Introduction:
Behavioral patterns of drug use have constituted a rising trend in overdose deaths in the United States (Ahmad et al., 2022). This effect is a result of societal and environmental factors and was exacerbated by the Covid-19 pandemic (Jones et al., 2022; Volkow, 2020). Drug overdose deaths are now the leading cause of accidental death (Ahmad et al., 2022; Hedegaard et al., 2021). Chronic exposure to drugs induces molecular changes in the brain that contribute to future drug seeking behavior (Nestler and Lüscher, 2019). Understanding the neurobiological changes that the brain undergoes in response to drugs is essential for the identification of pathways or targets that may be utilized for the development of therapies to reduce continued drug seeking events.
A significant proportion of the existing literature on molecular mechanisms of addiction has focused on drug-induced dysregulation of mRNA and protein species, including the involvement of transcription factors and epigenetic modifications to regulate downstream gene expression (Robison and Nestler, 2011). RNA sequencing, microarrays and proteomics profiling have identified mRNA and protein species that are regulated in discrete brain regions or blood samples following drug exposure in postmortem human samples as well as preclinical rodent models (Natividad et al., 2018; Zhou et al., 2014). However, mRNA and protein only account for a portion of the regulatory genetic machinery in the cell, and drug exposure induces regulation to other RNA species, including circular RNAs (circRNAs). circRNAs represent a unique class of RNAs with profound and diverse regulatory capabilities. circRNAs, unlike other non-coding RNAs, are generated from pre-mRNA through a back-splicing event which results in covalently bound 5’−3’ ends (Kristensen et al., 2019). This closed structure renders circRNAs resistant to many common RNA degradation processes, giving circRNAs more stability and a longer half-life than that of a typical mRNA (Enuka et al., 2016; Jeck and Sharpless, 2014; Jeck et al., 2013). As a single-stranded RNA sequence, circRNAs can bind microRNAs (miRNAs), RNA binding proteins and other complementary nucleic acid sequences (Li et al., 2018). Thus, their presence can disrupt the homeostatic environment within the cell and have wide-ranging effects on transcription, translation and protein location and function . Examples of circRNA functions including modulation of transcriptional events, interference with RNA splicing and protein polypeptide synthesis and miRNA sponging to impact mRNA translation (Li et al., 2018; Piwecka et al., 2017).
The cellular processes regulated via circRNAs can have a vast impact on neurological and cognitive function. For instance, the number of circRNAs detected in the mammalian brain increases with age and humans express many more circRNAs compared to rodents, suggesting that circRNAs may contribute to cognitive ability (Mahmoudi and Cairns, 2019; Rybak-Wolf et al., 2015). The abundance of some circRNAs exceeds that of its linear parent gene, indicating that a cell may preferentially shift its mRNA processing to favor a circRNA isoform. This suggests that circRNAs have a meaningful presence in the cell that can affect biological processes in a sequence-specific manner. Furthermore, circRNAs are most abundantly expressed in the brain, conserved from humans to rodents and even further enriched in neuronal synaptosomes, suggesting a functional role in synaptic transmission and plasticity (Rybak-Wolf et al., 2015; You et al., 2015). Recent studies have demonstrated that opioids, psychostimulants, and alcohol can regulate circRNA expression in the brain, indicating that our current understanding of drug exposure on RNA regulation is incomplete. In addition to gene expression analyses following drug exposure, research is underway to define the functional contributions of individual circRNA species to drug seeking in preclinical models of drug exposure with promising results. Further studies are warranted, given that circRNAs can regulate drug seeking behavior (Shen et al., 2022; Yu et al., 2021) and cognition (Hafez et al., 2022; Zimmerman et al., 2020). While many more circRNAs have been detected in the human brain, ~20–25% are conserved from rodents to human at the structural or sequence level (Irie et al., 2019; Jeck et al., 2013; Rybak-Wolf et al., 2015). We hypothesize that the long-lasting nature of circRNAs, combined with their complex cellular functions, may impart additional modulation of continued, perseverative drug seeking behavior that endures over time in individuals with substance use disorder (SUD). In this review, we will discuss this hypothesis by first describing the detection and function of circRNAs as they relate to neuronal processes that can impact addictive behaviors. We will review the individual circRNAs that have been studied in preclinical addiction models and post-mortem tissues from subjects that used addictive substances. Lastly, we will provide insight into how researchers may adapt to study circRNAs and the limitations that are associated with the current technologies available to study circRNAs.
Formation of circRNAs:
CircRNAs are derived from pre-mRNA and are generated via back-splicing events. During circRNA biogenesis, the 3’ end of an exon attacks the 5’ end of an upstream exon, resulting in covalently bound 5’−3’ tails and a circular structure (Figure 1a) (Jeck and Sharpless, 2014; Kristensen et al., 2019; Ragan et al., 2019). CircRNA biogenesis exhibits much heterogeneity, and the composition of individual circRNAs varies with multiple isoforms of circRNAs for a given gene. circRNAs can be classified based on sequence composition, with a majority of circRNAs derived purely from exons. However, circRNAs can also be derived from intronic, mixed- exonic and intronic- and intergenic RNA sequences (Li et al., 2018). Although circRNAs were first identified in 1979 (Hsu and Coca-Prados, 1979) and hundreds of thousands of circRNAs have been detected in the mammalian brain (Glažar et al., 2014), only a handful of splicing enzymes have been linked to circRNA biogenesis. Regulators of circRNA formation include the circRNA biogenesis enzymes Adenosine Deaminase RNA Specific 1 (Adar1), FUS RNA Binding Protein (Fus), and quaking (Qki) (Conn et al., 2015; Errichelli et al., 2017; Rybak-Wolf et al., 2015; Shi et al., 2017). However, only one RNA-binding protein has been described to regulate circRNA biogenesis in the brain in a model of drug exposure: ELAV like RNA binding protein 4 (Elavl4), also referred to as HuD (Oliver et al., 2018). Elavl4/HuD is significantly elevated in the nucleus accumbens (NAc) in response to cocaine conditioned place preference (CPP) and overexpression of Elavl4/HuD further enhances CPP behavior (Oliver et al., 2018). Nearly one-third of circRNAs expressed in the mouse brain are estimated to contain a consensus motif that would allow for binding to Elavl4/HuD and manipulation of Elavl4/HuD expression induces drastic regulation of striatal circRNAs (Dell’Orco et al., 2021; Dell’Orco et al., 2020). By simultaneously measuring circRNA, miRNA and mRNA, Dell’Orco et al determined that linear mRNA counterparts of many circRNAs regulated by Elavl4/HuD manipulation are also regulated (Dell’Orco et al., 2020). Additionally, examining the list of miRNAs regulated by Elavl4/HuD indicated that the expression levels of many putative miRNA target mRNAs were regulated in the opposite direction compared to their predicted miRNA regulator (Dell’Orco et al., 2020). Because miRNAs bind to their target mRNA and function to prevent mRNA translation to protein, it would be expected that putative miRNA targets exhibit regulation in the opposite direction of a miRNA (Friedman et al., 2009), thus indicating that regulation of circRNAs is associated with disruption of miRNA signaling and downstream miRNA-mediated target gene expression. Furthermore, the Elavl4/HuD-regulated miRNA list included miRNAs that have previously been demonstrated to functionally regulate cocaine-seeking behaviors, such as mir-132–3p/212–3p (Hollander et al., 2010; Im et al., 2010). These data indicate that circRNA biogenesis in the nervous system is associated with integrated regulation of miRNA and mRNA networks which may ultimately impact downstream cellular function and organism behavior.
