Abstract
Beside the oral cavity, bitter taste receptors are expressed in several non-gustatory tissues. Whether extra-oral bitter taste receptors function as sensors for endogenous agonists is unknown. To address this question, we devised functional experiments combined with molecular modeling approaches to investigate human and mouse receptors using a variety of bile acids as candidate agonists. We show that five human and six mouse receptors are responsive to an array of bile acids. Moreover, their activation threshold concentrations match published data of bile acid concentrations in human body fluids, suggesting a putative physiological activation of non-gustatory bitter receptors. We conclude that these receptors could serve as sensors for endogenous bile acid levels. These results also indicate that bitter receptor evolution may not be driven solely by foodstuff or xenobiotic stimuli, but also depend on endogenous ligands. The determined bitter receptor activation profiles of bile acids now enable detailed physiological model studies.
Subject terms: Taste receptors, High-throughput screening
Using a combined experimental-computational approach, the activation profiles of human and mouse bitter taste receptors, TAS2Rs and Tas2rs, by bile acids is described, suggesting a physiological role of bile acids for taste receptors in non-gustatory tissues.
Introduction
The mammalian taste system is generally able to distinguish the five basic taste qualities salty, sour, sweet, umami and bitter1,2. Among the four different cell types housed in taste buds, the type II cells express the G protein-coupled receptors (GPCRs) mediating sweet, umami and bitter taste3. Taste GPCRs are divided into the two subfamilies TAS1Rs and TAS2Rs4. The three TAS1R members, TAS1R1, TAS1R2, and TAS1R3, form the heterodimers TAS1R1/TAS1R3 for the functional umami and TAS1R2/TAS1R3 for the functional sweet taste receptor5–10. In contrast, the group of TAS2Rs consists of ~25 known functional bitter taste receptors in human, but this number varies considerably among species11. Despite the relatively few bitter taste receptors, mammals are able to sense hundreds of bitter tasting compounds12–14. This is ensured by the presence of broadly tuned receptors, which can detect high numbers of chemically diverse agonists12. In humans, the three broadly tuned receptors TAS2R1015,16, TAS2R1417 and TAS2R4614 recognize together more than half of all tested bitter substances12. Narrowly and intermediately tuned bitter taste receptors were shown to have more restricted agonist profiles12. A physiological relevance for the observed differences in tuning breadths remains to be determined.
In general, strong bitter taste is perceived as unpleasant and as many noxious substances are known to taste bitter, it was thought to have its main function in warning mammals of the ingestion of toxic substances18, although no strict correlation between bitterness and toxicity has been observed19,20. Moreover, the expression of bitter taste receptors also in non-gustatory tissues has been confirmed. The detection in tissues like the gastrointestinal tract, the respiratory tract and the heart hints at further biological functions beyond taste21–23. Beside the gastrointestinal tract, where the swallowed food compounds may directly activate the expressed bitter taste receptors, research is ongoing to uncover agonists of extra-oral bitter taste receptors. Activation of TAS2Rs in airway epithelial cells for example, was already shown to induce increase in ciliary beat frequency to speed up mucociliary clearance with bacterial quorum-sensing molecules as suggested agonists22.
It has long been known, that the body fluid bile containing the endogenously produced compound class of bile acids tastes extremely bitter24. In human, bile acids are produced as the primary bile acids cholic acid and chenodeoxycholic acid in the liver from cholesterol as scaffold structure, secreted into the gallbladder after conjugation to taurine or glycine and released into the small intestine in response to food intake as they primarily fulfill nutritional functions, like solubilization of lipophilic food compounds25–27. In the intestinal lumen, they are exposed to the gut microbiota, which further modify them to secondary bile acids28,29. By reabsorption, bile acids are released into the portal venous blood and transported back to the liver to start the circulation again30,31. Only a small proportion of bile acids is excreted by the feces or enters the systemic circulation32.
Besides their role in digestion, bile acids are already known to fulfill further physiological functions by activation of receptors like the GPCR TGR533. The stimulation of this receptor by taurolithocholic acid in human macrophages induces the expression of anti-inflammatory cytokine IL-10 and reduces the expression of proinflammatory cytokines, indicating the importance of bile acids in immune responses34. Therefore, the question arises if also extra-oral bitter taste receptors can mediate biological functions by stimulation with bile acids.
In fact, recent studies demonstrated the bile acid taurocholic acid representing an agonist for bitter taste receptors of mouse, human and bony fish11,13. For bony fish furthermore, chenodeoxycholic acid, deoxycholic acid, glycocholic acid and taurolithocholic acid were shown to activate the T2R02 of Latimeria chalumnae with activation threshold concentration for taurolithocholic acid between 0.3 and 1 µM11. Up to now, there are no data about the activation thresholds for human receptors when stimulated with bile acids, however, to conclude a physiological function for endogenous bile acids activating non-gustatory bitter taste receptors (periodic) supra-threshold concentrations are essential.
The human metabolome database (HMDB) revealed the detection of several endogenously occurring bile acids and provides quantitative data for almost 30 of them in various body fluids35. To determine if they might be relevant for the activation of non-gustatory TAS2Rs, their bitter taste receptor activation profile has to be investigated in more detail. Therefore, we employed a set of eight bile acids that were detected and quantified in the human body35 to perform a complete functional characterization of the activation of human and mouse bitter taste receptors. The chosen bile acids covered a broad range, including primary, secondary and tertiary bile acids.
Results
Screening of human and mouse bitter taste receptors for their activation by bile acids
To gain a comprehensive insight in the activation profiles of human and mouse bitter taste receptors by bile acids, we tested a set of 8 different bile acids for their activation of 25 human (TAS2R1, −R3, −R4, −R5, −R7, −R8, −R9, −R10, −R13, −R14, −R16, −R19, −R20, −R30, −R31, −R38, −R39, −R40, −R41, −R42, −R43, −R45, −R46, −R50, −R60) and 34 mouse bitter taste receptors (Tas2r102, −r103, −r104, −r105, −r106, −107, −r108, −r109, −r110, −r113, −r114, −r115, −r117, −r118, −r119, −r120, −r121, −r122, −r123, −r124, −r125, −r126, −r129, −r130, −r131, −r134, −r135, −r136, −r137, −r138, −r139, −r140, −r143, −r144). The set of bile acids included the primary bile acids cholic and chenodeoxycholic acid, the secondary bile acids lithocholic and deoxycholic acid, the tertiary bile acid ursodeoxycholic acid, as well as the conjugated bile acids taurolithocholic, glycocholic and taurocholic acid (Fig. 1).
Fig. 1. Chemical structures of the investigated bile acids.
