Abstract
Allergic diseases are a major global health issue. Interleukin-9 (IL-9) producing T helper cells (Th9) promote allergic inflammation, yet Th9 effector functions are incompletely understood because their lineage instability makes them challenging to study. Here, we found that resting Th9 cells produced IL-9 independent of TCR restimulation, due to STAT5- and STAT6-dependent bystander activation. This mechanism was seen in circulating cells from allergic patients and was restricted to recently activated cells. STAT5-dependent Il9/IL9 regulatory elements underwent remodeling over time, inactivating the locus. A broader “allergic Th9” transcriptomic and epigenomic program was also unstable. In vivo, Th9 cells induced airway inflammation via TCR-independent, STAT-dependent mechanisms. In allergic patients, Th9 expansion was associated with responsiveness to JAK inhibitors. These findings suggest that Th9 instability is a negative checkpoint on bystander activation that breaks down in allergy, and that JAK inhibitors should be considered for allergic patients with Th9 expansion.
Introduction
Allergic diseases affect up to 40% of the global population and cause substantial morbidity and mortality; understanding the drivers of allergic pathology is critical to devising effective therapies1. T helper 9 (Th9) cells are implicated as major mediators of allergic disease through production of interleukin-9 (IL-9)2. IL9 polymorphisms are associated with allergic disease risk; Th9 expansion is seen in asthma, atopic dermatitis, and food allergy; and Th9 cells exacerbate murine models of allergic disease3-8. IL-9 promotes allergic inflammation by inducing mast cell maturation, enhancing immunoglobulin E production, recruiting eosinophils and neutrophils, promoting mucous cell metaplasia, and increasing epithelial permeability5,9-11. Additionally, Th9 cells have emerged as major drivers of autoimmunity and antitumor immunity2,10-14.
Despite the importance of Th9 cells to this broad cross-section of inflammatory diseases, they are incompletely characterized. This is partly because Th9 cells are extremely heterogeneous, but also because they display unique lineage plasticity and effector kinetics: their capacity for IL-9 production decreases over time9,10,15-18. The factors constraining Th9 stability are poorly understood, with most studies focusing on mechanisms of Th9 differentiation17,19-22. Because the “Th9” phenotype disappears shortly after in vitro T cell receptor (TCR) stimulation is withdrawn, it has been suggested that Th9 lineage stability requires sustained TCR signaling16,23. However, TCR-dependent transcription factors (TFs) have not been found to enhance Th9 stability20-22. Other Th9-promoting TFs include STAT5 and STAT6, which bind to critical cis-regulatory elements (REs) in the IL9 locus and increase accessibility during differentiation16,24-27. STAT5 signaling also enhances TCR-stimulation and activation of effector Th9 cells28. These observations suggest that STAT-dependent cytokines might contribute to the unique kinetics of Th9 lineage commitment and function.
Here, we found that paracrine IL-2 and IL-4 induced bystander activation of resting Th9 cells through STAT5 and STAT6, independent of TCR-restimulation. Critical STAT5-/STAT6-binding IL9 cis-regulatory elements (REs) became poised for transcriptional induction during Th9 differentiation. RE accessibility slowly decreased after withdrawal of TCR stimulation, repressing STAT5-target expression and causing instability of the Th9 phenotype. In vivo, pulmonary Th9 underwent TCR-independent, STAT-dependent activation to promote allergic lung inflammation. In patients with allergic disease, Th9 expansion was associated with STAT5 induction, STAT6 induction, and responsiveness to JAK inhibitors (jacanids). Our findings link the epigenetic mechanisms underlying Th9 instability to STAT-dependent bystander activation. These activation mechanisms may have a role in antihelminth responses, where antigen-specific responses are suppressed, while simultaneously providing a negative checkpoint that breaks down in patients with allergic diseases29. Therefore, STAT-dependent Th9 bystander activation may underlie a novel allergic endotype that predicts jakinib-responsiveness, and JAK inhibitors should be considered for allergic patients with a Th9 signature.
Results
Bystander Th9 activation drives persistent IL-9 secretion
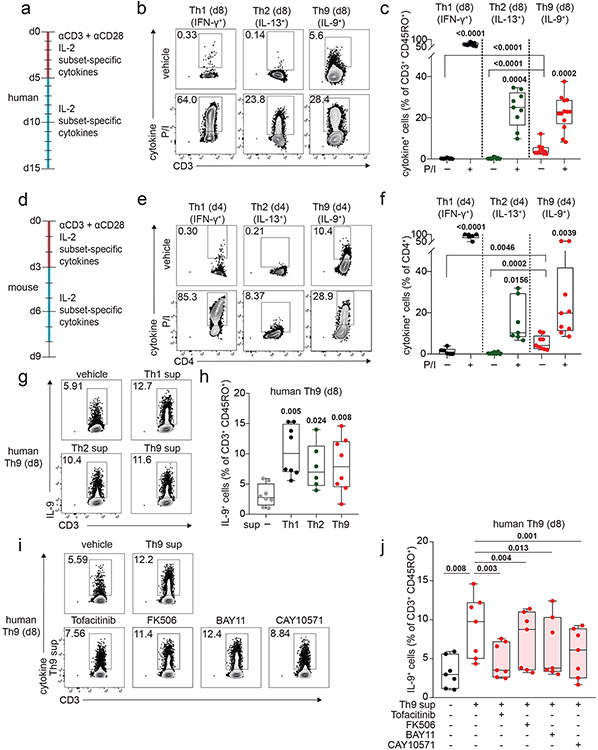
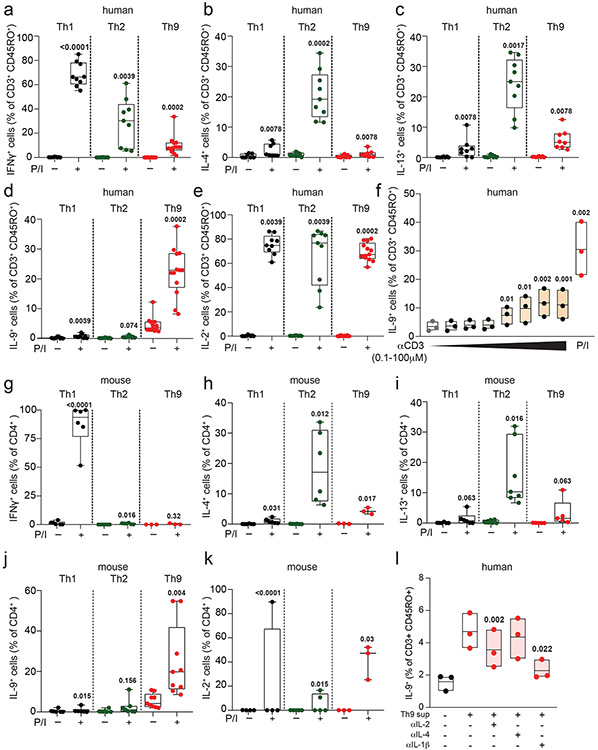
To determine how IL-9 kinetics might be affected by TCR activation, we purified naïve human CD4+ T cells and cultured them with TCR-stimulation (αCD3, αCD28), IL-2, and cytokines/neutralizing antibodies to promote Th1, Th2, or Th9 differentiation. After d5 of culture, we withdrew TCR-stimulation, resting the cells in IL-2 and subset-promoting cytokines/antibodies (Fig 1a). Resting Th1 and Th2 cells produced effector cytokines only upon short (6-hour) restimulation with TCR mimetics (Fig 1b-c, S1a-e). By contrast, resting Th9 cells produced IL-9 without restimulation, even after resting for 6 days (Fig 1b-c, S1f). In murine Th9 cells, IL-9 production was detectable without restimulation after resting for 2 days (Fig 1d-f, S1g-k).
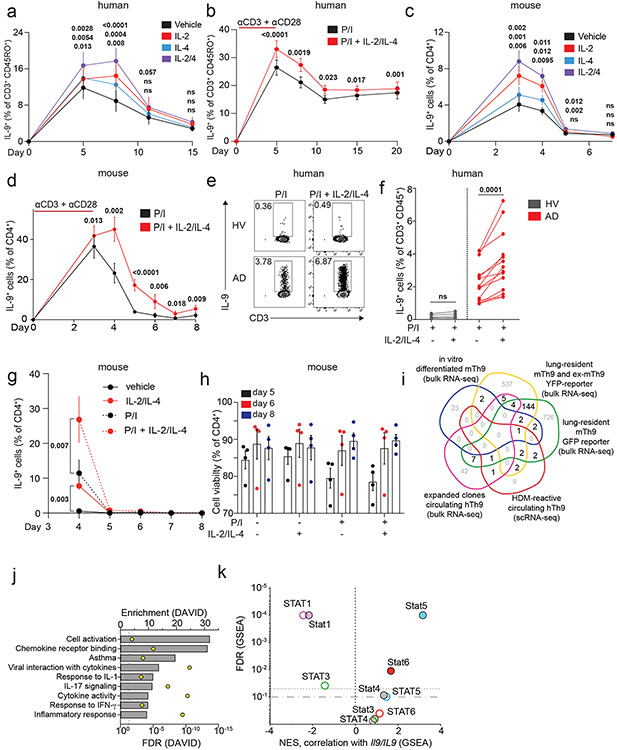
Figure 1. Resting Th9 cells uniquely produce IL-9 independent of T cell receptor (TCR) restimulation, downstream of paracrine STAT-dependent cytokines.
a. Timeline shows differentiation protocol for human T helper cells. Naïve CD4+ T cells from healthy volunteers were activated for 5 days with αCD3, αCD28, IL-2, and subset-promoting cytokines and antibodies. After 5 days, αCD3/αCD28 were withdrawn, and cells were cultured with IL-2 and subset-promoting cytokines and antibodies. b, c. Representative flow plot (b) and summary data (c) for % of cytokine production in human resting (d8, rested for 3 days) Th1 (IFN-γ+, n = 9), Th2 (IL-13+, n = 9), and Th9 (IL-9+, n = 13) with or without PMA and Ionomycin (P/I). d. Timeline shows differentiation protocol for murine T helper cells. Naïve CD4+ T cells from WT C57BL/6 mice were activated for 3 days with αCD3, αCD28, IL-2, and subset-promoting cytokines and antibodies. After 3 days, αCD3/αCD28 were withdrawn, and cells were cultured with IL-2 and subset-promoting cytokines and antibodies. e, f. Representative flow plot (e) and summary data (f) for % of cytokine production in resting (d4) murine Th1 (IFN-γ+, n = 3), Th2 (IL-13+, n = 4), and Th9 (IL-9+, n = 9) cells restimulated with vehicle control vs. P/I. g, h. Representative flow plots (g) and summary data (h) show % IL-9+ cells of resting (d8) human Th9 cells restimulated with supernatants from Th1 (n = 8), Th2 (n = 6), and Th9 (n = 8) cells. i, j. Representative flow cytometry plot (i) and summary data (j) show % IL-9+ cells of resting (d8) human Th9 cells restimulated with Th9 supernatants in the presence of vehicle, tofacitinib (JAK-STAT inhibitor), FK506 (calcineurin/NFAT inhibitor), BAY11 (NF-κB inhibitor), or CAY10571 (p38 MAPK inhibitor) (n = 7). For all experiments: paired or unpaired t-test, normally distributed data; Wilcoxon (paired) or Mann-Whitney (unpaired), non-normally distributed data. Box plots show all data points (min to max, lines at median). All statistical tests are 2-sided, all replicates are biologically independent samples.
These findings led us to hypothesize that paracrine cytokines might promote bystander activation of Th9 cells. To test this, we stimulated in vitro-differentiated human Th1, Th2, and Th9 cells with αCD3, collected supernatants, and used supernatants to stimulate d8-resting Th9 cells. All three supernatants induced IL-9, suggesting that resting Th9 cells undergo bystander activation downstream of paracrine cytokines (Fig 1g-h). Bystander activation of Th1 and Th2 cells is induced by IL-1-family cytokines through NF-κB and p38MAPK and enhanced by STAT-dependent cytokines30. We hypothesized that similar mechanisms might induce IL-9: IL-1β and IL-2 blockade reduced supernatant-induced IL-9 induction (Fig S1l). Because IL-1β enhances Th9 responsiveness to IL-231, we also treated resting Th9 cells with IL-1 family cytokines alone. IL-1β, IL-18, IL-33, IL-36α, and IL-36γ failed to induce IL-9, suggesting that these cytokines do not induce bystander Th9 activation (Fig S2a). Moreover, NF-κB (BAY11) and p38MAPK (CAY10571) inhibition had only a moderate effect on supernatant-induced IL-9 production (Fig 1i-j). NFAT signaling blockade (FK506) also had a modest effect, but JAK-STAT inhibition (tofacitinib) profoundly repressed supernatant-induced IL-9 production (Fig 1i-j). Together, these results indicated that STAT-dependent paracrine cytokines induce bystander activation of resting Th9 cells.
STAT5 and STAT6 induce Th9 bystander activation
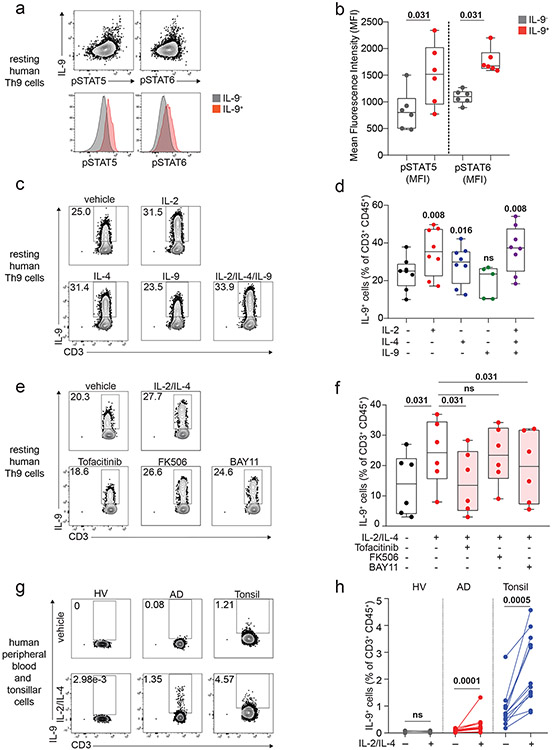
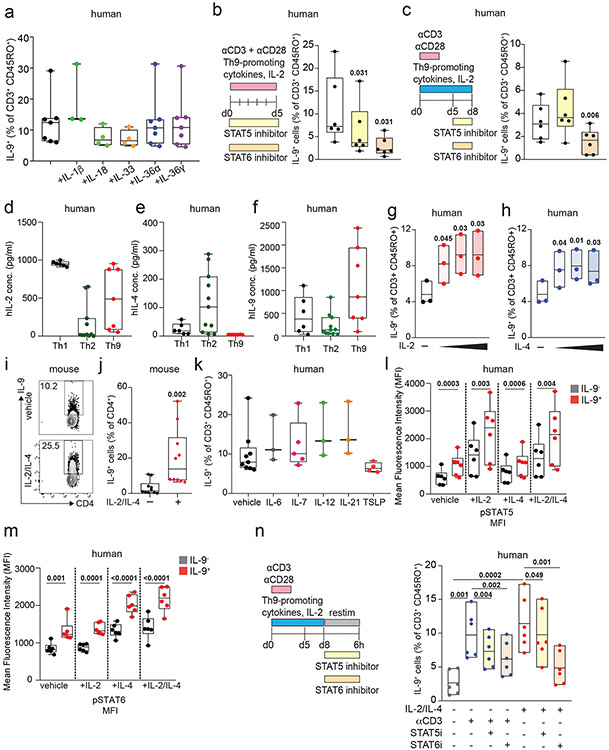
We next sought to identify the specific cytokines and TFs underlying Th9 bystander activation. STAT5 and STAT6 are critical for Th9 differentiation and function16,25; d8 IL-9+ cells exhibited high STAT5 and STAT6 phosphorylation (Fig 2a-b), and STAT5 and STAT6 inhibitors reduced IL-9 production in d8 Th9 cells (Fig S2b-c). Th9-promoting STAT5- and STAT6-activating cytokines include TSLP, IL-7, IL-2, IL-4, and IL-97,17,21,25,27. Because TSLP and IL-7 derive from non-hematopoietic cells7,32, we measured IL-2, IL-4, and IL-9 concentrations in Th1, Th2, and Th9 supernatants. We detected all three cytokines (Fig S2d-f), leading us to hypothesize that they might promote Th9 bystander activation. IL-2 and IL-4 induced IL-9, whereas IL-9 and other STAT-dependent cytokines had no effect (Fig 2c-d, S2g-k). STAT5 and STAT6 phosphorylation were induced in cytokine-treated IL-9+ cells (Fig Sl-m). JAK-STAT, STAT5, and STAT6 inhibition prevented cytokine-induced IL-9 production (Fig 2e-f, S2n).