Figure 1:
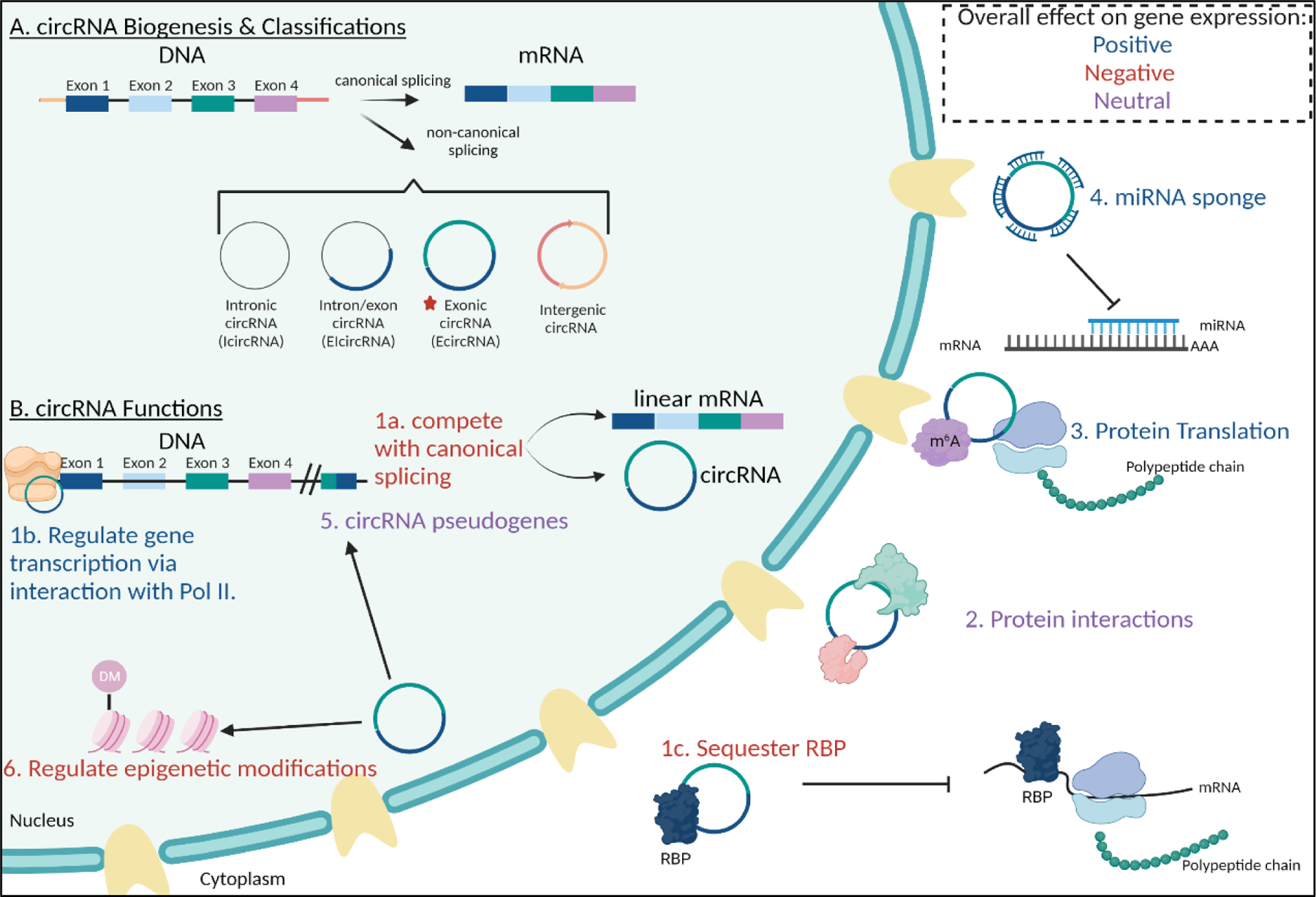
Biogenesis, classification, and regulatory functions of circRNAs.
(A) circRNAs can be classified into 4 subtypes: intronic circRNAs (icircRNA), exon-intron/sense overlapping circRNAs (ELcircRNA), exonic circRNAs (EcircRNA), and intergenic circRNAs with each group undergoing discrete biogenesis processes. (B) 6 circRNA functions identified to date. (1) Regulate gene expression through 3 discrete pathways (1a) compete with canonical splicing of linear counterpart (negative regulation); (1b) interact with polymerase II (Pol II) to promote transcription of parental gene (positive regulation); (1c) inhibit protein translation via sequestration of RBPs. (2) circRNAs have been shown to have dynamic protein interactions where they can function as transporters, scaffolding, decoys, and modulate protein activity. (3) Newer research has found some circRNAs can be translated into protein. (4) circRNAs may sponge miRNAs to allow translation of miRNA-repressed genes. (5) Through a process of reverse transcription and integration into host genomes, circRNAs can generate pseudogenes and thus physiological and pathological processes at the DNA, RNA, and protein levels; the mechanisms, and functions of circRNA derived pseudogenes is unknown. (6) circRNAs can also regulate epigenetic modifications via DNA and histone methylations.
RNA sequencing has been widely employed to measure mRNA expression levels in various brain regions after drug exposure. It is possible, depending on the method of library preparation, to extract information on circRNA expression from existing RNA-seq datasets (Chen et al., 2022). The caveat is that the RNA-seq library must have been prepared from total RNA using a ribosomal depletion strategy and not a poly-A enrichment strategy. It is highly likely that further analysis can be performed on existing datasets to yield useful information about the regulation of circRNAs in response to drugs. The two main technologies for unbiased profiling of circRNAs are microarrays and RNA-sequencing. Microarrays contain probes that hybridize with circRNA backsplice junctions and can give information about potential circRNA expression patterns. However, the backsplice probes do not indicate the size of the circRNA or its sequence content. Since circRNAs are formed as splice variants, they may include a variety of exons or introns and thus information about the sequences of the circRNA can only be obtained from RNA sequencing. Additionally, performing long-read RNA sequencing will allow the researcher to obtain more information about splice junctions that are part of circRNA backsplice junctions or indicative of other splicing events. Splice junction identification is essential to determine the composition of a circRNA, which can only be determined by sequencing and not by microarray.
circRNA functions in the nervous system:
The heterogeneity in circRNA composition indicates that circRNAs have a diverse range of functions attributed to their unique sequence arrangement even though multiple isoforms may be derived from the same gene. We have highlighted six proposed regulatory functions of circRNAs: (i) regulation of gene expression; (ii) interaction with proteins- circRNAs can function as transporters, scaffolding, or protein decoys and regulate protein activity; (iii) translation into protein; (iv) miRNA sponging; (v) creation of pseudogenes; and (vi) regulation of epigenetic processes including histone modifications and DNA methylation (Piwecka et al., 2017; Qu et al., 2017; You et al., 2015; Zhang et al., 2020). CircRNAs dually regulate gene expression through several discrete mechanisms (Figure 1b) including: (a) negative regulation of their parent gene by competing with canonical splicing; (b) promotion of gene transcription through interaction with DNA polymerase II (Pol II.); and (c) inhibition of gene translation by specific competitive binding of RNA binding proteins (RBP) (Qu et al., 2017; Zhang et al., 2020). The best characterized role of circRNAs is that of miRNA sponging (Hansen et al., 2013). miRNAs (miRNAs) are small ~18–24 nucleotide noncoding RNAs that inhibit mRNA translation to protein by binding ‘target’ mRNAs with sequence complementarity (Bartel, 2004). CircRNAs may retain miRNA binding sequences, thus serving to ‘sponge’ miRNAs by preventing them from interacting with their target mRNA and facilitating translation of miRNA-repressed genes (Sekar and Liang, 2019). However, computational analysis has determined that many circRNAs lack binding sites for miRNAs and miRNA sponging is likely not the main function of the majority of circRNAs (Guo et al., 2014). Alternatively, as circular isoforms of mRNAs, it seems clear that exonic circRNAs might retain their mRNA-parent gene’s capability to be translated into a protein product, and indeed several exonic circRNAs derived from synaptic encoding genes are capable of being translated into protein (Ragan et al., 2019). However, circRNAs lack the 5’ cap and 3’ poly-A tail requisite for mRNA translation. Instead, it appears circRNAs utilize cap independent mechanisms, primarily internal ribosome entry sites (IRES) elements and N6-methyladenosine (m6A) modified nucleotide sequences (Legnini et al., 2017; Pamudurti et al., 2017; Yang et al., 2017). To date, only a handful of circRNAs have been proven to code functional proteins, but other exonic circRNAs may also be capable of coding protein. Little is known on whether circ-proteins, proteins derived from circRNAs, function differently than proteins translated from mRNA. We suggest that it is unlikely that many circRNAs encode proteins and if they do, the proteins are expressed at very low levels because decades of mass-spectrometry data have failed to detect evidence of circ-proteins. Taken together, these findings demonstrate the profound regulatory capabilities of circRNAs across a plethora of biological processes. By impacting gene expression output in a variety of ways, drug-induced circRNAs are thus well-suited to impart regulatory modulation of cellular function.
One way in which drug-induced circRNAs may impact cellular neuroadaptations within the brain is by modulation of synaptic plasticity. A disproportionate percentage of circRNAs are derived from synaptic genes and published studies have implicated neuronally expressed circRNAs in regulation of synaptic plasticity (Piwecka et al., 2017; Ragan et al., 2019; Xu et al., 2020; You et al., 2015; Zhang et al., 2022). CircRNAs are dynamically expressed both spatially and temporally, with increased expression in neuronal regions participating in synaptic transmission and upregulation during periods of neural development and synaptogenesis (Rybak-Wolf et al., 2015; You et al., 2015). Similarly, artificial chemical induction of homeostatic plasticity induces a robust increase in circRNAs, many derived from well-characterized plasticity genes, locally at the neuronal synapse (You et al., 2015). Genetic deletion of a circRNA derived from a long-noncoding RNA was sufficient to alter neuronal function in brain cells (Piwecka et al., 2017). Therefore, by participating in regulation of synaptic function circRNAs are poised to contribute to drug-induced neuroplasticity and in turn, behavioral output that results from synaptic transmission.