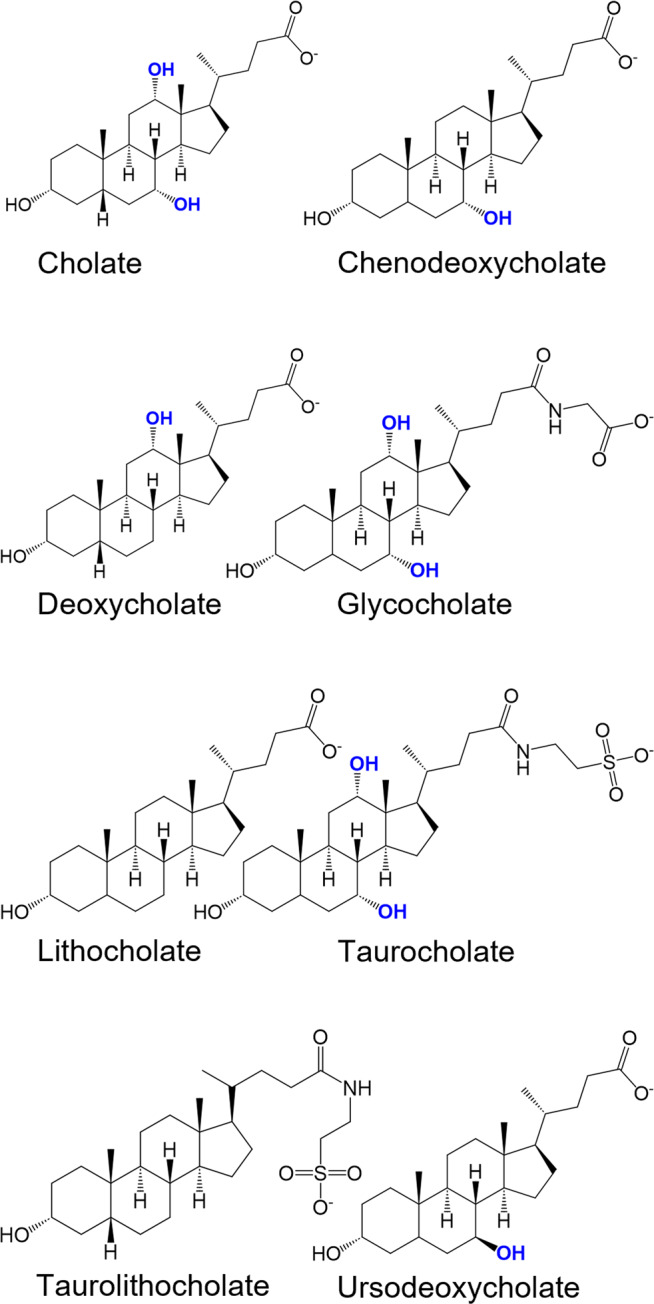
The structural formulas of the eight bile acids cholic acid, chenodeoxycholic acid, deoxycholic acid, glycocholic acid, lithocholic acid, taurocholic acid, thaurolithocholic acid, and ursodeoxycholic acid in their deprotonated form at physiological conditions (pH ~ 7) are presented. Hydroxyl groups at positions 7 and 12 are highlighted in bold and blue. Structures were generated with ChemDraw software.
By performing a screening using Ca2+-imaging assay, we were able to identify 5 human bitter taste receptors responding to bile acids (Fig. 2).
Fig. 2. Human bitter taste receptor responses to bile acids.

Fluorescence traces of HEK293T-Gα16gust44 cells transiently transfected with expression constructs of the human bitter taste receptors TAS2R1 (1), TAS2R4 (4), TAS2R14 (14), TAS2R39 (39), and TAS2R46 (46). Cells were exposed to cholic acid (CA), taurocholic acid (TCA), glycocholic acid (GCA), deoxycholic acid (DCA), lithocholic acid (LCA), taurolithocholic acid (TLCA), chenodeoxycholic acid (CDCA) and ursodeoxycholic acid (UDCA). Fluorescence changes were measured with an automated fluorometric imaging plate reader (FLIPRTETRA). Fluorescence traces are negative control corrected. Applied bile acid concentrations are given in µM in brackets. Only traces of responsive receptors are shown. A scale bar is provided at the bottom right.
The human bitter taste receptors TAS2R1, TAS2R4, TAS2R14, TAS2R39 and TAS2R46 responded to at least three of the tested bile acids. Of those, TAS2R1 was the least selective as it was stimulated by all eight bile acids. Additionally, TAS2R4 was activated by 6, TAS2R14 by 5, and both TAS2R39 and TAS2R46 by 3 of the bile acids.
To assess the potential phylogenetic conservation of the responses observed for the human receptors, we performed the identical screening procedure using mouse Tas2rs (Fig. 3).
Fig. 3. Mouse bitter taste receptor responses to bile acids.
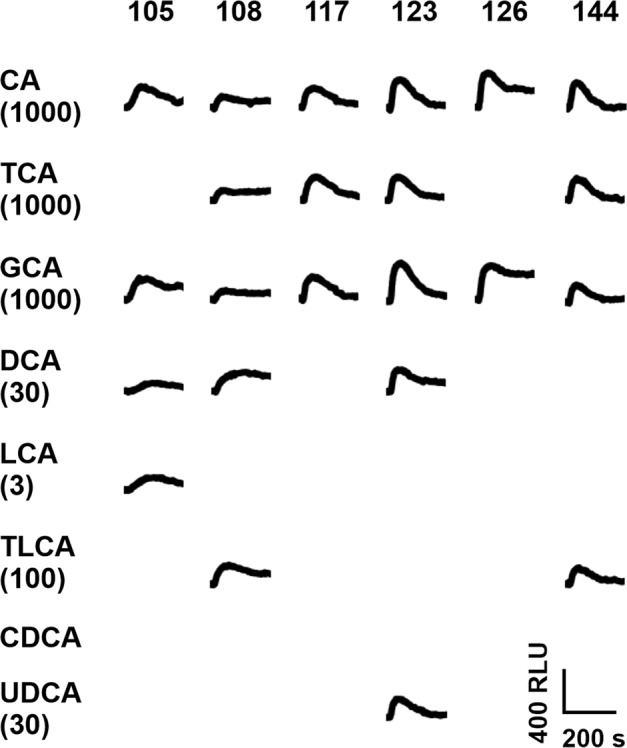
Fluorescence traces of HEK293T-Gα16gust44 cells transiently transfected with expression constructs of the mouse bitter taste receptors Tas2r105 (105), Tas2r108 (108), Tas2r117 (117), Tas2r123 (123), Tas2r126 (126), and Tas2r144 (144). Cells were exposed to cholic acid (CA), taurocholic acid (TCA), glycocholic acid (GCA), deoxycholic acid (DCA), lithocholic acid (LCA), taurolithocholic acid (TLCA), chenodeoxycholic acid (CDCA) and ursodeoxycholic acid (UDCA). Fluorescence changes were measured with an automated fluorescence plate reader (FLIPRTETRA). Fluorescence traces are negative control corrected. Applied bile acid concentrations are given in µM in brackets. Only traces of responsive receptors are shown. A scale bar is provided at the bottom right.
For mouse bitter taste receptors Tas2r105, Tas2r108, Tas2r117, Tas2r123, Tas2r126 and Tas2r144 responses to bile acids were observed. Compared to the human receptors, none of them responded to all tested bile acids and CDCA did not activate any of the tested mouse Tas2rs.
Establishment of dose-response relationships with activated bitter taste receptors
To elucidate the potency of bile acids to activate human bitter taste receptors and thus get a first hint if endogenous bile acid concentrations could suffice to activate bitter taste receptors, dose-response relationships were established (Fig. 4, Supplementary Figures 1–4). For this purpose, different concentrations of the bile acids were applied to the cells expressing the receptors. The lowest concentration eliciting a significantly increased fluorescence response (p < 0.01) compared to the empty vector control was defined as activation threshold concentration (Table 1). In case receptor saturation was reached with the highest applied bile acid concentration, we further calculated the EC50-value (Supplementary Table 1).
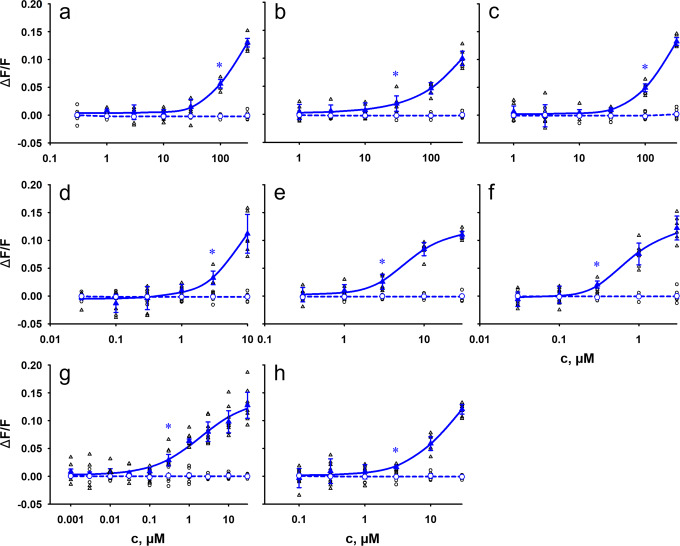
Fig. 4. Concentration-response relationships of eight tested bile acids with human TAS2R1.
