Abstract
The SARS-CoV-2 pandemic made evident that there are only a few drugs against coronavirus. Here we aimed to identify a cost-effective antiviral with broad spectrum activity and high safety profile. Starting from a list of 116 drug candidates, we used molecular modelling tools to rank the 44 most promising inhibitors. Next, we tested their efficacy as antivirals against α and β coronaviruses, such as the HCoV-229E and SARS-CoV-2 variants. Four drugs, OSW-1, U18666A, hydroxypropyl-β-cyclodextrin (HβCD) and phytol, showed in vitro antiviral activity against HCoV-229E and SARS-CoV-2. The mechanism of action of these compounds was studied by transmission electron microscopy and by fusion assays measuring SARS-CoV-2 pseudoviral entry into target cells. Entry was inhibited by HβCD and U18666A, yet only HβCD inhibited SARS-CoV-2 replication in the pulmonary Calu-3 cells. Compared to the other cyclodextrins, β-cyclodextrins were the most potent inhibitors, which interfered with viral fusion via cholesterol depletion. β-cyclodextrins also prevented infection in a human nasal epithelium model ex vivo and had a prophylactic effect in the nasal epithelium of hamsters in vivo. All accumulated data point to β-cyclodextrins as promising broad-spectrum antivirals against different SARS-CoV-2 variants and distant alphacoronaviruses. Given the wide use of β-cyclodextrins for drug encapsulation and their high safety profile in humans, our results support their clinical testing as prophylactic antivirals.
Keywords: SARS-CoV-2, COVID-19, Coronavirus, Antiviral, Drug repurposing, Cyclodextrin, β-cyclodextrin
Graphical Abstract
1. Introduction
Since the start of the SARS-CoV-2 pandemic novel vaccines have been developed that have changed the global landscape protecting from severe coronavirus disease (COVID-19) and decreasing the death toll during the early stages of the pandemic. Yet, it has taken more than a year for COVID-19 vaccines to reach 20 % of the population in low-income nations. Even in wealthy countries there is only about an 80 % of vaccination rate (https://ourworldindata.org/covid-vaccinations). Post- vaccination, vulnerable and immunocompromised individuals are still at high risk upon infection [2]. Unfortunately, all vaccines approved so far have failed to confer sterilizing immunity, and may not fully protect from infection.
Under this scenario, therapeutic approaches are still needed to protect those at higher risk after SARS-CoV-2 infection, which include individuals who lack access to vaccine regimens or those who are vaccinated but fail to mount adequate immune responses. Safe, affordable and effective antivirals could be of great help to mitigate these contingencies, offering treatment for those individuals not only at higher risk of developing severe COVID-19, but also for those with moderate or mild outcomes that could also benefit from reducing the time and disease-associated symptoms. WHO recommends different treatments that limit severe disease or risk of hospitalization [1]. Some of these antiviral treatments are based on neutralizing antibodies or intravenously administered drugs that are dispensed in the hospitals only. Therapeutic antibodies such as casirivimab-imdevimab have shown activity in clinical trials before the surge of new variants. However, pre-clinical studies suggest that this combination lacks neutralization activity against omicron [1]. In the case of sotrovimab, although activity might be retained, higher concentrations of the antibodies would be needed for neutralization of omicron [1]. Remdesivir targets the viral RNA polymerase but still requires intravenous treatment, although new orally-available formulations are about to be tested [44]. Molnupiravir, another inhibitor of the viral RNA polymerase, is administered orally and like remdesivir has shown clinical benefits when administered early upon infection [23]. WHO weakly recommends administration of Molnupiravir, and only for patients at high risk of being admitted to hospital with COVID-19 [1]. Paxlovid is taken orally, but is not widely accessible and imply costs that exceed those of vaccines [7]. Moreover, Paxlovid is only available for patients that tolerate Ritonavir as part of the treatment with the antiviral nirmatrelvir, and has also raised concerns as monotherapy due to associated viral resistance [59], which may limit efficacy against new variants of concern (VOC).
None of the antivirals currently approved have the cost-effective profile or the easy administration route needed to offer prophylaxis to those vulnerable individuals at higher risk of developing severe disease upon SARS-CoV-2 infection on a global scale. Nevertheless, the key to control the SARS-CoV-2 pandemic might follow an approach similar to HIV-1 pre-exposure prophylaxis (PrEP), which has been employed to decrease the HIV-1 infection rates [32]. With the success of this prior HIV-1 strategy, having an affordable antiviral with prophylactic potential and a broad-spectrum activity at the initial surges of novel VOCs could be key to decrease SARS-CoV-2 transmission rates.
Here we aimed to identify a cost-effective antiviral with broad spectrum activity and high safety and tolerability profiles. We began compiling a list of drugs previously used to treat other pathologies or characterized in preclinical studies with potential to treat coronavirus infections. We next employed molecular modelling tools to rank the most promising inhibitors and tested their efficacy as antivirals against two representative viruses from the α and β coronavirus genera: the HCoV-229E and SARS-CoV-2 viruses. With a combination of computational chemistry, virology, cell biology and electron microscopy methods, we studied 44 compounds. Four of them showed antiviral activity against HCoV-229E and SARS-CoV-2, β-cyclodextrins being the most promising candidates to treat the infection caused by SARS-CoV-2.
2. Material and methods
2.1. Molecular modelling
2.1.1. Viral targets
The characterization of the crystal structure of the Mpro [25], an enzyme recruited by the SARS-CoV-2 to complete the replication and transcription steps, was included in the protein data bank (PDB) library (code 6LU7) and allowed the use of virtual screening (VS) techniques [3]. Indeed, the same authors performed VS simulation searching for antiviral drugs in an in-house library of compounds [25]. Many other structures have been resolved since then, and there are several PDB entries with inhibitors located in the active site of the Mpro target (i.e. codes 5RG1, 5RGL, 7A1U, 7JU7) [10], [17]. Most of techniques aim for small molecules to reach the active site, which is characterized by a catalytic dyad of Cys145 and His41 residues [61].
The SARS-CoV-2 spike has been also extensively assessed through VS simulations as it governs host attachment and virus–cell membrane fusion upon infection [58]. In that framework, the characterization of the receptor-binding domain (RBD), deposited with PDB code 6M0J by Wang et al., 2020, allows for developing inhibitors by using molecular models [51]. It has been also proposed that the SARS-CoV-2 spike might bind two metabolites (biliverdin and bilirubin) avoiding the antibody recognition [43].
Within the last two years extensive research have led the discovery of other X-ray structures, for instance papain-like protease (PLpro), an enzyme that regulates SARS-CoV-2 viral spread and innate immunity [48]. Some of the more recent crystals with non-covalent inhibitors are associated to the PDB codes 7CMD, 7JIR, 7JIT, 7JIV, 7JIW, 7JN2, 7JRN, 7KOJ, 7KOK, 7KOL, 7KRX, 7LBR, 7LBS, 7LLF, 7LLZ and 7LOS [13], [33], [34], [40], [47]. In addition, the non-structural protein 16 (NSP16) has been shown to play an essential role for immune evasion by mimicking the human homolog, CMTr1. However, unlike CMTr1, NSP16 needs to form a heterodimer with NSP10 to activate its enzymatic activity. Although there are no available crystals for the inactive monomeric NSP16, Bowman and co-workers determined its activation mechanism and the location of a cryptic pocket [57], which if targeted with a small molecule, can be used to inhibit NSP16.
Niemann-Pick type C1 (NPC1), a lipid-transfer protein that regulates intracellular cholesterol traffic, has been shown to play a role in human cell infection [14]. Within that hypothesis, the regulation of cholesterol by targeting NPC1 might offer an additional therapeutic strategy to treat infected patients. The experimental data for NPC1 is still scarce, with only two X-ray structures available (PDB codes 6UOX and 5U73).
2.1.2. Library preparation, protein refinement and virtual screening
Our in-house library of 116 compounds were prepared for simulations by using LigPrep (Schrödinger Release 2021–3). Initial cartesian coordinates were retrieved from the PubChem database (Kim et al., 2021). Geometries were subsequently optimized by assessing protonation and tautomeric states at pH 7 ± 2 as predicted by Epik [15], [46]. All other parameters are set as default as implemented in the Schrödinger suite of programs (Schrödinger Release 2021–3). All abovementioned PDB structures were refined with the Protein Preparation Wizard module [30], a multistep protocol. During that structural refinement, missing hydrogen atoms are added to the protein, charges were assigned, protonation states were determined with Epik at pH 7.0 ± 2.0, and a final restricted minimization is performed with the OPLS4 force field [29]. A grid was created by considering co-crystal ligand or reference residues, as in the case of allosteric sites. Two docking runs were carried out, with single-precision (SP) or extra-precision (XP) scoring functions [12], [20]. For the records, ten poses per ligand were saved for each docking run. Binding energies are eventually computed with the molecular mechanics/generalized Born surface area (MMGBSA) implemented in Prime module [21], [22], a more refined method that accounts for the energies before and after ligand binding [39], [60]. The resulting free energies (ΔG) are therefore used for ranking compounds of our library. The last computational step uses molecular dynamics (MD) to assess the dynamic stability of the generated target-drug adducts. An orthorhombic box of TIP3P-waters was generated by using periodic boundary conditions with a buffer distance of 10 Å. Sodium cations are added to keep the system electronically neutral while additional ions were incorporated to mimic the physiological salt concentration of 0.15 M NaCl. The MD protocol includes minimization as implemented by default in the GPU-accelerated version of Desmond [5]. The MD simulation is completed by producing trajectories of 100 ns.
During our investigation several publications showed the antiviral activity of 10 compounds against SARS-CoV-2 (Simvastatin, Sirolimus, Everolimus, Itraconazole, Quercetin, Taxifolin, Resveratrol, Digoxin, Lanatoside C and SilibininA) that we found to inhibit 229E virus, and those were excluded from our study due to the lack of novelty [24], [27], [42], [55], [9]. Indeed, some of them (Simvastatin, Sirolimus, Itraconazole, Quercetin, Resveratrol and Silibinin A) have been or are currently included in clinical trials for COVID-19 (https://clinicaltrials.gov).
2.1.3. Reagents used
All the compounds and respective vendor origin are detailed in Table 1, and Table S1.
Table 1.
List of in vitro tested compounds against HCoV229E.