Additionally, newer studies into pathologically relevant circRNAs supports discrete roles of unique circRNA species on synaptic plasticity and glutamate homeostasis. For instance, circGria1, a circular isoform derived from Gria1, which encodes the GluR1 subunit of the AMPA receptor, was found to negatively regulate hippocampal synaptogenesis and GluR1 activity-dependent synaptic plasticity in male macaques (Xu et al., 2020). Glutamate homeostasis is adversely impacted by drug exposure and pharmacological manipulation of glutamate signaling regulates drug seeking phenotypes (D’Souza, 2015; Kalivas, 2009). Therefore drug-induced circRNA mechanisms that perturb glutamate signaling may also contribute to drug-induced neuropathologies.
Although limited information is available on circRNA function in drug seeking, insight into the flexibility of circRNA modulation of behavior can be gained from studies on other psychiatric conditions, including circRNA regulation of cognition and learning. For example, bidirectional manipulation of circHomer1a, a circRNA dysregulated in both human neuropsychiatric diseases (Cervera-Carles et al., 2020; Urdánoz-Casado et al., 2021) and preclinical rodent models, in the mouse orbitofrontal cortex regulates reversal learning behavior (Hafez et al., 2022; Zimmerman et al., 2020). Additionally, the brain’s repertoire of circRNAs increases from birth to adulthood (Gruner et al., 2016; Xu et al., 2020), and extensive expression profiling of circRNAs in both clinical samples from Alzheimer’s patients (Dube et al., 2019) and preclinical models of Alzheimer’s-like symptoms (Huang et al., 2018) have begun to identify candidate circRNAs (Lu et al., 2019) that may participate in cognitive decline or memory (Zajaczkowski and Bredy, 2021). A circulating peripheral blood circRNA, hsa_circ_0003391, was found to be associated with Alzheimer’s disease, suggesting a potential role of circRNAs as biomarkers for neurological disorders (Liu et al., 2020).
Because drugs impact striatal dopamine signaling (Nestler, 2005), additional insight into the putative function of drug-induced circRNAs can also be gained by examining circRNA regulation in Parkinson’s disease (PD), a condition in which striatal dopamine is lost. In a study of post-mortem substantia nigra (SN) tissue, researchers found that circRNAs accumulated in an age-dependent manner in control subjects, but not in individuals with PD, and PD patients had reduced total circRNA expression (Hanan et al., 2020). A clinical study of circulating blood circRNAs from PD patients implicated 4 circRNAs in predicting early-stage versus late-stage PD (Zhong et al., 2021). These studies demonstrate that circRNAs may have utility as biomarkers and are associated with a severe phenotype of loss of dopaminergic signaling. Lastly, changes in circRNA biogenesis are sufficient to alter animal behavior and cognition. In vivo rodent brain manipulation of circRNAs can rescue depression-like behavior in a model of chronic unpredictable stress (Zhang et al., 2018) as well as cognitive behavior and infarct volume in a cerebral focal ischemia model (Wu et al., 2019).
These studies highlight other potential avenues by which drug-induced circRNAs may contribute to drug seeking behavior through modulation of learning, stress responses or alteration of striatal signaling pathways. Drug self-administration in both preclinical models and human subjects is a result of extremely robust learned associates between drug-associated cues and drug administration (Kutlu and Gould, 2016; Lyons et al., 2013). Such drug-associated cues are potent initiators of relapse or reinstatement to drug seeking following periods of forced abstinence or extinction (Bossert et al., 2013). Likewise, stress and anxiety can promote a return to drug seeking behavior and are risk factors for vulnerability to relapse (Mantsch et al., 2016). CircRNA patterns associated with stress or learning phenotypes in other models may therefore by biologically relevant to drug seeking behaviors and should be thoroughly investigated.
Alcohol-induced circRNA regulation
Alcohol regulation of circRNAs has been examined in both human and preclinical samples, in a sex specific manner, in developing brain samples, as well as in circulating exosomes in blood in ethanol exposed tissues. In preclinical studies, circRNAs have been profiled in developing and adult animals after alcohol exposure. Paudel et al sought to identify circRNAs regulated by alcohol exposure in utero (Paudel et al., 2020). After exposing developing mice to 10% ethanol during the prenatal period, circRNA expression was profiled in whole brain samples using a microarray and identified sex-specific regulation of circRNAs (Paudel et al., 2020). No differentially regulated circRNAs were overlapping in male and female brains exposed to alcohol in utero, indicating that drugs may regulate brain circRNA expression in a sex-specific manner (Paudel et al., 2020). The lack of overlap of alcohol-induced circRNAs mirrors what has been described in mRNA studies for other drugs such as opioids: male and female animals display similar behavioral phenotypes but differ in their molecular response to drugs (Mayberry et al., 2022; Townsend et al., 2021). Because limited studies have investigated sex-specific regulation of genes in addiction models, it is possible that the reported sex-specific effects may be due to variability that cannot be quantified unless further studies are performed using multiple strains of rodents. Nevertheless, such findings emphasize the importance of performing studies with both male and female animals, as sex-specific circRNA mechanisms may underlie drug seeking behavior.
While the rodent microarray technology can measure expression of ~10,000–15,000 circRNAs (Paudel et al., 2020; You et al., 2015), high throughput sequencing detected expression of more than 50,000 circRNAs in the adult rodent brain following chronic ethanol exposure (Gong et al., 2022). CircRNA expression was compared in whole brain tissue from adult male mice that underwent a combination of chronic intermittent ethanol vapor exposure followed by voluntary ethanol intake using RNA sequencing technology (Gong et al., 2022). Nearly 400 circRNAs were differentially regulated in the brain following the ethanol exposure paradigm, with enrichment of circRNAs derived from genes involved in inhibitory synapses, cannabinoid signaling and signaling downstream of morphine exposure (Gong et al., 2022). A select subset of differentially regulated circRNAs was validated using quantitative PCR in both brain tissue as well as peripheral blood samples from ethanol-exposed animals (Gong et al., 2022). This latter finding suggests that the blood circRNA profile may be reflective of the brain circRNA profile, and that circRNAs from the brain may be released via extracellular vesicles (Lasda and Parker, 2016) or exosomes from the brain (Wang et al., 2022) into the circulating bloodstream.
Using human samples, circRNAs have been profiled in circulating exosomes for biomarker detection as well as in postmortem brain tissue. Liu et al profiled exosomal-derived circRNAs from serum samples of patients with Alcohol Use Disorder using RNA-sequencing technology and identified several hundred circRNAs that are associated with alcohol use (Liu et al., 2021). Correlation analysis determined that hsa_circ_0004771, which is derived from the linear mRNA for Nrip1, was associated with severity of alcohol use (Liu et al., 2021). Nrip1 is a nuclear receptor, sensitive to stress and gonadal hormone regulation, which has also been reported as a cocaine-induced gene in the NAc (Vallender et al., 2017). One possible mechanism of drug-induced circRNA biogenesis is through differential regulation of linear mRNA abundance first. However, further insight is required to determine if alcohol induces regulation of the linear mRNA that gives rise to the differentially regulated circRNAs described in the aforementioned studies. One strategy to address this issue is to perform parallel profiling of mRNA and circRNA simultaneously. Vornholt et al performed microarrays on NAc tissue from patients that were previously diagnosed with Alcohol Use Disorder to identify differentially expressed circRNAs as well as mRNAs and miRNAs in the postmortem samples (Vornholt et al., 2021). This innovative approach allows for the comparison of circRNA and mRNA expression simultaneously to determine if alcohol-induced circRNAs were regulated independent of their linear mRNA. Such a finding would suggest that alcohol, and other drugs, may induce differential activation or expression of splicing factors that then give rise to circRNAs; or of proteins that degrade circRNAs without impacting linear mRNA levels. Additionally, the study built putative circRNA-miRNA-mRNA networks associated with Alcohol Use Disorder to provide insight into how the sequence of a circRNA may sponge a miRNA to impact gene expression (Vornholt et al., 2021). One of the most significant interactions identified in subjects with Alcohol Use Disorder was a network that involves hsa_circ_406702 with miR-1200 and genes with well-characterized roles in synaptic plasticity and neurotransmission, such as HOMER1 (Vornholt et al., 2021). By performing RNA profiling of all three types of RNA simultaneously as well as extensive correlational analysis to identify patterns of genes that are co-regulated together, the study provides a framework for investigating the downstream impact of drug-induced circRNA regulation on gene expression and sequence-specific sponging of miRNA by circRNA. Furthermore, such analysis demonstrates the far-reaching consequences of drug-regulated circRNA dysregulation, as miRNA sponging may impact many of the 100’s-1000’s of targets for an individual miRNA (Friedman et al., 2009).
