm. (d) Primary cortical neurons transfected with Dsup-mApple, fixed 24 hours after transfection, and stained with the antibody against MAP2c and with the DAPI dye. Note that the Dsup-mApple construct is localized to the neuronal nucleus. Scale bar, 20 µm. (e) Primary cortical neurons were co-transfected with Dsup-BFP and mApple. 24 hours after transfection, neurons were fixed, stained the DCS1 dye and imaged. Note that Dsup-BFP is localized to the neuronal nucleus. Scale bar, 20 µm.
Next, primary rat cortical neurons were transfected with Dsup-mApple along with Wasabi, a marker of neuronal morphology. Live neurons were stained with the nuclear DAPI dye and imaged (Figure 1c). In parallel experiments, primary neurons were transfected with Dsup-mApple alone, fixed and stained with an antibody against MAP2c, a neuronal marker, and the DAPI dye (Figure 1d). Finally, neurons were transfected with Dsup-TagBFP (Dsup-BFP thereafter) and mApple to visualize neuronal morphology, fixed and stained a green nuclear dye, DCS1 (Figure 1e). Similarly to HEK 293 cells expressing Dsup, the Dsup constructs localized to the neuronal nucleus, illustrating the conserved mechanisms of nuclear import between the Tardigrades and mammalian cells including neurons (Figure 1b-e). These findings indicate that the murine-optimized Dsup protein, a DNA-binding protein from the Tardigrade Ramazzottius varieornatus, can be successfully expressed in murine neuronal cells with the protein being localized to the nucleus.
Dsup is toxic for primary neurons
While analyzing neurons that express Dsup-mApple, we noticed that many of these cells over time developed morphological signs of neurotoxicity such as neurite beading and blebbing. This was unexpected since the Dsup protein was in fact protective for HEK 293 cells that stably expressed the Dsup construct4. Therefore, we determined if the expression of Dsup-mApple contributes negatively to neuronal fate in our neuronal model. We used a microscopy system, automated imaging and longitudinal analysis, which enables following large cohorts of individual neurons over their lifetimes (Figure 2a and Supplemental Figure 1). By analyzing a sequence of images with individual neurons, we can apply statistical approaches to the survival data, and measure neurodegeneration or neuroprotection. We discovered that both Dsup-mApple and Dsup-BFP substantially increased the risk of neuronal death compared to neurons that expressed mApple or BFP (Figure 2b). Neurons expressing Dsup constructs often had their neurites retracted, which happens in neurodegenerative processes. We conclude that, unlike cancerous HEK 293 cells4, neurons are sensitive to the Dsup protein.
Figure 2.