HEK293T-Gα16gust44 cells were transiently transfected with human TAS2R1 (triangle, blue) and an empty vector control (circle, blue). Individual data points are depicted accordingly by black symbols. Receptor activation was recorded by increasing fluorescence intensities upon Ca2+ - release using an automated fluorometric imaging plate reader (FLIPRTETRA). For dose-response relationships, increasing concentrations of the bile acids cholic acid a), taurocholic acid b), glycocholic acid c), chenodeoxycholic acid d), deoxycholic acid e), lithocholic acid f), taurolithocholic acid g) and ursodeoxycholic acid h) were applied. The relative fluorescence intensities were mock subtracted and plotted against the bile acid concentration in µM (n = 3 biologically independent experiments). Data are presented as the mean ± standard deviation (STD). Beginning statistical significance (p < 0.01) is indicated by (*).
Table 1.
Activation threshold concentrations of bile acids with human bitter taste receptors.
| Cblood | Max c | TAS2R1 | TAS2R4 | TAS2R14 | TAS2R39 | TAS2R46 | |
|---|---|---|---|---|---|---|---|
| Cholic Acid | 0.1−1.761, 62 | 1000 | 100 | 30 | 300 | 300 | |
| Chenodeoxycholic Acid | 0.2−1.861, 62 | 30 | 3 | 30 | |||
| Lithocholic Acid | 0.08−0.3355, 62 | 3 | 0.3 | ||||
| Deoxycholic Acid | 0.33−0.5755, 63 | 30 | 3 | 3 | |||
| Taurocholic Acid | 0.1−0.3861, 63 | 1000 | 100 | 100 | 300 | 300 | 1000 |
| Glycocholic Acid | 0.6−0.8861, 64 | 1000 | 100 | 30 | 100 | 300 | 1000 |
| Taurolithocholic Acid | 0.61−1.8165 | 100 | 0.3 | 1 | 0.3 | 0.3 | |
| Ursodeoxycholic Acid | 0.1655 | 30 | 3 | 3 |
Presentation of TAS2Rs that were activated by bile acids. Determined threshold concentrations (p < 0.01) for receptor activation and maximum applied bile acid concentrations (Max c) are given in µM. Published bile acid concentration ranges in human blood (cblood;in µM) as summarized in the human metabolome database are listed.
In doing so, the secondary bile acid lithocholic acid and its taurine-conjugated form taurolithocholic acid were identified as the most potent bile acids. For lithocholic acid, the determined threshold concentration for the activation of the TAS2R1 was 0.3 µM. Same threshold values were detected for the activation of TAS2R1, TAS2R14, and TAS2R46 by taurolithocholic acid. For TAS2R1, also the EC50-values of these two bile acids were in the high nanomolar and low micromolar range, respectively (Supplementary Table 1). In total, the activation threshold concentrations of all tested bile acids varied considerably and ranged between high nanomolar values for the mentioned bile acids and low millimolar values for the activation of TAS2R46 by taurocholic acid and glycocholic acid (Table 1).
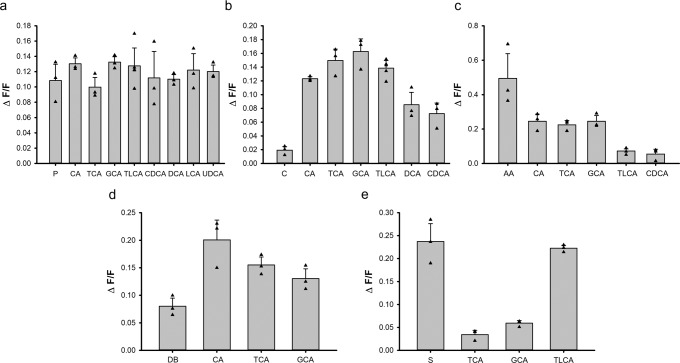
To evaluate the agonistic efficacy of bile acids for human TAS2Rs, we compared the signal amplitudes of well-known TAS2R agonists with bile acids activating TAS2R1, TAS2R4, TAS2R14, TAS2R39 and TAS2R46 (Fig. 5).
Fig. 5. Comparison of the efficacies of bile acids with prototypical TAS2R agonists.
Human bitter taste receptors TAS2R1 a), TAS2R4 b), TAS2R14 c), TAS2R39 d), and TAS2R46 e) activated by highest applied bile acid concentrations (cholic acid (CA), taurocholic acid (TCA), glycocholic acid (GCA), taurolithocholic acid (TLCA), chenodeoxycholic acid (CDCA), deoxycholic acid (DCA), lithocholic acid (LCA), ursodeoxycholic acid (UDCA)) are presented (n = 3). For comparison, maximal signal amplitudes (ΔF/F) obtained with control stimuli of the corresponding TAS2Rs were added. The control stimuli were: 1 mM picrotoxinin (P) for TAS2R1, 3 mM colchicine (C) for TAS2R4, 10 µM aristolochic acid (AA) for TAS2R14, 3 mM denatonium benzoate (DB) for TAS2R39 and 10 µM strychnine (S) for TAS2R46. Data are presented as the mean ± standard deviation (STD). Individual data points are depicted by black triangles.
In case of TAS2R1, measured efficacies of all 8 bile acids were comparable with the control stimulus picrotoxinin. For TAS2R14, the agonist aristolochic acid elicited responses about twice as high as the most effective bile acids and the response of TAS2R46 to taurolithocholic acid is about equal to that obtained for strychnine. Bile acids responses of TAS2R4 and TAS2R39 were considerably stronger than for the control stimuli colchicine and denatonium benzoate, respectively.
The same workflow for the evaluation of potencies was applied for the 34 mouse Tas2rs. Again, we calculated activation threshold concentrations (Table 2) and generate dose-response relationships (Fig. 6, Supplementary Figures 5–10).
Table 2.
Activation of mouse Tas2rs by bile acids.
| Max c (in µM) | Tas2r105 | Tas2r108 | Tas2r117 | Tas2r123 | Tas2r126 | Tas2r144 | |
|---|---|---|---|---|---|---|---|
| Cholic Acid | 1000 | 300 | 100 | 10 | 10 | 100 | 100 |
| Chenodeoxycholic Acid | 30 | ||||||
| Lithocholic Acid | 3 | 3 | |||||
| Deoxycholic Acid | 30 | 10 | 10 | 10 | |||
| Taurocholic Acid | 1000 | 100 | 30 | 100 | 300 | ||
| Glycocholic Acid | 1000 | 300 | 100 | 3 | 30 | 100 | 100 |
| Taurolithocholic Acid | 100 | 1 | 3 | ||||
| Ursodeoxycholic Acid | 30 | 10 |
Presentation of the mouse Tas2rs that were activated by bile acids. Determined threshold concentrations (p < 0.01) for receptor activation and maximum applied bile acid concentrations (Max c) are given in µM.
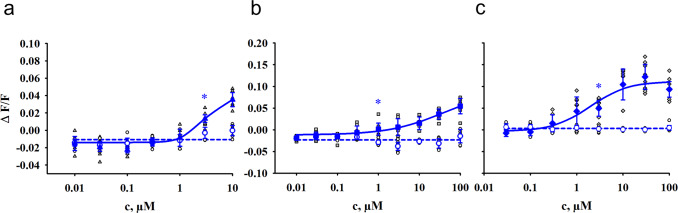
Fig. 6. Concentration-response relationships for the activation of mouse Tas2rs.
HEK293T-Gα16gust44 cells were transiently transfected with the murine Tas2r105 (triangle, blue), Tas2r108 (square, blue) or Tas2r144 (diamond, blue) and an empty vector control (circle, blue). Individual data points are depicted accordingly by black symbols. Receptor activation was recorded by increasing fluorescence intensities upon Ca2+ - release using an automated fluorometric imaging plate reader (FLIPRTETRA). For dose-response relationships, increasing concentrations of the bile acids lithocholic acid a) and taurolithocholic acid b) and c) were applied. The relative fluorescence intensities were mock subtracted and plotted against the bile acid concentration in µM (n = 3 biologically independent experiments). Data are presented as the mean ± standard deviation (STD). Beginning statistical significance (p < 0.01) is indicated by (*).