Figure 2. IL-2 and IL-4 rapidly induce bystander activation of resting Th9 cells through STAT5 and STAT6.
a, b. Representative flow plots (a) show pSTAT5, pSTAT6, and IL-9 expression in non-restimulated resting (d8 from start of culture) human cells differentiated from naïve T cells under Th9-promoting conditions (IL-2, IL-4, TGF-β, IL-1β, αIFN-γ). Representative histograms (a) and summary data (b) show mean fluorescence intensity (MFI) of pSTAT5 and pSTAT6 in resting human IL-9+ and IL-9− cells differentiated under Th9-promoting conditions. Bar graphs show pooled results (n = 6). c, d. Representative cytometry plot (c) and summary data (d) show % IL-9+ cells of resting (d8 from start of culture) human Th9 cells restimulated for 6 hours with vehicle, hIL-2 (n = 8), hIL-4 (n = 8),h IL-9 (n = 5), or a combination of the three (n = 8). e, f. Representative flow cytometric plot (e) and summary data (f) show % IL-9+ cells of resting (d8) human Th9 cells restimulated for 6 hours with IL-2 + IL-4 in the presence of vehicle, tofacitinib (JAK-STAT inhibitor), FK506 (calcineurin/NFAT inhibitor), BAY11 (NF-κB inhibitor), or CAY10571 (p38 MAPK inhibitor) (n = 6). g, h. Representative flow cytometric plot (g) and summary data (h) show % IL-9+ cells of circulating memory CD4+ T cells (CD3+/CD4+/CD8−/CD45RO+ cells) stimulated with vehicle vs. IL-2 + IL-4. Cells derive from healthy volunteer peripheral blood mononuclear cells (HV PBMC, n = 6), atopic dermatitis patient PBMC (AD, n = 14), and HV tonsils (n = 12). For all experiments: paired or unpaired t-test, normally distributed data; Wilcoxon (paired) or Mann-Whitney (unpaired), non-normally distributed data. Box plots show all data points (min to max, lines at median). All statistical tests are 2-sided, all replicates are biologically independent samples.
These findings led us to question whether similar mechanisms could activate in vivo-generated human memory Th9 cells 3,4,6. Treatment with IL-2 and IL-4 induced IL-9 in circulating CD45RO+CD4+ T cells from atopic dermatitis patients but not healthy volunteers (Fig 2g-h). Th9 cells can also be identified in lymphoid tissues, where they promote germinal center development33. Treatment with IL-2 and IL-4 induced IL-9 in human tonsil-derived CD4+CD45RO+ T cells (Fig 2g-h). Together, these results demonstrate that IL-2 and IL-4 induce bystander activation of resting human Th9 cells through STAT5 and STAT6.
Th9 bystander activation occurs via transcriptional induction
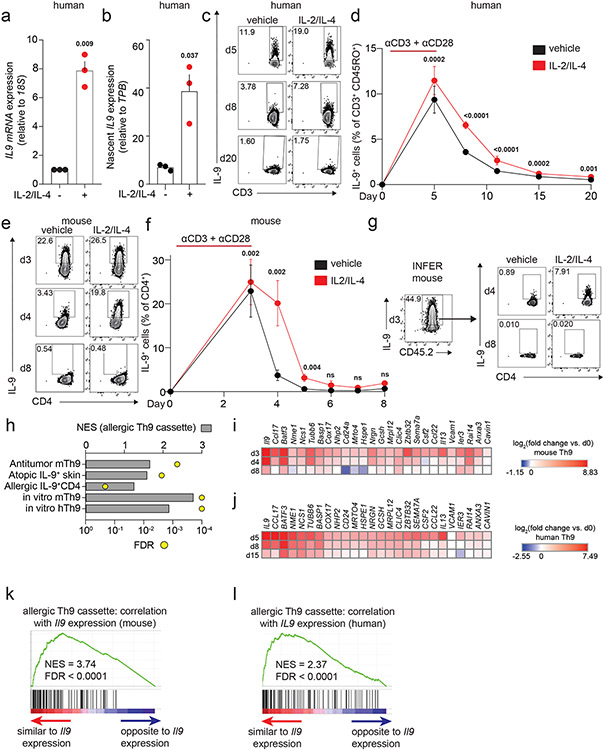
Acute cytokine induction can occur via transcriptional induction or post-transcriptional regulation34. Because STAT5 and STAT6 are Th9-promoting TFs16, we hypothesized that STAT-dependent Th9 bystander activation occurred through transcriptional induction. Accordingly, treatment with IL-2 and IL-4 induced IL9 total mRNA and nascent (newly synthesized) mRNA (Fig 3a-b). These results suggested that STAT5 and STAT6 rapidly and directly target the IL9 locus to promote bystander activation.
Figure 3. STAT-dependent cytokines induce IL9 transcription in recently activated Th9 cells.
a, b. Graphs show total (a) and nascent (b) IL9 expression in d8 human Th9 cells treated with vehicle or IL-2 + IL-4. (n = 3). c, d. Representative flow plots (c) and graphs (d) show % IL-9+ cells of human Th9 differentiated with αCD3/αCD28 for 5 days then continued with Th9-promoting cytokines/antibodies, but without αCD3/αCD28. On d5 (n = 14), d8/d11 (n = 17), d15 (n = 16), and d20 (n = 11), cells were restimulated with vehicle or IL-2 + IL-4. e, f. Representative flow plots (e) and graphs (f) show % IL-9+ cells of murine Th9 differentiated with αCD3/αCD28 for 3 days, then continued with other Th9-promoting cytokines/antibodies, but without αCD3/αCD28. On d3 (n = 12), d4 (n = 11), d5 (n = 9), d6 (n = 3), d7 (n = 3), and d8 (n = 6), cells were restimulated with vehicle or IL-2 + IL-4. g. Representative flow plots show % IL-9+ cells from IL-9 reporter (INFER) mice. IL-9+ cells were sorted on d3 and maintained in Th9-promoting conditions without αCD3/αCD28. Cells were restimulated on d4 and d8 with vehicle or IL-2 + IL-4. h. Graph shows normalized enrichment score (NES) and false discovery rate (FDR) of Th9 cassette (GSEA) in antitumor Th9 cells; IL9high atopic dermatitis skin; house-dust-mite-stimulated T cells from allergic subjects; d3-murine in vitro differentiated Th9; d5-human in vitro differentiated Th9. i,j. Heatmaps show average fold-change in gene expression of in vitro differentiated murine (n = 3, i) and human (n = 4, j) Th9 cells vs. naïve T cells at various time points, for “allergic Th9” genes with dynamic enrichment (FC > 2, FDR < 0.05) in murine (d3/4 vs. d0/8) and human (d5/8 vs. d0/15) cells. k,l. Enrichment plots show Pearson correlation of “allergic Th9” genes with Il9/IL9 expression over time in murine (k) and human (l) Th9 cells. For all experiments: paired or unpaired t-test, normally distributed data; Wilcoxon (paired) or Mann-Whitney (unpaired), non-normally distributed data. For boxplots/line graphs, error bars show ± SEM. All statistical tests are 2-sided, all replicates are biologically independent samples.
Bystander activation is restricted to recently activated Th9
Because STAT-dependent regulation of cytokine loci is also important for T helper lineage commitment, we next asked whether the mechanisms underlying Th9 lineage instability might prevent nonspecific Th9 bystander activation in healthy subjects. To address this, we followed human and murine in vitro Th9 differentiation through an extended time course (Fig 3c-f, S3a-d). Although murine Th9 cells lost IL-9-effector capacity by d7, 10-20% of human Th9 cells retained the ability to produce IL-9 (Fig S3a-d). IL-9 production was highest in cells restimulated with a combination of IL-2 + IL-4 (STAT-activators) and PMA + ionomycin (TCR-mimetics), suggesting that the mechanisms are additive (Fig S3b,d). We therefore hypothesized TCR and STAT agonists might additively induce IL-9 in circulating human Th9 cells. Stimulation of PBMCs from atopic dermatitis patients with STAT-dependent cytokines enhanced the effect of TCR-mimetics, suggesting bystander activation is required for optimal human Th9 function (Fig S3e-f).
Although some human Th9 cells exhibited lineage stability, cytokine-dependent IL-9 induction was limited to recently activated human and murine Th9 cells and could not be seen after prolonged resting (human d15, mouse d6, Fig 3c-f, S3a,c). This could be due to Th9 lineage instability or death of IL-9-producing clones. We therefore sorted d3-in vitro differentiated IL-9+CD4+ cells from IL-9 reporter (INFER) mice and rested them over time9. These remained viable but lost capacity to make IL-9, confirming that transient effector kinetics are related to lineage instability (Fig 3g, S3g-h). Taken together, these findings indicate that bystander IL-9 induction is limited to recently activated Th9 cells and that Th9 lineage instability may prevent nonspecific STAT-dependent bystander activation under homeostatic conditions.
The Th9 transcriptional program is unstable over time
To better understand the link between Th9 lineage instability and STAT-dependent bystander activation, we first set out to better delineate the transcriptional program characterizing Th9 cells. Prior transcriptomic analyses of IL-9-producing T cells have yielded conflicting results about the identity of these cells, with some studies indicating they are a distinct T helper subset and other studies suggesting they may be a group of early activated Th2 cells15-17,23. To resolve some of these questions, we integrated data from four transcriptomic datasets profiling in vivo Th9 cells15,16,23. Two datasets profiled lung-resident Th9 cells from IL-9-reporter mice exposed to intranasal papain, one profiled circulating Th9 clones from healthy volunteers, and one profiled house dust mite (HDM)-reactive Th9 cells from allergic asthma patients15,16,20,23. We also used one dataset comparing in vitro-differentiated Th9 with 4 other T helper subsets16. Overlap of DEGs between the various datasets was low, reflecting the heterogeneity of IL-9+ T cells in various contexts and models. Nonetheless, we defined a cassette of 175 “allergic Th9” genes that were differentially expressed in IL-9+ T cells in ≥2 datasets (Fig S3i, Extended Data Table 1). In addition to Il9, this cassette included several canonical Th2 genes (Il4, Il5, Il13, Ccr4) and “IL9+ Th2” genes linked to early/activated IL-9+ Th2 cells (Ccl17, Ccr8, Pparg). A group of “IL9+ non-Th2” genes was also seen, which are not typically expressed in the Th2 lineage (Spi1, Gzmb, Csf2). We further validated the “allergic Th9” cassette by testing for net enrichment (Gene Set Enrichment Analysis, GSEA) in multiple public datasets, including antitumor Th9 cells, IL9-expressing atopic dermatitis skin, and HDM-stimulated IL9-expressing T cells from allergic subjects (Fig 3h). Pathway analysis revealed enrichment for activation, chemotaxis, and cytokine signaling – particularly by IL-1, IL-17, and antiviral cytokines (Fig S3j).
We next probed Th9 transcriptomes over an extended time course of differentiation and resting, during which IL9/Il9 was first induced and then repressed (Fig 3c-f, i-j). The “allergic Th9” cassette was enriched in d3 (murine) and d5 (human) Th9 cells relative to naïve cells (Fig 3h). 589 genes were transiently induced in both murine Th9 (FC >2, FDR<0.05, d3-d4 vs. d0-d8), and human Th9 (FC>2, FDR<0.05, d5-d8 vs. d0-d15). 26 “allergic Th9” genes were transiently induced, including Il9/IL9, Il13/IL13, and Csf2/CSF2, but not Pparg/PPARG, Spi1/SPI1, Gzmb/GZMB, or Ccr8/CCR8 (Fig 3i,j). Net expression of the “allergic Th9” cassette correlated with IL9/Il9 expression, further suggesting that this program is unstable (Fig 3k,l). Net expression of STAT5-induced genes also correlated with IL9/Il9 expression; weaker enrichment was seen for STAT6-induced genes (Fig S3k). STAT1 and STAT3 are reported to antagonize Th9 differentiation by competing with each other and with STAT535-37; STAT1-induced genes inversely correlated with IL9/Il9 expression in murine and human cells, suggesting a potential role in Th9 instability (Fig S3k).
Chromatin remodeling promotes Th9 lineage instability
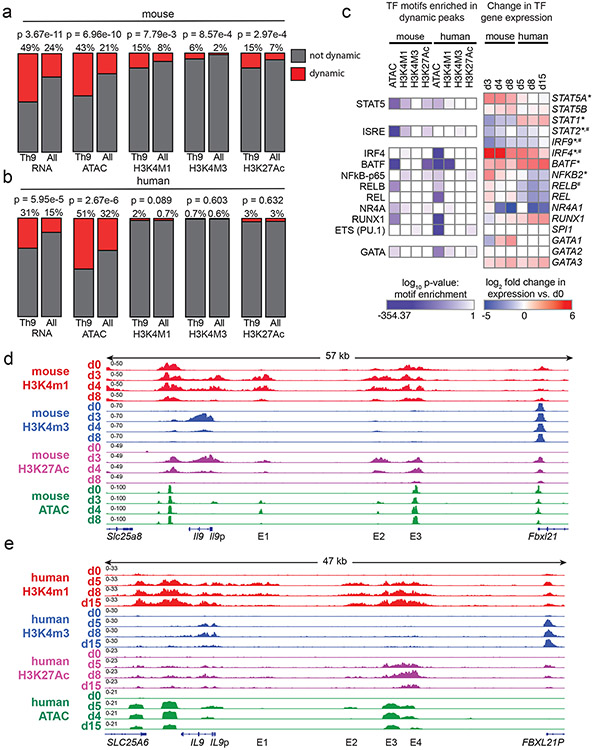
Global chromatin architecture provides an unbiased view of T helper identity that can be more stable than gene expression38. To determine how Th9 identity changes over time, we measured accessibility (ATAC-seq) and histone marks for cis-regulatory elements (REs): poised enhancers (H3K4Me1), active promoters (H3K4Me3) and active enhancers (H3K27Ac). We probed epigenomes over a time course of differentiation and resting in human and murine in vitro generated Th9 cells (Fig 3c-f). We identified a set of dynamic peaks that were gained on d3-4 and lost by d8 in murine Th9, or gained on d5-8 and lost by d15 in human Th9, and therefore specific to cells that could undergo bystander activation. “Allergic Th9” gene loci were enriched for dynamic ATAC-seq peaks, indicating that the Th9 lineage is unstable at an epigenetic level (Fig 4a,b). Enrichment for dynamic histone marks was seen in murine Th9 cells but not in human cells, suggesting that the murine Th9 epigenetic program is less stable than the human Th9 program (Fig 4a,b).