Opioid-induced circRNA regulation
Opioid-induced regulation of circRNA expression in the nervous system has been examined in preclinical models of opioid exposure in both whole brain tissue, as well as NAc and orbitofrontal cortex. Splice variants arising from the mu-opioid receptor (Oprm1) are diverse and indeed, circRNAs derived from Oprm1 are differentially regulated in whole brain tissue from mice that underwent chronic morphine exposure (Irie et al., 2019). This suggests that opioid interaction with the mu-opioid receptor may also induce post-transcriptional regulation of the receptor itself, and thus may impact mu-opioid receptor-mediated signaling cascades in the brain, as has been described in the heroin-exposed brain (Sillivan et al., 2013). Conservation of circRNAs derived from Oprm1 was also demonstrated by the detection of circOprm1 in brain and spinal cord tissue from humans, rats and mice (Irie et al., 2019).
In a rat model of heroin self-administration, chronic heroin exposure altered the profile of circRNAs in the orbitofrontal cortex (Floris et al., 2022), a brain region that modulates long-lasting heroin seeking behavior (Fanous et al., 2012). Among the circRNAs responsive to heroin was a circRNA derived from Grin2b (rno_circRNA_011731), the gene that encodes an NR2B subunit of the glutamate receptor, which was accompanied by an upregulation of linear Grin2b. This finding suggests that heroin either elevates levels of Grin2b to result in an increase in biogenesis of circGrin2b, or that heroin-induced upregulation of circGrin2b regulates the transcription of linear Grin2b, as some circRNAs are reported to regulate transcription of their parent gene. These findings are in accordance with previous studies that have demonstrated the utility of NR2B blockade for preventing reinstatement of heroin seeking and reversing heroin-induced synaptic remodeling (Shen et al., 2011). Thus, changes in the abundance of NR2B that are produced from circGrin2b biogenesis following heroin self-administration may impact aspects of drug seeking. Floris et al demonstrated that heroin-associated circRNAs regulated in the orbitofrontal cortex may be regulated independent of their linear counterparts, the majority are exonic, and their regulation can be specific to drug reward or general to appetitive reward (Floris et al., 2022).
Using a mouse model of morphine CPP, Yu et al identified 16 circRNAs in the NAc core that are associated with a phenotype of increased morphine seeking behavior following forced abstinence (Yu et al., 2021). Among these, circTmeff1, which is derived from the gene transmembrane protein with EGF like and two follistatin like domains 1 (Tmeff1), was the most significantly increased circRNA and manipulation of NAc core circTmeff1 expression bidirectionally modulated morphine CPP following 14 days of forced abstinence but not 1 day (Yu et al., 2021). These effects may be mediated by circTmeff1 sponging of mir-541–5p or mir-6934–3p and indicate that circTmeff1 may be involved in the functional modulation of long-lasting opioid seeking phenotypes (Yu et al., 2021). Additional insight into the pattern of circRNAs regulated by opioids may also be gained from studies of morphine-induced regulation of circRNAs in the spinal cord in preclinical models of morphine analgesia and tolerance (Bai et al., 2023; Weng et al., 2019; Xing et al., 2022).
Psychostimulant-induced circRNA regulation
Exposure to the psychostimulants methamphetamine or cocaine can regulate brain expression patterns of circRNAs as well as in vitro expression of circRNAs. In an experimenter-administered methamphetamine exposure study, circRNA expression was evaluated with RNA-sequencing in the cerebellum, a site of neuronal degeneration following methamphetamine (Boroujeni et al., 2020). While many cortical circRNAs are reported to contain only exons (You et al., 2015), the cerebellar profile of circRNAs contained mostly intronic circRNAs (~43%) that were over 100,000 base pairs in length (Boroujeni et al., 2020). Such large intronic circRNAs are believed to localize to the nucleus where they may interfere with transcription . Given the presence of degenerative markers also observed in cerebellar neurons after methamphetamine exposure, it is possible that methamphetamine-regulated intronic circRNAs may program cell death in cerebellar neurons. A separate study examined the impact of cocaine self-administration on circRNA expression in the dorsal striatum (Bu et al., 2019), a brain region critical for the formation of continued drug seeking behavior (Volkow et al., 2019), using microarray technology. Three qPCR validated circRNAs, mmu_circRNA_002381, mmu_circRNA_002520 and mmu_circRNA_003834, were regulated in the dorsal striatum by chronic cocaine self-administration as well as by experimenter administered injections of cocaine, indicating that these circRNAs respond robustly to cocaine (Bu et al., 2019). Using an siRNA knockdown approach, it was demonstrated that mmu_circRNA_002381 expression impacts both mRNA and protein levels of Limk1 and BDNF in N2A cultured cells, suggesting that mmu_circRNA_002381 may regulate expression of genes involved in neuroplasticity following cocaine exposure (Bu et al., 2019). A limitation of the aforementioned study is that the linear mRNA from which the drug-associated circRNAs are derived is not currently known and therefore, it is unclear whether these circRNAs are also regulated by other drug exposure paradigms.
Drug-induced circRNA regulation of Homer1 or circRNAs derived from Homer1 represent the most consistent finding in the literature to date. As detailed above, a circRNA from Homer1 can regulate cognition (Hafez et al., 2022; Zimmerman et al., 2020) and the Homer1 protein has a well-described role as a scaffolding protein that contributes to synaptic neurotransmission. Moreover, circHomer1 represents a translationally-relevant circRNA, as isoforms of circHomer1 are dysregulated in human disease (Cervera-Carles et al., 2020; Urdánoz-Casado et al., 2021), including alcohol use disorder (Vornholt et al., 2021), as we have described. However, primary cortical cells treated with methamphetamine also display upregulated circHomer1 (mmu_circ_0000491) very rapidly within 12 hours (Li et al., 2019; Li et al., 2020). No dopamine would presumably be released following methamphetamine exposure, due to the fact that the cultures no not contain dopaminergic synapses, suggesting that methamphetamine may regulate circHomer1 (mmu_circ_0000491) through a dopamine-independent pathway. Knockdown of circHomer1 (mmu_circ_0000491) in primary cultures reduced methamphetamine-induced cell damage, suggesting that drug-induced upregulation of circHomer1 may also contribute to drug toxicity. In another study of methamphetamine CPP, circHomer1 (mmu_circ_0000491) was significantly elevated in multiple brain regions, including the hippocampus, prefrontal cortex, ventral tegmental area, and NAc, suggesting that drug-induced biogenesis of this circRNA is widespread (Li et al., 2019). circHomer1 (mmu_circ_0000491) expression in the hippocampus and ventral tegmental area positively correlated with CPP scores (Li et al., 2020). This indicates that an animal’s individual response to, and perhaps motivation for, methamphetamine is related to production of this circRNA. Given Homer1’s role in synaptic plasticity and addiction (Ghasemzadeh et al., 2009; Gould et al., 2015; Okvist et al., 2011), regulation of the abundance of Homer1 in the cell may help to explain how circRNAs may regulate drug seeking behavior.
circTmeff1, which was identified as a circRNA associated with incubation of craving of morphine CPP, has also been evaluated functionally in a rodent cocaine CPP paradigm (Shen et al., 2022). In animals that previously underwent cocaine CPP, a single cocaine injection rapidly elevates levels of circTmeff1 in the NAc core within 30 minutes (Shen et al., 2022), suggesting that this circRNA may be responsive to dopamine signaling cascades that are produced by exposure to drugs. Elevated circTmeff1 is maintained in the core for at least 6 hours following cocaine. Moreover, cocaine-induced circTmeff1 expression correlates with a miRNA network of downregulated mir-206–3p and elevation of the mir-206–3p target brain-derived neurotrophic factor (BDNF) (Shen et al., 2022), a protein well studied for its role in learning and memory that supports incubation of opioid craving behavior (Theberge et al., 2012). Knockdown of circTmeff1 in the NAc core significantly impairs reconsolidation of cocaine CPP behavior, an effect blocked with a mir-206–3p antagomir (Shen et al., 2022). These findings suggest that circTmeff1 sponges mir-206–3p to disinhibit the translation of mir-206–3p genes, such as BDNF, allowing this pathway to support drug-associated memories.
