Abstract
The US National Institute of Allergy and Infectious Diseases hosted a two-day virtual workshop on skin microbial communities and their interactions with the host immune system in health and disease. The aim of the workshop was to evaluate the current state of knowledge in the field and identify gaps, challenges, and future directions.
The skin microbiome is composed of a diverse community of microorganisms and their associated products. These microorganisms interact directly with host cells and are affected by skin immune responses and external factors such as antibiotics. Benefits of the skin microbiome include the establishment of immunological tolerance in early life, production of antimicrobials and immunoregulating metabolites, facilitation of wound healing, enhancement of barrier function, and regulation of the migration, metabolism, and function of skin cells. By contrast, pathogens and pathobionts in the skin microbiome can cause disease and are associated with skin disorders (Fig. 1). The crosstalk between the skin microbiome and the host is highly complex and many knowledge gaps remain. Understanding the ‘rules’ that govern the microbial ecology of skin and the effect of its dysregulation on host immunity will be key to advancing this field and realizing the promise of using microorganisms and their metabolites for therapeutic purposes.
Fig. 1 |. The skin microbial kingdom.
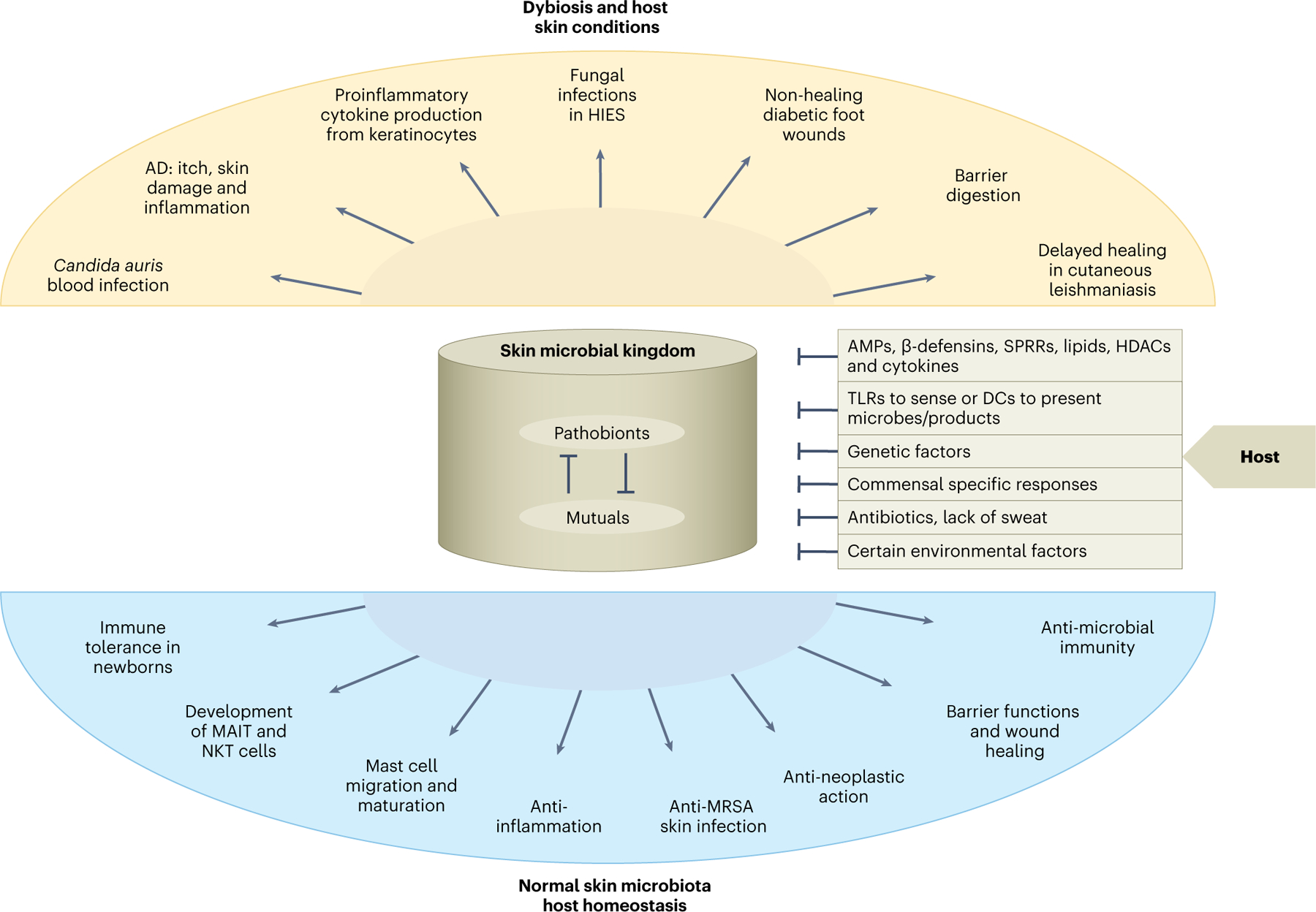
Pathobionts in the skin microbiome can cause disease and are associated with skin disorders. AD, atopic dermatitis; DCs, dendritic cells; HDAC, histone deacetylases; HIES, hyperimmunoglobulin E syndrome; SPRRs, induce small proline-rich proteins; TLRs, Toll-like receptors.
Host and environmental factors that influence the skin microbiome