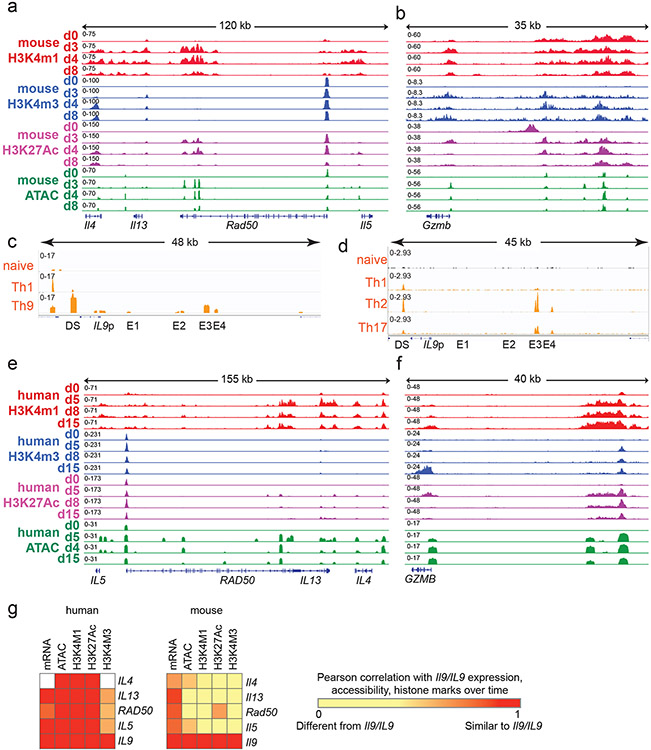
Figure 4. Prolonged resting of Th9 cells reduces accessibility of STAT5 binding sites and type 2 cytokine loci, including critical IL9 enhancers.
a,b. Bar graphs show enrichment (2-sided Fisher’s t-test) in murine (a) and human (b) Th9 cells of dynamically expressed genes, dynamic ATAC-seq (accessibility) peaks, dynamic H3K4Me1 (poised enhancer) peaks, dynamic H3K4Me3 (active promoter) peaks, and dynamic H3K27Ac (active enhancer) peaks in loci/genes mapping to the ‘allergic Th9” cassette vs. all coding gene loci. c. Heatmap (left) shows log (p-value, HOMER de novo motif discovery) for motif enrichment of transcription factors (TF) in human and murine dynamic peaks. All motifs with significant enrichment in at least 1 murine peak set and at least 1 human peak set are shown. Heatmap (right) shows log2 (average fold-change) in gene expression of TFs with motif enrichment in dynamic peaks. Values are shown for murine and human Th9 at various time points vs. naïve T cells. * transiently upregulated in murine Th9 (FC >2, FDR < 0.05, d3-d4 vs. d0-d8) # transiently upregulated in human Th9 (FC >2, FDR < 0.05, d5-d8 vs. d0-d15); d, e. Genome tracks show poised enhancer (H3K4Me1), active promoter (H3K4Me3), active enhancer (H3K27Ac) marks, and accessibility (ATAC) of the murine Il9 (d) and human IL9 (e) extended locus including the promoter (Il9p), downstream enhancer (DS) and upstream enhancers 1-4 (E1-E4), at different time points during Th9 differentiation and resting. All replicates are biologically independent samples.
We next sought to identify TFs that might preferentially induce gene expression in recently activated Th9 cells. Binding sites for these TFs should open and gain activating histone marks upon Th9 differentiation, then close and lose histone marks after prolonged resting39. We therefore searched for TF motifs with significantly enriched abundance (HOMER) in dynamic peaks. We identified 14 motifs that were enriched in human and murine dynamic peaks; 3 genes encoding for the associated TFs were also induced during Th9 differentiation – Stat5a/STAT5A, Irf4/IRF4, and Batf/BATF (Fig 4c). The STAT5 motif was enriched in 5 dynamic peak groups, representing the most conserved enriched motif, and Stat5a expression was dynamically induced in Th9 cells (Fig 4c). Thus, analysis of Th9 epigenomes suggested that chromatin remodeling reduces accessibility and activation of STAT5-binding cis-REs over time, even in the presence of IL-2.
Chromatin remodeling represses Il9/IL9 over time
Having found globally reduced accessibility and activation of STAT5-binding sites over time, we next investigated the Il9/IL9 locus16,40-42. The murine Il9 locus comprises five STAT5-binding cis-REs: a downstream enhancer (DS, also called CNS2, SEa), promoter (Il9p), and three upstream enhancers (E1, also cells CNS0, SEb; E2, also called CNS-25, SEc; and E3, also called CNS-25, SEc)16,40-42 (Fig 4d). Il9p, E1, and E2-E3 – previously defined as Th9-specific enhancers – gained accessibility and histone marks in murine d3-Th9 cells relative to naïve-T cells (Fig 4d). Il9p accessibility and H3K4me3 were reduced on d4 and absent by d8, whereas E1 and E2 were accessible and bore active enhancer marks on d4 but not d8; poised enhancer marks were also repressed on d8 (Fig 4d). Together, these changes caused the extended Il9 locus to revert to the naïve state in d8-cells that were refractory to bystander activation. By contrast, other “allergic Th9” loci, including the Th2 (Il4-Il13-Il5) and Gzmb loci exhibited sustained differences between d8 and d0 (naïve) (Fig S4a-b).
By contrast, the human IL9 locus contained six potential cis-REs: a downstream RE (DS), the promoter (IL9p), and four upstream cis-REs 5kb upstream of the promoter (Fig 4e). One of these REs was previously described as CNS-18, an enhancer homologous to murine E341,42. One RE (E1) previously described as CNS-4.5 was homologous to murine E141,42, and one RE (E2) located 14kb from the transcription start site was homologous to murine E2. One previously undescribed RE (E4) displayed Th9-specific accessibility relative to naïve cells. DS, IL9p, and E1-E4 also displayed Th9-specific accessibility compared to human naïve and in vitro differentiated Th1 cells (Fig S4c). In ex vivo human memory CD4+ cells (ENCODE), DS was accessible in multiple T helper subsets, whereas E3-E4 were specifically accessible in Th2 and Th17 cells, which can acquire IL-9-producing capacity23,25(Fig S4d), indicating that in vivo findings support the functional relevance of these human IL9-REs (Fig 4e).
We next compared IL9 locus architecture in naïve (d0), recently activated (d5-8) cells that can undergo bystander activation, and remotely activated (d15) human Th9 cells. As in murine Th9 cells, certain cis-REs within the extended IL9 locus lost accessibility by d15 (Fig 4e). By comparison, accessibility of the GZMB and Th2 (IL4-IL13-IL5) loci were sustained through late timepoints in human and murine cells (Fig S4e-f). Compared with the murine Il9 locus, dynamic regulation of the human IL9 locus was more stable and correlated more strongly with regulation of the Th2 locus (Fig S4g).
Together, these findings reveal that the extended Il9/IL9 locus undergoes epigenetic remodeling over time. During Th9 differentiation, the locus opens, allowing TCR-independent STAT5/STAT6-dependent bystander activation. After TCR stimulation is withdrawn, the Il9/IL9 locus loses accessibility over time, preventing TF binding and resulting in Th9 lineage instability. This mechanism is most pronounced in murine Th9 cells, consistent with the reduced stability of murine Th9 cells compared with human cells.
Pulmonary Th9 cells have an activated transcriptomic signature
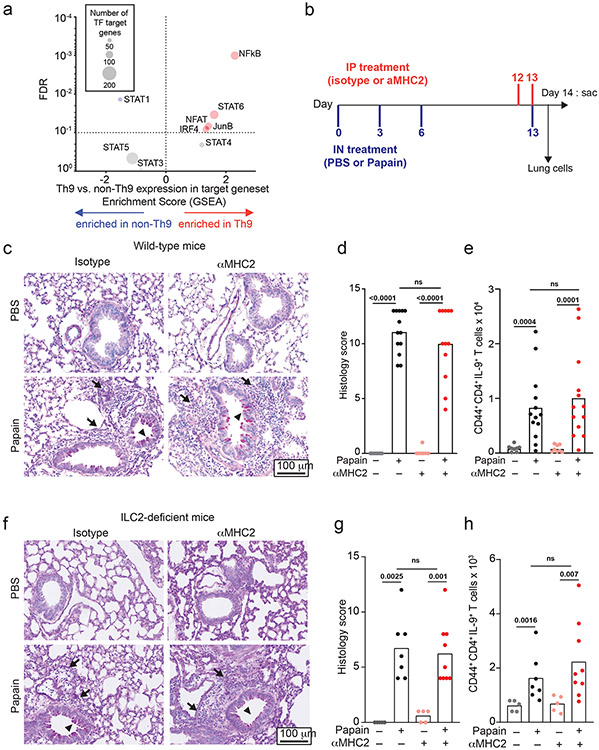
Like in vitro-generated and circulating Th9 cells, lung-resident Th9 cells have transient effector functions18,23. We therefore hypothesized that in vivo-generated Th9 cells might undergo STAT-dependent bystander activation to promote airway inflammation. Because bystander activation was restricted to recently activated in vitro-Th9 cells, we first asked whether lung-resident Th9 cells had an activated phenotype. We compared transcriptomes of lung-resident IL-9+ and IL-9− CD4+CD44+TCRβ+ cells from INFER mice exposed to the chronic papain model of airway inflammation, which induces expansion of IL-9-producing T-cells and is attenuated in the absence of IL-9 (Fig 5a-h, S5k-m, 6a-f, S6f-k)8,16,20. We identified four activation-induced TFs for which we could generate target genesets using public data (TF deletion or overexpression): NFAT, IRF4, JunB, and NF-κB. We compared net enrichment of TF-induced genes in Th9 vs. non-Th9 cells (GSEA), using STAT-target genesets as controls (Fig 5a). As a positive control, Th9 cells showed increased net expression of STAT6-induced genes. Lung-resident Th9 cells were also enriched for activation-dependent TF targets (Fig 5a), indicating that they had an activated transcriptomic signature.
Figure 5. Th9 cells promote TCR-independent airway pathology in vivo.
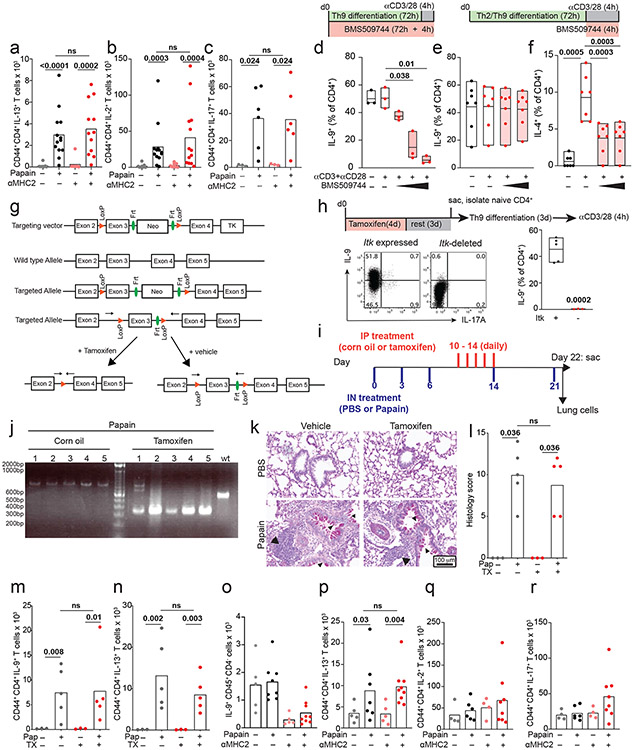
a. Enrichment plots show a normalized enrichment score and FDR (GSEA) for the average, or net, transcriptome of Th9 vs. non-Th9 cells for the following Th9-related and activation-dependent transcription factors (TFs): STAT1, STAT3, STAT4, STAT5, STAT6, IRF4, ,NF-kB16, NFAT, and JunB. A positive score indicates enrichment in Th9 cells, and a negative score indicates enrichment in non-Th9 cells. A neutral score indicates no significant enrichment. b. Timeline shows model of papain-induced airway inflammation with MHC2 blockade. Mice were sensitized and rechallenged with intranasal papain, and treated with isotype vs. αMHC2 during rechallenge. c-e. Representative periodic acid-Schiff (PAS) stained images (c), pooled histology scores (d), and live CD45+TCRβ+CD4+CD44+IL-9+ (Th9) cell counts (h) from lungs of mice exposed to papain and treated with isotype vs. αMHC II as in (b) (arrows, inflammatory infiltrate; triangles, mucus; n= 8, PBS; n = 13, Papain) f-h. Representative periodic acid-Schiff (PAS) stained images (e), pooled histology scores (f), and live CD45+TCRβ+CD4+CD44+IL-9+ (Th9) cell counts (h) from lungs of Type 2 innate lymphoid cell (ILC2) deficient mice exposed to papain and treated with αMHC II or isotype as in (b) (arrows, inflammatory infiltrate; triangles, mucus; n= 5, PBS; n = 7, Papain-iso; n = 9, Papain-αMHCII. For all in vivo experiments, statistical tests are 2-sided Mann-Whitney; all data are biologically independent samples from ≥ 2 replicate experiments.
Pulmonary Th9 cells undergo bystander activation in vivo
Having determined that lung-resident Th9 cells from papain-treated mice have an activated signature, we next assessed for evidence of bystander activation in vivo. To block TCR-activation, mice were treated between papain sensitization and challenge with an antibody against major histocompatibility complex class II (αMHCII) that was previously shown to prevent TCR-activation in the airway (Fig 5b)43. αMHCII-treated mice developed peribronchial leukocyte infiltration, goblet hyperplasia, and mucus production indistinguishable from isotype-treated mice. In lung-resident CD4+ T cells, αMHCII had no effect on expression of IL-9 or other effector cytokines (Fig 5c-e, S5a-c). These findings demonstrate that Th9 cells and Th2 cells can undergo MHCII/TCR-independent activation and tissue infiltration in vivo.
We next sought to confirm TCR-independent activation of differentiated Th9 cells using a genetic approach. The Tec family kinase Itk is required for full TCR signaling and is critical for Th9 differentiation; we confirmed this by differentiating Th9 cells in vitro in the presence of a pharmacologic Itk inhibitor (Fig S5d)20. We hypothesized that Itk signaling was dispensable for restimulation of Th9 cells in vitro and in vivo. Pharmacologic Itk inhibition did not affect IL-9 production from in vitro differentiated Th9 cells; as a positive control, inhibition reduced TCR-stimulated IL-4 production from Th2 cells (Fig S5e-f). To study acute TCR-Itk signaling in vivo, we crossed mice bearing exon 3 of Itk flanked by two loxP sites with mice expressing ERT-Cre to generate ItkERT mice (Fig S5g). We confirmed that deletion of Itk inhibited in vitro Th9 differentiation, recapitulating germline Itk deficiency (Fig S5h). To determine whether in vivo ablation of TCR-Itk signaling affected Th9 reactivation after in vivo differentiation of Th9 cells, mice were treated with tamoxifen for 5 days between papain sensitization and challenge, to delete exon 3 of Itk in CD4+ and CD8+ T cells (Fig S5i-j). Itk ablation had no significant effect on lung pathology or IL-9- and IL-13-producing pulmonary CD4+ T cells (Fig S5k-n), confirming that tissue-infiltrating Th9 cells can undergo TCR-independent activation in vivo.
While Th9 cells are important drivers of airway inflammation in vivo, type 2 innate lymphoid cells (ILC2) also induce lung pathology by producing type 2 cytokines independent of antigenic stimulation8,38. To eliminate this confounding factor, we treated ILC2-deficient (Gata3ΔKLRG1) mice with αMHCII between papain-sensitization and challenge (Fig 5a). Treatment with papain induced IL-9 in CD4+ T cells but had no effect on IL-9 production in other cell types (Figure 5f-h, S5o). Although airway inflammation was attenuated in ILC2-deficient mice, αMHCII had no significant effect on airway pathology, IL-9-producing pulmonary CD4+ T cells, or IL-13-producing CD4+ T cells (Fig 5f-h, S5p-r). This established that TCR-independent restimulation was sufficient to induce pulmonary Th9 infiltration, Th9 activation, and allergic lung pathology, even in the absence of ILC2.