| Num. | Drug | IC50 ± SD/CC50 ± SD | Mode of action | Previous clinical use | Vendor Origin | |
|---|---|---|---|---|---|---|
| A | 1 | OSW-1 | 0.5 ± 0.1 nM / > 100 nM | OSBP inhibitor | Pre-clinical | Cayman Chemical |
| 2 | HβCD | 4.3 ± 4.0 / 43.6 ± 5.7 mM | Extracts cholesterol from cellular membranes | Vaccine adjuvant Drug carrier | Trappsol Cyclo | |
| 3 | U18666A | 2.6 ± 1.1 / 74.4 ± 14.8 μM | NPC1 inhibitor | Pre-clinical | Millipore | |
| 4 | Phytol | 18.6 ± 6.9 / 56.2 ± 7.7 μM | Activator of the PPARs pathway and inhibitor of NF-κB | Pre-clinical | Sigma Aldrich | |
| B | 5 | Mdivi-1 | 11.7 ± 0.7 / 95.4 ± 1.8 µM | Inhibitor of Drp1-dependent mitochondrial fission and Bax/Bak-dependent cytochrome c release during apoptosis | Pre-clinical | Sigma Aldrich |
| 6 | FLI 06 | 0.4 ± 0.3 / > 50 μM | Inhibitor of Notch signaling | Pre-clinical | Sigma Aldrich | |
| 7 | Gingenoside RB1 | 48.3 ± 2.1 / > 1000 μM | Reversible inhibitor of PIK93γ and PI4KIIIβ | Pre-clinical | Sigma Aldrich | |
| 8 | Digitoxin | 0.021 ± 3.1 / 6.9 ± 6.3 nM | Inhibitor of the Na-K-ATPase membrane pump | Cystic fibrosis | Sigma Aldrich | |
| 9 | PIK93 | 4.2 ± 1.4 / > 50 μM | Reversible inhibitor of PIK93γ and PI4KIIIβ | Pre-clinical | Sigma Aldrich | |
| 10 | Bortezomib (PS-341) | 0.6 ± 0.3 / > 100 nM | Inhibitor of proteasome | Multiple myeloma Mantle cell lymphoma | Selleckchem | |
| 11 | Aspirin | 1.0 ± 9.8 / 1.1 ± 1.7 mM | Inhibitor of prostaglandin synthesis and platelet aggregation | Pain Fever Inflammation | Sigma Aldrich | |
| 12 | Wortmannin | 21.6 ± 15.6 / > 50 μM | Inhibitor of PI3Ks | Pre-clinical | Millipore | |
| 13 | Alpha-terpineol | 381.7 ± 107.1 / 1058.6 ± 332.6 μM | Inhibitor of NF-κB activity, COX-2 and IKK2 | Adjuvant in treatment for upper respiratory tract congestion | Sigma Aldrich | |
| 14 | Dehydrocostus lactone | 4.2 ± 1.8 / 39.1 ± 3.7 uM | Inhibitor of the IKKβ/NF-κB/COX-2 and PI3K/Akt/Bad signaling pathways | Pre-clinical | Sigma Aldrich | |
| 15 | Valinomycin | 2.5 ± 0.2 / 12.5 ± 3.5 μM | Binds and exchanges potassium across membranes | Pre-clinical | Sigma Aldrich | |
| 16 | Terpinolene | 8 ± 2.2 mM/ 69 ± 5 mM | ND | Pre-clinical | Sigma-Aldrich | |
| C | 17 | Baclogen | NA / > 1000 μM | GABA-B receptor agonist activity | Multiple sclerosis Hepatocellular carcinoma Liver cirrhosis | Sigma Aldrich |
| 18 | CI976 | NA / 50.7 ± 12.1 μM | Inhibitor of sterol O-acyltransferase and acyl-coenzyme A: cholesterol acyltransferase. Inhibitor of Golgi-associated LPL acyltransferase (LPAT) | Pre-clinical | Sigma Aldrich | |
| 19 | Sulfacetamide | NA / > 1 mM | Inhibitor of the dihydropteroate synthase. Inbibitor of bacterial para-aminobenzoic acid | Antibiotic | Sigma Aldrich | |
| 20 | Squalene | NA / 135.4 ± 5.4 mM | Intermediate metabolite in the synthesis of cholesterol | Pre-clinical | Sigma Aldrich | |
| 21 | Sodium 4-phenylbutyrate | NA / > 500 mM | Interacts with hydrophobic regions in unfolded proteins and reduces ER stress | Urea cycle disorders | Sigma Aldrich | |
| 22 | Oleanolic acid | NA / 25.2 ± 7.4 mM | ND | Pre-clinical | Sigma Aldrich | |
| 23 | MitoTEMPO | NA / NA | Mimetic of the mitochondria-targeted superoxide dismutase and scavenger of the superoxide and alkyl radicals | Pre-clinical | Sigma Aldrich | |
| 24 | N-acetyl-L-cystein | NA / NA | Reductant of disulfide bonds and scavenger of reactive oxygen species | Mucolytic agent Chronic and acute bronchitis Pulmonary fibrosis and emphysema Acetaminophen intoxication | Sigma Aldrich | |
| 25 | Camphene | NA / 72 ± 9.2 mM | Reduces plasma cholesterol and triglycerides | Pre-clinical | Sigma Aldrich | |
| 26 | Myricetin | NA / 433.6 ± 214.1 μM | Inhibitor of the cyclooxygenase 1 | Pre-clinical | Sigma-Aldrich | |
| 27 | Aminohipurric | NA / < 10 mM | ND | Test of renal plasma flow | Sigma-Aldrich | |
| 28 | L-Phenilalanine | NA / < 10 mM | ND | Depression Vitiligo | Sigma-Aldrich | |
| 29 | (-)β-Pinene | NA / 293.6 ± 103.8 μM | Inhibitor of acetylcholinesterase and positive modulator of GABA receptors | Treatment for bladder, kidney, and urinary stones | Sigma Aldrich | |
| 30 | R-(+)-Limonene | NA / 1908.4 ± 194.2 μM | Angiogenesis inhibitor and TNF and IL-6 suppressor | Genital Human Papilloma Virus infection Parotid Gland Tumor Submandibular Gland Tumor | HWI group | |
| 31 | (-)-Trans-Caryophyllene | NA / 1835 ± 173 μM | Agonist of the CB2, inhibitor of p38 MAPK/NF-κB and reductor of the cytokine release | Clinical trial for analgesia | Sigma Aldrich | |
| 32 | Esential Oil Pinus sylvestris | NA / 533.3 ± 57.7 μg/mL* | Down-regulator of Cdc25C and cell cycle. Enhancer of calcium channels activation | Pre-clinical | Terpenic | |
| 33 | Hydrosol Pinus sylvestris | NA / > 10 mg/mL* | ND | Pre-clinical | Le Gattilier |
The Table is divided in three sections: 1) Compounds with antiviral activity against HCoV229E and SARS-CoV-2 (A-white, 1–4); 2) compounds with antiviral activity against HCoV229E but not against SARS-CoV-2 (B-light grey, 5–16) and 3) compounds without antiviral activity against HCoV229E (C-dark grey, 17–33). For each compound, IC50 and CC50 values for HCoV229E, known mode of action, previous clinical use and vendor origin are indicated. ND, non-determined.
*These are complex mixtures of compounds (molarity unknown)
2.2. Assays with HCoV-229E
2.2.1. Cells and virus
MRC5 cells (ATCC CCL-171) were grown in Dulbecco’s minimal essential medium (DMEM) supplemented with 10 % inactivated fetal bovine serum (FBS, Biological Industries), 4 mM glutamine (Sigma-Aldrich), 1 x non-essential amino acid solution (Sigma-Aldrich), 100 U/mL penicillin and 100 μg/mL streptomycin (both from Sigma-Aldrich). For preparing virus stocks, human coronavirus 229E (HCoV-229E; ATCC VR-740) was propagated in MRC-5 cells as described [31] with a modification that is maintaining cell cultures at 35ºC. Viral titer was calculated as 50 % tissue culture infective dose (TCID50). Briefly, MRC-5 cells in a 96-well plate at 80 % confluency were inoculated with a serial dilution of the viral stock, from 10-1 to 10-8. The plate was incubated at 33 °C and with 5 % CO2 for 5 days and then fixed with 4 % paraformaldehyde (PFA) at room temperature (RT) for 20 min. Monolayers were processed by indirect immunofluorescence (IF) using specific antibodies against the HCoV-229E nucleocapsid (N) protein (see below), and titer calculated as described [38], [41].
2.2.2. Cytotoxicity assay
A stock solution of each compound was prepared. Hydrophobic compounds were dissolved in dimethyl sulfoxide (DMSO) or ethanol (as recommended by the manufacturer) and stored at −20 °C. To evaluate the viability of cell cultures when treated with the compounds, we performed an MTT (3-[4,5-dimethylthiazol-2-yl]-2,5 diphenyl tetrazolium bromide) assay, which measures the mitochondrial dehydrogenase activity of living cells [45], [54]. MRC-5 cells were cultured in a 96-well plate until 80 % confluency was reached. Serial dilutions of the compounds in DMEM with 10 % FBS were added in triplicate and in a final volume of 100 μL/well. After 24 h, 5 mg/mL of MTT reagent (Sigma-Aldrich) was added to a final concentration of 0.5 mg/mL. Incubation was maintained during 4 h at 37ºC with 5 % CO2 for metabolization of the reagent and then 100 μL of lysis buffer (10 % SDS, 0.01 M HCl and 85 % isopropanol) was added and incubation maintained for 30 min on an orbital shaker protected from light. Plates were read at 570 nm and the background (measured at 690 nm) was subtracted. Data ware calculated from three independent replicates.
2.2.3. Antiviral activity
The effect of the compounds on viral infection was determined by IF as follows: 20,000 MRC5 cells/well were seeded in duplicates per drug and condition in 96-wells flat-bottom plates. When 80 % confluency was reached, cells were adsorbed with HCoV-229E in DMEM media without FBS at MOI= 0.1 PFU/cell, and incubated for 1 h at 37 °C. The inoculum was then removed and a serial dilution of the compounds in DMEM supplemented with 1 % FBS was added. A duplicate of the infected cells without any drug treatment served as control. After 24 h, cells were fixed with 4 % PFA for 20 min and washed three times with PBS. Cells were permeabilized with 0.25 % saponin in PBS for 10 min and then treated 30 min with blocking buffer (1xPBS with 0.25 % saponin and 2 % FBS). Cells were then incubated 1 h with a rabbit antibody specific for the HCoV-229E nucleocapsid (N) protein (Ingenasa) diluted 1:200 in blocking buffer. After 3 washes with PBS, cells were incubated 45 min with an anti-rabbit secondary antibody conjugated with Alexa fluor 488 (Invitrogen) diluted 1:500 in blocking buffer and washed three times with PBS. Finally, cell nuclei were labeled 20 min with 4′,6-diamidino-2-phenylindole (DAPI) diluted 1:200 in blocking buffer and cells then washed 3 times with PBS. Images were obtained with a Leica DMi8 S widefield epifluorescence microscope and processed with Image J software. Data were normalized by setting the positive infection control as 100 % of infection. Inhibition data were plotted as dose-effect curves fitted to a nonlinear regression model in GraphPad Prism v 9.4 software. The IC50 was calculated with Quest Graph™ IC50 Calculator (https://www.aatbio.com/tools/ic50-calculator). All experiments were replicated three times.
2.3. Antiviral activity in cells infected with SARS-CoV-2
2.3.1. Biosafety statement
The biosafety committee of the Institute Germans Trias i Pujol approved the execution of SARS-CoV-2 experiments at the BSL3 laboratory of the Center for Bioimaging and Comparative Medicine (CSB-20–015-M7).
2.3.2. Cells
Vero E6 cells (ATCC CRL-1586) were cultured in Dulbecco’s modified Eagle medium, (DMEM) supplemented with 10 % fetal bovine serum (FBS), 100 U/mL penicillin, 100 μg/mL streptomycin, and 2 mM glutamine (all from Invitrogen). HEK-293 T cells (ATCC repository) were maintained in DMEM with 10 % FBS, 100 IU/mL penicillin and 100 μg/mL streptomycin (all from Invitrogen). HEK-293 T cells overexpressing the human ACE2 (293 T-ACE2) were kindly provided by Integral Molecular Company and maintained in DMEM with 10 % FBS, 100 IU/mL penicillin and 100 μg/mL streptomycin, and 1 μg/mL of puromycin (Invitrogen). CaLu-3 cells were kindly provided by the laboratory of Dr. Sanchez Cespedes and maintained in DMEM with 10 % FBS, 100 IU/mL penicillin and 100 μg/mL streptomycin.
2.3.3. Virus isolation and sequencing
SARS-CoV-2 variants were isolated from clinical nasopharyngeal swabs in Vero E6 cells, as previously described [42]. Viral stocks were grown in Vero E6 cells and supernatants were collected and stored at −80ºC until use. The following SARS-CoV-2 variants with deposited genomic sequence at the GISAID repository (http://gisaid.org) were tested: B.1 (D614G) isolated in Spain in March 2020 (EPI_ISL_510689); and 4 variants isolated in Spain from January to February 2021: Alpha or B.1.1.7 (EPI_ISL_1663569), β or B.1.351 (originally detected in South Africa; EPI_ISL_1663571), Zeta or P.2 (originally detected in Brazil; EPI_ISL_1831696), Delta or B.1.617.2 (originally detected in India; EPI_ISL_3342900), and Omicron or B.1.1.529 (EPI_ISL_8151031). Genomic sequencing was performed from viral supernatant by using standard ARTIC v3 or v4 based protocols followed by Illumina sequencing [dx.doi.org/10.17504/protocols.io.bhjgj4jw]. Raw data analysis was performed by viralrecon pipeline [https://github.com/nf-core/viralrecon] while consensus sequence was called using samtools/ivar at the 75 % frequency threshold. Viral variants were titrated at 1/3 dilutions on Vero E6 cells using the same luminometric assay described for antiviral testing. Thus, for all VOCs, we used equivalent infectious units inducing 50 % of viral induced cytopathic effect.
