Abstract
Background
The efficacy of monotherapy of AMG-510 is limited. This study explored whether the AMG-510 and cisplatin combination increases the anti-tumor effect in lung adenocarcinoma with the mutation of Kirsten rat sarcoma viral oncogene (KRAS) G12C.
Methods
Patients’ data were used to analyze the proportion of KRAS G12C mutation. Besides, the next-generation sequencing data was used to uncover information about co-mutations. The cell viability assay, the concentration inhibiting 50% of cell viability (IC50) determination, colony formation, and cell-derived xenografts were conducted to explore the anti-tumor effect of AMG-510, Cisplatin, and their combination in vivo. The bioinformatic analysis was conducted to reveal the potential mechanism of drug combination with improved anticancer effect.
Results
The proportion of KRAS mutation was 2.2% (11/495). In this cohort with KRAS mutation, the proportion of G12D was higher than others. Besides, KRAS G12A mutated tumors had the likelihood of concurrent serine/threonine kinase 11 (STK11) and kelch-like ECH-associated protein 1 (KEAP1) mutations. KRAS G12C and tumor protein p53 (TP53) mutations could appear at the same time. In addition, KRAS G12D mutations and C-Ros oncogene 1 (ROS1) rearrangement were likely to be present in one tumor simultaneously. When the two drugs were combined, the respective IC50 values were lower than when used alone. In addition, there was a minimum number of clones among all wells in the drug combination. In in vivo experiments, the tumor size reduction in the drug combination group was more than twice that of the single drug group (p < 0.05). The differential expression genes were enriched in the pathways of phosphatidylinositol 3 kinase-protein kinase B (PI3K-Akt) signaling and extracellular matrix (ECM) proteoglycans compared the combination group to the control group.
Conclusions
The anticancer effect of the drug combination was confirmed to be better than monotherapy in vitro and in vivo. The results of this study may provide some information for the plan of neoadjuvant therapy and the design of clinical trials for lung adenocarcinoma patients with KRAS G12C mutation.
Supplementary Information
The online version contains supplementary material available at 10.1007/s12672-023-00698-z.
Keywords: KRAS G12C mutation, AMG-510, Cisplatin, In vivo, Translational medicine
Introduction
Lung cancer is still the leading cause of worldwide malignancies-related death, which mainly contains two histological types, small cell lung cancer and non-small cell lung cancer (NSCLC) [1, 2]. The proportion of NSCLC is over 80%, and the 5-year overall survival (OS) rate is less than 30% [3, 4]. Squamous cell carcinoma and lung adenocarcinoma account for most NSCLCs [5]. In addition, some somatic driver events were found to have the ability to provide treatment targets, such as epidermal growth factor receptor (EGFR) mutation, Kirsten rat sarcoma viral oncogene (KRAS) mutation, and CAP-Gly domain-containing linker protein 1- leukocyte tyrosine kinase (CLIP1-LTK) fusion [6–8]. Those findings improve the OS of NSCLC patients significantly [6, 7]. However, for patients of KRAS mutation, it is still a challenge to form the targeted drug because of the narrow hydrophobic pocket until some outstanding structural and mechanistic research constructed the theoretical basis for the clinical development of KRAS G12C inhibitors [9, 10].
Based on those findings, a phase 2 clinical trial (NCT03600883) of AMG-510 (Sotorasib) was performed in 126 enrolled patients. Moreover, the objective response rate reached 37.1% in this phase 2 trial [7]. According to the significant outcomes, the Food and Drug Administration approved the utilization of AMG-510 in NSCLC patients with KRAS G12C mutation as subsequent therapy [11]. Therefore, there is no doubt that the use of AMG-510 provides improved survival outcomes for NSCLC patients with KRAS G12C mutation. However, we found that some patients in this phase 2 trial did not have satisfactory outcomes—26 patients did not get disease control.
In addition, in a phase 2 trial of another targeted drug Erlotinib (EGFR inhibitor, NCT01407822), neoadjuvant targeted therapy did not further make tumor regression to elevate the complete resection rate and reduce resection extent [12]. Thus, the effect of tumor regression and the anticancer ability using monotherapy of targeted drugs are limited according to the results of the abovementioned studies [7, 12]. To consider those findings, some researchers investigated the antitumor effect of the drug combination in NSCLC patients with EGFR mutation and suggested that the EGFR inhibitor and chemotherapy combination further improved the objective response rate [13]. Therefore, this study aimed to explore whether the AMG-510 and cisplatin combination increases the antitumor effect in lung adenocarcinoma with the mutation of KRAS G12C by experiments in vitro and in vivo. In addition, we used bioinformatic analysis to uncover the potential mechanism of drug combination with improved anticancer effect. We anticipated that these results would provide some key information for designing clinical trials and using clinical drugs in the NSCLC population with KRAS G12C mutation.
Methods
Patients and samples
Patients with NSCLC after surgery were recruited from July 2018 to July 2019 in Sun Yat-sen University Cancer Center. Mian inclusion criteria for this study included: (1) histologically confirmed stage IA-IIIA NSCLC; (2) without distant metastasis; (3) without neoadjuvant therapy for NSCLC. Patients were excluded according to following standards: (1) age < 18 years old; (2) did not receive KRAS gene test. Tumor tissues which were collected during surgery were performed the next-generation sequencing (NGS). The pathological stage was based on the 8th staging system [14].
Cell lines, cell culture, and drug treatment
Lung adenocarcinoma cell lines harbor KRAS G12C mutations were obtained from American Type Culture Collection. NCI-H23 (H23) and NCI-H358 (H358) cells were cultured in Roswell Park Memorial Institute 1640 medium (WISENT Inc., Nanjing, China) and supplemented with 10% fetal bovine serum (Excell Bio Inc., Taicang, China) and 50 µg/ml penicillin–streptomycin (Invitrogen). All cells used in this study were maintained in a 5% CO2 cell culture incubator at 37 °C. AMG-510 and Cisplatin (Targetmol Co. Ltd., Shanghai, China) were dissolved in dimethyl sulfoxide and H2O, respectively, then stored at -80 °C until use. Untreated cell lines were subdued until cell viability assays for determining the concentration inhibiting 50% of cell viability (IC50).