Bystander Th9 activation promotes airway inflammation in vivo
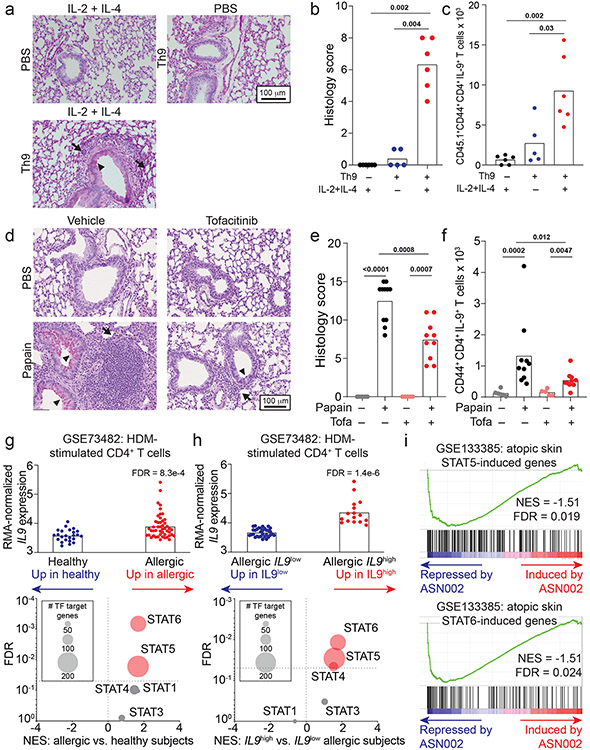
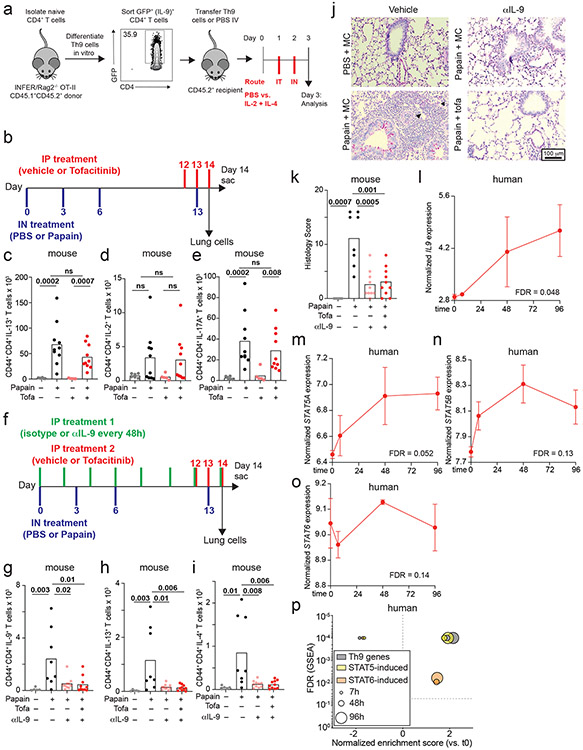
Although Th9 cells are important mediators of papain-induced airway inflammation, other cell types like eosinophils and mast cells can also produce IL-9, while other cytokines like IL-13 and IL-17A can promote airway inflammation. We therefore used a Th9-specific model to determine whether STAT-dependent activation of Th9 cells induces airway inflammation. We differentiated ovalbumin (OVA)-specific (OT-II) Th9 cells from INFER mice in vitro, transferred sorted-GFP+ (IL-9+) CD4+ cells into congenic hosts, and rechallenged recipient mice with intranasal IL-2 + IL-4 (Fig S6a). This promoted lung inflammation and induced IL-9 production from donor OT-II Th9 cells, revealing that Th9 cells can induce in vivo pathology in the absence of cognate antigen if they are stimulated with IL-2 + IL-4 (Fig 6a-c). Thus, STAT5- and STAT6-dependent Th9 bystander activation can promote airway inflammation in vivo, in the absence of acute MHC-II/antigen-TCR interactions.
Figure 6. STAT-dependent Th9 bystander activation promotes allergic disease and is associated with responsiveness to JAK inhibitors.
a-c. Representative periodic acid-Schiff (PAS) stained images (a), pooled histology scores (b), and live CD45.1+TCRβ+CD4+CD44+IL-9+ (adoptively transferred Th9) cell counts (c) in lungs of WT mice who underwent adoptive transfer of PBS or Th9 cells from congenic donors, followed by intratracheal and intranasal challenge with PBS or IL-2 + IL-4; 3 replicates, n = 2 per replicate; arrows, inflammatory infiltrate; triangles, mucus. d-f. Representative PAS-stained images (d), pooled histology scores (e), and live CD45+TCRβ+CD4+CD44+IL-9+ (Th9) cell counts (f) from lungs of mice exposed to chronic papain-induced airway inflammation and treated with methylcellulose (MC) or tofacitinib; n = 6 (PBS/MC), n = 5 (PBS/tofa), n = 11 (Papain/MC), n = 10 (Papain/tofa); arrows, inflammatory infiltrate; triangles, mucus. In vivo data are shown as mean ± SEM, Mann-Whitney. g. Bar graph shows RMA-normalized IL9 expression in house dust mite (HDM) treated CD4+ T cells from healthy volunteers vs. patients with allergic disease. Scatterplot shows normalized enrichment scores (NES; Gene Set Enrichment Analysis, GSEA) and FDR for genes induced by the following TFs in allergic vs. healthy CD4+ cells: STAT1; STAT3; STAT4; STAT5; STAT6. h. Bar graph shows RMA-normalized IL9 expression in HDM-treated CD4+ T cells from allergic patients in which IL9 expression is lower than mean expression for the dataset (IL9low) vs. higher than mean for the dataset (IL9high). Scatterplot shows NES (GSEA) and FDR for genes induced by STAT1, STAT3, STAT4, STAT5, and STAT6 in IL9low vs. IL9high CD4+ cells. i. GSEA plots show NES and FDR for STAT5- and STAT6-induced genes in d29 vs. d0 skin from AD patients treated with the JAK-SYK inhibitor ASN002, 80 mg daily. All statistical tests are 2-sided; all replicates are biologically independent samples.
Th9 expansion correlates with responsiveness to JAK inhibitors
The importance of STAT-dependent bystander activation to Th9 effector function suggested that JAK-STAT inhibitors (jakinibs), which are efficacious for allergic diseases32, might prevent acute Th9-driven pathology. We therefore treated C57BL/6 mice with the pan-jakinib tofacitinib to block JAK-STAT signaling between papain sensitization and challenge (Fig S6b). Tofacitinib-treated mice had significantly less peribronchial leukocyte infiltration, goblet hyperplasia, and mucus production than vehicle-treated mice (Fig 6d-e). In pulmonary CD4+ T cells, tofacitinib significantly reduced production of IL-9 but not of other cytokines (Fig 6f, S6c-e,. Tofacitinib did not affect airway inflammation in mice treated with IL-9-blocking antibodies (Fig S6f-k). Together, these findings suggest that blocking Th9 activation with JAK-STAT inhibitors could ameliorate allergic disease flares.
This led us to hypothesize that peripheral Th9 expansion, which can be seen in allergic patients (Fig 2h, S2f) and is associated with STAT5 activation36, might correlate with responsiveness to jakinibs. We therefore analyzed transcriptomes of HDM-treated CD4+ T cells from allergic patients, which had high IL9 and “allergic Th9” gene expression (Fig 6g, S3j)16. STAT5- and STAT6- induced genes were enriched in allergic CD4+ T cells relative to healthy volunteer cells (GSEA, Fig 6g). Moreover, STAT5- and STAT6- induced genes were enriched in allergic cells with high IL9 expression (> mean expression, Fig 6h) relative to cells with low IL9 expression (< mean expression, Fig 6h). In a separate dataset (GSE6281), nickel-allergic patients were exposed to nickel for 96 hours, inducing inflammation, IL9 expression (Fig S6l), and STAT5- and STAT6-target genes (Fig S6m-p). This suggested that Th9 cells from allergic patients are poised to respond to jakinibs, and that Th9 expansion might be associated with jakinib-responsiveness. We therefore investigated skin transcriptomes of atopic dermatitis patients with enriched “allergic Th9” genes (Fig S3j), in whom IL9 expression correlated with response to a JAK-SYK inhibitor44. Expression of STAT5- and STAT6- induced genes decreased in post-treatment lesional skin relative to pre-treatment skin (Fig 6i). Hence, in patients with allergic disease, Th9 expansion is associated with STAT5 activation, STAT6 activation, and responsiveness to jakinibs. This suggests that Th9 expansion may predict a jakinib-responsive endotype, further increasing the relevance of STAT-dependent Th9 bystander activation to human allergic disease.
Discussion
Th9 cells are critical inducers of allergy, autoimmunity, and antitumor immunity6,7,11,12,14,45. However, major questions surround the regulation and function of Th9 cells. These include the mechanisms underlying transient dynamics of IL-9 production, the physiological significance of Th9 lineage instability, and the molecular mechanisms regulating human Th9 effector function in patients with inflammatory diseases. Herein, we addressed these questions using genetic and functional approaches in human primary cells and murine models.
STAT5 and STAT6 are both critical for Th9 differentiation24-26,37. Here, we provide evidence that IL-9 is also induced by STAT5- and STAT6- dependent bystander activation, indicating a broader role for these TFs in Th9 effector function. This mechanism is unique to Th9 cells: Th1, Th2, and Th17 bystander activation are induced by IL-1 family members, although STATs can enhance NF-κB-dependent activation30. Moreover, Th1, Th2, and Th17 bystander activation requires prolonged stimulation whereas Th9-bystander activation is rapid30. The reasons for these Th9-specific mechanisms and kinetics may be related to the role of Th9 cells in anti-helminth immunity, where antigen-specific responses are suppressed29. While antigen-independent IL-9 production can also derive from non-T-cell sources like ILC2 and mast cells, Th9 cells serve important nonredundant functions because of their capacity for circulation, plasticity, and expansion40,42,46. Bystander activation may also promote Th9 function in immune-privileged sites like the central nervous system and tumor microenvironment45,47,48. The transient kinetics of bystander activation may constrain prolonged nonspecific Th9-mediated inflammation. The responsiveness of circulating IL-9-producing T cells from allergic patients – but not healthy volunteers – to STAT-dependent cytokines suggests that this negative checkpoint breaks down in allergic disease. This could promote Th9 stability in allergic patients, suggesting a novel potential driver and therapeutic target.
In addition to providing major insights about Th9 effector function, this mechanism of bystander activation is also translationally useful. Th9 expansion is associated with STAT5- and STAT6- induction and responsiveness to jakinibs in murine models and allergic patients. This suggests that Th9 expansion might be a biomarker for a jakinib-responsive endotype. Moreover, adding IL-2 and IL-4 to TCR mimetics improves detection of rare circulating human IL-9+ cells, and therefore of patients with Th9-expansion. Thus, STAT-dependent Th9 bystander activation not only defines a druggable allergic endotype but can also be leveraged to detect patients with this endotype. Further studies can determine how peripheral Th9/IL-9 induction can be used to best target biologic therapies for patients with allergic diseases.
Our findings also advance the understanding of Th9 identity. Previous studies have yielded conflicting results about the identity of the “Th9” subset, possibly due to disease- and tissue-specific differences in IL-9+ T cells. We resolve several of these questions using a transcriptional program of IL-9+ T cells developed using multiple independent datasets. While overlap of DEGs from individual datasets was low, net expression of this cassette was enriched in several additional independent datasets obtained from IL9+ allergic subjects, indicating that this cassette is characteristic of “allergic Th9” cells. Enrichment of the “allergic Th9” cassette in antitumor Th9 cells also demonstrates a conserved identity for IL-9+-T cells in different immunological contexts. Enrichment of “allergic Th9” genes in in vitro-generated Th9 cells, indicates that these cells are an appropriate model to study Th9 lineage identity. Within the “allergic Th9” cassette, co-expression of genes encoding conventional Th2 cytokines, activation markers, Granzyme B, and GM-CSF suggests that Th9 cells are a distinct group of highly activated cells that produce a unique combination of proinflammatory cytokines. Analysis of the “allergic Th9” cassette indicates responsiveness to antiviral cytokines like TL1A and to IL-1β, which potentiates IL-2/STAT5 signaling during Th9 differentiation and enhances the differentiation of “inflammatory Th2” cells21,31,49. Both cytokines are elevated during allergic inflammation and could promote Th9 activation, enhancing bystander IL-9 induction. Finally, following “allergic Th9” gene expression over time suggests that Th9 cells do not simply differentiate into conventional Th2 cells but rather maintain a distinct inflammatory phenotype.
Apart from the transcriptome, Th9 identity can also be studied by profiling the epigenome, including human IL9 locus structure and regulation. Th9 cells are regulated by a broad network of TFs including PU.1/ETV5, GATA3, STATs, TAK1, and RARα that directly target the Il9 locus13,16,17,19,25,27. These mechanisms have largely been investigated in mice, yet there are fundamental differences in human and murine Th9/IL-9 regulation. By integrating human and murine data, our work provides a highly translational model of IL9 regulation that identifies and utilizes the most relevant murine models of human Th9/IL-9 biology. For example, we find that IL9 regulation correlates with Th2 locus regulation in human Th9 cells more closely than murine cells. This may relate to fundamental differences in chromatin structure: human IL9 is located 3kb from the Th2 locus whereas the murine Il9 and Th2 loci are on different chromosomes. Hence, it may be necessary to study IL9-associated topologically associated domains (TADs), Th2 locus interactions, and long-range interactions primarily in human cells. Combining transcriptomic and epigenomic data reveals dynamic STAT5-target induction and STAT1-target repression in murine and human cells, but inconsistent results for STAT3. This is consistent with reports that STAT3 represses murine Th9 development by restraining STAT5 while promoting human Th9 differentiation by restraining STAT135,37. Our findings suggest that future investigations of STAT5-STAT1 interactions may resolve some of these inconsistencies.
Our epigenetic data also uncover a novel mechanism underlying the unstable Th9 phenotype. In murine and human cells, the Il9/IL9 locus gains accessibility during differentiation, permitting effector function. However, critical cis-REs close over time, rendering Il9/IL9 refractory to transcriptional induction. Because T effector function depends on the chromatin state of cytokine loci50, these epigenetic changes prevent Th9 lineage commitment. The transient nature of changes is more pronounced in murine Th9 cells than in human cells, and circulating IL-9+ memory T cells are reliably identified in allergic patients but not in healthy volunteers. The determinants of human Th9 stability will require further studies using cells from subjects with IL-9-associated diseases. Because Th9 cells are implicated in a broad cross-spectrum of diseases, studying human Th9 in diverse cohorts should uncover factors promoting lineage commitment in the context of these varied pathologies.
In summary, post-differentiation murine and human Th9 cells become transiently susceptible to TCR-independent, STAT-dependent bystander activation upon stimulation with IL-2 and IL-4. This mechanism is restricted to recently activated cells because of dynamic Il9/IL9 locus remodeling. Epigenetic changes reduce accessibility of STAT5-binding sites, rendering Il9/IL9 refractory to transcriptional induction and causing Th9 instability. Finally, STAT-dependent activation promotes TCR-independent Th9-mediated pathology in vivo and is associated with responsiveness to jakinibs in patients with Th9-expansion. These results not only advance our understanding of the basic mechanisms of allergic inflammation and T helper lineage commitment, but also define a novel potential allergic endotype.
Methods
Human samples
De-identified buffy coats for healthy volunteers were obtained from the National Institutes of Health Blood Bank. Whole blood was obtained from idiopathic atopic dermatitis (AD) patients who were evaluated at NIH Clinical Center under the IRB-approved protocol NCT01164241. Human tonsil samples from noninflamed volunteers with sleep-disordered breathing were obtained from Children’s National Hospital in Washington, DC under a multi-site IRB-approved protocol in which NIH was the IRB of record (94-HG-0105). All subjects provided written informed consent, and all experiments were done with male and female subjects. Subjects were not compensated.