Extensive RNA sequencing has been performed on tissue sets from postmortem brain of individuals with cocaine use disorder as well as preclinical models of cocaine or methamphetamine exposure and many of these datasets are publicly available on repositories such as Gene Expression Omnibus. Information on backsplice junction reads from these datasets may provide insight into putative psychostimulant-induced circRNA expression in the brain but requires additional analysis. Chen et al performed such analysis on an RNA-sequencing dataset of dorsal lateral prefrontal cortex tissue from individuals with cocaine use disorder (Ribeiro et al., 2017) and reported differential expression of 41 circRNAs (Chen et al., 2022). Among the regulated circRNAs was a 172bp circGrin2b (chr12:13708789|13708961), a circRNA derived from the gene that encodes the NR2B subunit (Chen et al., 2022). A separate circGrin2b was also regulated by heroin in a rat model (Floris et al., 2022), as described above, indicating that circRNA biogenesis from the Grin2b gene may be general to multiple types of drugs. Chen et al identified miRNA response element sequences and RNA binding protein sequences for cocaine-associated circRNAs, demonstrating that there are extensive sites through which circRNAs may interact with other RNA and proteins (Chen et al., 2022). Further studies with RNA- or protein-immuneprecipitation are required to understand the RNA-binding proteins that interact with circRNAs or the miRNAs that may be sponged by circRNAs.
Studying circRNAs in addiction
Currently RNA sequencing in preclinical models is limited by the fact that many companies require a large input of total RNA (e.g.- >5ug) that is not compatible with individual brain areas from the mouse brain. Working with rat models may provide more utility for circRNA researchers as the rat brain is significantly larger in size compared to the mouse brain and models of self-administration are very well-characterized for rats. Following detection of dysregulated circRNAs associated with drug exposure, quantitative PCR (qPCR) should be performed, using primers that span the circRNA backsplice junction , to confirm either RNA-sequencing or microarray findings (Nielsen et al., 2022). Both RNA sequencing and microarray detection of circRNAs are semi-quantitative and therefore, validation of these methodologies with high-quality qPCR is essential.
Because circRNAs are cargo that can be packaged into exosomes (Li et al., 2015), circRNAs may provide insight into drug seeking phenotypes by functioning as biomarkers. Neuronal exosomes can be released from the brain and detected in peripheral blood samples. By analyzing the content of neuronal exosomes, it is possible to obtain a window into the regulation of neuronal circRNAs. In preclinical models of drug exposure and self-administration, blood sampling measurements that correspond to drug seeking events can be collected from animals to correlate behavioral data to circRNA expression levels. Such measurements may reveal insight into brain circRNA expression levels that are indicative of enhanced drug seeking or provide insight into molecular mechanisms through which therapies may reduce drug seeking behavior. An analogous approach may also be applied to blood samples from human subjects that are in recovery from addiction to identify circRNA biomarker profiles that correspond to enhanced drug craving, relapse, or treatment responsiveness.
A major barrier to performing translationally relevant circRNA research on the neurobiology of addiction lies in the fact that no central database of circRNAs currently exists. This limitation makes it challenging to determine when a species-specific analog of a circRNA exists. A website that functions similar to miRBase, which tracks known miRNAs and allows a user to determine conservation status of a given miRNA, would be extremely useful for circRNA researchers. More annotated databases of circRNAs can be found for human and mouse circRNAs (Dori and Bicciato, 2019), such as circBase (http://circrna.org). circBank (http://www.circbank.cn) and circRNADb (http://reprod.njmu.edu.cn/circrnadb) are exclusive to human circRNAs. circRNADb also contains information about protein-coding circRNAs and such a resource would be extremely valuable to basic scientists seeking to understand the biological mechanisms of circRNAs in preclinical models, yet this information is not currently available for mouse, rat, worm, or fly circRNAs. In addition, web-based tools to investigate putative circRNA interactions, comparable to TargetScan or mirDB for miRNAs, are a much-needed resource for the circRNA field. Because circRNAs may interact with miRNA, mRNA or protein, sequence-based analysis of RNA-binding or protein-binding domains of a circRNA would allow researchers to identify potential interactions for a given circRNA to begin to delineate the cellular mechanisms of individual circRNAs. Like miRNAs, circRNAs have been more extensively studied in cancer biology and a well-curated database called CircNet exists to allow researchers to mine circRNA data associated with a particular cancer (https://awi.cuhk.edu.cn/~CircNet). CircRNA studies in the nervous system can benefit from databases such as CircNet by using its framework to identify circRNAs associated with neuropsychiatric conditions. At present, the existing circRNA databases use 3 separate strategies for circRNA names. Until there is a universally accepted circRNA nomenclature, comparing drug-associated circRNA results across species may still be challenging. To circumvent these challenges, researchers should consider employing well-characterized technologies that are readily available to study the consequences of a circRNA or its potential binding partners. Examples include RNA sequencing or mass-spectrometry to identify changes in RNA or protein expression following a circRNA knockdown or overexpression; as well as RNA-immuneprecipitation assays followed by mass-spectrometry to identify RNA-binding proteins that associate with a circRNA of interest.
To date, no study has examined circRNA expression in models of nicotine or cannabis exposure. Given the regulation of circRNAs in the brain after exposure to psychostimulants, opioids, or alcohol, it is likely that circRNAs are also regulated by nicotine or cannabis exposure. Future studies should examine circRNA contribution to drug seeking at various stages, including self-administration, extinction, reinstatement, and relapse. The limited studies that have examined the functional relevance of circRNAs to drug seeking demonstrate that this species of RNA may possess profound regulatory control over synaptic plasticity associated with drug exposure as well modulation of drug seeking behavior. Therefore, exploration into the role of drug-associated circRNAs and their vast modulatory networks represents a critical neurobiological process to study to further understand the neurobiology of addiction.
Table 1:
Published studies reporting drug-induced circular RNA regulation.
| First author | Year | PMID | Drug | Brain region |
|---|---|---|---|---|
| Paudel | 2020 | 33304235 | Alcohol | Whole brain |
| Liu | 2021 | 33821559 | Alcohol | Serum |
| Vornholt | 2021 | 34164896 | Alcohol | Nac |
| Gong | 2022 | 35838410 | Alcohol | Whole brain |
| Li | 2019 | 30797870 | Meth | NAc, PFC, VTA, HPC |
| Boroujeni | 2020 | 32576439 | Meth | Cerebellum |
| Li | 2020 | 32450188 | Meth | NAc, PFC, VTA, HPC |
| Bu | 2019 | 31434869 | Cocaine | Striatum |
| Yun Chen | 2019 | 35493321 | Cocaine | dorsolateral PFC |
| Shen | 2022 | 35489671 | Cocaine | NAc |
| Weng | 2019 | 31533844 | Morphine | Spinal cord |
| Irie | 2019 | 31243060 | Morphine | Whole brain, Spinal cord |
| Hailei yu | 2021 | 34116208 | Morphine | NAc |
| Floris & Gillespie | 2022 | 35163373 | Heroin | OFC |
| 2022 | 35992914 | Morphine | Spinal cord | |
| Bai | 2022 | 36202171 | Morphine | Spinal cord |
Table 2:
Potential mechanisms of drug-induced circRNAs.