As it was already shown for the human receptors, lithocholic acid and taurolithocholic acid are also the most potent bile acids for mouse Tas2rs (Table 2).
By generating dose-response relationships, the threshold concentration for the activation of the Tas2r108 by taurolithocholic acid was determined as 1 µM. With 3 µM the activation threshold concentration of lithocholic acid for Tas2r105 and taurolithocholic acid for Tas2r144 were in a similar range (Fig. 6). Compared to the mouse bitter taste receptors, the human receptors are more sensitive for these two bile acids.
Predicted binding modes of bile acids within the TAS2R1 binding site
The 3D structure of TAS2R1 was obtained with homology modeling using the recently solved structure of TAS2R4636 as a template (sequence identity = 27%). Interestingly, the first solved structures of TAS2R46 suggest a high flexibility of the EC loops, specifically of the ECL2 domain, which is not resolved in the bound state conformation of the receptor, and for which the folding obtained in the unbound states overlaps with the ligand position. The ECL2 connects transmembrane helices 4 and 5 and is diverse in length and composition in currently solved GPCRs37. It was demonstrated that docking performance could be insensitive to or even improved by excluding ECL2 from the calculations16,38,39 Therefore, because of the uncertainness of the ECL2 folding, we modelled TAS2R1 without the ECL2.
To predict the binding modes of bile acids within the orthosteric TAS2R1 binding site, we ran molecular docking simulations, and generated thirty different poses for each compound. Among all poses, we selected a consensus binding mode, namely the most frequent pose observed for all ligands that is also the best pose for lithocholic acid. The ligands insert into the orthosteric binding site by anchoring to TM3 and TM5. These poses have docking scores that correlate well with activation thresholds, but to further optimize ligand-receptor interactions, the poses were rescored with MM/GBSA minimization. The resulting binding modes were not affected but the scoring improved even more (Table 3, Supplementary Figures 11 and 12), suggesting that the model can capture key ligand-receptor interactions and supporting the assumption that bile acids bind to the orthosteric binding site.
Table 3.
Docking and MM/GBSA scores of analyzed bile acids within the TAS2R1 binding site.
| Activation thresholds [µM] | Docking scores [kcal/mol] | MM/GBSA dG Bind [kcal/mol] | |
|---|---|---|---|
| Taurolithocholic Acid | 0.30 | −5.61 | −86.45 |
| Lithocholic Acid | 0.30 | −5.28 | −70.11 |
| Chenodeoxycholic Acid | 3.00 | −5.08 | −69.75 |
| Ursodeoxycholic Acid | 3.00 | −5.78 | −69.60 |
| Deoxycholic Acid | 3.00 | −5.43 | −68.36 |
| Cholic Acid | 100.00 | −4.57 | −65.75 |
| Glycocholic Acid | 100.00 | −3.36 | −65.27 |
| Taurocholic Acid | 100.00 | −3.33 | −48.76 |
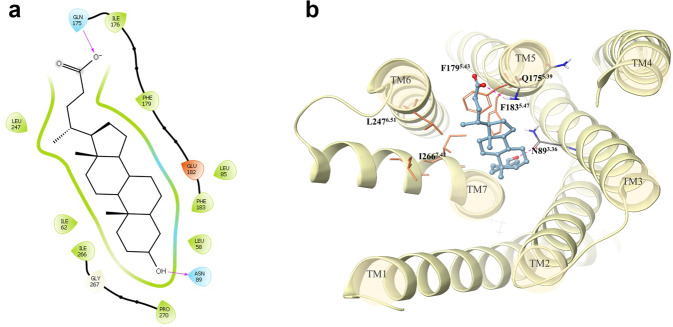
In Fig. 7, we show the 2D and 3D representations of the binding mode of lithocholic acid within TAS2R1. The ligand forms hydrogen bonds with the side chain of N893.36 and the main chain of Q1755.39 and it is accommodated in a hydrophobic cavity generated by F1795.43, F1835.47, L2476.51 and I2667.42.
Fig. 7. 2D and 3D representations of the putative binding mode of lithocholic acid in the TAS2R1 binding site obtained with MM/GBSA refinement.
The 2D plot a) was generated using the Ligand Interaction Diagram tool available in Maestro (Schrödinger Release 2022-3) showing residues at 4 Å from the ligand. In the 3D representation b), the ligand is shown as blue ball&stick, polar residues in CPK-colored sticks and hydrophobic residues as orange sticks. Hydrogen bonds are shown as dashed magenta lines in both representations.
The binding modes of all other ligands are reported in Supplementary Figure 11. Lithocholic, chenodeoxycholic, ursodeoxycholic, deoxycholic and cholic acids differ mostly in the presence/absence of hydroxyl groups in position 7 and 12. Interestingly, we found that these hydroxyl groups do not affect the pose of binding but point to F1795.43 and F1835.47, affecting the hydrophobic interactions observed for lithocholic acid (Fig. 7). Taurolithocholic acid, with a similar activity threshold to lithocholic acid, also misses the hydroxyl groups in positions 7 and 12, suggesting that hydrophobic complementarity is important for receptor binding and activation. Glycocholic and taurocholic acids are the ligands that differ most in the binding mode. The methyl groups of glycocholic acid push the ligand higher in the binding site, we lose the interaction with N893.36, and the ligand anchors instead to Y9 in TM1.
Discussion
The detection of bitter taste receptors in tissues beside the gustatory system has resulted in an increased interest in the biological role(s) of these receptors in non-gustatory tissues and in the nature and putative origins of the bitter substances that activate the receptors outside the oral cavity. Therefore, one hypothesis is the existence of endogenous agonists40 and previous studies already confirmed the activation of human, mouse and bony fish bitter taste receptors by bile acids11,13. In general, the group of bile acids is a very complex compound class. As they are released into the small intestine upon food uptake, they are exposed to the gut microbiota. Thereby bile acids are modified and finally a mixture of hundreds of different bile acids is present28,29. To get a deeper look into the activation of bitter taste receptors by bile acids, we investigated a diverse group of bile acids, including primary, secondary, and tertiary bile acids. For human and mouse receptors activated by taurocholic acid, we could confirm previously published data according to activation threshold concentrations and EC50-values in this study13. Besides, we further identified the human TAS2R1, TAS2R14, TAS2R39 and TAS2R46, as well as mouse Tas2r108 as receptors for taurocholic acid. The different outcomes of both studies may occur due to the rather high activation threshold concentrations of this bile acid (30–1000 µM), which is near the highest applied concentration (1000 µM), whereby a weak activation could have been missed in the previous study in which the concentration used for the screening was limited to 300 µM. Moreover, all newly identified taurocholic acid responsive receptors (mouse Tas2r108, human TAS2R1, −R14, −R39, and –R46) exhibited quite high threshold concentrations of 100–1000 µM and, in case of Tas2r108 and TAS2R1, also low signal amplitudes (cf. Figures 2 and 3). For mouse Tas2r105, the lack of taurocholic acid responsiveness reported by Lossow et al.13. has been confirmed, although other, previously not tested bile acids, were able to elicit responses of this receptor.
We further compared the efficacies of bile acids with cognate agonists of the identified human TAS2Rs to evaluate the relative strength of TAS2R activation by bile acids. The results demonstrated that for TAS2R1, bile acid responses are on a similar level with the agonist picrotoxinin indicating that bile acids represent full agonists of this receptor. In contrast, the bile acid agonists of TAS2R14 were not able to trigger a response similar to aristolochic acid, which is one of the most efficient agonists for this receptor. Therefore, the tested bile acids represent only partial agonists of TAS2R14. Responses to control stimuli of TAS2R4 and TAS2R39 were found to be less effective than the activating bile acid agonists, indicating that those may represent full agonists for both receptors. As indicated already by the determined dose-response relationships, the TAS2R46 seems to be a receptor specialized to detect distinct bile acids. Beside the differences in activation threshold concentrations, also the efficacy of TLCA is significantly higher than that of TCA and GCA and comparable to the control stimulus strychnine. Hence, TCLA can be judged as another full agonist of this receptor.