Animal experiments
Experiments were done on 8-14-week-old mice that were age- and sex- matched within experiments. All experiments were done in male and female mice and were performed in an AAALAC-accredited animal facility, approved by the NIAID Institutional Animal Care and Use Committee, in accordance with NIH guidelines for the use and care of live animals, under protocol LAD12E. Mice were maintained in a specific pathogen free facility on a 12h light/dark cycle, in open cages with food and water continuously available. Mice were checked periodically to ensure normal health and more often if adverse effects were anticipated. All mice were on a C57BL/6 background. Gata3fl/fl and ERTCre mice were purchased from Jackson. Cd45.1 x Rag−/− x OT-ii mice were purchased from Taconic. INFER and Klrg1Cre51 mice were generously provided by Drs. Richard Flavell and Paola Licona-Limon. ILC2-deficient mice were generated by crossing Klrg1Cre to Gata3fl/fl mice52. Detailed characterization of ILC2-deficient mice has been reported53.
To generate ItkERT mice, two loxP sites flanking exon 3 of Itk were introduced into the genome. A targeting vector (pL253) containing exon 3 flanked by two LoxP sites was generated by recombineering54; a neomycin resistance gene (Neo) flanked by two FRT sites was inserted before the second LoxP site for drug selection. The vector was linearized and electroporated into HGTC-8 ES cells; genomic DNA was isolated from G418-resistant cell clones and screened by long-range PCR. Correct integration of targeted ES clones was confirmed by Southern blots, and successfully targeted ES clones were microinjected into C57BL/6 blastocysts. Germline transmission from generated chimeric offspring was confirmed by genotyping. Mice carrying the targeted allele were bred to Flp recombinase transgenic mice (NHGRI Transgenic Core) to remove the FRT-flanked Neo cassette and to generate Itkfl/fl mice. To delete Itk, Itkfl/fl mice were bred to B6(129S4)-Et(cre/ERT2)6691Rdav/J (Jackson), which express the tamoxifen-inducible Cre-ERT2 fusion protein (Cre-ERT2). Genomic DNA was genotyped by PCR (gen Itk in3 F: 5’ TGCTTACTCTAGGAGAACAAGG 3’, gen Itk in3 R: 5’ GCTACTTAGACAATTGGAGGC 3’) which generated a 600 bp band from the wild type allele, a 785 bp band from the floxed allele and a 342 bp band from the knockout allele.
Reagents
All recombinant cytokines were obtained from Peprotech except for hIL-12, mIL-12, TSLP, GM-CSF, TL1-A, and hTGF-β (R&D). Details of antibody clones and validation are provided in Source Data. Tofacitinib, FK506, BAY11, CAY10571, STAT5-INHIBITOR, and STAT6 inhibitor AS1517499 were purchased from Cayman and reconstituted in DMSO to 100 mM, 10 mM, 50 μM, 100 mM, 50 μM, and 50 nM stock concentrations, respectively.
In vitro human T cell differentiation
Human peripheral blood mononuclear cells (PBMC) were isolated by density centrifugation (Ficoll-Paque). Naïve human CD4+ T cells were magnetically enriched to >95% purity (StemCell) and plated at a density of 0.5 x106 in 1 mL in a 24-well plate for Th1 and Th2 differentiation, and 1 x106 in 2 mL in a 24-well plate for Th9 differentiation on plate-bound αCD3 (1 μg/mL, OKT3). Cells were cultured in complete RPMI-1640 with glutamine (10% FBS, 100 IU/mL penicillin, 0.1 mg/mL streptomycin) in the presence of cytokines and antibodies to promote the differentiation of Th1 (1 μg/mL αCD28; 2.5 μg/mL αIL-4; 10 ng/mL hIL-12, 1 ng/mL hIL-2); Th2 (1 μg/mL αCD28, 2.5 μg/mL αIFN-γ, 2.5 μg/mL αIL-10, 30 ng/mL hIL-4, 10 ng/mL hIL-2), Th9 (1 μg/mL αCD28, 2.5 μg/mL αIFN-γ, 5 ng/mL hTGFβ, 10 ng/mL hIL-2, 30 ng/mL hIL-4, 10 ng/mL hIL-1β. For supernatant collection experiments, Th1 and Th9 cells were cultured for 5 days and Th2 cells were cultured for 9 days prior to restimulation. For all other experiments, after 5 days cells were washed and cultured without αCD3 or αCD28 but with all polarizing cytokines and antibodies for an additional 3-15 days (media, cytokines, and antibodies changed every 2 days). Cells were collected on day 5-20 for stimulation and analysis; resting cells were defined as cells not being cultured on αCD3 or αCD28 (days 6 and onward).
In vitro mouse T cell differentiation
Murine naïve CD4+CD62L+ cells were isolated from spleen and lymph nodes using negative selection (Miltenyi) followed by flow sorting to >95% purity. Cells were cultured in complete IMDM (10% FBS, 100 IU/mL penicillin, 0.1 mg/mL streptomycin, 55 μM 2-Mercaptoethanol) at a density of 0.5 x106 in 1mL in a 24 well plate for Th1, Th2, and Th9 differentiation. Cells were activated with plate bound αCD3Ɛ (10 μg/mL) and αCD28 (10 μg/mL) for 3 days with polarizing cytokine and antibodies to promote the differentiation of Th1 (10 μg/mL αIL-4, 10 ng/mL mIL-12,), Th2 (10 μg/mL αIFN-γ, 20 ng/mL mIL-4), Th9 (10 μg/mL αIFN-γ, 20 ng/mL mIL-4, 10 ng/mL hIL-2, 5 ng/mL hTGF-β). After 3 days, cells were washed and cultured without αCD3 or αCD28 but with polarizing cytokines and antibodies, and with 10 ng/mL hIL-2, in fresh media. Media, cytokines, and antibodies were changed every 2 days. Cells were collected on day 3-10 for stimulation and analysis; resting cells were defined as cells not being cultured on αCD3 or αCD28 (days 4 and onward).
For Itk inhibitor experiments, CD4+ T cells were purified by magnetic cell separation (Miltenyi) from pooled lymph nodes and spleens. Naive CD4+ T cells (1.2-2 × 105) were co-cultured in complete IMDM media, at a ratio of 1:5 with mitomycin C treated T-depleted splenocytes as APCs in 48-well plates under Th9 conditions (1μg/ml αCD3, 3μg/ml αCD28, 30ng/ml IL-4, 7.5ng/ml TGFβ1, 15ng/ml mTL1A, 15μg/ml αIFN-γ, 15μg/ml αIL-12) or Th2 conditions (1μg/ml αCD3, 3μg/ml αCD28, 60ng/ml IL-4, 30μg/ml αIL-12) for 3 days20. In some experiments Itk inhibitor (BMS 509744) was used at the indicated concentrations, either during T helper differentiation or during restimulation. In some experiments, mice were treated with intraperitoneal tamoxifen 100 μg IP daily for 4 days before isolation of naïve T cells, to delete Itk.
Restimulation
All cells were stimulated in 96-well plates. For ex vivo stimulations, PBMC from AD patients, PBMC from healthy volunteers, and tonsil samples were thawed and washed x2 with complete RPMI-1640 with 10 U/mL DNase I (Roche). PBMCs and tonsil cells were stimulated with 20 ng/mL hIL-2; 40 ng/mL mIL-4 for 2h, followed by brefeldin A (10 μg/mL) for 4h (total 6h) at 37°C. In vitro differentiated T cells, cells were washed x3 prior to restimulation. For supernatant collection experiments, Th1, Th2, and Th9 cells were stimulated in fresh media with plate-bound αCD3 for 48h. For other experiments, murine T cells were stimulated with the following agents alone or in combination, as detailed in individual experiments: 100 ng/mL phorbol 12-myristate 13-acete (PMA, Calbiochem); 1 mg/mL ionomycin (Sigma); 20 ng/mL hIL-2; 40 ng/mL mIL-4. Human T cells were stimulated with (alone or in combination) 20 ng/mL PMA; 1 mg/mL ionomycin; 20 ng/mL hIL-2; 60 ng/mL hIL-4. In some experiments, cells were stimulated with escalating doses of IL-2 and IL-4 as indicated. In some experiments, cells were stimulated with escalating doses of plate bound αCD3 as indicated. In some experiments, cells were restimulated with 30 ng/mL IL-9, 10 μg/mL LIGHT, 10 μg/mL ICOS, 10 μg/mL H7-2, 200 ng/mL OX40L, 10 ng/mL hIL-12 , 100 ng/mL hIL-21, 50 ng/mL IL-7, 10 ng/mL IL-6, 100 ng/mL IL-1β, 50 ng/mL TSLP, 100 ng/mL TL1-A, 100 ng/mL IL-18, 100 ng/mL IL-17A, 100 ng/mL IL-33, 100 ng/mL IL-36α, 100 ng/mL IL-36γ, or 100 ng/mL GM-CSF. For flow cytometric measurement of intracellular cytokines, cells were treated with brefeldin A (10 μg/mL) after 1.5 hours and stimulated for an additional 4.5h (6h total) at 37 °C. In some experiments, cells were treated with tofacitinib (0.3 μM), FK506 (10 nM), BAY11 (5 μM), CAY10571 (25 μM), STAT5-INHIBITOR (Cayman, CAS 285986-31-4, 50 μM), or the STAT6 inhibitor AS1517499 (Cayman, 50 nM) for 20 min before restimulation or during differentiation as indicated.
For supernatant-stimulation experiments, d8 human Th9 cells were washed and resuspended in 200 μL of supernatants for up to 6 hours. Brefeldin was added after 2 hours. In some experiments the following antibodies were added at 20 μg/mL: isotype, αIL-2, αIL-4, or αIL-1β.
For Itk inhibitor experiments, in vitro differentiated cells were stimulated for 4h with plate bound αCD3(1ug/ml) + αCD28(3ug/ml) in presence of Golgistop (BD) in absence or presence of 1μM or 500 nM Itk inhibitor (BMS-509744, Calbiochem).
For pulmonary T cell analysis, cells were stimulated with PMA (100 ng/mL), Ionomycin (1mg/mL), hIL-2 (20 ng/mL) and mIL-4 (40 ng/mL) for 2h followed by brefeldin A (10 μg/mL) for 3h (total 5h) at 37°C.
Flow cytometry
In vitro differentiated cells were stained with Zombie Aqua™ dye (BioLegend) for 15min in 4 °C, then fixed with Cytofix/Cytoperm (BD Bioscience) and underwent intracellular staining. For Itk inhibitor experiments, in vitro differentiated Th9 cells underwent surface staining for CD4 prior to fixation. For phospho-STAT staining, after viability staining cells were fixed with 1.6% PFA for 10 min in dark at RT then treated with ice-cold methanol overnight at −20 °C. Cells were washed with FACS buffer and intracellular staining antibodies were added. Cells isolated from lung tissue were stained with Zombie Aqua for 10 min in the presence of mouse Fc block (20 μg/ml, BioXCell, #BE0307), rat serum (1:50, ThermoFisher, #10710C), hamster serum (1:50, Abcam, #ab7483). After 10 minutes, surface staining antibodies were added for 20 minutes. Cells were then fixed (BD Cytofix/Cytoperm) and stained with intracellular antibodies in the presence of Fc block.
The following antibodies were used for intracellular staining of human cells (full panels available in Source Data): Human Fc block/IgG (10 μg/ml, MP Biomedicals, LLC, #82310) , CD3 APC-Cy7 (SK7) or CD3 AF700 (UCHT1), CD4 BUV395 (RPA-T4), CD8 APC-Cy7 (SK1), CD45RO PE-Texas-red (ECD), IL-2 PerCp-Cy5.5 (MQ1-17H12), IL-4 FITC or BV605 or PE-Cy7 (MP4-25D2), IL-5 BV421 (TRFK5), IL-9 PE (MH9A4), IL-10 PE-Cy7 (JES3-9D7), IL-13 BV711 (JES 10-5A2), IL-17A APC (ebio64CAP17), IFNγ AF700 or V450 (B27), p Y694-STAT5 v450 (47), pY641-STAT6 PerCp-Cy5.5 (18-1P-stat6). The following antibodies were used for murine cells (full panels available in Source Data): CD45.1 PerCp-Cy5.5 (A20), CD45.2 FITC or PE-Cy7 (104), CD44 AF700 (IM7), TCRβ APC-Cy7 (H57-597), CD4 PerCP-Cy5.5 (RM4-5) or CD4 V450 or BUV737 (GK1.5), IL-2 PE-Cy7 (MQ1-17H12) or IL-2 PE (JES-6-5H4), IL-4 PE-Texas-red or AF488 (11B11), IL-9 APC or PE or BV421 (RM9A4), IL-13 V450 or PE-Cy7 (eBio13A), IL-17A FITC (TC11-18H10.1), IFNγ PE or BV605 (XMG1.2).
Data were collected on an LSR Fortessa (BD Biosciences) or FACS Symphony A5-4 (BD Biosciences) and analyzed with FlowJo (Treestar). Gating strategies for murine experiments are shown in Figure S7, and gating strategies for human experiments are shown in Figure S8.
Cell sorting
Human naïve CD4+ T cells were enriched by magnetic separation (StemCell), then stained with Zombie Aqua, CD19 AF488 (HIB19), CD16 FITC (3G8), CD14 FITC (M5E2), CD4 APC (S3.5), CD8 APC-Cy7 (SK1), CD45RA BV711 (HI100). Aqua−/CD4+/CD45RA+/CD19−/CD16−/CD14− cells were sorted on a FACS Aria. For ATAC-seq experiments, in vitro differentiated Th9 cells were stained with DAPI and CD45RO PE-Texas-red and sorted for DAPI−/CD45RO+ cells.
Mouse naïve CD4+ T cells were enriched using magnetic separation (Miltenyi), then stained with DAPI, CD62L PE-Cy7 (MEL-14), CD25 PE (PC 61.5), CD4 V450 (GK1.5), and CD44 AF700 (IM7). DAPI−/CD4+/CD25−/CD44lo/CD62Lhi cells were sorted on a FACS Aria. For ATAC-seq experiments, in vitro differentiated Th9 cells were stained with DAPI and sorted for DAPI− cells. For plasticity experiments (Fig 3), d3 Th9 cells from INFER mice were stained with DAPI, and DAPI−/GFP+ cells were sorted on a FACS Aria.
Papain induced Asthma
All in vivo studies were randomized. Mice were anesthetized with isoflurane and exposed intranasally (IN) to 25 μg papain (Calbiochem) in 30 μl PBS on day 0, 3, 6, and 13. 10-12 hours after the last IN injection, mice were euthanized. In some experiments, 100 μg αMHC2 (clone Y-3P) or isotype (BioXCell) were administered daily by intraperitoneal injection (IP) on day 12 and 13. We validated activity of this antibody in vivo (see below). In some experiments, 50 mg/kg tofacitinib in 0.5% methylcellulose was administered twice daily IP on day 12 and 13. In some experiments, mice were treated with 100 μg αIL-9 (clone 9C1) or isotype antibody (BioXCell) every other day by intraperitoneal injection (IP) on day 0-14. For Itk deletion experiments, Mice were anesthetized with isoflurane and exposed intranasally (IN) to 25 μg papain (Calbiochem) in 30 μl PBS on day 0, 3, 6, 14, and 21, then euthanized 10-12 hours after the last IN injection as above. On days 10-14, mice were treated with tamoxifen 100 μg IP daily. After euthanasia, lung tissues were fixed in 10% neutral buffered formalin solution. Paraffin embedded sections (5 μm) were stained with hematoxylin and eosin (H&E) or Periodic Acid-Schiff (PAS). Cells from lung were isolated and digested in collagenase Type IV (100 unit/mL; Gibco) for 70 min at 37 °C. After digestion, cells were lysed with ACK buffer (Gibco) for 30-60 sec and strained through a 40 μM strainer, then stimulated and stained as above.