Workshop speakers highlighted how the microbial ecology of the skin can be shaped by local and systemic host immune factors, including the nature and composition of the skin microbiota. Tamia Harris-Tryon (University of Texas Southwestern) reported that sebaceous glands generate antimicrobial peptides and lipids that shape an individual’s skin microbial composition. The excretion of sebum can selectively support certain microorganisms that grow in a hypoxic environment. In this context, lipopolysaccharides from skin resident Gram-negative bacteria can induce small proline-rich proteins, which are bactericidal1. Host innate immune cells sense and take up skin microorganisms and/or products. Anna Di Nardo (University of California, San Diego) described how keratinocytes sense Gram-positive bacteria through TLR22, and Tiffany Scharschmidt (University of California, San Francisco) discussed how skin dendritic cells can present antigens from commensal bacteria3. Host systemic immune factors also markedly affect the composition of microorganisms on the skin. Workshop speakers cited examples of primary immunodeficiencies or inborn errors of immunity where there are clear changes in the skin microbiome. Richard Gallo (University of California, San Diego) reported that the skin lesions of individuals with Netherton’s syndrome with mutations in SPINK5 are predominated by Staphylococcus aureus and S. epidermidis. Heidi Kong (National Institute of Arthritis and Musculoskeletal and Skin Diseases) reported that individuals with DOCK8 deficiency, a rare monogenic immune disorder, experience an expansion of their skin virome4. Bone marrow transplantation can resolve skin lesions caused by viral infections in DOCK8 deficiency5, which highlights the crucial role of the systemic immune system in the establishment of the skin microbiome.
Composition of the skin microbiome is affected by other host factors such as age, sex, hormones, and certain lifestyle and external factors including topical skin products, antibiotics, environment, hygiene, diet, and drugs. Kong showed that the use ofsystemic antibiotics can have a long-lasting effect on skin microbiota6. Not surprisingly, these changes can coincide with the emergence of antimicrobial-resistant bacterial strains that have the potential to cause hard-to-treat infections.