| circRNA | Parent Gene | Species | Drug-Induced Expression | Drug/Model | Sample type | Citation | Potential Mechanism |
|---|---|---|---|---|---|---|---|
| circHomer1_(mmu_circ_0000491) | Homer | mouse | Upregulated | Methamph etamine exposure | Primary Cortical Neurons | Li et al.,2019 | Upregulation is associated with methamphet amine-induced toxicity and CPP score. Reduction may protect from methamphet amine toxicity |
| Methamph etamine CPP | Hippocampus, VTA, NAc, PFC | Li et al.,2020 | |||||
| mmu_circRNA_002381 | Unknown | mouse/N2A cultured cells | Upregulated | Cocaine self-administration | Whole Brain | Bu et al., 2019 | Positively regulates genes associated with drug-induced neuroplasticity and may sponge miR-138 |
| mmu_circRNA_002520 | Unknown | mouse | Downregulated | Cocaine self-administration | Whole Brain | Bu et al., 2019 | May function as a miRNA sponge: contains 33 putative binding sites for miR-138 and 29 for miR-212 |
| mmu_circRNA_003834 | Unknown | mouse | Downregulated | Cocaine self-administration | Whole Brain | Bu et al., 2019 | Contains 13 putative binding sites to sponge miR-138 |
| circTmeff-1 | Tmeff | mouse | Upregulated | Cocaine CPP | NAc Core | Shen et., 2022 | Induced during reconsolidation of cocaine-paired memory; may disrupt miR-206-mediated regulation of BDNF |
| Morphine CPP | NAc Core | Yu et al., 2021 | Induced by abstinence from morphine CPP and sponges miR-541-5p and miR-6934-3p to positively regulate Nfasc and Vamp1 | ||||
| circGrin2b chr12:13708789|13708961 | Grin 2b | Human | Upregulated | Cocaine | Postmortem Dorsal Lateral Prefrontal Cortex | Chen et al., 2022 | Induced by exposure to cocaine or heroin in both humans and rodents and may be generally responsive to drug reward; may positively regulate expression of its linear parent gene |
| circGrin2b_011731 | Grin 2b | Rat | Upregulated | Heroin | Orbitofrontal Cortex | Floris & Gillespie et al., 2022 | |
| hsa_circ_0004771 | Nrip1 | Human | Upregulated | Alcohol | Serum Exosome | Liu et al., 2021 | Positively associated with severity of Alcohol Use Disorder |
| hsa_circ_406702 | Unknown | Human | N/A | Alcohol | Postmortem NAc | Vornholt et al., | Interaction network significantly associated with miR-1200; may regulate miR-1200 and HRAS, PRKCB, HOMER1 and PCLO either directly or indirectly |
| circOprm1 | Oprm1 | Mouse | Upregulated | Morphine | Whole Brain | Irie et al., 2019 | Morphine may facilitate Oprm1 circR NA formation; Oprm1 circRNAs may sponge miRNAs |
Highlights:
Overview of circRNA functions that may contribute to drug-associated neuroadaptations
Review of existing studies that have identified drug-associated circRNAs and their functional contribution to drug-seeking behaviors
Identification of strategies that may be implemented to further identify drug-associated circRNAs and critical challenges for studying circRNAs in addiction
Funding:
This work was supported by NIDA/NIH grants T32DA007237 (AG) and DP1DA051550 (SS).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of Interest: The authors declare that we have no conflicts of interest.
References
- Ahmad FB, Cisewski JA, Rossen LM, Sutton P 2022. Provisional drug overdose death counts. National Center for Health Statistics. [Google Scholar]
- Bai X, Huang Y, Zhang K, Huang W, Mu Y, Li Y, Ouyang H, 2023. CircNf1-mediated CXCL12 expression in the spinal cord contributes to morphine analgesic tolerance. Brain Behav Immun 107, 140–151. [DOI] [PubMed] [Google Scholar]
- Bartel DP, 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297. [DOI] [PubMed] [Google Scholar]
- Boroujeni ME, Nasrollahi A, Boroujeni PB, Fadaeifathabadi F, Farhadieh M, Tehrani AM, Nakhaei H, Sajedian AM, Peirouvi T, Aliaghaei A, 2020. Exposure to methamphetamine exacerbates motor activities and alters circular RNA profile of cerebellum. J Pharmacol Sci 144, 1–8. [DOI] [PubMed] [Google Scholar]
- Bossert JM, Marchant NJ, Calu DJ, Shaham Y, 2013. The reinstatement model of drug relapse: recent neurobiological findings, emerging research topics, and translational research. Psychopharmacology (Berl) 229, 453–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu Q, Long H, Shao X, Gu H, Kong J, Luo L, Liu B, Guo W, Wang H, Tian J, Zhao Y, Cen X, 2019. Cocaine induces differential circular RNA expression in striatum. Transl Psychiatry 9, 199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervera-Carles L, Dols-Icardo O, Molina-Porcel L, Alcolea D, Cervantes-Gonzalez A, Muñoz-Llahuna L, Clarimon J, 2020. Assessing circular RNAs in Alzheimer’s disease and frontotemporal lobar degeneration. Neurobiol Aging 92, 7–11. [DOI] [PubMed] [Google Scholar]
- Chen Y, Li X, Meng S, Huang S, Chang S, Shi J, 2022. Identification of Functional CircRNA-miRNA-mRNA Regulatory Network in Dorsolateral Prefrontal Cortex Neurons of Patients With Cocaine Use Disorder. Front Mol Neurosci 15, 839233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conn SJ, Pillman KA, Toubia J, Conn VM, Salmanidis M, Phillips CA, Roslan S, Schreiber AW, Gregory PA, Goodall GJ, 2015. The RNA binding protein quaking regulates formation of circRNAs. Cell 160, 1125–1134. [DOI] [PubMed] [Google Scholar]
- D’Souza MS, 2015. Glutamatergic transmission in drug reward: implications for drug addiction. Front Neurosci 9, 404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dell’Orco M, Elyaderani A, Vannan A, Sekar S, Powell G, Liang WS, Neisewander JL, Perrone-Bizzozero NI, 2021. HuD Regulates mRNA-circRNA-miRNA Networks in the Mouse Striatum Linked to Neuronal Development and Drug Addiction. Biology (Basel) 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dell’Orco M, Oliver RJ, Perrone-Bizzozero N, 2020. HuD Binds to and Regulates Circular RNAs Derived From Neuronal Development- and Synaptic Plasticity-Associated Genes. Front Genet 11, 790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dori M, Bicciato S, 2019. Integration of Bioinformatic Predictions and Experimental Data to Identify circRNA-miRNA Associations. Genes (Basel) 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dube U, Del-Aguila JL, Li Z, Budde JP, Jiang S, Hsu S, Ibanez L, Fernandez MV, Farias F, Norton J, Gentsch J, Wang F, Salloway S, Masters CL, Lee JH, Graff-Radford NR, Chhatwal JP, Bateman RJ, Morris JC, Karch CM, Harari O, Cruchaga C, (DIAN), D.I.A.N., 2019. An atlas of cortical circular RNA expression in Alzheimer disease brains demonstrates clinical and pathological associations. Nat Neurosci 22, 1903–1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enuka Y, Lauriola M, Feldman ME, Sas-Chen A, Ulitsky I, Yarden Y, 2016. Circular RNAs are long-lived and display only minimal early alterations in response to a growth factor. Nucleic Acids Res 44, 1370–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errichelli L, Dini Modigliani S, Laneve P, Colantoni A, Legnini I, Capauto D, Rosa A, De Santis R, Scarfò R, Peruzzi G, Lu L, Caffarelli E, Shneider NA, Morlando M, Bozzoni I, 2017. FUS affects circular RNA expression in murine embryonic stem cell-derived motor neurons. Nat Commun 8, 14741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanous S, Goldart EM, Theberge FR, Bossert JM, Shaham Y, Hope BT, 2012. Role of orbitofrontal cortex neuronal ensembles in the expression of incubation of heroin craving. J Neurosci 32, 11600–11609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floris G, Gillespie A, Zanda MT, Dabrowski KR, Sillivan SE, 2022. Heroin Regulates Orbitofrontal Circular RNAs. Int J Mol Sci 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman RC, Farh KK, Burge CB, Bartel DP, 2009. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 19, 92–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghasemzadeh MB, Windham LK, Lake RW, Acker CJ, Kalivas PW, 2009. Cocaine activates Homer1 immediate early gene transcription in the mesocorticolimbic circuit: differential regulation by dopamine and glutamate signaling. Synapse 63, 42–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glažar P, Papavasileiou P, Rajewsky N, 2014. circBase: a database for circular RNAs. RNA 20, 1666–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Z, Rong X, Li X, Wang H, Liu D, He L, Pan J, Shen Q, Peng Y, 2022. Male mice exposed to chronic intermittent ethanol exposure exhibit significant upregulation or downregulation of circular RNAs. Am J Drug Alcohol Abuse 48, 562–572. [DOI] [PubMed] [Google Scholar]