2.3.4. Pseudovirus production
HIV-1 reporter pseudovirus expressing SARS-CoV-2 Spike protein and luciferase were generated using two plasmids. pNL4–3. Luc.R-.E- was obtained from the NIH AIDS repository. SARS-CoV-2. SctΔ19 was generated (Geneart) from the full protein sequence of SARS-CoV-2 spike with a deletion of the last 19 amino acids in C-terminal, human-codon optimized and inserted into pcDNA3.4-TOPO [35]. Spike plasmid was transfected with X-tremeGENE HP Transfection Reagent (Merck) into HEK-293 T cells, and 24 h post-transfection, cells were transfected with pNL4–3. Luc.R-.E-. Supernatants were harvested 48 h later, filtered with 0.45 μM (Millex Millipore) and stored at −80ºC until use. The p24gag content of all viruses was quantified using an ELISA (Perkin Elmer) and viruses were titrated in HEK-293 T overexpressing the human ACE2.
2.3.5. Pseudoviral entry inhibition assay
HEK-293 T overexpressing the human ACE2 were used to test the indicated compounds. A constant pseudoviral titer was used to pulse cells in the presence of the drugs. At 48 h post-inoculation, cells were lysed with the Bright Glo Luciferase system (Promega). Luminescence was measured with an EnSight Multimode Plate Reader (Perkin Elmer).
2.3.6. Antiviral activity against SARS-CoV-2
Increasing concentrations of the indicated antiviral compounds were added to Vero E6 cells and immediately after, we added equivalent infectious units of SARS-CoV-2 variants that induced a 50 % cytopathic effect. Untreated non-infected cells and untreated virus-infected cells were used as negative and positive controls of infection, respectively. To detect any drug-associated cytotoxic effect, Vero E6 cells were equally cultured in the presence of increasing drug concentrations, but in the absence of virus. Cytopathic or cytotoxic effects of the virus or drugs were measured 3 days after infection, using the CellTiter-Glo luminescent cell viability assay (Promega). Luminescence was measured in a Fluoroskan Ascent FL luminometer (ThermoFisher Scientific).
Viral replication of SARS-CoV-2 was also assessed on Calu-3 cells in the presence of the indicated antiviral compounds. Compounds were incubated with cells before adding the SARS-CoV-2 virus at MOI = 0.3. After 24 h of incubation at 37ºC and 5 % CO2, cells were washed with PBS and compounds were added in fresh media at the same concentration for 48 h. The amount of SARS-CoV-2 nucleoprotein released to the supernatant was measured with SARS-CoV-2 nucleocapsid protein High-Sensitivity Quantitative ELISA (ImmunoDiagnostics) according to the manufacturer’s protocol. The cytopathic effect on Calu-3 cells was assessed with Cell Titer-Glo Assay with a Fluoroskan Ascent FL luminometer at the time of supernatant collection.
2.3.7. IC50 calculation and statistical analysis
Response curves of compounds or their mixes were adjusted to a non-linear fit regression model, calculated with a four-parameter logistic curve with variable slope. Cells not exposed to the virus were used as negative controls of infection and were set as 100 % of viability to normalize data and calculate the percentage of cytopathic effect. All analyses and Figures were generated with the GraphPad Prism v8.0b Software, Adobe Photoshop or BioRender.
2.4. Transmission electron microscopy (TEM)
Mock- or SARS-CoV-2 – infected (MOI = 0.02 PFU/cell) cell monolayers were incubated in the absence or presence of two different concentrations of the compounds and fixed at 48 hpi with 4 % paraformaldehyde and 1 % glutaraldehyde in phosphate buffered saline (PBS) for 2 h at room temperature (RT). Cells were removed from the plates in the fixative, pelleted by centrifugation and washed three times with PBS. Post-fixation of cell pellets was done on ice with 1 % osmium tetroxide + 0.8 % potassium ferrocyanide in water. Afterwards, pellets were dehydrated on ice with increasing concentrations of acetone and processed for embedding in the epoxy resin EML-812 (Taab Laboratories), as previously described [52]. Infiltration with epoxy resin was performed at RT. All samples were polymerized at 60 °C for 48 h. Ultrathin sections (50–70 nm) were cut with a Leica UC6 microtome and placed on uncoated 300 mesh copper grids. Sections were contrasted with 4 % uranyl acetate and Reynold's lead citrate. Images were taken with a Tecnai G2 TEM operated at 120 kV with a Ceta camera or with a Jeol 1400 operated at 120 kV with a Gatan Rio camera. At least 100 cells per condition were studied by TEM.
2.5. SARS-CoV-2 Mpro inhibition assay
A SARS-CoV-2 3CL main protease (MBP-tagged) assay kit (BPS Bioscience) was used following the kit instructions. In brief, the assay buffer was prepared by adding dithiothreitol, and then the protease was diluted to a 3–5 ng/μL. Next, diluted protease solution was added to the test samples and the positive controls. The MβCD and the GC376 control solutions were prepared with the assay buffer. The reaction was started by adding the substrate solution to each well and incubated overnight. Fluorescence was read in an EnSight multimode plate reader (Perkin Elmer) at a 360 nm/460 nm excitation/ detection wavelength. Percentage of Mpro activity inhibition is calculated as follows: (positive control – test inhibitor) / positive control.
2.6. Lipidomic analysis of plasma membranes of Calu-3 cells after MβCD treatment
To assess the lipid composition, 3 million Calu-3 cells/well were seeded in a 6-well plate and treated with 0, 0.6 or 2.5 mM of MβCD for 2 h at 37 ºC and 5 % CO2. After extensive washing with PBS, cells were detached with trypsin and stored at −80 ºC until its analysis. A total of 750 μL of a methanol-chloroform (1:2, vol/vol) solution containing internal standards (16:0 D31_18:1 phosphocholine, C17:0 cholesteryl ester, stigmasterol and N-dodecanoylsphingosylphosphorylcholine, 0.2 nmol each, from Avanti Polar Lipids) were added to 300,000 cells. Samples were vortexed and sonicated until they appeared dispersed and extracted at 48 °C overnight. The samples were then evaporated and transferred to 1.5 mL Eppendorf tubes after the addition of 0.5 mL of methanol. Samples were evaporated to dryness and stored at −80 °C until analysis. Before analysis, 150 μL of methanol were added to the samples, centrifuged at 13,000 g for 3 min, and 130 μL of the supernatants was transferred to ultra-performance liquid chromatography (UPLC) vials for injection and analysis with an Acquity UPLC system (Waters) connected to a time-of-flight (TOF; LCT Premier XE, Waters) detector (Simbari et al., 2016). Experiments were performed in duplicate, and each sample was analyzed in triplicate.
2.7. SARS-CoV-2 antiviral activity in a human nasal epithelial model
To assess the MβCD antiviral activity in a physiological model, we used a human nasal airway epithelium (HNE) model (Mucilair™, Epithelix) with the specific medium provided. Inserts were cultured at the air liquid interface, mimicking in vivo nasal tissue. In this model we added 2.5 mM MβCD on the apical side or on the basal medium, or 2 µM Remdesivir on the basal medium, or left cultures untreated. SARS-CoV-2 D614G was added at an MOI= 0.01 to the apical side of each insert for 1 h at 37ºC and 5 % CO2. The apical side was then extensively washed with PBS and incubated for 24 h. MβCD on the apical side was washed and not replaced thereafter. Drugs added on the basal medium were present during the whole experiment and replaced at 48 hpi. Viral content was measured collecting the apical side of inserts after adding 300 μL/well of PBS at 24-, 48- and 72-hours pi. SARS-CoV-2 nucleocapsid concentration was measured by ELISA. The cytopathic effect was measured 72 hpi using the Cell Titer-Glo Assay to measure ATP released by living cells on a DL Ready Luminoskan (ThermoScientific).
2.8. SARS-CoV-2 antiviral activity in an in vivo hamster model
All animal experiments were approved by the IRTA committee on animal experimentation and were authorized by the Generalitat de Catalunya (11964). The researchers adhered to the 3Rs principle and assured animal welfare throughout the study. The study was evaluated by the Biosecurity committee of IRTA and was performed under BSL-3 conditions. Nine-week-old golden Syrian hamsters (Janvier) were divided into 3 groups in a sex-balanced ratio (50 % females): G0 (n = 4; untreated and uninfected animals), G1 (n = 8; untreated and SARS-CoV-2-NanoLuc infected animals), G2 (n = 8; MβCD-treated and SARS-CoV-2-NanoLuc infected animals). Animals were inoculated by intranasal instillation with 50 mM MβCD or PBS (100 μL/individual, 50 μL for each nostril), for the treated and untreated groups, respectively. Then, animals were inoculated by intranasal instillation with 103 SARS-CoV-2 NanoLuc replicative competent reporter virus (n = 16) resuspended in 50 mM MβCD (for the treated animals) or PBS (for the untreated animals; 100 μL/individual, 50 μL for each nostril). The four remaining hamsters were intranasally inoculated with PBS (100 μL/individual, 50 μL for each nostril) and used as negative controls. For virological examinations, 4 hamsters per group (G1-G2) and 2 control hamsters (G0) were euthanized on days 1 and 2 post-infection.
Oral swabs were collected from all animals before the challenge and at the euthanasia. At necropsy, samples from nasal turbinate and lung were taken and placed in individual microfuge tubes containing 500 μL of DMEM (GIBCO) supplemented with 1 % penicillin–streptomycin (PS) (GIBCO) and a single zinc-plated, steel, 4.5-mm bead. Samples were homogenized at 30 Hz for 2 min using a TissueLyser II (QIAGEN GmbH, Hilden, Germany) and centrifuged for 30 s at 11,000 rpm. Supernatants were analyzed for Nanoluciferase content with the Nano-Glo Luciferase Assay System (Promega) at a 1:1 sample:luciferase ratio and assessed on a Fluoroskan Ascent FL luminometer (ThermoFisher). Samples were stored at −70 °C for further analysis.
Viral RNA was extracted from samples using the IndiMag pathogen kit (Indical Bioscience) on a Biosprint 96 workstation (QIAGEN) according to the manufacturer’s instructions. RT–PCR used to detect viral gRNA is based on the one published by Corman et al. Eurosurveillance; 2020;25:2000045, with minor modifications to adapt it to the AgPath-ID One- Step RT–PCR Kit (Life Technologies). The primers and probes used, and their final concentration are the following: forward: 5′-ACAGGTACGTTAATAGTTAATAGCGT-3′ [400 nM], reverse: 5′-ATATTGCAGCAGTACGCACACA-3′ [400 nM] probe: 5′-FAM ACACTAGCCATCCTTA CTGCGC TTCG-TAMRA-3′ [200 nM]. Thermal cycling was performed at 55 °C for 10 min for reverse transcription, followed by 95 °C for 3 min and then 45 cycles of 94 °C for 15 s, 58 °C for 30 s
Upon euthanasia, animals were necropsied, and lungs, nasal turbinate and brain samples were fixed with formalin and routinely stained with haematoxylin & eosin. Slides were examined under an optical microscope and analysed in a blinded fashion. Lesions were scored following a semi-quantitative approach based on the inflammation severity (none, mild, moderate, or severe), as previously described [56], [6].
SARS-CoV-2 nucleoprotein (NP) antigen was detected in target tissues with an immunohistochemistry protocol using a rabbit monoclonal antibody (40143-R019, Sino Biological, Beijing, China) at a 1:15,000 dilution. The amount of NP was scored semi-quantitatively in a blinded fashion (no detection, low amount, moderate amount or high amount of antigen [56], [6]).
Viral titration of replicative SARS-CoV-2-nanoLuc was performed in VeroE6 cells. Briefly, homogenized tissue was serially diluted 10-fold and transferred to an opaque (white) 96-well plate with 70 % confluence of VeroE6 cells and incubated for 48 h at 37 °C and 5 % CO2. Then, samples were incubated for 3 min with Nano-Glo® reagent (Promega) at a 1:1 ratio in absence of light. Read-out was performed with a Fluoroskan Ascent FL (Thermo Fisher Scientific). Viral titration of replicative SARS-CoV-2 virus was performed as described elsewhere [19], [42] and quantified using the Reed-Muench method.