Cell viability assay and IC50 determination
To determine the IC50 value of AMG-510 and Cisplatin in H23 and H358 cell lines, we used Cell Count Kit8 (CCK-8, Targetmol Co. Ltd., Shanghai, China) after 72 h of drug treatment. In this assay, 3000 cells were seeded on 96-well plates and incubated for cell apposition in their complete medium and subsequently for 72 h with scalar doses of AMG-510 (0–50 µM) or Cisplatin (0–5µM) as single agents or as a constant combination. After 72 h of drug treatment, we aspirated the old medium, added a new complete medium containing 10% CCK8 reagent, and placed it in a 37° thermostat for 2 h. It was worth noting that the process was kept dark throughout. Infinite M Plex (Tecan Trading Co., Ltd., Shanghai, China) and Tecan. At.XFluor software was used for signal acquisition with a detection wavelength of 450 nm. The dose-effect curves were drawn, and IC50 values were calculated by GraphPad Prism software version 9.0 (https://www.graphpad.com/) and analyzed by two-way ANOVA with Bonferroni’s correction.
Colony formation
The cells were seeded into 6-well plates (2000 cells/well). Then medium changes were performed every other day, as well as medication changes. Each well was treated differently in the following four ways: control, 0.1µM AMG-510, 1µM Cisplatin, and 0.1µM AMG-510 plus 1µM Cisplatin. After 14–21 days of culture, the colonies were fixed with 4% paraformaldehyde and stained with 0.5% crystal violet (Yeasen Biotech Co., Ltd., Shanghai, China). The number of colonies was then counted.
Mice experiments
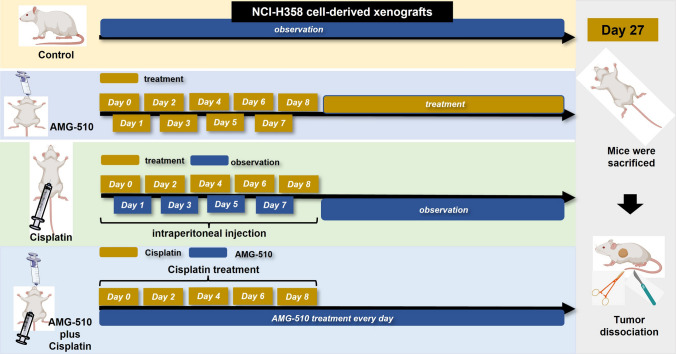
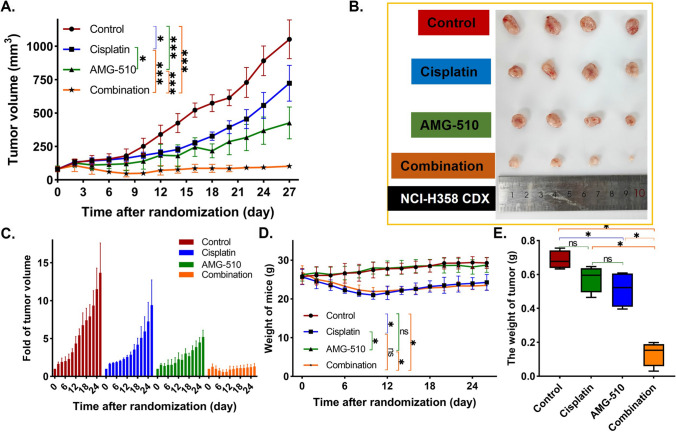
We constructed cell-derived xenografts (CDXs) to explore the anti-tumor effect of AMG-510, Cisplatin, and their combination in Balb/C nude mice (Yaokang Biotechnology Co., Ltd, Guangzhou, China, License No. SCXX2020-0054). The H358 cells (2*106/150µL) were injected under the skin after one-week feeding. The mice were classified into four groups randomly (control, AMG-510, Cisplatin, and combination) when the tumor size was about 53-111mm3. Mice in the control group did not receive any drug; mice in the AMG-510 group were administered by gavage at a dose of 30 mg/kg per day (100 µl suspension); mice in the cisplatin group were administered by intraperitoneal injection at 4 mg/kg per mouse on alternate days, with follow-up observation after five injections; the administration of the combination drug group combined the AMG-510 and cisplatin groups. The specific dosing flow chart is shown in Fig. 1. The 0.5% sodium carboxymethylcellulose (Targetmol Co. Ltd., Shanghai, China) was used as the resuspension medium for AMG-510, forming a stable homogeneous mixture. Cisplatin was dissolved in 0.9% saline into a working solution of 4 mg/kg. Mice were weighed regularly. After 28 days from the first drug injection observation, the difference in tumor size between the four groups was evident, and the animal experiment was ended. After anesthetizing the mice, the mice were executed using the cervical dislocation method, then the tumors were dissected, the tumors were weighed, and the tumor tissue was preserved.
Fig. 1.
Flow chart of animal experiments
mRNA sequence
Total RNA was extracted using the TRIzol reagent (Invitrogen, CA, USA) according to the manufacturer’s protocol. In this study, we used resected tumor tissue to perform this experiment. RNA purity and quantification were evaluated using the NanoDrop 2000 spectrophotometer (Thermo Scientific, USA). RNA integrity was assessed using the Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). Then the libraries were constructed using VAHTS Universal V6 RNA-seq Library Prep Kit according to the manufacturer’s instructions.
The libraries were sequenced on a llumina Novaseq 6000 platform and 150 bp paired-end reads were generated. About 50 M raw reads for each sample were generated. Raw reads of fastq format were firstly processed using fastp and the low-quality reads were removed to obtain the clean reads [15]. Then about 40 M clean reads for each sample were retained for subsequent analyses. Finally, the clean reads were mapped to the reference genome using HISAT2 [16].