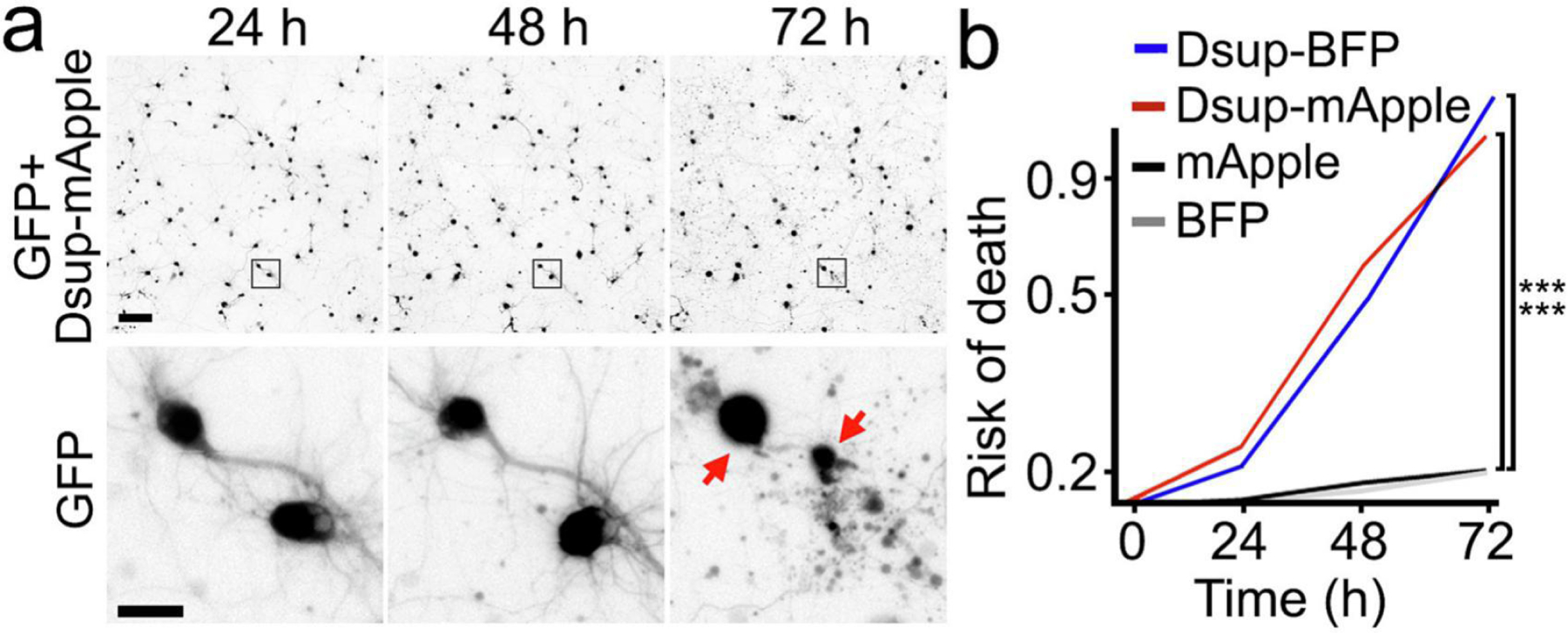
The Dsup constructs are neurotoxic. (a) An example of longitudinal imaging of primary neurons. Cultured cortical neurons were co-transfected with Dsup-mApple and a green fluorescent protein (GFP) to visualize neuronal morphology. 24 hours after transfection, the same group of neurons were imaged longitudinally with an automated microscope at different time points. The top panel is a montage of non-overlapping images captured in one well of a 24-well plate. Scale bar is 100 µm. The bottom panel is zoomed onto two neurons (the GFP channel) to demonstrate longitudinal single-cell tracking. Scale bar is 10 µm. Note that both neurons died before the last imaging event (dead cells are depicted with red arrows). (b) Neurons were transfected with mApple and GFP or Dsup-mApple and GFP or with BFP and GFP or with Dsup-BFP and GFP, and tracked with an automated microscope for 72 hours. Risk of death curves demonstrate that Dsup-mApple and Dsup-BFP are toxic for neurons. ***p (mApple vs Dsup-mApple) < 0.0001, ***p (BFP vs Dsup-mBFP) < 0.0001 (log-rank test, JMP statistical software). More than one hundred neurons per group were analyzed from three independent experiments.
Dsup induces DNA DSBs in neurons
We hypothesized that we could still detect protection from DNA damage before significant neurotoxicity occurs in neurons. We cultured primary neurons, transfected them with Dsup-BFP and mApple, fixed and stained with an antibody against phosphorylated histone H2A variant X (γH2A.X), which is commonly used as a read-out of DNA DSBs13. Unexpectedly, we discovered that, under control conditions with no stressors applied to neurons, the Dsup protein promoted DNA DSBs in the neuronal nuclei (Figure 3a, Supplemental Figure 2). Indeed, neurons that expressed the Dsup-BFP construct were positive for γH2A.X in the nuclei and were positive for annexin V, when compared to neurons that expressed BFP (Figure 3b, Supplemental Figure 3, Supplemental Figure 4). Our data demonstrate that Dsup promotes DNA damage in neuronal cells.
Figure 3. Dsup induces DNA DSBs in neurons.
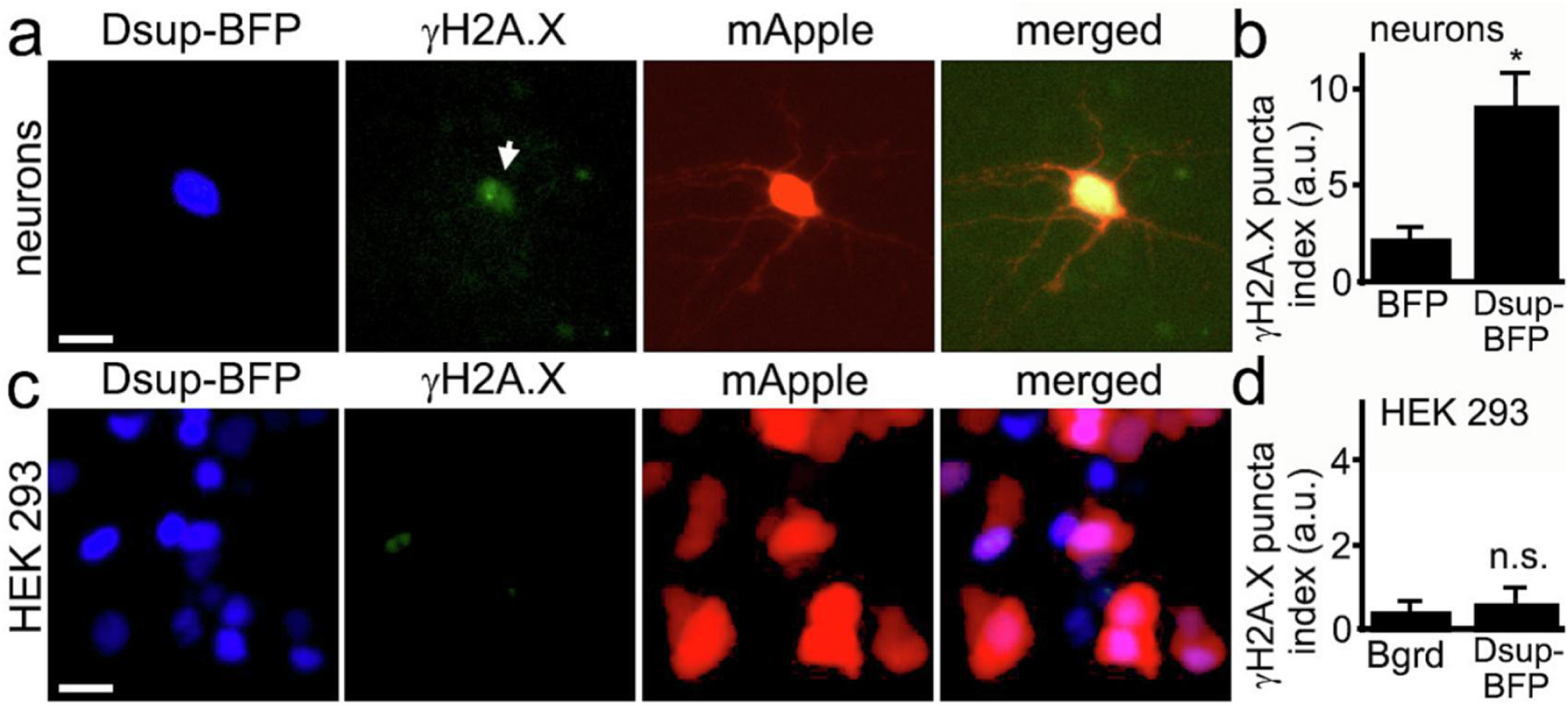
(a) Primary neurons transfected with Dsup-BFP and mApple, fixed 48 hours after transfection, and immunostained with antibodies against γH2A.X. Note that the Dsup-BFP-positive neuron exhibits γH2AX puncta, indicating DNA DSBs (depicted with the arrow). Scale bar is 20 µm. (b) Images of fixed neurons from (a) were analyzed with ImageJ and the puncta index was estimated by measuring the standard deviation of γH2A.X fluorescence intensity. The puncta index of γH2A.X staining is increased in neurons that express Dsup-BFP. BFP, BFP-transfected neurons from Supplemental Figure 3. p= 0.0137 (t-test), a.u., arbitrary units. 