This in vivo experiment was also performed using a wildtype SARS-CoV-2 virus (D614G variant, GISAID ID EPI_ISL-47147).
3. Results
3.1. Molecular modeling for antiviral selection
We first screened the bibliography to identify: (i) molecules targeting RNA viral polymerases or cellular factors used by viruses; (ii) hydro-soluble extracts and essential oils from plants used in medicine and with antiviral activity; (iii) commercially available drugs that could have antiviral activity. These studies produced a library of 116 compounds that were all further analyzed with molecular models to identify potential inhibitors of SARS-CoV-2 proteins or cell proteins used by the virus. The designed computational strategy is summarized in Fig. 1, including the selected viral targets (Fig. 1 A) as well as the chemical structures of the best-ranked compounds (Fig. 1 B). The performed calculations suggested that OSW-1 is efficiently anchored at the active site of the Mpro (Fig. 1 C). Indeed, that hit exhibited the largest binding energy (ΔG = −108.79 kcal/mol) among all the small molecules included in our library. Additional large interactions are predicted for the binding of U18666A, wortmannin and phytol to NPC1 (ΔG in the range of –90 to −80 kcal/mol). Unfortunately, only moderate interactions are associated with other viral targets as Spike, NSP16 and PLPro, with ΔG values under the threshold of −66 kcal/mol. The complete list of the predicted binding-free energies for the best-ranked compounds is provided in Tables S2 and S3.
Fig. 1.
Virtual screening and molecular dynamics for selecting antiviral drugs. (A) Schematic representation of our library of compounds; experimental structures for all selected viral targets displayed in cartoons. (B) Chemical structures for the best-ranked compounds. (C) Poses adopted by the best-ranked compounds (targets are sketched in grey cartoons and surface); the associated root mean-square deviation (RMSD) in angstroms (Å). For the sake of clarity, the same color scheme has been adopted: black for OSW-1, red for U18666A, green for wortmannin, blue for phytol and purple for β-CD.
The adopted poses by the best-ranked ligands is illustrated in Fig. 1 C once reached the target. In the case of cyclodextrins, β-CD was selected as a representative macrocycle. The analysis of the produced structures confirms that both OSW-1 and β-CD are in the catalytic pocket of the Mpro, while U18666A, phytol and wortmannin reach the central pocket of the NPC1. To ensure that ligands remain in their pockets under biological conditions, MD trajectories of 100 nano-seconds (ns) were performed. The stability was monitored through the so-called root means-square deviation (RMSD) (Fig. 1 C, right panel). A close inspection of RMSD values revealed that both OSW-1 (black line) and βCD (purple line) are stabilized with a RMSD variation of ca. 2 Å. On the contrary, U18666A, phytol and wortmannin yielded an average RMSD of ca. 4 Å. This difference is probably due to the fact that NCP1 has an internal pocket while the Mpro active site is localized on its surface, which induces a larger conformational change upon ligand binding. However, despite such dissimilarity, all five drugs were quickly equilibrated, with a variation of less than 1 Å after the first 20 ns of trajectory, suggesting the drug-target adducts are stable with time, a prerequisite to exhibit biological activity.
The energetic analysis offers an unbiased criterium for ligand selection. However, a large interaction with these targets does not guarantee an efficient activity against the virus, given that many other mechanisms may affect the inhibitory ability in the biological scenario [8]. Consequently, selection criteria for testing compounds in cell culture against coronaviruses HCoV-229E and SARS-CoV-2 was completed with additional parameters including their low toxicity and commercial availability. With this information, we selected 44 compounds to be tested first against HCoV-229E. In addition, we also tested the hydro-soluble extract and the essential oil from Pinus sylvestris in cells infected with HCoV-229E, given its described antiviral properties [18], [37].
3.2. Antiviral activity against HCoV-229E
To test the antiviral activity of selected compounds, MRC-5 cells were infected with HCoV-229E and treated in parallel with increasing concentrations of the selected compounds. Drug cytotoxicity was first measured, and safe drug-doses were used to determine the percentage of infected cells by immunofluorescence using an antibody specific for the HCoV-229E nucleocapsid protein ( Fig. 2 A). With these assays, we calculated the concentration of compound required to inhibit 50 % of the virus (IC50), and the concentration for the 50 % cytotoxic effect (CC50) (Fig. 2 B). Thirteen compounds, including OSW-1, HP-β-cyclodextrin (HβCD), U18666A and Phytol inhibited HCoV-229E infection at non-toxic concentrations (Table 1, Fig. 2 B top graphs). Mdivi-1, FLI06 and CI976 showed an inhibitory effect but at toxic concentrations, and Baclogen did not have an inhibitory effect on HCoV-229E (Fig. 2 B bottom graphs). In the case of BMS309403, Dimercaprol, and alpha-cedrol, they inhibited 229E but their CC50 was close to the IC50 and they were discarded for further analysis due to toxicity issues. As indicated in Table 1 , the most potent antiviral was OSW-1 (IC50 of 0.5 nM), followed by U18666A, phytol and HβCD (IC50 of 2.6 µM, 18.6 µM and 4.3 mM, respectively).
Fig. 2.
Antiviral activity of compounds tested against HCoV-229E. MRC-5 cells were absorbed with HCoV-229E at MOI of 0.1 PFUs/cell for 1 h, exposed to increasing concentrations of the drug for 24 h and processed by immunofluorescence with antibodies specific for the HCoV-229E N protein and with an anti-rabbit secondary antibody conjugated with Alexa fluor 488 (green). Nuclei were labeled with DAPI (blue). Images were collected with an epifluorescence microscope. (A) Representative pictures of the immunofluorescence assay. Scale bars, 100 µm. (B) Dose-response curves (red lines) of OSW-1, HβCD, U18666A, Phytol, Mdivi-1, FLI 06, Baclogen and CI976 were determined by nonlinear regression. Data is shown as mean ± S.E.M. of 3 biological replicates. Cytotoxic effect on MRC-5 cells exposed to increasing concentrations of drugs in the absence of virus is also shown (black lines).
3.3. OSW-1, HβCD, U18666A and Phytol show anti- SARS-CoV-2 activity in Vero E6 cells
Compounds that revealed antiviral activity against HCoV-229E were next evaluated against SARS-CoV-2. Vero E6 cells were exposed to SARS-CoV-2 in the presence of increasing concentrations of the different drugs. After three days, the cytopathic effect of the virus and the cytotoxic effect of the drugs on cells were analyzed and the IC50 value calculated for each drug at non-cytotoxic concentrations. Results showed that OSW-1, HβCD, U18666A and Phytol had antiviral activity on these cells ( Fig. 3 A), while the rest of tested compounds were not active or were cytotoxic (Fig. S1 and Table 2). The antiviral activity of these four drugs was similar when tested for different variants of concern of SARS-CoV-2 and comparable to the control Remdesivir (Fig. 3 B). The antiviral activity found for tested compounds with both HCoV-229E and SARS-CoV-2 is summarized in Table 1, Table 2.
Fig. 3.
Antiviral activity of drugs against SARS-CoV-2. (A) Cytopathic effect on Vero E6 cells exposed to 200 TCID50/mL of SARS-CoV-2 in the presence of increasing concentrations of OSW-1 (1 – 1,3 ×10-5 µM), HβCD (20 – 0,00026 mM), U18666A (20 – 2,6 ×10-4 µM) and Phytol (100 – 0,78 µM). Non-linear fit to a variable response curve from one representative experiment out of three with two replicates is shown (red lines), excluding data from drug concentrations with associated toxicity. Cytotoxic effect with the same drug concentrations in the absence of virus is also shown (black lines). The IC50 value is indicated on each graph. IC50 for OSW-1 could not be calculated because 100 % inhibition was not obtained with this compound. (B) Cytopathic effect on Vero E6 cells exposed to 200 TCID50/mL of different variants of concern of SARS-CoV-2 as described in A, and in presence of Remdesivir.
Table 2.
List of in vitro tested compounds against SARS-CoV-2.
| Drugs | IC50 ± SD (µM) | First cytotoxic conc. (µM) |
|---|---|---|
| Remdesivir | 0.51 ± 0.71 | 100 |
| OSW-1 | ∼ 0.002 ± 68.41 | 0.007 |
| Hβ-CD | 847.09 ± 2240.10 | 20000 |
| U18666A | ∼ 0.17 ± 539.43 | 20 |
| Phytol | ∼ 140.90 ± 402.29 | 100–200 |
| Cytotoxic drugs | ||
| Bortezomib | ∼ 0.4958 | 0.8 |
| FLI06 | 0.0151 | 4–6.25 |
| Mdivi-1 | ∼ 0.8317 | 20 |
| PIK93 | ∼ 0.005508 | 20 |
| CI976 | 0.2997 | 100 |
| Valinomycin | NA | 0.32 |
| Dehydrocostus lactone | ∼ 0.8439 | 100 |
| Digitoxin | ∼ 1.524 | 1.56 |
| Wortmanin | 0.941 | 25 |
| Terpinolene | ∼ 2.305 | 10000 |
| Gingenoside | ∼ 1.403 | 50 |
| No active drugs | ||
| L-arginine | ∼ 0.000 | NA |
| α-Terpineol | ∼ 23.67 | NA |
The IC50 median and SD of the viral cytopathic effect from three independent experiments is provided, and the first drug-cytotoxic concentration is provided for uninfected cells. SD, standard deviation; NA, non-applicable.
Then we investigated the mechanism of action of the four selected antivirals active against α and β coronavirus. We first performed morphological studies of Vero E6 cells infected with SARS-CoV-2 by transmission electron microscopy (TEM). These studies showed characteristic structures assembled by SARS-CoV-2, which are: double-membrane vesicles (DMVs) where the virus replicates its genome ( Fig. 4 A-C), viral particles inside single-membrane vesicles (SMVs) (Fig. 4 D), complex vacuoles (CV) with viruses (Fig. 4 E) and extracellular viral particles (VPs) (Fig. 4 F).
Fig. 4.
TEM of Vero E6 cells infected with SARS-CoV-2. Ultrathin sections of cells infected with SARS-CoV-2 at MOI of 0.02 PFU/cell and 48 hpi. (A) Overview of a cell with clusters of DMVs (asterisks) near the nucleus (N). (B) High magnification of a group of early DMVs with electron-dense content. (C) Cluster of late DMVs with fibrillar content. (D) Single membrane vesicles with viral particles in their lumen (white arrowheads). (E) Complex vacuole (CV) with viral particles (black arrowheads) in their lumen. The inset shows a higher magnification of one of the viral particles inside the CV. (F) Viral particles (black arrowheads) at the plasma membrane (P). M, mitochondrion, Scale bars, 1 µm in A, 200 in B-F.
TEM of SARS-CoV-2 infected cells treated with two different concentrations of OSW-1 (Fig. S2 and Table S4) and of U18666A (Fig. S3 and Table S4) suggested that in Vero E6 cells, these compounds block DMVs’ assembly and function, which would affect subsequent steps of viral morphogenesis. Mock-infected cells incubated with OSW-1 exhibited normal morphology and no alterations were detected in any of the cell organelles compared to mock-infected cells in the absence of the drug (Fig. S4 and S5). Incubation with the lower dose of U18666A caused no appreciable alterations in mock-infected cells (Fig. S6A -C) but swelling of Golgi cisternae and lysosomes were observed when the higher dose is applied (Fig. S6D -F). For phytol analysis, infected cells were incubated also with two different concentrations. Cells treated with the highest concentration showed clear signs of cytopathic effect and only the cells treated with the lowest concentration were processed for TEM. In phytol-treated cells, DMVs were altered to some extent but treatment had minor effects in virus assembly and egress (Fig. S7A -E and Table S4). In mock-infected cells, phytol did not cause significant alterations, with the only exception of mild increase in the amount of cytosolic glycogen granules and lipid droplets (Fig. S7F -I).