- Gould AT, Sacramento AD, Wroten MG, Miller BW, von Jonquieres G, Klugmann M, Ben-Shahar O, Szumlinski KK, 2015. Cocaine-elicited imbalances in ventromedial prefrontal cortex Homer1 versus Homer2 expression: implications for relapse. Addict Biol 20, 148–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruner H, Cortés-López M, Cooper DA, Bauer M, Miura P, 2016. CircRNA accumulation in the aging mouse brain. Sci Rep 6, 38907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JU, Agarwal V, Guo H, Bartel DP, 2014. Expanded identification and characterization of mammalian circular RNAs. Genome Biol 15, 409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafez AK, Zimmerman AJ, Papageorgiou G, Chandrasekaran J, Amoah SK, Lin R, Lozano E, Pierotti C, Dell’Orco M, Hartley BJ, Alural B, Lalonde J, Esposito JM, Berretta S, Squassina A, Chillotti C, Voloudakis G, Shao Z, Fullard JF, Brennand KJ, Turecki G, Roussos P, Perlis RH, Haggarty SJ, Perrone-Bizzozero N, Brigman JL, Mellios N, 2022. A bidirectional competitive interaction between circHomer1 and Homer1b within the orbitofrontal cortex regulates reversal learning. Cell Rep 38, 110282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanan M, Simchovitz A, Yayon N, Vaknine S, Cohen-Fultheim R, Karmon M, Madrer N, Rohrlich TM, Maman M, Bennett ER, Greenberg DS, Meshorer E, Levanon EY, Soreq H, Kadener S, 2020. A Parkinson’s disease CircRNAs Resource reveals a link between circSLC8A1 and oxidative stress. EMBO Mol Med 12, e11942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J, 2013. Natural RNA circles function as efficient microRNA sponges. Nature 495, 384–388. [DOI] [PubMed] [Google Scholar]
- Hedegaard H, Miniño AM, Warner M, 2021. Co-involvement of opioids in drug overdose deaths involving cocaine and psychostimulants. NCHS Data Brief 406, 1–8. [PubMed] [Google Scholar]
- Hollander JA, Im HI, Amelio AL, Kocerha J, Bali P, Lu Q, Willoughby D, Wahlestedt C, Conkright MD, Kenny PJ, 2010. Striatal microRNA controls cocaine intake through CREB signalling. Nature 466, 197–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu MT, Coca-Prados M, 1979. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature 280, 339–340. [DOI] [PubMed] [Google Scholar]
- Huang JL, Qin MC, Zhou Y, Xu ZH, Yang SM, Zhang F, Zhong J, Liang MK, Chen B, Zhang WY, Wu DP, Zhong ZG, 2018. Comprehensive analysis of differentially expressed profiles of Alzheimer’s disease associated circular RNAs in an Alzheimer’s disease mouse model. Aging (Albany NY) 10, 253–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im HI, Hollander JA, Bali P, Kenny PJ, 2010. MeCP2 controls BDNF expression and cocaine intake through homeostatic interactions with microRNA-212. Nat Neurosci 13, 1120–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irie T, Shum R, Deni I, Hunkele A, Le Rouzic V, Xu J, Wilson R, Fischer GW, Pasternak GW, Pan YX, 2019. Identification of Abundant and Evolutionarily Conserved Opioid Receptor Circular RNAs in the Nervous System Modulated by Morphine. Mol Pharmacol 96, 247–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeck WR, Sharpless NE, 2014. Detecting and characterizing circular RNAs. Nat Biotechnol 32, 453–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE, 2013. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 19, 141–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones CM, Compton WM, Han B, Baldwin G, Volkow ND, 2022. Methadone-Involved Overdose Deaths in the US Before and After Federal Policy Changes Expanding Take-Home Methadone Doses From Opioid Treatment Programs. JAMA Psychiatry 79, 932–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalivas PW, 2009. The glutamate homeostasis hypothesis of addiction. Nat Rev Neurosci 10, 561–572. [DOI] [PubMed] [Google Scholar]
- Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J, 2019. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet 20, 675–691. [DOI] [PubMed] [Google Scholar]
- Kutlu MG, Gould TJ, 2016. Effects of drugs of abuse on hippocampal plasticity and hippocampus-dependent learning and memory: contributions to development and maintenance of addiction. Learn Mem 23, 515–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasda E, Parker R, 2016. Circular RNAs Co-Precipitate with Extracellular Vesicles: A Possible Mechanism for circRNA Clearance. PLoS One 11, e0148407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legnini I, Di Timoteo G, Rossi F, Morlando M, Briganti F, Sthandier O, Fatica A, Santini T, Andronache A, Wade M, Laneve P, Rajewsky N, Bozzoni I, 2017. Circ-ZNF609 Is a Circular RNA that Can Be Translated and Functions in Myogenesis. Mol Cell 66, 22–37.e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Shi Q, Wang Q, Tan X, Pang K, Liu X, Zhu S, Xi K, Zhang J, Gao Q, Hu Y, Sun J, 2019. Profiling circular RNA in methamphetamine-treated primary cortical neurons identified novel circRNAs related to methamphetamine addiction. Neurosci Lett 701, 146–153. [DOI] [PubMed] [Google Scholar]
- Li J, Sun Q, Zhu S, Xi K, Shi Q, Pang K, Liu X, Li M, Zhang Y, Sun J, 2020. Knockdown of circHomer1 ameliorates METH-induced neuronal injury through inhibiting Bbc3 expression. Neurosci Lett 732, 135050. [DOI] [PubMed] [Google Scholar]
- Li X, Yang L, Chen LL, 2018. The Biogenesis, Functions, and Challenges of Circular RNAs. Mol Cell 71, 428–442. [DOI] [PubMed] [Google Scholar]
- Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J, Chen D, Gu J, He X, Huang S, 2015. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res 25, 981–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Chen X, Chen YH, Zhang K, 2020. Identification of Circular RNA hsa_Circ_0003391 in Peripheral Blood Is Potentially Associated With Alzheimer’s Disease. Front Aging Neurosci 12, 601965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Li J, Bu H, Wang H, Zhang Y, Shen Q, Li M, Lu Z, Rong X, Zheng D, Peng Y, 2021. Circular RNA expression alteration identifies a novel circulating biomarker in serum exosomal for detection of alcohol dependence. Addict Biol 26, e13031. [DOI] [PubMed] [Google Scholar]
- Lu Y, Tan L, Wang X, 2019. Circular HDAC9/microRNA-138/Sirtuin-1 Pathway Mediates Synaptic and Amyloid Precursor Protein Processing Deficits in Alzheimer’s Disease. Neurosci Bull 35, 877–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons D, de Jaeger X, Rosen LG, Ahmad T, Lauzon NM, Zunder J, Coolen LM, Rushlow W, Laviolette SR, 2013. Opiate exposure and withdrawal induces a molecular memory switch in the basolateral amygdala between ERK1/2 and CaMKIIα-dependent signaling substrates. J Neurosci 33, 14693–14704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmoudi E, Cairns MJ, 2019. Circular RNAs are temporospatially regulated throughout development and ageing in the rat. Sci Rep 9, 2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantsch JR, Baker DA, Funk D, Lê AD, Shaham Y, 2016. Stress-Induced Reinstatement of Drug Seeking: 20 Years of Progress. Neuropsychopharmacology 41, 335–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayberry HL, Bavley CC, Karbalaei R, Peterson DR, Bongiovanni AR, Ellis AS, Downey SH, Toussaint AB, Wimmer ME, 2022. Transcriptomics in the nucleus accumbens shell reveal sex- and reinforcer-specific signatures associated with morphine and sucrose craving. Neuropsychopharmacology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natividad LA, Buczynski MW, McClatchy DB, Yates JR, 2018. From Synapse to Function: A Perspective on the Role of Neuroproteomics in Elucidating Mechanisms of Drug Addiction. Proteomes 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nestler EJ, 2005. Is there a common molecular pathway for addiction? Nat Neurosci 8, 1445–1449. [DOI] [PubMed] [Google Scholar]
- Nestler EJ, Lüscher C, 2019. The Molecular Basis of Drug Addiction: Linking Epigenetic to Synaptic and Circuit Mechanisms. Neuron 102, 48–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen AF, Bindereif A, Bozzoni I, Hanan M, Hansen TB, Irimia M, Kadener S, Kristensen LS, Legnini I, Morlando M, Jarlstad Olesen MT, Pasterkamp RJ, Preibisch S, Rajewsky N, Suenkel C, Kjems J, 2022. Best practice standards for circular RNA research. Nat Methods 19, 1208–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okvist A, Fagergren P, Whittard J, Garcia-Osta A, Drakenberg K, Horvath MC, Schmidt CJ, Keller E, Bannon MJ, Hurd YL, 2011. Dysregulated postsynaptic density and endocytic zone in the amygdala of human heroin and cocaine abusers. Biol Psychiatry 69, 245–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver RJ, Brigman JL, Bolognani F, Allan AM, Neisewander JL, Perrone-Bizzozero NI, 2018. Neuronal RNA-binding protein HuD regulates addiction-related gene expression and behavior. Genes Brain Behav 17, e12454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pamudurti NR, Bartok O, Jens M, Ashwal-Fluss R, Stottmeister C, Ruhe L, Hanan M, Wyler E, Perez-Hernandez D, Ramberger E, Shenzis S, Samson M, Dittmar G, Landthaler M, Chekulaeva M, Rajewsky N, Kadener S, 2017. Translation of CircRNAs. Mol Cell 66, 9–21.e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paudel P, Pierotti C, Lozano E, Amoah SK, Gardiner AS, Caldwell KK, Allan AM, Mellios N, 2020. Prenatal Alcohol Exposure Results in Sex-Specific Alterations in Circular RNA Expression in the Developing Mouse Brain. Front Neurosci 14, 581895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piwecka M, Glažar P, Hernandez-Miranda LR, Memczak S, Wolf SA, Rybak-Wolf A, Filipchyk A, Klironomos F, Cerda Jara CA, Fenske P, Trimbuch T, Zywitza V, Plass M, Schreyer L, Ayoub S, Kocks C, Kühn R, Rosenmund C, Birchmeier C, Rajewsky N, 2017. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science 357. [DOI] [PubMed] [Google Scholar]