271 Dsup-BFP-expressing and 321 BFP-expressing neurons were analyzed. Results were pooled from four experiments. (c) HEK 293 cells transfected with mApple and Dsup-BFP, fixed 48 hours after transfection, and immunostained with antibodies against γH2A.X. Virtually no nuclear γH2A.X puncta were observed in Dsup-BFP expressing cells. Scale bar is 10 µm. (d) Images of fixed HEK 293 cells from (c) were analyzed with ImageJ and the puncta index was estimated by measuring the standard deviation of γH2A.X fluorescence intensity and compared to background. Bgrd., background; n.s., non-significant, p=0.35 (t-test), a.u., arbitrary units. Results were pooled from three experiments.
In their elegant work, Hashimoto et al., demonstrated that Dsup prevents the formation of DNA DSBs in HEK 293 cells. We decided to confirm that Dsup does not promote DNA damage in HEK 293 cells in our hands. We cultured HEK 293 cells and transfected them with the Dsup-BFP and mApple constructs, then fixed and stained these cells with an antibody against γH2A.X (Figure 3c). As expected, Dsup did not induce DNA DSBs in HEK 293 cells (Figure 3c, d). These data illustrate the important differences in DNA homeostatic mechanisms between primary neurons and cancer cells14.
Dsup promotes condensation of chromatin
Our finding that Dsup itself promotes DNA DSBs in neurons was surprising. We, therefore, decided to use transmission electron microscopy (TEM) and look at cultures that express the BFP or Dsup-BFP construct. For that, we generated two viruses, the BFP virus and the Dsup-BFP virus. Primary cultured neurons were infected with these constructs (Supplemental Figure 5). At 72 hours after infection, the expression plateaued and >70% of cells were expressing the constructs (Supplemental Figure 6). The expression of BFP and Dsup-BFP was also confirmed with western blotting (Supplemental Figure 6). Cultures were then fixed and analyzed with TEM. Some neurons in control cultures had neurons with condensed chromatin; however, neurons in Dsup-BFP-infected cultures often exhibited nuclei with significantly rearranged, condensed chromatin (Figure 4a, b). Intriguingly, the cytoplasm of neurons from the Dsup-BFP-infected cohort did not exhibit any major abnormalities (Figure 4c). For example, mitochondria appeared to be healthy, indicating that we likely caught the moment before chromatin condensation translated into overall neurotoxicity. We conclude that, unlike cancerous cells, neurons do not tolerate the Tardigrade protein likely due to Dsup’s interference with chromatin physiological structure and function.
Figure 4. Dsup alters the structure of chromatin in primary neurons.