When SARS-CoV-2 exposed cells were treated with HβCD at the suboptimal concentration of 0.16 mM, DMVs showed alterations of the inner membrane ( Fig. 5 A and B). Viruses inside SMVs appear normal, but large groups of distorted viral particles were seen inside complex vacuoles (Fig. 5 C). Extracellular virions attached to the plasma membrane looked normal (Fig. 5 D). At inhibiting concentrations using 20 mM of HβCD, all viral structures were reduced (Fig. 5 E and Table S4) and a few detected DMVs showed membrane alterations (Fig. 5 F and G). Intracellular viral particles inside SMVs (Fig. 5 F), CVs (Fig. 5 H) and extracellular virions (Fig. 5 I) were seen in low numbers compared with infected non-treated cells (Table S4). These results show that HβCD affects the biogenesis of DMVs and the subsequent morphogenesis of new viral particles, an observation that could be linked to the capacity of this compound to reduce viral fusion and alter cellular membranes. At the lowest concentration, HβCD did not produce appreciable changes in cell compartments of mock-infected cells (Fig. S8A -C). However, non-infected cells with 20 mM of HβCD had increased number of vacuoles (Fig. S8D), swelling of ER, Golgi and lysosomes (Fig. S8E and F).
Fig. 5.
TEM of Vero E6 cells infected with SARS-CoV-2 and effects of HβCD. Cells infected with SARS-CoV-2 at MOI of 0.02 PFU/cell were incubated with 0.16 mM (A-D) or 20 mM HβCD (E-I) and prepared for TEM at 48 hpi. (A) Overview of a cell with a cluster of DMVs (asterisk). (B) Group of DMVs with alteration of the inner membrane (arrows) and a single membrane vesicle with a viral particle (white arrowhead) in close vicinity. (C) Complex vacuole (CV) with viral particles in the lumen. The inset shows a viral particle at higher magnification. (D) Viral particles (black arrowheads) at the plasma membrane (P). (E) Overview of a cell. (F) DMV with a group of single membrane vesicles containing viral particles (white arrowheads) in close vicinity. (G) Group of DMVs with alteration of the inner membrane (arrow). (H) Complex vacuole with viral particles (arrowheads) in the lumen. (I) Viral particles (black arrowheads) at the plasma membrane (P). N, nucleus. Scale bars, 1 µm in A and E; 200 nm in B-D, F-I.
3.4. HβCD and U18666A block viral fusion, but only HβCD inhibits infection in pulmonary cells
We therefore assessed the capability of these four drugs to inhibit the entry of SARS-CoV-2 pseudovirus in ACE2 expressing HEK-293T cells. Cells were exposed to fixed amounts of lentiviruses pseudotyped with SARS-CoV-2 spike in the presence of decreasing drug concentrations. Results showed that at drug concentrations without any cytotoxic effect, HβCD and U18666A inhibited the entry of pseudovirus, while OSW-1 and Phytol did not ( Fig. 6 A). Cytopathic effect of the drugs in the absence of virus is shown Fig. S9A. These findings with the functional pseudoviral assay suggest that the mechanism of action of cyclodextrins is to reduce viral fusion.
Fig. 6.
Drug inhibition of pseudovirus entry in ACE2–293 T cells and of SARS-CoV-2 infection in pulmonary cells. (A) Relative viral entry of SARS-CoV-2 pseudoviruses in the presence of the indicated drugs in ACE2 expressing HEK-293 T cells. Cells were exposed to fixed amounts of SARS-CoV-2 Spike lentiviruses in the presence of decreasing drug concentrations. Values show luciferase expression of the reporter lentiviruses pseudotyped with SARS-CoV-2, normalized to the luciferase expression of mock-treated cells (set at 100 %). Mean and standard deviation from two experiments with two replicates each are represented, excluding cytotoxic values. (B) Relative viral replication of SARS-CoV-2 assessed on CaLu-3 cells in the presence of the indicated drugs. After 24 h of adding virus and drugs at the indicated concentrations, cells were washed and compounds were added at the same final concentration for an additional 48 h. Then supernatants were tested for viral release by detecting SARS-CoV-2 nucleocapsid concentration by ELISA. Values are normalized to the nucleocapsid concentration by mock-treated cells (set at 100 %), which reached 5716 ± 2237 pg/mL (mean ± SD). Mean and standard deviation from four experiments are represented, excluding cytotoxic values.
We next tested SARS-CoV-2 antiviral activity in pulmonary Calu-3 cells as a more physiological and relevant cellular model. After incubating these cells with the drugs and the virus for 24 h, the virus was washed away and compounds were added at the same concentration for 48 h. The amount of SARS-CoV-2 nucleoprotein released to the supernatant was then measured by ELISA (Fig. 6 B) and the cytopathic effect of the drugs in the absence of virus in Calu-3 cells was assessed by luminescence (Fig. S9B). Only HP-β-CD at 10 mM effectively inhibited SARS-CoV-2 viral release into the supernatant of Calu-3 cells as observed for control remdesivir, while no inhibition was observed for the other compounds (Fig. S9D) at non-cytotoxic concentrations. Hence, HP-β-CD resulted as the most promising candidate to block SARS-CoV-2 replication on pulmonary cells.
3.5. Other members of the Cyclodextrin family also inhibit SARS-CoV-2 infection in pulmonary cells
Given the well-known safety profiles of different types of β-cyclodextrins, that have been used as excipients in the pharmaceutical industry for decades [26], we next aimed to test whether other cyclodextrins could hold the potential to inhibit viral replication. To test if other cyclodextrins could inhibit SARS-Cov-2 pseudoviral entry, we performed studies in ACE2 expressing HEK-293 T cells. At no cytotoxic concentrations (Fig. S9C), seven cyclodextrins inhibited the pseudoviral SARS-CoV-2 entry, being the most potent β, HP-β, HP-β and methyl-β-CDs ( Fig. 7 A).
Fig. 7.
Members of the Cyclodextrin family inhibit SARS-CoV-2 infection in pulmonary cells. (A) Relative viral entry of SARS-CoV-2 pseudoviruses in the presence of the indicated cyclodextrins in ACE2 expressing HEK-293 T cells. Cells were exposed to fixed amounts of SARS-CoV-2 Spike lentiviruses in the presence of decreasing drug concentrations. Values show luciferase expression of the reporter lentiviruses pseudotyped with SARS-CoV-2, normalized to the luciferase expressi % on of mock-treated cells (set at 100 %). Mean and standard deviation from two experiments with two replicates each are represented, excluding cytotoxic values. (B) Relative viral replication of D614G (C) or Omicron (D) SARS-CoV-2 variant was assessed on CaLu-3 cells in the presence of the indicated cyclodextrins. After 24 h of adding virus and drugs at the indicated concentrations, cells were washed and compounds were added at the same final concentration for additional 48 h. Then supernatants were tested for viral release by detecting SARS-CoV-2 nucleocapsid concentration by ELISA. Values are normalized to the nucleocapsid concentration by mock-treated cells (set at 100 %), which reached 5716 ± 2237 pg/mL (mean ± SD). Mean and standard deviation from three experiments are represented, excluding cytotoxic values.
Next, we assayed if the most active cyclodextrins inhibiting pseudoviral fusion could also block SARS-CoV-2 replication in pulmonary Calu3 cells. The quantification of SARS-CoV-2 nucleoprotein released to the supernatant detected by ELISA revealed that β-CDs, HP-β-CDs, and methyl-β-CDs inhibited the SARS-CoV-2 viral activity on Calu-3 cells, for the D614G (Fig. 7 B) and the Omicron BA.1 variant of concern (Fig. 7 C) at no cytotoxic concentrations. These results further highlight the potential of the β cyclodextrin family as antivirals against SARS-CoV-2.
3.6. Cyclodextrins inhibit SARS-CoV-2 replication by interfering with viral fusion via cholesterol depletion
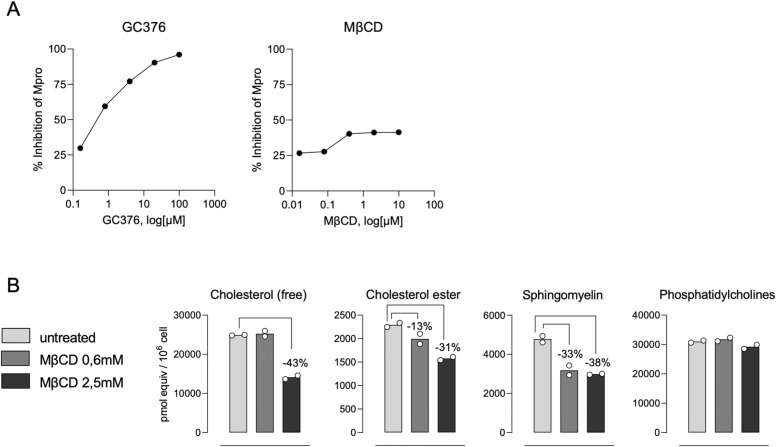
We had previously found that β-CD are in the catalytic pocket of the Mpro of SARS-CoV-2 by molecular modeling (Fig. 1 C). This finding pointed to a possible explanation for the antiviral activity detected. All cyclodextrins, including α, β, β, HP- α, HP-β, HP-β and methyl-β-CDs, were screened by molecular modeling against the SARS-CoV-2 Mpro. Since it is possible to inhibit the action of Mpro by targeting two allosteric sites -PDB codes 7AXM and 7AGA- [17]), we expanded the chemical space of search into the designed models. Theory predicts that cyclodextrins can bind both active and allosteric Mpro sites, although significant dissimilarities appear depending on the size and nature of the macrocycle (Table S3). To test whether this interaction occurred in vitro, the Mpro activity was measured in presence of increasing concentrations of methyl-β-CD (MβCD). While MβCD did not show activity, active GC376 control inhibited Mpro in a dose dependent manner ( Fig. 8 A).
Fig. 8.
Cyclodextrins inhibit SARS-CoV-2 replication by interfering with viral fusion via cholesterol depletion. (A) Mpro activity measured in presence of increasing concentrations of methyl-β-CD (MβCD) or GC376 as positive inhibitor. Results are represented as the percentage of inhibition of Mpro activity in the absence of drugs. (B) Lipidomic measurement of plasma membranes from Calu3 cells treated or not with MβCD for 2 h at 37ºC and 5 % CO2.
We next explored alternative antiviral mechanisms of action of cyclodextrins. Given the well-known capacity of cyclodextrins to extract cholesterol from biological membranes [28], we performed a lipidomic analysis focusing on different lipids associated to cholesterol enriched domains in biological membranes. Calu-3 cells treated with increasing concentrations of MβCD showed a reduction of free and ester cholesterol along with sphingomyelin, and did not affect phosphatidylcholine (Fig. 8 B). These results further confirm the capacity of cyclodextrins to alter the composition of cholesterol enriched domains actively involved in viral fusion processes. Taken together, these experiments along the pseudoviral fusion assays highlight the potential of cyclodextrins to inhibit SARS-CoV-2 replication by interfering with viral fusion via cholesterol depletion.
3.7. Methyl-β-cyclodextrin inhibits SARS-CoV-2 replication in a human nasal epithelial model
We aimed to test the antiviral effect of MβCD in a more physiologically relevant model. We used a human nasal epithelial (HNE) model which mimics the characteristics of in vivo nasal tissue. In this HNE model we added MβCD, either on the apical side or the basal medium of the culture ( Fig. 9 A), just before adding SARS-CoV-2 to the apical side for 1 h. Apical side was then extensively washed and inserts were incubated for 72 h. Remdesivir was added to the basal medium, while other inserts were left untreated. Viral replication was monitored at 48 and 72 hpi by measuring SARS-CoV-2 nucleocapsid concentration by ELISA. SARS-CoV-2 released to the apical supernatant picked at 48 hpi and decreased at 72 hpi (Fig. 9 B-C). Initial MβCD addition to the apical side reduced viral replication by 74 % at 48 h pi (Fig. 9 B), while constant presence of MβCD in the basal side almost had no effect (11 %) as opposed to the condition with remdesivir that completely blocked it (Fig. 9 B-C). The cytopathic effect measured at 72 hpi indicated the HNE cells were alive with all the tested conditions (Fig. S9E). These results further emphasize the potential use of MβCD as a prophylactic antiviral in nasal sprays against SARS-CoV-2.
Fig. 9.
