Abstract
The HIV RT and Protease Sequence Database is an online relational database that catalogs evolutionary and drug-related human immunodeficiency virus (HIV) reverse transcriptase (RT) and protease sequence variation (http://hivdb.stanford.edu ). The database contains a compilation of nearly all published HIV RT and protease sequences including International Collaboration database submissions (e.g., GenBank) and sequences published in journal articles. Sequences are linked to data about the source of the sequence sample and the antiretroviral drug treatment history of the individual from whom the isolate was obtained. The database is curated and sequences are annotated with data from >230 literature references. Users can retrieve additional data and view alignments of sequence sets meeting specific criteria (e.g., treatment history, subtype, presence of a particular mutation). A gene-specific sequence analysis program, new user-defined queries and nearly 2000 additional sequences were added in 1999.
MEDICAL AND BIOLOGICAL RELEVANCE
Human immunodeficiency virus type 1 (HIV) reverse transcriptase (RT) and protease enzymes are the molecular targets of the 14 currently licensed antiretroviral drugs. Sequence changes in the genes coding for these enzymes are directly responsible for phenotypic resistance to RT and protease inhibitors. Individuals infected with drug-susceptible HIV isolates experience reductions in morbidity and mortality with appropriate antiretroviral drug therapy. In contrast, individuals infected with drug-resistant isolates usually do not respond to drug therapy (1). Assays for sequencing HIV RT and protease are commercially available and are widely used in clinical settings. However, the optimal means of interpreting RT and protease sequences is not known and the potential utility of such sequence results in clinical settings is an area of intense clinical investigation.
HIV RT and protease are model proteins for studying genotypic and phenotypic correlations for the following reasons: (i) there is a wide range of genetic variability among HIV isolates and the RT and protease genes have been sequenced more frequently than those of any other virus; (ii) HIV evolution can be observed in vivo and in vitro in a matter of weeks; (iii) there are >120 published protease structures and >30 published RT structures in the Protein Data Bank; and (iv) there are numerous reports correlating sequence results with phenotypic drug susceptibility changes and with exposure to different drugs either in vitro or in vivo.
HIV RT AND PROTEASE
HIV RT is a heterodimer composed of p51 and p66 subunits. It is responsible for RNA-dependent DNA polymerization, RNase H activity and DNA-dependent DNA polymerization. The p51 subunit is composed of the first 450 amino acids of the RT gene. The p66 subunit is composed of all 560 amino acids of the RT gene. Although the p51 and p66 subunits share 450 amino acids, their relative arrangements are significantly different. The p66 subunit contains the DNA-binding groove and the active site; the p51 subunit appears to function as a scaffold for the enzymatically active p66 subunit. The three dimensional structure of HIV RT, bound to a double-stranded nucleic acid, to a non-nucleoside RT inhibitor, and unbound have been determined by X-ray crystallography (2).
HIV protease is responsible for the post-translational processing of the viral gag and gag-pol polyproteins to yield the structural proteins and enzymes of the virus. The enzyme is an aspartic protease composed of two non-covalently associated, structurally identical monomers 99 amino acids in length. The protease has a binding cleft that specifically recognizes and cleaves at least 10 different sequences on the viral precursor polyproteins. The three dimensional structure of wild-type HIV protease and of several drug-resistant mutant forms bound to various inhibitors have been determined crystallographically (3).
HIV SEQUENCE VARIATION
Factors contributing to HIV genetic variation include (i) the lack of proofreading capability by HIV RT; (ii) the high in vivo rate of HIV replication; (iii) the accumulation of proviral HIV variants during the course of infection; and (iv) recombination. The likelihood of developing drug resistance depends on the size and heterogeneity of the HIV population within an individual, the ease of acquisition of a particular mutation (or set of mutations), and the effect of drug-resistance mutations on changes in drug susceptibility and virus fitness (4). Some mutations selected during drug therapy confer resistance by themselves, other mutations produce measurable resistance only when present in combination.
Genetic analysis of HIV-1 isolates has revealed at least 10 distinct group M (main) subtypes (A–J) as well as several highly divergent group O (outlier) isolates. Differences between group M subtypes are based on the ~30% intersubtype genetic divergence in the env region and 14% intersubtype divergence in the gag region (5,6). The world-wide HIV pandemic is caused by group M HIV-1 virus. In North America, Europe and Australia, most HIV-1 isolates belong to subtype B and the available anti-HIV drugs have been developed by drug screening and susceptibility testing using subtype B isolates. However, subtype B accounts for only a small proportion of HIV-1 isolates worldwide and, even in industrialized countries, non-B isolates are being identified with increasing frequency.
