Abstract
EcoCyc is an organism-specific Pathway/Genome Database that describes the metabolic and signal-transduction pathways of Escherichia coli, its enzymes, and—a new addition—its transport proteins. MetaCyc is a new metabolic-pathway database that describes pathways and enzymes of many different organisms, with a microbial focus. Both databases are queried using the Pathway Tools graphical user interface, which provides a wide variety of query operations and visualization tools. EcoCyc and MetaCyc are available at http://ecocyc.PangeaSystems. com/ecocyc/
INTRODUCTION
The EcoCyc and MetaCyc databases (DBs) are online reference sources for metabolic data. They are similar in describing metabolic pathways, reactions, enzymes and substrate compounds. Both DBs use the same DB schema. Both are accessed using the same software environment, called the Pathway Tools. Both are review-level DBs in that a given entry in either DB often integrates information from multiple literature citations. There is also overlap in the content of the DBs—both contain all pathways of Escherichia coli small-molecule metabolism.
The DBs do differ significantly in content. EcoCyc aims to describe the full biochemical network of E.coli. As well as describing the metabolism of E.coli, EcoCyc describes its signal-transduction pathways, its transporters, and all E.coli genes. MetaCyc does not focus on a single organism as EcoCyc does. It describes pathways from a wide variety of species, with a focus on microorganisms, but includes some human and other mammalian pathways as well.
Intended uses of the DBs include the following.
• MetaCyc is a general reference source on metabolic pathways for the scientific community. It also serves as a reference pathway DB for prediction of the pathway complement of an organism from the annotated genome of the organism (1).
• EcoCyc is a resource for analysis of microbial genomes at the level of individual genes. Because the E.coli genome has a high fraction of genes whose functions were determined experimentally, it is an accurate reference for inferring gene function by sequence similarity.
• Because of its links to sequence DBs such as SWISS-PROT, EcoCyc can be used to perform function-based retrieval of DNA or protein sequences, for example to prepare datasets for studies of protein structure–function relationships.
• Both EcoCyc and MetaCyc are used as an aid in teaching biochemistry.
This article describes recent enhancements to EcoCyc and MetaCyc, and how to access them. We request that users of these DBs cite this article in publications related to their use.
Version 5.0 of EcoCyc was released in June, 1999; version 5.0 of MetaCyc was released in September 1999. In the future the same version number will be used for both DBs, and their releases will be synchronized.
PATHWAY TOOLS SOFTWARE AND DATABASE ENVIRONMENT
The Pathway Tools software that underlies EcoCyc and MetaCyc provides query, editing and visualization operations for Pathway/Genome DBs (1–3). The Pathway Tools is an environment for functional bioinformatics—for managing, curating and computing with a functional genome annotation. The Pathway Tools utilize a frame knowledge representation system (FRS) called Ocelot (3,4). FRSs use an object-oriented data model that organizes information within classes: collections of objects that share similar properties and attributes.
Recent software enhancements now allow users to issue structured queries to EcoCyc and MetaCyc using a new HTML query form that is available at http://ecocyc.pangeasystems. com:1555/query.html . This query interface allows the user to specify the class of objects they wish to query, plus a set of constraints that select the objects of interest, plus a set of attributes to be returned for each object. For example, the user might query the class of all genes to select all genes whose chromosomal location is within a specific location, and ask that the results include the unique identifier of each gene, its name, the function of its product and its map position.
The EcoCyc/MetaCyc web site was upgraded and reorganized for the 5.0 release. Although the location of the EcoCyc home page has not changed, the URL of the page used to formulate most queries to EcoCyc and MetaCyc has changed to http://ecocyc.pangeasystems.com:1555/server.html
EcoCyc
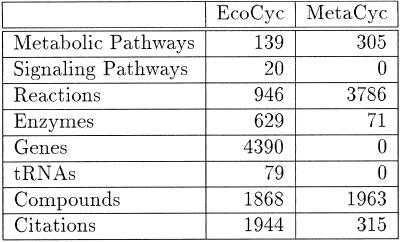
Table 1 shows the current size of the principal EcoCyc and MetaCyc classes.
Table 1. The number of objects in version 5.0 of EcoCyc and MetaCyc.
One of the recent enhancements to EcoCyc is the inclusion of membrane transport systems. Metabolic pathways are dependent on membrane transport processes for the uptake of exogenous substrates and for the export of metabolic end products. Membrane transporters also play a role in other important cellular processes such as energy interconversion, ion homeostasis and resistance to drugs and toxic compounds. Therefore, we believe that membrane transport systems represent a significant addition to EcoCyc and will help provide an understanding of the interplay between metabolic pathways and their corresponding transport systems.
A complete list of known and putative E.coli cytoplasmic membrane transporters has previously been compiled, and each of these transporters was classified on the basis of amino acid sequence similarities and transport function (5) (see also http://www-biology.ucsd.edu/~ipaulsen/transport/ and http://www-biology.ucsd.edu/~msaier/transport/ ). This compilation of over 300 transporters was imported into EcoCyc, and each transporter is now being annotated using the same detailed, literature-based approach that EcoCyc uses for E.coli enzymes and pathways. For each transporter, a comprehensive literature search of MEDLINE and Current Contents is performed for relevant keywords. Citations in relevant journal papers are used to identify further pertinent articles. Additionally, the background knowledge of transport systems of the two transport editors proved invaluable, and in some instances experts working on particular transport systems are contacted to clarify their findings.