As there are several evidences for the functional conservation of bitter taste receptors between species like the metal ion response of human and vampire bats or the overlapping agonist profiles of coelacanth and zebrafish T2R1, we further investigated mouse bitter taste receptors for their bile acid response11,41,42. To compare responses of one-to-one orthologous receptors of both species, we consulted the phylogenetic tree that was generated in a former study13. Here, we identified the two receptors TAS2R1 and TAS2R39 responding to bile acids, but their corresponding orthologues Tas2r119 and Tas2r139 do not. In contrast, the orthologue of TAS2R4, called Tas2r108 is activated by bile acids. They share common agonists among the tested bile acids, but there are also significant differences. The human receptor is very sensitive to ursodeoxycholic acid with an activation threshold concentration of 3 µM, whereas the mouse receptor does not respond (Table 1, Table 2). We have analyzed the sequences of the orthologous receptors TAS2R4 and Tas2r108. Among the considerable number of differences between the two receptors (98 positions (~33%) are not identical, see Supplementary Figures 13 – 15), only few minor differences occur in positions which have been demonstrated previously to be important for agonist binding (Supplementary Figures 14 and 15). As ursodeoxycholic acid is the only compound in which the C7 hydroxylgroup is positioned above the plane of ring B, we speculate that sterical hindrance between Tas2r108 residues and the C7 hydroxylgroup could be responsible for the lack of activation.
For the remaining responding receptors, no one-to-one orthologues were identified as they are organized in species-specific gene expansion groups. Therefore, we can confirm that a general conservation of the functionality of orthologous receptors between mice and human does not exist, as it was already proposed elsewhere13. We did not obtain any response of mouse receptors to CDCA, whereas human TAS2R1 and TAS2R14 were activated by this bile acid. It is known that mice possess an enzyme called Cyp2c70 in the liver, which is converting CDCA into muricholic acid (MCA)43. This keeps CDCA levels low, and hence might be the reason why mouse bitter taste receptors did not exhibit responsiveness to CDCA. Whether mouse Tas2rs instead are more specialized to the mouse-specific MCAs has to be clarified.
Furthermore, we demonstrated that the secondary bile acid lithocholic acid and its taurine conjugated form taurolithocholic acid are the most potent tested bile acids in activating human as well as mouse bitter taste receptors. These bile acids are activating the already known bile acid receptor TGR5 with EC50 values of 600 nM for lithocholic and 300 nM for taurolithocholic acid44. For the human TAS2R1 we measured EC50 values of 900 nM and 1.9 µM, respectively, concluding the TGR5 receptor to be slightly more sensitive to lithocholic acid and taurolithocholic acid. Related to these data, it can be assumed that the activation of bitter taste receptors by bile acids is only of biological relevance in tissues, cells or at subcellular localizations where the TGR5 is not expressed, respectively if bile acid concentration is increasing, the activation of bitter taste receptors might be additive to the TGR5 signal. According to literature, there are some tissues with overlapping TGR5 and TAS2R expression profiles, including the small intestine and the testis45,46. In particular in testis, TAS2R1 mRNA was detected in late spermatids at quite high levels, whereas TAS2R4 and TAS2R14 mRNAs were found at lower levels in several cell types of the small (TAS2R4: enteroendocrine cells, Paneth cells, goblet cells, enterocytes; TAS2R14: enterocytes) and large (TAS2R4: undifferentiated cells, enterocytes, goblet cells, enteroendocrine cells; TAS2R14: T-cells, enterocytes, undifferentiated cells, goblet cells, enteroendocrine cells) intestine47,(https://www.proteinatlas.org/search/TAS2R). It is a matter of future research to elucidate the function of TAS2Rs in these tissues and conclude the interplay between both receptor types. Furthermore, we only tested a subset of the variety of all bile acids. Therefore, we might have missed the best bile acid agonist for human bitter taste receptors, which is more potent to TAS2Rs than to TGR5. However, there are differences in published EC50-values of TGR5 activation by bile acids observed and a recent study reported EC50-concentrations of 20 µM for LCA and 2.3 µM for TLCA48. These results suggest, that occasionally TAS2Rs can be more sensitive to bile acids and fulfill their function independent from TGR5 or that the additive effect is mediated by TGR5.
To answer the question of potential biological relevance, we compared if the quantified concentrations of bile acids in human body fluids listed in the Human Metabolome Database35 match with our measured data. As already expected, concentrations in bile are in the millimolar range, for which reason a biological relevance of bitter taste receptors in tissues like the gallbladder, the liver or the small intestine is questionable as these concentrations would lead to a permanent activation of the receptors. A previous study, which showed the absence of the known bile acid bitter taste receptors Tas2r117, Tas2r123 and Tas2r144 in mouse small intestine therefore concluded that the absence of the bile acid-sensitive Tas2rs is due to the fact that such receptors are useless if physiological bile acid concentrations exceed threshold concentrations at all times and hence would constantly signal or remain constantly desensitized49. As already mentioned, we were able to identify the Tas2r105, the Tas2r108 and the Tas2r126 as further bile acid bitter taste receptors. Published expression data reveals high intestinal expression levels of the Tas2r108 and the Tas2r12649. For human TAS2R expression in the small intestine, it is reported that TAS2R4, TAS2R14, TAS2R39 and TAS2R46, which are all responding to bile acids, are expressed in jejunal crypts. In this context, some functions of these receptors in the small intestine are suggested. The activation of TAS2R4 by taurocholic acid was reported to increase the release of molecules that have a positive impact on E. coli growth50. Therefore, the ingestion of food, which results in the release of bile acids into the small intestine may have positive effects on E. coli growth and consequently for the process of digestion. Furthermore, activation of TAS2R14 in a human colorectal cancer cell line is supposed to result in increased GDF15 levels, which is involved in several biological functions like anti-inflammatory and apoptotic pathways50–52. To clarify, if these receptors play a role in bile acid detection in the small intestine, further research is necessary.
Beside high concentrations in bile, blood serum bile acid levels increase from 0.2 – 0.7 µM to 4 – 5 µM postprandial53,54 and for lithocholic acid, which is one of the most potent identified bile acids, a serum concentration of 0.33 µM was measured previously in healthy children subjects55. As blood is the main transporting unit in the body, this bile acid will be distributed in concentrations that were shown to be sufficient to activate the human TAS2R1, which is highly expressed in the human brain and testis46, in particular in late spermatids 47,(https://www.proteinatlas.org/search/TAS2R). It was further shown that the lithocholic acid serum concentration is decreased in children with cystic fibrosis and it is known that men with cystic fibrosis go later through puberty than healthy subjects55,56. As brain and testis are important players in puberty a role of bile acids in development from child to adult is conceivable, but further research is required.
Interestingly, the potency of activation of the human TAS2R1 seemed to depend a lot on the presence of hydroxyl groups at positions 7 and 12 of the steroid scaffold structure in our experiments. Docking simulations of bile acids investigated in this paper highlighted the main interactions established with TAS2R1. We found that hydroxyl groups at positions 7 and 12 can affect hydrophobic interactions between the ligands and two hydrophobic patches in the receptor binding site: one made by Leu853.32, Phe1835.47, and Phe1795.43 and the other one made by Ile2667.42 and Leu2476.51. We also suggest that the H-bond between the ligands and N893.36 is highly important for the ligand-receptor interaction, and it is supposed to be a key interaction for receptor selectivity. In fact, position 3.36 is highly conserved among the investigated TAS2Rs, but the residue in the close position 3.32 can influence the access to this interaction. In TAS2R1, a leucine occupies this position, but we have bulkier residues (F or W) in TAS2R4, TAS2R14, TAS2R39 and TAS2R46. This difference causes a change of pose in other receptors (we report the predicted binding mode for taurolithocholic acid within the TAS2R46 binding site in Supplementary Figure 16).