Statistical analysis
Fragments Per Kilobase of exon model per Million mapped fragments (FPKM) of each gene were calculated, and the read counts of each gene were obtained by HTSeq-count [17, 18]. Principal component analysis (PCA) analysis was performed using R 4.2.2 software (https://cloud.r-project.org/) to evaluate the biological duplication of samples. Differential expression analysis was performed using the DESeq2 [19]. Q value < 0.05 and foldchange > 2 or foldchange < 0.5 was set as the threshold for significantly differential expression genes (DEGs). Hierarchical cluster analysis of DEGs was performed using R 4.2.2 software to demonstrate the expression pattern of genes in different groups and samples. The radar map of the top 30 genes was drawn to show the expression of up-regulated or down-regulated DEGs using the R package “ggradar.” Based on the hypergeometric distribution, Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG), Wikipathway, and Reactome pathway enrichment analysis of DEGs were performed to screen the significantly enriched term using R 4.2.2 software, respectively [20–22]. The GO entries corresponding to the number of differential genes greater than 2 in the three classifications (biological process [BP], cellular component [CC], and molecular function [MF]) were screened, and ten entries were sorted by the -log10p-value of each entry in descending order. R 4.2.2 software was used to draw the column, chord, and bubble diagram of the significant enrichment term. Gene Set Enrichment Analysis (GSEA) was performed using GSEA software. Besides, we used the STRING database to conduct Protein-Protein Interaction Networks (PPI) analysis and screened the top 30 DEGs with a high combined score [23]. The analysis used a predefined gene set, and the genes were ranked according to the degree of differential expression in the two samples. Then, whether the predefined gene set was enriched at the top or bottom of the ranking list is tested. The Mann-Whitney U test was used to compare the difference of tumor weight between two groups. The standard deviation (SD) was conducted to evaluate the stability of data. A two-sided p-value < 0.05 was considered statistically significant.
Results
Patient characteristics
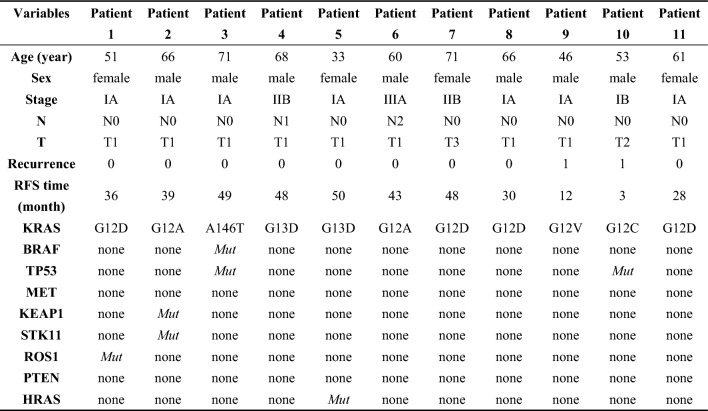
We reviewed the medical records of 495 NSCLC patients. The proportion of KRAS mutation was 2.2% (11/495). Those patients underwent surgical resection, and the majority of them belonged to stage IA disease (63.6%, 7/11). Two patients developed tumor recurrence at postoperative month three and month 12. In this cohort with KRAS mutation, the proportion of G12D was higher than others. Besides, KRAS G12A mutated tumors had the likelihood of concurrent serine/threonine kinase 11 (STK11) and kelch-like ECH-associated protein 1 (KEAP1) mutations. KRAS G12C and tumor protein p53 (TP53) mutations could appear at the same time. In addition, KRAS G12D mutations and C-Ros oncogene 1 (ROS1) rearrangement were likely to be present in one tumor at the same time. The detailed clinical characteristics are shown in Table 1.
Table 1.
Clinical characteristics of non-small-cell lung cancer patients with KRAS mutation
RFS recurrence-free survival; BRAF vrafmurine sarcoma viral oncegene homolog B; TP53 tumor protein p53; MET: mesenchymal epithelial transition factor; KEAP1: Kelch-1ike ECH- associated protein l; STK11: serine/threonine Kinase 11; ROS1: ROS proto-oncogene 1; PTEN phosphatase and tensin homolog; HRAS v-Ha-ras Harvey rat sarcoma viral oncogene homolog; Mut: mutation
Cell viability and colony formation
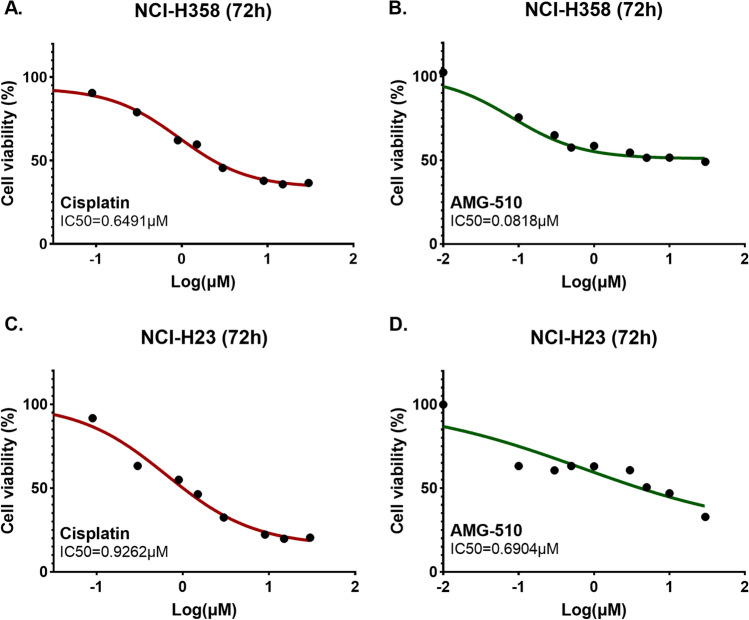
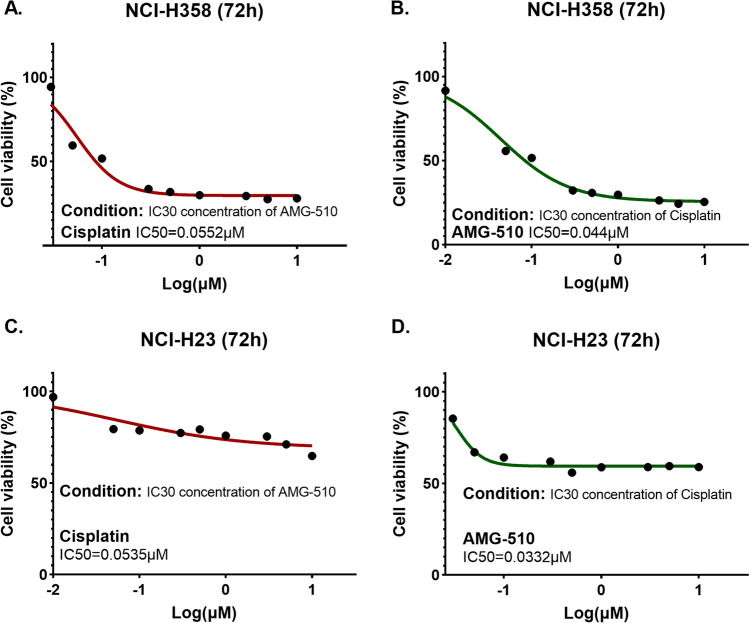
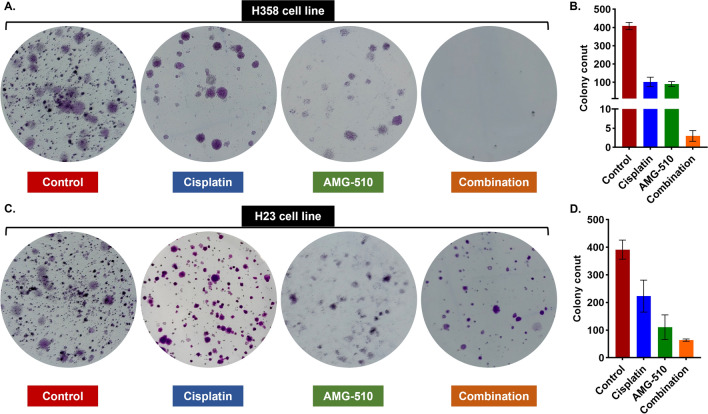