Positive regulation of host immune system and immunity by the skin microbiome
The skin microbiota are crucial in establishing immune tolerance and shaping development of host immune cells in early life. Scharschmidt reported that microbial colonization of skin in neonates helps to establish immune tolerance by inducing commensal-specific regulatory T (Treg) cells via interactions with CD301+ type 2 conventional dendritic cells. The dendritic cells present bacterial antigens that support the development of commensal-specific Treg cells and possess the highest immunotolerance capability4. An additional example of important immune–microbiome crosstalk described by Michael Constantinides (Scripps Research) involves innate-like T cells, including natural killer T (NKT) cells and mucosal-associated invariant T (MAIT) cells. The development of both cell types depends on early-life exposure to non-peptidic metabolites from the skin microbiome presented by monomorphic MHC class Ib molecules. NKT cells recognize bacterial glycolipids presented by CD1d, whereas MAIT cells recognize bacterial and fungal riboflavin through MR17.
The skin microbiome also regulates the function of immune cells and keratinocytes and maintenance of the cutaneous barrier. Di Nardo demonstrated that the recruitment and maturation of mast cells into the skin requires lipoteichoic acid from Gram-positive bacteria and TLR2 on keratinocytes, as well as stem cell factor from keratinocytes3. Gallo reported that S. epidermidis not only produces lipoteichoic acid to inhibit TLR3-triggered skin inflammation through keratinocytes in a TLR2-dependent manner, but some strains also produce 6-N-hydroxyaminopurine to selectively suppress tumor cell growth8. In addition, skin bacteria can induce host cells to secrete factors such as cathelicidins, β-defensins, lipids, and hyaluronan to boost antimicrobial immunity and histone deacetylases to reduce tissue-specific inflammation. Tom Dawson (A*STAR Skin Research Labs) discovered that the fungus Malassezia induces species-specific, enzymatically driven cytokine and prostaglandin E2 production from human keratinocytes. Elizabeth Grice (University of Pennsylvania) showed that human skin commensals promote barrier function and accelerate skin repair by activating the keratinocyte aryl hydrocarbon receptor (AHR)9. Such functions of the skin microbiota can have immediate therapeutic relevance.
Negative regulation of host immunity during skin conditions by skin microbiome
Several speakers discussed the pathogenic role of certain skin microorganisms in the progression of atopic dermatitis. Gallo noted that some gene products from S. epidermidis, including extracellular cysteine protease (EcpA), promote inflammation and correlate with disease severity. Kong found that in a cohort of pediatric individuals with atopic dermatitis, S. aureus and S. epidermidis predominate in acute exacerbations or ‘flares’, with S. aureus colonizing individuals with severe flares and S. epidermidis dominating in those with less severe disease. Similarly, Gurjit Khurana Hershey (Cincinnati Children’s Hospital) found that persistent S. aureus skin colonization is associated with increased severity of atopic dermatitis, allergen sensitization, and decreased filaggrin levels in infants. These data suggest that colonization by certain skin-associated microorganisms can drive inflammation in individuals with atopic dermatitis.
Dysbiosis of the skin microbiome also worsened other skin conditions. As reported by Fernanda Novais (Ohio State University), the clinical course of the parasitic skin infection, cutaneous leishmaniasis, was microbiome-dependent, with lesional S. aureus linked to delayed lesion resolution. Dawson demonstrated that hydrolases expressed by Malassezia on human skin converted sebum triglycerides into free fatty acids, and increased hydrophobicity at the surface of Malassezia species corresponded with boosted proinflammatory cytokine production from human keratinocytes. Julie Segre (National Human Genome Research Institute) discussed how fungal dysbiosis was associated with genetically defined primary immune deficiencies, such as STAT3 hyper IgE syndrome. Grice showed that S. aureus isolates from diabetic foot ulcers produced more staphyloxanthin, which promoted resistance to oxidative stress and enhanced bacterial survival in human neutrophils.
Pathogenic bacteria and fungi can migrate beyond the skin to cause systemic infections. Victor Nizet (University of California, San Diego) found that low platelet counts were associated with a high mortality rate in S. aureus bloodstream infections due to S. aureus α-toxin reduction in platelet viability and acceleration of platelet clearance. Notably, the drug ticagrelor (Brilinta), which inhibits α-toxin-mediated platelet injury, protected mice from lethal S. aureus infection. Candida auris, an emerging multidrug-resistant fungal pathogen that caused several recent outbreaks, triggered difficult-to-treat bloodstream infections10. Segre examined the underlying skin mycobiota during persistent colonization that lead to environmental shedding, which promoted hospital transmissions and predisposed patients to subsequent infections.