In conclusion, we were able to show the activation profile of human and mouse bitter taste receptors by bile acids. We identified five human and six mouse receptors, which are responsive to subsets of tested bile acids. Comparing the determined activation threshold concentrations with physiological bile acid concentrations in the human body, this compound class is very promising as endogenous agonists of bitter taste receptors. The comparative investigation of primary cell lines, intestinal organoids, or mouse models derived from TGR5-knockout57 and wildtype mice can provide further insights into the activation mechanism and downstream signaling of bitter taste receptors activated by bile acids. It is a future task to experimentally clarify, the exact biological functions of bitter taste receptor activation by bile acids.
Methods
Bile acids
Functional characterization of 25 human12 and 34 out of 35 mouse bitter taste receptors13 was performed using a set of 8 different bile acids, including the primary bile acids cholic (Calbiochem, San Diego, United States) and chenodeoxycholic acid (Sigma-Aldrich, Steinheim, Germany), the secondary bile acids lithocholic (AcrosOrganics, Geel, Belgium) and deoxycholic acid (Sigma-Aldrich, Steinheim, Germany), the conjugated bile acids taurocholic (Biochemika), taurolithocholic (Sigma-Aldrich, Steinheim, Germany), glycocholic acid (Sigma-Aldrich, Steinheim, Germany), as well as the tertiary bile acid ursodeoxycholic acid (Alfa Aesar, Kandel, Germany). This set of bile acids was chosen because of their commercial availability in high purities, their diversity within the class of bile acids and their previous detection and quantitation in human blood. Stock solutions were prepared in DMSO. For the prevention of unspecific cellular responses, the stock solutions are diluted in the assay buffer C1 (130 mM NaCl, 10 mM HEPES pH 7.4, 5 mM KCl, 2 mM CaCl2, 0.18 % glucose) to reduce the DMSO concentration to a maximum of 0.5 % in the final experiments. Maximal applied concentrations are due to solubility problems or receptor-independent artefacts during measurement at high bile acid concentrations (Table 1, Table 2)12,58.
Cell lines
As basal growth medium for the HEK293T-Gα16gust44 cell line17,59 served Dulbecco’s modified eagle medium (DMEM) (Thermo Fisher Scientific, Darmstadt, Germany) supplemented with 10 % fetal bovine serum (Sigma-Aldrich, Steinheim, Germany), 2 mM L-glutamine (Sigma-Aldrich, Steinheim, Germany), 100 units/ml penicillin (Sigma Aldrich, Steinheim, Germany) and 100 µg/ml streptomycin (Sigma Aldrich, Steinheim, Germany). Growth conditions were 37 °C, 5 % CO2 and saturated air-humidity60.
Transient transfection
HEK293T-Gα16gust44 cells were seeded on poly-D-lysine (10 µg/ml) coated 96-well plates to reach a confluence of 40-60 % the next day. For transient transfection 150 ng of plasmid DNA containing the receptor of interest and 0.3 µl lipofectamine 2000 (Thermo Fisher Scientific, Darmstadt, Germany) per well were used and transfection took place according to the manual of lipofectamine 2000. Mouse Tas2r116 was excluded because successful cloning was not possible in a previous work13. The empty vector DNA (mock) was transfected as negative control14,41.
Calcium imaging assay
Cells were loaded using the calcium-sensitive fluorescent dye Fluo-4-AM (Abcam, Cambridge, Great Britain) in the presence of 2.5 mM probenecid (Sigma-Aldrich, Steinheim, Germany) the day after transfection13,41. 1 h after loading, cells were washed with C1 buffer using a BioTek Cell Washer, incubated in the dark for half an hour and washed again. For automated agonist application and measurement of fluorescence changes, a FLIPRTETRA device (Molecular Devices, San José, United States) was used. Viability of cells was tested by application of 100 nM Somatostatin 14 (Bachem, Bubendorf, Switzerland)42.
Data analysis
Measured data were negative control corrected by subtracting the signal of the mock-transfected cells and exported to Microsoft Excel using the FLIPR software ScreenWorks 4.2. In Microsoft Excel software, standardization of maximum fluorescence intensities to the basal fluorescence and normalization to the buffer-only control was done to calculate the relative fluorescence changes (ΔF/F).
Statistics and reproducibility
Initial screening experiments performed in duplicate wells were confirmed by at least one replication and representative traces were selected for display. All dose-response relationships were determined by three independent experiments (biological replicates) performed in duplicates (technical replicates). Threshold concentrations, defined as lowest substance concentrations leading to statistically significant elevated fluorescence changes in receptor-transfected cells compared with empty vector-transfected cells, were determined using SigmaPlot with Student’s t-test to evaluate statistical significance (p < 0.01).
Molecular modeling
2D structures of bile acids investigated in this work were downloaded from PubChem. Ligprep (Schrödinger Release 2022-3: LigPrep, Schrödinger, LLC, New York, NY, 2022) was used to generate 3D structures and protonation states of all ligands at pH 7 ± 1.
The currently released receptor structure of TAS2R46 (PDB ID: 7XP6) was used as a template for modeling the structures of TAS2R1, -R4, -R14, and -R39 using Prime (Schrödinger Release 2022-3). The sequence identities between TAS2R1, -R4, -R14, and –R39 and the template are 27%, 24%, 43%, and 25%, respectively. All models are available at https://github.com/dipizio/TAS2R-models.
Glide Standard Precision (Schrödinger Release 2022-3) was used for docking studies on TAS2R1. The receptor binding site was prepared using the “Receptor Grid Generation” tool, the grid box was the centroid of the ligand in the experimental structure of TAS2R46. We saved 30 poses per ligand. The docking pose of lithocholic acid with the lowest Glide score was used as a selection filter for the docking poses of all bile acids. MM/GBSA minimization (Prime, Schrödinger, LLC, New York, NY, USA, 2022) was used to rescore the poses. The same procedure was applied to predict the binding mode of taurolithocholic acid within the TAS2R46 binding site. The 2D and 3D representations of lithocholic acid/TAS2R1 binding mode were generated with Maestro 13.2 (Schrödinger Release 2022-3).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
We thank Catherine Delaporte and Eva Boden for excellent technical assistance.
Author contributions
M.B. and F.Z. contributed to the study conception and design. F.Z. performed experimental work. M.B. and F.Z. analyzed the data. A.S. and A.D.P. performed and analyzed modeling studies. All authors contributed to the written manuscript. All authors have read and agreed to the published version of the manuscript.
Peer review
Peer review information
Communications Biology thanks Simon Foster, Hiroo Imai and the other, anonymous, reviewer for their contribution to the peer review of this work. Primary Handling Editors: Ross Bathgate and Joao Valente.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Data availability
All data of this study are provided in the main text and supplementary information. Source data for graphs shown in Figs. 2 to 6 are provided as Supplementary data files (Supplementary data 1 for Figs. 2 and 3; Supplementary data 2 for Figs. 4 and 6; Supplementary data 3 for Fig. 5).
Code availability
Receptor models and docking results are available at https://github.com/dipizio/TAS2R-models.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s42003-023-04971-3.
References
- 1.Yarmolinsky DA, Zuker CS, Ryba NJ. Common sense about taste: from mammals to insects. Cell. 2009;139:234–244. doi: 10.1016/j.cell.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kinnamon SC, Finger TE. Recent advances in taste transduction and signaling. F1000Res. 2019;8:F1000. doi: 10.12688/f1000research.21099.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang Y, et al. Coding of sweet, bitter, and umami tastes: different receptor cells sharing similar signaling pathways. Cell. 2003;112:293–301. doi: 10.1016/S0092-8674(03)00071-0. [DOI] [PubMed] [Google Scholar]
- 4.Behrens, M. & Meyerhof, W. in Chemosensory Transduction (eds Frank Zufall & S. D. Munger) 227–244 (Academic Press, 2016).