Methyl-β-cyclodextrin inhibit SARS-CoV-2 replication in a human nasal epithelial (HNE) model. (A) Schematic representation of the HNE model used, showing the apical side, the basal medium and the cells cultured in the air-liquid interphase. (B) SARS-CoV-2 replication in the HNE model in the presence of 2.5 mM MβCD either on the apical side (red) or on the basal medium (orange), or 2.5 uM Remdesivir (green) on the basal medium, without drugs (blue), or in the absence of virus (black). SARS-CoV-2 was added to the apical side for 1 h, extensively washed afterwards, and nucleocapsid concentration was measured by ELISA at 24, 48 and 72 hpi. (C) The results of two independent experiments following the same procedure explained in ‘B’ and measured at 72 hpi.
3.8. Prophylaxis treatment with Methyl-β-cyclodextrin protects animals from SARS-CoV-2 infection
To test the capacity of MβCD to prevent SARS-CoV-2 infection in vivo, we tested its prophylactic activity in hamsters. Animals were pre-treated with MβCD or PBS intra-nasally before challenging via the same route with a SARS-CoV-2 reporter virus that produces nano-luciferase upon infection. Animals were euthanized on day 1 and 2 post infection and compared to uninfected controls, measuring the amount of nanoluciferase content in nasal turbinates and lungs ( Fig. 10 A-B). On day 1, the four infected untreated animals had detectable nanoluciferase levels both at the nasal turbinates and lungs, while no nanoluciferase signal was detected on MβCD treated animals. Similar results were obtained on day 2 post-infection, where higher levels of nanoluciferase were detected in the nasal turbinates and lungs of untreated infected animals, and no sign of infection was detected on MβCD treated animals (Fig. 10 A-B). These results were further confirmed by qPCR on days 1 and 2 post-infection in nasal turbinates and lungs (Fig. 10 C-D), and by viral infectivity (Fig. 10 E-F). Similar protection trend was observed in the histopathology and haematoxylin and eosin-stained sections of nasal turbinates and lungs of MβCD-treated animals as compared to controls (Fig. 10 G-J). Protective trend was also observed with a replicative SARS-CoV-2 isolate specially in the lung tissue (Fig. S10), but with a less potent prophylactic effect as compared to the Nanoluciferase virus. These results further confirm the prophylactic capacity of MβCD to protect from SARS-CoV-2 infection in the nasal epithelium of hamsters.
Fig. 10.
Methyl-β-cyclodextrin nasal application inhibits SARS-CoV-2 replication in a hamster model. Hamsters were intranasally administered with or without MβCD at 50 mM and challenged with a SARS-CoV-2 Nanoluciferase reporter virus. Control uninfected animals were also assayed. (A-B) At 1- or 2-days post-infection, nasal turbinates and lungs collected from euthanized animals were lysed with the Nano-glo Luciferase system (Promega) and luminescence was measured with a plate reader in relative light units (RLUs). (C-J) At 1- or 2-days post-infection, nasal turbinates and lungs collected from euthanized animals were analysed for (C-D) viral RNA presence in inverted CTs by qPCR, (E-F) by viral infectivity, (G-H) by immunohistochemistry to detect the NP of SARS-CoV-2 and (I-J) by histopathological observation in haematoxylin and eosin-stained sections.
4. Discussion
Although pathogenic viruses pose a real and growing threat to public health, we have few medicines to prevent and treat viral infections. Here, a library of potential inhibitors of coronavirus infection was elaborated with a “based-on-knowledge” strategy. Examining the available information about what viruses use to complete their life cycle in cells and the description of the mechanism of action of drugs, we found compounds with potential to be used as antivirals to treat coronavirus infections. The main advantage of this strategy compared to high-throughput analysis is that the list of candidates is limited, and that different protocols of infection and drug treatment can be tested, which increases the probabilities of identifying molecules with antiviral activity. We have used a workflow involving biocomputational analysis and several biological assays to carefully select potential antivirals against different coronaviruses, including HCoV-229E and SARS-CoV-2. From our list of 116 compounds that target cell factors and pathways, 4 showed antiviral activity against both coronaviruses. Results with β-CDs were particularly relevant as showed consistent efficacy in different cellular models including human pulmonary cells and the nasal epithelium of hamsters in vivo. Particularly promising is our finding suggesting that the mechanism of action of β-CDs is interfering with viral fusion. Results from lipidomic analysis and transmission electron microscopy (TEM) showed that β-CDs may interfere with coronavirus infection by altering cholesterol and sphingomyelin content and disrupting the organization of membranes used by the virus. This is supported by our TEM results that showed dose-dependent effects of β-CDs in all SARS-CoV-2 structures in infected cells. The integrity of DMVs was compromised and viral morphogenesis was impaired, with the production of abnormal viral particles that are sometimes trapped in DMVs and inside large vacuoles that could represent degradation compartments. Our results confirm and broaden recent findings focused on HP-β-CDs [4], and with different methods further expand these results encompassing other members of the cyclodextrin family and SARS-CoV2 VOCs, including Omicron.
Given that cyclodextrins are suited for oral, nasal or nebulized solutions, these results open avenues to test diverse drug formulations known to be safe in humans [50], [53]. The reduced infection after MβCD prophylactic addition on a human nasal airway epithelium and in a physiologically relevant hamster model shown herein further supports the clinical testing of these compounds. To the best of our knowledge this is the first set of data that shows the efficacy of MβCD in inhibiting viral infection in the nasal epithelium and upper respiratory tract of an animal model. The well-known safety profiles of β-CDs render these molecules as ideal candidates to develop affordable prophylactic compounds against coronaviruses [11], [49]. Such drugs, which are already approved for clinical use in nasal spray devices [16], [36], may be easier to deploy in low income countries compared to vaccines, which often require cold storage and must be administered by trained personnel. Given that β-CDs are widely used for compound encapsulation, they could be easily combined with other antivirals to potentiate activity and avoid viral resistance. Finally, the broad-spectrum mechanism of action shown herein, which inhibits viral fusion with cellular membranes, could help to counteract other respiratory viruses, providing an arsenal to deploy in front of new variants of concern or future novel coronaviruses with pandemic potential. Broad-spectrum antivirals such as β-CDs could be ultimately applied to counteract unknown emergent viruses yet to appear, but will need rigorous clinical assessment for further development.
Funding
This work has been funded by grant RTI2018-094445-B100 (MCIU/AEI/FEDER, UE) from the Ministry of Science and Innovation of Spain (C.R.), by Palex Medical S.A., Sika S.A.U. and 7 more companies, and by Ms. Raquel Casaus Alvarez, Mr. Miguel Pardo Gil, Mr. Jacques Noguès and a total of 2916 citizens through the Precipita crowdfunding platform of Fecyt (Fundación Española para la Ciencia y la Tecnología). NI-U is supported by the Spanish Ministry of Science and Innovation (grant PID2020-117145RB-I00), EU HORIZON-HLTH-2021-CORONA-01 (grant 101046118) and by institutional funding of Grifols, Pharma Mar, HIPRA, Amassence and Palobiofarma. This work used the computational resources of the Centro de Supercomputación de Galicia (CESGA) supported by the Partnership for Advanced Computing in Europe (PRACE) COVID-19 Fast Track Call for Proposals – Allocation Decision – Proposal COVID19-85.
CRediT authorship contribution statement
Dalia Raïch-Regué: Conceptualization, Investigation, Validation, original draft preparation. Raquel Tenorio: Conceptualization, Investigation, Validation, original draft preparation. Isabel Fernández de Castro: Conceptualization, Investigation, Validation, original draft preparation. Ferrán Tarrés: Conceptualization, Investigation. Martin Sachse: Conceptualization, Investigation, Validation, original draft preparation. Daniel Perez-Zsolt: Conceptualization, Investigation. Jordana Muñoz-Basagoiti: Conceptualization, Investigation. Sara Y. Fernández-Sánchez: Investigation, Validation. Marçal Gallemi: Conceptualization, Investigation. Paula Ortega-González: Conceptualization, Investigation. Alberto Fernández-Oliva: Conceptualization, Investigation. José A. Gabaldón: Conceptualization, Resources. Estrella Núñez-Delicado: Conceptualization, Resources. Josefina Casas: Investigation, Formal analysis. Núria Roca: Methodology, Investigation. Guillermo Cantero: Methodology, Investigation. Mónica P é rez: Methodology, Investigation. Carla Usai: Methodology, Investigation. Cristina Lorca-Oró: Methodology, Investigation. Júlia Vergara-Alert: Conceptualization. Joaquim Segalés: Conceptualization. Jorge Carrillo: Conceptualization. Julià Blanco: Conceptualization. Bonaventura Clotet Sala: Conceptualization, Resources, Funding acquisition. José P. Cerón-Carrasco: Conceptualization, Investigation, Formal analysis, Funding acquisition, Writing – review & editing. Nuria Izquierdo-Useros: Conceptualization, Supervision, Funding acquisition, Writing – review & editing. Cristina Risco: Conceptualization, Supervision, Funding acquisition, Writing – review & editing.
Declaration of Competing Interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: CR, IFC, MS, RT, PO-G, AF-O, SF-S, NI-U, DP-Z, JM-B, DR-R, JPC-C, JAG and EN-D are inventors of a patent application for use of cyclodextrins to treat viral infection (PCT/EP2023/057735). The authors declare no other related competing interest.
Acknowledgements
Special thanks to Dr. Mª Carmen Martínez Jiménez, from the Hospital Nuestra Señora de Sonsoles de Avila for sharing knowledge about the use of plant extracts and their active compounds to treat viral infections. We are grateful to Jorge Díaz for his constant support at the CMCIB facility. We thank the animal facility technicians A. Pou and Ò. García, the pathological anatomy technicians A. Barceló and R. López and the laboratory management unit I. Cordón, M. Mora an X. Abad for their support with the in vivo experiments at IRTA-CReSA. We thank Rytis Boreika for editing the manuscript.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.biopha.2023.114997.
Appendix A. Supplementary material
Supplementary material.
.
Supplementary material.
.
Supplementary material.
.
Supplementary material.
.