In the cell viability experiments, H358 and H23 cell lines were sensitive to drug treatment. The IC50 values of Cisplatin and AMG-510 in H358 and H23 cell lines were 0.6491µM vs. 0.0818µM and 0.9262µM vs. 0.6904µM, respectively (Fig. 2). To explore the anticancer effect of combination with two drugs in both cell lines, we fixed the concentration of one drug (setting the concentration of IC30) and increased the concentration of the other drug in a gradient. The IC30 concentration of Cisplatin in H358 and H23 cell lines were 0.27µM and 0.216µM, respectively. Besides, the IC30 concentration of AMG-510 in H358 and H23 cell lines were 0.035µM and 0.044µM, respectively. Under the condition with the IC30 concentration of AMG-510 pre-treatment, the IC50 values of Cisplatin were 0.0552µM and 0.0535µM in the H358 and H23 cell lines (Fig. 3). Similarly, with the combination of IC30-concentration Cisplatin, the IC50 values of AMG-510 were 0.044µM and 0.0332µM in the H358 and H23 cell lines (Fig. 3). Overall, when the two drugs were combined, the respective IC50 values were lower than when the drugs were used alone. As showed results in Fig. 4, the inhibition effect of drugs presents an increasing trend. The combination of AMG-510 and Cisplatin had the best anticancer ability among different use of drugs. In addition, in wells of the drug combination, there was the minimum number of clones among all wells (Fig. 4B and D).
Fig. 2.
Cell viability experiment for monotherapy in different cell lines
Fig. 3.
Cell viability experiment for drug combination in different cell lines
Fig. 4.
Colony formation experiment and colony count for H358 (A and B) and H23 (C and D) cell lines
Confirmation of anticancer effect in vivo
Animal experiments were performed to validate the anticancer effect of the drug combination in vivo. The flow chart is presented in Fig. 1. After the tumors were measurable, we randomized the mice to groups. Animal experiments were performed for 28 days, and the changes in tumor volume in each group can be seen in Fig. 5A, B. The mean tumor volume were 1052.56mm3 (SD = 142.54mm3), 723.39mm3 (SD = 133.88mm3), 426.66mm3 (SD = 118.72mm3), and 102.51mm3 (SD = 20.14mm3) in control, Cisplatin, AMG-510, and combination groups on Day 27, respectively. The size reduction of tumor in the drug combination group was more than twice that of the single drug group. At the end of the animal experiments, the tumor volume in each group was the smallest in the combined drug group (AMG-510 group vs. combination group, p < 0.001; Cisplatin group vs. combination group, p < 0.001). The picture to fold of tumor volume showed that the tumor growth in the combination group was evidently inhibited (Fig. 5C). Besides, the mean weight of mice was 29.28 g (SD = 1.42 g), 24.25 g (SD = 2.05 g), 28.73 g (SD = 1.87 g), and 23.50 g (SD = 1.17 g) in control, Cisplatin, AMG-510, and combination groups on Day 27, respectively. Compared to the control and AMG-510 groups, Cisplatin and combination groups had more severe weight loss (all p < 0.05, Fig. 5D). Overall, however, the weight of the combination group remained comparable to that of the Cisplatin group (p = 0.549). Among control (0.69 g, SD = 0.05 g), Cisplatin (0.58 g, SD = 0.08 g), AMG-510 (0.51 g, SD = 0.11 g), and combination groups (0.13 g, SD = 0.07 g), the tumor weight was the smallest in the combination groups (Fig. 5E).
Fig. 5.
The growth of the tumor volume over time (A). The photo of tumor removed from mice (B). The fold change of tumor volume over time (C). The weight of mice over time (D). The wight of tumor in the end (E)
The landscape of DEGs
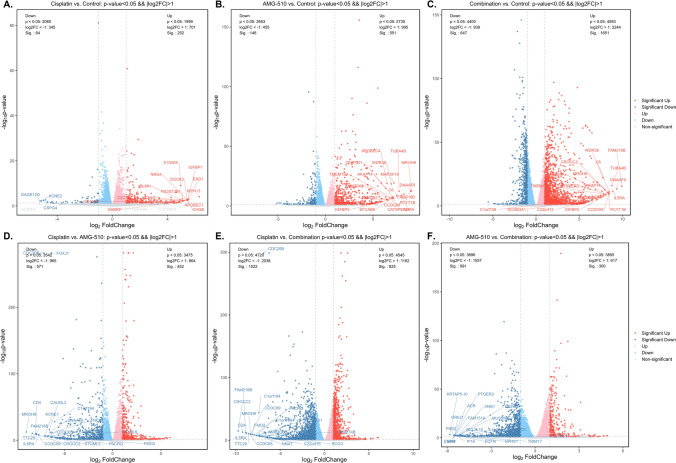
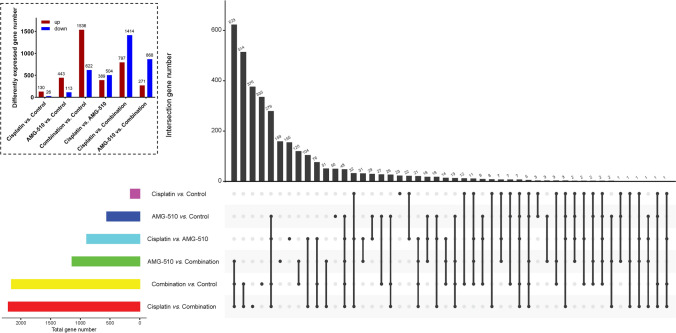
After differential expression analysis in the data of RNA seq among resected tumor tissues from mice of control, Cisplatin, AMG-510, and combination groups, up-regulated and downregulated DEGs were obtained. In the different comparison groups, the DEGs were different. Detailed information about DEGs could see in Figs. 6 and 7. There were 504 down-regulated and 389 up-regulated DEGs in the comparison between Cisplatin and AMG-510 (Figs. 6D and 7). Besides, in comparing the Cisplatin and Combination, 797 up-regulated genes and 1414 down-regulated genes were demonstrated (Fig. 6E). As for AMG-510 vs. Combination, 271 up-regulated and 868 down-regulated were included in this analysis (Fig. 6F).