- Qu S, Zhong Y, Shang R, Zhang X, Song W, Kjems J, Li H, 2017. The emerging landscape of circular RNA in life processes. RNA Biol 14, 992–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragan C, Goodall GJ, Shirokikh NE, Preiss T, 2019. Insights into the biogenesis and potential functions of exonic circular RNA. Sci Rep 9, 2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro EA, Scarpa JR, Garamszegi SP, Kasarskis A, Mash DC, Nestler EJ, 2017. Gene Network Dysregulation in Dorsolateral Prefrontal Cortex Neurons of Humans with Cocaine Use Disorder. Sci Rep 7, 5412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robison AJ, Nestler EJ, 2011. Transcriptional and epigenetic mechanisms of addiction. Nat Rev Neurosci 12, 623–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybak-Wolf A, Stottmeister C, Glažar P, Jens M, Pino N, Giusti S, Hanan M, Behm M, Bartok O, Ashwal-Fluss R, Herzog M, Schreyer L, Papavasileiou P, Ivanov A, Öhman M, Refojo D, Kadener S, Rajewsky N, 2015. Circular RNAs in the Mammalian Brain Are Highly Abundant, Conserved, and Dynamically Expressed. Mol Cell 58, 870–885. [DOI] [PubMed] [Google Scholar]
- Sekar S, Liang WS, 2019. Circular RNA expression and function in the brain. Noncoding RNA Res 4, 23–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H, Moussawi K, Zhou W, Toda S, Kalivas PW, 2011. Heroin relapse requires long-term potentiation-like plasticity mediated by NMDA2b-containing receptors. Proc Natl Acad Sci U S A 108, 19407–19412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Q, Xie B, Galaj E, Yu H, Li X, Lu Y, Zhang M, Wen D, Ma C, 2022. CircTmeff-1 in the nucleus accumbens regulates the reconsolidation of cocaine-associated memory. Brain Res Bull 185, 64–73. [DOI] [PubMed] [Google Scholar]
- Shi L, Yan P, Liang Y, Sun Y, Shen J, Zhou S, Lin H, Liang X, Cai X, 2017. Circular RNA expression is suppressed by androgen receptor (AR)-regulated adenosine deaminase that acts on RNA (ADAR1) in human hepatocellular carcinoma. Cell Death Dis 8, e3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sillivan SE, Whittard JD, Jacobs MM, Ren Y, Mazloom AR, Caputi FF, Horvath M, Keller E, Ma’ayan A, Pan YX, Chiang LW, Hurd YL, 2013. ELK1 transcription factor linked to dysregulated striatal mu opioid receptor signaling network and OPRM1 polymorphism in human heroin abusers. Biol Psychiatry 74, 511–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theberge FR, Pickens CL, Goldart E, Fanous S, Hope BT, Liu QR, Shaham Y, 2012. Association of time-dependent changes in mu opioid receptor mRNA, but not BDNF, TrkB, or MeCP2 mRNA and protein expression in the rat nucleus accumbens with incubation of heroin craving. Psychopharmacology (Berl) 224, 559–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend EA, Kim RK, Robinson HL, Marsh SA, Banks ML, Hamilton PJ, 2021. Opioid withdrawal produces sex-specific effects on fentanyl-vs.-food choice and mesolimbic transcription. Biol Psychiatry Glob Open Sci 1, 112–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urdánoz-Casado A, Sánchez-Ruiz de Gordoa J, Robles M, Acha B, Roldan M, Zelaya MV, Blanco-Luquin I, Mendioroz M, 2021. Gender-Dependent Deregulation of Linear and Circular RNA Variants of HOMER1 in the Entorhinal Cortex of Alzheimer’s Disease. Int J Mol Sci 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallender EJ, Goswami DB, Shinday NM, Westmoreland SV, Yao WD, Rowlett JK, 2017. Transcriptomic profiling of the ventral tegmental area and nucleus accumbens in rhesus macaques following long-term cocaine self-administration. Drug Alcohol Depend 175, 9–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkow ND, 2020. Collision of the COVID-19 and Addiction Epidemics. Ann Intern Med 173, 61–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkow ND, Michaelides M, Baler R, 2019. The Neuroscience of Drug Reward and Addiction. Physiol Rev 99, 2115–2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vornholt E, Drake J, Mamdani M, McMichael G, Taylor ZN, Bacanu SA, Miles MF, Vladimirov VI, 2021. Identifying a novel biological mechanism for alcohol addiction associated with circRNA networks acting as potential miRNA sponges. Addict Biol 26, e13071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZY, Wen ZJ, Xu HM, Zhang Y, Zhang YF, 2022. Exosomal noncoding RNAs in central nervous system diseases: biological functions and potential clinical applications. Front Mol Neurosci 15, 1004221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng Y, Wu J, Li L, Shao J, Li Z, Deng M, Zou W, 2019. Circular RNA expression profile in the spinal cord of morphine tolerated rats and screen of putative key circRNAs. Mol Brain 12, 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F, Han B, Wu S, Yang L, Leng S, Li M, Liao J, Wang G, Ye Q, Zhang Y, Chen H, Chen X, Zhong M, Xu Y, Liu Q, Zhang JH, Yao H, 2019. Circular RNA TLK1 aggravates neuronal injury and neurological deficits after ischemic stroke via miR-335–3p/TIPARP. J Neurosci. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing M, Deng M, Shi Y, Dai J, Ding T, Song Z, Zou W, 2022. Identification and characterization of N6-methyladenosine circular RNAs in the spinal cord of morphine-tolerant rats. Front Neurosci 16, 967768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K, Zhang Y, Xiong W, Zhang Z, Wang Z, Lv L, Liu C, Hu Z, Zheng YT, Lu L, Hu XT, Li J, 2020. CircGRIA1 shows an age-related increase in male macaque brain and regulates synaptic plasticity and synaptogenesis. Nat Commun 11, 3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Fan X, Mao M, Song X, Wu P, Zhang Y, Jin Y, Chen LL, Wang Y, Wong CC, Xiao X, Wang Z, 2017. Extensive translation of circular RNAs driven by N. Cell Res 27, 626–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You X, Vlatkovic I, Babic A, Will T, Epstein I, Tushev G, Akbalik G, Wang M, Glock C, Quedenau C, Wang X, Hou J, Liu H, Sun W, Sambandan S, Chen T, Schuman EM, Chen W, 2015. Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat Neurosci 18, 603–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Xie B, Zhang J, Luo Y, Galaj E, Zhang X, Shen Q, Liu Y, Cong B, Wen D, Ma C, 2021. The role of circTmeff-1 in incubation of context-induced morphine craving. Pharmacol Res 170, 105722. [DOI] [PubMed] [Google Scholar]
- Zajaczkowski EL, Bredy TW, 2021. Circular RNAs in the Brain: A Possible Role in Memory? Neuroscientist 27, 473–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Hu J, Yu Y, 2020. CircRNA Is a Rising Star in Researches of Ocular Diseases. Front Cell Dev Biol 8, 850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Gao Y, Li Y, Geng D, Liang Y, He Q, Wang L, Cui H, 2022. Nrf2 improves hippocampal synaptic plasticity, learning and memory through the circ-Vps41/miR-26a-5p/CaMKIV regulatory network. Exp Neurol 351, 113998. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Du L, Bai Y, Han B, He C, Gong L, Huang R, Shen L, Chao J, Liu P, Zhang H, Gu L, Li J, Hu G, Xie C, Zhang Z, Yao H, 2018. CircDYM ameliorates depressive-like behavior by targeting miR-9 to regulate microglial activation via HSP90 ubiquitination. Mol Psychiatry. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong L, Ju K, Chen A, Cao H, 2021. Circulating CircRNAs Panel Acts as a Biomarker for the Early Diagnosis and Severity of Parkinson’s Disease. Front Aging Neurosci 13, 684289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Enoch MA, Goldman D, 2014. Gene expression in the addicted brain. Int Rev Neurobiol 116, 251–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman AJ, Hafez AK, Amoah SK, Rodriguez BA, Dell’Orco M, Lozano E, Hartley BJ, Alural B, Lalonde J, Chander P, Webster MJ, Perlis RH, Brennand KJ, Haggarty SJ, Weick J, Perrone-Bizzozero N, Brigman JL, Mellios N, 2020. A psychiatric disease-related circular RNA controls synaptic gene expression and cognition. Mol Psychiatry 25, 2712–2727. [DOI] [PMC free article] [PubMed] [Google Scholar]