Supplementary material.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
References
- 1.Agarwal A., Rochwerg B., Lamontagne F., Siemieniuk R.A.C., Agoritsas T., Askie L., Lytvyn L., Leo Y.-S., Macdonald H., Zeng L., Amin W., Barragan F.A.J., Bausch F.J., Burhan E., Calfee C.S., Cecconi M., Chanda D., Dat V.Q., De Sutter A., Du B., Geduld H., Gee P., Harley N., Hashmi M., Hunt B., Jehan F., Kabra S.K., Kanda S., Kim Y.-J., Kissoon N., Krishna S., Kuppalli K., Kwizera A., Lisboa T., Mahaka I., Manai H., Mino G., Nsutebu E., Preller J., Pshenichnaya N., Qadir N., Sabzwari S., Sarin R., Shankar-Hari M., Sharland M., Shen Y., Ranganathan S.S., Souza J.P., Stegemann M., Swanstrom R., Ugarte S., Venkatapuram S., Vuyiseka D., Wijewickrama A., Maguire B., Zeraatkar D., Bartoszko J.J., Ge L., Brignardello-Petersen R., Owen A., Guyatt G., Diaz J., Kawano-Dourado L., Jacobs M., Vandvik P.O. A living WHO guideline on drugs for covid-19. BMJ. 2020:m3379. doi: 10.1136/bmj.m3379. [DOI] [PubMed] [Google Scholar]
- 2.Benet S., Blanch-Lombarte O., Ainsua-Enrich E., Pedreño-Lopez N., Muñoz-Basagoiti J., Raïch-Regué D., Perez-Zsolt D., Peña R., Jiménez E., de la Concepción M.L.R., Ávila C., Cedeño S., Escribà T., Romero-Martín L., Alarcón-Soto Y., Rodriguez-Lozano G.F., Miranda C., González S., Bailón L., Blanco J., Massanella M., Brander C., Clotet B., Paredes R., Esteve M., Izquierdo-Useros N., Carrillo J., Prado J.G., Moltó J., Mothe B. Limited humoral and specific T-cell responses after SARS-CoV-2 vaccination in PLWH with poor immune reconstitution. J. Infect. Dis. 2022 doi: 10.1093/infdis/jiac406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berman H.M., Battistuz T., Bhat T.N., Bluhm W.F., Bourne P.E., Burkhardt K., Feng Z., Gilliland G.L., Iype L., Jain S., Fagan P., Marvin J., Padilla D., Ravichandran V., Schneider B., Thanki N., Weissig H., Westbrook J.D., Zardecki C. The protein data bank. Acta Crystallogr. Sect. D. Biol. Crystallogr. 2002;58:899–907. doi: 10.1107/S0907444902003451. [DOI] [PubMed] [Google Scholar]
- 4.Bezerra B.B., Silva G.P.D., da, Coelho S.V.A., Correa I.A., Souza M.R.M., de, Macedo K.V.G., Matos B.M., Tanuri A., Matassoli F.L., Costa L.J., da, Hildreth J.E.K., Arruda L.B. de. Hydroxypropyl-beta-cyclodextrin (HP-BCD) inhibits SARS-CoV-2 replication and virus-induced inflammatory cytokines. Antivir. Res. 2022;205 doi: 10.1016/j.antiviral.2022.105373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bowers K.J., Chow E., Xu H., Dror R.O., Eastwood M.P., Gregersen B.A., Klepeis J.L., Istvan Kolossvary M., Moraes A., Sacerdoti F.D., Salmon J.K., Shan Y., Shaw D.E. Proceedings of the 2006 ACM/IEEE Conference on Supercomputing (SC ’06) Association for Computing Machinery; 2006. Scalable algorithms for molecular dynamics simulations on commodity clusters. [DOI] [Google Scholar]
- 6.Brustolin M., Rodon J., Rodríguez de la Concepción M.L., Ávila-Nieto C., Cantero G., Pérez M., Te N., Noguera-Julián M., Guallar V., Valencia A., Roca N., Izquierdo-Useros N., Blanco J., Clotet B., Bensaid A., Carrillo J., Vergara-Alert J., Segalés J. Protection against reinfection with D614- or G614-SARS-CoV-2 isolates in golden Syrian hamster. Emerg. Microbes Infect. 2021;10:797–809. doi: 10.1080/22221751.2021.1913974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buxeraud J., Faure S., Fougere É. Le nirmatrelvir-ritonavir (Paxlovid®), un traitement contre la Covid-19. Actual. Pharm. 2022;61:10–12. doi: 10.1016/j.actpha.2022.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cerón‐Carrasco J.P. When virtual screening yields inactive drugs: dealing with false theoretical friends. ChemMedChem. 2022:17. doi: 10.1002/cmdc.202200278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cho J., Lee Y.J., Kim J.H., Kim S., il, Kim S.S., Choi B.-S., Choi J.-H. Antiviral activity of digoxin and ouabain against SARS-CoV-2 infection and its implication for COVID-19. Sci. Rep. 2020;10:16200. doi: 10.1038/s41598-020-72879-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Douangamath A., Fearon D., Gehrtz P., Krojer T., Lukacik P., Owen C.D., Resnick E., Strain-Damerell C., Aimon A., Ábrányi-Balogh P., Brandão-Neto J., Carbery A., Davison G., Dias A., Downes T.D., Dunnett L., Fairhead M., Firth J.D., Jones S.P., Keeley A., Keserü G.M., Klein H.F., Martin M.P., Noble M.E.M., O’Brien P., Powell A., Reddi R.N., Skyner R., Snee M., Waring M.J., Wild C., London N., von Delft F., Walsh M.A. Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease. Nat. Commun. 2020;11:5047. doi: 10.1038/s41467-020-18709-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fatmi S., Taouzinet L., Skiba M., Iguer-Ouada M. The use of cyclodextrin or its complexes as a potential treatment against the 2019 novel coronavirus: a mini-review. Curr. Drug Deliv. 2021;18:382–386. doi: 10.2174/1567201817666200917124241. [DOI] [PubMed] [Google Scholar]
- 12.Friesner R.A., Murphy R.B., Repasky M.P., Frye L.L., Greenwood J.R., Halgren T.A., Sanschagrin P.C., Mainz D.T. Extra precision glide: docking and scoring incorporating a model of hydrophobic enclosure for protein−ligand complexes. J. Med. Chem. 2006;49:6177–6196. doi: 10.1021/jm051256o. [DOI] [PubMed] [Google Scholar]
- 13.Gao X., Qin B., Chen P., Zhu K., Hou P., Wojdyla J.A., Wang M., Cui S. Crystal structure of SARS-CoV-2 papain-like protease. Acta Pharm. Sin. B. 2021;11:237–245. doi: 10.1016/j.apsb.2020.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.García-Dorival I., Cuesta-Geijo M.Á., Barrado-Gil L., Galindo I., Garaigorta U., Urquiza J., Puerto A., del, Campillo N.E., Martínez A., Gastaminza P., Gil C., Alonso C. Identification of Niemann-Pick C1 protein as a potential novel SARS-CoV-2 intracellular target. Antivir. Res. 2021;194 doi: 10.1016/j.antiviral.2021.105167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Greenwood J.R., Calkins D., Sullivan A.P., Shelley J.C. Towards the comprehensive, rapid, and accurate prediction of the favorable tautomeric states of drug-like molecules in aqueous solution. J. Comput. Aided Mol. Des. 2010;24:591–604. doi: 10.1007/s10822-010-9349-1. [DOI] [PubMed] [Google Scholar]
- 16.Guard S., Watling K.J., Watson S.P. Characterisation of [3H]-senktide binding to NK3 tachykinin receptors in guinea-pig ileum and cerebral cortex. Br. J. Pharmacol. 1989;98:798P. [PubMed] [Google Scholar]
- 17.Günther Sebastian, Reinke P.Y.A., Fernández-García Y., Lieske J., Lane T.J., Ginn H.M., Koua F.H.M., Ehrt C., Ewert W., Oberthuer D., Yefanov O., Meier S., Lorenzen K., Krichel B., Kopicki J.-D., Gelisio L., Brehm W., Dunkel I., Seychell B., Gieseler H., Norton-Baker B., Escudero-Pérez B., Domaracky M., Saouane S., Tolstikova A., White T.A., Hänle A., Groessler M., Fleckenstein H., Trost F., Galchenkova M., Gevorkov Y., Li C., Awel S., Peck A., Barthelmess M., Schlünzen F., Lourdu Xavier P., Werner N., Andaleeb H., Ullah N., Falke S., Srinivasan V., França B.A., Schwinzer M., Brognaro H., Rogers C., Melo D., Zaitseva-Doyle J.J., Knoska J., Peña-Murillo G.E., Mashhour A.R., Hennicke V., Fischer P., Hakanpää J., Meyer J., Gribbon P., Ellinger B., Kuzikov M., Wolf M., Beccari A.R., Bourenkov G., von Stetten D., Pompidor G., Bento I., Panneerselvam S., Karpics I., Schneider T.R., Garcia-Alai M.M., Niebling S., Günther C., Schmidt C., Schubert R., Han H., Boger J., Monteiro D.C.F., Zhang L., Sun X., Pletzer-Zelgert J., Wollenhaupt J., Feiler C.G., Weiss M.S., Schulz E.-C., Mehrabi P., Karničar K., Usenik A., Loboda J., Tidow H., Chari A., Hilgenfeld R., Uetrecht C., Cox R., Zaliani A., Beck T., Rarey M., Günther Stephan, Turk D., Hinrichs W., Chapman H.N., Pearson A.R., Betzel C., Meents A. X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease. Science (80–.) 2021;372:642–646. doi: 10.1126/science.abf7945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ha T.K.Q., Lee B.W., Nguyen N.H., Cho H.M., Venkatesan T., Doan T.P., Kim E., Oh W.K. Antiviral Activities of Compounds Isolated from Pinus densiflora (Pine Tree) against the Influenza A Virus. Biomolecules. 2020;10:711. doi: 10.3390/biom10050711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haagmans B.L., van den Brand J.M.A., Raj V.S., Volz A., Wohlsein P., Smits S.L., Schipper D., Bestebroer T.M., Okba N., Fux R., Bensaid A., Solanes Foz D., Kuiken T., Baumgärtner W., Segalés J., Sutter G., Osterhaus A.D.M.E. An orthopoxvirus-based vaccine reduces virus excretion after MERS-CoV infection in dromedary camels. Science. 2016;80-(351):77–81. doi: 10.1126/science.aad1283. [DOI] [PubMed] [Google Scholar]
- 20.Halgren T.A., Murphy R.B., Friesner R.A., Beard H.S., Frye L.L., Pollard W.T., Banks J.L. Glide: a new approach for rapid, accurate docking and scoring. 2. Enrichment factors in database screening. J. Med. Chem. 2004;47:1750–1759. doi: 10.1021/jm030644s. [DOI] [PubMed] [Google Scholar]
- 21.Jacobson M.P., Friesner R.A., Xiang Z., Honig B. On the role of the crystal environment in determining protein side-chain conformations. J. Mol. Biol. 2002;320:597–608. doi: 10.1016/S0022-2836(02)00470-9. [DOI] [PubMed] [Google Scholar]
- 22.Jacobson M.P., Pincus D.L., Rapp C.S., Day T.J.F., Honig B., Shaw D.E., Friesner R.A. A hierarchical approach to all-atom protein loop prediction. Proteins Struct. Funct. Bioinforma. 2004;55:351–367. doi: 10.1002/prot.10613. [DOI] [PubMed] [Google Scholar]
- 23.Jayk Bernal A., Gomes da Silva M.M., Musungaie D.B., Kovalchuk E., Gonzalez A., Delos Reyes V., Martín-Quirós A., Caraco Y., Williams-Diaz A., Brown M.L., Du J., Pedley A., Assaid C., Strizki J., Grobler J.A., Shamsuddin H.H., Tipping R., Wan H., Paschke A., Butterton J.R., Johnson M.G., De Anda C. Molnupiravir for oral treatment of Covid-19 in nonhospitalized patients. N. Engl. J. Med. 2022;386:509–520. doi: 10.1056/NEJMoa2116044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jeon S., Ko M., Lee J., Choi I., Byun S.Y., Park S., Shum D., Kim S. Identification of antiviral drug candidates against SARS-CoV-2 from FDA-Approved Drugs. Antimicrob. Agents Chemother. 2020:64. doi: 10.1128/AAC.00819-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., Zhang B., Li X., Zhang L., Peng C., Duan Y., Yu J., Wang L., Yang K., Liu F., Jiang R., Yang Xinglou, You T., Liu Xiaoce, Yang Xiuna, Bai F., Liu H., Liu Xiang, Guddat L.W., Xu W., Xiao G., Qin C., Shi Z., Jiang H., Rao Z., Yang H. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582:289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 26.Lachowicz M., Stańczak A., Kołodziejczyk M. Characteristic of cyclodextrins: their role and use in the pharmaceutical technology. Curr. Drug Targets. 2020;21:1495–1510. doi: 10.2174/1389450121666200615150039. [DOI] [PubMed] [Google Scholar]