Fig. 6.
The differential expressed genes (DEGs) in different comparisons
Fig. 7.
The number of the differential expressed genes (DEGs) in different comparisons
Functional enrichment and PPI analyses of DEGs
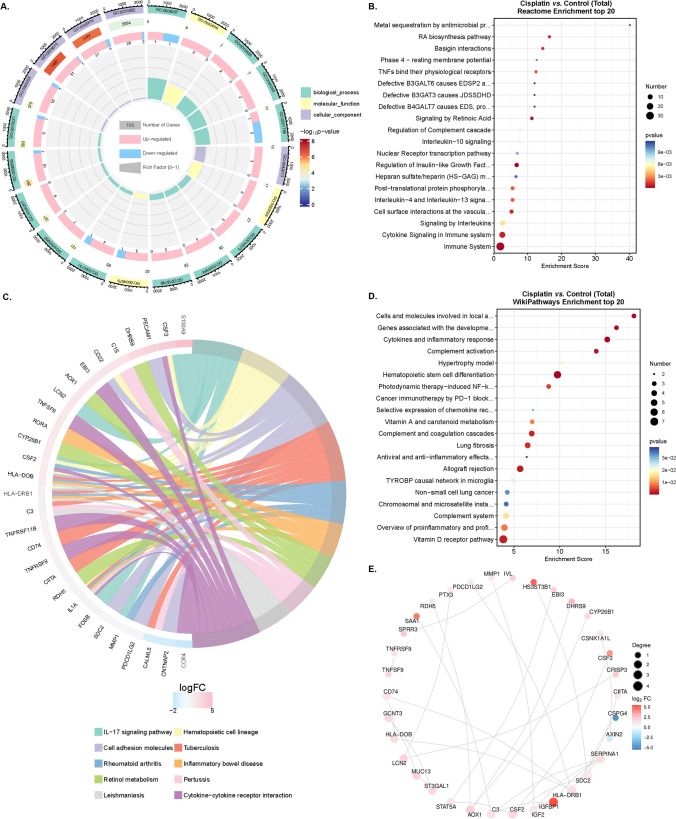
To study the relationship among DEGs and BPs, MFs, CCs, biological pathways, and diseases in comparing the Cisplatin and Control group, we first performed a functional enrichment analysis of DEGs (Fig. 8A). DEGs were the most abundant in BPs, such as biological adhesion, cell killing, and immune system processing (Supplementary Figure 1A). Further, the DEGs obtained in the comparison of Cisplatin vs. control was enriched in the pathway of the immune system after KEGG and Reactome analyses (Supplementary Figure 1B and Fig. 8B–C). Besides, the pathways of hematopoietic stem cells, cytokines, and inflammatory response were founded in the DEGs enrichment according to the results of Wikipathway analysis (Fig. 8D). According to the analysis of PPI, serum amyloid A1 (SAA1), heparan sulfate 3-O-sulfotransferase 3B1 (HS3ST3B1), and insulin-like growth factor binding protein 1 (IFGBP1), as up-regulated DEGs, had more related genes than other genes (Fig. 8E).
Fig. 8.
The GO enrichment (A), Reactome enrichment (B), KEGG pathway (C), WikiPathway (D), and Protein-Protein Interaction Networks (E) analyses in the comparison between Cisplatin and Control groups
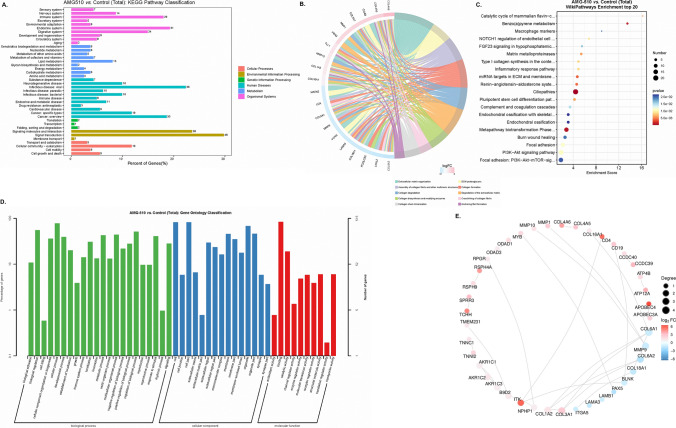
For the comparison between AMG-510 vs. control, the DEGs were enriched in the pathways of signal transduction and signaling molecules and interaction in the KEGG analysis (Fig. 9A). Besides, Reactome analysis showed there were many DEGs enriched in the pathway of extracellular matrix organization (Fig. 9B). After WikiPathway analysis, the results presented that the ciliopathies gathered many DEGs (Fig. 9C). GO enrichment analysis suggested that the DEGs were rich in the BP function (Fig. 9D). In addition, the PPI analysis suggested that four down-regulated DEGs (collagen, type VI, alpha 1 [COL6A1], matrix metallopeptidase 9 [MMP9], collagen, type VI, alpha 2 [COL6A2], and B-cell linker protein [BLNK]) had the largest number of related genes among all DEGs (Fig. 9E).
Fig. 9.