For each transporter, where possible, the annotation includes: (i) functional information such as the energy-coupling mechanism, substrate specificity and relative affinity of each transporter; (ii) a brief description of the experimental evidence for function, e.g. cloning and expression data, knockout mutants, protein purification and functional reconstitution, vesicle or whole cell transport assays; (iii) sequence similarity to other known transporters; (iv) the inferred physiological role of the transporter and/or the role of its substrate in metabolism; and (v) in some cases additional relevant information is provided, such as structure/function studies, domain structure, details of transcriptional regulation.
Version 5.0 of EcoCyc contains all of the E.coli phosphotransferase system (PTS) transporters. Subsequent versions will include detailed descriptions of all of the other transporters present in E.coli (primary and secondary transporters, and channels). The PTS functions by transporting and concomitantly phosphorylating its sugar substrates using phosphoenolpyruvate as a phosphate donor. Each PTS transporter characteristically consists of a sugar-specific multidomain Enzyme II complex with between one and four protein subunits and two general energy coupling proteins, Enzyme I and HPr. The Version 5.0 release describes each of the 20 Enzyme II complexes and the two general energy coupling proteins of the PTS.
A representative example of an EcoCyc transporter entry is available at http://ecocyc.PangeaSystems.com:1555/ECOLI/new-image?type=ENZYME&object=CPLX-166 . This entry describes the mannitol PTS transporter Enzyme IIMtl, encoded by the mtlA gene. Mannitol is imported and concomitantly phosphorylated by Enzyme IIMtl to yield mannitol-1-phosphate. Phosphoenolpyruvate is used as energy source and phosphate donor. The phosphoryl transfer reaction involves Enzyme I and HPr in addition to Enzyme IIMtl.
The inclusion of transport systems in the EcoCyc DB not only provides a valuable resource for researchers, but also provides the opportunity for probing the interrelationships between transport and metabolism at a global level. For instance, in preliminary analyses we have compared the complete inventory of compounds transported into the cell with the starting reactants of all of the metabolic pathways of E.coli, as well as with the complete lists of metabolites and cofactors present in E.coli (I.T.Paulsen and P.D.Karp, unpublished data). This analysis provides an idea of the transporters which may be present in E.coli, but have not yet been experimentally identified, as well as highlighting the physiological role of the transported substrates.
Our schema for transport data builds upon our representations of enzyme function. Each transporter is represented by one or more DB objects that encode the transporter and its monomer subunits, if any. The transporter is linked to one or more (in the case of multifunctional transporters) objects that describe its function as a biochemical reaction. We have extended our schema for reactions to allow each substrate to be tagged with a cellular compartment, which if omitted defaults to the cytoplasm. For example, the representation of the mannitol PTS transporter tags the substrate mannitol with the compartment periplasm; the other substrates default to the cytoplasm. The PTS provides a strong example of why the reaction-based representation is a suitable one, since these transport systems both translocate and chemically alter their substrates.
THE EVOLVING ANNOTATION OF THE E.coli GENOME
We seek to make the annotation of the E.coli genome within EcoCyc as complete and up-to-date as possible based on new functional characterizations of E.coli genes in the literature, and based on an ongoing sequence analysis of the E.coli genome by the EcoCyc project.
In the initial annotation of the E.coli genome published by Blattner et al. in September 1997, 38% of the open reading frames had no attributed function. In EcoCyc 4.5, there are 1400 ORFs with no attributed function (32%), and 939 genes whose attributed function is marked as putative (21%).
MetaCyc
MetaCyc is a meta-metabolic pathway database that contains pathways from a variety of different organisms, which are listed in Table 2. MetaCyc describes metabolic pathways, reactions, enzymes and substrate compounds. The MetaCyc data were gathered from a variety of literature and on-line sources. ‘MetaCyc’ is pronounced ‘met-a-sike’. It sounds like ‘encyclopedia’.
Table 2. MetaCyc contains pathways from these species.
MetaCyc has two major goals: to provide a reference source on metabolic pathways that is accessible to scientists through the WWW, and to serve as a reference for computational prediction of metabolic pathways for sequenced genomes (6). The philosophy of MetaCyc is to encode pathways that have been reported in the experimental literature, and to label each pathway with the organism(s) in which it is known to occur, based on wet-lab experiments. Thus, unlike EcoCyc, which aims to describe the complete metabolic map of a single organism, MetaCyc aims to provide a smorgasbord of pathways from many organisms, none of whose metabolic maps have been studied in as much detail in the laboratory as has that of E.coli.
MetaCyc employs the same database schema as does EcoCyc. It aims to provide the same rich literature-based annotation for each pathway as does EcoCyc, although a minority of pathways currently lack the extensive commentary and literature citations that we plan to provide. Each MetaCyc pathway contains a citation to the source from which it was obtained. Unlike EcoCyc, MetaCyc does not provide genomic data such as genomic maps or sequences.