- 5.Hoon MA, et al. Putative mammalian taste receptors: a class of taste-specific GPCRs with distinct topographic selectivity. Cell. 1999;96:541–551. doi: 10.1016/S0092-8674(00)80658-3. [DOI] [PubMed] [Google Scholar]
- 6.Nelson G, et al. Mammalian sweet taste receptors. Cell. 2001;106:381–390. doi: 10.1016/S0092-8674(01)00451-2. [DOI] [PubMed] [Google Scholar]
- 7.Li X, et al. Human receptors for sweet and umami taste. Proc. Natl. Acad. Sci. 2002;99:4692. doi: 10.1073/pnas.072090199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nelson G, et al. An amino-acid taste receptor. Nature. 2002;416:199–202. doi: 10.1038/nature726. [DOI] [PubMed] [Google Scholar]
- 9.Montmayeur JP, Liberles SD, Matsunami H, Buck LB. A candidate taste receptor gene near a sweet taste locus. Nat. Neurosci. 2001;4:492–498. doi: 10.1038/87440. [DOI] [PubMed] [Google Scholar]
- 10.Max M, et al. Tas1r3, encoding a new candidate taste receptor, is allelic to the sweet responsiveness locus Sac. Nat. Genet. 2001;28:58–63. doi: 10.1038/ng0501-58. [DOI] [PubMed] [Google Scholar]
- 11.Behrens M, Di Pizio A, Redel U, Meyerhof W, Korsching SI. At the Root of T2R Gene Evolution: Recognition Profiles of Coelacanth and Zebrafish Bitter Receptors. Genome Biol. Evol. 2021;13:evaa264. doi: 10.1093/gbe/evaa264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meyerhof W, et al. The molecular receptive ranges of human TAS2R bitter taste receptors. Chem. Senses. 2010;35:157–170. doi: 10.1093/chemse/bjp092. [DOI] [PubMed] [Google Scholar]
- 13.Lossow K, et al. Comprehensive Analysis of Mouse Bitter Taste Receptors Reveals Different Molecular Receptive Ranges for Orthologous Receptors in Mice and Humans. J. Biol. Chem. 2016;291:15358–15377. doi: 10.1074/jbc.M116.718544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brockhoff A, Behrens M, Massarotti A, Appendino G, Meyerhof W. Broad tuning of the human bitter taste receptor hTAS2R46 to various sesquiterpene lactones, clerodane and labdane diterpenoids, strychnine, and denatonium. J. Agric. Food Chem. 2007;55:6236–6243. doi: 10.1021/jf070503p. [DOI] [PubMed] [Google Scholar]
- 15.Bufe B, Hofmann T, Krautwurst D, Raguse JD, Meyerhof W. The human TAS2R16 receptor mediates bitter taste in response to beta-glucopyranosides. Nat. Genet. 2002;32:397–401. doi: 10.1038/ng1014. [DOI] [PubMed] [Google Scholar]
- 16.Born S, Levit A, Niv MY, Meyerhof W, Behrens M. The human bitter taste receptor TAS2R10 is tailored to accommodate numerous diverse ligands. J. Neurosci. 2013;33:201–213. doi: 10.1523/JNEUROSCI.3248-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Behrens M, et al. The human taste receptor hTAS2R14 responds to a variety of different bitter compounds. Biochem. Biophys. Res. Commun. 2004;319:479–485. doi: 10.1016/j.bbrc.2004.05.019. [DOI] [PubMed] [Google Scholar]
- 18.Lindemann B. Taste reception. Physiol. Rev. 1996;76:719–766. doi: 10.1152/physrev.1996.76.3.719. [DOI] [PubMed] [Google Scholar]
- 19.Glendinning JI. Is the bitter rejection response always adaptive? Physiol. Behav. 1994;56:1217–1227. doi: 10.1016/0031-9384(94)90369-7. [DOI] [PubMed] [Google Scholar]
- 20.Nissim I, Dagan-Wiener A, Niv MY. The taste of toxicity: A quantitative analysis of bitter and toxic molecules. IUBMB Life. 2017;69:938–946. doi: 10.1002/iub.1694. [DOI] [PubMed] [Google Scholar]
- 21.Hofer D, Puschel B, Drenckhahn D. Taste receptor-like cells in the rat gut identified by expression of alpha-gustducin. Proc. Natl. Acad. Sci. USA. 1996;93:6631–6634. doi: 10.1073/pnas.93.13.6631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shah AS, Ben-Shahar Y, Moninger TO, Kline JN, Welsh MJ. Motile cilia of human airway epithelia are chemosensory. Science. 2009;325:1131–1134. doi: 10.1126/science.1173869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Foster SR, et al. Expression, regulation and putative nutrient-sensing function of taste GPCRs in the heart. PLoS One. 2013;8:e64579. doi: 10.1371/journal.pone.0064579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heaton KW, Morris JS. Bitter humour: the development of ideas about bile salts. J. R. Coll Phys. Lond. 1971;6:83–97. [PMC free article] [PubMed] [Google Scholar]
- 25.Falany CN, Johnson MR, Barnes S, Diasio RB. Glycine and taurine conjugation of bile acids by a single enzyme. Molecular cloning and expression of human liver bile acid CoA:amino acid N-acyltransferase. J. Biol. Chem. 1994;269:19375–19379. doi: 10.1016/S0021-9258(17)32178-6. [DOI] [PubMed] [Google Scholar]
- 26.Hofmann AF, Hagey LR. Bile acids: chemistry, pathochemistry, biology, pathobiology, and therapeutics. Cell Mol. Life Sci. 2008;65:2461–2483. doi: 10.1007/s00018-008-7568-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alnouti Y. Bile Acid sulfation: a pathway of bile acid elimination and detoxification. Toxicol. Sci. 2009;108:225–246. doi: 10.1093/toxsci/kfn268. [DOI] [PubMed] [Google Scholar]
- 28.Ridlon JM, Kang DJ, Hylemon PB. Bile salt biotransformations by human intestinal bacteria. J. Lipid. Res. 2006;47:241–259. doi: 10.1194/jlr.R500013-JLR200. [DOI] [PubMed] [Google Scholar]
- 29.Jones BV, Begley M, Hill C, Gahan CG, Marchesi JR. Functional and comparative metagenomic analysis of bile salt hydrolase activity in the human gut microbiome. Proc. Natl. Acad. Sci. U.S.A. 2008;105:13580–13585. doi: 10.1073/pnas.0804437105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alrefai WA, Gill RK. Bile acid transporters: structure, function, regulation and pathophysiological implications. Pharm. Res. 2007;24:1803–1823. doi: 10.1007/s11095-007-9289-1. [DOI] [PubMed] [Google Scholar]
- 31.Dawson PA, et al. The heteromeric organic solute transporter alpha-beta, Ostalpha-Ostbeta, is an ileal basolateral bile acid transporter. J. Biol. Chem. 2005;280:6960–6968. doi: 10.1074/jbc.M412752200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mertens KL, Kalsbeek A, Soeters MR, Eggink HM. Bile Acid Signaling Pathways from the Enterohepatic Circulation to the Central Nervous System. Front Neurosci. 2017;11:617. doi: 10.3389/fnins.2017.00617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kawamata Y, et al. A G protein-coupled receptor responsive to bile acids. J. Biol. Chem. 2003;278:9435–9440. doi: 10.1074/jbc.M209706200. [DOI] [PubMed] [Google Scholar]
- 34.Haselow K, et al. Bile acids PKA-dependently induce a switch of the IL-10/IL-12 ratio and reduce proinflammatory capability of human macrophages. J. Leukoc. Biol. 2013;94:1253–1264. doi: 10.1189/jlb.0812396. [DOI] [PubMed] [Google Scholar]
- 35.Wishart DS, et al. HMDB 5.0: the Human Metabolome Database for 2022. Nucleic Acids Res. 2022;50:D622–D631. doi: 10.1093/nar/gkab1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu W, et al. Structural basis for strychnine activation of human bitter taste receptor TAS2R46. Science. 2022;377:1298–1304. doi: 10.1126/science.abo1633. [DOI] [PubMed] [Google Scholar]
- 37.Nicoli A, Dunkel A, Giorgino T, de Graaf C, Di Pizio A. Classification Model for the Second Extracellular Loop of Class A GPCRs. J. Chem. Inform. Model. 2022;62:511–522. doi: 10.1021/acs.jcim.1c01056. [DOI] [PubMed] [Google Scholar]