The KEGG pathway (A), Reactome enrichment (B), WikiPathway (C), GO enrichment (D), and Protein-Protein Interaction Networks (E) analyses in the comparison between AMG-510 and Control groups
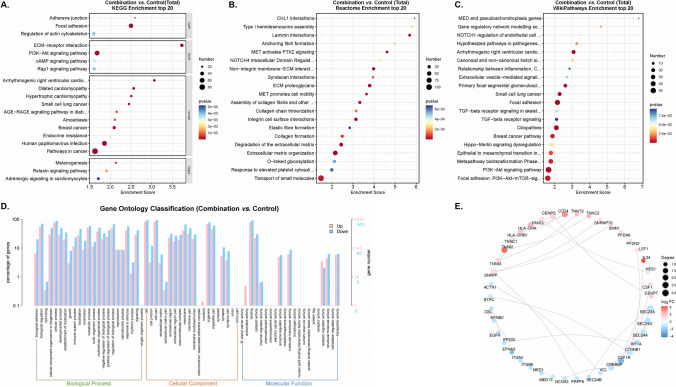
We further focused on the DEGs obtained in the comparison between the combination and control groups. We found that the DEGs were enriched in the pathways of phosphatidylinositol 3 kinase-protein kinase B (PI3K-Akt) signaling and extracellular matrix (ECM) proteoglycans after analyses of KEGG enrichment, Reactome enrichment, and WikiPathway (Fig. 10A–C). GO classification uncovered that the DEGs were mainly located in the BP function, and were enriched in the cilium movement, cell adhesion, and calcium ion binding function pathways (Fig. 10D). Of note, the up-regulated DEGs were different from the abovementioned two comparisons, of which three DEGs (CD74 molecule [CD74], troponin I2, fast skeletal type [TNNI2], and interleukin 34 [IL34]) had the most number of related genes among genes (Fig. 10E).
Fig. 10.
The KEGG pathway (A), Reactome enrichment (B), WikiPathway (C), GO enrichment (D), and Protein-Protein Interaction Networks (E) analyses in the comparison between combination and Control groups
Discussion
In this study, we used the data from our hospital to explore the proportion of KRAS in all patients and found that KRAS G12C and TP53 mutations could appear at the same time, and KRAS G12D mutations and ROS1 rearrangement were likely to be present in one tumor at the same time. Besides, we explored the anticancer effect of the combination of Cisplatin and AMG-510 and suggested that the combination treatment had a better ability to regress the tumor than the use of the single drug in vitro and in vivo (Figs. 3, 4 and 5). To further explore the possible mechanism of the good effect of the dual drug combination, we used some methods of bioinformatics for the analysis of the RNA-seq data. In addition, we found that the DEGs obtained in the comparison between the combination and control groups were enriched in the pathways of PI3K-Akt signaling, cilium movement, cell adhesion, and calcium ion binding function pathways and ECM proteoglycans after analyses of KEGG enrichment, Reactome enrichment, GO classification, and WikiPathway (Fig. 10). Interestingly, CD74, TNNI2, and IL34, as the up-regulated DEGs, had the largest number of related genes in all DEGs (Fig. 10E).
Neoadjuvant therapy is essential for local advanced NSCLC patients. The mood of preoperative drug treatment plus surgical resection provides more survival benefits than surgery or drug treatment alone in those patients [24]. Accordingly, a number of clinical trials focused on NSCLC patients with locally advanced staging disease [25, 26]. In a previous study, however, the use of targeted therapy alone did not increase the rate of R0 (complete resection) compared with chemotherapy. Even more, the data showed that patients treated with targeted agents had (8.1%, n = 3) a higher rate of R2 resection (visible tumor under the naked eye) than those treated with chemotherapy (2.9%, n = 1) [12]. R2 resection means that patients have higher recurrence rates and shorter disease-free survival. Thus, it is important to increase the rate of tumor regression for patients with locally advanced staging disease. As for translational and preclinical research, the results of this study may provide some information for the plan of neoadjuvant therapy and the design of clinical trials for NSCLC patients with KRAS G12C mutation. First, the anticancer effect of the use of drug combination was confirmed to be better than monotherapy in vitro and in vivo. Interestingly, the reduction of tumor size in the drug combination group was more than twice that of the single drug group. Besides, as seen in Fig. 5 of the fold of tumor volume, the tumors in the control group showed a growing trend over time, and the monotherapy group only inhibited the growing trend, while the combination group interrupted the tumor growth trend. Therefore, the combination of AMG-510 and Cisplatin may be a good trial for KRAS G12C mutation patients with locally advanced staging disease in phase I/II clinical trials. Second, although the weights of the mice did not differ between the cisplatin and combination groups, one of the mice in the combination group died during the first cycle of Cisplatin. Accordingly, it is still important to pay attention to the side effects during the drug combination, especially the tolerance of patients during the first cycle of chemotherapy and targeted drugs.
The proportion of KRAS G12C mutation is varied in NSCLC patients among different cohorts. A previous report suggested that among NSCLC patients, White (13.0%) and Black patients (10.9%) had more proportion of KRAS G12C mutations than Asians (3.6%) [27]. Other studies also confirmed this distribution phenomenon of KRAS G12C mutation [28–30]. The proportion of KRAS G12C mutation was low in the present study, like the abovementioned research. Thus, the low mutation rate of KRAS G12C in Asian populations reminds to researchers to consider the proper use of resources and fit when conducting research. Besides, as reported in previous studies, our study also found that STK11 can be co-mutated with KRAS G12A [31]. Interestingly, we found that ROS1 rearrangements could coexist with KRAS G12D, which was not common in other studies. Although in our study, two patients with KRAS G12C and KRAS G12V recurred, because of the small sample size, it cannot be said that patients with these two mutations have a poor prognosis. It’s essential for NSCLC patients to perform gene tests because exact gene status means personalized medicine and survival benefit [32]. Recently, a phase III clinical trial (NCT04303780) uncovered that AMG-510 significantly increased progression-free survival compared with docetaxel in patients with advanced NSCLC with the KRAS G12C mutation [33]. Patients need to be confirmed for KRAS G12C mutation prior to AMG-510 treatment. Therefore, molecular testing is necessary for the design of clinical trials.