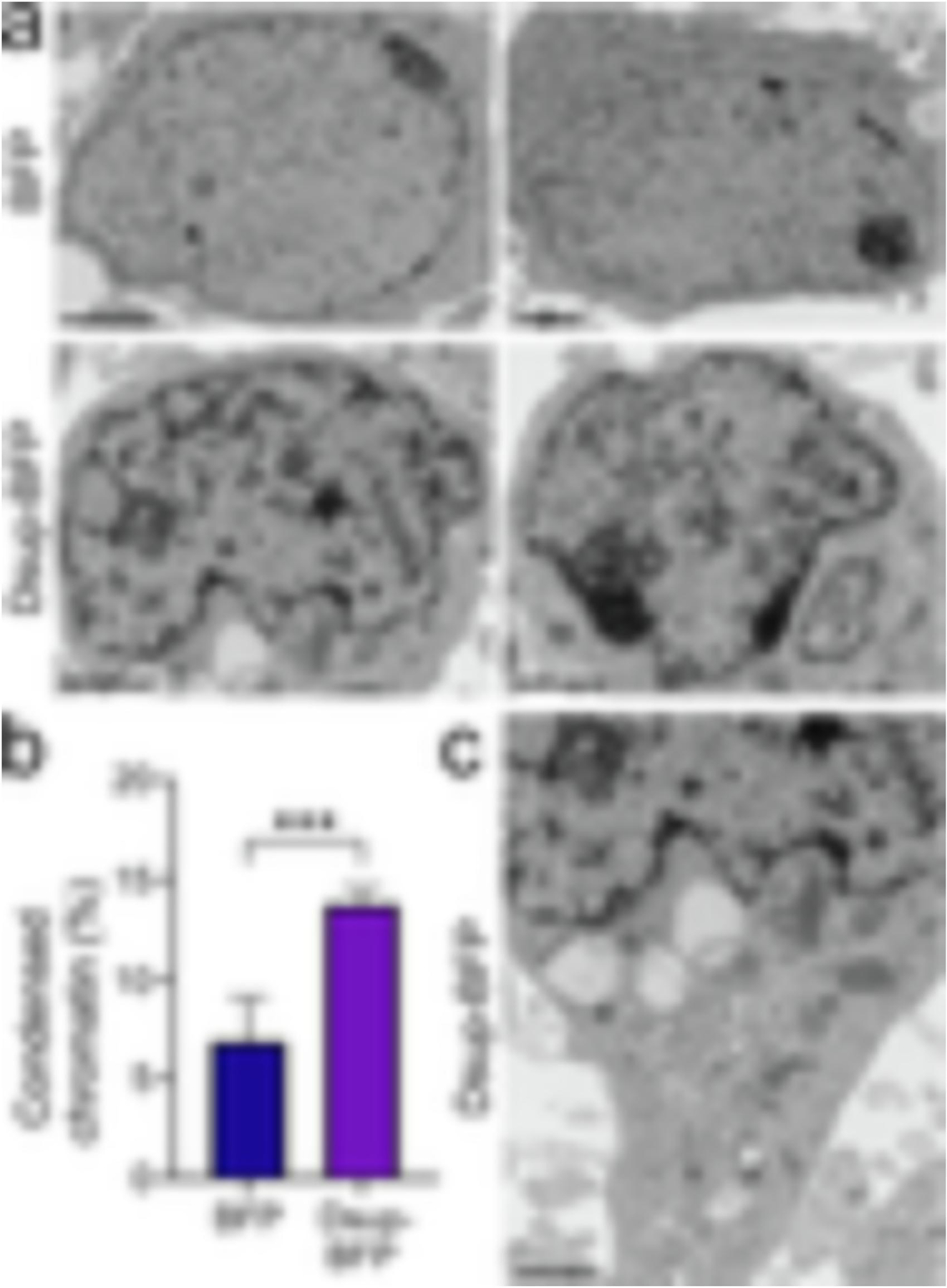
(a) Representative electron micrograph of cultured cortical neurons infected with the BFP lentivirus (BFP) or the Dsup-BFP lentivirus. Primary cultures were infected, fixed 72 hours when the expression plateaued and >70% of cells were expressing the constructs, and processed for electron microscopy imaging. Results were pooled from two experiments. Bar (BFP), 2 μm; bar (Dsup-BFP), 1 μm. (b) The percentage of condensed chromatin was calculated in the BFP-infected and Dsup-BFP-infected cohorts. P=0.0002 (t-test). (c) The cytoplasm of the neuron from the Dsup-BFP-infected cohort from (a) does not exhibit major abnormalities. Bar, 1 μm.
Discussion
Here, we demonstrate that Dsup, a protein from the Tardigrade, localizes to the nucleus and promotes neurotoxicity, induces the formation of DNA DSBs and rearranges chromatin. Unlike cancerous cells, neurons are sensitive to Dsup, and Dsup is a questionable surrogate for DNA protection in neurons.
Tardigrades (from Latin tardigradus, slow-stepper) is a model organism in medicine10,15 and astrobiology16–18, as they can survive extreme low and high temperatures (from −273°C to +150°C), vacuum, high pressure and high doses of ionizing radiation19 and exposure to outer space1,20. Radiation causes severe DNA damage, results in genome instability and death of the organism, but Tardigrades can withstand almost a thousand times more radiation than other animals9. An effective DNA repair system, DNA condensation, and the levels of cellular hydration (e.g., a desiccated state) contribute to radiotolerance in radiotolerant organisms9. A protein that protects DNA from radiation was discovered in Tardigrades, the Dsup protein4. Importantly, Dsup protects DNA from damage rather than facilitates DNA damage repair4. When expressed in human cancerous cells, Dsup considerably reduces DNA damage associated with X-ray irradiation and treatment with hydrogen peroxide4. Dsup binds to the nucleosomes directly and protects DNA from hydroxyl radicals8. These unique properties of Dsup, therefore, suggested that Dsup itself or small molecules with Dsup-like features could be used in human cells to increase tolerance to radiation and ROS10,21, even to extend cell longevity8, and for use in space exploration6. Our data demonstrate, however, that expression of Dsup in primary cultured neurons, post-mitotic cells, itself alters chromatin and promotes DNA DSBs, indicating that recapitulating protective Dsup mechanisms in all cell types is not always possible. Nevertheless, there is a possibility that differences between the prior studies and our study could be due the methodology. We cannot completely rule out that imaging of fluorescently tagged Dsup could potentially affect Dsup’s protective properties and induce neurotoxicity. However, it is unlikely because Dsup fused to a fluorescent protein (e.g., mApple and BFP) induces DNA damage in neurons and dramatically alters chromatin structure in neurons. The expression of fluorescent proteins alone does not promote DNA damage or alters chromatin structure. Future studies will investigate if methodology affects Dsup’s protective properties and/or toxicity and whether Dsup is toxic for other post-mitotic cells.