IDENTIFYING DRUG RESISTANCE MUTATIONS
Drug resistance mutations have traditionally been identified during the pre-clinical and early clinical evaluation of a new anti-HIV drug. During these studies, drug-resistant HIV-1 isolates are identified, sequenced and tested for drug susceptibility. Site-directed mutagenesis experiments are then done to confirm the role of specific mutations introduced into a wild-type virus. This experimental approach, however, has limitations because many different combinations of mutations are associated with HIV-1 drug resistance and because the effect of a mutation often depends on the genetic context in which it develops (7). The HIV RT and Protease Sequence Database was developed on the premise that sequences of clinical HIV-1 isolates are experiments of nature that should be cataloged and examined methodically to help prioritize clinical investigations and further in vitro experimental work.
DATABASE SCHEMA
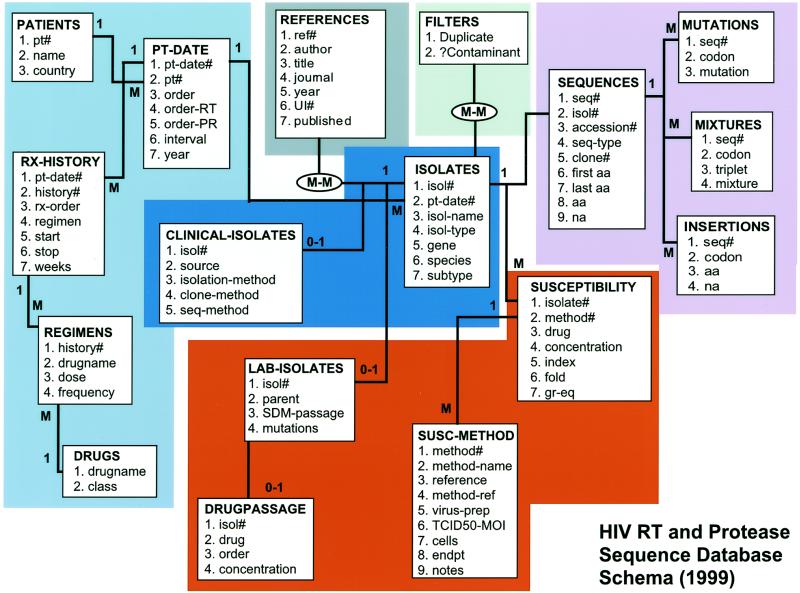
The web site is built around a relational database containing the text of each sequence, data about the person from whom the sequence was obtained (e.g., country, treatment history) and data about the methods of sequencing and sample isolation (e.g., year of isolation, body source, cloning method) (Fig. 1). Sequences are stored in a virtual alignment with the subtype B consensus sequence (5); thus, amino acid sequences are also represented as lists of amino acid differences from the consensus sequence (MUTATIONS table). The number of the start and stop residue of each sequence is maintained along with a table containing insertions, thereby avoiding the need for a multiple sequence alignment algorithm when a set of sequences is retrieved.
Figure 1.
Schema of HIV RT and Protease Sequence Database. The 17 rectangles (entities) depict 12 of the base tables in the database. Within the tables, the attributes (fields) are listed. The multiplicities of the linkages are demonstrated by either a 1 or an M (many). The schema demonstrates the hierarchical relationship between patients, patient-dates, isolates and sequences.
There is a hierarchical relationship linking four of the entities in the database: patient, patient-date, isolate and sequence (Fig. 1). An individual (PATIENT table) may have isolates obtained at different times (PATIENT-DATE table). A patient-date will have more than one isolate (ISOLATE table) if samples are obtained from more than one source (e.g., peripheral blood mononuclear cells, plasma, lymph node). Finally, if multiple clones are sequenced from an individual isolate then each clone is considered a different sequence (SEQUENCE table).
In 1999, the database schema was revised to accommodate drug susceptibility data. The database has been populated with >2000 published drug susceptibility results. These data are being curated and will be available on the site by early 2000.