The potential mechanism of the anticancer effect from a combination of AMG-510 and Cisplatin is complex, and it is not a simple superposition of the cancer-inhibiting mechanisms of chemotherapeutic agents or AMG-510. The DEGs differed among three comparisons (Fig. 6, Cisplatin vs. Control, AMG-510 vs. Control, and Combination vs. Control). Accordingly, the DEGs enrichment’s function pathway was inconsistent (Figs. 8, 9 and 10). After Cisplatin treatment, the DEGs were mainly located in the pathway of immune system. Besides, the use of AMG-510 inhibited the expression level of MMP9, which could promote tumor metastasis through the PI3K/Akt pathway [34]. The DEGs obtained from the comparison between AMG-510 and control groups were enriched in the function pathway of signal transduction. The extracellular matrix (ECM) pathway-related genes, like COL16A1 and COL4A6, were up-regulated after AMG-510 treatment. Notably, the ECM pathway may be related to tumor progression and drug resistance [35]. In addition, the phenomenon that the DEGs were enriched in the pathway of PI3K-Akt after treatment combination of AMG-510 and Cisplatin may indicate one of the drug resistance mechanisms.
This study still has some shortcomings. First, we only performed simple, functional experiments and did not further investigate the mechanism. However, the functional validation experiments have provided useful information for conducting subsequent clinical trials of neoadjuvant therapy. Second, the clinical data in this study were partially from a single hospital with a small sample size, and further expansion of the sample size is still needed to confirm our findings. Third, because the drug AMG-510 is not yet available in China, we could not obtain samples from patients after dosing them for genetic testing. We hope that AMG-510 will be available in China soon to benefit more lung cancer patients.
Conclusions
The anticancer effect of the use of drug combination was confirmed to be better than monotherapy in vitro and in vivo. The results of this study may provide some information for the plan of neoadjuvant therapy and the design of clinical trials for lung adenocarcinoma patients with KRAS G12C mutation.
Supplementary Information
Supplementary material 1: The GO enrichmentand KEGG pathwayclassifications.
Acknowledgements
We appreciate the help in bioinformatic analysis from the staff of OE Biotech Co., Ltd, Shanghai, China.
Author contributions
LLW, WMJ, GWM, and DX contributed to the study design, data collection, data analyses, data interpretation, and manuscript drafting. ZYL, YYZ, YEL, JYQ, and KL contributed to data analyses and manuscript review. ZXL, YYH, and DX contributed to data interpretation and manuscript review. All authors contributed to the final approval of the manuscript. All authors read and approved the final manuscript.
Funding
This study was supported by the Shanghai ShenKang Hospital Development Centre (SHDC22020218), Science and Technology Commission of Shanghai Municipality, (21Y11913400), and Outstanding Young Medical Talent of Rising Star in Medical Garden of Shanghai Municipal Health Commission “Dong Xie”. The funding bodies played no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Data availability
Any researchers interested in this study could contact corresponding author for requiring data.
Declarations
Ethics approval and consent to participate
The Clinical Research Ethic Committee of Sun Yat-sen University Cancer Center approved this study (No. B2020-255-01), and the human data was in accordance with the Declaration of Helsinki in the manuscript, with the need for informed consent waived for this retrospective study. All procedures involving animals were in compliance with the European Community Council Directive of 24 November 1986, and ethical approval was granted by the Ethical Review Committee and Laboratory Animal Welfare Committee of Shanghai Pulmonary Hospital (No. K22-390). The maximal tumor size/burden approved by our IRB was not exceeded.
Consent for publication
Not applicable.
Competing interests
There are no competing interests to declare.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Lei-Lei Wu and Wen-Mei Jiang have contributed equally to this work.
Contributor Information
Guo-Wei Ma, Email: magw@sysucc.org.cn.
Dong Xie, Email: xiedong@tongji.edu.cn.
References
- 1.Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin. 2022;72:7–33. doi: 10.3322/caac.21708. [DOI] [PubMed] [Google Scholar]
- 2.Wu LL, Li CW, Lin WK, Qiu LH, Xie D. Incidence and survival analyses for occult lung cancer between 2004 and 2015: a population-based study. BMC Cancer. 2021;21:1009. doi: 10.1186/s12885-021-08741-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 4.Miller KD, Nogueira L, Mariotto AB, Rowland JH, Yabroff KR, Alfano CM, Jemal A, Kramer JL, Siegel RL. Cancer treatment and survivorship statistics, 2019. CA Cancer J Clin. 2019 doi: 10.3322/caac.21565. [DOI] [PubMed] [Google Scholar]
- 5.Wu L-L, Li C-W, Li K, Qiu L-H, Xu S-Q, Lin W-K, Ma G-W, Li Z-X, Xie D. The difference and significance of parietal pleura invasion and Rib invasion in pathological T classification with non-small cell lung cancer. Front Oncol. 2022 doi: 10.3389/fonc.2022.878482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu YL, Tsuboi M, He J, John T, Grohe C, Majem M, Goldman JW, Laktionov K, Kim SW, Kato T, et al. Osimertinib in Resected EGFR-Mutated non-small-cell Lung Cancer. N Engl J Med. 2020 doi: 10.1056/NEJMoa2027071. [DOI] [PubMed] [Google Scholar]
- 7.Skoulidis F, Li BT, Dy GK, Price TJ, Falchook GS, Wolf J, Italiano A, Schuler M, Borghaei H, Barlesi F, et al. Sotorasib for Lung cancers with KRAS p.G12C mutation. N Engl J Med. 2021;384:2371–81. doi: 10.1056/NEJMoa2103695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Izumi H, Matsumoto S, Liu J, Tanaka K, Mori S, Hayashi K, Kumagai S, Shibata Y, Hayashida T, Watanabe K, et al. The CLIP1-LTK fusion is an oncogenic driver in non-small-cell lung cancer. Nature. 2021;600:319–23. doi: 10.1038/s41586-021-04135-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ostrem JM, Peters U, Sos ML, Wells JA, Shokat KM. K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature. 2013;503:548–51. doi: 10.1038/nature12796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lito P, Solomon M, Li LS, Hansen R, Rosen N. Allele-specific inhibitors inactivate mutant KRAS G12C by a trapping mechanism. Science. 2016;351:604–8. doi: 10.1126/science.aad6204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.National Comprehensive Cancer Network. Non-small cell lung cancer (Version 2.2023; Accessed February 17, 2023).