A conserved domain in the Dsup proteins, which enables binding to nucleosomes, is homologous to the nucleosome-binding domain of HMGN proteins in vertebrates8. HMGN proteins are non-histone architectural proteins that bind to nucleosomes and regulate the structure of chromatin12. Being ubiquitous in vertebrate cells, five members of the HMGN family are involved in many chromatin-dependent processes such as transcription, replication, and DNA repair12. Intriguingly, individuals with Down syndrome overexpress HMGN1 protein; however, it is not clear if HMGN1 levels contribute to Down syndrome phenotypes12. Overexpression of HMGN4 leads to altered transcription and increases the levels of a DNA DSB marker, γH2AX22. It has been suggested that functions of Dsup and HMGN proteins may overlap at least in part8,12.
In neuronal cells, DNA damage induced by endogenous or exogenous processes, if not effectively repaired, promotes genome instability and results in premature neuronal aging and age-associated neurodegenerative diseases. Neurons are post-mitotic and often last for a lifetime, and that adds complexity to guarding their genome integrity23. Accumulation of DNA damage has been linked to many age-associated neurodegenerative diseases24–26. It is not clear how Dsup induces DNA DSBs. Theoretically, Dsup bound to DNA may stall DNA polymerase during transcription, and then transcription–coupled–repair poisoning with the action of endonucleases could result in DNA damage27. Another possibility is the disruption of DNA repair complexes in the nucleus, leading to DNA damage, chromatin condensation, and apoptosis. There are many possibilities and it is likely that a combination of several mechanisms leads to neurotoxicity. Future studies in other cell types, mitotic and post-mitotic, will demonstrate if Dsup and Dsup-like proteins can be practically used to understand DNA protection mechanisms.
Methods
Chemicals, antibodies, plasmids, and viruses
The DAPI dye was from Santa Cruz Biotechnology (sc-394039). The Nuclear Green DCS1 dye was from Cayman Chemical (25171). Mouse antibodies against MAP2c were from Santa Cruz Biotechnology (1:500; sc-74421). Rabbit antibodies against annexin V were from Proteintech (1:500; 11060–1-AP). Rabbit antibodies against gH2AX were from Abcam (1:500; ab11174). The TagRFP polyclonal antibody were from ThermoFisher Scientific (1:1000; R10367). According to the manufacturer, this antibody recognizes fluorescent proteins derived from Entacmaea and Discosoma sea anemones. Rabbit antibodies against HRP were from Abcam (1:2000; ab97023) Anti-mouse Alexa Fluor 488-labeled (1:1000 #A-11001) and anti-rabbit Alexa Fluor 488-labeled (1:1000 #A-11008) secondary antibodies were from Thermo Fisher Scientific. pGW1-mApple and pGW1-GFP were described28. pGW1-Wasabi was cloned from pmWasabi-C (Allele Biotechnology). The pLV[Exp]-EF1A>Dsup:3xGGGGS:TagBFP2 lentivirus (>108 TU/ml) and pLV[Exp]-EF1A>TagBFP2 lentivirus (>108 TU/ml) were constructed and prepared by Vectorbuilder.
The protein sequence was previously published9. pCAG-Dsup-mApple, pCAG-Dsup-TagBFP, and pCAG-TagBFP were synthesized by VectorBuilder (the CAG promoter is the CMV early enhancer fused to modified chicken β-actin promoter). There are no studies that analyzed the codon system of the Tardigrades. However, there are papers on closely related to Arthropoda and Onychophora characterized by a specific coding system for particular codons29–33. The Zhang lab optimized the Dsup’s cDNA for the expression in the tobacco plant7. The Kadonaga lab optimized the Dsup’s cDNA for the expression in E. coli8. The Goldstein lab optimized Tardigrade proteins for the expression in E. coli as well2. The Dsup cDNA was, therefore, optimized for the expression in mammalian cells with VectorBuilder.
Cell cultures
Cortices from rat embryos (E17–18) were dissected and dissociated. Cells were plated on 24-well tissue-culture plates (2×106/well), coated with poly-D-lysine (Sigma-Aldrich, P6407), as described34. Primary cortical neurons were grown in Neurobasal Plus Medium (Gibco, 21103049) supplemented with B-27 (Gibco, 17504001), and penicillin-streptomycin (Gibco, 15140122). Some primary cultures were transfected after 4 days in vitro with Lipofectamine 2000 (Thermo Fisher Scientific) and with a total of 1 μg of plasmid DNA per well, as described34.
HEK 293 cells were a gift by Carmen Dessauer (Integrative Biology and Pharmacology, the University of Texas McGovern Medical School at Houston). Cells were maintained in Dulbecco’s Modified Eagle Medium (Thermo Fisher Scientific) supplemented with 10% fetal bovine serum (Sigma), and penicillin-streptomycin (Life Technologies). Before an experiment, HEK 293 cells were plated into 24-well tissue-culture plates. After 24 h, cells were transfected with Lipofectamine 2000 (Thermo Fisher Scientific), with a total of 1 μg of plasmid DNA per well, according to the manufacturer's protocol.