Th9 Adoptive Transfer and OVA asthma
All in vivo studies were randomized. To validate αMHC2 in vivo, mice were exposed IP ovalbumin (OVA) on day 0 and 7, followed by IN OVA on day 15 and 16. 100 μg αMHC2 (clone Y-3P) was administered IP on day 15 and 16, and mice were euthanized on day 18. To generate ovalbumin (OVA)-specific Th9 cells, INFER (GFP IL-9 reporter) mice were crossed to Rag2−/− OT-II CD45.1+ mice. Resulting INFER/Rag2−/− OT-II CD45.1+ CD45.2+ naïve T cells were co-cultured with antigen presenting cells (APCs) under Th9 promoting conditions as previously described16. On day 3, cells were collected and stained with Zombie Aqua and antibodies to CD4, CD45.1, and OT-ii as above. IL-9+ Th9 cells were sorted (DAPI−/CD4+/OT-ii+/CD45.1+/GFP+) and washed x3 with PBS. 1x106 Th9 cells were injected retroorbitally into congenic (CD45.2) C57BL/6 mice. After 24h, mice were anesthetized as previously described16 and exposed intratracheally to mIL-2 (100 ng) and mIL-4 (500 ng). After 48h, mice were anaesthetized with isoflurane, challenged intranasally with mIL-2 (100 ng) and mIL-4 (500 ng), and euthanized 12h after the last challenge.
Scoring Papain induced asthma and OVA-asthma
Lung histology was scored blindly by a pathologist as previously described based on perivascular cuffing, peribronchial cuffing, interstitial inflammation, and goblet cell hyperplasia16.
Enzyme-linked immunosorbent assay (ELISA)
ELISA for human IL-2 (BioLegend), IL-4 (BioLegend), and IL-9 (R&D system) were performed according to the manufacturer’s protocol. Absorbance readings (450 nm and 570 nm) were collected on a TECAN plate reader.
mRNA extraction and reverse transcription quantitative PCR (RT-qPCR)
Cells were suspended in Trizol LS (Invitrogen), and total RNA extracted according to the manufacturer instruction. 250 ng RNA was reverse transcribed using cDNA synthesis kit (Applied Biosystems™) and qPCR performed in triplicate using TaqMan master mix (Applied Biosystems™) and primers for IL9 (Hs00174125_m1) and 18S (Hs99999901_s1). Fold change in transcript level was calculated by normalizing threshold values to 18S.
Nascent RNA extraction and digital droplet PCR (ddPCR)
To measure nascent RNA level in human Th9 cells, total RNA was extracted as above. EU-labeled cellular RNAs were separated using the Click-it Nascent RNA Capture kit (ThermoFisher) according to the manufacturer’s protocol. Nascent IL9 expression was measured relative to TBP (BioRad cat 10031257).
RNA-seq library preparation
mRNA was extracted as above. Libraries for mRNA-sequencing were prepared using the NEBNext Ultra II Directional RNAseq Library kit, per the manufacturer’s instructions. Samples were sequenced on a NovaSeq 6000, paired end, 50 cycles, and processed with bcl2fastq v2.20.0.422.
RNAseq analysis
Reads of 50 bases were processed using the rna-seek pipeline (https://ccbr.github.io/RNA-seek/). Briefly, adapter sequences were trimmed with cutadapt 1.18 and mapped to the mouse transcriptome and genome mm10 or human transcriptome and genome GRCh38 (hg38) using STAR. Gene expression values (FPKM: Fragments Per Kilobase exon Per Million mapped reads; TPM, Tags Per Million mapped reads; counts) were generated using RSEM. Differential gene expression was calculated using DESeq2. To calculate fold-change cutoffs, datasets were normalized based on FPKM and purged of micro-RNAs, sno-RNAs and sca-RNAs. To minimize fold-change artifact from low abundance transcripts, a small offset was added to all RPKM values (0.37; equal to the averaged second quartiles of each dataset). When multiple transcripts were detected for one gene, only the most abundant (highest average RPKM across all replicates) was analyzed. Transcripts with FPKM < 1 were excluded. Downstream analyses were performed with R 4.2.2, Morpheus (https://software.broadinstitute.org/morpheus/), and DAVID (https://david.ncifcrf.gov/).
ChIP-seq sample and library preparation
ChIP-seq sample preparation was performed with Covaris kit (PN 520154) as previously described16. Naïve T cells were cultured in vitro under Th9 promoting conditions and treated with chemical cross-linking (1% formaldehyde) on days 0, 3, 4, and 8 (murine) or days 0, 5, 8, and 15 (human). At least 0.5 million cells were used for each immunoprecipitation. After chemical chromatin cross-linking (1% formaldehyde), cells were washed and frozen at −80C. Cells were resuspended in Lysis Buffer B (Covaris) with protease inhibitors, and DNA fragmentation was performed on a Covaris Sonicator (ME220) for 8 min to an average length of 200-700 bp. After sonication, cells were immunoprecipitated with anti-H3K27Ac (ab4729; Abcam), anti-H3K4Me1 (ab8895; Abcam), and anti-H3K4Me3 (ab8580; Abcam). Genomic DNA (input) was prepared by treating aliquots of chromatin with proteinase K and heating to de-crosslink, followed by purification (28106, Qiagen). An aliquot of chromatin underwent immunoprecipitation, after which decrosslinking was performed. After recovering purified DNA, 5ng or more of DNA was used to generate libraries (NEBNext Ultra Directional II DNA sequencing kit). Samples were sequenced on a NovaSeq 6000, paired end, 50 cycles, and processed with bcl2fastq v2.20.0.422.
Chip-Seq analysis
Reads of 50 bases were processed using the ccbr pipeliner tool (https://ccbr.github.io/pipeliner-docs/). Briefly, adapter sequences were trimmed with cutadapt 1.18 and mapped to the mouse genome mm10, human genome hg19 (for bigwig files/visualization, to harmonize with ATAC-seq data), or human genome GRCh38 (hg38) (for peak calling) using BWAmem 0.7.17. Uniquely mapped and nonredundant reads were generated using Picard 2.17.11 and used for downstream analysis. The aligned files were converted to bam format using SAMtools 1.6, and bigwig files were created using deepTools 3.0.1. Genome browser files are displayed with IGV.
For downstream analysis, peaks were called using MACS v2.1.1 using FDR 0.01 and with the input sample for background correction. In the case of H3K4me1, H3K4me3, and H3K27Ac samples, “—broad” setting was used. Peaks were annotated using Uropa 4.2.1 and the annotatePeaks module in HOMER. Only regions called in both replicates were used in downstream analysis. After calling peaks with MACS, the union peaks of replicate samples were created using mergePeaks module in HOMER and divided into shared and dynamic peaks using the same utility. Motif enrichment analysis of dynamic peaks was done using the findMotifsGenome module in HOMER, de novo motif analysis.
ATAC-seq transposition and library preparation
Omni-ATAC-seq sample preparation was performed as previously described55. After flow cytometric sorting, 50,000 cells were lysed with 50 μl cold ATAC-Resuspension buffer (RSB) (10 mM Tris-HCl pH 7.4, 10 mM NaCl, 3 mM MgCl2, 0.1% Tween 20) with 0.1% NP40, and 0.01% Digitonin on ice for 3 min. Lysis was stopped with 1 mL cold ATAC-RSB. After centrifugation (500×g for 10 min), nuclei were resuspended in 50μl transposition reaction containing 2.5μl Tn5 transposase (FC-121-1030; Illumina) to tag and fragmentalize accessible chromatin. The reaction was incubated at 37°C, 400rpm for 30 min; DNA was then purified using a MinElute kit (QIAGEN) and amplified with 8-12 cycles of PCR based on the amplification curve. Samples were purified using a MinElute kit, sequenced on a NovaSeq 6000, paired end, 50 cycles, and processed with bcl2fastq v2.20.0.422.
ATAC-sequencing
ATAC-seq reads from two biological replicates were used for each sample and analyzed as previously described16. Briefly, redundant paired end (PE) reads were removed using fastquniq. PE reads of 50 bases were aligned to the mouse genome build mm10 or human build hg19 with Bowtie 0.12.8, following the guidelines presented by Buenrostro et al56. Customized python scripts were used to calculate fragment length of each pair of uniquely mapped PE reads for size distribution analysis, and to group uniquely mapped reads into bins of 0 to 175 bases and 180 to 250 bases, respectively38. UCSC Genome Browser viewable and normalized BigWig files were generated with the Hypergeometric Optimization of Motif EnRichment program (HOMER) version 4.8. Genome browser files are displayed with IGV.
Only one mapped read to each unique region of the genome that was less than 175 bp was kept and used in peak calling. Regions of open chromatin were identified by MACS 1.4.2 using a p-value of 1 × 10−5. Only regions called in both replicates were used in downstream analysis. Peak intensities (“tags” column) were normalized as tags per 10 million reads (RP10M) in the original library. Signal across all sites (i.e., all annotated genes and all accessible chromatin regions) was analyzed to eliminate potential bias by pre-selection. After calling ATAC-seq peaks with MACS, the union peaks of replicate samples were created using mergePeaks module in HOMER and divided into shared and dynamic peaks using the same utility.
Correlation analysis
For analysis of Il9 and Th2 locus correlations (Fig S4g), chromatin accessibility and epigenetic marks, peaks were annotated based on proximity to the nearest gene and checked manually for Il9/IL9, Il4/IL4, Rad50/RAD50, Il13/IL13, and Il5/IL5. Tag densities (normalized reads) were calculated using HOMER. For each sample at each timepoint, the total number of tags at each gene locus were combined to obtain the total tag density over the entire locus. For RNA-seq data, FPKM-normalized gene expression for each gene was used. Pearson correlation values were calculated with R 4.2.2 for Il9/IL9 with Il4/IL4, Rad50/RAD50, Il13/IL13, and Il5/IL5. Other downstream analysis and heatmap generation were performed as described above.
Selection and analysis of public gene expression datasets
For GSEA analysis57,58, enrichment score curves and member ranks were generated by GSEA software (Broad Institute). For Figure 6, lists of TF-induced and TF-repressed genes were obtained using a previously published analysis of public datasets16. These included STAT1 (GSE40666), STAT3 (GSE65621); STAT4 (GSE40463); STAT5 (GSE77656); STAT6 (GSE40463), IRF4 (GSE39756); NF-kB16; NFAT (GSE64409); JunB (GSE98413). For Figures 3-6, public datasets were selected based on significantly increased IL9 expression during allergic response as previously described or based on isolation and analysis of Th9 cells for datasets describing in vivo and in vitro generated Th9 cells16. The following datasets were selected: GSE97087, antitumor Th9 vs. Th1/Th17; GSE133385, lesional IL9high vs. nonlesional IL9low atopic dermatitis skin; GSE73482, house dust mite (HDM) stimulated T cells from allergic subjects vs. healthy volunteers; GSE133385, AD patients treated with the JAK-SYK inhibitor ASN002; GSE146170, IL-9-expressing HDM-reactive T helper cells from allergic patients; MTAB-5739, IL-9-expressing T helper clones from healthy volunteers; GSE123501, in vitro differentiated Th9 cells and Th9 cells from IL-9-reporter mice treated with papain.
Additionally, the Gene Expression Omnibus was searched for datasets or series examining time-course gene expression in atopic or allergic inflammation, treated with JAK inhibitors and demonstrating significantly increased IL9 expression during allergic response. Although no datasets were identified that detected IL9 expression on transcriptome analysis, one dataset (GSE133385) was identified in which IL9 expression measured by RT-qPCR increased during allergic response44. A list of public datasets used can be found in Extended Data Table 2.
For analysis of ex vivo human memory CD4+ cell epigenetics, ENCODE tracks were analyzed directly in the igv browser 59.
Statistical analysis and reproducibility of PCR, flow cytometry, ELISA, and in vivo experiments
All in vitro experiments contained ≥ 3 biological replicates, and all data were expressed as mean ± SEM (bar/line graphs) or as min-to-max with lines at median (box plots). All in vivo experiments included 2-3 independent experiments with 1-6 animals per group per experiment, comprising ≥ 3 biological replicates. No statistical methods were used to pre-determine sample sizes but our sample sizes are similar to those reported in previous publications7,16,18. No animals/data points were excluded. Statistics were analyzed with GraphPad Prism 8.0. Normality testing was performed to determine whether samples were normally distributed. Normally distributed samples were analyzed using unpaired or paired Student’s t-test. Nonparametrically distributed samples were analyzed using Wilcoxon test (paired) or Mann-Whitney test (unpaired). All statistical testing was 2-sided.
Extended Data
Extended Data Figure 1. Corresponds to Figure 1.
a-e. Naïve CD4+ T cells from healthy volunteers were activated for 5 days with αCD3, αCD28, IL-2, and subset-promoting cytokines and antibodies. After 5 days, αCD3/αCD28 were withdrawn, and cells were cultured with IL-2 and subset-promoting cytokines and antibodies. Pooled results show production of IFN-γ (a), n=9 (Th1, Th2) or 10 (Th9); IL-4 (b), n=8 (Th1), 9 (Th2), or 4 (Th9); IL-13 (c) n=8 (Th1), 9 (Th2), or 4 (Th9); IL-9 (d) n=9 (Th1, Th2), or 10 (Th9); and IL-2 (e) n=9 (Th1, Th2), or 10 (Th9), with or without PMA and Ionomycin (P/I). f. Bar graphs show % IL-9 positive cells differentiated to d8 Th9 as above and restimulated with vehicle vs. plate bound αCD3 at escalating doses (n=3) g-k. Naïve CD4+ T cells from WT C57BL/6 mice were activated for 3 days with αCD3, αCD28, IL-2, and subset-promoting cytokines and antibodies. After 3 days, αCD3/αCD28 were withdrawn, and cells were cultured with IL-2 and subset-promoting cytokines and antibodies. Pooled results show production of IFN-γ (f), n=6 (Th1, Th2) or 3 (Th9); IL-4 (g), n=6 (Th1, Th2) or 5 (Th9); IL-13 (h), n=6 (Th1), 7 (Th2), or 5 (Th9); IL-9 (i), n=6 (Th1), 7 (Th2), or 10 (Th9); and IL-2 (j), n=4 (Th1), 5 (Th2), or 3 (Th9), with or without P/I. l. Bar graphs show % IL-9+ cells of resting (d8) human Th9 cells restimulated with Th9 supernatants and vehicle (n=3), αIL-2 (20 μg/mL), αIL-4 (20 μg/mL), or αIL-1β (20 μg/mL). For all line/bar graphs, error bars show ± S.E.M. Box plots show all data points (min to max, lines at median). For all experiments: paired or unpaired t-test, normally distributed data, Wilcoxon (paired) or Mann-Whitney (unpaired), non-normally distributed data. All statistical tests are 2-sided, all replicates are biologically independent samples.
Extended Data Figure 2. Corresponds to Figure 2.
a. Plot shows % IL-9+ of d8 human Th9 cells restimulated with vehicle (n=7), IL-1β (n=3), IL-18 (n=4), IL-33 (n=4), IL-36α (n=7), or IL-36γ (n=7). b-c. Timelines show experimental design; graphs shows % IL-9+ (n=6) for human non-restimulated T cells differentiated as in Fig 1a, with STAT5 inhibitor or STAT6 inhibitor from d0-5 (b) or d5-8 (c). d-f. Box plot shows concentration (pg/mL, ELISA) of IL-2 (d), IL-4 (e), and IL-9 (f) in supernatants of human in vitro differentiated Th1- or Th9 (d5), or Th2 (d14) cells, after 48h restimulation.. g,h. d8 human Th9 cells were restimulated with escalating doses of IL-2 (b) (2, 20, or 200 ng/mL) or IL-4 (c) (6, 60, or 500 ng/mL) (n=3). i,j. Representative flow plots (i) and box plot (j) show % IL-9+ cells in resting (d4) murine Th9 cells restimulated with IL-2 + IL-4 (n=10) k. Box plot shows pooled results d8 human Th9 cells restimulated with vehicle (n=9), IL-6 (n=3), IL-7 (n=5), IL-12 (n=3), IL-21 (n=3), or TSLP (n=4). l, m. Box plots show pooled (n=6) mean fluorescence intensity (MFI) of pSTAT5 (l) and pSTAT6 (m) in d8 human IL-9+ and IL-9− Th9 cells restimulated with IL-2, IL-4, or IL-2 + IL-4. n. Box plots show pooled (n=3) % IL-9+ d8 human Th9 cells stimulated with IL-2 and IL-4 in the presence of STAT5 or STAT6 inhibitor. For all experiments: paired or unpaired t-test, normally distributed data; Wilcoxon (paired) or Mann-Whitney (unpaired), non-normally distributed data. Box plots show all data points (min to max, lines at median). All statistical tests are 2-sided, all replicates are biologically independent samples.