- 12.Zhong WZ, Chen KN, Chen C, Gu CD, Wang J, Yang XN, Mao WM, Wang Q, Qiao GB, Cheng Y, et al. Erlotinib Versus Gemcitabine Plus Cisplatin as Neoadjuvant Treatment of Stage IIIA-N2 EGFR-Mutant non-small-cell Lung Cancer (EMERGING-CTONG 1103): a randomized phase II study. J Clin Oncol. 2019;37:2235–45. doi: 10.1200/jco.19.00075. [DOI] [PubMed] [Google Scholar]
- 13.Chen Z, Shen S, Shi W, Jiang G, Wang X, Jian H, Zhou Z, Ding Z, Lu S. Intercalated combination of chemotherapy and erlotinib for stage IIIA non-small-cell lung cancer: a multicenter, open-label, single-arm, phase II study. Cancer Manag Res. 2019;11:6543–52. doi: 10.2147/cmar.S189287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rami-Porta R, Asamura H, Travis WD, Rusch VW. Lung cancer - major changes in the american Joint Committee on Cancer eighth edition cancer staging manual. Cancer J Clin. 2017;67:138–55. doi: 10.3322/caac.21390. [DOI] [PubMed] [Google Scholar]
- 15.Chen S, Zhou Y, Chen Y, Gu J. Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinf. 2018;34:i884–90. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12:357–60. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roberts A, Trapnell C, Donaghey J, Rinn JL, Pachter L. Improving RNA-Seq expression estimates by correcting for fragment bias. Genome Biol. 2011;12:R22. doi: 10.1186/gb-2011-12-3-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anders S, Pyl PT, Huber W. HTSeq–a python framework to work with high-throughput sequencing data. Bioinf. 2015;31:166–9. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014 doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.The Gene Ontology Resource 20 years and still going strong. Nucleic Acids Res. 2019;47:D330–d338. doi: 10.1093/nar/gky1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36:D480–484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li K, Wu LL, Wang H, Cheng H, Zhuo HM, Hao Y, Liu ZY, Li CW, Qian JY, Li ZX, et al. The characterization of tumor microenvironment infiltration and the construction of predictive index based on cuproptosis-related gene in primary lung adenocarcinoma. Front Oncol. 2022;12:1011568. doi: 10.3389/fonc.2022.1011568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rosner S, Reuss JE, Zahurak M, Zhang J, Zeng Z, Taube J, Anagnostou V, Smith KN, Riemer J, Illei PB, et al. Five-year clinical outcomes after Neoadjuvant Nivolumab in Resectable Non-Small Cell Lung Cancer. Clin Cancer Res. 2023;29:705–10. doi: 10.1158/1078-0432.Ccr-22-2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang B, Zhong H, Han B. Neoadjuvant immunotherapy for patients with Non-Small Cell Lung Cancer-Is a new era coming? JAMA Oncol. 2023 doi: 10.1001/jamaoncol.2022.6898. [DOI] [PubMed] [Google Scholar]
- 26.Godoy LA, Chen J, Ma W, Lally J, Toomey KA, Rajappa P, Sheridan R, Mahajan S, Stollenwerk N, Phan CT, et al. Emerging precision neoadjuvant systemic therapy for patients with resectable non-small cell lung cancer: current status and perspectives. Biomark Res. 2023 doi: 10.1186/s40364-022-00444-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nassar AH, Adib E, Kwiatkowski DJ. Distribution of KRAS (G12C) somatic mutations across race, sex, and Cancer Type. N Engl J Med. 2021;384:185–7. doi: 10.1056/NEJMc2030638. [DOI] [PubMed] [Google Scholar]
- 28.Wahl SGF, Dai HY, Emdal EF, Berg T, Halvorsen TO, Ottestad AL, Lund-Iversen M, Brustugun OT, Førde D, Paulsen EE, et al. The prognostic effect of KRAS mutations in non-small cell lung carcinoma revisited: a norwegian multicentre study. Cancers. 2021 doi: 10.3390/cancers13174294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tamiya Y, Matsumoto S, Zenke Y, Yoh K, Ikeda T, Shibata Y, Kato T, Nishino K, Nakamura A, Furuya N, et al. Large-scale clinico-genomic profile of non-small cell lung cancer with KRAS G12C: results from LC-SCRUM-Asia study. Lung Cancer. 2023;176:103–11. doi: 10.1016/j.lungcan.2022.12.019. [DOI] [PubMed] [Google Scholar]
- 30.Loong HH, Du N, Cheng C, Lin H, Guo J, Lin G, Li M, Jiang T, Shi Z, Cui Y, et al. KRAS G12C mutations in Asia: a landscape analysis of 11,951 chinese tumor samples. Transl Lung Cancer Res. 2020;9:1759–69. doi: 10.21037/tlcr-20-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ricciuti B, Son J, Okoro JJ, Mira A, Patrucco E, Eum Y, Wang X, Paranal R, Wang H, Lin M, et al. Comparative analysis and isoform-specific therapeutic vulnerabilities of KRAS mutations in non-small cell lung cancer. Clin Cancer Res. 2022;28:1640–50. doi: 10.1158/1078-0432.Ccr-21-2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.El-Deiry WS, Goldberg RM, Lenz HJ, Shields AF, Gibney GT, Tan AR, Brown J, Eisenberg B, Heath EI, Phuphanich S, et al. The current state of molecular testing in the treatment of patients with solid tumors, 2019. CA Cancer J Clin. 2019;69:305–43. doi: 10.3322/caac.21560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Langen AJ, Johnson ML, Mazieres J, Dingemans AC, Mountzios G, Pless M, Wolf J, Schuler M, Lena H, Skoulidis F, et al. Sotorasib versus docetaxel for previously treated non-small-cell lung cancer with KRAS(G12C) mutation: a randomised, open-label, phase 3 trial. Lancet. 2023 doi: 10.1016/s0140-6736(23)00221-0. [DOI] [PubMed] [Google Scholar]
- 34.Tian K, Du G, Wang X, Wu X, Li L, Liu W, Wu R. MMP-9 secreted by M2-type macrophages promotes Wilms’ tumour metastasis through the PI3K/AKT pathway. Mol Biol Rep. 2022;49:3469–80. doi: 10.1007/s11033-022-07184-9. [DOI] [PubMed] [Google Scholar]
- 35.Brown Y, Hua S, Tanwar PS. Extracellular matrix in high-grade serous ovarian cancer: advances in understanding of carcinogenesis and cancer biology. Matrix Biol. 2023 doi: 10.1016/j.matbio.2023.02.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material 1: The GO enrichmentand KEGG pathwayclassifications.
Data Availability Statement
Any researchers interested in this study could contact corresponding author for requiring data.