Immunocytochemistry
Primary cortical neurons and HEK 293 cells were fixed with 4% paraformaldehyde for 15 min at room temperature. Cells were then permeabilized with 0.1% Triton X-100 in PBS (5 min), and blocked with 1% bovine serum albumin in PBS for 1 h at room temperature. Cells were incubated with a primary antibody diluted in blocking buffer at 4°C overnight. Cells were washed and incubated with secondary antibodies diluted in a blocking buffer for 1 h at room temperature. Nuclei were stained with the DAPI dye in PBS for 2 min. Some cells were stained with the Nuclear Green DCS1 dye, according to the manufacturer's protocol. Cells were washed and analyzed.
Fluorescence microscopy and image analysis
Live and fixed cell imaging was performed with the EVOS microscopy system (Thermo Fisher Scientific) as described34–40. γH2AX fluorescence was analyzed by the puncta index, which is the standard deviation of the intensities measured among pixels within the neuronal nuclei, as described38. Low puncta index represents diffuse localization, whereas a high puncta index represents punctate localization. Condensation of chromatin was analyzed with ImageJ.
Survival analysis
Survival analysis was described36,38,39,41. Briefly, transfected neurons were imaged every 24 h for three days. An image of the fiducial field with neurons on the plate was collected at the first time-point and used as a reference image for tracking the same neurons over time. Each time the same plate was imaged thereafter, the fiducial image was aligned with the reference image. the survival time is measured for each neuron from the moment a transfected neuron is visualized until its death, indicated by the abrupt disappearance of the transfected marker. Survival time for each neuron in a cohort is tabulated, and a survival curve for the neuronal cohort is constructed with software. Neurons that survived the entire experiment are weighted differently to account for an indeterminate survival time. Survival functions are fit with Kaplan-Meier analysis, and differences between the cohorts are assessed with the log-rank test with JMP software (SAS Institute) as described41,42. A hazard function, which describes the instantaneous risk that a neuron from the cohort will reach the endpoint of interest, is generated from the survival function (JMP software).
Immunoblotting
Neuronal cultures were lysed in RIPA buffer (150 mM NaCl, 1% P40, 0.5% sodium deoxycholate, 0.1% SDS and 50 mM Tris/HCl (pH 8.0), with phosphatase and protease inhibitors) on ice. Lysates were vortexed and cleared by centrifugation (14000 g, 10 min, 4°C). Supernatants were collected, and protein concentrations were determined by the Bicinchoninic Acid Protein Assay Kit (Thermo Scientific). Samples were analyzed by SDS/PAGE (4–12% gradient gels), and proteins were transferred on to nitrocellulose membranes. Membranes were blocked with 5% skimmed milk and were incubated with the primary antibodies (the TagRFP antibody) overnight at 4°C. Membranes were then washed with TBS (Tris-buffered saline; 10 mM Tris/HCl and 150 mM NaCl (pH 7.4)) and incubated with secondary antibodies (HRP) for 1 hr at room temperature. Chemiluminescent signal was visualized with Prometheus ProSignal Pico (Genesee Scientific) on Blue Devil autoradiography films (Genesee Scientific).
Statistical analyses
For longitudinal survival analysis, neurons that died during the imaging interval were assigned a survival time. These event times were used to generate exponential cumulative survival curves in JMP statistical software. Survival curves describe the risk of death for single cells in the group being longitudinally imaged. To determine differences in the survival curves, they were then analyzed for statistical significance by the log-rank test as described36,38,39,41. To compare differences across two groups, the groups were analyzed with student’s t-test. Differences across multiple groups were analyzed with ANOVA.
Transmission electron microscopy
Neurons were infected with the BFP or Dsup-BFP viruses, cultured for 3 days, and fixed overnight in Karnovsky’s fixative. Neurons were then post-fixed in 1% osmium tetroxide for one hour, dehydrated using a graded series of ethanol, embedded in epoxy resin and heat-polymerized. Ultra-thin sections were cut at 100 nm on a Leica EM UC7 ultramicrotome (Leica, Buffalo Grove, IL) and stained with saturated methanolic uranyl acetate and Reynold’s lead citrate. Sections were examined using a JEOL JEM-1230 TEM (JEOL, Peabody, MA) equipped with an AMT (AMT, Woburn, MA) NanoSprint15 sCMOS camera. TEM images were analyzed with ImageJ. The total amount of condensed DNA (heterochromatin) was calculated as a percentage of the nuclear area occupied by condensed DNA.
Ethics statement
Rats were maintained in accordance with guidelines and regulations of the University of Texas McGovern Medical School at Houston (the protocol number #AWC-16–0081). All experimental protocols were approved by the University of Texas McGovern Medical School at Houston. The methods were carried out in accordance with the approved guidelines.
Supplementary Material
Highlights.
A Tardigrade protein, Dsup (Damage Suppressor), protects the Tardigrade’s DNA during harsh environmental stress and X-rays. These unusual properties of Dsup suggested that understanding how the protein functions may help in the design of small molecules that could protect humans during radiotherapy or space travel.
In cancer cells, Dsup protects DNA from single and double-strand breaks (DSBs) induced by radiation and increases survival of irradiated cells.