Extended Data Figure 3. Corresponds to Figure 3.
a b. Line graph shows % IL-9+ cells of human Th9 differentiated as in Fig 1a. Cells were restimulated with vehicle, IL-2, IL-4, or IL-2 + IL-4 (a, n=6); b. on d5 (n=14), d8/d11 (n=17), d15 (n=16), and d20 (n=11), cells were restimulated with PMA + ionomycin (P/I) or P/I + IL-2 + IL-4. c,d. Line graph shows % IL-9+ cells of murine Th9 differentiated as in Fig 1d. Cells were restimulated with vehicle, IL-2, IL-4, or IL-2 + IL-4 (c, n=4). d. On d3 (n=12), d4 (n=11), d5 (n=9), d6 (n=3), d7 (n=3), and d8 (n=6), cells were restimulated P/I or P/I + IL-2 + IL-4. e, f. Representative flow plots (e) and graphs (f) show % IL-9+ of circulating Th9 cells stimulated with P/I or P/I + IL-2 + IL-4, from healthy volunteer (HV, n=5) or atopic patients (AD, n=14). g. Line graphs shows pooled results (n=5) for % IL-9+ cells from IL-9 reporter (INFER) mice, sorted on d3 and maintained as in Fig 1d. Cells were restimulated with vehicle, IL-2 + IL-4, P/I or P/I + IL-2 + IL-4, p-value is for d4. h. Bar graphs show viability (n=4) for Th9 cells. i. Venn diagram shows generation of “allergic Th9” cassette using: HDM-reactive circulating Th9 cells from allergic patients1, Th9 clones from healthy subjects2, pulmonary Th9 cells from IL-9 reporter mice, and in vitro differentiated Th9 cells3. Genes differentially expressed in >1 dataset were selected. j. Bar graph shows pathway enrichment scores and false discovery rates (FDR, DAVID) for the “allergic Th9” cassette. k. Scatterplot shows enrichment score (GSEA) and FDR for correlation (Pearson) of STAT1, STAT3, STAT4, STAT5, and STAT6 target genes with murine Il9 expression and human IL9 expression. For all experiments: paired or unpaired t-test, normally distributed data; Wilcoxon (paired) or Mann-Whitney (unpaired), non-normally distributed data. Bar/line graphs show mean ± SEM; box plots show all data points (min to max, lines at median). All statistical tests are 2-sided, all replicates are biologically independent samples.
Extended Data Figure 4. Corresponds to Figure 4.
a, b. Genome tracks show poised enhancer (H3K4m1), active promoter (H3K4m3), active enhancer (H3K27Ac) marks, and accessibility (ATAC) of the murine Th2 (a, Il4-Il13-Rad50-Il5) and Gzmb loci at different time points during Th9 differentiation and resting. c, d. ATAC-seq tracks show the human extended IL9 locus including the promoter (Il9p), downstream enhancer (DS) and upstream enhancers 1-4 (E1-E4), in naïve T cells, Th1 cells (d5), and Th9 cells (d5) (c), and in ex vivo naïve, Th1, Th2, and Th17 cells from ENCODE (d). e, f. Genome tracks show poised enhancer (H3K4m1), active promoter (H3K4m3), active enhancer (H3K27Ac) marks, and accessibility (ATAC) of the human Th2 (e, IL4-IL13-RAD50-IL5) and GZMB loci at different time points during Th9 differentiation and resting. g. Heatmap shows Pearson correlation of Il9/IL9 expression, ATAC-seq tag density, H3K4me1 tag density, H3K4me3 tag density, and H3K27Ac tag density with those of Il4/IL4, Il13/IL13, Rad50/RAD50, and Il5/IL5.
Extended Data Figure 5. Corresponds to Figure 5.
a-c. Pooled counts of pulmonary CD45+TCRβ+CD4+CD44+IL-13+ cells (a, n=8, PBS; n=13, papain), CD45+TCRβ+CD4+CD44+IL-17A+ cells (b, n=3, PBS; n=6, papain), and CD45+TCRβ+CD4+CD44+IL-2+ cells (c, n=8, PBS; n=13, papain) from mice treated with isotype vs. αMHC2 between papain sensitization and challenge. d. Th9 cells were differentiated in the presence of vehicle vs. BMS509744 (Itk inhibitor; 0.5 μM, 1 μM, 1.5 μM) and restimulated with αCD3 + αCD28 (n=3) e-f. Bar graphs (n=7) show %IL-9+ (e) of Th9 and %IL-4+ (f) of Th2 cells restimulated with αCD3 + αCD28 and vehicle vs. BMS509744 (0.5 μM, 1 μM), g. Design of inducible Itk-deleted mouse. Itkflox/flox were crossed ERT-Cre mice to generate ItkERT mice. h. Representative flow plot and bar graph show %IL-9+ cells of Th9 cells from TAM-treated (Itk-deleted, n = 3) vs. TAM-untreated (n=5) mice. i. Timeline shows model of papain-induced airway inflammation with Itk deletion. Mice were treated with vehicle vs. tamoxifen between sensitization and challenge. j. Gel shows that injection of tamoxifen results in deletion of WT Itk gene in mice from (i). k-n. Representative periodic acid-Schiff (PAS) stained images (k), pooled histology scores (l, n=3, PBS; n=5, Papain), pulmonary CD45+TCRβ+CD4+CD44+IL-9+ (Th9) (m, n=5, PBS; n=10, Papain), and pulmonary CD45+TCRβ+CD4+CD44+IL-13+ (Th2) cell counts (n, n=3, PBS; n=5, Papain), from Itkf/f treated with vehicle vs. tamoxifen as in (i). o-r. Pooled counts of pulmonary CD45+CD4− IL-9+ cells (o, n=5, PBS; n=8, Papain), CD45+TCRβ+CD4+CD44+IL-13+ cells (Th2) (p, n=5, PBS; n=8, Papain), CD45+TCRβ+CD4+CD44+IL-17A+ cells (Th17) (q, n=4, PBS; n=7, Papain), and CD45+TCRβ+CD4+CD44+IL-2+ cells (r n=4, PBS; n=7, Papain) ILC2-deficient mice treated with isotype vs. αMHC2 between papain sensitization and challenge. For all experiments: paired or unpaired t-test, normally distributed data; Wilcoxon (paired) or Mann-Whitney (unpaired), non-normally distributed data. Bar graphs show mean ± SEM; box plots show all data points (min to max, lines at median). All statistical tests are 2-sided, all replicates are biologically independent samples from ≥ 2 independent experiments.
Extended Data Figure 6. Corresponds to Figure 6.
a. Timeline shows Th9 adoptive transfer-induced airway inflammation. Ovalbumin (OVA)-specific OT-ii Th9 cells were differentiated in vitro, sorted for IL-9+ cells, and adoptively transferred. Recipient mice were treated with IL-2 and IL-4 intratracheally and intranasally. b. Timeline shows papain-induced airway inflammation with tofacitinib; mice were treated with vehicle vs. tofacitinib between sensitization and challenge. c-e. Pooled counts of pulmonary CD45+TCRβ+CD4+CD44+IL-13+ (c), CD45+TCRβ+CD4+CD44+IL-17A+ (d), and CD45+TCRβ+CD4+CD44+IL-2+ cells (e) mice treated with vehicle vs. tofacitinib between sensitization and challenge. n=6 (PBS/PBS), 5 (PBS/tofa), 10 (Papain/MC and Papain/tofa) f. Timeline shows papain-induced airway inflammation with tofacitinib and αIL-9. Mice were treated as in (b) but also received isotype or αIL-9 every other day. g-k. Pooled counts of pulmonary CD45+TCRβ+CD4+CD44+IL-9+ (g), CD45+TCRβ+CD4+CD44+IL-13+ (h), and live CD45+TCRβ+CD4+CD44+IL-4+ cells (i), representative periodic acid-Schiff (PAS) stained images (j) and pooled histology scores (k) of mice treated as in Fig S6f; n=5 (PBS/isotype/MC), 8 (Papain/isotype/MC), and 10 (Papain/αIL-9/MC and Papain/αIL-9/tofa). For all in vivo experiments: Bar graphs show mean ± SEM; 2-sided Mann-Whitney (unpaired). All replicates are biologically independent samples from ≥ 2 independent experiments. l-o. Line graphs show expression (mean ± SEM) over 96 hours of IL9 (l), STAT5A (m), STAT5B (n), and STAT6 (o), in skin of nickel-allergic patients exposed to nickel over 96 hours (n=7) p. Graph shows normalized enrichment scores and false discovery rate (GSEA) for expression of “allergic Th9” cassette, STAT5-induced genes, and STAT6-induced genes in 7h-nickel-exposed, 48h-nickel-exposed, and 96h-nickel-exposed skin relative to pre-exposed/unexposed skin.
Extended Data Figure 7. Reporting of gating strategy and validation for murine cell studies.
a-f. Representative flow cytometric plots show gating strategies for naïve T cell sorting (a), Th9 sorting for transfer experiments (b), for in vitro differentiated Th9, Th1, and Th2 cells shown in Figures 1-3 and S1-S3 (c), for in vivo memory T helper subsets in Figures 5-6 and S5-S6 (d-e), and for adoptively transferred CD45.1+ Th9 cells analyzed in murine lung two days after transfer (f). g. Timeline shows protocol for validation of MHC2 blocking antibody in vivo. Mice were sensitized to ovalbumin (OVA) intraperitoneally on d0 and d7, then challenged on d13 and d14. MHC blocking antibody was administered with each dose of OVA. Bar graphs show results of MHC2 validation experiment. Blockade of MHC2 reduced histology scores, BAL Eosinophil counts, and lung-infiltrating IL-4+ and IL-13+ (Th2) cell counts. Mean ± SEM, 2-sided Mann Whitney, all data points represent biological replicates.
Extended Data Figure 8. Reporting of gating strategy for human cell studies.
Representative flow cytometric plots show gating strategies for naïve T cell sorting (a), gating for in vitro differentiated Th9, Th1, and Th2 cells shown in Figures 1-3 and S1-S3 (b), and for ex vivo circulating memory Th9 cells in stimulated PBMC samples (c).
Extended Data Table 1.
Th9 cassette genes
| Murine | Human |
|---|---|
| Adam15 | ADAM15 |
| Anpep | ANPEP |
| Anxa3 | ANXA3 |
| Anxa4 | ANXA4 |
| App | APP |
| Arg1 | ARG1 |
| Arg2 | ARG2 |
| Atf3 | ATF3 |
| Atp6v0c | ATP6V0C |
| Basp1 | BASP1 |
| Batf3 | BATF3 |
| Bcl2a1d | BCL2A1 |
| AA467197 | C15ORF48 |
| C4b | C4A |
| Cav1 | CAV1 |
| Cavin1 | CAVIN1 |
| Ccl11 | CCL11 |
| Ccl12 | CCL17 |
| Ccl17 | CCL2 |
| Ccl22 | CCL22 |
| Ccl24 | CCL24 |
| Ccl3 | CCL3 |
| Ccl7 | CCL7 |
| Ccr1 | CCR1 |
| Ccr4 | CCR4 |
| Ccr8 | CCR8 |
| Cd24a | CD24 |
| Cd300c2 | CD300C |
| Cd63 | CD63 |
| Cdc42ep4 | CDC42EP4 |
| Cfh | CFH |
| Cish | CISH |
| Clec4a3 | CLEC4C |
| Clec4d | CLEC4D |
| Clic4 | CLIC4 |
| Clu | CLU |
| Col6a1 | COL6A1 |
| Cox17 | COX17 |
| Crabp2 | CRABP2 |
| Creg1 | CREG1 |
| Creld2 | CRELD2 |
| Csf2 | CSF2 |
| Csf2ra | CSF2RA |
| Cst3 | CST3 |
| Cstb | CSTB |
| Ctsh | CTSH |
| Ctsz | CTSZ |
| Cx3cr1 | CX3CR1 |
| Cxcl16 | CXCL16 |
| Cxcl2 | CXCL3 |
| Cxcl3 | CXCL3 |
| Cxcl5 | CXCL6 |
| Cyp11a1 | CYP11A1 |
| Cyp4f18 | CYP4F2 |
| Dohh | DOHH |
| Dstn | DSTN |
| Ece1 | ECE1 |
| Egfl7 | EGFL7 |
| Eif4ebp1 | EIF4EBP1 |
| Emp2 | EMP2 |
| Fabp5 | FABP5 |
| Fam110a | FAM110A |
| Fkbp11 | FKBP11 |
| Fkbp2 | FKBP2 |
| Ftl1 | FTL |
| G0s2 | G0S2 |
| Gabarapl1 | GABARAPL1 |
| Galk1 | GALK1 |
| Gcm1 | GCM1 |
| Gcsh | GCSH |
| Gpx3 | GPX3 |
| Gsn | GSN |
| Gstm1 | GSTM5 |
| Gzmb | GZMB |
| Havcr2 | HAVCR2 |
| Hdc | HDC |
| Hk2 | HK2 |
| Hlf | HLF |
| Hmgn3 | HMGN3 |
| Hpgd | HPGD |
| Hspb1 | HSPB1 |
| Hspe1 | HSPE1 |
| Idi2 | IDI2 |
| Ier3 | IER3 |
| Ifi30 | IFI30 |
| Ifitm3 | IFITM3 |
| Igfbp7 | IGFBP7 |
| Il10 | IL10 |
| Il13 | IL13 |
| Il1r2 | IL1R2 |
| Il1rl1 | IL1RL1 |
| Il4 | IL4 |
| Il5 | IL5 |
| Il9 | IL9 |
| Kctd14 | KCTD14 |
| Lat2 | LAT2 |
| Lcn2 | LCN2 |
| Lgmn | LGMN |
| Litaf | LITAF |
| Manf | MANF |
| Map3k8 | MAP3K8 |
| Marcks | MARCKS |
| Mcfd2 | MCFD2 |
| Mgll | MGLL |
| Mgp | MGP |
| Mgst1 | MGST1 |
| Mif | MIF |
| Mrpl12 | MRPL12 |
| Mrpl36 | MRPL36 |
| Mrto4 | MRTO4 |
| Mt1 | MT1G |
| Mt2 | MT2A |
| Myo1e | MYO1E |
| Ncs1 | NCS1 |
| Ndufc2 | NDUFC2 |
| Nek6 | NEK6 |
| Nhp2 | NHP2 |
| Nipa2 | NIPA2 |
| Nkain1 | NKAIN1 |
| Nlrp3 | NLRP3 |
| Nme1 | NME1 |
| Nrgn | NRGN |
| Nucb2 | NUCB2 |
| Nupr1 | NUPR1 |
| Olr1 | OLR1 |
| Pdzk1ip1 | PDZK1IP1 |
| Perp | PERP |
| Phlda1 | PHLDA1 |
| Pla2g7 | PLA2G7 |
| Pld4 | PLD4 |
| Plin2 | PLIN2 |
| Plk2 | PLK2 |
| Pparg | PPARG |
| Ppic | PPIC |
| Pth | PTH |
| Pxdc1 | PXDC1 |
| Rai14 | RAI14 |
| Retnla | RETNLB |
| Rgl1 | RGL1 |
| Rpph1 | RPPH1 |
| Rps27l | RPS27L |
| S100a1 | S100A1 |
| S100a8 | S100A8 |
| Sap30 | SAP30 |
| Sdc1 | SDC1 |
| Sec61b | SEC61B |
| Sec61g | SEC61G |
| Sema7a | SEMA7A |
| Serpina1a | SERPINA1 |
| Slc15a3 | SLC15A3 |
| Slc9a3r2 | SLC9A3R2 |
| Spi1 | SPI1 |
| Sv2c | SV2C |
| Tfec | TFEC |
| Thbd | THBD |
| Thbs1 | THBS1 |
| Timm13 | TIMM13 |
| Timp1 | TIMP1 |
| Tmem220 | TMEM220 |
| Tnfsf10 | TNFSF10 |
| Tnip3 | TNIP3 |
| Trmt1 | TRMT1 |
| Trmt61a | TRMT61A |
| Tubb2a | TUBB2A |
| Tubb6 | TUBB6 |
| Txndc17 | TXNDC17 |
| Uqcc2 | UQCC2 |
| Uqcr10 | UQCR10 |
| Uqcr11 | UQCR11 |
| Vat1 | VAT1 |
| Vcam1 | VCAM1 |
| Ybx3 | YBX3 |
| Yif1a | YIF1A |
| Zbtb32 | ZBTB32 |
| Zeb2 | ZEB2 |
Extended Data Table 2.