- 27.Liesenborghs L., Spriet I., Jochmans D., Belmans A., Gyselinck I., Teuwen L.-A., ter Horst S., Dreesen E., Geukens T., Engelen M.M., Landeloos E., Geldhof V., Ceunen H., Debaveye B., Vandenberk B., Van der Linden L., Jacobs S., Langendries L., Boudewijns R., Do T.N.D., Chiu W., Wang X., Zhang X., Weynand B., Vanassche T., Devos T., Meyfroidt G., Janssens W., Vos R., Vermeersch P., Wauters J., Verbeke G., De Munter P., Kaptein S.J.F., Rocha-Pereira J., Delang L., Van Wijngaerden E., Neyts J., Verhamme P. Itraconazole for COVID-19: preclinical studies and a proof-of-concept randomized clinical trial. EBioMedicine. 2021;66 doi: 10.1016/j.ebiom.2021.103288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.López C.A., de Vries A.H., Marrink S.J. Molecular mechanism of cyclodextrin mediated cholesterol extraction. PLoS Comput. Biol. 2011;7 doi: 10.1371/journal.pcbi.1002020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu C., Wu C., Ghoreishi D., Chen W., Wang L., Damm W., Ross G.A., Dahlgren M.K., Russell E., Von Bargen C.D., Abel R., Friesner R.A., Harder E.D. OPLS4: improving force field accuracy on challenging regimes of chemical space. J. Chem. Theory Comput. 2021;17:4291–4300. doi: 10.1021/acs.jctc.1c00302. [DOI] [PubMed] [Google Scholar]
- 30.Madhavi Sastry G., Adzhigirey M., Day T., Annabhimoju R., Sherman W. Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments. J. Comput. Aided Mol. Des. 2013;27:221–234. doi: 10.1007/s10822-013-9644-8. [DOI] [PubMed] [Google Scholar]
- 31.Mesel-Lemoine M., Millet J., Vidalain P.-O., Law H., Vabret A., Lorin V., Escriou N., Albert M.L., Nal B., Tangy F. A human coronavirus responsible for the common cold massively kills dendritic cells but not monocytes. J. Virol. 2012;86:7577–7587. doi: 10.1128/JVI.00269-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Molina J.-M., Ghosn J., Assoumou L., Delaugerre C., Algarte-Genin M., Pialoux G., Katlama C., Slama L., Liegeon G., Beniguel L., Ohayon M., Mouhim H., Goldwirt L., Spire B., Loze B., Surgers L., Pavie J., Lourenco J., Ben-Mechlia M., Le Mestre S., Rojas-Castro D., Costagliola D. Daily and on-demand HIV pre-exposure prophylaxis with emtricitabine and tenofovir disoproxil (ANRS PREVENIR): a prospective observational cohort study. Lancet HIV. 2022;9:e554–e562. doi: 10.1016/S2352-3018(22)00133-3. [DOI] [PubMed] [Google Scholar]
- 33.Osipiuk J., Azizi S.-A., Dvorkin S., Endres M., Jedrzejczak R., Jones K.A., Kang S., Kathayat R.S., Kim Y., Lisnyak V.G., Maki S.L., Nicolaescu V., Taylor C.A., Tesar C., Zhang Y.-A., Zhou Z., Randall G., Michalska K., Snyder S.A., Dickinson B.C., Joachimiak A. Structure of papain-like protease from SARS-CoV-2 and its complexes with non-covalent inhibitors. Nat. Commun. 2021;12:743. doi: 10.1038/s41467-021-21060-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Osipiuk J., Wydorski P.M., Lanham B.T., Tesar C., Endres M., Engle E., Jedrzejczak R., Mullapudi V., Michalska K., Fidelis K., Fushman D., Joachimiak A., Joachimiak L.A. Dual domain recognition determines SARS-CoV-2 PLpro selectivity for human ISG15 and K48-linked di-ubiquitin. bioRxiv Prepr. Serv. Biol. 2022 doi: 10.1101/2021.09.15.460543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ou X., Liu Y., Lei X., Li P., Mi D., Ren L., Guo L., Guo R., Chen T., Hu J., Xiang Z., Mu Z., Chen X., Chen J., Hu K., Jin Q., Wang J., Qian Z. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020;11:1620. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paolacci S., Ergoren M.C., De Forni D., Manara E., Poddesu B., Cugia G., Dhuli K., Camilleri G., Tuncel G., Kaya Suer H., Sultanoglu N., Sayan M., Dundar M., Beccari T., Ceccarini M.R., Gunsel I.S., Dautaj A., Sanlidag T., Connelly S.T., Tartaglia G.M., Bertelli M. In vitro and clinical studies on the efficacy of α-cyclodextrin and hydroxytyrosol against SARS-CoV-2 infection. Eur. Rev. Med. Pharmacol. Sci. 2021;25:81–89. doi: 10.26355/eurrev_202112_27337. [DOI] [PubMed] [Google Scholar]
- 37.Patel N.K., Jaiswal G., Bhutani K.K. A review on biological sources, chemistry and pharmacological activities of pinostrobin. Nat. Prod. Res. 2016;30:2017–2027. doi: 10.1080/14786419.2015.1107556. [DOI] [PubMed] [Google Scholar]
- 38.Ramakrishnan M.A. Determination of 50% endpoint titer using a simple formula. World J. Virol. 2016;5:85–86. doi: 10.5501/wjv.v5.i2.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rastelli G., Rio A., Del, Degliesposti G., Sgobba M. Fast and accurate predictions of binding free energies using MM-PBSA and MM-GBSA. J. Comput. Chem. NA-NA. 2009 doi: 10.1002/jcc.21372. [DOI] [PubMed] [Google Scholar]
- 40.Ratia K., Kilianski A., Baez-Santos Y.M., Baker S.C., Mesecar A. Structural basis for the ubiquitin-linkage specificity and deISGylating activity of SARS-CoV papain-like protease. PLoS Pathog. 2014;10 doi: 10.1371/journal.ppat.1004113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reed L.J., Muench H. A simple method of estimating fifty per cent endpoint. Am. J. Hyg. 1938;27:493–497. [Google Scholar]
- 42.Rodon J., Muñoz-Basagoiti J., Perez-Zsolt D., Noguera-Julian M., Paredes R., Mateu L., Quiñones C., Perez C., Erkizia I., Blanco I., Valencia A., Guallar V., Carrillo J., Blanco J., Segalés J., Clotet B., Vergara-Alert J., Izquierdo-Useros N. Identification of plitidepsin as potent inhibitor of SARS-CoV-2-induced cytopathic effect after a drug repurposing screen. Front. Pharmacol. 2021;12 doi: 10.3389/fphar.2021.646676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rosa A., Pye V.E., Graham C., Muir L., Seow J., Ng K.W., Cook N.J., Rees-Spear C., Parker E., dos Santos M.S., Rosadas C., Susana A., Rhys H., Nans A., Masino L., Roustan C., Christodoulou E., Ulferts R., Wrobel A.G., Short C.-E., Fertleman M., Sanders R.W., Heaney J., Spyer M., Kjær S., Riddell A., Malim M.H., Beale R., MacRae J.I., Taylor G.P., Nastouli E., van Gils M.J., Rosenthal P.B., Pizzato M., McClure M.O., Tedder R.S., Kassiotis G., McCoy L.E., Doores K.J., Cherepanov P. SARS-CoV-2 can recruit a heme metabolite to evade antibody immunity. Sci. Adv. 7. 2021 doi: 10.1126/sciadv.abg7607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schäfer A., Martinez D.R., Won J.J., Meganck R.M., Moreira F.R., Brown A.J., Gully K.L., Zweigart M.R., Conrad W.S., May S.R., Dong S., Kalla R., Chun K., Du Pont V., Babusis D., Tang J., Murakami E., Subramanian R., Barrett K.T., Bleier B.J., Bannister R., Feng J.Y., Bilello J.P., Cihlar T., Mackman R.L., Montgomery S.A., Baric R.S., Sheahan T.P. Therapeutic treatment with an oral prodrug of the remdesivir parental nucleoside is protective against SARS-CoV-2 pathogenesis in mice. Sci. Transl. Med. 2022:14. doi: 10.1126/scitranslmed.abm3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shearman M.S., Ragan C.I., Iversen L.L. Inhibition of PC12 cell redox activity is a specific, early indicator of the mechanism of beta-amyloid-mediated cell death. Proc. Natl. Acad. Sci. U. S. A. 1994;91:1470–1474. doi: 10.1073/pnas.91.4.1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shelley J.C., Cholleti A., Frye L.L., Greenwood J.R., Timlin M.R., Uchimaya M. Epik: a software program for pK a prediction and protonation state generation for drug-like molecules. J. Comput. Aided Mol. Des. 2007;21:681–691. doi: 10.1007/s10822-007-9133-z. [DOI] [PubMed] [Google Scholar]
- 47.Shen Z., Ratia K., Cooper L., Kong D., Lee H., Kwon Y., Li Y., Alqarni S., Huang F., Dubrovskyi O., Rong L., Thatcher G.R., Xiong R. Potent, novel SARS-CoV-2 PLpro inhibitors block viral replication in monkey and human cell cultures. bioRxiv Prepr. Serv. Biol. 2021 doi: 10.1101/2021.02.13.431008. [DOI] [Google Scholar]
- 48.Shin D., Mukherjee R., Grewe D., Bojkova D., Baek K., Bhattacharya A., Schulz L., Widera M., Mehdipour A.R., Tascher G., Geurink P.P., Wilhelm A., van der Heden van Noort G.J., Ovaa H., Müller S., Knobeloch K.-P., Rajalingam K., Schulman B.A., Cinatl J., Hummer G., Ciesek S., Dikic I. Papain-like protease regulates SARS-CoV-2 viral spread and innate immunity. Nature. 2020;587:657–662. doi: 10.1038/s41586-020-2601-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sorice M., Misasi R., Riitano G., Manganelli V., Martellucci S., Longo A., Garofalo T., Mattei V. Targeting lipid rafts as a strategy against coronavirus. Front. Cell Dev. Biol. 2020;8 doi: 10.3389/fcell.2020.618296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stella V.J., He Q. Cyclodextrins. Toxicol. Pathol. 2008;36:30–42. doi: 10.1177/0192623307310945. [DOI] [PubMed] [Google Scholar]
- 51.Tai W., He L., Zhang X., Pu J., Voronin D., Jiang S., Zhou Y., Du L. Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: implication for development of RBD protein as a viral attachment inhibitor and vaccine. Cell. Mol. Immunol. 2020;17:613–620. doi: 10.1038/s41423-020-0400-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tenorio R., de Castro I.F., Knowlton J.J., Zamora P.F., Lee C.H., Mainou B.A., Dermody T.S., Risco C. Reovirus σNS and μNS proteins remodel the endoplasmic reticulum to build replication neo-organelles. MBio. 2018;9:15. doi: 10.1128/mBio.01253-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tian B., Hua S., Liu J. Cyclodextrin-based delivery systems for chemotherapeutic anticancer drugs: a review. Carbohydr. Polym. 2020;232 doi: 10.1016/j.carbpol.2019.115805. [DOI] [PubMed] [Google Scholar]
- 54.Tolosa L., Donato M.T., Gómez-Lechón M.J. General Cytotoxicity Assessment by Means of the MTT Assay. 2015. pp. 333–348. [DOI] [PubMed] [Google Scholar]
- 55.Van Damme E., De Meyer S., Bojkova D., Ciesek S., Cinatl J., De Jonghe S., Jochmans D., Leyssen P., Buyck C., Neyts J., Van Loock M. In vitro activity of itraconazole against SARS‐CoV‐2. J. Med. Virol. 2021;93:4454–4460. doi: 10.1002/jmv.26917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vidal E., López-Figueroa C., Rodon J., Pérez M., Brustolin M., Cantero G., Guallar V., Izquierdo-Useros N., Carrillo J., Blanco J., Clotet B., Vergara-Alert J., Segalés J. Chronological brain lesions after SARS-CoV-2 infection in hACE2-transgenic mice. Vet. Pathol. 2021 doi: 10.1177/03009858211066841. 030098582110668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vithani N., Ward M.D., Zimmerman M.I., Novak B., Borowsky J.H., Singh S., Bowman G.R. SARS-CoV-2 Nsp16 activation mechanism and a cryptic pocket with pan-coronavirus antiviral potential. Biophys. J. 2021;120:2880–2889. doi: 10.1016/j.bpj.2021.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu C., Liu Y., Yang Y., Zhang P., Zhong W., Wang Y., Wang Q., Xu Y., Li M., Li X., Zheng M., Chen L., Li H. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm. Sin. B. 2020;10:766–788. doi: 10.1016/j.apsb.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang K.S., Leeuwon S.Z., Xu S., Liu W.R. Evolutionary and Structural Insights about Potential SARS-CoV-2 Evasion of Nirmatrelvir. J. Med. Chem. 2022;65:8686–8698. doi: 10.1021/acs.jmedchem.2c00404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ylilauri M., Pentikäinen O.T. MMGBSA As a Tool To Understand the Binding Affinities of Filamin–Peptide Interactions. J. Chem. Inf. Model. 2013;53:2626–2633. doi: 10.1021/ci4002475. [DOI] [PubMed] [Google Scholar]
- 61.Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L., Becker S., Rox K., Hilgenfeld R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science (80–.) 2020;368:409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material.
Supplementary material.
Supplementary material.
Supplementary material.
Supplementary material.
.
.
.
.
.
.
.
.
.
.