In primary mammalian neurons, the codon-optimized Dsup localizes to the nucleus and promotes the formation of DNA DSBs.
With electron microscopy, we discovered that, in neurons, Dsup promotes chromatin condensation.
Mammalian neurons are sensitive to Dsup, and Dsup is a doubtful surrogate for DNA protection in neuronal cells.
Acknowledgments
We thank members of the A.S.T. laboratory and the BRAINS laboratory for useful discussions. Nur Compan provided administrative assistance. We thank Jeffrey Wefel (Neuro-Oncology, the University of Texas MD Anderson Cancer Center) for his thoughtful comments on the manuscript.
Funding Sources
This work was supported by 1R21AG067204 from the National Institute on Aging (A.S.T.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure of potential conflicts of interest
The authors declare that there is no conflict of interest.
References
- 1.Jonsson KI, Rabbow E, Schill RO, Harms-Ringdahl M & Rettberg P Tardigrades survive exposure to space in low Earth orbit. Curr Biol 18, R729–R731, doi: 10.1016/j.cub.2008.06.048 (2008). [DOI] [PubMed] [Google Scholar]
- 2.Boothby TC et al. Tardigrades Use Intrinsically Disordered Proteins to Survive Desiccation. Mol Cell 65, 975–984 e975, doi: 10.1016/j.molcel.2017.02.018 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Horikawa DD et al. Establishment of a rearing system of the extremotolerant tardigrade Ramazzottius varieornatus: a new model animal for astrobiology. Astrobiology 8, 549–556, doi: 10.1089/ast.2007.0139 (2008). [DOI] [PubMed] [Google Scholar]
- 4.Hashimoto T et al. Extremotolerant tardigrade genome and improved radiotolerance of human cultured cells by tardigrade-unique protein. Nat Commun 7, 12808, doi: 10.1038/ncomms12808 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ricci C et al. The Tardigrade Damage Suppressor Protein Modulates Transcription Factor and DNA Repair Genes in Human Cells Treated with Hydroxyl Radicals and UV-C. Biology (Basel) 10, doi: 10.3390/biology10100970 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Puig J, Knödlseder N, Quera J, Algara M & Güell M DNA Damage Protection for Enhanced Bacterial Survival Under Simulated Low Earth Orbit Environmental Conditions in Escherichia coli. Front Microbiol 12, 789668, doi: 10.3389/fmicb.2021.789668 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kirke J, Jin XL & Zhang XH Expression of a Tardigrade Dsup Gene Enhances Genome Protection in Plants. Mol Biotechnol 62, 563–571, doi: 10.1007/s12033-020-00273-9 (2020). [DOI] [PubMed] [Google Scholar]
- 8.Chavez C, Cruz-Becerra G, Fei J, Kassavetis GA & Kadonaga JT The tardigrade damage suppressor protein binds to nucleosomes and protects DNA from hydroxyl radicals. eLife 8, e47682, doi: 10.7554/eLife.47682 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hashimoto T & Kunieda T DNA Protection Protein, a Novel Mechanism of Radiation Tolerance: Lessons from Tardigrades. Life (Basel, Switzerland) 7, 26, doi: 10.3390/life7020026 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jönsson KI Radiation Tolerance in Tardigrades: Current Knowledge and Potential Applications in Medicine. Cancers 11, 1333, doi: 10.3390/cancers11091333 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klomchitcharoen S et al. MINERVA: A CubeSat for demonstrating DNA damage mitigation against space radiation in C. elegans by using genetic modification. Heliyon 8, e10267, doi: 10.1016/j.heliyon.2022.e10267 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nanduri R, Furusawa T & Bustin M Biological Functions of HMGN Chromosomal Proteins. Int. J. Mol. Sci 21, doi: 10.3390/ijms21020449 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moruno Manchon JF et al. Levetiracetam mitigates doxorubicin-induced DNA and synaptic damage in neurons. Sci. Rep 6, 25705, doi: 10.1038/srep25705 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iyama T & Wilson DM 3rd. DNA repair mechanisms in dividing and non-dividing cells. DNA repair 12, 620–636, doi: 10.1016/j.dnarep.2013.04.015 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goldstein B The Emergence of the Tardigrade Hypsibius exemplaris as a Model System. Cold Spring Harb Protoc 2018, 10.1101/pdb.emo102301, doi: 10.1101/pdb.emo102301 (2018). [DOI] [PubMed] [Google Scholar]
- 16.Weronika E & Łukasz K Tardigrades in Space Research - Past and Future. Orig Life Evol Biosph 47, 545–553, doi: 10.1007/s11084-016-9522-1 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jagadeesh MK, Roszkowska M & Kaczmarek Ł Tardigrade indexing approach on exoplanets. Life Sci Space Res (Amst) 19, 13–16, doi: 10.1016/j.lssr.2018.08.001 (2018). [DOI] [PubMed] [Google Scholar]
- 18.Jönsson KI Tardigrades as a potential model organism in space research. Astrobiology 7, 757–766, doi: 10.1089/ast.2006.0088 (2007). [DOI] [PubMed] [Google Scholar]
- 19.Horikawa DD et al. Radiation tolerance in the tardigrade Milnesium tardigradum. Int J Radiat Biol 82, 843–848, doi: 10.1080/09553000600972956 (2006). [DOI] [PubMed] [Google Scholar]