Technologies and tools for studying skin microbiome
Various in vivo models of skin conditions have been used to understand how microorganisms influence homeostasis. Several workshop speakers presented data from studies with germ-free and gnotobiotic mice to demonstrate the crucial role of the skin microbiome in regulating host immunity. Di Nardo used the NOD/SCID/IL2rγnull (NSG) mouse model engrafted with human hematopoietic stem cells to delineate how the skin microbiome affected human mast cell maturation. Using immunodeficient mice grafted with adult human skin, Rachael Clark (Brigham Women’s Hospital) demonstrated broad immunoregulatory effects of the topically applied AHR agonist tapinarof, which suggests that this mouse model was a useful tool to study the interaction between the human immune system and the skin microbiome.
Organoids are powerful in vitro tools to model the cellular composition, structure, complexity, and physiology of whole tissues and provide a unique opportunity to examine interactions of immune cells with skin commensals. Katherine Lemon (Baylor College of Medicine) discussed success in monocolonizing human nasal organoids with several human nasal microbionts (unpublished). Julia Oh ( Jackson Laboratories) described how a 3D air–liquid interface skin organoid model could recapture some human skin features, including the formation of stratum corneum, epidermis, and dermis. Meanwhile, Clark applied a 3D skin equivalent to study the human cutaneous response to radiation.
Shotgun metagenomics is a useful tool to capture microbial genomic content and resolve species- to strain-level microbiome diversity. By assembling metagenomic reads into metagenome-assembled genomes, Segre developed the skin microbial genome catalogue (http://ftp.ebi.ac.uk/pub/databases/metagenomics/genome_sets/skin_micro-biome/) that identifies 213 novel bacterial species, 14 bacterial genera, as well as Corynebacterium isolates. Christopher Dupont ( J. Craig Venter Institute) described the VEBA (viral eukaryotic bacterial archaeal) open-sourced software suite (https://github.com/jolespin/veba), the first end-to-end pipeline for in silico analysis that recovers, assesses quality, and classifies prokaryotic and viral genomes from metagenomes or metatranscriptomes11. Besides de novo assembly via the metatranscriptome, VEBA also enables the analysis of existing datasets to identify microorganisms without existing genome representatives.
Translational potential for therapeutic targets
Skin commensals offer unique opportunities to develop microorganism-based therapeutic agents or delivery tools to target infections or cancer. Interestingly, there are mechanisms in which skin commensals interact with each other, which can be exploited as targets to treat skin infections. Gallo provided evidence that some coagulase-negative Staphylococci (CoNS) commensals exhibited a selective anti-microbial function by producing phenol-soluble modulins γ and δ, antimicrobial peptides, and/or lantibiotics that inhibit group A Streptococcus (GAS) and S. aureus growth. Application of these CoNS strains reduced S. aureus abundance on the skin of individuals with atopic dermatitis12. Because the S. aureus serine protease V8 mediates itch by cleaving PAR1 in sensory neurons, PAR1 could also be a therapeutic candidate in atopic dermatitis. Isaac Chiu (Harvard Medical School) demonstrated that PAR1 antagonists such as vorapaxar, which was recently approved by the US Food and Drug Administration, block itch and skin damage in S. aureus-infected mice. Alexander Horswill (University of Colorado) discussed how CoNS bacteria produced several autoinducing peptides that interfered with the quorum-sensing system of S. aureus13. Oh described that engineered S. epidermidis that express antimicrobials inhibited methicillin-resistant S. aureus14. Michael Fischbach (Stanford University) showcased data on a tumor-derived neoantigen in S. epidermidis that elicited tumor-specific T cell responses resulting in protection against local and metastatic progression of a mouse melanoma in vivo15. Finally, Nizet reported that biomimetic toxin nanosponges coated with human red blood cell membranes sequestered GAS pore-forming streptolysin O to protect host cells and retain immune function.