MetaCyc was initialized to contain all metabolic pathways of EcoCyc. Many additional pathways were then added to the database. The majority of pathways have been added by Dr Riley’s group at the Marine Biological Laboratory, but some pathways have added by Dr Karp’s group. The pathways have been gathered from many different phyla, including micro-organisms, plants and humans.
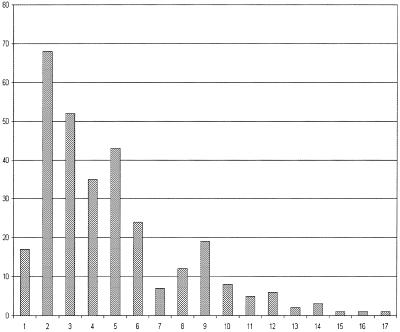
Each MetaCyc pathway has a slot (attribute) called ‘species distribution’ that lists the one or more species in which the experimental literature reports that this pathway has been observed. The fact that a given species is not listed in the species distribution of a pathway does not necessarily imply that the pathway is not present in that species—experiments may not yet have been performed, or the relevant literature may not have been located. Table 2 lists the species for which MetaCyc 5.0 contains pathways. Figure 1 shows the length distributions of MetaCyc pathways.
Figure 1.
The length distribution of MetaCyc pathways. We graph the number of MetaCyc pathways having a given length in reaction steps.
Each distinct pathway (where a pathway is defined as a set of reaction steps that are connected in a particular topology) is encoded only one time in MetaCyc—separate observations of the same pathway in new organisms do expand the species distribution recorded for that pathway, but those new observations do not result in new pathway records in MetaCyc unless the pathway differs in its component reactions. Different variants of a pathway that are observed in different organisms are recorded as distinct pathway objects in the DB. Different pathway variants might differ by the addition or removal of one or more reactions, or by substitution of one reaction for a similar reaction—such as substituting a reaction in which NAD is the hydride acceptor for a reaction in which NADP is the hydride acceptor.
The pathways within MetaCyc are annotated at different levels of detail. For all pathways, the pathway DB object is linked to objects for each reaction in the pathway. For some pathways, additional DB objects are provided for each enzyme and for each enzyme subunit in the pathway, and the pathway and enzyme include extensive commentary and literature citations.
MetaCyc contains all reactions in the enzyme-classification system devised by the Nomenclature Committee of the International Union of Biochemistry and Molecular Biology (NC-IUBMB). It also contains thousands of objects representing individual metabolites.
The MetaCyc data reside within the same software environment as used for EcoCyc: the Pathway Tools. The Pathway Tools run in both a WWW mode and an X-windows mode. Both EcoCyc and MetaCyc are accessed through a query form at http://ecocyc.PangeaSystems.com:1555/server.html . The user selects the DB to query using the dataset selector button near the top of the form. All query options in the remainder of the form can be applied to either DB. Put another way, all of the visualization and query tools available for EcoCyc are also available for MetaCyc (the exception being the genomic-map browser, which is not available for MetaCyc because MetaCyc contains no genomic maps).
DISTRIBUTION
EcoCyc and MetaCyc are available under license from Pangea Systems in two forms:
• EcoCyc and MetaCyc are accessible online through the WWW at http://ecocyc.PangeaSystems.com/ecocyc/ (this version supports a subset of the GUI functionality of the X-windows version).
• An X-windows version of EcoCyc and MetaCyc for the Sun workstation bundles together the Pathway/Genome Navigator software with the EcoCyc and MetaCyc DBs.
Both of the preceding forms of access are free to academic institutions for research use (contact pkarp@ai.sri.com ); a fee applies to other forms of use. The EcoCyc/MetaCyc WWW site provides background information about the DBs and software, and access to the publications produced by the EcoCyc project.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by grant 1-R01-RR07861-01 from the Comparative Medicine Program at the National Center for Research Resources. The contents of this article are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
REFERENCES
- 1.Karp P.D. (1999) Trends Biotechnol., 17, 275–281. [DOI] [PubMed] [Google Scholar]
- 2.Karp P.D. (1999) EcoCyc: The Resource and the Lessons Learned. Kluwer Academic Publishers, pp. 47–62.
- 3.Karp P.D. and Paley,S. (1996) J. Comp. Biol., 3, 191–212. [DOI] [PubMed] [Google Scholar]
- 4.Karp P.D., Chaudhri,V.K. and Paley,S.M. (1999) J. Intell. Inf. Sys., in press. [Google Scholar]
- 5.Sliwinski M.K., Paulsen,I.T. and Saier,M.H. (1998) J. Mol. Biol., 277, 573–592. [DOI] [PubMed] [Google Scholar]
- 6.Karp P.D., Ouzounis,C. and Paley,S.M. (1996) In States,D.J., Agarwal,P., Gaasterland,T., Hunter,L. and Smith,R. (eds), Proceedings of the Fourth International Conference on Intelligent Systems for Molecular Biology. AAAI Press, Menlo Park, CA, pp. 116–124. [PubMed]