DATABASE CONTENT
The HIV RT and Protease Database maintains a page with dynamically updated summaries of its contents (http://hivdb. stanford.edu/hiv/summary.asp ). As of October 1, 1999, the database contained 5464 sequences from 1450 individuals, including 2563 RT and 2901 protease sequences. Nearly 4200 sequences had GenBank accession numbers and nearly 1300 sequences were from published journal articles and were not in GenBank. The summary page also includes tables populated with dynamically updated numbers of sequences from individuals by country and subtype and by antiretroviral drug treatment.
DATABASE USE
In 1999 the user interface was updated to provide an increased number of user-defined queries. Users can now retrieve a large number of different sequences sets matching selection criteria based on drug treatment history, HIV subtype and specific mutations. Each query returns a new table and each record in the new table contains 8–12 columns of associated data. The data returned include (i) hyperlinks to the MEDLINE abstract, the GenBank record, and the complete nucleotide sequence and translation; (ii) a classification of the sequence by patient, patient-date and isolate; (iii) data on HIV-1 subtype; (iv) data on drug treatment including lists of drugs, duration of therapy and complete summaries of each drug regimen received by the patient from whom the isolate was obtained; and (v) technical data on method of virus isolation and sequencing. Following retrieval of a sequence set, users have the option of viewing or downloading complete sequence alignments, as well as, composite alignments summarizing the frequency and type of mutation at each position in the sequence.
In 1999, a program was added to the database to allow researchers to make sense of new HIV-1 RT and protease sequences in the context of previously published sequence data [HIV-1 RT and protease – a sequence analysis program (HRP-ASAP), http://hivdb.stanford.edu/hiv/programs.asp ]. The program compares a new HIV-1 sequence to a reference sequence and uses the differences or mutations as query parameters for the sequence database. The ability to examine new sequences in the context of previously published sequence data has two main advantages. First, unusual sequence results can be detected, allowing the person doing the sequencing to recheck the primary sequence output (e.g., electropherogram) while it is on his/her desktop (8). Second, unexpected associations can be uncovered because the person analyzing the sequence can immediately retrieve data on isolates sharing one or more mutations with the new sequence.
CITING THE DATABASE
Please refer to this article when citing the HIV RT and Protease Sequence Database.
REFERENCES
- 1.Hirsch M.S., Conway,B., D’Aquila,R.T., Johnson,V.A., Brun-Vezinet,F., Clotet,B., Demeter,L.M., Hammer,S.M., Jacobsen,D.M., Kuritzkes,D.R., Loveday,C., Mellors,J.W., Vella,S. and Richman,D.D. (1998) J. Am. Med. Assoc., 279, 1984–1991. [DOI] [PubMed] [Google Scholar]
- 2.Tantillo C., Ding,J., Jacobo-Molina,A., Nanni,R.G., Boyer,P.L., Hughes,S.H., Pauwels,R., Andries,K., Janssen,P.A. and Arnold,E. (1994) J. Mol. Biol., 243, 369–387. [DOI] [PubMed] [Google Scholar]
- 3.Erickson J.W. and Burt,S.K. (1996) Annu. Rev. Pharmacol. Toxicol., 36, 545–571. [DOI] [PubMed] [Google Scholar]
- 4.Coffin J.M. (1995) Science, 267, 483–489. [DOI] [PubMed] [Google Scholar]
- 5.Korber B., Kuiken,C., Foley,B., Hahn,B., McCutchan,F., Mellors,J.W. and Sodroski,J. (eds) (1998) Human Retroviruses and AIDS 1997: A Compilation and Analysis of Nucleic and Amino Acid Sequences. Theoretical Biology and Biophysics Group. Los Alamos National Laboratory, Los Alamos, NM, pp. II-A-20–II-A-31.
- 6.Hu D.J., Dondero,T.J., Rayfield,M.A., George,J.R., Schochetman,G., Jaffe,H.W., Luo,C.C., Kalish,M.L., Weniger,B.G., Pau,C.P., Schable,C.A. and Curran,J.W. (1996) J. Am. Med. Assoc., 275, 210–216. [PubMed] [Google Scholar]
- 7.Shafer R.W., Winters,M.A., Palmer,S. and Merigan,T.C. (1998) Ann. Intern. Med., 128, 906–911. [DOI] [PubMed] [Google Scholar]
- 8.Learn G.H.J., Korber,B.T., Foley,B., Hahn,B.H., Wolinsky,S.M. and Mullins,J.I. (1996) J. Virol., 70, 5720–5730. [DOI] [PMC free article] [PubMed] [Google Scholar]