Public datasets analyzed
| Dataset | Utilization |
|---|---|
| GSE97087 | antitumor Th9,Th1, and Th17 |
| GSE133385 |
|
| GSE73482 |
|
| GSE40666 | STAT1 target genes |
| GSE65621 | STAT3 target genes |
| GSE40463 |
|
| GSE77656 | STAT5 target genes |
| GSE39756 | IRF4 target genes |
| GSE64409 | NFAT target genes |
| GSE98413 | JunB target genes |
| GSE6281 | Pre-exposure vs. post-exposure skin from nickel-allergic patients exposed to nickel for 96 hours |
| GSE123501 |
|
| E-MTAB-5739 | IL-9+ T cell clones expanded from PBMC of healthy volunteers |
| GSE146170 | IL-9+ house dust mite reactive T helper cells isolated from asthmatic patients |
Acknowledgements
The authors would like to thank Richard Flavell and Paula Licona-Limon for generously providing INFER and Klrg1Cre mice. We would like to thank Franziska Petermann, Yvonne Baumer, and Shmuel Muallem for their thoughtful review of and comments on the manuscript. We would like to acknowledge the NIAID flow core for their assistance with flow sorting and analysis and the NIAMS sequencing core for assistance with sequencing and analysis. We would like to thank Julie Reilley and Jennifer Cannons for assistance with sample retrieval. The study was funded by the Intramural Research Programs of NIAID and NIAMS. The relevant grants are: ZIA-AI001251 (DMS), ZIA-AI001202 (PAF-G), ZIA-AI001098 (JDM), ZIA-AI001240 (PS), and ZIC-AR041181 (FM).
Footnotes
Competing Interests statement
J G-R is currently employed by TCR Therapeutics. All the other authors have no interests to declare.
Data and software availability
Raw and analyzed data are available on GEO, accession number GSE222910.
References
- 1.Immunology, A. A. o. A. A. a. Allergy Statistics, <https://www.aaaai.org/About/News/For-Media/Allergy-Statistics> (2021). [Google Scholar]
- 2.Angkasekwinai P & Dong C IL-9-producing T cells: potential players in allergy and cancer. Nat Rev Immunol 21, 37–48, doi: 10.1038/s41577-020-0396-0 (2021). [DOI] [PubMed] [Google Scholar]
- 3.Shimbara A et al. IL-9 and its receptor in allergic and nonallergic lung disease: increased expression in asthma. J Allergy Clin Immunol 105, 108–115, doi: 10.1016/s0091-6749(00)90185-4 (2000). [DOI] [PubMed] [Google Scholar]
- 4.Brough HA et al. IL-9 is a key component of memory TH cell peanut-specific responses from children with peanut allergy. J Allergy Clin Immunol 134, 1329–1338 e1310, doi: 10.1016/j.jaci.2014.06.032 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Abdelilah S et al. Functional expression of IL-9 receptor by human neutrophils from asthmatic donors: role in IL-8 release. J Immunol 166, 2768–2774, doi: 10.4049/jimmunol.166.4.2768 (2001). [DOI] [PubMed] [Google Scholar]
- 6.Liu J et al. IL-9 regulates allergen-specific Th1 responses in allergic contact dermatitis. J Invest Dermatol 134, 1903–1911, doi: 10.1038/jid.2014.61 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yao W et al. Interleukin-9 is required for allergic airway inflammation mediated by the cytokine TSLP. Immunity 38, 360–372, doi: 10.1016/j.immuni.2013.01.007 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wilhelm C et al. An IL-9 fate reporter demonstrates the induction of an innate IL-9 response in lung inflammation. Nat Immunol 12, 1071–1077, doi: 10.1038/ni.2133 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Licona-Limon P et al. Th9 Cells Drive Host Immunity against Gastrointestinal Worm Infection. Immunity 39, 744–757, doi: 10.1016/j.immuni.2013.07.020 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaplan MH, Hufford MM & Olson MR The development and in vivo function of T helper 9 cells. Nat Rev Immunol 15, 295–307, doi: 10.1038/nri3824 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gerlach K et al. TH9 cells that express the transcription factor PU.1 drive T cell-mediated colitis via IL-9 receptor signaling in intestinal epithelial cells. Nat Immunol 15, 676–686, doi: 10.1038/ni.2920 (2014). [DOI] [PubMed] [Google Scholar]
- 12.Xue G, Jin G, Fang J & Lu Y IL-4 together with IL-1beta induces antitumor Th9 cell differentiation in the absence of TGF-beta signaling. Nat Commun 10, 1376, doi: 10.1038/s41467-019-09401-9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nakatsukasa H et al. The DNA-binding inhibitor Id3 regulates IL-9 production in CD4(+) T cells. Nat Immunol 16, 1077–1084, doi: 10.1038/ni.3252 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schlapbach C et al. Human TH9 cells are skin-tropic and have autocrine and paracrine proinflammatory capacity. Sci Transl Med 6, 219ra218, doi: 10.1126/scitranslmed.3007828 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seumois G et al. Single-cell transcriptomic analysis of allergen-specific T cells in allergy and asthma. Sci Immunol 5, doi: 10.1126/sciimmunol.aba6087 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schwartz DM et al. Retinoic Acid Receptor Alpha Represses a Th9 Transcriptional and Epigenomic Program to Reduce Allergic Pathology. Immunity 50, 106–120 e110, doi: 10.1016/j.immuni.2018.12.014 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaplan MH The transcription factor network in Th9 cells. Semin Immunopathol 39, 11–20, doi: 10.1007/s00281-016-0600-2 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ulrich BJ et al. Allergic airway recall responses require IL-9 from resident memory CD4(+) T cells. Sci Immunol 7, eabg9296, doi: 10.1126/sciimmunol.abg9296 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee WH et al. BATF3 is sufficient for the induction of Il9 expression and can compensate for BATF during Th9 cell differentiation. Exp Mol Med 51, 1–12, doi: 10.1038/s12276-019-0348-6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gomez-Rodriguez J et al. Itk is required for Th9 differentiation via TCR-mediated induction of IL-2 and IRF4. Nat Commun 7, 10857, doi: 10.1038/ncomms10857 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Richard AC et al. The TNF-family ligand TL1A and its receptor DR3 promote T cell-mediated allergic immunopathology by enhancing differentiation and pathogenicity of IL-9-producing T cells. J Immunol 194, 3567–3582, doi: 10.4049/jimmunol.1401220 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xiao X et al. OX40 signaling favors the induction of T(H)9 cells and airway inflammation. Nat Immunol 13, 981–990, doi: 10.1038/ni.2390 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Micosse C et al. Human "TH9" cells are a subpopulation of PPAR-gamma(+) TH2 cells. Sci Immunol 4, doi: 10.1126/sciimmunol.aat5943 (2019). [DOI] [PubMed] [Google Scholar]
- 24.Liao W et al. Opposing actions of IL-2 and IL-21 on Th9 differentiation correlate with their differential regulation of BCL6 expression. Proc Natl Acad Sci U S A 111, 3508–3513, doi: 10.1073/pnas.1301138111 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fu Y et al. STAT5 promotes accessibility and is required for BATF-mediated plasticity at the Il9 locus. Nat Commun 11, 4882, doi: 10.1038/s41467-020-18648-6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harusato A et al. IL-36gamma signaling controls the induced regulatory T cell-Th9 cell balance via NFkappaB activation and STAT transcription factors. Mucosal Immunol 10, 1455–1467, doi: 10.1038/mi.2017.21 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dardalhon V et al. IL-4 inhibits TGF-beta-induced Foxp3+ T cells and, together with TGF-beta, generates IL-9+ IL-10+ Foxp3(−) effector T cells. Nat Immunol 9, 1347–1355, doi: 10.1038/ni.1677 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Olson MR et al. Paracrine IL-2 Is Required for Optimal Type 2 Effector Cytokine Production. J Immunol 198, 4352–4359, doi: 10.4049/jimmunol.1601792 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harnett W & Harnett MM Helminth-derived immunomodulators: can understanding the worm produce the pill? Nat Rev Immunol 10, 278–284, doi: 10.1038/nri2730 (2010). [DOI] [PubMed] [Google Scholar]
- 30.Guo L et al. IL-1 family members and STAT activators induce cytokine production by Th2, Th17, and Th1 cells. Proc Natl Acad Sci U S A 106, 13463–13468, doi: 10.1073/pnas.0906988106 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Canaria DA et al. IL-1beta promotes IL-9-producing Th cell differentiation in IL-2-limiting conditions through the inhibition of BCL6. Front Immunol 13, 1032618, doi: 10.3389/fimmu.2022.1032618 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luo Y et al. JAK-STAT signaling in human disease: From genetic syndromes to clinical inhibition. J Allergy Clin Immunol 148, 911–925, doi: 10.1016/j.jaci.2021.08.004 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Y et al. Germinal-center development of memory B cells driven by IL-9 from follicular helper T cells. Nat Immunol 18, 921–930, doi: 10.1038/ni.3788 (2017). [DOI] [PubMed] [Google Scholar]
- 34.Salerno F et al. Translational repression of pre-formed cytokine-encoding mRNA prevents chronic activation of memory T cells. Nat Immunol 19, 828–837, doi: 10.1038/s41590-018-0155-6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Y et al. Human TH9 differentiation is dependent on signal transducer and activator of transcription (STAT) 3 to restrain STAT1-mediated inhibition. J Allergy Clin Immunol 143, 1108–1118 e1104, doi: 10.1016/j.jaci.2018.06.036 (2019). [DOI] [PubMed] [Google Scholar]
- 36.Canaria DA et al. STAT5 Represses a STAT3-Independent Th17-like Program during Th9 Cell Differentiation. J Immunol 207, 1265–1274, doi: 10.4049/jimmunol.2100165 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Olson MR, Verdan FF, Hufford MM, Dent AL & Kaplan MH STAT3 Impairs STAT5 Activation in the Development of IL-9-Secreting T Cells. J Immunol 196, 3297–3304, doi: 10.4049/jimmunol.1501801 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shih HY et al. Developmental Acquisition of Regulomes Underlies Innate Lymphoid Cell Functionality. Cell 165, 1120–1133, doi: 10.1016/j.cell.2016.04.029 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Heinz S et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38, 576–589, doi: 10.1016/j.molcel.2010.05.004 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abdul Qayum A et al. The Il9 CNS-25 Regulatory Element Controls Mast Cell and Basophil IL-9 Production. J Immunol 203, 1111–1121, doi: 10.4049/jimmunol.1900272 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xiao X et al. Guidance of super-enhancers in regulation of IL-9 induction and airway inflammation. J Exp Med 215, 559–574, doi: 10.1084/jem.20170928 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Koh B et al. A conserved enhancer regulates Il9 expression in multiple lineages. Nat Commun 9, 4803, doi: 10.1038/s41467-018-07202-0 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guo L et al. Innate immunological function of TH2 cells in vivo. Nat Immunol 16, 1051–1059, doi: 10.1038/ni.3244 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pavel AB et al. Oral Janus kinase/SYK inhibition (ASN002) suppresses inflammation and improves epidermal barrier markers in patients with atopic dermatitis. J Allergy Clin Immunol 144, 1011–1024, doi: 10.1016/j.jaci.2019.07.013 (2019). [DOI] [PubMed] [Google Scholar]
- 45.Lu Y et al. Th9 Cells Represent a Unique Subset of CD4(+) T Cells Endowed with the Ability to Eradicate Advanced Tumors. Cancer Cell 33, 1048–1060 e1047, doi: 10.1016/j.ccell.2018.05.004 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chopp L, Redmond C, O'Shea JJ & Schwartz DM From thymus to tissues and tumors: A review of T-cell biology. J Allergy Clin Immunol, doi: 10.1016/j.jaci.2022.10.011 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Joyce JA & Fearon DT T cell exclusion, immune privilege, and the tumor microenvironment. Science 348, 74–80, doi: 10.1126/science.aaa6204 (2015). [DOI] [PubMed] [Google Scholar]
- 48.Elyaman W & Khoury SJ Th9 cells in the pathogenesis of EAE and multiple sclerosis. Semin Immunopathol 39, 79–87, doi: 10.1007/s00281-016-0604-y (2017). [DOI] [PubMed] [Google Scholar]
- 49.Caucheteux SM et al. IL-1beta enhances inflammatory TH2 differentiation. J Allergy Clin Immunol 138, 898–901 e894, doi: 10.1016/j.jaci.2016.02.033 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ansel KM, Djuretic I, Tanasa B & Rao A Regulation of Th2 differentiation and Il4 locus accessibility. Annu Rev Immunol 24, 607–656, doi: 10.1146/annurev.immunol.23.021704.115821 (2006). [DOI] [PubMed] [Google Scholar]
Methods-only References
- 51.Herndler-Brandstetter D et al. KLRG1(+) Effector CD8(+) T Cells Lose KLRG1, Differentiate into All Memory T Cell Lineages, and Convey Enhanced Protective Immunity. Immunity 48, 716–729 e718, doi: 10.1016/j.immuni.2018.03.015 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhu J et al. Conditional deletion of Gata3 shows its essential function in T(H)1-T(H)2 responses. Nat Immunol 5, 1157–1165, doi: 10.1038/ni1128 (2004). [DOI] [PubMed] [Google Scholar]
- 53.Gurram RK et al. Crosstalk between ILC2s and Th2 cells varies among mouse models. Cell Rep 42, 112073, doi: 10.1016/j.celrep.2023.112073 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu P, Jenkins NA & Copeland NG A highly efficient recombineering-based method for generating conditional knockout mutations. Genome Res 13, 476–484, doi: 10.1101/gr.749203 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Corces MR et al. An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat Methods 14, 959–962, doi: 10.1038/nmeth.4396 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Buenrostro JD, Wu B, Chang HY & Greenleaf WJ ATAC-seq: A Method for Assaying Chromatin Accessibility Genome-Wide. Curr Protoc Mol Biol 109, 21 29 21–21 29 29, doi: 10.1002/0471142727.mb2129s109 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Subramanian A et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102, 15545–15550, doi: 10.1073/pnas.0506580102 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mootha VK et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet 34, 267–273, doi: 10.1038/ng1180 (2003). [DOI] [PubMed] [Google Scholar]
- 59.Davis CA et al. The Encyclopedia of DNA elements (ENCODE): data portal update. Nucleic Acids Res 46, D794–D801, doi: 10.1093/nar/gkx1081 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
References
- 1.Seumois G et al. Single-cell transcriptomic analysis of allergen-specific T cells in allergy and asthma. Sci Immunol 5, doi: 10.1126/sciimmunol.aba6087 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Micosse C et al. Human "TH9" cells are a subpopulation of PPAR-gamma(+) TH2 cells. Sci Immunol 4, doi: 10.1126/sciimmunol.aat5943 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Schwartz DM et al. Retinoic Acid Receptor Alpha Represses a Th9 Transcriptional and Epigenomic Program to Reduce Allergic Pathology. Immunity 50, 106–120 e110, doi: 10.1016/j.immuni.2018.12.014 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]