Knowledge gaps, obstacles, and next steps
The skin microbiota engages in crosstalk with the host and these interactions have a vital role in maintaining skin function and regional homeostatic immunity. Changes to the microbiome can contribute to various skin conditions. Harnessing therapeutically beneficial microorganisms and their products will require a clear understanding of the functional microbiome at all levels, from specific strains to the collective output of their biosynthetic pathways. Scientific studies, clinical research, and interventional trials are underway to investigate the mechanisms and safety of bacteriotherapy and to fill existing knowledge gaps (Table 1).
Table 1 |.
Knowledge gaps in skin microbiome research
| Topics | Gaps |
|---|---|
| Microbial interactions |
|
| Interaction between microbiome and host |
|
| Microbiome transmission and dynamics |
|
| Microorganisms and skin conditions |
|
| Implementing integration of strains in vivo |
|
Despite recent advances in skin microbiome research, several obstacles remain, including: better models that accurately mimic the commensal colonization on human skin, within its niches such as the hair follicle, and interactions with human cells; expansion of research to include more mechanistic studies to define the role and function of the skin microbiota in regulating host homeostasis and diseases; identification of diagnostic and prognostic biomarkers from longitudinal sample collection from healthy individuals and patients, and making these resources publicly available; development of more robust bioinformatic capabilities for increasingly large human metagenomic and metatranscriptomic dataset; strengthening collaborations between experts in microbial genomics, microbiology, bioinformatics, immunology, and dermatology; and facilitation of translational capabilities and shortening the transition time from bench to bedside to promote therapeutic and drug development.
Future research directions should include the identification of new commensals and/or metabolites and their role in health and disease. Microorganism–microorganism and microorganism–host interactions, as well as the crosstalk between skin microbiota with remote organ systems must also be better understood. Continued efforts should focus on the development of innovative technologies and computational tools to integrate data for a human skin microbiota atlas. Multi-cohort and longitudinal studies are required to advance microorganism-based therapeutic agents and better understand the role of skin microbiota and their products in human disease.
Acknowledgements
We thank the invited speakers, and participants for their expertise and insights on crosstalk between skin microbiota and host immune systems in health and disease. We thank W. Leitner and A. Augustine for editing this manuscript.
Footnotes
Competing interests
The authors declare no competing interests.
References
- 1.Zhang C et al. Elife 11, 76729–76746 (2022). [Google Scholar]
- 2.Wang Z et al. J Allergy Clin Immunol 139, 1205–1216 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weckel A et al. Immunity 56, 1–16 (2023).36630909 [Google Scholar]
- 4.Tirosh O et al. Nat. Med 24, 1815–1821 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cuellar-Rodriguez J et al. Bio. Blood Marrow Transplant 21, 1037–1045 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.MacGibeny MA JAMA Dermatol 158, 989–991 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Constantinides MG & Belkaid Y Science 374, 6464–6477 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nakatsuji T et al. Sci Adv 4, 4502–4511 (2018). [Google Scholar]
- 9.Uberoi A et al. Cell Host Microbe 29, 1235–1248 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Proctor DM et al. Nat. Med 27, 1401–1409 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Espinoza JL BMC Bioinformatics 23, 419–455 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakatsuji T et al. Sci. Trans. Med 9, 378–388 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Severn MM et al. mBio 13, e0093033–e0093054 (2022). [Google Scholar]
- 14.Guan G et al. PLoS ONE 17, e0276795–e0076811 (2022).36520793 [Google Scholar]
- 15.Chen YE et al. Preprint at bioRxiv 10.1101/2021.02.17.431662 (2021). [DOI] [Google Scholar]