- 20.Neves RC et al. Differential expression profiling of heat stressed tardigrades reveals major shift in the transcriptome. Comp Biochem Physiol A Mol Integr Physiol 267, 111169, doi: 10.1016/j.cbpa.2022.111169 (2022). [DOI] [PubMed] [Google Scholar]
- 21.Jönsson KI, Holm I & Tassidis H Cell Biology of the Tardigrades: Current Knowledge and Perspectives. Results Probl Cell Differ 68, 231–249, doi: 10.1007/978-3-030-23459-1_10 (2019). [DOI] [PubMed] [Google Scholar]
- 22.Kugler J, Postnikov YV, Furusawa T, Kimura S & Bustin M Elevated HMGN4 expression potentiates thyroid tumorigenesis. Carcinogenesis 38, 391–401, doi: 10.1093/carcin/bgx015 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iyama T & Wilson DM 3rd. DNA repair mechanisms in dividing and non-dividing cells. DNA repair 12, 620–636, doi: 10.1016/j.dnarep.2013.04.015 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Canugovi C, Misiak M, Ferrarelli LK, Croteau DL & Bohr VA The role of DNA repair in brain related disease pathology. DNA Repair 12, 578–587, doi: 10.1016/j.dnarep.2013.04.010 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Penndorf D, Witte O & Kretz A DNA plasticity and damage in amyotrophic lateral sclerosis. Neural Regeneration Research 13, 173–180, doi: 10.4103/1673-5374.226377 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Massey TH & Jones L The central role of DNA damage and repair in CAG repeat diseases. Dis Model Mech 11, doi: 10.1242/dmm.031930 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Puget N, Miller KM & Legube G Non-canonical DNA/RNA structures during Transcription-Coupled Double-Strand Break Repair: Roadblocks or Bona fide repair intermediates? DNA Repair (Amst) 81, 102661, doi: 10.1016/j.dnarep.2019.102661 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moruno Manchon JF et al. Cytoplasmic sphingosine-1-phosphate pathway modulates neuronal autophagy. Sci. Rep 5, 15213, doi: 10.1038/srep15213 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Castresana J, Feldmaier-Fuchs G & Pääbo S Codon reassignment and amino acid composition in hemichordate mitochondria. Proc Natl Acad Sci U S A 95, 3703–3707, doi: 10.1073/pnas.95.7.3703 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barbhuiya RI, Uddin A & Chakraborty S Compositional properties and codon usage pattern of mitochondrial ATP gene in different classes of Arthropoda. Genetica 147, 231–248, doi: 10.1007/s10709-019-00067-1 (2019). [DOI] [PubMed] [Google Scholar]
- 31.Abascal F, Posada D, Knight RD & Zardoya R Parallel evolution of the genetic code in arthropod mitochondrial genomes. PLoS Biol 4, e127, doi: 10.1371/journal.pbio.0040127 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Whittle CA & Extavour CG Expression-Linked Patterns of Codon Usage, Amino Acid Frequency, and Protein Length in the Basally Branching Arthropod Parasteatoda tepidariorum. Genome Biology and Evolution 8, 2722–2736, doi: 10.1093/gbe/evw068 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rota-Stabelli O et al. Ecdysozoan mitogenomics: evidence for a common origin of the legged invertebrates, the Panarthropoda. Genome Biol Evol 2, 425–440, doi: 10.1093/gbe/evq030 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tabor N et al. Differential responses of neurons, astrocytes, and microglia to G-quadruplex stabilization. Aging (Albany NY) 13, doi: 10.18632/aging.203222 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lejault P et al. Regulation of autophagy by DNA G-quadruplexes. Autophagy, doi: 10.1080/15548627.2020.1769991 (2020). [DOI] [PMC free article] [PubMed]
- 36.Moruno-Manchon JF et al. Small-molecule G-quadruplex stabilizers reveal a novel pathway of autophagy regulation in neurons. eLife 9, e52283, doi: 10.7554/eLife.52283 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moruno-Manchon JF et al. Sphingosine kinase 1-associated autophagy differs between neurons and astrocytes. Cell Death Dis 9, 521, doi: 10.1038/s41419-018-0599-5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moruno-Manchon JF et al. The G-quadruplex DNA stabilizing drug pyridostatin promotes DNA damage and downregulates transcription of Brca1 in neurons. Aging (Albany NY) 9, 1957–1970, doi: 10.18632/aging.101282 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moruno-Manchon JF et al. Inhibiting sphingosine kinase 2 mitigates mutant Huntingtin-induced neurodegeneration in neuron models of Huntington disease. Hum Mol Genet 26, 1305–1317, doi: 10.1093/hmg/ddx046 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moruno-Manchon JF et al. Peroxisomes contribute to oxidative stress in neurons during doxorubicin-based chemotherapy. Mol Cell Neurosci 86, 65–71, doi: 10.1016/j.mcn.2017.11.014 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Arrasate M, Mitra S, Schweitzer ES, Segal MR & Finkbeiner S Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature 431, 805–810, doi: 10.1038/nature02998 (2004). [DOI] [PubMed] [Google Scholar]
- 42.Tsvetkov AS, Ando DM & Finkbeiner S Longitudinal imaging and analysis of neurons expressing polyglutamine-expanded proteins. Methods Mol. Biol 1017, 1–20, doi: 10.1007/978-1-62703-438-8_1 (2013). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