- 38.de Graaf C, Foata N, Engkvist O, Rognan D. Molecular modeling of the second extracellular loop of G-protein coupled receptors and its implication on structure-based virtual screening. Proteins. 2008;71:599–620. doi: 10.1002/prot.21724. [DOI] [PubMed] [Google Scholar]
- 39.Jaiteh M, Rodriguez-Espigares I, Selent J, Carlsson J. Performance of virtual screening against GPCR homology models: Impact of template selection and treatment of binding site plasticity. PLoS Comput. Biol. 2020;16:e1007680. doi: 10.1371/journal.pcbi.1007680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Behrens M, Ziegler F. Structure-Function Analyses of Human Bitter Taste Receptors-Where Do We Stand? Molecules. 2020;25:4423. doi: 10.3390/molecules25194423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Behrens M, Korsching SI, Meyerhof W. Tuning Properties of Avian and Frog Bitter Taste Receptors Dynamically Fit Gene Repertoire sizes. Mol. Biol. Evol. 2014;31:3216–3227. doi: 10.1093/molbev/msu254. [DOI] [PubMed] [Google Scholar]
- 42.Ziegler F, Behrens M. Bitter taste receptors of the common vampire bat are functional and show conserved responses to metal ions in vitro. Proc. Biol. Sci. 2021;288:20210418. doi: 10.1098/rspb.2021.0418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takahashi S, et al. Cyp2c70 is responsible for the species difference in bile acid metabolism between mice and humans. J. Lipid Res. 2016;57:2130–2137. doi: 10.1194/jlr.M071183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sato H, et al. Novel potent and selective bile acid derivatives as TGR5 agonists: biological screening, structure-activity relationships, and molecular modeling studies. J. Med. Chem. 2008;51:1831–1841. doi: 10.1021/jm7015864. [DOI] [PubMed] [Google Scholar]
- 45.Duboc H, Taché Y, Hofmann AF. The bile acid TGR5 membrane receptor: From basic research to clinical application. Digest. Liver Dis. 2014;46:302–312. doi: 10.1016/j.dld.2013.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Flegel C, Manteniotis S, Osthold S, Hatt H, Gisselmann G. Expression profile of ectopic olfactory receptors determined by deep sequencing. PLoS One. 2013;8:e55368. doi: 10.1371/journal.pone.0055368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Karlsson M, et al. A single-cell type transcriptomics map of human tissues. Sci. Adv. 2021;7:eabh2169. doi: 10.1126/sciadv.abh2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Leonhardt J, et al. Circulating Bile Acids in Liver Failure Activate TGR5 and Induce Monocyte Dysfunction. Cell Mol. Gastroenterol. Hepatol. 2021;12:25–40. doi: 10.1016/j.jcmgh.2021.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Prandi S, Voigt A, Meyerhof W, Behrens M. Expression profiling of Tas2r genes reveals a complex pattern along the mouse GI tract and the presence of Tas2r131 in a subset of intestinal Paneth cells. Cell. Mol. Life Sci. 2018;75:49–65. doi: 10.1007/s00018-017-2621-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liszt KI, et al. Human intestinal bitter taste receptors regulate innate immune responses and metabolic regulators in obesity. J. Clin. Invest. 2022;132:e144828. doi: 10.1172/JCI144828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Baek SJ, Kim KS, Nixon JB, Wilson LC, Eling TE. Cyclooxygenase inhibitors regulate the expression of a TGF-beta superfamily member that has proapoptotic and antitumorigenic activities. Mol. Pharmacol. 2001;59:901–908. doi: 10.1124/mol.59.4.901. [DOI] [PubMed] [Google Scholar]
- 52.Baek SJ, Wilson LC, Eling TE. Resveratrol enhances the expression of non-steroidal anti-inflammatory drug-activated gene (NAG-1) by increasing the expression of p53. Carcinogenesis. 2002;23:425–434. doi: 10.1093/carcin/23.3.425. [DOI] [PubMed] [Google Scholar]
- 53.Wang DQH, Neuschwander-Tetri BA, Portincasa P. The Biliary System. Coll. Series. Integr. Syst. Physiol. From Mol. Funct. 2012;4:1–148. [Google Scholar]
- 54.LaRusso NF, Korman MG, Hoffman NE, Hofmann AF. Dynamics of the enterohepatic circulation of bile acids. Postprandial serum concentrations of conjugates of cholic acid in health, cholecystectomized patients, and patients with bile acid malabsorption. N. Engl. J. Med. 1974;291:689–692. doi: 10.1056/NEJM197410032911401. [DOI] [PubMed] [Google Scholar]
- 55.Smith JL, et al. Endogenous ursodeoxycholic acid and cholic acid in liver disease due to cystic fibrosis. Hepatology. 2004;39:1673–1682. doi: 10.1002/hep.20238. [DOI] [PubMed] [Google Scholar]
- 56.Goldsweig B, Kaminski B, Sidhaye A, Blackman SM, Kelly A. Puberty in cystic fibrosis. J. Cyst. Fibros. 2019;18:S88–S94. doi: 10.1016/j.jcf.2019.08.013. [DOI] [PubMed] [Google Scholar]
- 57.Thomas C, et al. TGR5-mediated bile acid sensing controls glucose homeostasis. Cell Metab. 2009;10:167–177. doi: 10.1016/j.cmet.2009.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Behrens M, Gu M, Fan S, Huang C, Meyerhof W. Bitter substances from plants used in traditional Chinese medicine exert biased activation of human bitter taste receptors. Chem. Biol. Drug Des. 2018;91:422–433. doi: 10.1111/cbdd.13089. [DOI] [PubMed] [Google Scholar]
- 59.Kuhn C, et al. Bitter taste receptors for saccharin and acesulfame K. J. Neurosci. 2004;24:10260–10265. doi: 10.1523/JNEUROSCI.1225-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ueda T, Ugawa S, Yamamura H, Imaizumi Y, Shimada S. Functional interaction between T2R taste receptors and G-protein alpha subunits expressed in taste receptor cells. J. Neurosci. 2003;23:7376–7380. doi: 10.1523/JNEUROSCI.23-19-07376.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang L, et al. Impaired Bile Acid Homeostasis in Children with Severe Acute Malnutrition. PLoS One. 2016;11:e0155143. doi: 10.1371/journal.pone.0155143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gustafsson J, Alvelius G, Bjorkhem I, Nemeth A. Bile acid metabolism in extrahepatic biliary atresia: lithocholic acid in stored dried blood collected at neonatal screening. Ups J. Med. Sci. 2006;111:131–136. doi: 10.3109/2000-1967-017. [DOI] [PubMed] [Google Scholar]
- 63.Magos, L. C. Lentner (ed.). Geigy Scientific Tables, 8th edition. J. Appl. Toxicol.7, 413–413, (1987).
- 64.Matsui A, Psacharopoulos HT, Mowat AP, Portmann B, Murphy GM. Radioimmunoassay of serum glycocholic acid, standard laboratory tests of liver function and liver biopsy findings: comparative study of children with liver disease. J. Clin. Pathol. 1982;35:1011–1017. doi: 10.1136/jcp.35.9.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Burkard I, von Eckardstein A, Rentsch KM. Differentiated quantification of human bile acids in serum by high-performance liquid chromatography-tandem mass spectrometry. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2005;826:147–159. doi: 10.1016/j.jchromb.2005.08.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
All data of this study are provided in the main text and supplementary information. Source data for graphs shown in Figs. 2 to 6 are provided as Supplementary data files (Supplementary data 1 for Figs. 2 and 3; Supplementary data 2 for Figs. 4 and 6; Supplementary data 3 for Fig. 5).
Receptor models and docking results are available at https://github.com/dipizio/TAS2R-models.